Abstract
Analyzing vast textual data and summarizing key information from electronic health records imposes a substantial burden on how clinicians allocate their time. Although large language models (LLMs) have shown promise in natural language processing (NLP) tasks, their effectiveness on a diverse range of clinical summarization tasks remains unproven. Here we applied adaptation methods to eight LLMs, spanning four distinct clinical summarization tasks: radiology reports, patient questions, progress notes and doctor–patient dialogue. Quantitative assessments with syntactic, semantic and conceptual NLP metrics reveal trade-offs between models and adaptation methods. A clinical reader study with 10 physicians evaluated summary completeness, correctness and conciseness; in most cases, summaries from our best-adapted LLMs were deemed either equivalent (45%) or superior (36%) compared with summaries from medical experts. The ensuing safety analysis highlights challenges faced by both LLMs and medical experts, as we connect errors to potential medical harm and categorize types of fabricated information. Our research provides evidence of LLMs outperforming medical experts in clinical text summarization across multiple tasks. This suggests that integrating LLMs into clinical workflows could alleviate documentation burden, allowing clinicians to focus more on patient care.
Documentation plays an indispensable role in healthcare practice. Currently, clinicians spend a substantial amount of time summarizing vast amounts of textual information—whether it be compiling diagnostic reports, writing progress notes or synthesizing a patient’s treatment history across different specialists1–3. Even for experienced physicians with a high level of expertise, this intricate task naturally introduces the possibility for errors, which can be detrimental in healthcare where precision is paramount4–6.
The widespread adoption of electronic health records has expanded clinical documentation workload, directly contributing to increasing stress and clinician burnout7–9. Recent data indicate that physicians can expend up to 2 hours on documentation for each hour of patient interaction10. Similarly, documentation responsibilities for nurses can consume up to 60% of their time and account for considerable work stress11–13. These tasks divert attention from direct patient care, leading to worse outcomes for patients and decreased job satisfaction for clinicians2,14–16.
Large language models (LLMs) have gained remarkable traction, leading to widespread adoption of models such as ChatGPT17, which excel at information retrieval, nuanced understanding and text generation18,19. Although LLM benchmarks for general natural language processing (NLP) tasks exist20,21, they do not evaluate performance on relevant clinical tasks. Addressing this limitation presents an opportunity to accelerate the process of clinical text summarization, hence alleviating documentation burden and improving patient care.
Crucially, machine-generated summaries must be non-inferior to those of seasoned clinicians, especially when used to support sensitive clinical decision-making. Previous work has demonstrated potential across clinical NLP tasks22,23, adapting to the medical domain by training a new model24,25, fine-tuning an existing model26,27 or supplying task-specific examples in the model prompt27,28. However, adapting LLMs to summarize a diverse set of clinical tasks has not been thoroughly explored nor has non-inferiority to medical experts been achieved.
With the overarching objective of bringing LLMs closer to clinical readiness, we demonstrate here the potential of these models for clinical text summarization. Our evaluation framework (Fig. 1) follows a three-step process: (1) use quantitative NLP metrics to identify the best model and adaptation method across those selected on four summarization tasks; (2) conduct a clinical reader study with 10 physicians comparing the best LLM summaries to medical expert summaries across the key attributes of completeness, correctness and conciseness; and (3) perform a safety analysis of examples, potential medical harm and fabricated information to understand challenges faced by both models and medical experts. This framework aims to guide future enhancements of LLMs and their integration into clinical workflows.
Fig. 1 |. Framework overview.
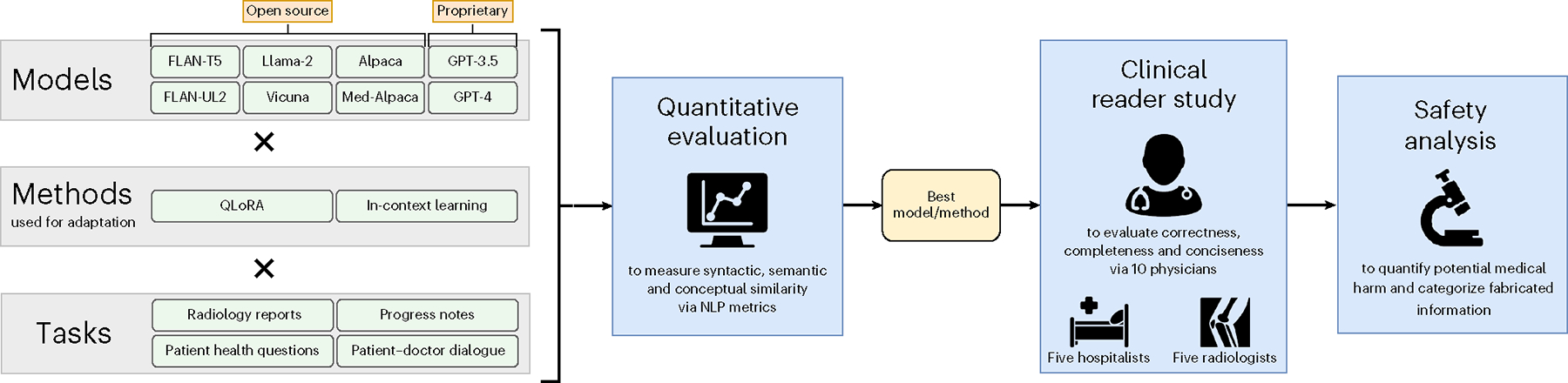
First, we quantitatively evaluated each valid combination (×) of LLM and adaptation method across four distinct summarization tasks comprising six datasets. We then conducted a clinical reader study in which 10 physicians compared summaries of the best model/method against those of a medical expert. Lastly, we performed a safety analysis to categorize different types of fabricated information and to identify potential medical harm that may result from choosing either the model or the medical expert summary.
Results
Constructing prompt anatomy
We structured prompts (Fig. 2) by following best practices29,30 and evaluating a handful of options for model expertise and task instructions. Figure 2 also illustrates the effect of model expertise on GPT-3.5. For example, we achieved better performance by nudging the model toward medical expertise compared to specializing in wizardry or having no specific expertise at all. This illustrates the value of relevant context in achieving better outcomes for the target task. We also explored the temperature hyperparameter, which adjusts the LLM’s conditional probability distributions during sampling, hence affecting how often the model will output less likely tokens or individual units of text. Higher temperatures lead to more randomness and ‘creativity’, whereas lower temperatures produce more deterministic outputs. After searching over temperature values {0.1, 0.5, 0.9} using GPT-3.5, Fig. 2 demonstrates that the lowest value, 0.1, performed best. We, thus, set temperature to this value for all models. Intuitively, a lower value seems appropriate given our goal of factually summarizing text with a high aversion to factually incorrect text.
Fig. 2 |. Model prompts and temperature.
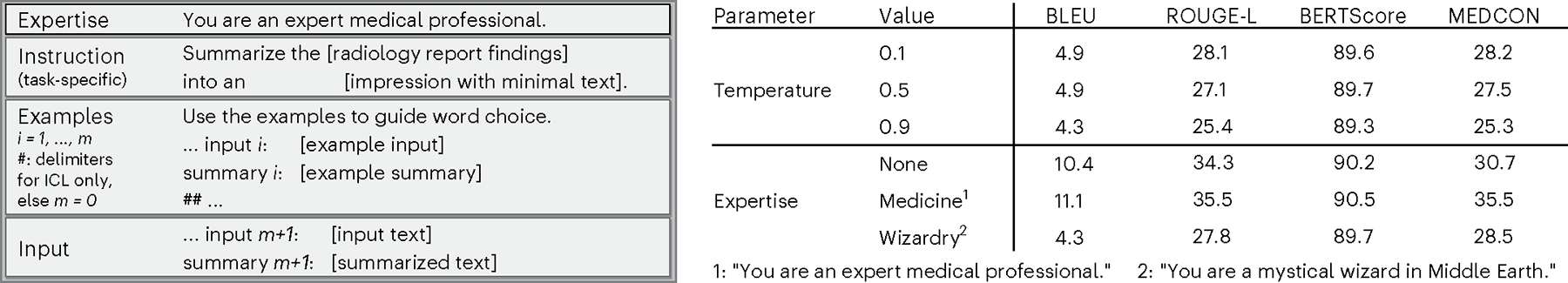
Left, prompt anatomy. Each summarization task uses a slightly different instruction. Right, model performance across different temperature values and expertise.
Identifying the best model/method
Following Fig. 1, we identified the best model/method across four summarization tasks comprising six datasets (Extended Data Table 1). This includes eight LLMs described in Extended Data Table 2: six open-source (FLAN-T5 (ref. 31), FLAN-UL2 (ref. 32), Alpaca33, Med-Alpaca34, Vicuna35 and Llama-2 (ref. 36)) and two proprietary (GPT-3.5 (ref. 37) and GPT-4 (ref. 38)). For adapting each model to a particular summarization task, we considered two proven adaptation strategies: in-context learning (ICL39), which adapts by including examples within the model prompt, and quantized low-rank adaptation (QLoRA40), which adapts by fine-tuning a subset of model weights on examples.
Impact of domain-specific fine-tuning.
When considering which open-source models to evaluate, we first assessed the benefit of fine-tuning open-source models on medical text. For example, Med-Alpaca is a version of Alpaca that was further instruction-tuned with medical question and answer (Q&A) text, consequently improving performance for the task of medical question–answering. Figure 3a compares these models for our task of summarization, showing that most data points are below the dashed lines, denoting equivalence. Hence, despite Med-Alpaca’s adaptation for the medical domain, it actually performed worse than Alpaca for our tasks of clinical text summarization, highlighting a distinction between domain adaptation and task adaptation. With this in mind, and considering that Alpaca is known to perform worse than our other open-source autoregressive models, Vicuna and Llama-2 (refs. 21,35), for simplicity we excluded Alpaca and Med-Alpaca from further analysis.
Fig. 3 |. Identifying the best model/method.
