Abstract
Helicobacter pylori (H. pylori) is a bacterial pathogen that exclusively colonizes the human gastric mucosa and can cause persistent infection. In this process, H. pylori employs various strategies to avoid recognition by the human immune system. These range from passive defense strategies (e.g., altered LPS or flagellin structures) that prevent recognition by pattern recognition receptors (PRRs) to more active approaches, such as inhibition of IL-2 secretion and proliferation of T cells via VacA. Despite the growing evidence that H. pylori actively manipulates the human immune system for its own benefit, the direct interaction of H. pylori with immune cells in situ is poorly studied. Here, we present a novel intravital imaging model of the murine stomach gastric mucosa and show for the first time the in situ recruitment of neutrophils during infection and a direct H. pylori-macrophage interaction. For this purpose, we applied multiphoton intravital microscopy adapted with live drift correction software (VivoFollow) on LysM-eGFP and CX3CR1-eGFP reporter mice strains in which specific subsets of leukocytes are fluorescently labeled. Multiphoton microscopy is proving to be an excellent tool for characterizing interactions between immune cells and pathogens in vivo.
Author summary
This study focuses on the dynamics of H. pylori infection and its interaction with myeloid immune cells, using an innovative in vivo multiphoton imaging approach to observe real-time events in the infected gastric mucosa. In contrast to conventional methods, this novel technique allows us to dynamically investigate the host immune response. Our results show the spatio-temporal distribution of neutrophils and the internalization of H. pylori by macrophages over time. Using intravital multiphoton microscopy enhanced with live drift correction software and utilizing LysM-eGFP and CX3CR1-eGFP reporter mice, we established an intravital imaging model for the gastric mucosa of mice. Notably, we observe a significant increase in neutrophil infiltration into the gastric submucosa during early infection, indicating a rapid response of the innate immune system within the gastric crypts. However, in the chronic infection phase, a contrasting decrease in neutrophil influx is seen, indicating an adaptive innate immune response with fewer neutrophils infiltrating the tissue at an increased rate. Importantly, our data suggest that H. pylori is dependent on macrophages to proliferate and colonize the gastric mucosa during chronic infection.
Introduction
The gastric pathogen Helicobacter pylori causes one of the most common chronic bacterial infections in humans. Diseases such as atrophic gastritis, peptic ulcers, and gastric cancer resulting from H. pylori infections are the leading causes of infection-related morbidity and mortality worldwide. Thus, it can be estimated that over 800000 new cases of gastric cancer per year are due to H. pylori infection [1]. Consequently, H. pylori has been classified as a definitive carcinogen (Class I) by the World Health Organization (WHO) [2]. Importantly, the immune system is unable to clear the infection, resulting in the characteristic gastric inflammation that persists for decades if untreated. Following infection, H. pylori induces activation of inflammatory cytokines, including interleukin-8 (IL-8) [3]. Production and secretion of IL-8 promotes neutrophil activation and monocyte recruitment, resulting in mucosal tissue damage. Thus, secretion of IL-8 is the critical mediator of neutrophil migration to sites of infection and requires direct contact of viable H. pylori with epithelial cells via its cag-type IV secretion system (cag-T4SS) [4]. Using the cag-T4SS, H. pylori is able to inject its CagA oncoprotein into gastric epithelial cells and various types of immune cells. For this purpose, H. pylori uses cell surface molecules of the carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) family as host cell receptors to interact with and inject CagA into epithelial cells as well as immune cells, e.g. neutrophils [5,6]. The initial inflammatory response to H. pylori infection in humans is studied mainly by histopathologic analysis, where a strong neutrophilic response has been observed. After eradication of the bacteria, the inflammatory reaction of the gastric mucosa subsides. It is well known that H. pylori actively interferes with the host immune system to prevent its elimination. Important factors include secretion of a vacuolating cytotoxin (VacA) that impedes IL-2 secretion by T lymphocytes [7]. In addition, VacA targets phagocytes to prevent proper phagosome maturation, antigen processing, and presentation [8]. Furthermore, the γ-glutamyl transpeptidase GGT contributes critically to H. pylori’s tolerizing effects on dendritic cells (DCs) [9]. In addition, the bacteria avoid host recognition by Toll-like receptors (TLRs), via binding of human annexin A5 to lipid A of the bacterial LPS preventing TLR4 signaling [10], and via its modulated flagellin subunit H. pylori avoids recognition via TLR5 [11]. In animal models, such as the conventional C57BL/6 H. pylori mouse model, the gastric inflammation is generally lower as compared to the human situation. In vivo studies to observe and quantify e.g. macrophages and neutrophil infiltration are still poorly reported. Thus, novel approaches need to be developed to study this phenomenon in vivo in more detail using appropriate animal models. Here, we developed for the first time in vivo imaging of the gastric mucosa in mice to visualize immune cell interactions with H. pylori in situ. Intravital multiphoton imaging combined with transgenic mouse lines expressing specific fluorescent labels is a powerful method that allows researchers to directly observe cell dynamics and biological processes in living organisms. The principle of multiphoton imaging permits visualization of organs with significant depth and minimal phototoxicity due to infrared laser light excitation, making it an excellent tool for animal model studies. The technique is suitable for real-time observation of cell-cell interactions in organs commonly used in immunology. It also facilitates visualization of cell movement and direct observation of immune cell interactions and dynamics such as intravascular migration and extravasation [12,13]. The influx of neutrophils into the tissue is an early hallmark of a bacterial infection. The major aim of this study is to visualize neutrophil and macrophage recruitment in the first 24 hours and compare it with long-term H. pylori infection conditions (up to 6 weeks post-infection). Our approach consists of combining intravital multiphoton microscopy with a transgenic mouse line expressing fluorescently labeled immune cells. The immune system recognizes the bacteria and attempts to eliminate them. Some phagocytic cells such as neutrophils can also take up and inactivate H. pylori [14]. Since a good in vivo model to analyze the role of immune cells during H. pylori infection is still lacking, we have developed an instrument to visualize labeled immune cells and H. pylori in vivo. By multiphoton microscopy, it is possible to produce three- or four-dimensional, deep tissue images over extended periods of time with minimal phototoxicity.
Generally, intravital imaging of organs localized in the abdominal cavity is quite challenging due to peristalsis, cardiac and respiratory motion, which compromises image quality and complicates quantification of cell tracking and cell-cell interaction in vivo. To circumvent these problems, in our multiphoton setup, we implemented a live-drift correction software named VivoFollow, which allows a real-time correction of the tissue drift by determining the offset between two image stacks acquired at different time points and minimize imaging drift during acquisition [15]. Using this approach, we studied neutrophil and macrophage recruitment during acute and chronic H. pylori infection. Our results showed a rapid influx of neutrophils during acute H. pylori infection compared to 6 weeks of infection, and an increased number of macrophages in the chronic stage of infection. H. pylori can be phagocytized by macrophages, and we show an increased number of bacteria inside macrophages in the chronic stage of infection. The depletion of macrophages resulted in a reduction in bacterial growth, suggesting a contribution of macrophage to bacterial proliferation in the later phase of infection. In conclusion, the imaging techniques we deployed prove to be a useful tool for the in vivo study of immune cell dynamics and their interaction with H. pylori.
