Abstract
Context
Insulin sensitivity (IS) is an important factor in type 2 diabetes (T2D) and can be estimated by many different indices.
Objective
We aimed to compare the genetic components underlying IS indices obtained from fasting and oral glucose-stimulated plasma glucose and serum insulin levels.
Methods
We computed 21 IS indices, classified as fasting, OGTT0,120, and OGTT0,30,120 indices, using fasting and oral glucose tolerance test (OGTT) data in 2 cohorts. We used data from a family cohort (n = 313) to estimate the heritability and the genetic and phenotypic correlations of IS indices. The population cohort, Inter99 (n = 5343), was used to test for associations between IS indices and 426 genetic variants known to be associated with T2D.
Results
Heritability estimates of IS indices ranged between 19% and 38%. Fasting and OGTT0,30,120 indices had high genetic (ρG) and phenotypic (ρP) pairwise correlations (ρG and ρP: 0.88 to 1) The OGTT0,120 indices displayed a wide range of pairwise correlations (ρG: 0.17-1.00 and ρP: 0.13-0.97). We identified statistically significant associations between IS indices and established T2D-associated variants. The PPARG rs11709077 variant was associated only with fasting indices and PIK3R rs4976033 only with OGTT0,30,120 indices. The variants in FAM63A/MINDY1, GCK, C2CD4A/B, and FTO loci were associated only with OGTT0,120 indices.
Conclusion
Even though the IS indices mostly share a common genetic background, notable differences emerged between OGTT0,120 indices. The fasting and OGTT-based indices have distinct associations with T2D risk variants. This work provides a basis for future large-scale genetic investigations into the differences between IS indices.
Keywords: insulin sensitivity, insulin sensitivity indices, type 2 diabetes, OGTT, fasting insulin, genetic variants
Type 2 diabetes (T2D) is a heterogeneous disease defined by deficient insulin secretion and reduced insulin sensitivity, which are major contributors to T2D etiology. Decreased insulin sensitivity (IS) is a major risk factor for T2D and cardiometabolic diseases (1-3). Understanding and quantifying IS and identifying its genetic components are essential for dissecting T2D heterogeneity, thereby paving the way for T2D subtyping for more personalized and effective treatment strategies tailored to individual genetic profiles (4).
Several methods for estimating IS have been developed and studied in various settings and populations (5-9). Most of these studies have focused on estimating the phenotypic correlation between IS estimated by the well-established hyperinsulinemic euglycemic clamp (HEC) method or frequently sampled intravenous glucose tolerance test with 1 of the fasting or oral glucose tolerance test (OGTT)-based methods. Most often the fasting and OGTT-based indices have high correlations (ρP: 0.6–0.7) with the HEC method (7). Although high phenotypic correlations between HEC and OGTT-based indices indicate that these indices can estimate IS almost as accurately as the HEC method, they do not provide insights into genetic factors underlying these IS indices. Only a few studies have investigated the genetic basis of the IS indices used in most clinical and epidemiological studies. These studies investigated the genetic overlap primarily between HEC or frequently sampled intravenous glucose tolerance tests and fasting-based indices, such as fasting insulin and Homeostatic Model Assessment of Insulin Resistance (HOMA-IR) (10-12). Interestingly, these studies have shown variability in the heritability and genetic correlations between HOMA-IR, fasting insulin, and HEC-based IS and between HOMA-IR and minimal model-based IS among different studies (10-12). This variability is partly due to differences in population size, ethnicity, and study design and likely also due to the inherent heterogeneity of IS as a trait and different methods used to estimate IS. However, for most IS indices, the understanding of their genetic underpinnings remains unclear. This requires further studies to investigate the genetic overlap between known IS indices and their usage as a tool to delineate the genetic determinants of IS contributing to other metabolic traits.
Given the inherent heterogeneity of IS and the multiple mathematical models employed to estimate it, we posed 3 critical questions: Are fasting and OGTT-based IS indices heritable traits, and to what extent does heritability vary between them? What is the degree of genetic overlap among these IS indices? Can we utilize these indices as a tool to dissect T2D-associated genetic variants and identify those that influence IS?
Using existing mathematical models in the literature, we computed 21 IS indices in both a family and a population cohort, categorizing them into fasting-, OGTT0,120-, and OGTT0,30,120-based indices. To answer the previously posed questions, we aimed to estimate and compare heritability, genetic, and phenotypic correlations among these IS indices. We also aimed to test associations between IS indices and 426 genetic variants known to be associated with T2D.
Materials and Methods
Study Populations
The family cohort included 533 individuals from 95 families with 1 parent diagnosed with T2D according to World Health Organization 1998 criteria (13) and 1 parent without known diabetes (14). All family members without diabetes, including spouses, offspring, and other relatives, were included, and a total of 366 individuals underwent an OGTT. Participants with incomplete data were excluded. As such, data from 313 individuals with complete information were available for analysis (Table 1).
Table 1.
Summary of clinical characteristics of study populations
| Traits | Family cohort | Inter99 |
|---|---|---|
| n | 313 | 5343 |
| Sex [female/male; n] | 175/138 | 2663/2680 |
| Age (years) | 45.1 [40.0, 50.2] | 40.7 [34.4, 50.6] |
| Weight (kg) | 77.0 [66.5, 88.0] | 75.5 [67.6, 86.5] |
| Height (cm) | 172.0 [165.5, 179.0] | 171.0 [165.0, 178.0] |
| BMI (kg/m2) | 25.6 [23.2, 28.6] | 25.8 [22.8, 28.9] |
| Waist to hip ratio | 0.86 [0.79, 0.93] | 0.86 [0.79, 0.92] |
| Waist circumference (cm) | 86 [77, 95] | 85 [77, 95] |
| Plasma glucose (mmol/L) | ||
| Fasting | 5.4 [5.1, 5.8] | 5.1 [4.8, 5.5] |
| 30 minutes | 8.6 [7.4, 9.8] | 8.4 [7.4, 9.3] |
| 120 minutes | 5.9 [5.0, 7.0] | 6.1 [5.2, 7.1] |
| Serum insulin (pmol/L) | ||
| Fasting | 35 [24, 52] | 36 [25, 53] |
| 30 minutes | 247 [176, 358] | 262 [178, 379] |
| 120 minutes | 157 [98, 257] | 183 [127, 317] |
Numbers presented in the table, from age and onwards, as medians and interquartile ranges (IQR: [IQ1, IQ3]).
Abbreviations: BMI, body mass index.
The Inter99 cohort is a Danish population-based cohort for the primary prevention of cardiovascular diseases (http://www.inter99.dk/) by nonpharmacological lifestyle intervention (15, 16). The baseline of the Inter99 cohort consists of 6784 individuals, and for this study, we analyzed baseline data of 6184 individuals with genotype data available. After excluding individuals with known diabetes, related individuals, and incomplete phenotype data for IS indices calculation, the final sample comprised 5343 individuals with complete genotype and phenotype data used in this study (Table 1).
