Abstract
The nonpolyadenylated mRNAs of rotavirus are templates for the synthesis of protein and the segmented double-stranded RNA (dsRNA) genome. During serial passage of simian SA11 rotaviruses in cell culture, two variants emerged with gene 5 dsRNAs containing large (1.1 and 0.5 kb) sequence duplications within the open reading frame (ORF) for NSP1. Due to the sequence rearrangements, both variants encoded only C-truncated forms of NSP1. Comparison of these and other variants encoding defective NSP1 with their corresponding wild-type viruses indicated that the inability to encode authentic NSP1 results in a small-plaque phenotype. Thus, although nonessential, NSP1 probably plays an active role in rotavirus replication in cell culture. In determining the sequences of the gene 5 dsRNAs of the SA11 variants and wild-type viruses, it was unexpectedly found that their 3′ termini ended with 5′-UGAACC-3′ instead of the 3′ consensus sequence 5′-UGACC-3′, which is present on the mRNAs of nearly all other group A rotaviruses. Cell-free assays indicated that the A insertion into the 3′ consensus sequence interfered with its ability to promote dsRNA synthesis and to function as a translation enhancer. The results provide evidence that the 3′ consensus sequence of the gene 5 dsRNAs of SA11 rotaviruses has undergone a mutation causing it to operate suboptimally in RNA replication and in the expression of NSP1 during the virus life cycle. Indeed, just as rotavirus variants which encode defective NSP1 appear to have a selective advantage over those encoding wild-type NSP1 in cell culture, it may be that the atypical 3′ end of SA11 gene 5 has been selected for because it promotes the expression of lower levels of NSP1 than the 3′ consensus sequence.
Rotavirus virions are icosahedral particles consisting of three layers of protein and containing 11 segments of double-stranded RNA (dsRNA) (8). The innermost protein layer has a T=2 arrangement and is formed by the core lattice protein VP2 (reviewed in reference 29). Associated with the interior side of each of the 12 pentamers of the VP2 lattice is believed to be one copy each of the viral RNA-dependent RNA polymerase (RdRP) VP1 and the mRNA-capping enzyme VP3 (19, 20). Based on structural studies of rotaviruses and other members of the family Reoviridae (29), it is thought that each genome segment exists as a tightly wound spiral around one of the 12 RdRP-capping complexes of the VP2 lattice (9). Collectively, the VP2 lattice, the RdRP-capping complexes, and the dsRNA genome make up the core of the virion. Double-layered particles consisting of cores surrounded by the intermediate protein VP6 have transcriptase activity and are responsible for the synthesis of the 11 viral mRNAs (1, 37). The dsRNA genome probably exists as a liquid crystal within the core and, in this form, has the fluidity necessary for the dsRNA segments to slide through the anchored RdRP-capping enzyme complex during transcription (9). Nascent transcripts produced by the viral RdRP are extruded through channels located at the vertices of double-layered particles (18).
Translation of the viral mRNAs yields six structural (VP1 to VP4, VP6, and VP7) and six nonstructural (NSP1 to NSP6) proteins (8). In the viral life cycle, the mRNAs are also used by the viral RdRP as templates for the synthesis of minus-strand RNAs, resulting in the formation of dsRNAs (3, 29). The rotavirus mRNAs are unique in that, although they possess methylated 5′ caps, they lack 3′ poly(A) tails (15, 23). Except for short sequences at their 5′ and 3′ termini, the 11 viral mRNAs exhibit no sequence similarity. The 3′-terminal sequence of the mRNAs of the group A rotaviruses has consistently been reported to be 5′-UGACC-3′ (8). Several studies have illustrated the importance of this 3′ consensus sequence in the viral life cycle. In particular, template-dependent cell-free replication systems have shown that the 3′ consensus sequence contains a cis-acting signal that is essential for the viral mRNA to function efficiently as a template for minus-strand synthesis (31, 42). Moreover, the last four nucleotides of the conserved sequence form a signal (3′ translation enhancer) that enhances translation of the viral mRNAs (4). The upregulation of gene expression is mediated by NSP3, a protein which is believed to cause circularization of viral mRNAs in polysomes by simultaneously binding to the 3′ consensus sequence and to the cap binding protein eIF4G (34, 35, 41).
Rotaviruses with atypical genome segments have been recovered from animals and immunocompromised children and have been generated by serial passage of the virus at high multiplicity of infection (MOI) in cell culture (7, 13, 40). In most cases, molecular analysis has shown that the atypical genome segments contain a head-to-tail duplication of sequences within the dsRNA (5, 14, 24, 32, 36). Such sequence rearrangements have been identified for segments 5 to 7, 10, and 11 (6). So far, the sequence duplications that have been described for segments 6, 7, 10, and 11 begin after the open reading frame (ORF) and, as a result, the rearrangement does not affect the ability of the RNAs to encode full-length proteins. In contrast, most of the sequence duplications described for segment 5 begin within the ORF and alter the coding capacity of the gene such that it no longer encodes wild-type NSP1 (10, 40). Other atypical genome segments 5 have been identified which, instead of containing duplicated sequences, contain sequence deletions or nonsense mutations within the ORF that also prevent the gene from encoding wild-type NSP1 (38, 40). The fact that rotaviruses can replicate in cell culture, even when unable to produce wild-type NSP1, has led to the suggestion that the protein is nonessential. The role of NSP1 in the viral life cycle is uncertain, but the protein is known to bind RNA and to be associated with the cytoskeleton fraction of infected cells (11).
In this study, we have characterized two variants of simian rotavirus SA11 whose gene 5 dsRNAs contain rearrangements stemming from duplication of sequences located within the NSP1 ORF. The rearrangements altered the ORFs such that they encoded C-truncated NSP1. Like other NSP1-defective isolates, the two variants displayed a small-plaque phenotype, suggesting that NSP1 contributes to the viral life cycle even in cell culture. Remarkably, sequencing also revealed that the 3′ termini of the segment 5 dsRNAs of the variants and of wild-type SA11 were atypical (UGAACC) in that they contained an A insertion relative to the expected 3′ consensus sequence (UGACC). The fact that the wild-type SA11 viruses grew to high titer and produced NSP1 demonstrated that the 3′ consensus sequence is not required for genome packaging, RNA replication, or viral gene expression. However, in vitro assays indicated that the A insertion altered the 3′ consensus sequence such that it operated suboptimally in replication and gene expression.
MATERIALS AND METHODS
Isolation of rotavirus variants 30-1A and 5S.
The simian SA11 strain SA11-FEM (26) was received from G. W. Gary (Centers for Disease Control and Prevention, Atlanta, Ga.), and the SA11-4F strain (22, 33) was received from R. L. Ward (Children's Hospital Medical Center, Cleveland, Ohio), who had obtained the strain from M. K. Estes (Baylor College of Medicine, Houston, Tex.). Both of these SA11 strains carry a bovine rotavirus-like VP4 gene, and both were originally provided by H. H. Marlherbe (21). The simian SA11 strains of rotavirus SA11-FEM and SA11-4F were serially passaged without dilution in MA104 cells. The gene 5 variant 30-1A and its wild-type correlate 30-19 were isolated by triple-plaque purification from the passage 30 lysate of SA11-FEM-infected cells, and the gene 5 variant 5S was obtained by limiting dilution from the passage 26 lysate of SA11-4F-infected cells. Viruses were propagated in the presence of 0.5 μg of trypsin per ml, and titers were determined by plaque assay on MA104 cells (30). The diameter of plaques was measured 5 days post-infection (p.i.).
Genomic dsRNAs were purified from rotavirus particles by phenol-chloroform extraction and ethanol precipitation. The genotypes of the viruses were examined by electrophoresis of purified dsRNA on 12% polyacrylamide gels containing sodium dodecyl sulfate (SDS–12% PAGE), followed by staining with ethidium bromide (30).
cDNA cloning and sequencing of gene 5 dsRNAs.
To prepare full-length gene 5 cDNAs, purified dsRNAs were denatured by resuspension in water and heating to 95°C for 5 min or by resuspension in 90% dimethyl sulfoxide and heating to 65°C for 10 min. First-strand cDNAs were synthesized by incubating the denatured RNA with Superscript II reverse transcriptase (Life Technologies) and the primers 5′-ggcttttttttgaaaagtcttgtgttagcc-3′ and 5′-ggttcacagtattttgccagctaggcgc-3′. The cDNAs were amplified by PCR using Elongase DNA polymerase (Life Technologies) and the same primers. PCR products of the appropriate size were gel purified and ligated into the vector pT7Blue (Novagen). Following transformation of Escherichia coli DH5α, bacteria containing the appropriate plasmids (pT7-gene 5) were identified based on antibiotic resistance, plasmid size, and restriction enzyme digestion. Plasmids containing full-length gene 5 cDNA inserts were purified with Concert maxiprep kits (Life Technologies). Due to the presence of duplicated sequences in the rearranged gene 5 cDNAs, subclones were prepared that contained fragments of the gene 5 cDNAs of 30-1A and 5S pT7-gene 5. The subclones were generated by digesting 30-1A and 5S pT7-gene 5 with EcoRV and with EcoRI, EcoRV, or EcoRV and PstI, respectively, and ligating the gene 5-specific fragments into plasmid SP72 or SP65 (Promega).
