Abstract
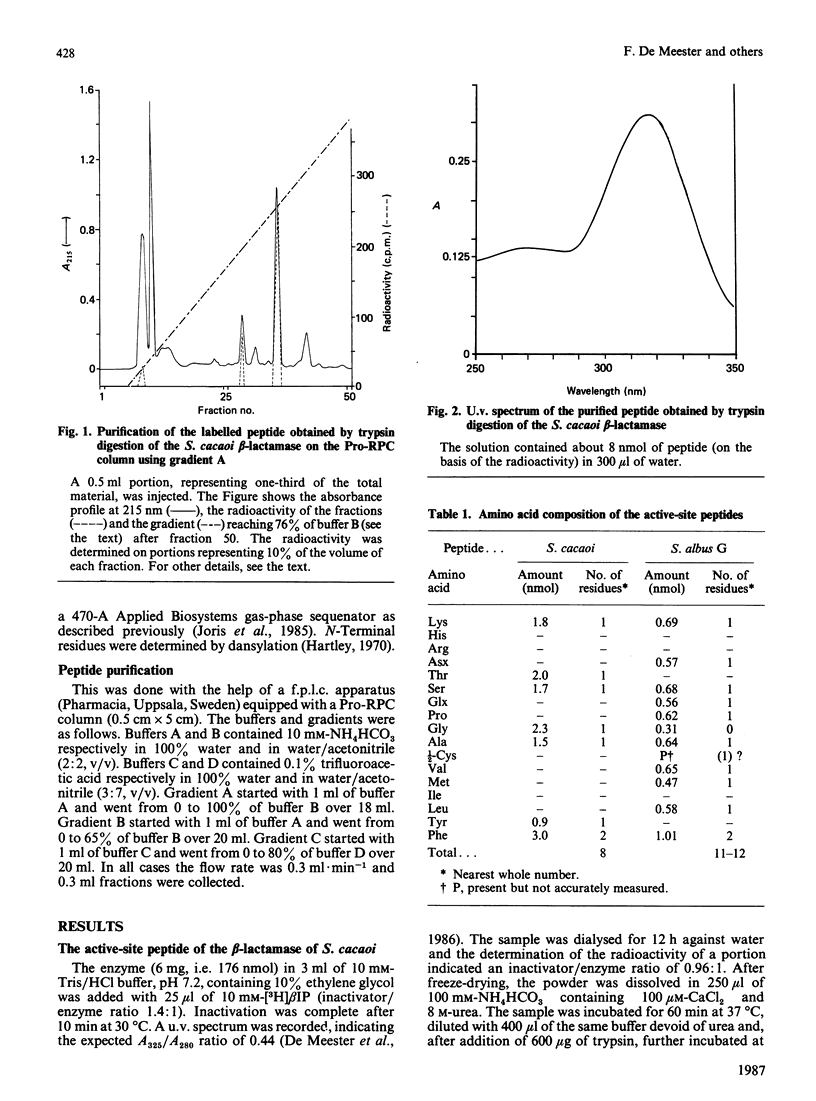
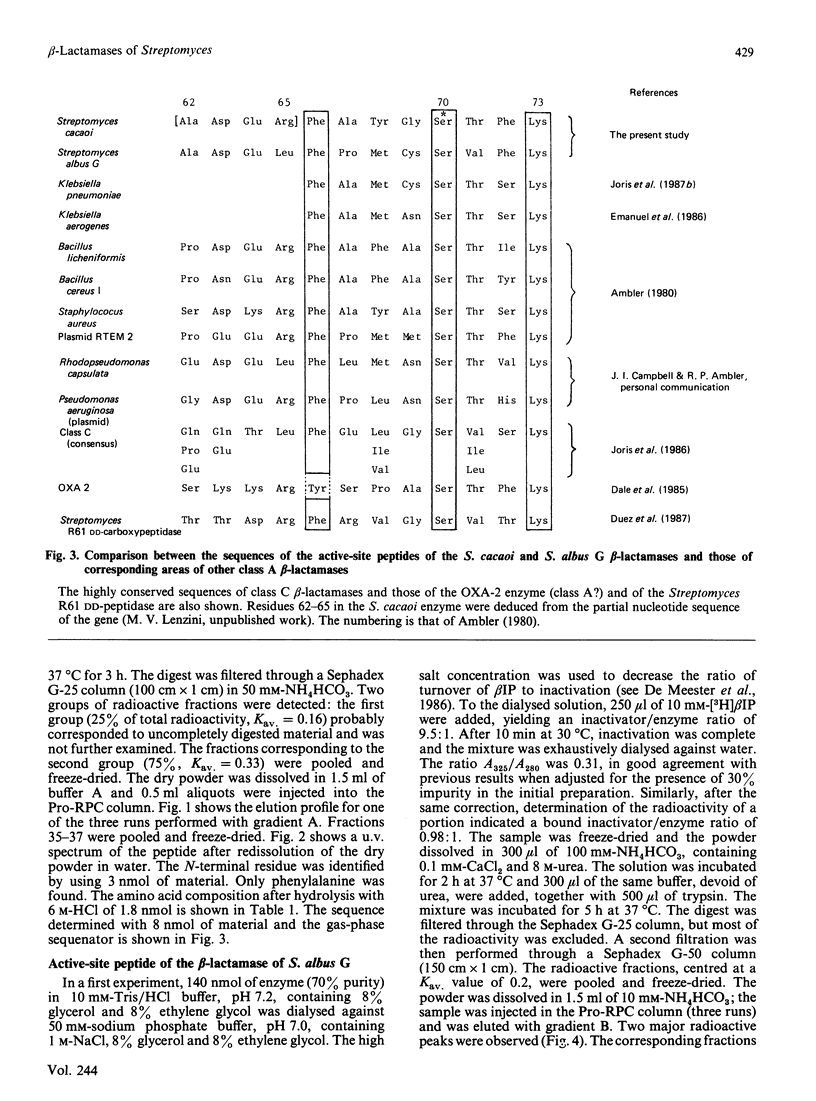
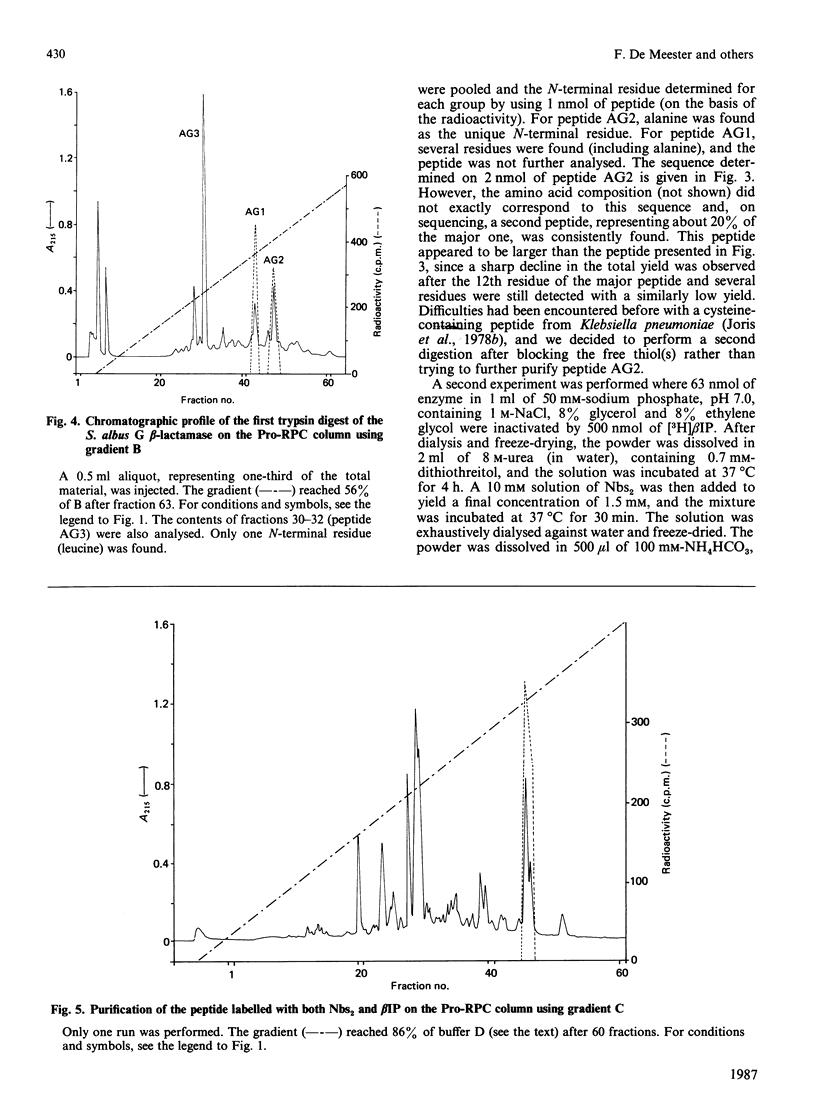
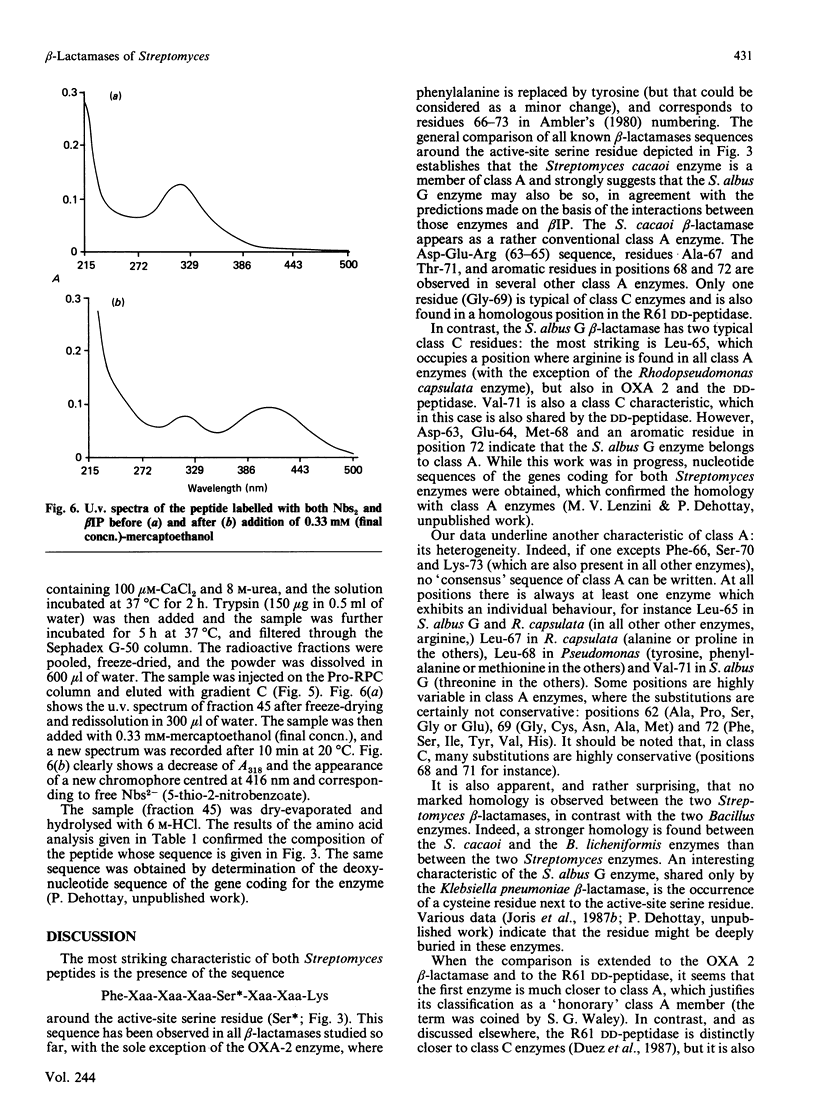
The active-site serine of the extracellular beta-lactamases of Streptomyces cacaoi and Streptomyces albus G has been labelled with beta-iodopenicillanate. The determination of the sequence of the labelled peptides obtained after trypsin digestion of the denatured proteins indicate both enzymes to be class A beta-lactamases. Surprisingly the two Streptomyces enzymes do not appear to be especially homologous, and none of them exhibited a high degree of homology with the Streptomyces R61 DD-peptidase. Our data confirm that, as a family of homologous enzymes, class A is rather heterogeneous, with only a small number of conserved residues in all members of the class.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Dale J. W., Godwin D., Mossakowska D., Stephenson P., Wall S. Sequence of the OXA2 beta-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985 Oct 21;191(1):39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- De Meester F., Frère J. M., Waley S. G., Cartwright S. J., Virden R., Lindberg F. 6-beta-Iodopenicillanate as a probe for the classification of beta-lactamases. Biochem J. 1986 Nov 1;239(3):575–580. doi: 