Results
Inner gastric mucosa preparation for multiphoton intravital imaging
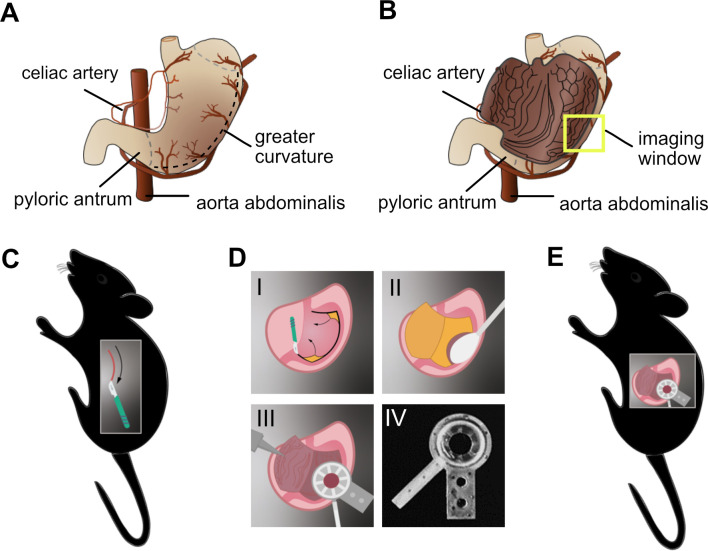
For our initial experiments on visualization of gastric mucosa by multiphoton microscopy, we started with the dissected intact stomach of mice. Due to the thick and rigid muscle layer of the stomach wall, direct microscopy on the intact stomach was very difficult and prevented high resolution imaging of the inner gastric mucosa. To address this issue, the stomach wall was cut laterally to expose the inner gastric mucosa (Fig 1A and 1B). Another obstacle we faced is the high peristaltic movement of the organ in the living animal. Therefore, an innovative gastric imaging system was designed in a way to effectively shield the gastric mucosa from the peristaltic movements and the neighboring bowel movement. After opening, the gastric mucosa was carefully cleaned with isotonic solution and food was removed with a moist cotton swab. Next, it was fixed with a suction ring holder attached to a cover slip with an imaging window of 8 mm, for imaging acquisition. The holder was designed and fabricated by the 3D printing technology. The detailed process steps are shown in Fig 1C–1E.
Fig 1. Fixation of murine gastric mucosal tissue for intravital microscopy imaging.
(A) Cartoon representing the closed (or intact) stomach. The dashed line delineates the area of cutting to open the stomach. (B) Open stomach showing the inner gastric mucosa where the yellow square represents the imaging window. (C) After sedation, the mouse is placed in the right lateral decubitus position. The stomach is exposed by making an incision of approximately 150 mm across the lateral side. (D) Steps demonstrating the gastric mucosa preparation and its fixation by the suction ring device with an imaging window of 8 mm, which was designed, and 3D printed. (D-I) After opening the skin, the stomach is opened with a cauterizing scalpel and the gastric mucosa is exposed. (D-II) Any remaining gastric contents are carefully removed with a moist cotton swab and irrigation with isotonic solution. Rinsing with isotonic solution, taking care not to injure the gastric mucosa. (D-III) The vacuum chamber can now be placed on to the lining of the stomach to permit multiphoton imaging of the gastric mucosa. (D-IV) 3D-printed suction ring with an imaging window of 8 mm. (E) Overview of the gastric mucosa preparation for multiphoton imaging.
VivoFollow real time drift correction software setup
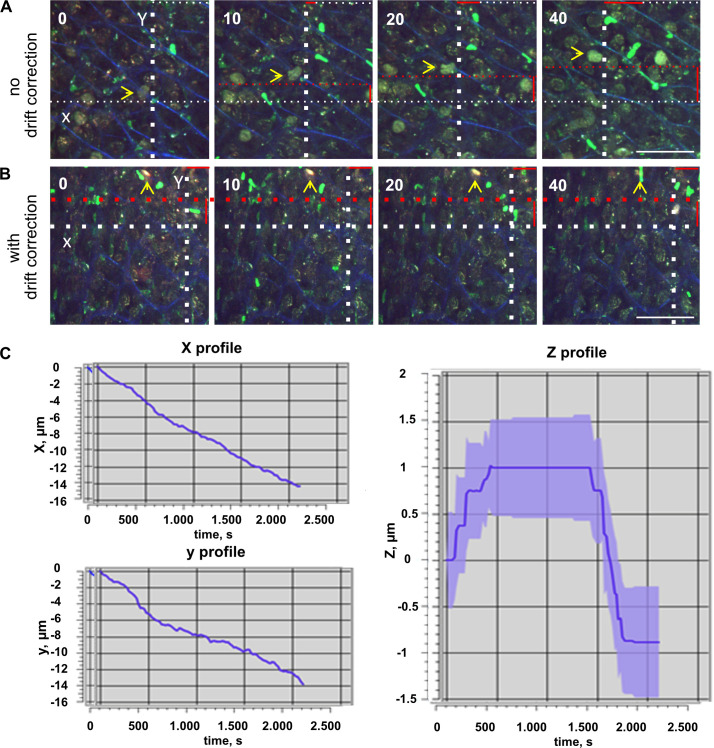
One of the most challenging tasks in performing intravital microscopy imaging is to avoid excessive vital movements during imaging acquisition, making imaging analyses not possible. A good tissue fixation is essential, however even with a good tissue holding, the longtime imaging acquisition leads to tissue drifting. Therefore, we applied and integrated a real time drift correction software, named VivoFollow, into our system to bypass this issue [15]. A detailed protocol of how to setup image acquisition using the VivoFollow software is described in [16]. The mouse with the fixed gastric mucosa was positioned inside a heated microscope chamber and the mucosa gastric window was placed under a 16x objective. To correct the drift movement during imaging acquisition, the live drift correction software (VivoFollow) was applied to successfully stabilize imaging acquisition in vivo (Fig 2 and S1 Video).
Fig 2. VivoFollow the real time drift correction software.
(A) Time-lapse sequence imaging of LysM-GFP mouse gastric mucosa showing drift along the X and Y axis. The white dotted lines are for reference; the red dotted lines show the drift, and the red solid lines indicate the magnitude of the shift. The yellow arrow indicates the followed immobilized reference structure showing the imaging drift in (A) and the stable corrected time series in (B). (B) Effects of the live drift correction stabilization. There is no major drift from the yellow arrow and white dotted lines serving as references (see also S1 Video). Numbers indicate minutes. Scale bars are 50 μm. (C) VivoFollow cartoon showing the path of image acquisition and correction in x, y and z, from microscope to GPU-equipped computer.
From the real-time stable videos acquired, it was possible to analyze the leukocyte dynamics in the gastric mucosa during H. pylori infection (Figs 3 and 4).
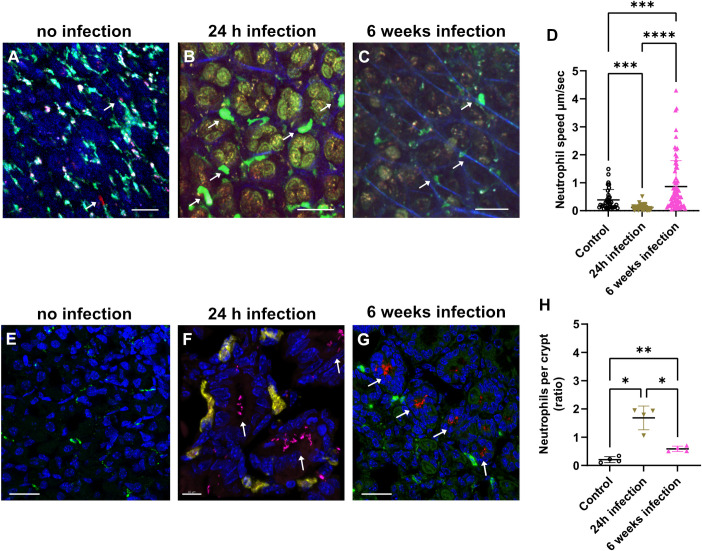
Fig 3. Neutrophil recruitment after H. pylori infection monitored by multiphoton microscopy in vivo.