Sequences of the 5′ and 3′ termini of the gene 5 dsRNAs were determined from cDNA clones obtained by rapid amplification of cDNA 5′ ends (5′ RACE system, version 2.0; Life Technologies). Terminal cDNAs were prepared by incubating reaction mixtures containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2.5 mM MgCl2, 10 mM dithiothreitol, the four deoxynucleoside triphosphates at 400 μM each, 0.5 μg of denatured dsRNA, the appropriate primer at 100 nM, and 200 U of Superscript II reverse transcriptase for 50 min at 45°C. The primers used to generate the gene 5 plus-strand cDNAs of 30-19 and 30-1A and of 4F and 5S were 5′-gcgcactttgaatgtagatagc-3′ and 5′-gggagcagcaaatgattatacc-3′, respectively. The primers used to generate the gene 5 minus-strand cDNAs of 30-19 and 30-1A and of 4F and 5S were 5′-attttctcatcaggaactgg-3′ and 5′-cacatggatcaaacaattgaag-3′, respectively. The cDNAs were tailed by incubation with 10 U of terminal deoxynucleotidyl transferase (Life Technologies) and 200 μM dATP. Subsequently, the tailed cDNAs were amplified by PCR using the poly(T)-containing anchor primer 5′-cggctcgagtttttttttttttttttttt-3′ (XhoI site underlined) and gene 5-specific primers (5′-taaggatccaatgtcaataaaatcaacggagg-3′ and 5′-taaggatccgcaaatgaggttcattgtcaagg-3′ for the plus-strand cDNAs of 30-19 and 30-1A and of 4F and 5S, respectively, or 5′-ctgggatccaaatagaatttgcaccaatgttgc-3′ and 5′-atcggatcccgccagtgtcagttgaaggta-3′ for the minus-strand cDNAs of 30-19 and 30-1A and of 4F and 5S, respectively (BamHI sites underlined). The cDNA products were purified, digested with XhoI and BamHI, and ligated into the XhoI/BamHI sites of pUC19.
Gene 5 sequences were obtained either with an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems) or with a Sequenase kit (Amersham Life Science) and suitable oligonucleotide primers.
Viral protein synthesis in infected cells.
Monolayers of MA104 cells were mock infected or infected with the 30-19, 30-1A, 4F, or 5S strain of rotavirus. Beginning at 1 h p.i., the cells were maintained in 80% Met- and Cys-free minimal essential medium containing 5 μg of actinomycin D per ml, and at 3 h p.i., 35S-Express (1,175 mCi per mmol; NEN) was added to the medium to a final concentration of 17 μCi/ml. At 8 h p.i., the monolayers were rinsed twice with cold, dilute reticulocyte standard buffer (RSB; 3 mM Tris-HCl [pH 8.5], 0.5 mM MgCl2, 3 mM NaCl), and then the cells were scraped into dilute RSB containing 1% Triton X-100. After gentle mixing, nuclei and the cytoskeleton fraction were pelleted from the lysates by centrifugation at 3,200 × g for 2 min. The pellets were resuspended in dilute RSB and, along with the supernatant (cytosol), stored at −70°C. Proteins in the pellets and supernatants were analyzed by SDS–10% PAGE and autoradiography.
For Western blot analysis, proteins in the soluble fraction were resolved by SDS–10% PAGE (10% NuGels; In Vitrogen) and then electroblotted onto a nitrocellulose sheet. The blot was blocked by soaking for 1 h in phosphate-buffered saline containing 5% skim milk. Subsequently, the blot was incubated overnight with a 1:1,000 dilution of rabbit anti-SA11 NSP1 C19 serum (11). Alkaline phosphatase-conjugated goat anti-rabbit antibody was used as the secondary antibody at a dilution of 1:5,000. The blots were developed with 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt and nitroblue tetrazolium chloride (Life Technologies).
Open-core replicase assays.
Open cores were prepared from DS1 × RRV virions and treated with micrococcal nuclease to remove endogenous dsRNA (2, 30). Reaction mixtures contained 50 mM Tris-HCl (pH 7.2), 5 mM MgCl2, 5 mM dithiothreitol, 20 U of RNasin (Promega), the four deoxynucleoside triphosphates at 200 μM each, 10 μCi of [α-32P]UTP, and the indicated amount of plus-strand template RNA and open cores. The reaction mixtures were incubated at 32°C for 2 h, and the 32P-labeled dsRNA products were resolved by SDS–12% PAGE and detected by autoradiography. A PhosphorImager was used to quantify the amount of 32P-labeled dsRNA.
Preparation of gene 5 mRNAs.
To generate first-strand gene 5 cDNAs, SA11-4F dsRNAs were denatured by heating to 100°C for 2 min and incubated with Superscript II reverse transcriptase, the plus-sense primer 5′-agctctagaccgcGGCTTTTTTTTGAAAAGTCTTGTG-3′, and the minus-sense primer 5′-agctctagaccgcGGTTCACAGTATTTTGCCAGC-3′ (XhoI sites are underlined, and viral sequences are in uppercase). The cDNAs were then amplified by PCR using the same primers and pfu DNA polymerase (Stratagene). Afterwards, the PCR product was digested with XbaI and ligated into the XbaI site of the plasmid pUC19, producing the construct pUC4F5. Templates for the synthesis of SA11 gene 5 mRNAs containing the 3′-terminal sequences UGACC and UGAACC were generated by PCR. The amplification mixtures included pfu DNA polymerase, the plus-sense primer 5′-cgcggatcctaatacgactcactataGGTTTTTTTTGAAAAGTCTTG-3′ (T7 promoter is underlined), and the minus-sense primer 5′-GGTCACAGTATTTTGCCAGC-3′(UGACC) or 5′-GGTTCACAGTATTTTGCCAGC-3′ (UGAACC). The mixtures also included the gene 5 cDNA insert released from pUC4F5 by digestion with XbaI. The PCR products were sequenced to ensure that they contained the desired terminal sequences. Transcripts were made from the PCR templates with an Ambion MEGAscript T7 transcription kit according to the protocol of the manufacturer.
Construction of the NSP3 expression vector.
Purified SA11 dsRNAs were denatured by resuspension in 90% dimethyl sulfoxide and heating to 95°C for 2 min. The NSP3 ORF in genome segment 7 was amplified from the denatured dsRNAs using the Titan One Tube Reverse Transcriptase-PCR System (Boehringer Mannheim), the plus-sense primer 5′-gcttttcagatcttgATGCTCAAGATGGAGTCT-3′, and the minus-sense primer 5′-gtatttgatagatctACATGTATCAAAATGGTT-3′ (BglII sites are underlined, and viral sequences are in uppercase). The amplified product was gel purified, digested with BglII, and ligated into isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pQE32 (Qiagen), which had been digested with BamHI and dephosphorylated with alkaline phosphatase (Life Technologies). Following transformation into E. coli DH5α, bacteria with the appropriate plasmid (pQE32g7) were identified based on antibiotic resistance, plasmid size, and restriction enzyme digestion. The expected sequence of pQE32g7 was confirmed by automated sequencing. The plasmid pQE32g7 was then electroporated into E. coli M15 carrying the pREP4 repressor plasmid. In pQE32g7, the NSP3 ORF is situated downstream of sequential codons for Arg, His, Ser, and six His residues.
Expression and purification of rNSP3.
