Abstract
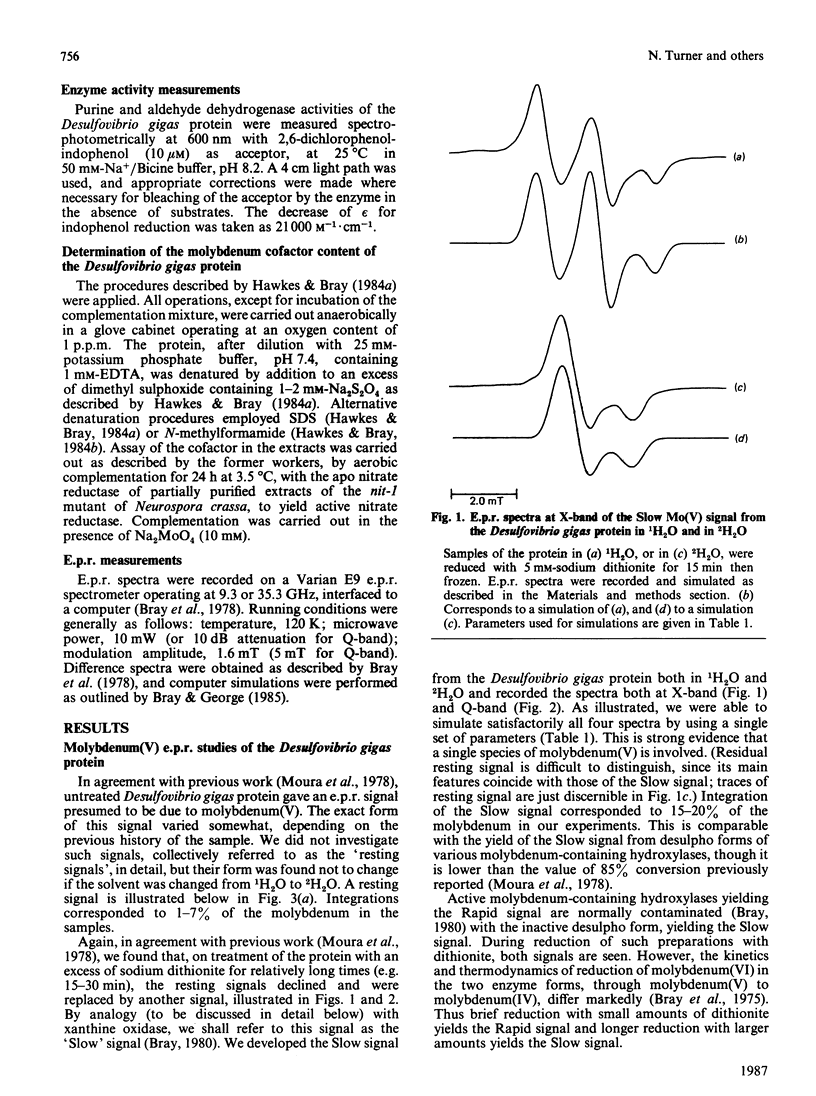
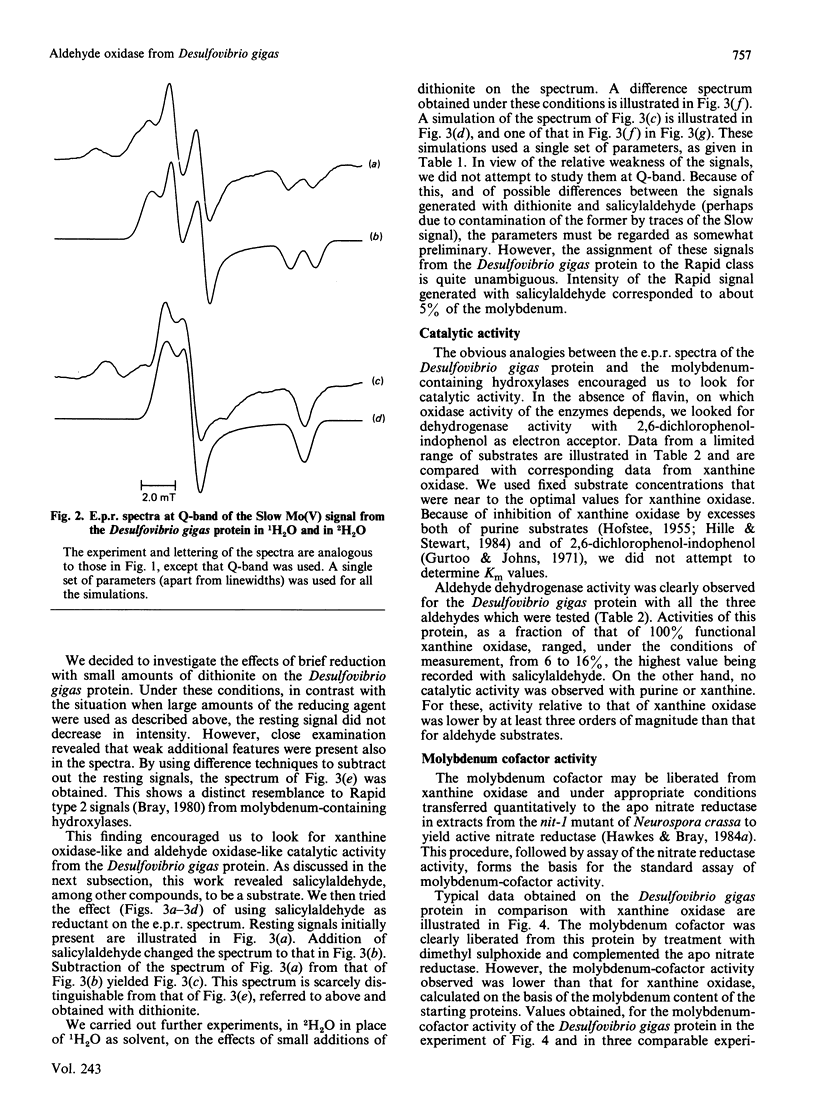
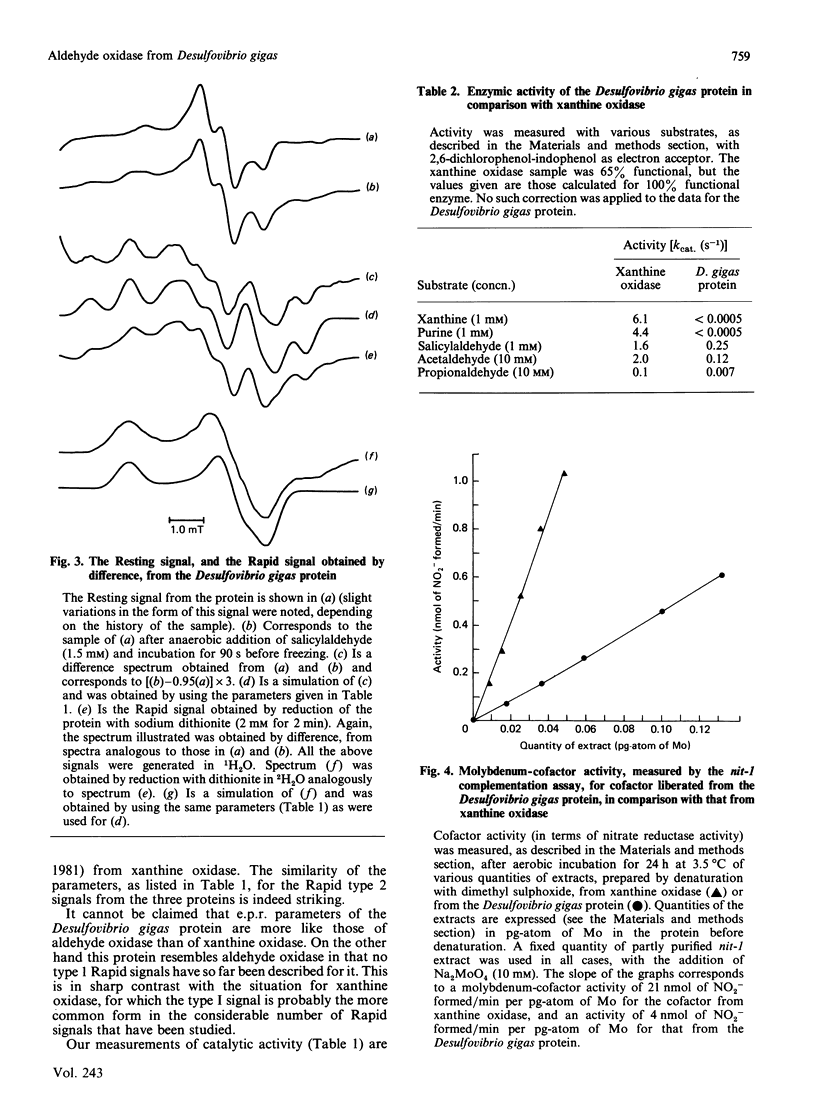
The molybdenum iron-sulphur protein originally isolated from Desulfovibrio gigas by Moura, Xavier, Bruschi, Le Gall, Hall & Cammack [(1976) Biochem. Biophys. Res. Commun. 72, 782-789] has been further investigated by e.p.r. spectroscopy of molybdenum(V). The signal obtained on extended reduction of the protein with sodium dithionite has been shown, by studies at 9 and 35 HGz in 1H2O and 2H2O and computer simulations, to have parameters corresponding to those of the Slow signal from the inactive desulpho form of various molybdenum-containing hydroxylases. Another signal obtained on brief reduction of the protein with small amounts of dithionite was shown by e.p.r. difference techniques to be a Rapid type 2 signal, like that from the active form of such enzymes. In confirmation that the protein is a molybdenum-containing hydroxylase, activity measurements revealed that it had aldehyde:2,6-dichlorophenol-indophenol oxidoreductase activity. No such activity towards xanthine or purine was observed. Salicylaldehyde was a particularly good substrate, and treatment of the protein with it also gave rise to the Rapid signal. Molybdenum cofactor liberated from the protein was active in the nit-1 Neurospora crassa nitrate reductase assay. It is concluded that the protein is a form of an aldehyde oxidase or dehydrogenase. From the intensity of the e.p.r. signals and from enzyme activity measurements, 10-30% of the protein in the sample examined appeared to be in the functional form. The evolutionary significance of the protein, which may represent a primitive form of the enzyme rather than a degradation product, is discussed briefly.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badwey J. A., Robinson J. M., Karnovsky M. J., Karnovsky M. L. Superoxide production by an unusual aldehyde oxidase in guinea pig granulocytes. Characterization and cytochemical localization. J Biol Chem. 1981 Apr 10;256(7):3479–3486. [PubMed] [Google Scholar]