The 2 studies received approval from the Ethical Committee of Copenhagen (reference numbers KA 93033 for family study and KA98155 and H-3-2012-155 for Inter99) and were conducted following the ethical guidelines and principles outlined in the Declaration of Helsinki II.
Clinical Examinations
All study participants underwent a comprehensive assessment of their demographics, and height and weight were measured to calculate body mass index (BMI). Those without a prior diagnosis of T2D underwent a standardized OGTT in accordance with World Health Organization guidelines (13). Fasting blood samples were collected from each participant after a 12-hour overnight fast prior to the OGTT. Subsequently, a 75-gram glucose load was administered, and additional blood samples were obtained at 30 and 120 minutes post-ingestion to evaluate plasma glucose and serum insulin levels.
Calculation of IS Indices
Twenty-one IS indices were calculated for both cohorts, using fasting and OGTT measures of plasma glucose (G0, G30, and G120) and serum insulin (I0, I30, and I120) using the formulas listed in Table 2.
Table 2.
Formulas for insulin sensitivity indices calculation
| IS Index | Formula (units) | Reference |
|---|---|---|
| Fasting indices | ||
| RaynaudSI | 40/I0 (µU/L) | Raynaud et al (17) |
| QUICKI | 1/(log I0 (µU/L) + log G0 (mg/dL)) | Katz et al (18) |
| ISIbasal | 104/(I0 (µU/L) × G0 (mg/dL)) | Sluiter et al (19) |
| FIns/FGlu | I0 (µU/L) /G0 (mmol/L) | Hanson et al (20) |
| BennettSI | 1/(log I0 (µU/L) × log G0 (mg/dL)) | Anderson et al (21) |
| Belfiorebasal | 2/((I0 (µU/L)/N × G0 (mmol/L)/N) + 1) | Belfiore et al (22) |
| AvignonSI0 | 108/(I0 (µU/L) × G0 (mg/dL) × VD(150mL/kg BW)) | Avignon et al (23) |
| HOMA-IR | (I0 (µU/L) × G0 (mmol))/22.5 | Matthews et al (24) |
| FIns | I0 (pmol/L) | Laakso (25) |
| OGTT0,120 indices | ||
| StumvollDem | 0.222 − 0.00333 × BMI − 0.0000779 × I120 (pmol/L) − 0.000422 × Age(years) | Stumvoll et al (26) |
| StumvollModi | 0.156 − 0.0000459 × I120 (pmol/L) − 0.000321 × I0 (pmol/L) − 0.00541 × G120 (mmol/L) | Stumvoll et al (26) |
| ISI120 | 104/(I120 (µU/L) × G120 (mg/dL)) | Sluiter et al (19) |
| Ins/Glu120 | I120 (µU/L)/G120 (mmol/L) | Hanson et al (20) |
| GuttIndex | 75 000 + (G0 (mg/dL) − G120 (mg/dL)) × 0.19 × BW(kg)/(120 × MPG(mg/dL) × log(MSI (µU/L))) | Gutt et al (27) |
| AvignonSI120 | 108/(I120 (µU/L) × G120 (mg/dL) × VD(150mL/kg BW)) | Avignon et al (23) |
| AvignonSIM | ((w(mean Si2h/mean Sib) × AvignonSI0) + AvignonSI120)/2 | Avignon et al (23) |
| Ins120 | I120 (pmol/L) | Laakso (25) |
| Glu120 | G120 (mmol/L) | |
| IFC | ln(I120 (pmol/L)/I0 (pmol/L)) | Williamson et al (28) |
| OGTT0,30,120 indices | ||
| Matsuda | 10 000/(sqrt(G0 (mg/dL) × I0 (µU/L) × mean G(mg/dL) × mean I(µU/L))) | Matsuda et al (29) |
| BIGTTSI | exp(4.90 − (0.00402 × I0 (pmol/L)) − (0.000556 × I30 (pmol/L)) − (0.00127 × I120 (pmol/L)) − (0.152 × G0 (mmol/L)) − (0.00871 × G30 (mmol/L)) − (0.0373 × G120 (mmol/L)) − (0.145 × sex) − (0.0376 × BMI (kg/m2))) | Hansen et al (30) |
Abbreviations: AvignonSI0, AvignonSI at fasting; AvignonSI120, AvignonSI at 120 minutes; AvignonSIM, average of AvignonSI120 and AvignonSI0; BW, body weight; BennettSI, Bennett fasting sensitivity index; Belfiorebasal, Belfiore fasting index; BIGTTSI, BIGTT insulin sensitivity index; FIns, fasting insulin; FIns/FGlu, ratio of fasting insulin and glucose; Glu120, glucose at 120 minutes; GuttIndex, Gutt Index; HOMA-IR, Homeostatic Model Assessment of Insulin Resistance; IFC, insulin fold change; Ins120, insulin at 120 minutes; Ins/Glu120, ratio of insulin and glucose at 120 minutes; ISI120, insulin sensitivity index at 120 minutes; ISIbasal, insulin sensitivity index at fasting; Matsuda, Matsuda calculated by using estimated mean over 3 time points (0, 30, 120 minutes glucose and insulin values); MPG, mean plasma glucose; MSI, mean serum insulin;, N, number of sampling points; OGTT, oral glucose tolerance test; QUICKI, quantitative insulin sensitivity check index; RaynaudSI, Raynaud's index; StumvollDem, Stumvoll with demographics (age, body mass index) at 120 minutes; StumvollModi, modified Stumvoll at 0 and 120 minutes; VD, glucose distribution volume of 150 mL per kilogram of body weight.
Normalization of IS Indices
To normalize IS indices, we employed inverse normal quantile transformation, as detailed in the supplementary materials (31). The transformation applied a rank-based normal transformation to raw IS indices to achieve a Gaussian-like distribution and to standardize all IS indices onto a consistent scale. This approach enhances both the statistical validity and comparability of the IS indices.
Genotyping and Quality Control
The participants from Inter99 were genotyped using Illumina Human OmniExpress-24 (v1.0A/v1.1A) and Genotyping module (version 1.9.4) of GenomeStudio software (version 2011.1; Illumina). We conducted quality control checks for genotype data, excluding samples with a call rate lower than 95%, heterozygosity exceeding the median +3 interquartile range, mismatched genetically inferred and reported sex, and duplicates. Variants with a call rate less than 98%, Hardy–Weinberg Equilibrium with a P-value less than 10−5, and monomorphic markers were excluded from the genotyped data. We used Michigan Imputation Server with the HRC reference panel (GRCh37 genome build, HRC r1.1 2016 release) for imputing variants.