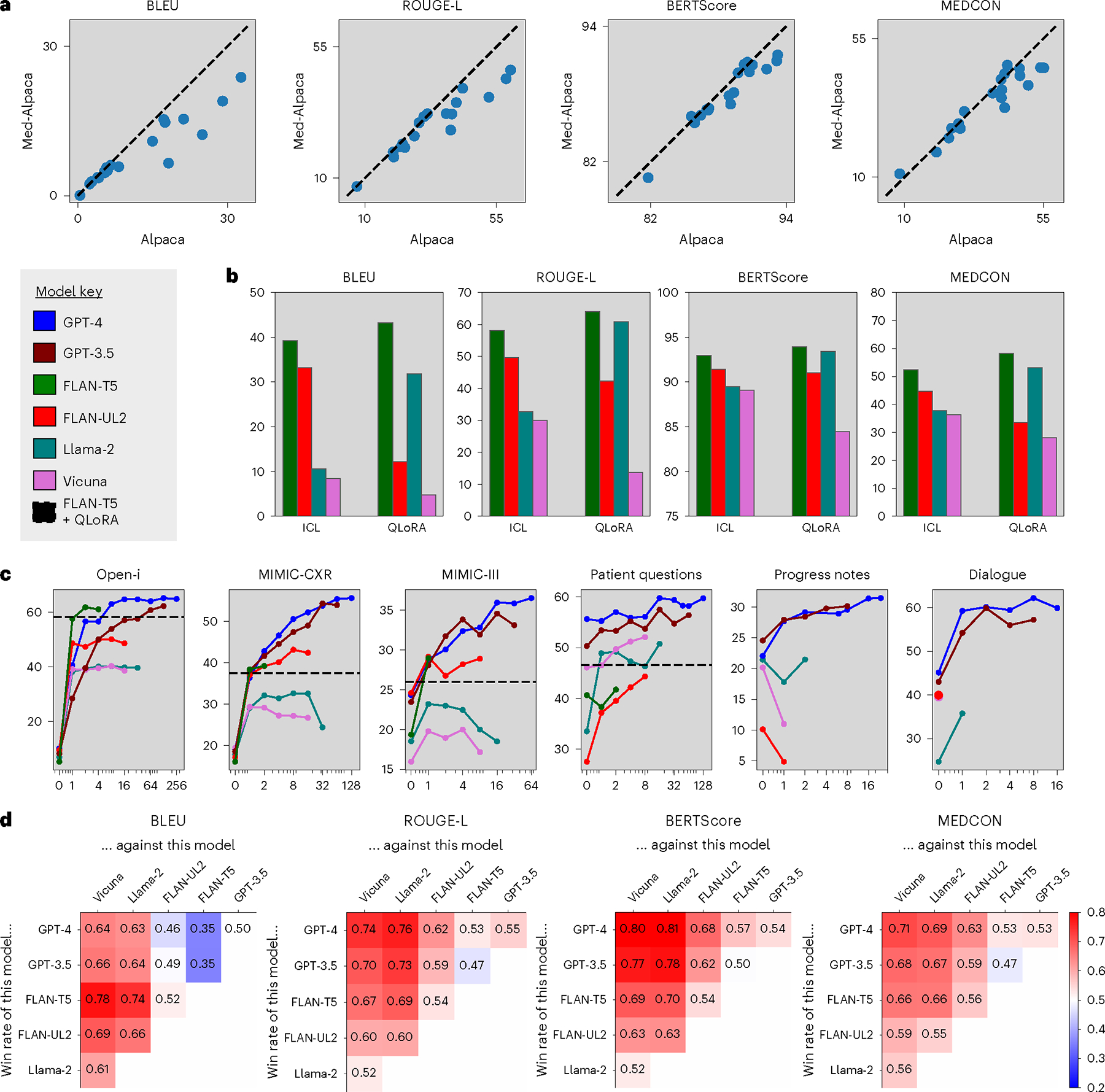
a, Impact of domain-specific fine-tuning. Alpaca versus Med-Alpaca. Each data point corresponds to one experimental configuration, and the dashed lines denote equal performance. b, Comparison of adaptation strategies. One in-context example (ICL) versus QLoRA across all open-source models on the Open-i radiology report dataset. c, Effect of context length for ICL. MEDCON scores versus number of in-context examples across models and datasets. We also included the best QLoRA fine-tuned model (FLAN-T5) as a horizontal dashed line for valid datasets. d, Head-to-head model comparison. Win percentages of each head-to-head model combination, where red/blue intensities highlight the degree to which models on the vertical axis outperform models on the horizontal axis.
Comparison of adaptation strategies.
Next, we compared ICL versus QLoRA across the remaining open-source models using the Open-i radiology reports dataset in Fig. 3b and the patient health questions dataset in Extended Data Fig. 1. We chose these datasets because their shorter context lengths allow for training with lower computational cost. FLAN-T5 emerged as the best-performing model with QLoRA. QLoRA typically outperformed ICL with the better models (FLAN-T5 and Llama-2); given a sufficient number of in-context examples, however, most models surpassed even the best QLoRA fine-tuned model, FLAN-T5 (Extended Data Fig. 2). FLAN-T5 (2.7B) eclipsed its fellow sequence-to-sequence (seq2seq) model, FLAN-UL2 (20B), despite being an older model with almost 10× fewer parameters.
Effect of context length for ICL.
Figure 3c displays MEDCON41 scores, which capture the quality of summaries with respect to medical concepts. These scores are plotted for all models against number of in-context examples, up to the maximum number of examples permitted by each model and dataset. This graph also includes the best-performing model (FLAN-T5) with QLoRA as a reference, depicted by a horizontal dashed line. Compared to prompting a model without examples (zero-shot prompting), adapting with even one example considerably improved performance in almost all cases, underscoring the importance of adaptation methods. Although ICL and QLoRA were competitive for open-source models, proprietary models GPT-3.5 and GPT-4 far outperformed other models and methods given sufficient in-context examples. For a similar graph across all metrics, see Extended Data Fig. 2.
Head-to-head model comparison.
Figure 3d compares models using win rates—that is, the head-to-head winning percentage of each model combination across the same set of samples. In other words, for what percentage of samples do model A’s summaries have a higher score than model B’s summaries? Although FLAN-T5 was more competitive for syntactic metrics, such as BLEU42, this model is constrained to a shorter context length of 512 (Extended Data Table 2).
Best model/method.
We deemed the best model and method to be GPT-4 (32,000 context length) with a maximum allowable number of in-context examples, hereon identified as the best-performing model.
Analyzing reader study results
Given our clinical reader study overview (Fig. 4a), pooled results across 10 physicians (Fig. 4b) demonstrate that summaries from the best-adapted model (GPT-4 using ICL) were more complete and contained fewer errors compared to medical expert summaries, which were created either by medical doctors during clinical care or by a committee of medical doctors and experts (Methods).
Fig. 4 |. Clinical reader study.
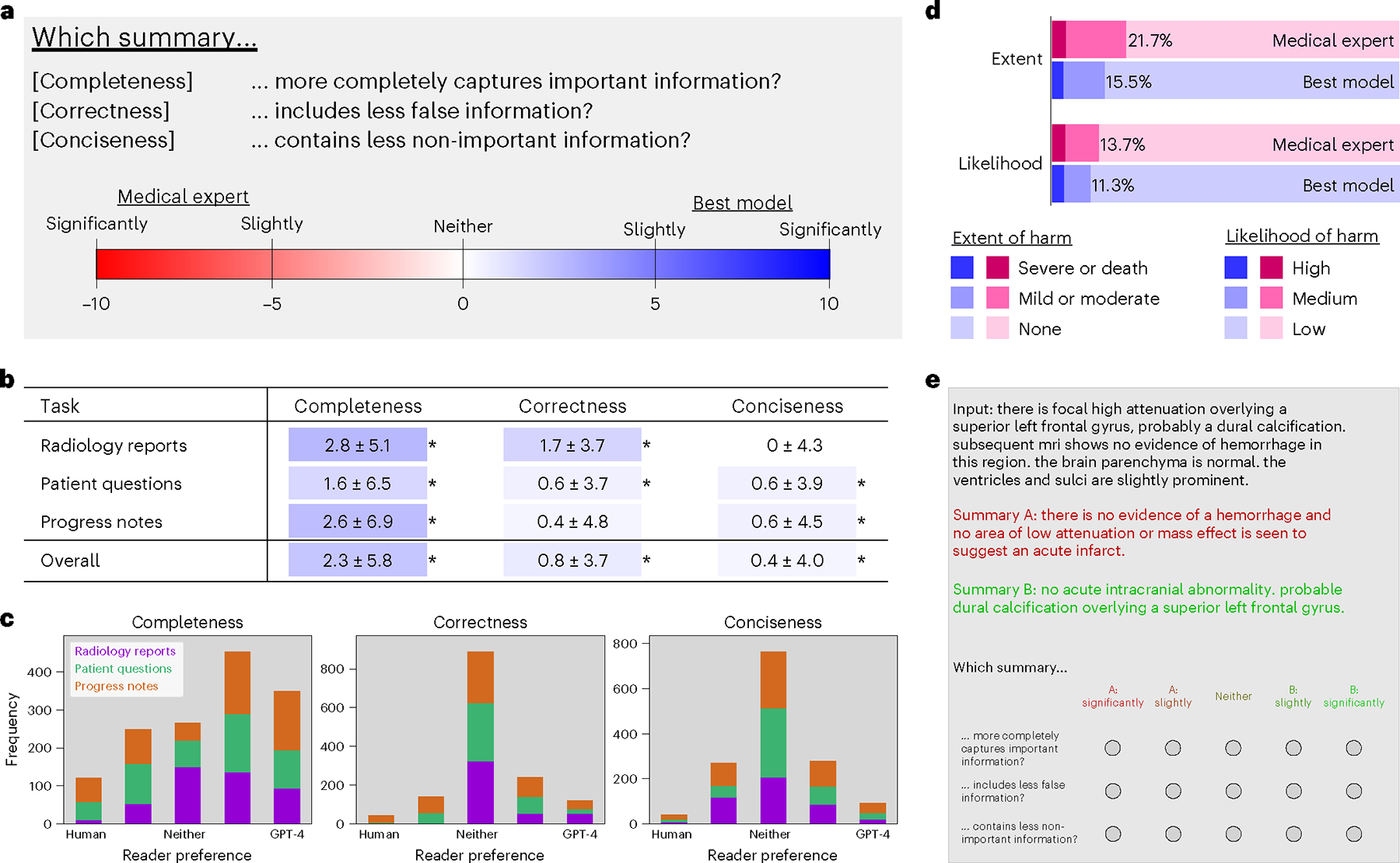
a, Study design comparing summaries from the best model versus that of medical experts on three attributes: completeness, correctness and conciseness. b, Results. Highlight colors correspond to a value’s location on the color spectrum. Asterisks (*) denote statistical significance by a one-sided Wilcoxon signed-rank test, P < 0.001. c, Distribution of reader scores for each summarization task across attributes. Horizontal axes denote reader preference as measured by a five-point Likert scale. Vertical axes denote frequency count, with 1,500 total cases for each plot. d, Extent and likelihood of possible harm caused by choosing summaries from the medical expert (pink) or best model (purple) over the other. e, Reader study user interface.
The distributions of reader responses in Fig. 4c show that medical expert summaries were preferred in only a minority of cases (19%), whereas, in a majority, the best model was either non-inferior (45%) or preferred (36%). Extended Data Table 3 contains scores separated by individual readers and affirms the reliability of scores across readers by displaying positive intra-reader correlation values. Based on physician feedback, we undertook a qualitative analysis to illustrate strengths and weaknesses of summaries by the model and medical experts (Fig. 5 and Extended Data Figs. 3 and 4).
Fig. 5 |. Annotation: radiology reports.
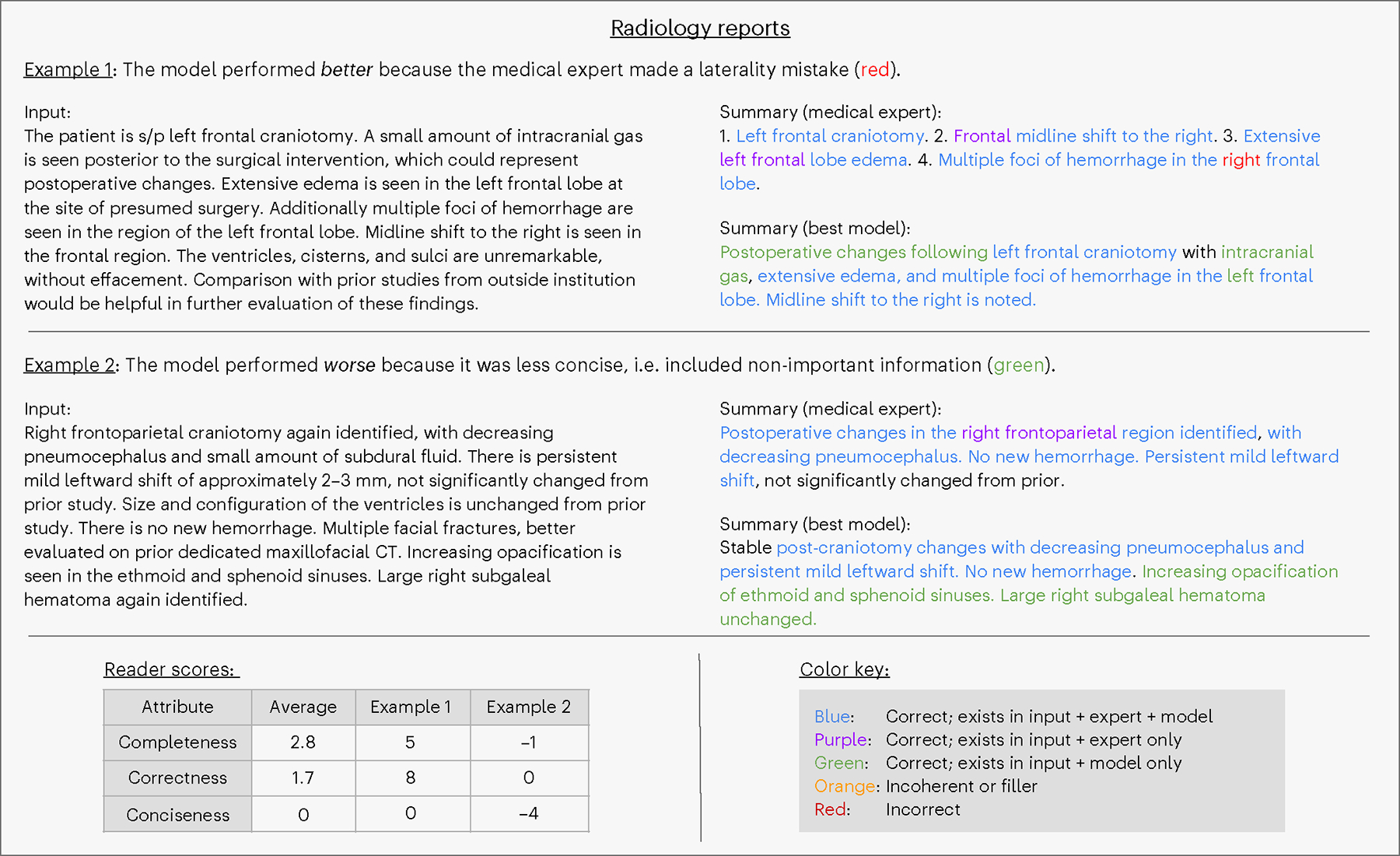
Qualitative analysis of two radiologist report examples from the reader study. The table (lower left) contains reader scores for these two examples and the task average across all samples.
