Abstract
Background
Somatic genetic alterations of the estrogen receptor 1 gene (ESR1) are enriched in endocrine therapy-resistant, estrogen receptor-positive (ER+) metastatic breast cancer (mBC). Herein, we investigated and compared the clinical and genomic landscape of ESR1-mutant (ESR1MUT) and ESR1 wild type (ESR1WT) ER+/ human epidermal growth factor receptor 2 (HER2)− mBCs.
Methods
Clinical and genomic data were retrieved from cBioPortal using the publicly-available MSK MetTropism dataset. Metastatic, ER+/HER2− mBC samples were included in the analysis. Only oncogenic and likely oncogenic alterations according to OncoKB were included. Statistical analyses were carried out using alpha level of 0.05, with a false discovery rate threshold of 10% for multiple comparisons using the Benjamini–Hochberg method.
Results
Among 679 samples, 136 ESR1MUT among 131 tumors were found (19.2%). The frequency of ESR1MUT was higher in ductal versus lobular mBC (21.2% versus 13.8%, P = 0.052) and enriched in liver metastasis compared with other sites (22.5% versus 12.7%; q = 0.02). Compared with ESR1WT mBC, ESR1MUT tumors showed higher fraction of genome altered (FGA) {[0.28 interquartile range (IQR), 0.15-0.43] versus 0.22 (0.11-0.38); P = 0.04} and tumor mutational burden (TMB) [4.89 (IQR 3.46-6.85) versus 3.92 (2.59-6.05) mut/Mb; P = 0.001]. Tumors harboring p.E380X alterations showed higher TMB compared with those with H11-12 alterations [8.24 (IQR 5.06-15.3) versus 4.89 (IQR 3.46-6.75) mut/Mb; P = 0.01]. Genetic alterations of TP53 were enriched in ESR1WT tumors (36% versus 14%) [odds ratio (OR) 3.17, 95% confidence interval (CI) 1.88-5.64, q = 0.001]. Considering signaling pathways, ESR1MUT tumors showed a lower occurrence of TP53 (OR 0.48, 95% CI 0.30-0.74; q = 0.003) and MAPK (OR 0.29, 95% CI 0.11-0.65; q = 0.009) alterations. TP53 (q < 0.001), CDH1 (q < 0.001), and ERBB2 (q < 0.001) demonstrated mutual exclusivity with ESR1MUT.
Conclusions
ER+/HER2− mBCs carrying ESR1MUT exhibit a divergent genomic background, characterized by a lower prevalence of TP53 and MAPK pathway alterations. Less common ESR1 alterations falling outside the H11-H12 region seem to occur in tumors with higher TMB, deserving further investigation to understand their potential actionability.
Key words: ESR1, breast cancer, genomic analysis, precision medicine, next-generation sequencing
Highlights
-
•
Distinct genomic mechanisms promote endocrine resistance among ESR1-mutant and ESR1 wild-type ER+/HER2− breast cancer.
-
•
ESR1-mutant ER+/HER2− breast cancer displays peculiar genetic landscape and genomic signatures.
-
•
Potentially actionable genetic alterations are differently distributed according to the presence of ESR1 alterations.
-
•
ER+/HER2− breast cancer carrying ESR1 p.E380Q might be particularly susceptible to the use of immunotherapy.
Introduction
Estrogen receptor-positive (ER+) and human epidermal growth factor receptor 2 (HER2)-negative breast cancer (BC) represents the most common BC subtype, accounting for ∼70% of diagnosed cases.1 Endocrine therapy (ET) represents the therapeutic cornerstone for ER+/HER2− BC, having been shown to result in improved survival in all settings of disease.1 ER signaling can be manipulated through different pharmacological strategies, including direct antagonism to estrogen receptor 1 (ESR1),2 by means of reduction of expression,3 or inhibition of estrogen production from the ovaries4 or from androgen aromatization into estrogens.5
Most of ER+ metastatic breast cancer (mBC) will ultimately develop biological resistance to ET, which may be promoted by several factors. Among them, the emergence of mutations in estrogen receptor 1 (ESR1MUT) represents one of the most well-characterized mechanisms, occurring in ∼30% of cases displaying ET resistance after prior exposure to antiestrogen therapy.1,6 Most genomic variants in ESR1 occur within the ligand-binding domain, promoting a conformational change of the protein resulting in an estrogen-independent constitutive transcription of ER-regulated genes.7, 8, 9
The impact of ESR1MUT on the activity of ET agents varies across different classes, being an established mechanism of resistance to aromatase inhibitors (AIs), while generally preserving variable sensitivity to selective estrogen receptor degrader (SERD).6,7,10, 11, 12 More recently, a novel generation of oral SERDs have been developed, demonstrating more favorable pharmacokinetic properties than fulvestrant and the ability to target a larger spectrum of hotspot ESR1MUT.10,13 Following preclinical studies, randomized clinical trials demonstrated the superior efficacy of oral SERD to AI and fulvestrant, and particularly among ET-resistant, ESR1MUT ER+ mBC, thus leading to the validation of ESR1MUT as a predictive biomarker for which diagnostic testing is routinely undertaken in clinical practice.14, 15, 16, 17
Nevertheless, other genomic mechanisms may account for ET resistance, whose actionability yielded likewise expanded therapeutic options, with most represented by agents targeting the phosphoinositide 3-kinase-protein kinase B-mammalian target of rapamycin (PI3K-AKT-mTOR) pathway.18,19 Accordingly, in light of the increasing genomically-driven therapeutic strategies for ER+ mBC, a critical task consists of elucidating the principal pathway promoting ET resistance, which is essential for tailoring treatments to restore sensitivity to ET and potentially delaying time to chemotherapy.