- Barber M. J., Coughlan M. P., Rajagopalan K. V., Siegel L. M. Properties of the prosthetic groups of rabbit liver aldehyde oxidase: a comparison of molybdenum hydroxylase enzymes. Biochemistry. 1982 Jul 20;21(15):3561–3568. doi: 10.1021/bi00258a006. [DOI] [PubMed] [Google Scholar]
- Bordas J., Bray R. C., Garner C. D., Gutteridge S., Hasnain S. S. X-ray absorption spectroscopy of xanthine oxidase. The molybdenum centres of the functional and the desulpho forms. Biochem J. 1980 Nov 1;191(2):499–508. doi: 10.1042/bj1910499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray R. C., Barber M. J., Lowe D. J. Electron-paramagnetic-resonance spectroscopy of complexes of xanthine oxidase with xanthine and uric acid. Biochem J. 1978 Jun 1;171(3):653–658. doi: 10.1042/bj1710653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray R. C., George G. N. Electron-paramagnetic-resonance studies using pre-steady-state kinetics and substitution with stable isotopes on the mechanism of action of molybdoenzymes. Biochem Soc Trans. 1985 Jun;13(3):560–567. doi: 10.1042/bst0130560. [DOI] [PubMed] [Google Scholar]
- Bray R. C. The reactions and the structures of molybdenum centers in enzymes. Adv Enzymol Relat Areas Mol Biol. 1980;51:107–165. doi: 10.1002/9780470122969.ch3. [DOI] [PubMed] [Google Scholar]
- Coughlan M. P., Betcher-Lange S. L., Rajagopalan K. V. Isolation of the domain containing the molybdenum, iron-sulfur I, and iron-sulfur II centers of chicken liver xanthine dehydrogenase. J Biol Chem. 1979 Nov 10;254(21):10694–10699. [PubMed] [Google Scholar]
- Coughlan M. P., Mehra R. K., Barber M. J., Siegel L. M. Optical and electron paramagnetic resonance spectroscopic studies on purine hydroxylase II from Aspergillus nidulans. Arch Biochem Biophys. 1984 Mar;229(2):596–603. doi: 10.1016/0003-9861(84)90192-9. [DOI] [PubMed] [Google Scholar]
- Cramer S. P., Moura J. J., Xavier A. V., LeGall J. Molybdenum EXAFS of the Desulfovibrio gigas Mo(2Fe-2S) protein--structural similarity to "desulfo" xanthine dehydrogenase. J Inorg Biochem. 1984 Apr;20(4):275–280. doi: 10.1016/0162-0134(84)85026-6. [DOI] [PubMed] [Google Scholar]