Mice were infected with H. pylori for 24 hours or 6 weeks. (A) Multiphoton 4D time-lapse images of uninfected CX3CR1-GFP+ mice, which were injected with anti-Ly6G-PE antibodies to visualize neutrophils in red. (B) LysM-eGFP mice show neutrophil recruitment (green) after H. pylori 24h weeks of infection (red), second harmonic generation (SHG) showing the vessels walls (blue) or 6 weeks after infection in (C). (D) Histogram showing neutrophil velocity at 24 hours and 6 weeks post-infection. Neutrophils show significantly reduced mobility in the gastric mucosa in vivo after the first 24 hours of H. pylori infection compared to control. After 6 weeks of H. pylori infection, neutrophil velocity is significantly increased compared to control 24-hours post infection. Immunostaining image of 6 weeks infected gastric mucosal section showing neutrophils stained with anti-Ly6G-488 (green) and H. pylori-mRFP (red), DAPI (blue). Immunohistology of non-infected gastric mucosa (E), scale bar is 20 μm. (F) 24h after infection, showing H. pylori (magenta) localized inside the crypts and neutrophils (yellow). Scale bar is 10 μm. (G) 6 weeks after infection, showing H. pylori (red) located inside the crypts and neutrophils (green), DAPI (nucleus), scale is 20 μm. The arrows indicate the localization of the H. pylori inside the crypts. (H) Quantification of neutrophil influx after 24 hours of H. pylori infection and decrease after 6 weeks of infection. n = 4 mice. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, or not significant (ns). One way ANOVA t-test. Mean ± S.D.
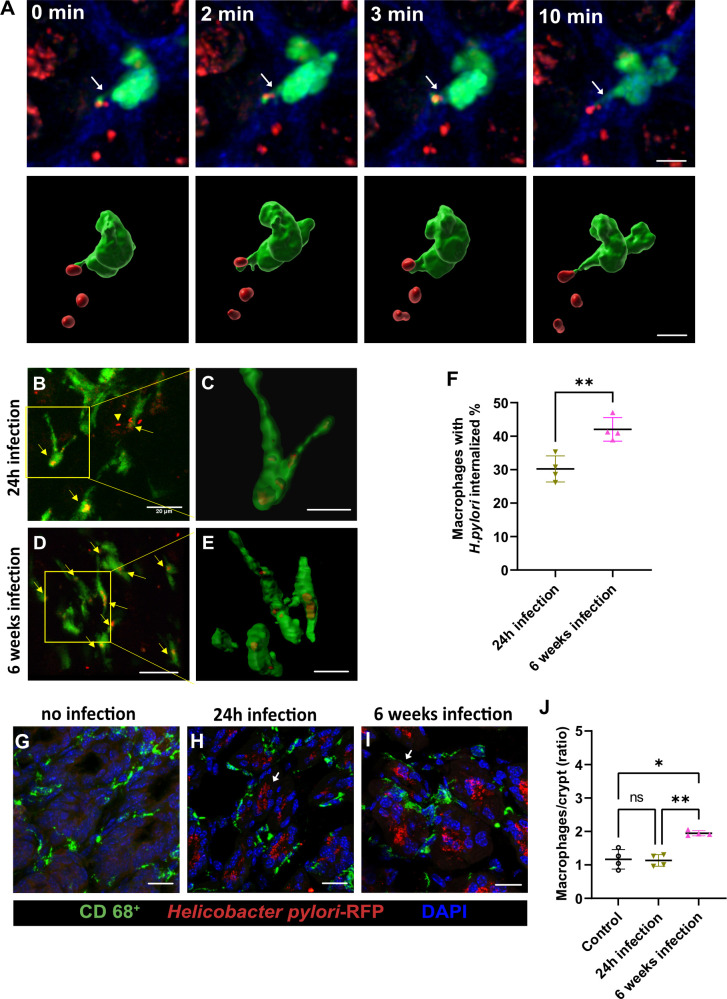
Fig 4. Interaction of macrophages with H. pylori monitored by multiphoton microscopy in vivo.
(A) Multiphoton imaging of CX3CR1-GFP mouse showing macrophages (green) promoting cytoplasmic extensions in closer contact with H. pylori, as indicated by the arrows in the upper panel. In the lower panel a 3D image reconstruction of the macrophages (green) and bacteria (red) in a closer proximity. Scale bar is 5 μm. (B-E) Multiphoton imaging of CX3CR1-GFP mouse showing H. pylori (red) localized inside macrophages (green) in vivo after 24 hours (B-C) or 6 weeks of infection (D-E), as indicated by the arrows. (C and D) the images represent the zoomed-out macrophages with H. pylori phagocytized. (F) Histogram showing the internalization of H. pylori by macrophages after 24 hours or 6 weeks of infection. (G-I) Immunohistology showing resident macrophages peripherally localized in crypts. Scale bars are 20 μm. (J) Quantification of macrophages in gastric mucosa after 24 hours and 6 weeks of H. pylori infection. n = 4 mice, ns (non-significant difference), ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05. One way ANOVA t test. Mean ± S.D.
Leukocyte dynamics measurement revealed by multiphoton microscopy in vivo
The dynamics of leukocytes during H. pylori gastric infection of the mouse stomach mucosa can be monitored in real time by using multiphoton microscopy. This high microscopy technology offers a deeper imaging penetration, with less photo-toxicity to the cells. LysM-eGFP mice [17] were infected with H. pylori for 24h (acute infection) or 6 weeks (chronic infection). The inner gastric mucosa was prepared for multiphoton intravital imaging (Fig 1). Gastric mucosa of non-infected, injected with anti-Ly6G-PE antibodies to label neutrophils (Fig 3A and S2 Video) and LysM-GFP infected mice for 24h (Fig 3B, see also S3 Video) or 6 weeks (Fig 3C and S4 Video), were imaged to track neutrophil speed and numbers (Fig 3D and 3H and S1 Data). Interestingly, the neutrophil mobility was affected by H. pylori infection of the gastric mucosa, with significant changes observed at different time points. Specifically, after the initial 24 hours of infection, there was a significant reduction in neutrophil mobility compared to uninfected controls (Fig 3D). However, after six weeks of infection, in the chronic infection phase, there was a significant increase in neutrophil velocity compared to both the control and the 24-hour time point. These observations indicate that the effects of H. pylori infection on neutrophil mobility are time-dependent and may vary during the course of the infection. The quantification of neutrophil influx in response to H. pylori infection reveals distinct patterns over time. Specifically, there is a significant increase in neutrophil infiltration into the gastric submucosa after the first 24 hours of infection, determined as neutrophils per crypt (Fig 3E–3H and S2 Data) representing an early response of the innate immune system to the pathogen localized inside the gastric mucosa crypts (Fig 3F and 3G and S8 Video). However, after 6 weeks of infection, a marked decrease in neutrophil influx was observed, suggesting that a smaller number of neutrophils is patrolling the tissue with an elevated speed indicating an adaptation of the innate immune system response to the ongoing infection (Fig 3H).
Multiphoton visualization of H. pylori internalized by macrophages in vivo in the murine gastric mucosa
In addition to studying the behavior of neutrophils under conditions of infection over time, the behavior of macrophages in the gastric mucosa was also investigated. The novel multiphoton setup gave us the possibility to visualize the interaction of H. pylori with macrophages in vivo during gastric mucosa infection. We primarily examined resident macrophages labeled with GFP (CX3CR1-GFP) [18]. With this approach, the macrophage cytoplasmic extension in direction to H. pylori was visualized in the first 24h of infection (Fig 4A and S5 Video), and the internalization of H. pylori by the macrophages in situ could be visualized after 24h and 6 weeks of infection (Fig 4B–4E) and quantified (Fig 4F and S6 and S7 Videos and S3 Data). Furthermore, immunohistology revealed resident macrophages localized in crypts in the periphery (Fig 4G–4I). Quantification of the data revealed a significant increase in H. pylori phagocytosis in the chronic phase (6 weeks) as compared to the acute phase (24 hours) (Fig 4F). Furthermore, the number of macrophages per crypt also increased significantly from the acute to the chronic phase of infection (Fig 4J and S4 Data). These findings suggest that macrophages play a crucial role in the immune response to H. pylori infection and the study highlights the use of multiphoton microscopy in vivo to monitor the interaction between immune cells and pathogens, which could provide a deeper understanding of the immune response to infections.