Inversion of the Insulin Resistance Indices to Represent IS
Prior to the association analysis between IS indices and T2D-associated variants, we reversed the direction of originally rank-based normalized insulin resistance indices [fasting insulin (FIns), ratio of fasting insulin and glucose (FIns/FGlu), HOMA-IR, ratio of insulin and glucose at 120 minutes (Ins/Glu120), insulin at 120 minutes (Ins120) insulin fold change (IFC), and glucose at 120 minutes (Glu120)] by multiplying the normalized index values by −1. Therefore, we used the “inv” prefix for the inverted indices (inv-FIns, inv-FIns/FGlu, inv-HOMA-IR, inv-Ins/Glu120, inv-Ins120, inv-IFC, and inv-Glu120) and used these indices for association analysis together with other IS indices. This step was done to ensure that the direction of these indices aligns with other IS indices.
Statistical Analyses
Heritability and correlation analysis
In the family cohort, we used Sequential Oligogenic Linkage Analysis Routines software (SOLAR version 8.5.1 2021) variance component analysis to estimate the pedigree-based heritability, genetic correlations, and phenotypic correlations for IS indices based on the pedigree data (32). Age, sex, and BMI were included as covariates. The BIGTT insulin sensitivity index (BIGTTSI) was not adjusted for sex and BMI, and Stumvoll with demographics (age, BMI) (StumvollDem) was not adjusted for age and BMI. The additive effects of shared genetics were calculated using the commands “polygen -testrhog -testrhop -rhopse” to determine the pairwise genetic and phenotypic correlations between the IS indices.
Associations with T2D-associated genetic variants
We selected 426 genome-wide significant variants (P < 5 × 10−8) with minor allele frequency greater than 1% from a previous genome-wide association study of T2D (Supplementary Table S1) (31) sourced from previous studies (33, 34). The majority of the variants were noncoding, intronic, or intergenic with subsets of 13 missense and 4 synonymous variants. We assessed the association between T2D-associated variants and IS indices in the cohort Inter99. We employed a linear regression model, adjusting for age, sex, and BMI along with 10 principal components (calculated with PLINK v2.00a3.7LM) to account for population structure. BIGTTSI was not adjusted for sex and BMI, and StumvollDem was not adjusted for age and BMI as these covariates are already included in their calculations. The association analysis was conducted using the RVTEST (version 20190205) genetic association testing software. The lead variants and nearest loci were assigned based on criteria defined by Mahajan et al (33) and Vujkovic et al (34). Values of P < .05 were considered nominally statistically significant. Correction for multiple testing was performed using a false discovery rate (FDR) of 5%. The variants associated with the IS indices were then soft clustered using the Bayesian information criterion method using package (mclust, version 6.0.1) to determine the optimal number of clusters in R (version 4.3.2, 10-31-2023). Complex Heatmap package (35) was used to generate heat maps, and a hierarchical clustering top-down approach was used to align the clusters both in rows and columns. The clustering was allowed both on IS indices and the associated variants to facilitate the identification of local patterns of association.
Phenotypic Clusters of Variants
T2D-associated variants without nominal association with IS indices or with an IS increasing effect were further intersected with a previously performed clustering of T2D variants (36) to investigate their T2D-related mechanistic endophenotype. Variants were grouped into 8 clusters based on association with 37 cardiometabolic phenotypes from 7 phenotype groups (glycemic traits, anthropometric measures, adipose, blood pressure, circulating plasma lipid levels, liver function, and biomarkers). Hereby 8 mechanistic T2D endophenotype clusters were assigned: beta cell +proinsulin, beta cell −proinsulin, residual glycemic, body fat, metabolic syndrome, obesity, lipodystrophy, and liver and lipid metabolism (36). We used 250 kb of window for matching the variants and creating an intersection with the variants to the reported clusters.
Results
Heritability, Genetic and Phenotypic Correlations
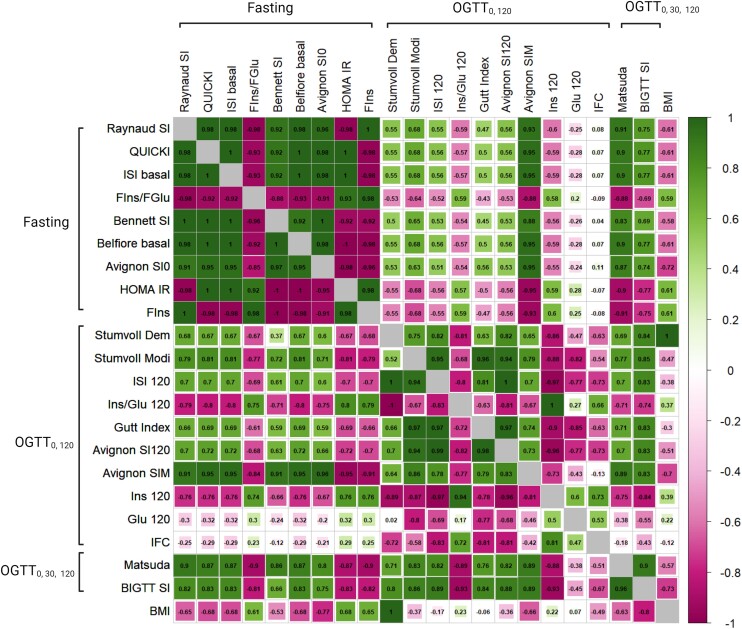
From the IS indices evaluated, the OGTT0,120 displayed a wide range of heritability values (0.31-0.38) in contrast to the fasting group, which showed a narrow range (0.23-0.29) with few exceptions, and the OGTT0,30,120 group, which exhibited confined ranges of values (0.19-0.24), as detailed in Table 3. In pairwise correlations, the OGTT0,120 indices displayed a wide range of genetic (ρG) and phenotypic (ρP) correlations (ρG:0.52-1.00 and ρP: 0.63-0.97), excluding Glu120 and IFC. Conversely, both the fasting and OGTT0,30,120 indices exhibited strong genetic and phenotypic correlations (ρG: 0.92-1.00 and ρP: 0.88-1.00). In the group-wise comparison, the OGTT0,120 group had ρG < 0.8 and ρP < 0.7 with the fasting group and ρG < 0.9 and ρP < 0.8 with OGTT0,30,120 group indices excluding Glu120 and IFC (Fig. 1). The fasting indices displayed notably moderate to high correlations with the OGTT0,30,120 indices (ρG: 0-66-0.90 and ρP: 0-69-0.91) (Supplementary Tables S2 and S3) (31). The environmental correlations aligned closely with the phenotypic correlations in the fasting and OGTT0,30,120 indices, although they were slightly lower in the OGTT0,120 indices (Supplementary Table S4) (31). Furthermore, our analysis also revealed a diverse range of phenotypic and genetic correlations between BMI and IS indices (Fig. 1 and Supplementary Tables S2-S4) (31).
Table 3.