We observed that the best model summaries were more complete, on average, than medical expert summaries, achieving statistical significance across all three summarization tasks with P < 0.001 (Fig. 4b). Lengths of summaries were similar between the model and medical experts for all three datasets: 47 ± 24 versus 44 ± 22 tokens for radiology reports, 15 ± 5 versus 14 ± 4 tokens for patient questions and 29 ± 7 versus 27 ± 13 tokens for progress notes. Hence, the model’s advantage in completeness is not simply a result of generating longer summaries. We provide intuition for completeness by investigating a specific example in progress notes summarization. In Extended Data Fig. 3, the model correctly identified conditions that were missed by the medical expert, such as hypotension and anemia. Although the model was more complete than the expert in generating its progress note summary, it also missed historical context (a history of hypertension).
Regarding correctness, the best model generated significantly fewer errors (P < 0.001) compared to medical expert summaries (Fig. 4b) overall and on two of three summarization tasks. As an example of the model’s superior correctness performance on the radiology report summarization task, we observe that it avoided common medical expert errors related to lateral distinctions (right versus left; Fig. 5). For the problem list summarization task, Extended Data Fig. 3 reveals an intriguing case: during the blinded study, the physician reader erroneously assumed that a hallucination—in this case, the incorrect inclusion of urinary tract infection—was made by the model. In this case, the medical expert was responsible for the hallucination. This instance underscores the point that even medical experts, not just LLMs, can hallucinate. Despite this promising performance, the model was not perfect across all tasks. We see a clear example in Extended Data Fig. 3 where the model mistakenly generated (hallucinated) several absent conditions, such as eosinophilia.
Regarding conciseness, the best model performed significantly better than medical experts (P < 0.001) overall and on two tasks, whereas, for radiology reports, it performed similarly to medical experts. We note that the model’s summaries are more concise while concurrently being more complete. Figure 5 provides an example in which the model’s summary includes correct information that readers deemed not important.
We then conducted a supplemental reader study connecting summarization errors to medical harm, inspired by the Agency for Healthcare Research and Quality (AHRQ)’s harm scale43. The results of this harm study (Fig. 4d) indicate that the medical expert summaries would have both a higher likelihood (14%) and a higher extent (22%) of possible harm compared to the summaries from the best model (12% and 16%, respectively).
Fabricated information
Fabricated information, or factually incorrect text, poses a substantial obstacle to the clinical integration of LLMs given the critical need for accuracy in medical applications. Our reader study results for correctness (Fig. 4b) indicate that the best model produces fewer instances of fabricated information than medical experts. Further, the need for accuracy motivates a more nuanced understanding of correctness for clinical text summarization. As such, we define three types of fabricated information: (1) misinterpretations of ambiguity; (2) factual inaccuracies: modifying existing facts to be incorrect; and (3) hallucinations: inventing new information that cannot be inferred from the input text. We found that the model committed misinterpretations, inaccuracies and hallucinations on 6%, 2% and 5% of samples, respectively, compared to 9%, 4% and 12%, respectively, by medical experts. Given the model’s lower error rate in each category, this suggests that incorporating LLMs could actually reduce fabricated information in clinical practice.
Connecting quantitative and clinical evaluations
Figure 6 captures the correlation between NLP metrics and physicians’ preferences. These values are calculated as the Spearman correlation coefficient between NLP metric scores and the magnitudes of reader scores. For correctness, the metrics BERTScore44 (measuring semantics) and MEDCON41 (measuring medical concepts) correlated most strongly with reader preference; meanwhile, the BLEU42 metric (measuring syntax) correlated most with completeness and least with conciseness. However, the low magnitude of correlation values (approximately 0.2) underscores the need to go beyond NLP metrics with a clinical reader study when assessing clinical readiness.
Fig. 6 |. Connecting NLP metrics and reader scores.
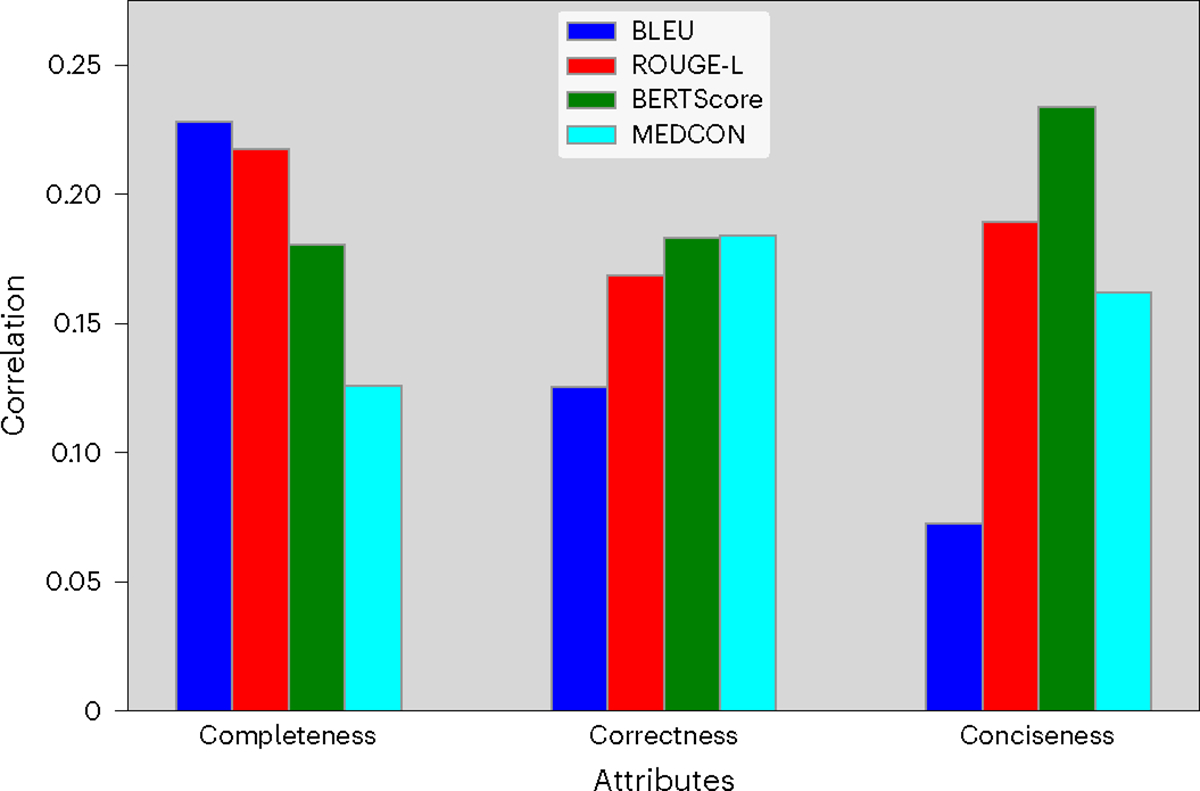
Spearman correlation coefficients between quantitative metrics and reader preference assessing completeness, correctness and conciseness.
Separately, we note the inclusion of additional results analyzing model size, demonstrating the dialogue task and comparing to summarization baselines in Extended Data Figs. 5 and 6 and Extended Data Table 4, respectively.
Discussion
In this research, we evaluated methods for adapting LLMs to summarize clinical text, analyzing eight models across a diverse set of summarization tasks. Our quantitative results underscore the advantages of adapting models to specific tasks and domains. The ensuing clinical reader study demonstrates that LLM summaries are often preferred over medical expert summaries due to higher scores for completeness, correctness and conciseness. The subsequent safety analysis explores qualitative examples, potential medical harm and fabricated information to demonstrate the limitations of both LLMs and medical experts. Evidence from this study suggests a potential avenue for LLMs to reduce documentation burden for clinicians.
We first highlight the importance of ‘prompt engineering’, or modifying and tuning the input prompt to improve model performance. This is well reflected in our evaluation of conciseness. We specified the desired summary length in the instruction—for example, with ‘one question of 15 words or less’ for summarizing patient questions (Extended Data Table 1). Without this instruction, the model might generate lengthy outputs, occasionally even longer than the input text. When considering conciseness scores (Fig. 4b), radiology reports were the only task in which physicians did not prefer the best model’s summaries to the medical experts. This could be attributed to the relatively vague length specification in the radiology reports instruction—that is, ‘…with minimal text’—whereas the other two task instructions quantify length.
Overall, our best-adapted model achieved non-inferior results to medical experts while performing a basic search across 1–2 options for each task instruction (Extended Data Table 1). Prompt phrasing and model temperature can have a considerable effect on LLM output, as demonstrated in the literature45,46 and in Fig. 2. This suggests that results could be further improved with additional prompt engineering and model hyperparameters, which is subject to future studies. In addition, beyond the scope of this manuscript, there is further potential to improve accuracy through incorporating checks by human operators and checks by other LLMs or using a model ensemble47,48.
Model performance generally improved with more context. Even one example provided considerable benefit compared to zero-shot prompting, underscoring the value of adaptation methods. Note that the number of allowable examples depends on the number of tokens per example and the model context length. This motivates future work to pursue more challenging tasks, such as summarizing longer documents or multiple documents of different types. Addressing these cases demands two key advancements: (1) extending model context length, potentially through multi-query aggregation or methods that increase context length49,50, and (2) introducing open-source datasets that include broader tasks and lengthier documents.
In terms of trade-offs between lightweight adaptation methods, while QLoRA fine-tuning performed similarly for some cases, ICL was the best overall, especially when including proprietary models GPT-3.5 and GPT-4. The proprietary nature of these models raises an interesting consideration for healthcare, where data and model governance are important, especially if summarization tools are cleared for clinical use by the Food and Drug Administration. This could motivate the use of fine-tuning methods on open-source models. Governance aside, ICL provides many benefits: (1) model weights are fixed, hence enabling queries of pre-existing LLMs, and (2) adaptation is feasible with even a few examples, whereas fine-tuning methods, such as QLoRA, typically require hundreds or thousands of examples.
We consider trade-offs of different model types: autoregressive and seq2seq. Seq2seq models (FLAN-T5 and FLAN-UL2) performed very well on syntactical metrics, such as BLEU, but worse on others (Fig. 3d), suggesting that these models excel more at matching word choice than matching semantic or conceptual meaning. Note that seq2seq models are often constrained to much shorter context length than autoregressive models, such as GPT-4, because seq2seq models require the memory-intensive step of encoding the input sequence into a fixed-size context vector. Among open-source models, seq2seq models performed better than autoregressive models (Llama-2 and Vicuna) on radiology reports but worse on patient questions and progress notes (Fig. 3c). Given that these latter datasets have higher lexical variance (Extended Data Table 1) and more heterogeneous formatting compared to radiology reports, we hypothesize that autoregressive models may perform better with increasing data heterogeneity and complexity.