E. coli M15 (pREP4) containing pQE32g7 was grown to an A600 of 0.5 in Terrific Broth (Quality Biologics), and the expression of NSP3 was induced by adding IPTG to a final concentration of 1 mM. After incubation for 4 h at 37°C, the bacteria were recovered by centrifugation at 4,000 × g for 10 min and lysed by resuspension in buffer containing 0.5 mM NaH2PO4 (pH 8), 300 mM NaCl, 5 mM imidazole, 1% Triton X-100, and 1 mg each of lysozyme, aprotinin, and leupeptin per ml. The lysate was sonicated and centrifuged at 10,000 × g for 30 min. Recombinant NSP3 (rNSP3) was purified from the pellet according to the protocol of Piron et al. (34). Briefly, the pellet was washed in 20 mM Tris-HCl (pH 8)–500 mM NaCl–5 mM imidazole and then solubilized overnight in the same buffer containing 8 M urea. rNSP3 was purified from the solubilized proteins with a Ni-nitrilotriacetic acid agarose column (Qiagen) and eluted with solubilization buffer, pH 4.5, containing 8 M urea. The NSP3 sample was then renatured by dialysis against 50 mM Tris-HCl (pH 8.0)–150 mM NaCl–10% glycerol–0.5 mM oxidized glutathione–5 mM reduced glutathione for 18 h at 4°C. The renatured rNSP3 was dialyzed against low-salt buffer (2 mM Tris-HCl [pH 7.2], 0.5 mM EDTA, 0.5 mM dithiothreitol) for 48 h at 4°C. The concentration of the purified protein was determined by Bradford assay using bovine serum albumin as the protein standard and by comparison with known amounts of bovine serum albumin coelectrophoresed on SDS-polyacrylamide gels and stained with Coomassie brilliant blue R-250.
Preparation of RNA probes.
The DNA templates for synthesis of the 32P-labeled RNA probes, v40-GACC and v40-GAACC, were generated by amplification with the Expand High Fidelity PCR System (Boehringer Mannheim). The amplification reaction mixtures contained the plasmid SP72-v40 (2), the plus-sense primer 5′-taatacgactcactataG-3′, and the minus-sense primer 5′-GGTCACATAAGCGCTTTC-3′ or 5′-GGTTCACATAAGCGCTTTC-3′ (T7 promoter is underlined, and viral sequences are in uppercase). SP72-v40 contains a sequence corresponding to the last 40 nucleotides (nt) of the SA11 gene 8 mRNA positioned downstream of the promoter for T7 RNA polymerase. The template for the synthesis of v40Δ3′-11 was made by digesting SP72-v40 with Eco47III, which cleaves the gene 8-specific sequence in the vector 11 residues upstream from its 3′ end. The 32P-labeled probes were made by runoff transcription using the Ambion MEGAshortscript kit according to the protocol of the manufacturer, except that the concentration of unlabeled UTP was reduced by one-fourth and 50 μCi of [α-32P]UTP was included per 20 μl of reaction mixture. After runoff transcription, the RNA products were purified by phenol-chloroform extraction and isopropanol precipitation. The 32P-labeled RNA probes were purified by electrophoresis and elution from 8% polyacrylamide gels containing 7 M urea (28). RNA concentrations were calculated from optical densities at 260 nm.
The sequence of the 72-nt v40-GACC RNA was 5′-GGGAGACCGGCAGAU C U GA UAUCAU C GAU GAAU U GA U G A U G G C U U A G C A A G A A U A G A AAGCGCUUAUGUGACC-3′ (2). The underlined portion of the sequence represents the 3′-terminal 40 nt of the SA11 gene 8 mRNA. Except for ending with GAACC instead of GACC, the sequence of the 73-nt v40-GAACC RNA was the same as that of the v40-GACC RNA. Although otherwise identical, the v40Δ3′-11 RNA lacks the last 11 nt of the v40-GACC RNA.
Gel shift assays.
The procedure used for analysis of rNSP3-RNA interactions by gel shift assay was similar to that described earlier (39). One pmol (∼24 ng) of the 32P-labeled RNA probe v40-GACC, v40-GAACC, or v40Δ3′-11 was incubated with 5 pmol of rNSP3 in low-salt buffer in a final volume of 10 μl for 30 min at room temperature. The reaction mixtures were analyzed by electrophoresis for 2.5 h at 175 V on nondenaturing 8% polyacrylamide gels containing 50 mM Tris-HCl and 50 mM glycine (pH 8.8). Protein-probe complexes were detected on the gel by autoradiography, and the intensities of the radiolabeled bands were measured with a PhosphorImager. The percent binding activities of rNSP3 for probes v40-GACC and v40-GAACC were calculated by dividing the intensity value obtained for the shifted probe with the combined intensity values obtained for the free probe and shifted probe and then multiplying the result by 100.
Preparation of g6-Fluc chimeric RNAs.
The plasmid pVec2.0g6-Fluc contains the ORF for firefly luciferase situated between the 5′ and 3′ untranslated regions (UTRs) of the SA11 gene 6 RNA (4). The T7 transcription templates used to produce the gene 6 analog RNAs g6-Fluc-UGACC, g6-Fluc-UGAACC, glo/g6-Fluc-UGACC, and glo/g6-Fluc-UGAACC were synthesized with the High Fidelity PCR System (Roche Molecular Biochemicals). The reaction mixtures contained pVec2.0g6-Fluc, the plus-sense primer 5′-cccaggtaccctaatacgactcactataGGCTTTTAAACGAAGTCTTCACCATG-3′ (gene 6 5′ UTR) or 5′-cccaggtaccctaatacgactcactatagacacttgcttctgacacacaccATGGAAGACGCCAAAAACATAAAG-3′ (β-globin 5′ UTR), and the minus-sense primer 5′-GGTCACATCCTCTCACTATACCATC-3′ (gene 6 3′ UTR, UGACC) or 5′-GGTTCACATCCTCTCACTATACCATC-3′ (gene 6 3′ UTR, GAACC). In the primers, the T7 promoters are underlined and the viral sequences are in uppercase. Capped mRNAs were generated from the PCR products using the Ambion mMESSAGE transcription system. The quality of the mRNAs was assessed by electrophoresis on 5% polyacrylamide gels containing 7 M urea (30).
To produce capped nv-Rluc RNA, the T7 transcription vector pRL-null (Promega) was linearized with BamHI and then transcribed as described above. The transcripts contain the ORF for Renilla luciferase.
Luciferase expression in infected cells.
Capped viral analog RNAs encoding firefly luciferase and nonviral RNAs encoding Renilla luciferase were cotransfected into rotavirus-infected MA104 cells at 1 h p.i. using lipofectAMINE (Life Technologies) (4). At 9 h p.i., the cells were lysed in 1× Passive Lysis Buffer (provided in the Dual Luciferase Kit [Promega]). The levels of firefly and Renilla luciferase activities in the lysates were assayed with a Turner TD-20E luminometer (4).
Nuclestide sequence accession numbers.
The GenBank accession numbers of the gene 5 cDNAs are AF290881 (30-19), AF290882 (30-1A), AF290883 (4F), and AF290884 (5S).
RESULTS
SA11 variants with novel genotypes.
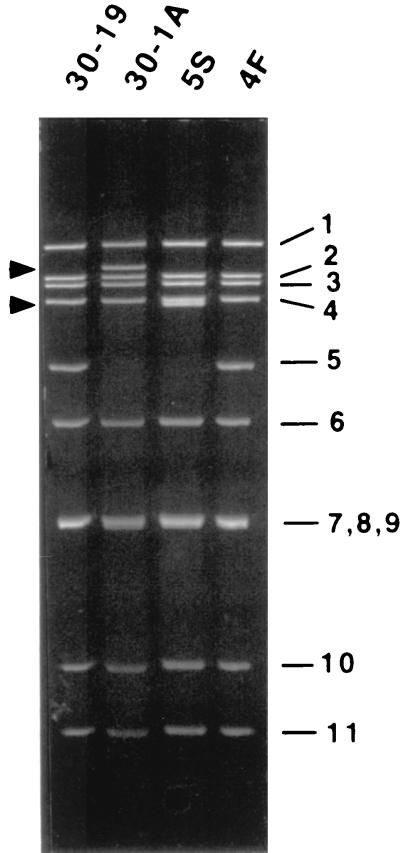
Rotavirus strains SA11-FEM and SA11-4F were serially passaged greater than 20 times at high MOI in MA104 cells. Following the emergence of one or more novel dsRNAs in the virus population, the variants 30-1A and 5S were isolated. 30-1A was derived from SA11-FEM, and its genome lacked a wild-type gene 5 dsRNA but contained a large novel dsRNA migrating between gene 1 and 2 dsRNAs upon PAGE (Fig. 1). In comparison, 5S was derived from SA11-4F and its genome, which also lacked a wild-type gene 5 dsRNA, contained a large, novel dsRNA migrating slightly faster than gene 4 dsRNA. Except for the absence of the wild-type gene 5 dsRNAs and the presence of the novel dsRNAs, the genome segments of the variants 30-1A and 5S comigrated precisely with the genome segments of their wild-type counterparts 30-19 and 4F, respectively (Fig. 1).
FIG. 1.