Heritability estimates of insulin sensitivity indices in the family cohort
| IS Index | Heritability (h2) |
|---|---|
| Fasting indices | |
| RaynaudSI | 0.29 ± 0.11 (2.8 × 10−04) |
| QUICKI | 0.24 ± 0.11 (1.7 × 10−03) |
| ISIbasal | 0.24 ± 0.11 (1.7 × 10−03) |
| FIns/FGlu | 0.35 ± 0.12 (2.1 × 10−05) |
| BennettSI | 0.33 ± 0.12 (7.8 × 10−05) |
| Belfiorebasal | 0.24 ± 0.11 (1.7 × 10−03) |
| AvignonSI0 | 0.23 ± 0.11 (3.9 × 10−03) |
| HOMA-IR | 0.24 ± 0.11 (1.7×10−03) |
| FIns | 0.29 ± 0.11 (2.8 × 10−04) |
| OGTT0,120 indices | |
| StumvollDem | 0.26 ± 0.14 (2.5 × 10−02) |
| StumvollModi | 0.34 ± 0.12 (2.5 × 10−04) |
| ISI120 | 0.34 ± 0.12 (2.6 × 10−04) |
| Ins/Glu120 | 0.38 ± 0.12 (3.5 × 10−05) |
| Gutt Index | 0.31 ± 0.12 (9.0 × 10−04) |
| AvignonSI120 | 0.34 ± 0.12 (3.0 × 10−04) |
| AvignonSIM | 0.36 ± 0.12 (9.6 × 10−05) |
| Ins120 | 0.35 ± 0.12 (1.1 × 10−04) |
| Glu120 | 0.33 ± 0.13 (1.2 × 10−03) |
| IFC | 0.21 ± 0.12 (2.1 × 10−02) |
| OGTT0,30,120 indices | |
| Matsuda | 0.24 ± 0.10 (4.9 × 10−04) |
| BIGTTSI | 0.19 ± 0.13 (4.0 × 10−02) |
Heritability estimates of IS indices in the Family cohort. The indices are categorized into 3 groups: fasting, OGTT0,120, and OGTT0,30,120 based indices. The first column lists the names of the indices, while the second column displays the heritability estimates (h2) with their corresponding SDs and P-values.
Abbreviations: AvignonSI0, AvignonSI at fasting; AvignonSI120, AvignonSI at 120 minutes; AvignonSIM, average of AvignonSI120 and AvignonSI0; BennettSI, Bennett fasting sensitivity index; Belfiorebasal, Belfiore fasting index; BIGTTSI, BIGTT insulin sensitivity index; FIns, fasting insulin; FIns/FGlu, ratio of fasting insulin and glucose; Glu120, glucose at 120 minutes; GuttIndex, Gutt Index; HOMA-IR, Homeostatic Model Assessment of Insulin Resistance; IFC, insulin fold change; ISI120, insulin sensitivity index at 120 minutes; ISIbasal, insulin sensitivity index at fasting; Ins/Glu120, ratio of insulin and glucose at 120 minutes; Matsuda, Matsuda calculated by using estimated mean over 3 time points (0, 30, 120 minutes glucose and insulin values); OGTT, oral glucose tolerance test; QUICKI, quantitative insulin sensitivity check index; RaynaudSI, Raynaud's index; StumvollDem, Stumvoll with demographics (age, body mass index) at 120 minutes; StumvollModi, modified Stumvoll at 0 and 120 minutes.
Figure 1.
Genetic and phenotypic correlations between 21 IS indices in the family study. The lower left triangle (genetic) and upper right triangle (phenotypic) represent the correlation between fasting, OGTT0,120, and OGTT0,30,120 indices, with values ranging from −1 to 1. The estimated correlation coefficients are noted within cells. The diagonal of the matrix shows the correlation of each trait with itself, which is always 1, shown by the empty grey box. Abbreviations: OGTT, oral glucose tolerance test; IS, insulin sensitivity.
Association Between IS Indices and T2D Genetic Variants
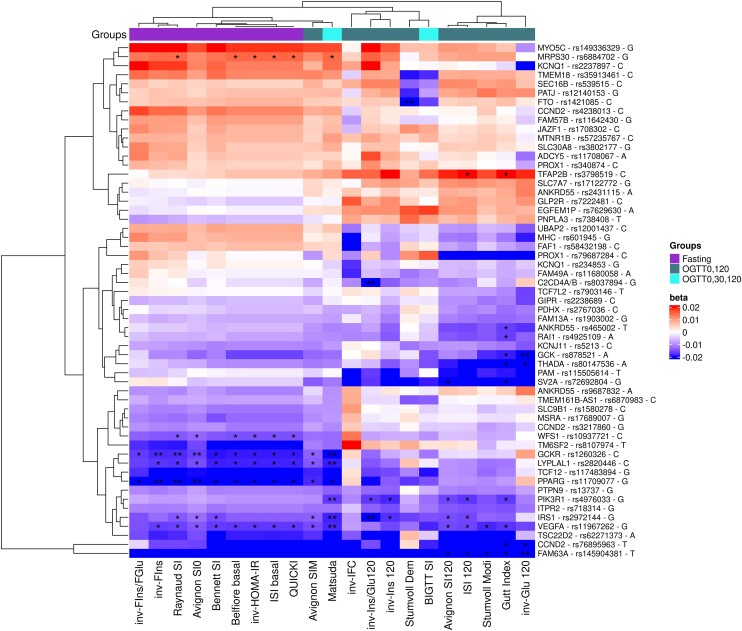
We tested the association between 21 IS indices and 426 known T2D-associated genetic variants with minor allele frequency >1% in Inter99. Of the 426 genetic variants, 163 were nominally associated with at least 1 IS index (P < .05). Three distinct clusters of IS indices emerged based on association with these 163 variants (Supplementary Fig. S1) (31). Cluster 1 included all fasting indices; cluster 2 was composed of a combination of indices from OGTT0,120 and OGTT0,30,120 groups including Matsuda insulin sensitivity index (Matsuda), AvignonSIM, inv-Ins/Glu120, invIns120, StumvollDem, BIGTTSI, inv-IFC; and cluster 3 contained OGTT0,120 indices including Avignon insulin sensitivity at 120 minutes, ISI120 StumvollModi, Gutt Index (GuttIndex), inv-Glu120 (Fig. 2, Supplementary Fig. S1) (31). Across the 3 clusters, we identified 2 groups; 1 of 114 T2D variants, where the T2D risk allele was associated with decreased IS, and 1 of 49 T2D variants, where the T2D risk allele was associated with increased IS (Supplementary Fig. S1) (31).
Figure 2.
Association between IS indices and 62 T2D-associated variants with MAF > 1% and with a P-value of <.05 with at least 1 index. All effects are for T2D-risk increasing allele. The x-axis displays the indices, and the y-axis represents the gene, variant, and T2D-risk allele. An asterisk (*) indicates an FDR of <.05, while 2 asterisks (**) denote an FDR of <.01.