10.1042/bj2390575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehottay P., Dusart J., Duez C., Lenzini M. V., Martial J. A., Frère J. M., Ghuysen J. M., Kieser T. Cloning and amplified expression in Streptomyces lividans of a gene encoding extracellular beta-lactamase from Streptomyces albus G. Gene. 1986;42(1):31–36. doi: 10.1016/0378-1119(86)90147-2. [DOI] [PubMed] [Google Scholar]
- Duez C., Frère J. M., Klein D., Noël M., Ghuysen J. M., Delcambe L., Dierickx L. The exocellular beta-lactamase of Streptomyces albus G. Purification, properties and comparison with the exocellular DD-carboxypeptidase. Biochem J. 1981 Jan 1;193(1):75–82. doi: 10.1042/bj1930075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez C., Piron-Fraipont C., Joris B., Dusart J., Urdea M. S., Martial J. A., Frère J. M., Ghuysen J. M. Primary structure of the Streptomyces R61 extracellular DD-peptidase. 1. Cloning into Streptomyces lividans and nucleotide sequence of the gene. Eur J Biochem. 1987 Feb 2;162(3):509–518. doi: 10.1111/j.1432-1033.1987.tb10669.x. [DOI] [PubMed] [Google Scholar]
- Emanuel E. L., Gagnon J., Waley S. G. Structural and kinetic studies on beta-lactamase K1 from Klebsiella aerogenes. Biochem J. 1986 Mar 1;234(2):343–347. doi: 10.1042/bj2340343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frère