Genome segments of wild-type viruses 30-19 and 4F and variants 30-1A and 5S. RNAs were recovered from the viruses by phenol-chloroform extraction and ethanol precipitation, resolved by SDS-PAGE, and detected by staining with ethidium bromide. The arrows denote the positions of aberrant genome segments for 30-1A and 5S.
Duplication of sequences in gene 5 dsRNAs.
Reverse transcription-PCR was used to prepare cDNAs of the gene 5 dsRNAs of the variants 30-1A and 5S and the wild-type viruses 30-19 and 4F. Nucleotide sequencing of the cDNAs showed that the gene 5 dsRNAs of the wild-type viruses 30-19 and 4F were 1614 and 1610 nt in length, respectively, and shared 98 to 99% identity with the previously reported sequences of the SA11 gene 5 dsRNA (12, 25). The wild-type gene 5 dsRNAs contained single long ORFs of 1,488 nt (residues 31 to 1518), 5′ UTRs of 30 nt, and 3′ UTRs of 96 nt for 30-19 and 92 nt for 4F. The protein product, NSP1, encoded by the wild-type gene 5 dsRNAs was predicted to be of 496 amino acids (aa) (Fig. 2), to have a molecular mass of 59 kDa and to share 97 to 98% identity with the NSP1 encoded by other SA11 rotaviruses (12, 25).
FIG. 2.
Alignment of the predicted amino acid sequences of NSP1 encoded by the gene 5 dsRNAs of wild-type viruses 30-19 and 4F and variants 30-1A and 5S. Sequence identity is indicated with dots.
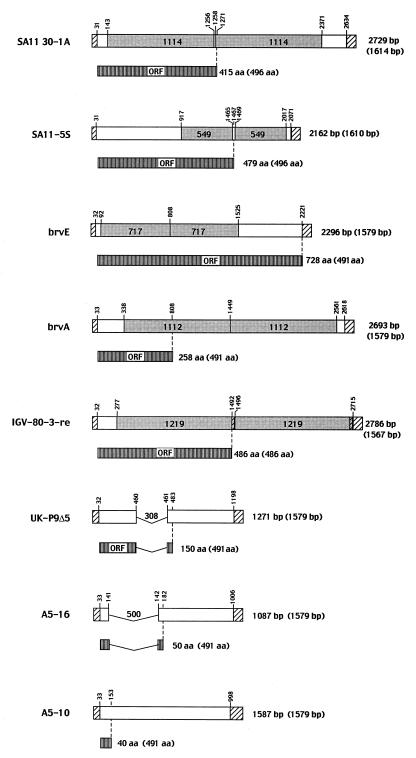
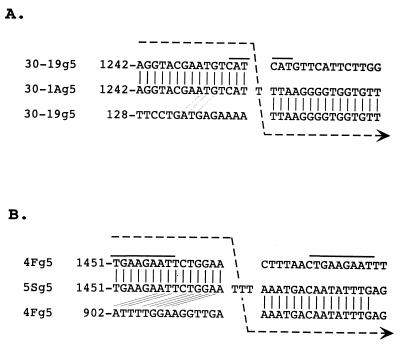
The gene 5 dsRNA of the 30-1A variant was 2,729 nt in length and contained a 1,114-nt duplication of the sequence from residues 143 to 1256 of the 30-19 gene 5 dsRNA (Fig. 3). In comparison to the three other gene 5 dsRNAs containing sequence duplications which have been molecularly described, i.e., brvA (10), brvE (40), and IGV-80-3-re (17) (Fig. 3), the overall length of the 30-1A gene 5 dsRNA is the second longest. In the 30-1A gene 5 dsRNA, the copies of the duplicated sequences are located at residues 143 to 1256 and 1258 to 2371 and are separated by a single T residue (Fig. 4). The origin of the T is not known, but based on comparison with the sequence of the 30-19 gene 5 dsRNA, the residue was not specified by the template strand during RNA synthesis and, therefore, was likely generated by an error of the RdRP. The presence of the T at the junction site causes a shift in the reading frame, which results in the addition of five incorrect amino acids to the growing NSP1 polypeptide and in its premature termination at nt 1271. The C-truncated NSP1 encoded by 30-1A gene 5 dsRNA is 414 aa in length (Fig. 2), has a molecular mass of 49 kDa, and lacks the last 82 aa of wild-type NSP1.
FIG. 3.
Sequence organization of aberrant gene 5 dsRNAs of rotavirus. The lengths of the aberrant gene 5 dsRNAs and their NSP1 products are given, as well as the corresponding values for wild-type gene 5 dsRNAs and their NSP1 products (in parentheses). Regions representing the 5′ and 3′ UTRs of the wild-type dsRNA are shown with diagonal lines. The sizes and positions of duplicated sequences in the gene 5 dsRNAs of SA11 30-1A, SA11-5S, brvE, brvA, and IGV-80-3-re and deleted sequences in the gene 5 dsRNAs of UK-P9Δ5 and A5-16 and the locations of nonsense mutations in the gene 5 dsRNAs of brvA and A5-10 are indicated. The accession numbers for the gene 5 homologs are AF290882 (SA11 30-1A), AF290884 (SA11-5S), Z24735 (brvE, junction sequence only), L12248 (brvA), AF190169 (IGV-80-3-re), Z24736 (UK-P9Δ5), D38149 (A5-16), and D38147 (A5-10).
FIG. 4.
Junction of the duplicated sequences in the gene 5 dsRNAs of SA11 30-1A and SA11-5S. The junction of the duplicated sequences includes one (30-1A) or three (5S) nontemplated T residues. Aligned with each junction are the sequences of the wild-type gene 5 dsRNAs of SA11 30-19 or SA11-4F. The alignment indicates the potential sites where the viral RdRP and nascent RNA dissociated from the wild-type RNA template and then, at an upstream site on the same template, reassociated and reinitiated RNA synthesis using the nascent RNA as a primer. The arrowed line illustrates the movement of the RdRP along the wild-type sequence and the synthesis of the nontemplated T residues required to generate the junction sequence during plus-strand synthesis. Repeated sequences near the site where the RdRP is proposed to have dissociated from the wild-type template are overlined. Identity between the 3′-terminal sequence of the nascent transcript and the sequence near where the RdRP reinitiated transcription is also shown.
Sequencing revealed that the gene 5 dsRNA of the variant 5S was 2,162 nt in length and contained a rearrangement resulting from a 549-nt duplication of the sequence between residues 917 and 1465 of the 4F gene 5 dsRNA (Fig. 3). The two copies of the duplicated sequences are located at residues 917 to 1465 and 1469 to 2017 and are separated by a stretch of three Ts (Fig. 4). As was the case for the single T detected between the duplicated sequences of the 30-1A gene 5 dsRNA, the origin of the TTT stretch is not apparent based on the sequence of the 4F gene 5 dsRNA and may represent the addition of nontemplated residues by the RdRP. The presence of TTT at the junction alters the NSP1 ORF such that threonine 479 becomes isoleucine and is followed by a termination codon (UAA). As a result, the 5S gene 5 dsRNA encodes a C-truncated NSP1 of 479 aa (Fig. 2) that has a molecular mass of 57 kDa and lacks the last 18 aa of the wild-type protein.
Lack of expression of full-length NSP1 by the 30-1A and 5S variants.
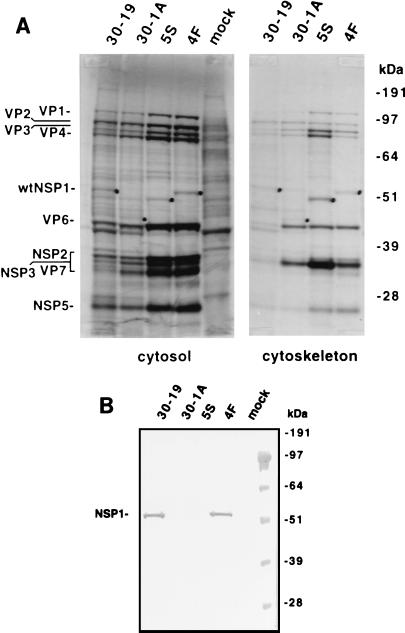
To verify that the gene 5 dsRNAs of the 30-1A and 5S variants lacked the ability to encode wild-type NSP1, MA104 cells were mock infected or infected with the variants or their wild-type counterparts and then maintained in the presence of 35S-labeled amino acids. After harvesting, the cytosol and cytoskeleton fractions of the cells were recovered. The radiolabeled proteins in the fractions were resolved by SDS-PAGE and detected by autoradiography. As shown in Fig. 5A (cytosol), a radiolabeled protein with the same molecular mass as NSP1 (59 kDa) was present in cells infected with wild-type viruses 30-19 and 4F but was absent in cells that were mock infected or infected with 30-1A or 5S. The locations of the C-truncated NSP1 proteins of 30-1A and 5S on the gel were predicted based on their expected molecular masses and the presence of novel proteins in the cytoskeleton fractions of 30-1A- and 5S-infected cells that were absent in the cytoskeletal fractions of mock-infected and 30-19- and 4F-infected cells (Fig. 5). The mock-infected and infected cells were also examined for the presence of NSP1 by Western blot assay using a monospecific polyclonal antiserum made against a peptide representing the C-terminal 19 aa of SA11 NSP1 (Fig. 5B). The analysis confirmed that full-length NSP1 was produced in cells infected with the wild-type viruses 30-19 and 4F but was not produced in mock-infected cells or cells infected with the 30-1A or 5S variant. The lack of availability of other antisera to SA11 NSP1 prevented us from using Western blot analysis to confirm the expression of truncated NSP1 in 30-1A- and 5S-infected cells.