Abbreviations: FDR, false discovery rate; IS, insulin sensitivity; MAF, minor allele frequency; T2D, type 2 diabetes.
The 269 variants, without nominal association with any IS index, mapped to 7 different clusters of the 8 mechanistic endophenotype clusters as recently defined by Suzuki et al (36). Forty-two percent were from the beta cell function +/– proinsulin or residual glycemic traits clusters, while 52% mapped to obesity or metabolic syndrome, body fat, or obesity clusters (Supplementary Fig. S2) (31). Conversely, of 49 variants showing an association with increased IS, 52% were from the beta cell function +/– proinsulin, or residual glycemic traits clusters, while the rest primarily were associated with obesity and metabolic syndrome clusters (Supplementary Fig. S3) (31).
We observed variants that increased the risk of T2D yet differed in their direction of association with the various IS indices (Fig. 2). For example, rs878521 (GCK locus), rs80147536 (THADA) rs115505614 (PAM), rs72692801 (SV2A), rs465002 (ANKRD55), and rs4925109 (RAI1) had a strong negative association with cluster 3, a heterogeneous pattern of association with cluster 2, and weak to no associations with cluster 1 (Fig. 2). Similarly, rs12001437 (UBAP2), rs601945 (MHC), rs58432198 (FAF1), rs7903146 (TCF7L2), rs234853 (KCNQ1), and rs11680058 (FAM49A) had negative associations with cluster 3, a mixed association with cluster 2, and positive or no associations with cluster 1. Conversely, rs3798519 (TFAP2B), rs17122772 (SLC7A7), rs2431115 (ANKRD55), rs7222481 (GLP2R), rs7629630 (EGFEM1P), and rs738408 (PNPLA3) had strong positive associations with cluster 3 and 2 and negative or no associations with cluster 1.
There were some unique associations with individual indices; for example, the rs149336329 (MYO5C), rs2207897 (KCNQ1), rs57235767 (MTNR1B), rs3802177 (SLC30A8), rs11708067 (ADCY5), and rs340874 (PROX1) variants had a positive association in all 3 clusters except with inv-Glu120 and inv-IFC indices. Similarly, the rs35913461 (TMEM18), rs539515 (SEC16B), rs12140153 (PATJ), and rs1421085 (FTO) variants had a positive association in all 3 clusters except with BIGTTSI and StumvollDem indices (Fig. 2).
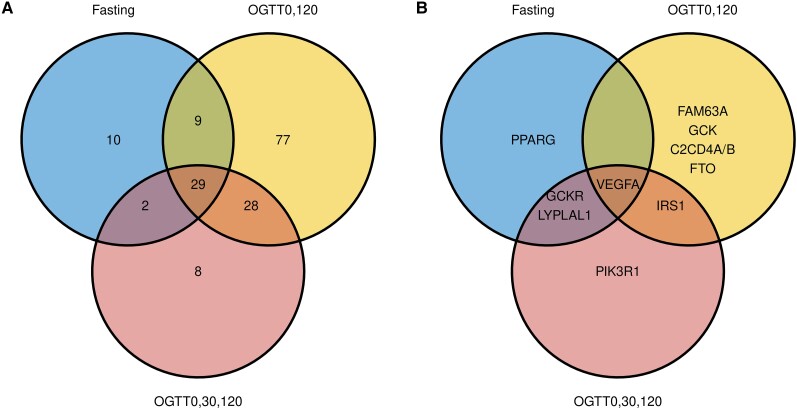
Further analysis of the 163 genetic variants revealed that some were only associated with indices from a single group, and some were associated with indices across groups (Fig. 3A, Supplementary Table S5) (31). Of 11 variants showing associations with indices at FDR < 0.05, 4 variants in the FAM63A, GCK, C2CD4A/B, and FTO loci were exclusively associated with OGTT0,120 indices, PPARG was uniquely associated with fasting indices and PIK3R1 solely with OGTT0,30,120 indices. Four variants in the GCKR, LYPLAL1, IRS1, and VEGFA loci were associated with 2 or all 3 groups of indices (Fig. 3B, Supplementary Table S6) (31). The rs145904381 variant in the FAM63A locus had significant (P < .05) negative association with all 3 groups but most significantly (P < .0001, FDR < 0.02) with OGTT0,120-based indices.
Figure 3.
(A) Unique and common T2D-associated variants significantly associated with IS indices, with a P-value of <.05. (B) Gene loci containing variants associated with IS indices at an FDR of <.05.
Abbreviations: FDR, false discovery rate; IS, insulin sensitivity; T2D, type 2 diabetes.
The comprehensive association chart, showcasing the relationship between 426 T2D genetic variants and 21 indices, is accessible as a resource at (https://sufyansuleman.github.io/isi_t2d/).
Discussion
In this study, we investigated the genetic architecture underlying the fasting, OGTT0,120, and OGTT0,30,120 indices of IS. We also examined their association with variants known to be associated with T2D. We observed that the heritability of OGTT0,120 indices is higher compared to both fasting and OGTT0,30,120 indices. Conversely, the genetic and phenotypic correlations were lower within the OGTT0,120 indices group when compared with the fasting and OGTT0,30,120 indices groups. Most T2D variants have the expected association with IS indices with the T2D-increasing alleles being associated with lower IS, whereas for some variants, the T2D-increasing allele is associated with higher IS.
The fasting state provides a stable milieu wherein both insulin and glucose levels remain relatively constant, offering an opportunity for precise measurement of insulin and glucose concentrations, which are the basis of most fasting-based IS indices. The persistent genetic correlations observed in our study among fasting indices imply a common genetic influence on the underlying physiology of fasting state IS as well as a mathematical similarity between them. Nonetheless, the simplicity and cost-effectiveness of fasting IS indices substantiate their utility in clinical and epidemiological estimation of IS.
Fasting indices are inherently unable to estimate postprandial IS. In contrast, OGTT-based indices considering data up to 120 minutes offer a better reflection of postprandial IS. Given the distinct models used by OGTT0,120 indices compared to fasting, a varied pattern of genetic correlations was expected. The observed pattern suggests that these indices are somewhat influenced by unique genetic factors, implying that they can capture different genetic effects on IS in genetic association studies. The observed variation may, in part, stem from differences in mathematical models. However, this variability cannot be solely attributed to mathematical models as evidenced by prior studies showing distinct genetic correlations between IS indices (10, 11) In this context, fasting indices are well studied in genetic research (37-39), whereas OGTT indices like StumvollMod (40) and Glu120 (38), along with others like GuttIndex and Avignon insulin sensitivity at 120 minutes, are underexplored in large-scale genetic studies.