- Dalton H., Lowe D. J., Pawlik T., Bray R. C. Studies by electron-paramagnetic-resonance spectroscopy on the mechanism of action of xanthine dehydrogenase from Veillonella alcalescens. Biochem J. 1976 Feb 1;153(2):287–295. doi: 10.1042/bj1530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton H., Lowe D. J., Pawlik T., Bray R. C. Studies by electron-paramagnetic-resonance spectroscopy on the mechanism of action of xanthine dehydrogenase from Veillonella alcalescens. Biochem J. 1976 Feb 1;153(2):287–295. doi: 10.1042/bj1530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fháolain I. N., Coughlan M. P. Effects of limited proteolysis on the structure and activity of turkey liver xanthine dehydrogenase [proceedings]. Biochem Soc Trans. 1977;5(6):1705–1707. doi: 10.1042/bst0051705. [DOI] [PubMed] [Google Scholar]
- Gurtoo H. L., Johns D. G. On the interaction of the electron acceptor 2,6-dichlorophenolindophenol with bovine milk xanthine oxidase. J Biol Chem. 1971 Jan 25;246(2):286–293. [PubMed] [Google Scholar]
- Gutteridge S., Tanner S. J., Bray R. C. Comparison of the molybdenum centres of native and desulpho xanthine oxidase. The nature of the cyanide-labile sulphur atom and the nature of the proton-accepting group. Biochem J. 1978 Dec 1;175(3):887–897. doi: 10.1042/bj1750887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFSTEE B. H. On the mechanism of inhibition of xanthine oxidase by the substrate xanthine. J Biol Chem. 1955 Sep;216(1):235–244. [PubMed] [Google Scholar]
- Hart L. I., McGartoll M. A., Chapman H. R., Bray R. C. The composition of milk xanthine oxidase. Biochem J. 1970 Mar;116(5):851–864. doi: 10.1042/bj1160851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes T. R., Bray R. C. Quantitative transfer of the molybdenum cofactor from xanthine oxidase and from sulphite oxidase to the deficient enzyme of the nit-1 mutant of Neurospora crassa to yield active nitrate reductase. Biochem J. 1984 Apr 15;219(2):481–493. doi: 10.1042/bj2190481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes T. R., Bray R. C. Studies by electron-paramagnetic-resonance spectroscopy of the environment of the metal in the molybdenum cofactor of molybdenum-containing enzymes. Biochem J. 1984 Sep 15;222(3):587–600. doi: 10.1042/bj2220587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R., Hagen W. R., Dunham W. R. Spectroscopic studies on the iron-sulfur centers of milk xanthine oxidase. J Biol Chem. 1985 Sep 5;260(19):10569–10575. [PubMed] [Google Scholar]