FIG. 5.
Expression of truncated NSP1 in cells infected with the variants 30-1A and 5S and wild-type viruses 30-19 and 4F. Mock- or virus-infected MA104 cells which had been maintained in medium containing 35S-labeled amino acids were lysed at 8 h p.i. with Triton X-100, and the cytosol and cytoskeleton fractions of the cell lysates were recovered. (A) 35S-labeled proteins in the fractions were resolved by SDS-PAGE and detected by autoradiography. The suspected positions of wild-type (30-19 and 4F) and C-truncated (30-1A and 5S) NSP1 are indicated with dots. (B) The cytosol fractions described in panel A were resolved by SDS-PAGE, and the proteins were blotted onto nitrocellulose. The blot was probed with antiserum raised against a peptide corresponding to the last 19 aa of SA11 NSP1. Molecular masses were determined by coelectrophoresis of prestained protein markers.
Small-plaque phenotype of the NSP1-defective variants.
Propagation of the wild-type virus 30-19 and its variant 30-1A showed that each grew to a titer of ∼1 × 107 in MA104 cells. Likewise, the wild-type virus 4F and its variant 5S grew equally well in MA104 cells but they each reached a final titer that was at least 10-fold greater than that of 30-19 and 30-1A (data not shown). The basis for the difference between the titers of the 30-19 and 30-1A pair and the 4F and 5S pair is not known. However, the fact that each of the wild-type viruses and its variant counterpart grew to the same titer indicated that the inability to encode full-length NSP1 did not affect virus yield.
In contrast, the diameters of the plaques produced by the 30-1A and 5S variants were approximately one-half of those of the plaques produced by the corresponding wild-type viruses (Table 1). Except for P9Δ5, whose plaque phenotype has not been reported (38), a decrease in plaque size has also been observed for all variants that encode defective NSP1. This is the case regardless of whether the gene 5 dsRNA encoding the defective protein contains a sequence duplication (30-1A, 5S, brvA, and brvE [40]), a sequence deletion (A5-16 [(38)]), or a nonsense mutation within the NSP1 ORF (A5-10 [38]) (Fig. 3). Thus, although NSP1-defective rotaviruses can grow to the same final titer as rotaviruses producing wild-type NSP1, the inability to produce wild-type NSP1 apparently confers a relatively small-plaque phenotype on the virus. The small-plaque phenotype cannot be correlated with the size of the gene 5 dsRNA. Notably, the plaque size of the IGV-80-3-re variant, which contains a 1.2-kb duplication but encodes full-length NSP1 (17), does not differ significantly from IGV isolates containing a wild-type gene 5 dsRNA (K. Kojima, personal communication).
TABLE 1.
Plaque sizes of wild-type SA11 rotavirus and SA11 variants encoding C-truncated NSP1
| Strain | No. of plaques | Plaque diam (mm)a
|
|
|---|---|---|---|
| Range | Mean (SD) | ||
| SA11-5N (wt)b | 10 | 2.5–5.5 | 4.3 (0.88) |
| SA11-5S | 10 | 1.0–3.0 | 2.2 (0.79) |
| SA11-30-19 (wt) | 10 | 2.0–3.5 | 2.7 (0.71) |
| SA11-1A | 10 | 0.5–2.0 | 1.3 (0.48) |
Diameters were determined at 5 days p.i.
wt, wild type.
Absence of the 3′ consensus sequence in SA11 gene 5 dsRNAs.
While the 3′-terminal sequence of the plus-strand RNAs of nearly all group A rotaviruses is 5′-UGACC-3′, sequencing of multiple cDNA clones obtained by rapid amplification of cDNA 5′ ends of the 3′ termini of the gene 5 dsRNAs of the wild-type viruses 30-19 and 4F and variants 30-1A and 5S showed that they ended with the sequence 5′-UGAACC-3′ (data not shown). Similar analysis of the 3′ end of gene 5 dsRNA of an SA11-4F isolate obtained from R. F. Ramig (Baylor College of Medicine) showed that it too contained the nonconsensus sequence UGAACC (data not shown). Given that Mitchell and Both (25) determined that the gene 5 dsRNAs of their SA11 isolate ended with this same unusual sequence, we conclude that a feature common to the gene 5 segments of all SA11 strains of rotavirus is the presence of the atypical 3′ -terminal sequence UGAACC. Sequencing of the gene 5 dsRNAs of the human KU and canine K9 strains have revealed that they too end with the atypical sequence UGAACC (27). But sequencing of the gene 5 dsRNAs of the bovine A5 (38), RF (34), UK (data not shown), rhesus RRV (date not shown), and avian PO-13 (GenBank accession no. AB009633) strains of rotavirus has shown that they end with the consensus sequence UGACC. Thus, the atypical 3′ end found for the SA11 gene 5 dsRNA appears to be a feature that is found for the gene 5 dsRNAs of some, but not all, strains of rotavirus. While there are many gene 5 sequences in the GenBank database, the 3′-terminal sequences of most of these were not directly determined. Instead, the reported 3′-terminal sequence of the gene 5 RNAs represents the primer that was used to prepare cDNAs of the gene by reverse trancription-PCR. Therefore, it remains unclear whether the atypical 3′ sequence is a common or a rare feature of rotavirus gene 5 dsRNAs.
Atypical 3′-terminal sequence reduces efficiency of gene 5 dsRNA synthesis in vitro.
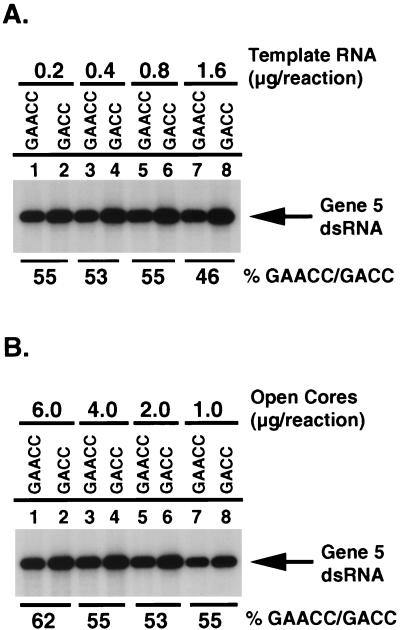
In vitro assays performed with the open-core replication system have shown that the 3′ consensus sequence contains a cis-acting signal that promotes the synthesis of minus-strand RNA (31, 42). Site-specific mutagenesis has revealed that modifications made to the 3′ consensus sequence can reduce the efficiency of minus-strand synthesis (2). To determine whether the atypical 3′ end of SA11 gene 5 dsRNA differed from the 3′ consensus sequence in the ability to promote minus-strand synthesis, two types of SA11 gene 5 mRNAs were prepared which were identical in sequence except that one ended with UGAACC and the other ended with UGACC. The two types of mRNAs were separately incubated in the open-core system in the presence of [32P]UTP, and the radiolabeled gene 5 dsRNA products of the assays were resolved by PAGE and quantified by phosphorimaging. The results showed that the gene 5 mRNA ending with UGAACC was replicated to approximately one-half of the level of the gene 5 mRNA ending with the 3′ consensus sequence UGACC (Fig. 6). This same difference was observed regardless of the amount of gene 5 template RNA or open cores added to the reaction mixtures. Thus, based on this assay, the presence of GAACC at the end of the gene 5 mRNAs of SA11 rotavirus significantly reduces the efficiency by which the viral RdRP can replicate the RNA. However, the fact that the atypical sequence is present in the genome of a nondefective rotavirus which can grow to high titer in cell culture indicates that the change in replication efficiency does not have a marked impact on the ability of the virus to propagate.
FIG. 6.