The OGTT0,30,120 indices such as Matsuda and BIGTTSI show strong genetic and phenotypic correlations with fasting indices. This could be due to the inclusion of the 30-minute time point that makes the index more akin to fasting-based indices compared to OGTT0,120 indices (41). The minimal variation observed between the OGTT0,30,120 indices and other groups may be due to the limited number of analyzed indices in this category.
We observed that 49 (30%) of the 163 T2D-associated genetic variants were nominally associated with at least 1 index and displayed a positive association with 1 or more IS indices. Interestingly, many of the variants exhibiting a positive association with IS indices were located in or near the genes known to be involved in beta cell function such as KCNQ1, MTNR1B, SLC30A8, JAZF1, and ADCY5 (42-46). This observation was concurred when we assigned mechanistic endophenotype clusters (36) to these variants showing that a high fraction mapped to beta cell-related clusters, indicating positive associations with fasting glucose and hemoglobin A1C with positive or negative associations with circulating proinsulin. Physiologically, the positive association of these T2D risk alleles with IS makes sense given the intricate relationship between beta cell function and IS.
T2D-associated variants without an at least nominal association with IS predominantly mapped to mechanistic clusters related to obesity, underscoring the BMI adjustment in the current association analysis to identify the genetic impact on IS independent of BMI.
We noted a nuanced association between some genetic loci and IS indices. The loci SLC7A7, ANKRD55, GLP2R, EGFEM1P, and PNPLA3 were nominally negatively associated with fasting indices and positively with OGTT-based indices. In contrast, UBAP2, MHC, FAF1, and TCF7L2 were positively associated with fasting indices and negatively with OGTT-based indices, suggesting that these genes may influence insulin signaling differently during fasting and postprandial states. The differential roles of these genes during fasting and postprandial states may offer insights into the pathophysiology of insulin resistance and T2D.
The PPARG genetic variant was only associated with the fasting indices group. Peroxisome proliferator-activated receptor (PPAR) γ is a nuclear receptor transcription factor that both lipid (47, 48) and glucose metabolism (49). Fasting-induced PPARG activation affects IS differently across tissues, enhancing it in adipose by decreasing fat synthesis and increasing breakdown but reducing it in muscles by promoting fat storage and inhibiting fat breakdown (50). This differential modulation of fat storage and utilization by PPARG contributes to IS variations during fasting states in muscles and adipose tissue.
In the liver, PPARγ plays a key role by regulating fatty acid oxidation and gluconeogenesis (49). It influences fat metabolism through interactions with nuclear receptors like PPARα and SREBP-1c (51). Furthermore, it activates enzymes involved in gluconeogenesis such as glucose-6-phosphatase, fructose-1,6-bisphosphatase, and pyruvate carboxylase and inhibits glucose uptake enzymes like GLUT2 and glucokinase, reducing glucose absorption and usage by the liver (52). The intronic variant in the PPARG locus may influence PPARγ expression, potentially leading to a decreased IS in the fasting state.
The PIK3R1 rs4976033 variant was uniquely associated with OGTT0,30,120 indices. As a regulatory component of phosphoinositide 3-kinase, it is activated by insulin and prominently expressed in skeletal muscles, brain, liver, and adipose tissues (53). (Supplementary Fig. S4A and S4B) (31). PIK3R1 binds to phosphorylated insulin receptor substrate proteins and facilitates AKT recruitment and activation, which is essential for glucose uptake in muscle and fat cells and for glycogen synthesis in the liver. Variants in PIK3R1 that modify its expression can notably influence phosphoinositide 3-kinase activity and the insulin signaling pathway. The pathogenic human PIK3R1 Y657* loss-of-function mutation knocked into mice confers severe insulin resistance also observed in humans (54) showing its role in decreased IS. The PIK3R1 rs4976033-G allele has previously been associated with lower IS estimated by the Matsuda (55). Interestingly, we found that the PIK3R1 locus associated significantly (FDR < 0.05) only with an OGTT0,30,10 index (Matsuda), which implies that the index can identify loci with a role in postprandial insulin action.
The variant in the FAM63A locus was strongly associated with the OGTT0,120 indices. This genetic variant was previously reported not to associate with IS estimated by the StumvollMod index (n = 55,000, P = .2) (28). However, in our analysis, it shows a statistically significant (FDR < 0.05) association with decreased IS with the OGTT0,120 indices group. Furthermore, this variant also shows significant associations with other cardiometabolic traits, as shown by the PheWAS analysis (Supplementary Fig. S5) (31). FAM63A encodes a deubiquitinating enzyme (56), vital for insulin-mediated breakdown of insulin receptor substrate 1 via the ubiquitin pathway, thereby regulating phosphoinositide 3-kinase activity essential for normal insulin function (57). However, given the conflicting evidence of association, we are not able to designate this locus to the list of loci inflicting insulin resistance.
Limitations of our study constitute the small family study sample, potentially affecting the accuracy of estimates of heritability and pairwise correlations. Limited in sample size, our association analysis between T2D risk variants and IS indices may have been underpowered, potentially leading to type 2 error. A larger-scale genome-wide association study could reveal more and novel genetic associations with IS.
In summary, this study revealed that fasting and OGTT0,30,120 indices generally exhibit similar genetic underpinnings, whereas OGTT0,120 indices showed notably distinct genetic bases. Despite overall moderate to high genetic correlations across IS indices, they may capture different genetic facets of T2D. Additionally, the study suggests the need for large-scale genome-wide association studies to comprehensively uncover additional genetic factors associated with IS using multiple IS indices.
Acknowledgments
The authors thank C.F. Rundsten, A.P. Gjesing, and J. Bork-Jensen for their technical assistance with the project. We also thank L. Ryborg and K.N. Kaadtmann for their administrative and research support.
Abbreviations
- AvignonSI0
AvignonSI at fasting
- AvignonSI120
AvignonSI at 120 minutes
- AvignonSIM
average of AvignonSI120 and AvignonSI0
- BennettSI
Bennett fasting sensitivity index
- Belfiorebasal
Belfiore fasting index
- BennettSI
Peter Bennett fasting insulin sensetivity
- BIGTTSI
BIGTT insulin sensitivity index
- BMI
body mass index
- Fins
fasting insulin
- FIns/FGlu
fasting insulin and glucose ratio
- FDR
false discovery rate
- Glu120
glucose at 120 minutes
- HEC
hyperinsulinemic euglycemic insulin clamp
- GuttIndex
Gutt Index
- HOMA-IR
homeostatic model assessment of insulin resistance
- IFC
insulin fold change
- Ins120
insulin at 120 minutes
- IS
insulin sensitivity
- ISI120
insulin sensitivity index at 120 minutes
- ISIbasal
insulin sensitivity index basal
- Ins/Glu120
insulin-glucose ratio at 120 minutes
- Matsuda
Matsuda insulin sensitivity index
- OGTT
oral glucose tolerance test
- PPAR
peroxisome proliferator-activated receptor
- StumvollDem
Stumvoll with demographics (age, body mass index)
- StumvollModi
modified Stumvoll
- T2D
type 2 diabetes
- ρG
genetic correlation
- ρP
phenotypic correlation
Contributor Information
Sufyan Suleman, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Anne L Madsen, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Lars H Ängquist, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Mikkel Schubert, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Allan Linneberg, Center for Clinical Research and Prevention, Copenhagen University Hospital—Bispebjerg and Frederiksberg, Copenhagen 2000, Denmark; Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Ruth J F Loos, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Torben Hansen, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Niels Grarup, Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2200, Denmark.