Macrophages contribute to H. pylori proliferation over time
To investigate if macrophages in the gastric mucosa are able to actively interfere with H. pylori proliferation, e.g. by restricting the number of colonizing bacteria due to phagocytosis, or by facilitating the bacteria proliferation in macrophages, we made use of PLX5622, an inhibitor of the colony-stimulating factor 1 receptor (CSF1R), which was originally used to eliminate microglia from the brain [19]. We show that feeding of mice with PLX5622 chow for 7 days (Fig 5A–5C and S5 Data), 2 or 3 weeks (Fig 6B), strongly reduces or eliminates resident CX3CR1 macrophages from the stomach mucosa. Furthermore, we show that the decrease in macrophage number was accompanied by an increase in neutrophil counts (Fig 5D and S6 Data).
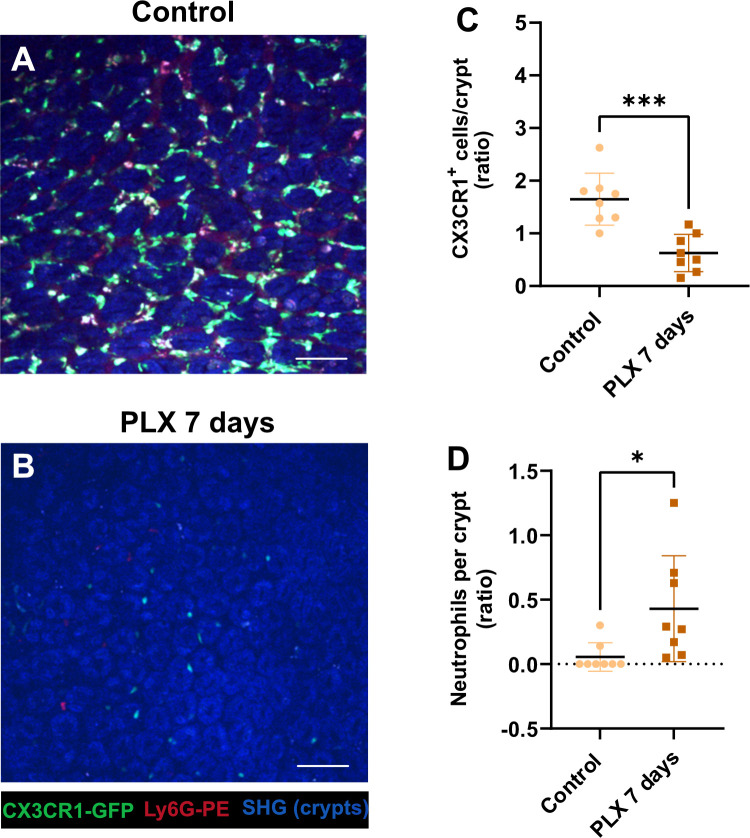
Fig 5. Macrophage depletion and neutrophils response under PLX 5622 treatment.
(A-B) Multiphoton imaging of CX3CR1-GFP mouse gastric mucosa showing macrophages (green) and neutrophils, labelled by the injection of anti-Ly6G-PE antibodies (red), treated with PLX 5622 chow (B) and control chow treated (A), SHG (blue), scale bar is 20 μm. (C) Quantification of the CX3CR1-GFP positive cells after 7 days of PLX 5622 treatment. (D) Quantification of neutrophils in gastric mucosa after PLX treatment versus untreated (control). n = 8 mice, ns (non-significant difference), ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05. One way ANOVA t-test. Mean ± S.D.
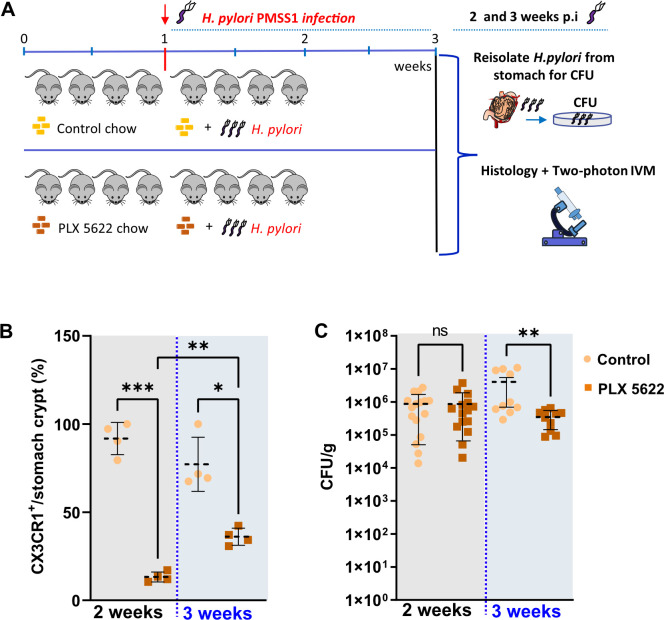
Fig 6. Continuous macrophage depletion is associated with reduced H. pylori infection.
(A) Schematic representation of H. pylori infection (2–3 weeks) and PLX 5622 food treatment (3–4 weeks) to reduce macrophage replication. (B) Quantification of CX3CR1+ cells in the gastric mucosa after PLX treatment versus untreated (control) for 2 or 3 weeks of infection. n = 4 mice for each treatment. (C) Determination of colony forming units (CFU) of H. pylori infected CX3CR1+ mice after PLX treatment (PLX 7 days) or untreated (control). ns (non-significant difference), ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05. One way ANOVA t-test. Mean ± S.D. Mouse cartoon by lemmling, sourced from Open Clip Art, licensed under CC0 1.0.
Macrophages contribute to the multiplication of H. pylori as demonstrated by PLX 5622 treatment and infection for 2–3 weeks (Fig 6A–6C and S7 Data). Reducing the number of macrophages on the gastric mucosa by approximately 80% has no significant effect on bacterial colonization efficiency after 2 weeks of infection, but does affect bacterial growth after 3 weeks of infection, as determined by the re-isolation of bacteria (Fig 6C and S8 Data). This indicates that H. pylori might rely on macrophages to proliferate and colonize the gastric mucosa during chronic infection.
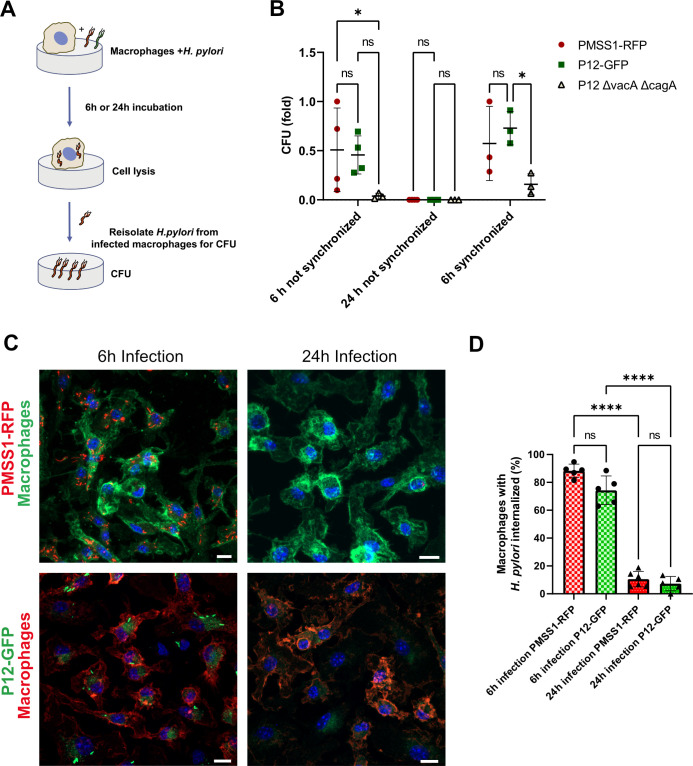
To investigate whether H. pylori can replicate within macrophages, we conducted a survival assay using isolated primary mouse bone marrow-derived macrophages infected with the bacteria. After incubating the primary macrophages and bacteria for 6 or 24 hours, we lysed the macrophages and re-isolated the bacteria via agar plating to determine bacterial survival (Fig 7A and 7B). We used two H. pylori strains for the infections: PMSS1-RFP and P12-GFP [10], along with a P12 strain lacking the vacA and cagA genes (P12ΔvacAΔcagA) (Fig 7B and S9 Data), as these virulence factors might influence intracellular survival [20]. Both PMSS1-RFP and P12-GFP strains successfully infected macrophages (Fig 7C and 7D and S10 Data), whereas the P12ΔvacAΔcagA mutant showed a very low infection rate (Fig 7B). To ensure this was not due to reduced physical interaction between the mutant strain and macrophages, we included an additional centrifugation step during the 6-hour synchronized infection (see Methods for details), which resulted in better infection, but still at a significantly reduced level, suggesting that the bacteria use an active mechanism for intracellular survival. The localization of the PMSS1-RFP and P12-GFP strains inside macrophages with higher resolution is displayed in Fig 8 and S9 and S10 Videos and S1 Fig.