The evidence from our reader study suggests that adapting LLMs can outperform medical experts in terms of completeness, correctness and conciseness. When qualitatively analyzing summaries, we notice a few general trends. As implied by the completeness scores, the best-adapted model (GPT-4 using ICL) excelled at identifying and understanding the most relevant information from the source text. However, both the model and the medical experts faced challenges interpreting ambiguity, such as user queries in patient health questions. Consider example 1 in Extended Data Fig. 4, in which the input question mentioned ‘diabetes and neuropathy’.
The model mirrored this phrasing verbatim, whereas the medical expert interpreted it as ‘diabetic neuropathy’. This highlights the model’s tendency toward a literal approach without interpretation, which may be either advantageous or limiting. In example 2 of Extended Data Fig. 4, the model simply reformulated the input question about tests and their locations, whereas the medical expert inferred a broader query about tests and treatments. In both cases, the model’s summaries leaned toward literalness, a trait that readers sometimes favored and sometimes did not. In future work, a systematic exploration of model temperature could further illuminate this trade-off.
Regarding general trends for our clinical NLP metrics, the syntactic metric BLEU provided the highest correlation with physician preference for completeness. Given that BLEU measures sequence overlap, this result seems reasonable, as more text provides more ‘surface area’ for overlap; more text also reduces the brevity penalty that BLEU applies on generated sequences, which are shorter than the reference42. In addition, the metrics BERTScore and MEDCON correlated most strongly with physician preference for correctness. This implies that the semantics (BERTScore) and concepts (MEDCON) measured by these metrics correspond to correctness more effectively than syntactic metrics BLEU and ROUGE-L51.
Many previous clinical NLP studies rely primarily on quantitative metrics for evaluation41,52–54. Given the critical nature of nuanced medical tasks, such as summarization, that typically have no objective solutions, including human experts in the evaluation process beyond quantitative metrics is crucial to demonstrate clinical readiness. To address this, there have been recent releases of expert evaluations for adjacent clinical NLP tasks3,55. Other studies employ human experts to evaluate synthesized abstracts, demonstrating that NLP metrics are not sufficient to measure summary quality56. Aside from the low correlation values in Fig. 6, our reader study results in Fig. 4 also highlight another limitation of NLP metrics, especially as model-generated summaries become increasingly viable. These metrics rely on a reference (in our case, the medical expert), which we have demonstrated may contain errors. Hence, we suggest that human evaluation is essential when assessing the clinical feasibility of new methods. When human evaluation is not feasible, Fig. 6 suggests that syntactic metrics are better at measuring completeness, whereas semantic and conceptual metrics are better at measuring correctness.
The present study has several limitations that must be addressed in future research. First, we did not consider the inherently context-specific nature of summarization. For example, a gastroenterologist, a radiologist and an oncologist may have different preferences for summaries of a cancer patient with liver metastasis. Or perhaps an abdominal radiologist will want a different summary than a neuroradiologist. Furthermore, individual clinicians may prefer different styles or amounts of information. Although we did not explore such a granular level of adaptation, this may not require much further development: because the best model and method uses a handful of examples via ICL, one could plausibly adapt using examples curated for a particular specialty or clinician. Another limitation is that radiology report summaries from medical experts occasionally recommend further studies or refer to prior studies—for example, ‘… not significantly changed from prior’ in Fig. 5. These instances are out of scope for the LLM, as it does not have access to prior studies nor the purview to make recommendations. Hence, for our clinical reader study, physicians were told to disregard these phrases. However, future work can explore providing more context via prior reports and allow the LLM to make a treatment suggestion. An additional consideration for our study and other LLM studies, especially with proprietary models, is that it is not possible to verify whether a particular open-source dataset was included in model training. Although three of our datasets (MIMIC-CXR, MIMIC-III and ProbSum) require PhysioNet57 access to ensure safe data usage by third parties, this is no guarantee against data leakage. This complication highlights the need for validating results on internal data when possible. We further note the potential for LLMs to be biased58,59. Although our datasets do not contain demographic information, we advocate for future work to consider whether summary qualities have any dependence upon group membership.
The findings from our study demonstrate that adapting LLMs can outperform medical experts for clinical text summarization across the diverse range of documents that we evaluated. This suggests that incorporating LLM-generated candidate summaries could reduce documentation load, potentially leading to decreased clinician strain and improved patient care. Testing this hypothesis requires future prospective studies in clinical environments.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-024-02855-5.
Methods
LLMs
We investigated a diverse collection of transformer-based LLMs for clinical summarization tasks. This included two broad approaches to language generation: seq2seq models and autoregressive models. Seq2seq models use an encoder–decoder architecture to map the input text to a generated output, often requiring paired datasets for training. These models have shown strong performance in machine translation60 and summarization61. In contrast, the autoregressive models typically use only a decoder. They generate tokens sequentially—where each new token is conditioned on previous tokens—thus efficiently capturing context and long-range dependencies. Autoregressive models are typically trained with unpaired data, and they are particularly useful for various NLP tasks, such as text generation, question–answering and dialogue interactions17,35.
We included prominent seq2seq models owing to their strong summarization performance61 and autoregressive models owing to their state-of-the-art performance across general NLP tasks21. As shown in Extended Data Table 2, our choice of models varied widely with respect to number of parameters (2.7 billion to 175 billion) and context length (512 to 32,000)—that is, the maximum number of input tokens a model can process. We organized our models into three categories:
Open-source seq2seq models.
The original T5 ‘text-to-text transfer transformer’ model62 demonstrated excellent performance in transfer learning using the seq2seq architecture. A derivative model, FLAN-T5 (refs. 31,63), improved performance via instruction prompt tuning. This T5 model family has proven effective for various clinical NLP tasks27,64. The FLAN-UL2 model32,31 was introduced recently, which features an increased context length (fourfold that of FLAN-T5) and a modified pre-training procedure called unified language learning (UL2).
Open-source autoregressive models.
The Llama family of LLMs36 has enabled the proliferation of open-source instruction-tuned models that deliver similar performance to GPT-3 (ref. 17) on many benchmarks despite their smaller sizes. Descendants of this original model have taken additional fine-tuning approaches, such as fine-tuning via instruction-following (Alpaca33), medical Q&A data (Med-Alpaca34), user-shared conversations (Vicuna35) and reinforcement learning from human feedback (Llama-2 (ref. 36)). Llama-2 allows for twofold longer context lengths (4,096) relative to the aforementioned open-source autoregressive models.
Our focus was primarily on the 7B-parameter tier of these models despite some models, such as Llama-2, having larger versions. The benefit of larger models is explored in Extended Data Fig. 5, which found this improvement marginal for Llama-2 (13B) compared to Llama-2 (7B). Although other open-source models might have slightly outperformed our selections, this likely would not have substantially changed our analysis, especially because the clinical reader study employed a state-of-the-art proprietary model21.
Proprietary autoregressive models.
We include GPT-3.5 (ref. 37) and GPT-4 (ref. 38), the latter of which has been regarded as state of the art on general NLP tasks21 and has demonstrated strong performance on biomedical NLP tasks, such as medical examinations65–67. Both models offer significantly higher context length (16,384 and 32,768) than open-source models. We note that, since sharing our work, GPT-4’s context length has been increased to 128,000.
Adaptation methods
We considered two proven techniques for adapting pre-trained, general-purpose LLMs to domain-specific tasks:
ICL.
ICL is a lightweight adaptation method that requires no altering of model weights; instead, one includes a handful of in-context examples directly within the model prompt62. This simple approach provides the model with context, enhancing LLM performance for a particular task or domain27,28. We implemented this by choosing, for each sample in our test set, the m nearest neighbors training samples in the embedding space of the PubMedBERT model68. Note that choosing ‘relevant’ in-context examples has been shown to outperform choosing examples at random69. For a given model and dataset, we used m = 2x examples, where x ∈ {0, 1, 2, 3,…, M} for M such that no more than 1% of the s = 250 samples were excluded due to prompts exceeding the model’s context length. Hence, each model’s context length limited the allowable number of in-context examples.
To demonstrate the benefit of adaptation methods, we included the baseline zero-shot prompting—that is m = 0 in-context samples.
QLoRA.
Low-rank adaptation (LoRA)70 has emerged as an effective, lightweight approach for fine-tuning LLMs by altering a small subset of model weights, often less than 0.1% (ref. 27). LoRA inserts trainable matrices into the attention layers; then, using a training set of samples, this method performs gradient descent on the inserted matrices while keeping the original model weights frozen. Compared to training model weights from scratch, LoRA is much more efficient with respect to both computational requirements and the volume of training data required. Recently, QLoRA40 was introduced as a more memory-efficient variant of LoRA, employing 4-bit quantization to enable the fine-tuning of larger LLMs given the same hardware constraints. This quantization negligibly impacts performance40; as such, we used QLoRA for all model training. Note that QLoRA could not be used to fine-tune proprietary models on our consumer hardware, as their model weights are not publicly available. Fine-tuning of GPT-3.5 via API was made available after our internal model cutoff date of 31 July 202371.
Data
To robustly evaluate LLM performance on clinical text summarization, we chose four distinct summarization tasks, comprising six open-source datasets. As depicted in Extended Data Table 1, each dataset contained a varying number of samples, token lengths and lexical variance. Lexical variance is calculated as the ratio of unique words to total words across the entire dataset; hence, a higher ratio indicates less repetition and more lexical diversity. We describe each task and dataset below. For examples of each task, see Fig. 5 and Extended Data Figs. 3, 4 and 6.
Radiology reports.
Radiology report summarization takes as input the findings section of a radiology study containing detailed examination analysis and results. The goal is to summarize these findings into an impression section that concisely captures the most salient, actionable information from the study. We considered three datasets for this task, where both reports and findings were created by attending physicians as part of routine clinical care. Open-i72 contains de-identified narrative chest x-ray reports from the Indiana Network for Patient Care 10 database. From the initial set of 4,000 studies, Demner-Fushman et al.72 selected a final set of 3,400 reports based on the quality of imaging views and diagnostic content. MIMIC-CXR73 contains chest x-ray studies accompanied by free-text radiology reports acquired at the Beth Israel Deaconess Medical Center between 2011 and 2016. For this study, we used a dataset of 128,000 reports69 pre-processed by the RadSum23 shared task at BioNLP 2023 (refs. 74,75). MIMIC-III76 contains 67,000 radiology reports spanning seven anatomies (head, abdomen, chest, spine, neck, sinus and pelvis) and two modalities: magnetic resonance imaging (MRI) and computed tomography (CT). This dataset originated from patient stays in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. For this study, we used a pre-processed version via RadSum23 (refs. 74,75). Compared to x-rays, MRIs and CT scans capture more information at a higher resolution. This usually leads to longer reports (Extended Data Table 1), rendering MIMIC-III a more challenging summarization dataset than Open-i or MIMIC-CXR.
Patient questions.
Question summarization consists of generating a condensed question expressing the minimum information required to find correct answers to the original question77. For this task, we employed the MeQSum dataset77. MeQSum contains (1) patient health questions of varying verbosity and coherence selected from messages sent to the US National Library of Medicine and (2) corresponding condensed questions created by three medical experts such that the summary allows retrieving complete, correct answers to the original question without the potential for further condensation. These condensed questions were then validated by a medical doctor and verified to have high inter-annotator agreement. Due to the wide variety of these questions, MeQSum exhibited the highest lexical variance of our datasets (Extended Data Table 1).
Progress notes.
The goal of this task was to generate a ‘problem list’, or a condensed list of diagnoses and medical problems using the provider’s progress notes during hospitalization. For this task, we employed the ProbSum dataset78. This dataset, generated by attending internal medicine physicians during the course of routine clinical practice, was extracted from the MIMIC-III database of de-identified hospital intensive care unit (ICU) admissions. ProbSum contains (1) progress notes averaging more than 1,000 tokens and substantial presence of unlabeled numerical data—for example, dates and test results—and (2) corresponding problem lists created by attending medical experts in the ICU. We accessed these data via the BioNLP Problem List Summarization shared task75,79,80 and PhysioNet81.