Funding
The study was supported by a grant from Independent Research Fund Denmark (1030-00280B) to N.G. and S.S. Additionally, S.S., A.L.M., L.A., M.S., R.J.F.L., T.H., and N.G. were also supported by a Novo Nordisk Foundation grant to Center for Basic Metabolic Research (https://cbmr.ku.dk/) University of Copenhagen, Copenhagen 2200, Denmark (Grant No. NNF18CC0034900).
Disclosures
N.G. is currently employed at Novo Nordisk. All other authors do not have any disclosures to make. There are no conflicts of interest that could potentially influence the objectivity or conclusions of the present study.
Data Availability
The genotype and phenotype data analyzed in this study are not publicly available but are available from the corresponding authors on reasonable request. The family study and Inter99 data sets may be obtained by a third party by contacting Allan Linneberg at allan.linneberg@regionh.dk and/or Torben Hansen at torben.hansen@sund.ku.dk.
References
- 1. Weyer C, Tataranni PA, Bogardus C, Pratley RE. Insulin resistance and insulin secretory dysfunction are independent predictors of worsening of glucose tolerance during each stage of type 2 diabetes development. Diabetes Care. 2001;24(1):89‐94. [DOI] [PubMed] [Google Scholar]
- 2. Lillioja S, Mott DM, Spraul M, et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med. 1993;329(27):1988‐1992. [DOI] [PubMed] [Google Scholar]
- 3. Lorenzo C, Haffner SM, Stancáková A, Laakso M. Relation of direct and surrogate measures of insulin resistance to cardiovascular risk factors in nondiabetic Finnish offspring of type 2 diabetic individuals. J Clin Endocrinol Metab. 2010;95(11):5082‐5090. [DOI] [PubMed] [Google Scholar]
- 4. DiCorpo D, LeClair J, Cole JB, et al. Type 2 diabetes partitioned polygenic scores associate with disease outcomes in 454,193 individuals across 13 cohorts. Diabetes Care. 2022;45(3):674‐683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trikudanathan S, Raji A, Chamarthi B, Seely EW, Simonson DC. Comparison of insulin sensitivity measures in south Asians. Metabolism. 2013;62(10):1448‐1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moura FA, Carvalho LSF, Cintra RMR, et al. Validation of surrogate indexes of insulin sensitivity in acute phase of myocardial infarction based on euglycemic-hyperinsulinemic clamp. Am J Physiol Endocrinol Metab. 2014;306(4):E399‐E403. [DOI] [PubMed] [Google Scholar]
- 7. Otten J, Ahrén B, Olsson T. Surrogate measures of insulin sensitivity vs the hyperinsulinaemic-euglycaemic clamp: a meta-analysis. Diabetologia. 2014;57(9):1781‐1788. [DOI] [PubMed] [Google Scholar]
- 8. da Silva C, Zambon MP, Vasques ACJ, et al. Homeostatic model assessment of adiponectin (HOMA-Adiponectin) as a surrogate measure of insulin resistance in adolescents: comparison with the hyperglycaemic clamp and homeostatic model assessment of insulin resistance. PLoS One. 2019;14(3):e0214081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. So A, Sakaguchi K, Okada Y, et al. Relation between HOMA-IR and insulin sensitivity index determined by hyperinsulinemic-euglycemic clamp analysis during treatment with a sodium-glucose cotransporter 2 inhibitor. Endocr J. 2020;67(5):501‐507. [DOI] [PubMed] [Google Scholar]
- 10. Bergman RN, Zaccaro DJ, Watanabe RM, et al. Minimal model-based insulin sensitivity has greater heritability and a different genetic basis than homeostasis model assessment or fasting insulin. Diabetes. 2003;52(8):2168‐2174. [DOI] [PubMed] [Google Scholar]
- 11. Rasmussen-Torvik LJ, Pankow JS, Jacobs DR, et al. Heritability and genetic correlations of insulin sensitivity measured by the euglycaemic clamp. Diabet Med. 2007;24(11):1286‐1289. [DOI] [PubMed] [Google Scholar]
- 12. Almgren P, Lehtovirta M, Isomaa B, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia study. Diabetologia. 2011;54(11):2811‐2819. [DOI] [PubMed] [Google Scholar]
- 13. Alberti KGMM, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1997;15(7):539‐553. [DOI] [PubMed] [Google Scholar]
- 14. Gjesing AP, Hornbak M, Allin KH, et al. High heritability and genetic correlation of intravenous glucose- and tolbutamide-induced insulin secretion among non-diabetic family members of type 2 diabetic patients. Diabetologia. 2014;57(6):1173‐1181. [DOI] [PubMed] [Google Scholar]
- 15. Jørgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glümer C, Pisinger C. A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99 (1). Eur J Cardiovasc Prev Rehabil. 2003;10(5):377‐386. [DOI] [PubMed] [Google Scholar]
- 16. Kolberg JA, Jørgensen T, Gerwien RW, et al. Development of a type 2 diabetes risk model from a panel of serum biomarkers from the inter99 cohort. Diabetes Care. 2009;32(7):1207‐1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raynaud E, Perez-Martin A, Brun JF, Benhaddad AA, Mercier J. Revised concept for the estimation of insulin sensitivity from a single sample. Diabetes Care. 1999;22(6):1003‐1004. [DOI] [PubMed] [Google Scholar]
- 18. Katz A, Nambi SS, Mather K, et al. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab. 2000;85(7):2402‐2410. [DOI] [PubMed] [Google Scholar]
- 19. Sluiter WJ, Erkelens DW, Terpstra P, Reitsma WD, Doorenbos H. Glucose tolerance and insulin release, a mathematical approach. II. Approximation of the peripheral insulin resistance after oral glucose loading. Diabetes. 1976;25(4):245‐249. [DOI] [PubMed] [Google Scholar]
- 20. Hanson RL, Pratley RE, Bogardus C, et al. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am J Epidemiol. 2000;151(2):190‐198. [DOI] [PubMed] [Google Scholar]
- 21. Anderson RL, Hamman RF, Savage PJ, et al. Exploration of simple insulin sensitivity measures derived from frequently sampled intravenous glucose tolerance (FSIGT) tests. The insulin resistance atherosclerosis study. Am J Epidemiol. 1995;142(7):724‐732. [DOI] [PubMed] [Google Scholar]
- 22. Belfiore F, Iannello S, Volpicelli G. Insulin sensitivity indices calculated from basal and OGTT-induced insulin, glucose, and FFA levels. Mol Genet Metab. 1998;63(2):134‐141. [DOI] [PubMed] [Google Scholar]
- 23. Avignon A, Bøegner C, Mariano-Goulart D, Colette C, Monnier L. Assessment of insulin sensitivity from plasma insulin and glucose in the fasting or post oral glucose-load state. Int J Obes. 1999;23(5):512‐517. [DOI] [PubMed] [Google Scholar]