Fig 7. H. pylori survival in infected macrophages.
(A) Bone marrow-derived mouse macrophages were incubated with H. pylori PMSS1-RFP, P12-GFP or P12ΔvacAΔcagA. After 6h or 24h infected macrophages were lysed, and H. pylori were re-isolated for colony forming unit (CFU) assay. (B) CFU quantification indicating the bacterial survival after 6h of internationalization by macrophages. (C) Infected macrophages were fixed and stained with anti-F4/80-Alexa-488 (green) or Alexa-594 (red) for macrophages, and DAPI for nucleus (blue). Images show H. pylori internalized by macrophages. Scale bar is 10 μm. (D) Quantification of macrophages infected with H. pylori strains PMSS1-RFP (red) or P12-GFP (green) for 6h or 24h. n = 5–6, ****p<0.0001, ***p<0.001, **p<0.01,*p<0.05, or not significant (ns). Two-way ANOVA t-test. Mean ± S.D.
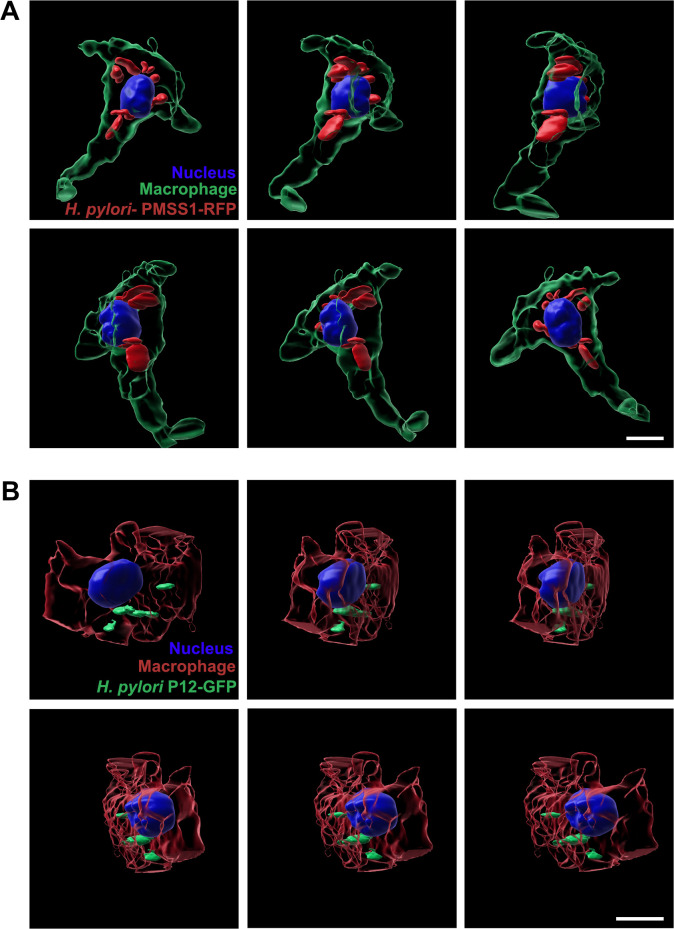
Fig 8. H. pylori internalization by macrophages.
3D rendering image sequences showing the localization of PMMS1-RFP (A) or P12-GFP (B) after 6h of infection inside macrophages. The images series are displayed from different angles and were extracted from S9 and S10 Videos. Scale bar is 10μm. DAPI is labelling the nucleus. Anti-F4/80 antibodies were used to label macrophages.
Discussion
Our study presents a novel gastric imaging system that was designed to allow for the first time a direct view into the stomach mucosa of the cellular events, such as neutrophil dynamics and H. pylori–macrophage interaction in an infected animal. The novel system shields the gastric mucosa from peristaltic movements and neighboring bowel movements, allowing for high-resolution imaging of the inner gastric mucosa. In addition, the use of the VivoFollow live drift correction software successfully stabilizes imaging acquisition in vivo, enabling analysis of leukocyte dynamics in the gastric mucosa during H. pylori infection. The combination of these innovative techniques provides a powerful tool for studying the immune response in the gastric mucosa.
Furthermore, our results show that H. pylori infection affects the mobility of neutrophils at different time points, with a reduction in mobility after 24 hours and an increase after six weeks. It appears that during the acute phase of H. pylori infection neutrophils arriving in the gastric submucosa lower their speed to monitor the tissue for the bacterial invaders. Immunohistology of the inner gastric mucosa revealed distinct patterns over time. A significant increase in neutrophil infiltration was observed during the first 24 hours of H. pylori infection, but a marked decrease of neutrophils was seen after 6 weeks, although the bacteria were still present having established the chronic phase of infection. Our earlier data using neutrophils in vitro showed that H. pylori contact strongly enhanced the oxidative burst of neutrophils and stimulated the process of H. pylori phagocytosis. However, the expression of human CEACAM3/6 on these murine neutrophils allowed the extended survival of H. pylori within neutrophils [6], suggesting that the conventional H. pylori mouse model does not adequately mirror the human infection conditions. Nevertheless, the novel in vivo multiphoton setup provides first in vivo data concerning the adaptation of the innate immune system response to the ongoing H. pylori infection. Further investigations are needed to fully understand the underlying mechanisms and implications of these findings.
Our study also aimed to investigate the behavior of macrophages in the gastric mucosa during H. pylori infection using the novel multiphoton setup, which allowed for visualization of the RFP-labeled H. pylori with resident macrophages labeled with GFP. The experimental data showed an increase in H. pylori phagocytosis by macrophages in the chronic phase of infection compared to the acute phase, highlighting a crucial role of macrophages in the immune response to this pathogen. Data on phagocytosis of H. pylori PMSS1-RFP by CX3CR1-GFP macrophages in vivo were also previously reported by fluorescence-activated cell sorting (FACS) [21]. Mice that had been neonatally infected and therefore possessed high colonization rates exhibited as adults RFP signals in approximately 4% of the CD11b+ myeloid cell population [21]. Our multiphoton microscopy tool revealed the in vivo dynamics of macrophage forming protrusions in presence of the bacteria in the early period of infection (Fig 4A and S5 Video) and the internalized H. pylori inside the macrophages in the early and increased number in the later stage of the infection (Figs 4B–4D and S6 and S7 Videos). To further understand the functional role of resident macrophages in the control of the bacteria we used the PLX5622 inhibitor of the colony-stimulating factor 1 receptor (CSF1R) to eliminate macrophages from the stomach mucosa. Interestingly, the reduced number of macrophages had a significant effect on bacterial colonization efficiency in the later stage of bacterial proliferation, however, it has no effect in acute infection, suggesting that macrophages, despite their phagocytosis activity shown under in vivo conditions, are not able to contribute to the bacterial proliferation or elimination of H. pylori in acute phase of infection. We show here that elimination of macrophages for long term, causes a reduction of bacterial growth, suggesting that macrophages may serve as a host for H. pylori proliferation. Earlier work using murine and human macrophage cell lines show that H. pylori strains expressing the vacuolating cytotoxin interrupt phagosome maturation in macrophages by recruiting and retaining coronin 1A protein [20]. Other studies show that the internalization of H. pylori by macrophages induces autophagosome formation and these autophagic vesicles are adapted to allow bacterial replication inside macrophages [22]. In another work, it was found that the transient absence of macrophages in the early period of H. pylori infection reduced the gastric pathology of the disease but did not affect the bacterial load in the stomach [23].