J. M., Dormans C., Duyckaerts C., De Graeve J. Interaction of beta-iodopenicillanate with the beta-lactamases of Streptomyces albus G and Actinomadura R39. Biochem J. 1982 Dec 1;207(3):437–444. doi: 10.1042/bj2070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley B. S. Strategy and tactics in protein chemistry. Biochem J. 1970 Oct;119(5):805–822. doi: 10.1042/bj1190805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., De Meester F., Galleni M., Frère J. M., Van Beeumen J. The K1 beta-lactamase of Klebsiella pneumoniae. Biochem J. 1987 Apr 15;243(2):561–567. doi: 10.1042/bj2430561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., De Meester F., Galleni M., Masson S., Dusart J., Frère J. M., Van Beeumen J., Bush K., Sykes R. Properties of a class C beta-lactamase from Serratia marcescens. Biochem J. 1986 Nov 1;239(3):581–586. doi: 10.1042/bj2390581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., De Meester F., Galleni M., Reckinger G., Coyette J., Frere J. M., Van Beeumen J. The beta-lactamase of Enterobacter cloacae P99. Chemical properties, N-terminal sequence and interaction with 6 beta-halogenopenicillanates. Biochem J. 1985 May 15;228(1):241–248. doi: 10.1042/bj2280241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Jacques P., Frère J. M., Ghuysen J. M., Van Beeumen J. Primary structure of the Streptomyces R61 extracellular DD-peptidase. 