Dialogue.
The goal of this task was to summarize a doctor–patient conversation into an ‘assessment and plan’ paragraph. For this task, we employed the ACI-Bench dataset41,82–84, which contains (1) 207 doctor–patient conversations and (2) corresponding patient visit notes, which were first generated by a seq2seq model and subsequently corrected and validated by expert medical scribes and physicians. Because ACI-Bench’s visit notes include a heterogeneous collection of section headers, we chose 126 samples containing an ‘assessment and plan’ section for our analysis. Per Extended Data Table 1, this task entailed the largest token count across our six datasets for both the input (dialogue) and the target (assessment).
As we are not the first to employ these datasets, Extended Data Table 4 contains quantitative metric scores from other works25,27,41,52–54 that developed methods specific to each individual summarization task.
Experimental setup
For each dataset, we constructed test sets by randomly drawing the same s samples, where s = 250 for all datasets except dialogue (s = 100), which included only 126 samples in total. After selecting these s samples, we chose another s as a validation set for datasets, which incorporated fine-tuning. We then used the remaining samples as a training set for ICL examples or QLoRA fine-tuning.
We leveraged PyTorch for our all our experiments, which included the parameter-efficient fine-tuning84 and the generative pre-trained transformers quantization85 libraries for implementing QLoRA. We fine-tuned models with QLoRA for five epochs using the Adam optimizer with weight decay fix86. An initial learning rate of 1 × 10−3 was decayed linearly to 1 × 10−4 after a 100-step warm-up; we determined this configuration after experimenting with different learning rates and schedulers. To achieve an effective batch size of 24 on each experiment, we adjusted both individual batch size and number of gradient accumulation steps to fit on a single consumer GPU, a NVIDIA Quadro RTX 8000. All open-source models are available on HuggingFace87.
Quantitative metrics
We used well-known summarization metrics to assess the quality of generated summaries. BLEU42, the simplest metric, calculates the degree of overlap between the reference and generated texts by considering 1-gram to 4-gram sequences. ROUGE-L51 evaluates similarity based on the longest common subsequence; it considers both precision and recall, hence being more comprehensive than BLEU. In addition to these syntactic metrics, we employed BERTScore, which leverages contextual BERT embeddings to evaluate the semantic similarity of the generated and reference texts44. Lastly, we included MEDCON41 to gauge the consistency of medical concepts. This employs QuickUMLS88, a tool that extracts biomedical concepts via string-matching algorithms89. MEDCON was restricted to relvant UMLS semantic groups (Anatomy, Chemicals & Drugs, Device, Disorders, Genes & Molecular Sequences, Phenomena and Physiology). All four metrics ranged from [0, 100] with higher scores indicating higher similarity between the generated and reference summaries.
Reader study
After identifying the best model and method via NLP quantitative metrics, we performed a clinical reader study across three summarization tasks: radiology reports, patient questions and progress notes. The dialogue task was excluded owing to the unwieldiness of a reader parsing many lengthy transcribed conversations and paragraphs; see Extended Data Fig. 6 for an example and Extended Data Table 1 for the token count.
Our readers included two sets of physicians: (1) five board-certified radiologists to evaluate summaries of radiology reports and (2) five board-certified hospitalists (internal medicine physicians) to evaluate summaries of patient questions and progress notes. For each task, each physician viewed the same 100 randomly selected inputs and their A/B comparisons (medical expert versus the best model summaries), which were presented in a blinded and randomized order. An ideal summary would contain all clinically important information (‘completeness’) without any errors (‘correctness’) or superfluous information (‘conciseness’). Hence, we posed the following three questions for readers to evaluate using a five-point Likert scale.
Completeness: ‘Which summary more completely captures important information?’ This compares the summaries’ recall—that is, the amount of clinically important detail retained from the input text.
Correctness: ‘Which summary includes less false information?’ This compares the summaries’ precision—that is, instances of fabricated information.
Conciseness: ‘Which summary contains less non-important information?’ This compares which summary is more condensed, as the value of a summary decreases with superfluous information.
To obfuscate any formatting differences between the model and medical expert summaries, we applied simple post-processing to standardize capitalization, punctuation, newline characters, etc. Figure 4e demonstrates the user interface for this study, which we created and deployed via Qualtrics.
Statistical analysis
Given these non-parametric, categorical data, we assessed the statistical significance of responses using a Wilcoxon signed-rank test with type 1 error rate = 0.05, adjusted for multiple comparisons using Bonferroni correction. We estimated intra-reader correlation based on a mean-rating, fixed-agreement, two-may mixed-effects model90 using the Pingouin package91. Additionally, readers were provided comment space to make observations for qualitative analysis.
Connecting errors to medical harm
We then conducted a supplemental reader study connecting summarization errors to medical harm, inspired by the AHRQ harm scale43. We selected radiology reports (nr = 27) and progress notes (nn = 44) samples that contained disparities in completeness and/or correctness between the best model and medical expert summaries. Here, disparities occur if at least one physician significantly preferred, or at least two physicians slightly preferred, one summary to the other. These summary pairs were then randomized and blinded. For each sample, we asked the following multiple-choice questions: ‘Summary A is more complete and/or correct than Summary B. Now, suppose Summary B (worse) is used in the standard clinical workflow. Compared to using Summary A (better), what would be the…’ (1) ‘… extent of possible harm?’ options: {none, mild or moderate harm, severe harm or death} and (2) ‘… likelihood of possible harm?’ options: {low, medium, high}. The percentages displayed in Fig. 4d were computed with respect to all samples, such that the subset of samples with similar A/B summaries (in completeness and correctness) were assumed to contribute no harm.
Connecting quantitative and clinical evaluations
We next outlined our calculation of correlation values between NLP metrics and clinical reader scores in Fig. 6. Note that, in this work, these tools measured different quantities: NLP metrics measured the similarity between two summaries, whereas reader scores measured which summary is better. Consider an example where two summaries are exactly the same: NLP metrics would yield the highest possible score (100), whereas clinical readers would provide a score of 0 to denote equivalence. As the magnitude of a reader score increases, the two summaries are increasingly dissimilar, yielding a lower quantitative metric score. Hence, the correlation values are calculated as the Spearman correlation coefficients between NLP metric scores and the magnitudes of the reader scores. Because these features are inversely correlated, for clarity we display the negative correlation coefficient values.
Statistics and reproducibility
When determining sample size, we used our best judgment based on the size of datasets and clinician time. Note that these models are new, and they were applied to a new set of tasks. Thus, there was no prior effect size available to guide our sample size estimates. For the quantitative experiments, our sample size of 250 was chosen to maximize the number of samples given constraints of dataset size and cost of computing resources. We deem this sufficient as it enabled reproducible results when running the same experiments with a different set of samples. Furthermore, our use of six datasets enhances the robustness of our experiments. Note that we reduced the temperature parameter of the LLMs to be near zero, hence reducing randomness in the generated output summaries. For the clinical reader study, our sample size of 100 comparisons per reader per task was chosen to maximize the number of comparisons that could be made in a reasonable amount of time, which we estimated as 10 h. We deem this sufficient as it enabled statistically significant results for many combinations of tasks and attributes. No data were excluded from the analyses.
Data, models and code are all publicly available (https://github.com/StanfordMIMI/clin-summ). Internally, we verified reproducibility by achieving similar results across all six datasets. Because the LLM temperature parameter is near zero, outputs have very little randomness and are, thus, reproducible. Additionally, we chose the smallest dataset––which consequently required the lowest cost of computational resources––and re-ran a set of experiments that rendered very similar quantitative metric scores. This provided confidence moving forward with datasets that required higher computational resources.
Regarding randomization, for the quantitative experiments, we randomly selected 250 test samples and then randomly divided remaining samples into training and validation sets. For the clinical reader study, we randomly selected 100 samples for comparison; these samples and the A/B comparisons within each sample are displayed in random order. Regarding blinding, for the clinical reader study, we presented, in a blinded manner, the A/B comparison of summaries from the model and human experts. To obfuscate any formatting differences between A and B, we applied simple post-processing to standardize capitalization, punctuation, newline characters, etc.
Ethics approval
The clinical reader study component of this research involved the participation of physicians. This study adhered to the principles outlined in the Declaration of Helsinki. Informed consent was obtained from each physician before their participation. This study used only retrospective, de-identified data that fell outside the scope of institutional review board oversight.
Extended Data
Extended Data Fig. 1 |. ICL vs. QLoRA.
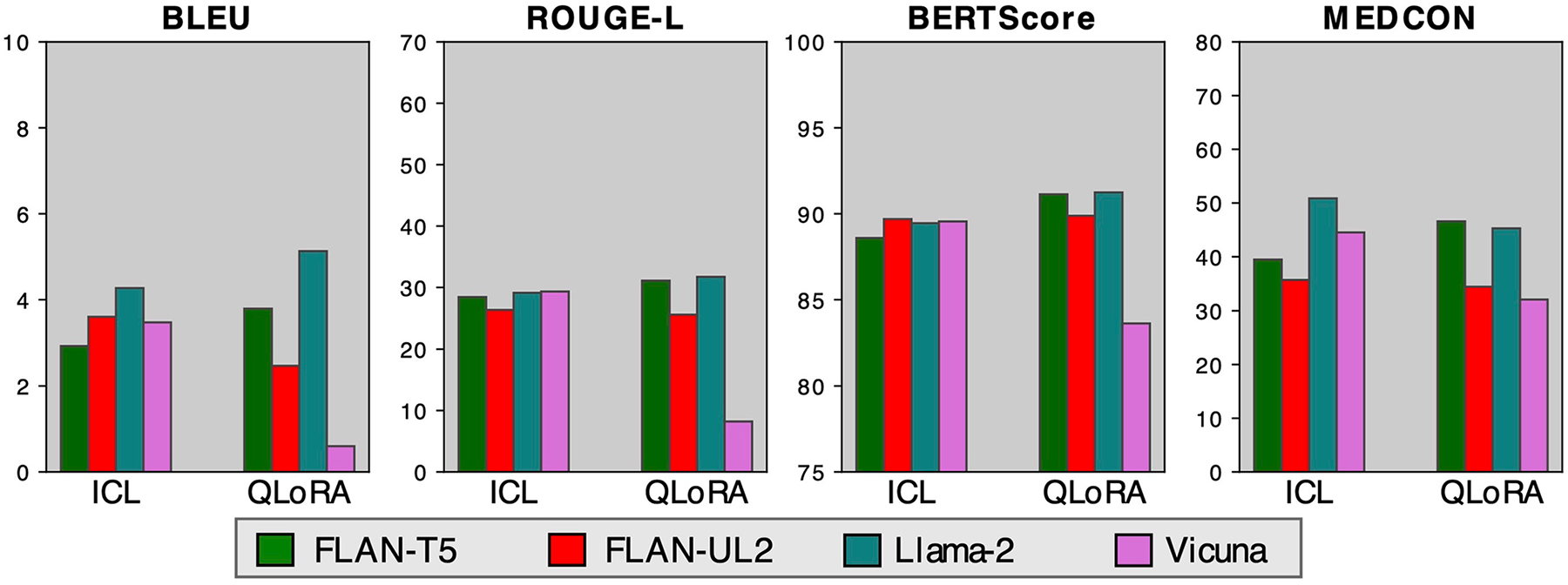
Summarization performance comparing one in-context example (ICL) vs. QLoRA across all open-source models on patient health questions.
Extended Data Fig. 2 |. Quantitative results across all metrics.