Our primary mouse macrophage infection experiments support the in vivo data, indicating that H. pylori can infect and survive within primary mouse bone marrow-derived macrophages. The P12 strain lacking vacA and cagA genes exhibited a significantly lower infection rate, even after improving physical interaction with macrophages, suggesting these virulence factors are crucial for efficient intracellular survival. Our data imply that macrophages may facilitate H. pylori proliferation over the course of infection, playing a role in long-term bacterial growth rather than in the acute stage. This mechanism of bacterial residence inside macrophages may contribute to increased resistance to antimicrobial treatment and protection against humoral antibody responses.
Materials and methods
Ethics statement
The animal study protocol was approved by the local regulatory agency (Regierung von Oberbayern, Munich, Germany, record number ROB-55.2-2532.Vet_02-18-189 and ROB-55.2-2532.Vet_02-18-183.
Mice
C57BL/6, CX3CR1-GFP (B6.129P2(Cg)-Cx3cr1tm1Litt/J) (Strain #:005582), LysM-GFP were obtained from The Jackson Laboratory (Boston, MA, USA). The mice were maintained and bred under specific-pathogen free and fed standard mouse diet. All animals undergoing procedures were over 10 weeks old.
Bacterial strains
H. pylori strain PMSS1-RFP [21], P12-GFP and P12ΔvacAΔcagA [10] were grown on GC agar plates (Oxoid) supplemented with vitamin mix (1%) and horse serum (8%) (serum plates) and cultured in a microaerobic atmosphere (85% N2, 10% CO2, 5% O2) at 37°C.
Mice infection and bacterial culture for CFU experiments
Mice were fed with PLX 5622 chow (D19101002i, AIN-76A), or control chow (D10001i, AIN-76A) from Research Diets, for 7 consecutive days prior to infection and continued for 2 or 3 weeks after the infection with H. pylori PMSS1-RFP. Frozen stock of H. pylori PMSS1-RFP strain was thawed from -80°C and subcultured on GC agar plates (with 1% vitamin mix and 8% horse serum) for five days under microaerophilic conditions (85% N2, 10% CO2, 5% O2). On the sixth day, H. pylori was taken into liquid culture in Brucella Broth. Mice were infected twice every other day with approximately 6 x 107 CFUs in Brucella Broth using an oral gavage needle. H. pylori was re-isolated at 2- or 3-weeks post infection from the antrum and corpus following stomach excision and homogenization using a Wheaton homogenizer and selected on DENT plates (Oxoid H. pylori selective supplement–SR0147E containing Vancomycin, Cefsulodin Trimethoprim, Amphotericin B with Polymyxin B (Simga: P9602), Nalidixin acid (Roche), Bacitracin and 1% vitamin mix and 8% horse serum) for the CFU experiments. The stomach was collected for immunohistology analysis.
Mice and organ preparation for multiphoton intravital imaging
CX3CR1-GFP or LysM-GFP mice were anesthetized with 5.0 Vol. % Isofluran (cp-pharma, Burgdorf, Germany) and 0.25 l/l oxygen. Mice were injected intraperitoneally with one dose of anesthetic mixture containing Fentanyl (0.05mg/kg body weight, Albrecht GmbH, Aulendorf, Germany), Midazolam (5mg/kg body weight, B. Braun, Melsungen, Hessen, Germany) and Medetomidine (0.5mg/kg body weight, Zoetis, Germany). Mice under narcosis were placed on a warming pad, the eyes were covered with eye cream (Bepanthene) and mice were positioned in right lateral cubic. The skin incision (about 1 cm) was made on the median right lateral position, after skin disinfection. The stomach was pulled out carefully from the abdominal cavity, to allow the visualization of the complete organ. The lateral curvature of the stomach corpus was open using a cautery pen (Bovie Medical Corporation) and the upper part was fleted out as shown in Fig 1, to expose the stomach inner mucosa. The food inside the stomach was carefully removed with a wet swab cotton without touching the tissue. The inner gastric mucosa was rinsed with NaCl (0.9%) to remove food residues. The open stomach was fixed by a suction ring, attached with a cover glass of 8 mm diameter, connected with a small vacuum pump, set up with a pressure of 0.2 PA. The mouse was positioned on the microscope stage within an incubator chamber to maintain a constant temperature of 37°C. Ultrasonic gel, acting as an immersion medium, was applied to the cover slip prior to contacting the microscope objective. Gastric mucosa imaging was conducted using a multiphoton LaVision Biotech (Bielefeld, Germany) TrimScope II system connected to an upright Olympus microscope, equipped with a Ti;Sa Chameleon Ultra II laser (Coherent) tunable in the range of 680 to 1080 nm and additionally an optical parametric oscillator (OPO) compact to support the range of 1000 to 1600 nm and a 16 × water immersion objective (numerical aperture 0.8, Nikon). Live images were acquired from 30–40 μm depth, with z-interval of 2–3 μm. 870 nm was used as an excitation wavelength, with 350 × 350 pixels. The signal was detected by Photomultipliers (G6780-20, Hamamatsu Photonics, Hamamatsu, Japan). ImSpector Pro (LaVision) version 275 was used as acquisition software.
Imaging stabilization and analysis
During multiphoton intravital imaging acquisition, VivoFollow live drift correction software tool was applied to correct for the tissue motion in real-time by maintaining the region of interest in focus over the course of imaging (15–16).
Customized VivoFollow drift correction software (15) was connected and run into the ImSpector Pro version 275. A customized chamber was made to cover the microscope automatic stage, to maintain a stable 37°C warm environment.
Immunohistology and imaging
Mice were orally infected with H. pylori strain and after the determinate period of infection, were anesthetized and euthanized by cervical dislocation. The blood was collected by cardiac puncture and the stomachs were perfused with PBS and collected in cold PBS. The stomachs were fixed in 4% PFA overnight followed by overnight infiltration in sucrose at 4°C. Subsequently, the samples were infiltrated in tissue teck and frozen in dry ice, before storage at -80°C. Cryosectioning of the samples with a thickness of 10 μm was performed using a cryotome (Cryostar NX70 from Epredia) in preparation for immunohistological analysis. The sliced tissue was fixed with 4%PFA for 15 min at RT, washed with PBS 3x and blocked with BSA 3% overnight. Next day the primary antibodies were added overnight at 4°C, washed and incubated with the secondary antibodies for 2h at RT. After washing DAPI (1 μg/ml, 1:1000) was added for 30 min, before the tissue mounting with fluorescence mounting medium DAKO. Images were taken using a confocal laser scanning microscope LSM 880 or 980 (Carl Zeiss) with Airyscan super-resolution module, using 63x objective (Nikon) and a 415 x 415 μm or 312 x 312 μm field of views and step size 1.0 μm. Macrophages were labeled with anti-CD68 (clone FA-11, BioRad # MCA1957GA) and neutrophils with anti-ly6G primary antibodies (clone 1A8, Biolegend # 127608).
Bone marrow-derived macrophages culture
Bone marrow-derived macrophages were generated as previously described [24]. Briefly, bone marrow was obtained by flushing murine femur and tibia with ice cold 1x PBS through a 70 μm filter. The suspension was centrifiged with 350 g, 4°C for 5 min, and the cell pellet was resuspended in RPMI1640 medium (Sigma-Aldrich), supplemented with 10% fetal bovine serum (FBS) (BioSell), 1x Pen/Strep (Sigma-Aldrich) and 20 ng/mL macrophage colony-stimulating factor (M-CSF) from (ImmunoTools). Medium was changed every other day. At day 6 of differentiation, macrophages were detached using Accutase (Sigma-Aldrich), and plated in 24-well non-tissue culture treated plates at a density of 2.0x105 macrophages per well.