In this setting, limited data exist to examine the genomic background of ESR1MUT ER+/HER2− mBC, and particularly to determine the presence of additional potentially actionable pathways, and to assess whether distinct molecular mechanisms may account for ET resistance compared with ESR1 wild-type (ESR1WT) tumors. Within this context, the present study aimed to assess whether the presence of ESR1MUT dictates or is dictated by a distinct genomic background, as well as to examine further potential biomarkers suitable for clinical actionability among ER+ mBC carrying or not ESR1MUT.
Methods
Data retrieval
Clinical and genomic data were retrieved from cBioPortal20,21 using the cBioPortalR package.22 For our analysis, publicly available data were queried from the MSK MetTropism dataset, an integrated pan-cancer cohort study of >25 000 patients affected by different primary tumors, including about 2500 BC.23 Metastatic samples from patients affected by ER+/HER2−, which were previously exposed to ET, were considered for the analysis. All samples were subjected to genomic profiling by the FDA-cleared, tumor-normal MSK-IMPACT assay.24
Genomic analysis
Oncogenic and likely oncogenic alterations by OncoKB annotation25 were included for downstream analysis. In the ESR1MUT group, we excluded tumors carrying ESR1 copy number gains while we considered for inclusion single nucleotide variants (SNV), small insertion/deletions (indels), rearrangement, or fusion within the ESR1 gene, having copy number positive changes of the ESR1 gene a less clarified pathogenetic significance and not deemed as responsive to SERDs.26 We selected genes to include in the pathway-level analysis according to those included in The Cancer Genome Atlas Program (TCGA) project.27 ESR1MUT were categorized according to the functional protein domain affected, as previously reported.28 Tumor mutational burden (TMB) was calculated as the proportion of nonsynonymous mutations to the total number of base pairs sequenced per sample. Fraction of genome altered (FGA) was defined as the fraction of genome analyzed for copy number changes presenting with log2 copy number gain >0.2 or loss less than −0.2. The MSIsensor score with a threshold of ≥10 was used to define microsatellite instability (MSI-high).29
Statistical analysis
Categorical variables were reported as absolute numbers and proportions, and continuous variables as median and interquartile range (IQR). Associations of categorical variables were carried out using the Fisher’s exact test, or logistic regression model, as appropriate. The Bartlett test and Shapiro–Wilk test were used to assess variances and normal distributions, respectively. Nonparametric tests for group comparisons for continuous variables included the Wilcoxon rank sum test and Kruskal–Wallis test. Dunn’s test was used for multiple pairwise comparisons after a significant Kruskal–Wallis test. Statistical tests were carried out using a two-sided significance level of <0.05. The Benjamini and Hochberg method was used for multiple comparisons, whenever appropriate, using a false discovery rate threshold of 10%. Statistical analyses were carried out using R Software version 4.3.2.30 Genomic co-occurrence and mutual exclusivity analyses were carried out using the DISCOVER31 and Rediscover32 R packages.