Effect of the UGACC→UGAACC mutation on the efficiency of minus-strand synthesis. SA11 gene 5 mRNAs were prepared that ended with the 3′ consensus sequence UGACC (GACC) or the atypical sequence UGAACC (GAACC) and used as templates for the synthesis of minus-strand RNA in the open-core replication system. Reaction mixtures contained various amounts of template RNA (A) or open cores (B) and included [32P]UTP to radiolabel RNA products. 32P-labeled dsRNA products were detected by SDS-PAGE and autoradiography, and the relative levels of dsRNA products were quantified with a PhosphorImager. The percent GAACC/GACC was calculated by dividing the amount of dsRNA product made in reactions containing template RNA ending with UGAACC by the amount of that ending with UGACC and multiplying the result by 100.
NSP3 recognizes the atypical 3′-terminal sequence.
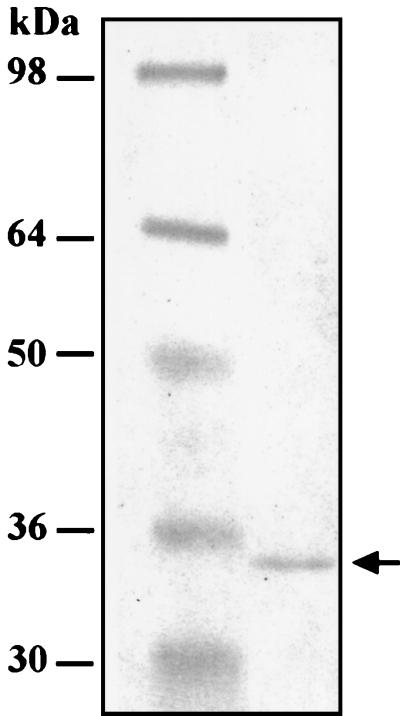
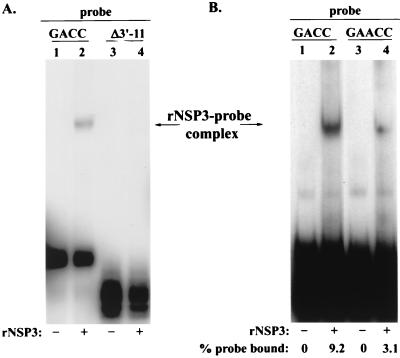
In addition to containing a cis-acting signal that promotes minus-strand synthesis, the 3′ consensus sequence also contains an element that enhances translation of rotavirus mRNAs (4). In particular, the last 4 nt of the consensus sequence, GACC, constitute a translation enhancer that is specifically recognized by NSP3, a nonstructural protein that upregulates gene expression by promoting circularization of viral mRNAs (34, 35). The fact that MA104 cells infected with the SA11 wild-type viruses 30-19 and 4F produced NSP1 (Fig. 5) demonstrates that viral mRNAs which lack the precise NSP3 recognition element can be translated. To determine whether NSP3 was able to recognize the atypical 3′ end of the SA11 gene 5 mRNA (UGAACC), rNSP3 containing an N-terminal His tag was expressed in bacteria and purified to homogeneity with a Ni affinity column (Fig. 7). The rNSP3 was then incubated with the 32P-labeled RNA probe v40-GACC, v40Δ3′-11, or v40-GAACC, and the mixtures were analyzed for the formation of rNSP3-probe complexes by gel mobility shift assay. The 3′ end of the v40-GACC probe corresponds to the last 40 nt of the SA11 gene 8 mRNA and ends with the 3′ consensus sequence UGACC. The v40Δ3′-11 probe is the same, except that it lacks the last 11 nt of the v40-GACC probe. Instead of ending with UGACC, the v40-GAACC probe ends with UGAACC. The gel mobility shift analysis showed that rNSP3 formed a complex with v40-GACC but not with v40Δ3′-11 (Fig. 8A). This is consistent with earlier studies showing that the recognition element for NSP3 is located at the 3′ end of viral mRNAs (35). The analysis also showed that rNSP3 bound not only to v40-GACC but to v40-GAACC as well (Fig. 8B). However, quantitation of the results revealed that the level of NSP3-probe complexes formed with v40-GAACC was approximately one-third of that formed with v40-GACC (data not shown). These data suggest that while the 3′ GAACC sequence of SA11 gene 5 mRNAs does not contain the precise recognition element for NSP3, the protein retains the ability to bind specifically to the 3′ end of the mRNA but with decreased affinity.
FIG. 7.
Isolation of His-tagged rNSP3 expressed in bacteria. Protein markers (lane 1) and His-tagged rNSP3, purified with a Ni-nitrilotriacetic acid agarose column (lane 2), were resolved by SDS-PAGE and detected by staining with Coomassie blue R-250.
FIG. 8.
Impact of the UGACC→UGAACC mutation on the RNA-binding activity of NSP3. Complexes formed by incubating purified rNSP3 with the 32P-labeled RNA probes v40-GACC and v40Δ3′-11 (A) or v40-GACC and v40-GAACC (B) were resolved by electrophoresis on a nondenaturing 6% polyacrylamide gel and detected by autoradiography. The intensity of bands representing rNSP3-probe complexes and free probe were quantified with a PhosphorImager. The values were used to calculate the percentage of probe binding to rNSP3.
Atypical 3′-terminal sequence reduces efficiency of gene expression.
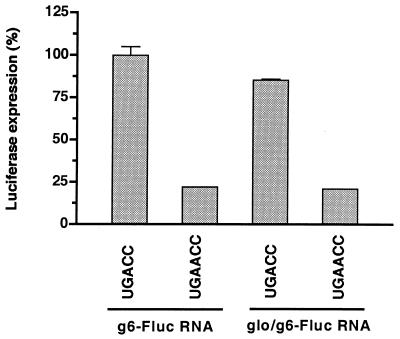
Chimeric RNAs, which contained the ORF for firefly luciferase and the UTRs either of SA11 gene 6 mRNA or of nonviral mRNAs were used previously to identify and characterize the rotavirus 3′ translation enhancer (4). We employed this same system to address the question of whether the translational efficiency of the SA11 gene 5 mRNAs was reduced because they ended with GAACC instead of the 3′ translation enhancer GACC. To perform this analysis, 5′-capped chimeric reporter RNAs were made that contained the ORF for firefly luciferase and either the 5′ and 3′ UTRs of SA11 gene 6 mRNA (g6-Fluc-UGACC) or the 5′ UTR of β-globin mRNA and the 3′ UTR of SA11 gene 6 mRNA (glo/g6-Fluc-UGACC). Two derivatives of these chimeric RNAs were also prepared in which the 3′-terminal UGACC sequences were replaced with UGAACC (g6-Fluc-UGAACC and glo/g6-Fluc-UGAACC). Finally, a capped but nonpolyadenylated reporter RNA (nv-Rluc) was prepared that encodes Renilla luciferase and contains UTRs of nonviral origin. Each of the g6-Fluc and glo/g6-Fluc RNAs was then individually cotransfected with nv-Rluc RNA into SA11 rotavirus-infected MA104 cell at 1 h p.i. At 9 h posttransfection, the levels of firefly and Renilla luciferases expressed in the cells were determined with a luminometer. To compensate against possible variations in transfection efficiency, the values obtained for the expression of firefly luciferase were then normalized against the values obtained for the expression of Renilla luciferase. The results showed that cells transfected with g6-Fluc-GACC and glo/g6-Fluc-GACC produced approximately four times as much firefly luciferase as cells transfected with g6-Fluc-GAACC and glo/g6-Fluc-GAACC, respectively (Fig. 9). Thus, mutation of the terminal sequence from GACC to GAACC caused a significant decrease in the expression of the ORF of the chimeric RNAs, suggesting that the mutation partially inactivated the 3′-terminal enhancer. Consistent with an earlier report (4), the results also showed that the effect of the 3′-terminal sequences of the chimeric RNAs on gene expression was not influenced by the 5′ UTR. In summary, these data provide indirect evidence that the atypical 3′-terminal sequence on the mRNAs of SA11 gene 5 dsRNA probably has a negative effect on the expression of NSP1 from these mRNAs.
FIG. 9.
Effect of the UGACC→UGAACC mutation on protein expression in infected cells. The chimeric reporter RNAs g6-Fluc-UGACC and -UGAACC and glo/g6-Fluc-UGACC and -UGAACC were separately cotransfected with nv-Rluc into SA11-infected MA104 cells at 1 h p.i. At 9 h p.i., the levels of firefly and Renilla luciferases per milligram of cell lysate were determined and the expression of firefly luciferase was normalized to the expression of Renilla luciferase. To ease the comparison of values, the expression of firefly luciferase for the g6-Fluc-UGACC and glo/g6-Fluc-UGACC RNAs was set to 100%.