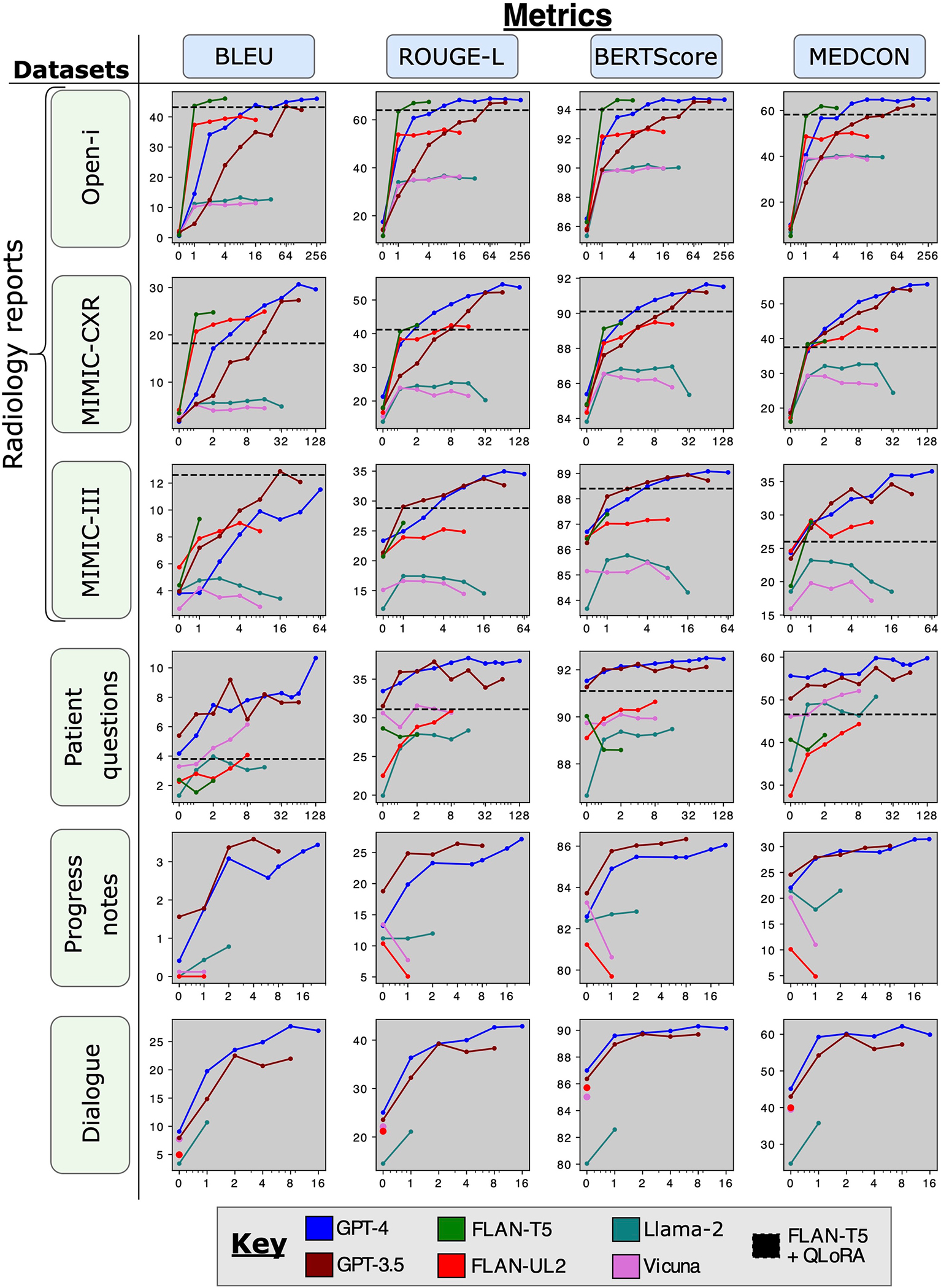
Metric scores vs. number of in-context examples across models and datasets. We also include the best model fine-tuned with QLoRA (FLAN-T5) as a horizontal dashed line.
Extended Data Fig. 3 |. Annotation: progress notes.

Qualitative analysis of two progress notes summarization examples from the reader study. The table (lower right) contains reader scores for these examples and the task average across all samples.
Extended Data Fig. 4 |. Annotation: patient questions.
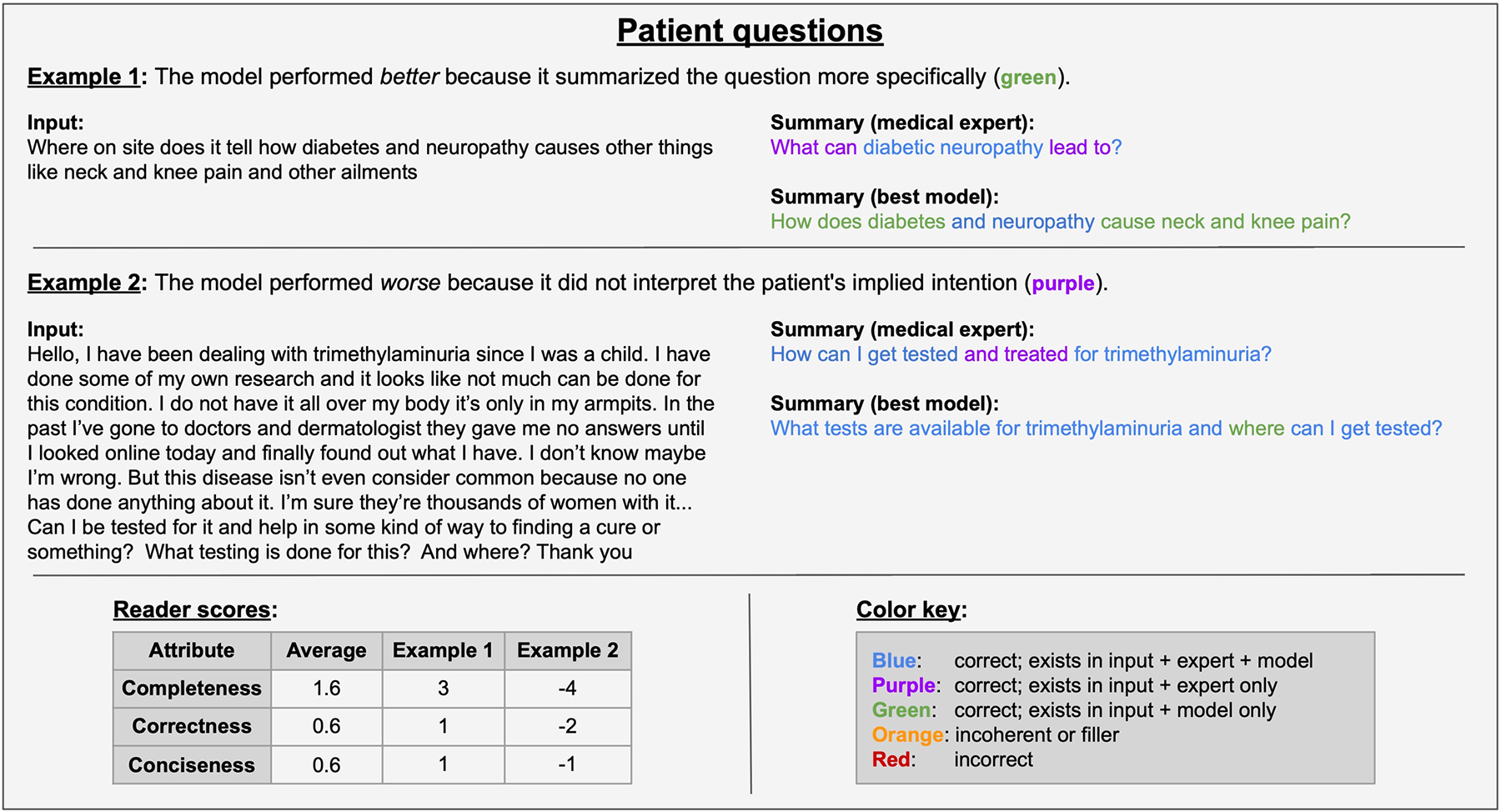
Qualitative analysis of two patient health question examples from the reader study. The table (lower left) contains reader scores for these examples and the task average across all samples.
Extended Data Fig. 5 |. Effect of model size.
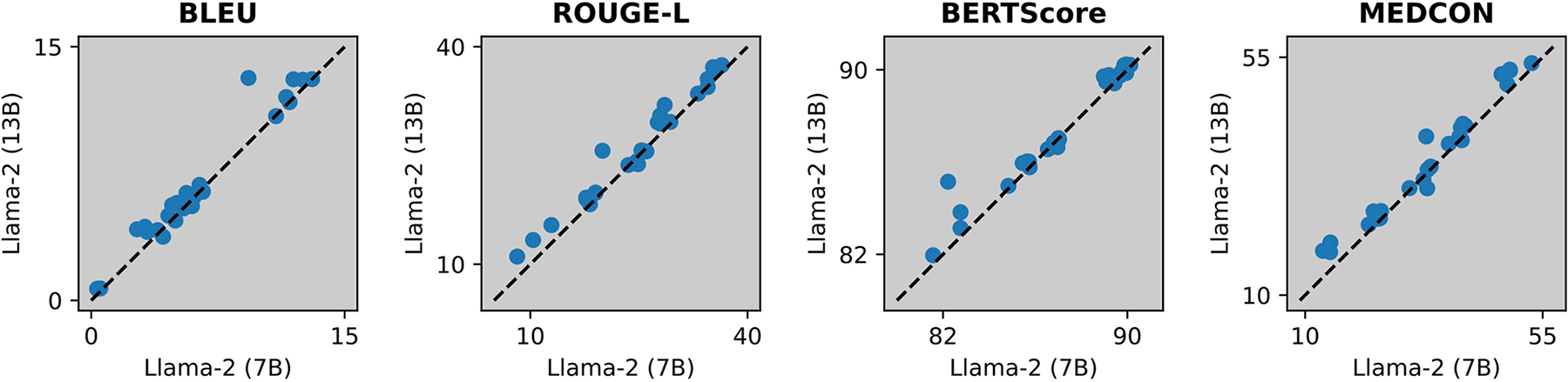
Comparing Llama-2 (7B) vs. Llama-2 (13B). The dashed line denotes equivalence, and each data point corresponds to the average score of s = 250 samples for a given experimental configuration, that is {dataset x m in-context examples}.
Extended Data Fig. 6 |. Example: dialogue.

Example of the doctor-patient dialogue summarization task, including ‘assessment and plan’ sections generated by both a medical expert and the best model.
Extended Data Table 1 |.
Datasets, task instructions
| Model | Context | Parameters | Proprietary? | Seq2seq | Autoreg. |
|---|---|---|---|---|---|
|
| |||||
| FLAN-T5 | 512 | 2.7B | - | ✔ | - |
| FLAN-UL2 | 2,048 | 20B | - | ✔ | - |
| Alpaca | 2,048 | 7B | - | - | ✔ |
| Med-Alpaca | 2,048 | 7B | - | - | ✔ |
| Vicuna | 2,048 | 7B | - | - | ✔ |
| Llama-2 | 4,096 | 7B, 13B | - | - | ✔ |
| GPT-3.5 | 16,384 | 175B | ✔ | - | ✔ |
| GPT-4 | 32,768* | unknown | ✔ | - | ✔ |
The context length of GPT-4 has since been increased to 128,000.
We quantitatively evaluated eight models, including state-of-the-art seq2seq and autoregressive models. Unless specified, models are open source (versus proprietary).
Extended Data Table 2 |.
Models
| Dataset descriptions | |||||
|---|---|---|---|---|---|
|
| |||||
| Dataset | Task | Number of samples | Avg. number of tokens | Lexical variance | |
| Input | Target | ||||
|
| |||||
| Open-i | Radiology reports | 3.4K | 52 ± 22 | 14 ± 12 | 0.11 |
| MIMIC-CXR | Radiology reports | 128K | 75 ± 31 | 22 ± 17 | 0.08 |
| MIMIC-III | Radiology reports | 67K | 160 ± 83 | 61 ± 45 | 0.09 |
| MeQSum | Patient questions | 1.2K | 83 ± 67 | 14 ± 6 | 0.21 |
| ProbSum | Progress notes | 755 | 1,013 ± 299 | 23 ± 16 | 0.15 |
| ACI-Bench | Dialogue | 126 | 1,512 ± 467 | 211 ± 98 | 0.04 |
| Task Instructions | |
|---|---|
|
| |
| Task | Instruction |
|
| |
| Radiology reports | “Summarize the radiology report findings into an impression with minimal text.” |
| Patient questions | “Summarize the patient health query into one question of 15 words or less.” |
| Progress notes | “Based on the progress note, generate a list of 3–7 problems (a few words each) ranked in order of importance.” |
| Dialogue | “Summarize the patient/doctor dialogue into an assessment and plan.” |
Top, description of six open-source datasets with a wide range of token Length and Lexical variance, or the ratio of unique words to total words. Bottom, instructions for each of the four summarization tasks.