Mouse macrophage survival assays
Macrophages were setled into each 24 well (2x105 cells/well) and were washed twice with RPMI medium + 10% fetal calf serum (FCS) to remove antibiotics and kept in 200 μl RPMI + 10% FCS + M-CSF. For macrophage infection, H. pylori PMSS1-RFP, P12-GFP, and P12ΔvacAΔcagA were removed from a serum plate with a sterile cotton swab and resuspended in PBS + 10% FCS. The optical density (OD) at 600 nm was measured to determine the concentration, with an OD of 1 corresponding to 2.5 x 108 bacteria/ml. Bacteria were resuspended in RPMI + 10% FCS + M-CSF to a concentration of 5 x 107 bacteria/ml. 100 μl of this suspension (5 x 106 bacteria, multiplicity of infection: 25) were added per well to the macrophages and incubated at 37°C and 5% CO2 for 5 h (6 h time point) or 23 h (24 h time point). Alternatively, to obtain synchronization of infection, plates were centrifuged at 400 g for 6 min at 4°C. Immediately afterwards the cells (H. pylori infected macrophages) were washed three times with cold RPMI + 10% FCS before addition of pre-warmed RPMI + 10% FCS + M-CSF and incubation at 37°C and 5% CO2 for 1 h. In case of syncronized as well as non-synchronized infection assays the cells were washed twice with RPMI + 10% FCS before addition of RPMI + 10% FCS + M-CSF containing 100 μg/ml gentamicin and incubated for 1 h at 37°C and 5% CO2. Afterwards, the cells were washed twice with PBS and per well 200 μl 0.1% saponin in PBS was added and incubated for 5 min at room temperature for cell lysis. Different volumes were plated on serum plates and incubated at 37°C and 10% CO2 for at least 96 h before colonies were counted.
Infected macrophages were plated on round cover slip of 13 mm diameter, coated with poly-L-lysin (Merck), fixed with 2% PFA for 15 min RT, blocked with 2% BSA for 30 min, followed by adding anti-mouse F4/80-Alexa-488 (Biolegend, #123120) or Alexa 594 (Biolegend, #123140) antibodies, for 1h RT. Washed 3x with PBS, added DAPI (1μg/ml) and mounted with fluorescence mounting medium DAKO.
Statistical analysis
Visualization and statistical analyses were performed with GraphPad Prism software (San Diego, USA) version 9.0. In all Figs, the data are presented as mean ± SD. An unpaired student‘s t-test was used for comparison between two groups. For comparison of multiple groups with one variable, one-way ANOVA testing was performed. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, or not significant (ns).
Supporting information
LysM-GFP mouse was infected with H. pylori-RFP (red), vessels are visible with the blue color (second harmonic generation; SHG). After 40 min of recording the tissue has drift from the field of view without the drift correction, when the VivoFollow was applied the image was in focus.
(MP4)
There is a reduced number of neutrophils in the non-infected stomach in comparison with the infected mice (S3 Video). CX3CR1-eGFP (macrophages), neutrophils are labelled by the Ly6G-PE antibodies injected (i.v.) into the mice 15 min before imaging. Stomach crypts are labeled blue by the SHG effect.
(MP4)
There is an increased number of neutrophils in the 24h infected stomach in comparison with the non-infected mice (S2 Video). LysM-eGFP (neutrophils), H. pylori-RFP (red) inside the crypts. Vessels are labelled by the SHG.
(AVI)
There is an increased speed of the neutrophils in comparison with 24h of infection (see S3 Video for comparison). LysM-eGFP (neutrophils), H. pylori-RFP (red). Vessels are labelled by the SHG.
(AVI)
Multiphoton imaging of the CX3CR1-GFP gastric mucosa was infected with H. pylori-RFP (red) for 24h, macrophage (green), vessels are visible with the blue color (second harmonic generation; SHG), second part of the video show the 3D rendering of macrophage and H. pylori.
(MP4)
CX3CR1-GFP mouse infected with H. pylori for 24h. Macrophages (green) show cytoplasmic extensions and H. pylori phagocytized (red). The second part of the movie show the imaging 3D reconstruction and rendering.
(MP4)
CX3CR1-GFP mouse infected with H. pylori for 6 weeks. Macrophages (green) show cytoplasmic extensions and H. pylori phagocytized (red). The second part of the movie show the imaging 3D reconstruction and rendering.
(AVI)
LysM-eGFP (yellow) mice gastric mucosa infected with H. pylori for 24h (magenta). 3D projection shown neutrophils localized at the periphery of the crypts and H. pylori are located inside the crypts (magenta). Nucleus are displayed in blue.
(AVI)
Bone marrow isolated macrophages were co-cultivated with PMSS1-RFP H. pylori strain (red) and after 6h of incubation the infected macrophages were fixed and labelled with anti-F4/80- Alexa 488 (green). 3D rendering projection shows the internalization of the bacteria by macrophages. Scale bar is 10 μm.
(MP4)
Bone marrow isolated macrophages were co-cultivated with P12-GFP H. pylori strain (green) and after 6h of incubation the infected macrophages were fixed and labelled with anti-F4/80- Alexa 594 (red). 3D rendering projection shows the internalization of the bacteria by macrophages. Scale bar is 10 μm.
(MP4)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
3D projection image sequences showing the localization of PMMS1-RFP (A) or P12-GFP (B) after 6h of infection inside macrophages. The images series are displayed from different angles representing the raw and unprocessed data used for 3D rendering of Fig 8 and S9 and S10 Videos. Scale bar is 10μm. DAPI is labelling the nucleus. Anti-F4/80 antibodies were used to label macrophages.
(TIFF)
Acknowledgments
We thank Susanne Sauer for the support in animal experiments and Wolfgang Fischer for providing H. pylori strains. Many thanks to Anna Titova, Fillip Prica and Simona Nasiscionyte for excellent technical support. Our special thanks also go to Karen M. Ottemann for the generous gift of the H. pylori strain PMSS1-RFP.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was funded by the German Research Foundation (DFG), SFB914 projects B05 (R.H.), SFB914 project Z01 (H.I-A. and S.M.) and A10 (C.S.), TRR359 “PILOT – Perinatal Development of Immune Cell Topology” Project ID 491676693, (C.S.), Else Kröner-Fresenius-Stiftung, German Centre for Cardiovascular Research (DZHK) - Start-Up Grant, (L.T.) and TRR332 project C3 (B.W. and D.M.B) as well as the ERC AdvGrant “Immunothrombosis” #833440 (S.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8(2):e180–e90. Epub 2019/12/22. doi: 10.1016/S2214-109X(19)30488-7 . [DOI] [PubMed] [Google Scholar]
- 2.Herrero R, Park JY, Forman D. The fight against gastric cancer—the IARC Working Group report. Best Pract Res Clin Gastroenterol. 2014;28(6):1107–14. Epub 2014/12/03. doi: 10.1016/j.bpg.2014.10.003 . [DOI] [PubMed] [Google Scholar]
- 3.Fischer W, Püls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Molecular Microbiology. 2001;42(5):1337–48. [DOI] [PubMed] [Google Scholar]
- 4.Rieder G, Hatz RA, Moran AP, Walz A, Stolte M, Enders G. Role of adherence in interleukin-8 induction in Helicobacter pylori-associated gastritis. Infection and Immunity. 1997;65(9):3622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Königer V, Holsten L, Harrison U, Busch B, Loell E, Zhao Q, et al. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat Microbiol. 2016;2:16188–99. [pii]; doi: 10.1038/nmicrobiol.2016.188 [DOI] [PubMed] [Google Scholar]
- 6.Behrens IK, Busch B, Ishikawa-Ankerhold H, Palamides P, Shively JE, Stanners C, et al. The HopQ-CEACAM Interaction Controls CagA Translocation, Phosphorylation, and Phagocytosis of Helicobacter pylori in Neutrophils. mBio. 2020;11(1). Epub 2020/02/06. doi: 10.1128/mBio.03256-19 ; PubMed Central PMCID: PMC7002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301(5636):1099–102. [DOI] [PubMed] [Google Scholar]