DISCUSSION
Role of NSP1 in the viral life cycle.
With the completion of this study, a total of eight aberrant gene 5 dsRNAs of rotavirus have been molecularly described. Of these, five contain sequence duplications (SA11 30-1A, SA11-5S, brvA, brvE, and IGV-80-3-re), two contain sequence deletions (UK-P9Δ5 and A5-16), and two contain nonsense mutations (brvA and A5-10). Of the eight aberrant gene 5 dsRNAs, only that of the variant IGV-80-3-re encodes full-length NSP1. In contrast, analysis of the plaques produced by the variants encoding defective NSP1 has so far revealed that they are significantly smaller than those produced by the corresponding wild-type viruses, regardless of whether the variant contained a large (e.g., SA11 30-1A) or a small (e.g., A5-16) gene 5 dsRNA. These results indicate that although NSP1 is nonessential, the protein does affect the plaque phenotype of the virus and therefore does have a role in the biology of the virus when it is propagated in cell culture. Notably, the variant SA11-5S produced a C-truncated NSP1 that was missing only the last 17 aa of the full-length protein. The fact that the plaque size of this variant was one-half of that of its wild-type counterpart suggests that even deletion of a relatively few amino acids from the C terminus of NSP1 alters the protein's function and results in a change in the plaque phenotype of the virus. Thus, the complete NSP1 protein, or nearly all of it, is apparently required to confer the normal plaque phenotype on the virus. It was also observed that although SA11 30-1A and SA11-5S produced smaller plaques than their wild-type counterparts, MA104 cells infected with these variants grew to the same final titer as cells infected with the wild-type viruses. Given the growth and plaque characteristics of the variants, it may be proposed that defects in NSP1 increase the length of the viral life cycle (and thereby produce slower-growing plaques) but do not affect the number of progeny virions generated in the infected cell.
Mechanism of sequence rearrangement.
Previous analysis of a rotavirus variant containing a sequence duplication in the gene 11 dsRNA has provided evidence that intragenic rearrangements can occur during viral transcription (16). The molecular events leading to rearrangements are not known. However, it has been proposed that during plus-strand synthesis, the RdRP and nascent transcript detach from the dsRNA template and then move together to a distant site on the same template where the RdRP reinitiates plus-strand synthesis using the nascent transcript as a primer (6, 16).
Inspection of the sequences of the atypical gene 5 dsRNAs suggests that during plus-strand synthesis, the RdRP can disengage from all regions of the dsRNA template, including near the 5′ end (A5-16), the middle (brvE), and the 3′ end (IGV-80-3-re) (Fig. 3). The RdRP is able to re-engage and reinitiate RNA synthesis at sites that are upstream (e.g., A5-16) or downstream (e.g., SA11-5S) of the sites at which the RdRP disengaged, and thereby, the RdRP is able to create deletions or duplications, respectively, within the product RNA. The distances between the sites where the polymerase disengages and re-engages the template are quite variable, ranging from 0.3 kb for UK-P9Δ5 to 1.2 kb for IGV-80-3-re. As revealed by the analysis of the site at which the RdRP disengaged from the gene 5 dsRNA of 30-19, in generating the rearranged gene 5 dsRNA of 30-19 (Fig. 4A), repeated sequences of significant length need not be present for the polymerase to release from the RNA template. Similarly, for the disengaged RdRP to reinitiate RNA synthesis elsewhere on the dsRNA template using the nascent RNA as a primer, the 3′ end of the nascent RNA need not have extensive complementarity with sequences of the template at or near the site of reinitiation (Fig. 4A).
During the rearrangement of the gene 5 dsRNAs of the variants 30-1A and 5S, one or three nontemplated T residues, respectively, were introduced at the junction site of the duplicated sequences (Fig. 4). An important but unanswerable question is whether the T residues were added before or after the RdRP disengaged from the template. If before, then the presence of the nontemplated residues may have destabilized the interaction between the nascent RNA and the RNA template at the transcription fork, causing the release of the nascent strand and RdRP. However, the fact that gene 5 rearrangements have been described which lack any notable nontemplated nucleotides at the junction site suggests that other events are also involved in causing the RdRP to disengage from the RNA template.
Basis for selection of RNAs containing sequence duplications.
All of the variants shown in Fig. 3 that contain duplicated gene 5 sequences emerged during passage of wild-type rotaviruses at high MOI. The increasing predominance of the rearranged gene 5 dsRNA as the virus lysates were serially passaged indicates that there was a selective advantage in their use in the viral life cycle over the wild-type gene 5 dsRNA. Indeed, analysis of cells coinfected with equal amounts of wild-type virus and variants with duplicated sequences in gene 5 or 11 has also suggested that dsRNAs with sequence duplications have a selective advantage over those that do not (5, 13). Since rotavirus mRNAs are believed to undergo selection (assortment) and packaging into cores prior to replication into dsRNAs, it is reasonable to propose that, in fact, the sequence rearrangement alters the mRNA in a way that, directly or indirectly, increases the frequency by which the mRNA is assorted and/or packaged. The nature of the advantage is unclear but simply may be related to an increase in the overall size of the transcripts made from gene 5 dsRNAs containing sequence duplications. Alternatively, it is possible that the increase in the length of the 3′ UTR generated by the sequence rearrangement in itself provides a selective advantage. The increased size of the transcript or its 3′ UTR may make it more difficult for NSP3 to cause the circularization of the mRNAs and, thereby, for the mRNAs to be recruited into polysomes. As a consequence, the probability may increase that the transcripts interact with those viral RNA-binding proteins that target their movement to viroplasms and their introduction into replication intermediates.
Importance of the 3′ consensus sequence for RNA replication and gene expression.
It was found that the 3′-terminal sequence of the gene 5 dsRNAs of all isolates of SA11 rotaviruses was UGAACC and not the expected consensus sequence UGACC. Our analyses indicate that the A insertion into the consensus sequence affects the ability of rotavirus mRNAs to serve as templates for minus-strand synthesis, to be recognized by NSP3, and to be efficiently translated. Based on assays performed with the open-core replication system, the UGAACC sequence is only 50% as efficient in promoting dsRNA synthesis as the UGACC sequence. However, since SA11 viruses grow to high titer (>108) and have relatively large-plaque phenotypes, the negative impact that the atypical 3′ end has on the replication of the gene 5 mRNA is not necessarily significant to the biology of these viruses. In terms of using the open-core system as a tool for identifying sequences in viral mRNAs that promote minus-strand synthesis, our results indicate that mutations made in viral mRNAs that reduce minus-strand synthesis in vitro by 50% cannot be interpreted as representing changes that would render a virus nonviable.
Gel shift assays demonstrated that SA11 NSP3 specifically recognizes the atypical 3′ sequence UGAACC of SA11 gene 5 dsRNA, although less efficiently than the 3′ consensus sequence UGACC. This result is contrary to previous results which indicated that the four-base sequence GACC was necessary and sufficient for interaction with NSP3. In particular, Poncet et al. (35) reported that NSP3 was not able to form complexes with UAACC but was able to form complexes with GGACC, suggesting that in their studies the G residue at −4 was critical for recognition by NSP3. In their studies, the probe UGAACC was not tested. The difference between our results and those of Poncet et al. (35) may stem from minor differences in specificity between SA11 NSP3 and bovine NSP3. Even though the gene 5 dsRNA of SA11 lacked the 3′ consensus sequence, cells infected with the virus expressed NSP1. Thus, despite the proposed importance of the 3′ consensus sequence in viral mRNA circularization and gene expression, the sequence, in fact, is not required for translation of viral mRNAs.
If the 3′ sequence UGAACC functions suboptimally in promoting dsRNA synthesis and gene expression, then why does it exist in SA11 gene 5 dsRNAs? The simplest answer is that it leads to lower expression of NSP1 in infected cells. The frequency at which gene 5 rearrangements occur and then dominate over wild-type gene 5 RNAs during serial passage of the rotaviruses in cell culture indicates that viruses which no longer encode wild-type NSP1 may have a selective advantage over those that do. If this is true, then it is reasonable to presume that an A insertion into the gene 5 3′ consensus sequence may confer an advantage on the SA11 virus because it partially inactivates the 3′ translation enhancer and, as a result, decreases the expression of wild-type NSP1. In this scenario, the selective advantage in favor of inactivating the 3′ translation enhancer would be dominant over any deleterious effect that the A insertion would have on the ability of the 3′ sequence to promote minus-strand synthesis.