- 24. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412‐419. [DOI] [PubMed] [Google Scholar]
- 25. Laakso M. How good a marker is insulin level for insulin resistance? Am J Epidemiol. 1993;137(9):959‐965. [DOI] [PubMed] [Google Scholar]
- 26. Stumvoll M, Van Haeften T, Fritsche A, Gerich J. Oral glucose tolerance test indexes for insulin sensitivity and secretion based on various availabilities of sampling times [11]. Diabetes Care. 2001;24(4):796‐797. [DOI] [PubMed] [Google Scholar]
- 27. Gutt M, Davis CL, Spitzer SB, et al. Validation of the insulin sensitivity index (ISI0,120): comparison with other measures. Diabetes Res Clin Pract. 2000;47(3):177‐184. [DOI] [PubMed] [Google Scholar]
- 28. Williamson A, Norris DM, Yin X, et al. Genome-wide association study and functional characterization identifies candidate genes for insulin-stimulated glucose uptake. Nat Genet. 2023;55(6):973‐983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22(9):1462‐1470. [DOI] [PubMed] [Google Scholar]
- 30. Hansen T, Drivsholm T, Urhammer SA, et al. The BIGTT test: a novel test for simultaneous measurement of pancreatic beta-cell function, insulin sensitivity, and glucose tolerance. Diabetes Care. 2007;30(2):257‐262. [DOI] [PubMed] [Google Scholar]
- 31. Suleman S, Madsen AL, Ängquist LH, et al. Supplementary material for “Genetic Underpinnings of Fasting and Oral Glucose-stimulated Based Insulin Sensitivity Indices”. GitHub. https://github.com/sufyansuleman/isi_t2d. Date accessed 13 December 2023. [DOI] [PMC free article] [PubMed]
- 32. Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198‐1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vujkovic M, Keaton JM, Lynch JA, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52(7):680‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847‐2849. [DOI] [PubMed] [Google Scholar]
- 36. Suzuki K, Hatzikotoulas K, Southam L, et al. Genetic drivers of heterogeneity in type 2 diabetes pathophysiology. Nature. 2024;19:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen J, Spracklen CN, Marenne G, et al. The trans-ancestral genomic architecture of glycemic traits. Nat Genet. 2021;53(6):840‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Downie CG, Dimos SF, Bien SA, et al. Multi-ethnic GWAS and fine-mapping of glycaemic traits identify novel loci in the PAGE study. Diabetologia. 2022;65(3):477‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walford GA, Gustafsson S, Rybin D, et al. Genome-wide association study of the modified stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes. 2016;65(10):3200‐3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abdul-Ghani MA, Matsuda M, Balas B, DeFronzo RA. Muscle and liver insulin resistance indexes derived from the oral glucose tolerance test. Diabetes Care. 2007;30(1):89‐94. [DOI] [PubMed] [Google Scholar]
- 42. Grarup N, Andersen G, Krarup NT, et al. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57(9):2534‐2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boesgaard TW, Zilinskaite J, Vänttinen M, et al. The common SLC30A8 Arg325Trp variant is associated with reduced first-phase insulin release in 846 non-diabetic offspring of type 2 diabetes patients–the EUGENE2 study. Diabetologia. 2008;51(5):816‐820. [DOI] [PubMed] [Google Scholar]
- 44. Dimas AS, Lagou V, Barker A, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63(6):2158‐2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Torekov SS, Iepsen E, Christiansen M, et al. KCNQ1 long QT syndrome patients have hyperinsulinemia and symptomatic hypoglycemia. Diabetes. 2014;63(4):1315‐1325. [DOI] [PubMed] [Google Scholar]
- 46. Prasad RB, Kristensen K, Katsarou A, Shaat N. Association of single nucleotide polymorphisms with insulin secretion, insulin sensitivity, and diabetes in women with a history of gestational diabetes mellitus. BMC Med Genomics. 2021;14(1):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879‐887. [DOI] [PubMed] [Google Scholar]
- 48. Francque S, Szabo G, Abdelmalek MF, et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat Rev Gastroenterol Hepatol. 2021;18(1):24‐39. [DOI] [PubMed] [Google Scholar]
- 49. Kim HI, Ahn YH. Role of peroxisome proliferator-activated receptor-γ in the glucose-sensing apparatus of liver and β-cells. Diabetes. 2004;53(suppl_1):S60‐S65. [DOI] [PubMed] [Google Scholar]
- 50. Tsuchida A, Yamauchi T, Takekawa S, et al. Peroxisome proliferator–activated receptor (PPAR)α activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: comparison of activation of PPARα, PPARγ, and their combination. Diabetes. 2005;54(12):3358‐3370. [DOI] [PubMed] [Google Scholar]
- 51. Pettinelli P, Videla LA. Up-regulation of PPAR-γ mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J Clin Endocrinol Metab. 2011;96(5):1424‐1430. [DOI] [PubMed] [Google Scholar]
- 52. Schuit FC, Huypens P, Heimberg H, Pipeleers DG. Glucose sensing in pancreatic β-cells: a model for the study of other glucose-regulated cells in gut, pancreas, and hypothalamus. Diabetes. 2001;50(1):1‐11. [DOI] [PubMed] [Google Scholar]
- 53. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 54. Kwok A, Zvetkova I, Virtue S, et al. Truncation of Pik3r1 causes severe insulin resistance uncoupled from obesity and dyslipidaemia by increased energy expenditure. Mol Metab. 2020;40:101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prokopenko I, Poon W, Mägi R, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10(4):e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abdul Rehman SA, Kristariyanto YA, Choi SY, et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 2016;63(1):146‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhande R, Mitchell JJ, Wu J, Sun XJ. Molecular mechanism of insulin-induced degradation of insulin receptor substrate 1. Mol Cell Biol. 2002;22(4):1016‐1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genotype and phenotype data analyzed in this study are not publicly available but are available from the corresponding authors on reasonable request. The family study and Inter99 data sets may be obtained by a third party by contacting Allan Linneberg at allan.linneberg@regionh.dk and/or Torben Hansen at torben.hansen@sund.ku.dk.