- 8.Djekic A, Muller A. The Immunomodulator VacA Promotes Immune Tolerance and Persistent Helicobacter pylori Infection through Its Activities on T-Cells and Antigen-Presenting Cells. Toxins (Basel). 2016;8(6). Epub 2016/06/21. doi: 10.3390/toxins8060187 ; PubMed Central PMCID: PMC4926153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oertli M, Noben M, Engler DB, Semper RP, Reuter S, Maxeiner J, et al. Helicobacter pylori gamma-glutamyl transpeptidase and vacuolating cytotoxin promote gastric persistence and immune tolerance. Proc Natl Acad Sci U S A. 2013;110(8):3047–52. 1211248110 [pii]; doi: 10.1073/pnas.1211248110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidinger B, Petri K, Lettl C, Li H, Namineni S, Ishikawa-Ankerhold H, et al. Helicobacter pylori binds human Annexins via Lipopolysaccharide to interfere with Toll-like Receptor 4 signaling. PLoS Pathog. 2022;18(2):e1010326. Epub 2022/02/18. doi: 10.1371/journal.ppat.1010326 ; PubMed Central PMCID: PMC8890734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102(26):9247–52. doi: 10.1073/pnas.0502040102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada T, Takahashi S, Ishida A, Ishigame H. In vivo multiphoton imaging of immune cell dynamics. Pflugers Arch. 2016;468(11–12):1793–801. Epub 20160922. doi: 10.1007/s00424-016-1882-x ; PubMed Central PMCID: PMC5138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147(5):983–91. doi: 10.1016/j.cell.2011.11.004 ; PubMed Central PMCID: PMC3824153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen LA. The role of the neutrophil and phagocytosis in infection caused by Helicobacter pylori. Curr Opin Infect Dis. 2001;14(3):273–7. [DOI] [PubMed] [Google Scholar]
- 15.Vladymyrov M, Abe J, Moalli F, Stein JV, Ariga A. Real-time tissue offset correction system for intravital multiphoton microscopy. Journal of immunological methods. 2016;438:35–41. Epub 2016/09/04. doi: 10.1016/j.jim.2016.08.004 . [DOI] [PubMed] [Google Scholar]
- 16.Lasch M, Vladymyrov M, van den Heuvel D, Gotz P, Deindl E, Ishikawa-Ankerhold H. Multiphoton Intravital Imaging for Monitoring Leukocyte Recruitment during Arteriogenesis in a Murine Hindlimb Model. J Vis Exp. 2021;(175). Epub 2021/10/19. doi: 10.3791/62969 . [DOI] [PubMed] [Google Scholar]
- 17.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96(2):719–26. Epub 2000/07/11. . [PubMed] [Google Scholar]
- 18.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20(11):4106–14. Epub 2000/05/11. doi: 10.1128/MCB.20.11.4106-4114.2000 ; PubMed Central PMCID: PMC85780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dagher NN, Najafi AR, Kayala KM, Elmore MR, White TE, Medeiros R, et al. Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J Neuroinflammation. 2015;12:139. Epub 2015/08/02. doi: 10.1186/s12974-015-0366-9 ; PubMed Central PMCID: PMC4522109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng PY, Jones NL. Helicobacter pylori strains expressing the vacuolating cytotoxin interrupt phagosome maturation in macrophages by recruiting and retaining TACO (coronin 1) protein. Cell Microbiol. 2003;5(1):25–40. [DOI] [PubMed] [Google Scholar]
- 21.Arnold IC, Zhang X, Urban S, Artola-Boran M, Manz MG, Ottemann KM, et al. NLRP3 Controls the Development of Gastrointestinal CD11b(+) Dendritic Cells in the Steady State and during Chronic Bacterial Infection. Cell Rep. 2017;21(13):3860–72. Epub 2017/12/28. doi: 10.1016/j.celrep.2017.12.015 . [DOI] [PubMed] [Google Scholar]
- 22.Wang YH, Wu JJ, Lei HY. When Helicobacter pylori invades and replicates in the cells. Autophagy. 2009;5(4):540–2. Epub 20090513. doi: 10.4161/auto.5.4.8167 . [DOI] [PubMed] [Google Scholar]
- 23.Kaparakis M, Walduck AK, Price JD, Pedersen JS, van Rooijen N, Pearse MJ, et al. Macrophages are mediators of gastritis in acute Helicobacter pylori infection in C57BL/6 mice. Infect Immun. 2008;76(5):2235–9. Epub 20080310. doi: 10.1128/IAI.01481-07 ; PubMed Central PMCID: PMC2346689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toda G, Soeda K, Okazaki Y, Kobayashi N, Masuda Y, Arakawa N, et al. Insulin- and Lipopolysaccharide-Mediated Signaling in Adipose Tissue Macrophages Regulates Postprandial Glycemia through Akt-mTOR Activation. Mol Cell. 2020;79(1):43–53 e4. Epub 20200527. doi: 10.1016/j.molcel.2020.04.033 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LysM-GFP mouse was infected with H. pylori-RFP (red), vessels are visible with the blue color (second harmonic generation; SHG). After 40 min of recording the tissue has drift from the field of view without the drift correction, when the VivoFollow was applied the image was in focus.
(MP4)
There is a reduced number of neutrophils in the non-infected stomach in comparison with the infected mice (S3 Video). CX3CR1-eGFP (macrophages), neutrophils are labelled by the Ly6G-PE antibodies injected (i.v.) into the mice 15 min before imaging. Stomach crypts are labeled blue by the SHG effect.
(MP4)
There is an increased number of neutrophils in the 24h infected stomach in comparison with the non-infected mice (S2 Video). LysM-eGFP (neutrophils), H. pylori-RFP (red) inside the crypts. Vessels are labelled by the SHG.
(AVI)
There is an increased speed of the neutrophils in comparison with 24h of infection (see S3 Video for comparison). LysM-eGFP (neutrophils), H. pylori-RFP (red). Vessels are labelled by the SHG.
(AVI)
Multiphoton imaging of the CX3CR1-GFP gastric mucosa was infected with H. pylori-RFP (red) for 24h, macrophage (green), vessels are visible with the blue color (second harmonic generation; SHG), second part of the video show the 3D rendering of macrophage and H. pylori.
(MP4)
CX3CR1-GFP mouse infected with H. pylori for 24h. Macrophages (green) show cytoplasmic extensions and H. pylori phagocytized (red). The second part of the movie show the imaging 3D reconstruction and rendering.
(MP4)
CX3CR1-GFP mouse infected with H. pylori for 6 weeks. Macrophages (green) show cytoplasmic extensions and H. pylori phagocytized (red). The second part of the movie show the imaging 3D reconstruction and rendering.
(AVI)
LysM-eGFP (yellow) mice gastric mucosa infected with H. pylori for 24h (magenta). 3D projection shown neutrophils localized at the periphery of the crypts and H. pylori are located inside the crypts (magenta). Nucleus are displayed in blue.
(AVI)
Bone marrow isolated macrophages were co-cultivated with PMSS1-RFP H. pylori strain (red) and after 6h of incubation the infected macrophages were fixed and labelled with anti-F4/80- Alexa 488 (green). 3D rendering projection shows the internalization of the bacteria by macrophages. Scale bar is 10 μm.
(MP4)
Bone marrow isolated macrophages were co-cultivated with P12-GFP H. pylori strain (green) and after 6h of incubation the infected macrophages were fixed and labelled with anti-F4/80- Alexa 594 (red). 3D rendering projection shows the internalization of the bacteria by macrophages. Scale bar is 10 μm.
(MP4)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
3D projection image sequences showing the localization of PMMS1-RFP (A) or P12-GFP (B) after 6h of infection inside macrophages. The images series are displayed from different angles representing the raw and unprocessed data used for 3D rendering of Fig 8 and S9 and S10 Videos. Scale bar is 10μm. DAPI is labelling the nucleus. Anti-F4/80 antibodies were used to label macrophages.
(TIFF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.