ACKNOWLEDGMENTS
V.G. was supported in part by a fellowship from CNPq, Brasilia, Brazil, and by a grant from FAPERJ, Rio de Janeiro, Brazil.
REFERENCES
- 1.Bican P, Cohen J, Charpilienne A, Scherrer R. Purification and characterization of bovine rotavirus cores. J Virol. 1982;43:1113–1117. doi: 10.1128/jvi.43.3.1113-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen D, Patton J T. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J Virol. 1998;72:7387–7396. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Zeng C Q-Y, Wentz M J, Gorziglia M, Estes M K, Ramig R F. Template-dependent, in vitro replication of rotavirus RNA. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chizhikov V, Patton J T. A four-nucleotide translation enhancer in the 3′-terminal terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA. 2000;6:814–825. doi: 10.1017/s1355838200992264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chnaiderman J, Diaz J, Magnusson G, Liprandi F, Spencer E. Characterization of a rotavirus rearranged gene 11 by gene reassortment. Arch Virol. 1998;143:1711–1722. doi: 10.1007/s007050050411. [DOI] [PubMed] [Google Scholar]
- 6.Desselberger U. Genome rearrangements of rotaviruses. Arch Virol Suppl. 1996;12:37–51. doi: 10.1007/978-3-7091-6553-9_5. [DOI] [PubMed] [Google Scholar]
- 7.Eiden J, Losonski G A, Johnson J, Yolken R. Rotavirus RNA variation during chronic infection of immunocompromised children. Pediatr Infect Dis J. 1985;4:632–637. doi: 10.1097/00006454-198511000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 731–761. [Google Scholar]
- 9.Gouet P, Grimes J M, Malby R, Burroughs J N, Zientara S, Stuart D I, Mertens P P C. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 10.Hua J, Patton J T. The carboxyl-half of the rotavirus nonstructural protein NS53 (NSP1) is not required for virus replication. Virology. 1994;198:567–576. doi: 10.1006/viro.1994.1068. [DOI] [PubMed] [Google Scholar]
- 11.Hua J, Chen X, Patton J T. Deletion mapping of the rotavirus metalloprotein NS53 (NSP1): the conserved cysteine-rich region is essential for virus-specific RNA binding. J Virol. 1994;68:3990–4000. doi: 10.1128/jvi.68.6.3990-4000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua J, Mansell E A, Patton J T. Comparative analysis of the rotavirus NS53 gene: conservation of basic and cysteine-rich regions in the protein and possible stem-loop structures in the RNA. Virology. 1993;196:372–378. doi: 10.1006/viro.1993.1492. [DOI] [PubMed] [Google Scholar]
- 13.Hundley F, Biryahwaho B, Gow M, Desselberger U. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology. 1985;143:88–103. doi: 10.1016/0042-6822(85)90099-6. [DOI] [PubMed] [Google Scholar]
- 14.Hundley F, McIntyre M, Clark B, Beards G, Wood D, Chrystie L, Desselberger U. Heterogeneity of genome rearrangements in rotaviruses isolated from a chronically infected immunodeficient child. J Virol. 1987;61:3365–3372. doi: 10.1128/jvi.61.11.3365-3372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai M, Akatani K, Ikegami N, Furuichi Y. Capped and conserved terminal structures in human rotavirus double-stranded RNA segments. J Virol. 1983;47:125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojima K, Taniguchi K, Urasawa T, Urasaw S. Sequence analysis of normal and rearranged NSP5 genes from human rotavirus strains isolated from nature: implications for the occurrence of the rearrangement at the step of plus strand synthesis. Virology. 1996;224:446–452. doi: 10.1006/viro.1996.0551. [DOI] [PubMed] [Google Scholar]
- 17.Kojima K, Taniguchi K, Kawagishi-Kobayashi M, Matsuno S, Urasaw S. Rearrangement generated in double genes, NSP1 and NSP3, of viable progenies from a human rotavirus strain. Virus Res. 2000;67:163–171. doi: 10.1016/s0168-1702(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 18.Lawton J A, Estes M K, Prasad B V V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 19.Lawton J A, Zeng C Q-Y, Mukherjee S K, Cohen J, Estes M K, Prasad B V V. Three dimensional analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M, Offit P A, Estes M K. Identification of the siman rotavirus SA11 genome segment 3 product. Virology. 1988;162:26–32. doi: 10.1016/0042-6822(88)90230-9. [DOI] [PubMed] [Google Scholar]
- 21.Malherbe H H, Strickland-Cholmley M. Simian virus SA11 and the related O agent. Arch Gesamte Virusforsch. 1967;22:235–245. doi: 10.1007/BF01240518. [DOI] [PubMed] [Google Scholar]
- 22.Mattion N M, Estes M K. Sequence of a rotavirus gene 4 associated with unique biologic properties. Arch Virol. 1991;120:109–113. doi: 10.1007/BF01310953. [DOI] [PubMed] [Google Scholar]
- 23.McCrae M A, McCorquodale J G. Molecular biology of rotaviruses. V. Terminal structure of viral RNA species. Virology. 1983;126:204–212. doi: 10.1016/0042-6822(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 24.Mendez E, Arias C F, Lopez S. Genomic rearrangements in human rotavirus strain WA: analysis of rearranged RNA segment 7. Arch Virol. 1992;125:331–338. doi: 10.1007/BF01309651. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell D B, Both G W. Conservation of a potential metal binding motif despite extensive sequence diversity in the rotavirus nonstructural protein NS53. Virology. 1990;174:618–621. doi: 10.1016/0042-6822(90)90117-a. [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa K, Taniguchi K, Torres A, Hoshino Y, Green K, Kapikian A Z, Chanock R M, Gorziglia M. Comparative analysis of the VP3 gene of divergent strains of the rotaviruses simian SA11 and bovine Nebraska calf diarrhea virus. J Virol. 1988;62:4022–4026. doi: 10.1128/jvi.62.11.4022-4026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada J, Kobayashi N, Taniguchi K, Shiomi H. Functional analysis of the heterologous NSP1 genes in the genetic background of simian rotavirus SA11. Arch Virol. 1999;144:1439–1449. doi: 10.1007/s007050050600. [DOI] [PubMed] [Google Scholar]
- 28.Patton J T. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton J T, Spencer E. Differences and similarities in RNA replication of viruses with segmented double-stranded RNA genomes. Virology. 2000;277:217–225. doi: 10.1006/viro.2000.0645. [DOI] [PubMed] [Google Scholar]
- 30.Patton J T, Taraporewala Z, Chizhikov V, Chen D. Virus replication. Methods Mol Med. 1999;34:33–66. doi: 10.1385/1-59259-078-0:33. [DOI] [PubMed] [Google Scholar]
- 31.Patton J T, Wentz M, Xiaobo J, Ramig R F. cis-acting signals that promote genome replication in rotavirus mRNA. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedley S, Hundley F, Chrystie I, McCrae M A, Desselberger U. The genomes of rotaviruses isolated from chronically infected immunodeficient children. J Gen Virol. 1984;65:1141–1150. doi: 10.1099/0022-1317-65-7-1141. [DOI] [PubMed] [Google Scholar]
- 33.Pereira H G, Azeredo R S, Fialho A M, Vidal M N P. Genomic heterogeneity of simian rotavirus SA11. J Gen Virol. 1984;65:815–818. doi: 10.1099/0022-1317-65-4-815. [DOI] [PubMed] [Google Scholar]
- 34.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poncet D, Laurent S, Cohen J. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J. 1994;13:4165–4173. doi: 10.1002/j.1460-2075.1994.tb06734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen S, Burke B, Desselberger U. Rearrangement of the VP6 gene of a group A rotavirus in combination with a point mutation affecting trimer stability. J Virol. 1994;68:1682–1688. doi: 10.1128/jvi.68.3.1682-1688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer E, Arias M L. In vitro transcription catalyzed by heat-treated human rotavirus. J Virol. 1981;40:1–10. doi: 10.1128/jvi.40.1.1-10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi K, Kojima K, Urasawa S. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153 to 155. J Virol. 1996;70:4125–4130. doi: 10.1128/jvi.70.6.4125-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taraporewala Z, Chen D, Patton J T. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian Y, Tarlow O, Ballard A, Desselberger U, McCrae M A. Genomic concatemerization/deletion in rotaviruses: a new mechanism for generating rapid genetic change of potential epidemiological importance. J Virol. 1993;67:6625–6632. doi: 10.1128/jvi.67.11.6625-6632.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vende P, Piron M, Castagne N, Poncet D. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J Virol. 2000;74:7064–7071. doi: 10.1128/jvi.74.15.7064-7071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wentz M J, Patton J T, Ramig R F. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]