- Hille R., Stewart R. C. The inhibition of xanthine oxidase by 8-bromoxanthine. J Biol Chem. 1984 Feb 10;259(3):1570–1576. [PubMed] [Google Scholar]
- Komai H., Massey V., Palmer G. The preparation and properties of deflavo xanthine oxidase. J Biol Chem. 1969 Apr 10;244(7):1692–1700. [PubMed] [Google Scholar]
- Krenitsky T. A., Neil S. M., Elion G. B., Hitchings G. H. A comparison of the specificities of xanthine oxidase and aldehyde oxidase. Arch Biochem Biophys. 1972 Jun;150(2):585–599. doi: 10.1016/0003-9861(72)90078-1. [DOI] [PubMed] [Google Scholar]
- Malthouse J. P., Gutteridge S., Bray R. C. Rapid type 2 molybdenum(V) electron-paramagnetic resonance signals from xanthine oxidase and the structure of the active centre of the enzyme. Biochem J. 1980 Mar 1;185(3):767–770. doi: 10.1042/bj1850767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R. K., Coughlan M. P. Purification and properties of purine hydroxylase II from Aspergillus nidulans. Arch Biochem Biophys. 1984 Mar;229(2):585–595. doi: 10.1016/0003-9861(84)90191-7. [DOI] [PubMed] [Google Scholar]
- Moura J. J., Xavier A. V., Bruschi M., Le Gall J., Hall D. O., Cammack R. A molybdenum-containing iron-sulphur protein from Desulphovibrio gigas. Biochem Biophys Res Commun. 1976 Oct 4;72(3):782–789. doi: 10.1016/s0006-291x(76)80201-x. [DOI] [PubMed] [Google Scholar]
- Moura J. J., Xavier A. V., Cammack R., Hall D. O., Bruschi M., Le Gall J. Oxidation-reduction studies of the Mo-(2Fe-2S) protein from Desulfovibrio gigas. Biochem J. 1978 Aug 1;173(2):419–425. doi: 10.1042/bj1730419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R. C., Hageman R. V., Rajagopalan K. V. The relationship of Mo, molybdopterin, and the cyanolyzable sulfur in the Mo cofactor. Arch Biochem Biophys. 1984 Apr;230(1):264–273. doi: 10.1016/0003-9861(84)90107-3. [DOI] [PubMed] [Google Scholar]
- Wahl R. C., Warner C. K., Finnerty V., Rajagopalan K. V. Drosophila melanogaster ma-l mutants are defective in the sulfuration of desulfo Mo hydroxylases. J Biol Chem. 1982 Apr 10;257(7):3958–3962. [PubMed] [Google Scholar]