Extended Data Table 3 |.
Individual reader scores
| Task | Reader | Completeness | Correctness | Conciseness |
|---|---|---|---|---|
|
| ||||
| Radiology reports | 1 | 3.5 ± 5.6 | 1.7 ± 3.6 | 1.2 ± 4.8 |
| 2 | 3.6 ± 6.6 | 2.5 ± 4.7 | −0.3 ± 5.4 | |
| 3 | 0.8 ± 2.9 | 0.6 ± 3.2 | −1.7 ± 3.0 | |
| 4 | 4.7 ± 4.7 | 2.9 ± 3.9 | 1.2 ± 3.8 | |
| 5 | 1.4 ± 4.0 | 0.6 ± 2.2 | −0.6 ± 3.4 | |
| Pooled | 2.8 ± 5.1 * | 1.7 ± 3.7 * | 0.0 ± 4.3 | |
| ICC | 0.45 | 0.58 | 0.48 | |
|
| ||||
| Patient questions | 1 | 1.7 ± 7.2 | 0.6 ± 3.4 | 0.3 ± 3.4 |
| 2 | 1.0 ± 5.6 | −0.1 ± 3.6 | 0.1 ± 3.6 | |
| 3 | 2.3 ± 7.2 | 2.0 ± 5.3 | 2.2 ± 5.9 | |
| 4 | 1.9 ± 6.7 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| 5 | 0.9 ± 5.7 | 0.4 ± 3.6 | 0.4 ± 3.6 | |
| Pooled | 1.6 ± 6.5 * | 0.6 ± 3.7 * | 0.6 ± 3.9 * | |
| ICC | 0.67 | 0.31 | 0.21 | |
|
| ||||
| Progress notes | 1 | 3.4 ± 7.5 | 0.5 ± 2.5 | 0.1 ± 4.5 |
| 2 | 2.3 ± 6.5 | 0.6 ± 4.4 | 0.4±4.2 | |
| 3 | 2.7 ± 6.3 | 1.0 ± 4.4 | 0.9 ± 3.7 | |
| 4 | 2.5 ± 7.2 | 0.5 ± 6.8 | 1.7 ± 6.9 | |
| 5 | 2.0 ± 6.8 | −0.8 ± 4.5 | −0.1 ± 1.2 | |
| Pooled | 2.6 ± 6.9 * | 0.4 ± 4.8 | 0.6 ± 4.5 * | |
| ICC | 0.77 | 0.74 | 0.42 | |
|
| ||||
| Overall | Pooled | 2.3 ± 5.8 * | 0.8 ± 3.7 * | 0.4 ± 4.0 * |
| ICC | 0.63 | 0.56 | 0.38 | |
Reader study results evaluating completeness, correctness and conciseness (columns) across individual readers and pooled across readers. Scores are on the range [−10, 10], where positive scores denote that the best model is preferred to the medical expert. Asterisks (*) on pooled rows denote statistical significance by a one-sided Wilcoxon signed-rank test, P < 0.001. Intra-class correlation (ICC) values across readers are on a range of [−1, 1] where −1, 0 and +1 correspond to negative, no and positive correlations, respectively.
Extended Data Table 4 |.
Summarization baselines
| Dataset | Baseline | BLEU | ROUGE-L | BERTScore | MEDCON |
|---|---|---|---|---|---|
|
| |||||
| Open-i | Ours | 46.0 | 68.2 | 94.7 | 64.9 |
| ImpressionGPT [52] | - | 65.4 | - | - | |
|
| |||||
| MIMIC-CXR | Ours | 29.6 | 53.8 | 91.5 | 55.6 |
| RadAdapt [27] | 18.9 | 44.5 | 90.0 | - | |
| ImpressionGPT [52] | - | 47.9 | - | - | |
|
| |||||
| MIMIC-III | Ours | 11.5 | 34.5 | 89.0 | 36.5 |
| RadAdapt [27] | 16.2 | 38.7 | 90.2 | - | |
| Med-PaLM M [25] | 15.2 | 32.0 | - | - | |
|
| |||||
| Patient questions | Ours | 10.7 | 37.3 | 92.5 | 59.8 |
| ECL° [53] | - | 50.5 | - | - | |
|
| |||||
| Progress notes | Ours | 3.4 | 27.2 | 86.1 | 31.5 |
| CUED [54] | - | 30.1 | - | - | |
|
| |||||
| Dialogue | Ours | 26.9 | 42.9 | 90.2 | 59.9 |
| ACI-Bench° [41] | - | 45.6 | - | 57.8 | |
Comparison of our general approach (GPT-4 using ICL) against baselines specific to each individual dataset. We note that the focal point of our study is not to achieve state-of-the-art quantitative results, especially given the discordance between NLP metrics and reader study scores. A dash (-) indicates that the metric was not reported; a ° indicates that the dataset was pre-processed differently.
Acknowledgements
Microsoft provided Azure OpenAI credits for this project via both the Accelerate Foundation Models Academic Research (AFMAR) program and also a cloud services grant to Stanford Data Science. Additional compute support was provided by One Medical, which A.A. used as part of his summer internship. C.L. is supported by National Institute of Health (NIH) grants R01 HL155410 and R01 HL157235, by Agency for Healthcare Research and Quality grant R18HS026886, by the Gordon and Betty Moore Foundation and by the National Institute of Biomedical Imaging and Bioengineering under contract 75N92020C00021. A.C. receives support from NIH grants R01 HL167974, R01 AR077604, R01 EB002524, R01 AR079431 and P41 EB027060; from NIH contracts 75N92020C00008 and 75N92020C00021; and from GE Healthcare, Philips and Amazon.
Footnotes
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41591-024-02855-5.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-024-02855-5.
Peer review information Nature Medicine thanks Kirk Roberts and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ming Yang, in collaboration with the Nature Medicine team.
Reprints and permissions information is available at www.nature.com/reprints.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
All code used for experiments in this study can be found in a GitHub repository (https://github.com/StanfordMIMI/clin-summ), which also contains links to open-source models hosted by HuggingFace87.
Data availability
This study used six datasets that are all publicly accessible at the provided references. Three of those datasets require PhysioNet81 access due to their terms of use: MIMIC-CXR73 (radiology reports), MIMIC- III76 (radiology reports) and ProbSum79 (progress notes). For the other three datasets not requiring PhysioNet access—Open-i72 (radiology reports), MeQSum77 (patient questions) and ACI-Bench41 (dialogue)—researchers can access original versions via the provided references, in addition to our data via the following GitHub repository: https://github.com/StanfordMIMI/clin-summ. Note that any further distribution of datasets is subject to the terms of use and data-sharing agreements stipulated by the original creators.
References
- 1.Golob JF Jr, Como JJ & Claridge JA The painful truth: the documentation burden of a trauma surgeon. J. Trauma Acute Care Surg. 80, 742–747 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Arndt BG et al. Tethered to the EHR: primary care physician workload assessment using EHR event log data and time–motion observations. Ann. Fam. Med. 15, 419–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleming SL et al. MedAlign: a clinician-generated dataset for instruction following with electronic medical records. Preprint at 10.48550/arXiv.2308.14089 (2023). [DOI]
- 4.Yackel TR & Embi PJ Unintended errors with EHR-based result management: a case series. J. Am. Med. Inform. Assoc. 17, 104–107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman S Impact of electronic health record systems on information integrity: quality and safety implications. Perspect. Health Inf. Manag. 10, 1c (2013). [PMC free article] [PubMed] [Google Scholar]
- 6.Gershanik EF, Lacson R & Khorasani R Critical finding capture in the impression section of radiology reports. AMIA Annu. Symp. Proc. 2011, 465–469 (2011). [PMC free article] [PubMed] [Google Scholar]
- 7.Gesner E, Gazarian P & Dykes P The burden and burnout in documenting patient care: an integrative literature review. Stud. Health Technol. Inform. 21, 1194–1198 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Ratwani RM et al. A usability and safety analysis of electronic health records: a multi-center study. J. Am. Med. Inform. Assoc. 25, 1197–1201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrenfeld JM & Wanderer JP Technology as friend or foe? Do electronic health records increase burnout? Curr. Opin. Anaesthesiol. 31, 357–360 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Sinsky C et al. Allocation of physician time in ambulatory practice: a time and motion study in 4 specialties. Ann. Intern. Med. 165, 753–760 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Khamisa N, Peltzer K & Oldenburg B Burnout in relation to specific contributing factors and health outcomes among nurses: a systematic review. Int. J. Environ. Res. Public Health 10, 2214–2240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy WJ, Kharasch MS & Du H Point of care documentation impact on the nurse–patient interaction. Nurs. Adm. Q. 34, E1–E10 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Chang C-P, Lee T-T, Liu C-H & Mills ME Nurses’ experiences of an initial and reimplemented electronic health record use. Comput. Inform. Nurs. 34, 183–190 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Shanafelt TD et al. Relationship between clerical burden and characteristics of the electronic environment with physician burnout and professional satisfaction. Mayo Clin. Proc. 91, 836–848 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Robinson KE & Kersey JA Novel electronic health record (EHR) education intervention in large healthcare organization improves quality, efficiency, time, and impact on burnout. Medicine (Baltimore) 97, e12319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toussaint W et al. Design considerations for high impact, automated echocardiogram analysis. Preprint at 10.48550/arXiv.2006.06292 (2020). [DOI]
- 17.Brown T et al. Language models are few-shot learners. In Advances in Neural Information Processing Systems 33 https://proceedings.neurips.cc/paper/2020/file/1457c0d6bfcb4967418bfb8ac142f64a-Paper.pdf (NeurIPS, 2020). [Google Scholar]
- 18.Zhao WX et al. A survey of large language models. Preprint at 10.48550/arXiv.2303.18223 (2023). [DOI]
- 19.Bubeck S et al. Sparks of artificial general intelligence: early experiments with GPT-4. Preprint at 10.48550/arXiv.2303.12712 (2023). [DOI]
- 20.Liang P et al. Holistic evaluation of language models. Transact. Mach. Learn. Res. (in the press). [Google Scholar]
- 21.Zheng L et al. Judging LLM-as-a-Judge with MT-Bench and Chatbot Arena. Preprint at 10.48550/arXiv.2306.05685 (2023). [DOI]
- 22.Wornow M et al. The shaky foundations of large language models and foundation models for electronic health records. NPJ Digit. Med. 6, 135 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thirunavukarasu AJ et al. Large language models in medicine. Nat. Med. 29, 1930–1940 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Singhal K et al. Large language models encode clinical knowledge. Nature 10.1038/s41586-023-06291-2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu T et al. Towards generalist biomedical AI. Preprint at 10.48550/arXiv.2307.14334 (2023). [DOI]
- 26.Toma A et al. Clinical Camel: an open-source expert-level medical language model with dialogue-based knowledge encoding. Preprint at 10.48550/arXiv.2305.12031 (2023). [DOI] [Google Scholar]
- 27.Van Veen D et al. RadAdapt: radiology report summarization via lightweight domain adaptation of large language models. In 22nd Workshop on Biomedical Natural Language Processing and BioNLP Shared Tasks 449–460 (Association for Computational Linguistics, 2023). [Google Scholar]
- 28.Mathur Y et al. SummQA at MEDIQA-Chat 2023: in-context learning with GPT-4 for medical summarization. Preprint at 10.48550/arXiv.2306.17384 (2023). [DOI] [Google Scholar]
- 29.Saravia E Prompt engineering guide. https://github.com/dair-ai/Prompt-Engineering-Guide (2022).
- 30.Best practices for prompt engineering with OpenAI API. https://help.openai.com/en/articles/6654000-best-practices-for-prompt-engineering-with-openai-api (2023).