2. Amino acid sequence data. Eur J Biochem. 1987 Feb 2;162(3):519–524. doi: 10.1111/j.1432-1033.1987.tb10670.x. [DOI] [PubMed] [Google Scholar]
- Kelly J. A., Dideberg O., Charlier P., Wery J. P., Libert M., Moews P. C., Knox J. R., Duez C., Fraipont C., Joris B. On the origin of bacterial resistance to penicillin: comparison of a beta-lactamase and a penicillin target. Science. 1986 Mar 21;231(4744):1429–1431. doi: 10.1126/science.3082007. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lenzini M. V., Frère J. M. The beta-lactamase of Streptomyces cacaoi: interaction with cefoxitin and beta-iodopenicillanate. J Enzyme Inhib. 1985;1(1):25–34. doi: 10.3109/14756368509031279. [DOI] [PubMed] [Google Scholar]
- Ogawara H., Mantoku A., Shimada S. beta-lactamase from Streptomyces cacaoi. Purification and properties. J Biol Chem. 1981 Mar 25;256(6):2649–2655. [PubMed] [Google Scholar]
- Samraoui B., Sutton B. J., Todd R. J., Artymiuk P. J., Waley S. G., Phillips D. C. Tertiary structural similarity between a class A beta-lactamase and a penicillin-sensitive D-alanyl carboxypeptidase-transpeptidase. 1986 Mar 27-Apr 2Nature. 320(6060):378–380. doi: 10.1038/320378a0. [DOI] [PubMed] [Google Scholar]