- 31.Chung H et al. Scaling instruction-finetuned language models. Preprint at 10.48550/arXiv.2210.11416 (2022). [DOI]
- 32.Tay Y et al. UL2: unifying language learning paradigms. Preprint at 10.48550/arXiv.2205.05131 (2023). [DOI]
- 33.Taori R et al. Stanford Alpaca: an instruction-following LLaMA model. https://github.com/tatsu-lab/stanford_alpaca (2023).
- 34.Han T et al. MedAlpaca—an open-source collection of medical conversational AI models and training data. Preprint at 10.48550/arXiv.2304.08247 (2023). [DOI]
- 35.The Vicuna Team. Vicuna: an open-source chatbot impressing GPT-4 with 90%* ChatGPT quality. https://lmsys.org/blog/2023-03-30-vicuna/ (2023).
- 36.Touvron H et al. Llama 2: open foundation and fine-tuned chat models. Preprint at 10.48550/arXiv.2307.09288 (2023). [DOI]
- 37.OpenAI. ChatGPT https://openai.com/blog/chatgpt (2022).
- 38.OpenAI. GPT-4 technical report. Preprint at 10.48550/arXiv.2303.08774 (2023). [DOI]
- 39.Lampinen AK et al. Can language models learn from explanations in context? In Findings of the Association for Computational Linguistics: EMNLP 2022 https://aclanthology.org/2022.findings-emnlp.38.pdf (Association for Computational Linguistics, 2022). [Google Scholar]
- 40.Dettmers T, Pagnoni A, Holtzman A & Zettlemoyer L QLoRA: efficient finetuning of quantized LLMs. Preprint at 10.48550/arXiv.2305.14314 (2023). [DOI]
- 41.Yim WW et al. Aci-bench: a novel ambient clinical intelligence dataset for benchmarking automatic visit note generation. Sci. Data 10.1038/s41597-023-02487-3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papineni K, Roukos S, Ward T & Zhu W-J Bleu: a method for automatic evaluation of machine translation. In Proc. of the 40th Annual Meeting of the Association for Computational Linguistics. 10.3115/1073083.1073135 (Association for Computing Machinery, 2002). [DOI] [Google Scholar]
- 43.Walsh KE et al. Measuring harm in healthcare: optimizing adverse event review. Med. Care 55, 436–441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang T, Kishore V, Wu F, Weinberger KQ & Artzi Y BERTScore: evaluating text generation with BERT. International Conference on Learning Representations. https://openreview.net/forum?id=SkeHuCVFDr (2020). [Google Scholar]
- 45.Strobelt H et al. Interactive and visual prompt engineering for ad-hoc task adaptation with large language models. IEEE Trans. Vis. Comput. Graph. 29, 1146–1156 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Wang J et al. Prompt engineering for healthcare: methodologies and applications. Preprint at 10.48550/arXiv.2304.14670 (2023). [DOI]
- 47.Jozefowicz R, Vinyals O, Schuster M, Shazeer N & Wu Y Exploring the limits of language modeling. Preprint at 10.48550/arXiv.1602.02410 (2016). [DOI]
- 48.Chang Y et al. A survey on evaluation of large language models. A CM Trans. Intell. Syst. Technol. 10.1145/3641289 (2023). [DOI] [Google Scholar]
- 49.Poli M et al. Hyena hierarchy: towards larger convolutional language models. In Proceedings of the 40thInternational Conference on Machine Learning 202, 1164 (2023). [Google Scholar]
- 50.Ding J et al. LongNet: scaling transformers to 1,000,000,000 tokens. Preprint at 10.48550/arXiv.2307.02486 (2023). [DOI]
- 51.Lin C-Y ROUGE: a package for automatic evaluation of summaries. In Text Summarization Branches Out 74–81 (Association for Computational Linguistics, 2004). [Google Scholar]
- 52.Ma C et al. ImpressionGPT: an iterative optimizing framework for radiology report summarization with chatGPT. Preprint at 10.48550/arXiv.2304.08448 (2023). [DOI]
- 53.Wei S et al. Medical question summarization with entity-driven contrastive learning. Preprint at 10.48550/arXiv.2304.07437 (2023). [DOI]
- 54.Manakul P et al. CUED at ProbSum 2023: Hierarchical ensemble of summarization models. In The 22nd Workshop on Biomedical Natural Language Processing and BioNLP Shared Tasks 516–523 (Association for Computational Linguistics, 2023). [Google Scholar]
- 55.Yu F et al. Evaluating progress in automatic chest x-ray radiology report generation. Patterns (N Y) 4, 100802 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang L et al. Evaluating large language models on medical evidence summarization. NPJ Digit. Med. 6, 158 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson A, Pollard T & Mark R MIMIC-III Clinical Database Demo (version 1.4). PhysioNet 10.13026/C2HM2Q (2019). [DOI] [Google Scholar]
- 58.Omiye JA, Lester JC, Spichak S, Rotemberg V & Daneshjou R Large language models propagate race-based medicine. NPJ Digit. Med. 6, 195 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zack T et al. Assessing the potential of GPT-4 to perpetuate racial and gender biases in health care: a model evaluation study. Lancet Digit. Health 6, e12–e22 (2024). [DOI] [PubMed] [Google Scholar]
- 60.Chen MX et al. The best of both worlds: combining recent advances in neural machine translation. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) 10.18653/v1/P18-1008 (Association for Computational Linguistics, 2018). [DOI] [Google Scholar]
- 61.Shi T, Keneshloo Y, Ramakrishnan N & Reddy CK Neural abstractive text summarization with sequence-to-sequence models. ACM Trans. Data Sci. 10.1145/3419106 (2021). [DOI] [Google Scholar]
- 62.Raffel C et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 5485–5551 (2020). [Google Scholar]
- 63.Longpre S et al. The Flan collection: designing data and methods for effective instruction tuning. Preprint at 10.48550/arXiv.2301.13688 (2023). [DOI]
- 64.Lehman E et al. Do we still need clinical language models? In Proceedings of Machine Learning Research 209, 578–597 (Conference on Health, Inference, and Learning, 2023). [Google Scholar]
- 65.Lim ZW et al. Benchmarking large language models’ performances for myopia care: a comparative analysis of ChatGPT-3.5, ChatGPT-4.0, and Google Bard. EBioMedicine 95, 104770 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosoł M, Gąsior JS, Łaba J, Korzeniewski K & Młyńczak M Evaluation of the performance of GPT-3.5 and GPT-4 on the Medical Final Examination. Preprint at medRxiv 10.1101/2023.06.04.23290939 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brin D et al. Comparing ChatGPT and GPT-4 performance in USMLE soft skill assessments. Sci. Rep. 13, 16492 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deka P et al. Evidence extraction to validate medical claims in fake news detection. In Lecture Notes in Computer Science. 10.1007/978-3-031-20627-6_1 (Springer, 2022). [DOI] [Google Scholar]
- 69.Nie F, Chen M, Zhang Z & Cheng X Improving few-shot performance of language models via nearest neighbor calibration. Preprint at 10.48550/arXiv.2212.02216 (2022). [DOI]
- 70.Hu E et al. LoRA: low-rank adaptation of large language models. Preprint at 10.48550/arXiv.2106.09685 (2021). [DOI]
- 71.Peng A, Wu M, Allard J, Kilpatrick L & Heidel S GPT-3.5 Turbo fine-tuning and API updates https://openai.com/blog/gpt-3-5-turbo-fine-tuning-and-api-updates (2023).
- 72.Demner-Fushman D et al. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 23, 304–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson A et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 6, 317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delbrouck J-B, Varma M, Chambon P & Langlotz C Overview of the RadSum23 shared task on multi-modal and multi-anatomical radiology report summarization. In Proc. of the 22st Workshop on Biomedical Language Processing 10.18653/v1/2023.bionlp-1.45 (Association for Computational Linguistics, 2023). [DOI] [Google Scholar]
- 75.Demner-Fushman D, Ananiadou S & Cohen KB The 22nd Workshop on Biomedical Natural Language Processing and BioNLP Shared Tasks. https://aclanthology.org/2023.bionlp-1 (Association for Computational Linguistics, 2023). [Google Scholar]
- 76.Johnson A et al. Mimic-iv. https://physionet.org/content/mimiciv/1.0/ (2020).
- 77.Ben Abacha A & Demner-Fushman D On the summarization of consumer health questions. In Proc. of the 57th Annual Meeting of the Association for Computational Linguistics 10.18653/v1/P19-1215 (Association for Computational Linguistics, 2019). [DOI] [Google Scholar]
- 78.Chen Z, Varma M, Wan X, Langlotz C & Delbrouck J-B Toward expanding the scope of radiology report summarization to multiple anatomies and modalities. In Proc. of the 61st Annual Meeting of the Association for Computational Linguistics 10.18653/v1/2023.acl-short.41 (Association for Computational Linguistics, 2023). [DOI] [Google Scholar]
- 79.Gao Y et al. Overview of the problem list summarization (ProbSum) 2023 shared task on summarizing patients’ active diagnoses and problems from electronic health record progress notes. In Proceedings of the Association for Computational Linguistics. Meeting 10.18653/v1/2023.bionlp-1.43 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao Y, Miller T, Afshar M & Dligach D BioNLP Workshop 2023 Shared Task 1A: Problem List Summarization (version 1.0.0). PhysioNet. 10.13026/1z6g-ex18 (2023). [DOI] [Google Scholar]
- 81.Goldberger AL et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101, e215–e220 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Abacha AB, Yim W-W, Adams G, Snider N & Yetisgen-Yildiz M Overview of the MEDIQA-Chat 2023 shared tasks on the summarization & generation of doctor–patient conversations. In Proc. of the 5th Clinical Natural Language Processing Workshop 10.18653/v1/2023.clinicalnlp-1.52 (2023). [DOI] [Google Scholar]
- 83.Yim W, Ben Abacha A, Snider N, Adams G & Yetisgen M Overview of the MEDIQA-Sum task at ImageCLEF 2023: summarization and classification of doctor–patient conversations. In CEUR Workshop Proceedings https://ceur-ws.org/Vol-3497/paper-109.pdf (2023). [Google Scholar]
- 84.Mangrulkar S, Gugger S, Debut L, Belkada Y & Paul S PEFT: state-of-the-art parameter-efficient fine-tuning methods https://github.com/huggingface/peft (2022).
- 85.Frantar E, Ashkboos S, Hoefler T & Alistarh D GPTQ: accurate post-training quantization for generative pre-trained transformers. Preprint at 10.48550/arXiv.2210.17323 (2022). [DOI]
- 86.Loshchilov I & Hutter F Decoupled weight decay regularization. In International Conference on Learning Representations https://openreview.net/forum?id=Bkg6RiCqY7 (2019) [Google Scholar]
- 87.Wolf T et al. Transformers: state-of-the-art natural language processing. In Proc. of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations 10.18653/v1/2020.emnlp-demos.6 (Association for Computational Linguistics, 2020). [DOI] [Google Scholar]
- 88.Soldaini L & Goharian N QuickUMLS: a fast, unsupervised approach for medical concept extraction. https://ir.cs.georgetown.edu/downloads/quickumls.pdf (2016).
- 89.Okazaki N & Tsujii J Simple and efficient algorithm for approximate dictionary matching. In Proc. of the 23rd International Conference on Computational Linguistics https://aclanthology.org/C10-1096.pdf (Association for Computational Linguistics, 2010). [Google Scholar]
- 90.Koo TK & Li MY A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 15, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vallat R Pingouin: statistics in Python. J. Open Source Softw. 3, 1026 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study used six datasets that are all publicly accessible at the provided references. Three of those datasets require PhysioNet81 access due to their terms of use: MIMIC-CXR73 (radiology reports), MIMIC- III76 (radiology reports) and ProbSum79 (progress notes). For the other three datasets not requiring PhysioNet access—Open-i72 (radiology reports), MeQSum77 (patient questions) and ACI-Bench41 (dialogue)—researchers can access original versions via the provided references, in addition to our data via the following GitHub repository: https://github.com/StanfordMIMI/clin-summ. Note that any further distribution of datasets is subject to the terms of use and data-sharing agreements stipulated by the original creators.