Results
Clinical characteristics
A total of 679 ET-pre-treated ER+/HER2− mBC cases were retrieved. Among them, 99.26% (674 of 679) were females, with a median age of 58.8 years (IQR 50.6-67.0 years). One-hundred thirty-one (19.29%) samples exhibited ESR1MUT, while the ESR1WT group included 80.71% (548 of 679) of cases. Patients whose tumors had ESR1MUT exhibited a significantly higher median age [62.5 years (IQR 54.5-67.8 years) versus 57.9 years (IQR 49.6-66.8 years); P = 0.004], while a numerically lower proportion of ESR1MUT was observed among Asian patients (Asian 3 of 31, 9.68%; versus black 10 of 55, 18.2%; n = 55; white 111 of 544, 20.4%; P = 0.329) (Table 1).
Table 1.
Clinical and pathological characteristics among ESR1 mutant and ESR1 wild-type tumors
| Characteristic | ESR1MUT, N = 131 | ESR1WT, N = 548 | P value |
|---|---|---|---|
| Sex | 0.6 | ||
| Female | 131 (100%) | 543 (99%) | |
| Male | 0 (0%) | 5 (0.9%) | |
| Age, years | 63 (IQR 55-68) | 58 (IQR 50-67) | 0.004 |
| Self-reported ethnicity | 0.3 | ||
| Asian | 3 (2.4%) | 28 (5.5%) | |
| Black or African American | 10 (8.1%) | 45 (8.9%) | |
| White | 111 (90%) | 433 (86%) | |
| Unknown | 7 | 42 | |
| Histology subtype | 0.026 | ||
| Ductal | 110 (84%) | 410 (75%) | |
| Lobular | 21 (16%) | 138 (25%) |
Significant P values (P < 0.05) are indicated in bold.
IIQR, interquartile range; MUT, mutant; WT, wild-type.
Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) represented 76.58% (520 of 679) and 23.42% (159 of 679) of cases, respectively. IDC exhibited a numerically higher proportion of ESR1MUT compared with ILC (21.2% versus 13.8%, P = 0.052).
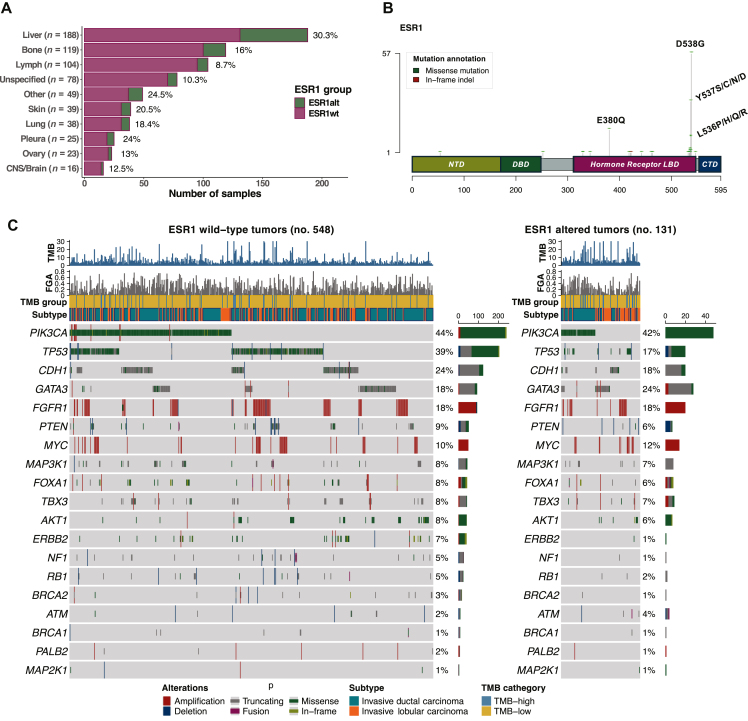
Liver represented the most commonly sampled site on which comprehensive genomic profiling (CGP) had been carried out (27.69%, 188 of 679), followed by bone (17.53%, 119 of 679), and lymph nodes (15.32%, 104 of 679). Considering the proportion of ESR1MUT according to the anatomical site sampled, an enrichment of ESR1MUT was observed in liver lesions compared with non-liver sites (22.5% versus 12.7%; q < 0.001) (Figure 1A).
Figure 1.
Proportion of ESR1MUTaccording to metastatic site, type of ESR1 mutations, and driver genes among ESR1-mutated and ESR1 wild-type tumors. (A) Sampled sites with proportion of ESR1MUT detected by each site (shown in red). (B) Spectrum of ESR1MUT detected among 679 ER+ mBC. (C) Oncoprints of driver genes in breast cancer among ESR1WT and ESR1MUT tumors. CTD, C-terminal domain; DBD, DNA-binding domain; ER, estrogen receptor; FGA, fraction of genome altered; LBD, ligand binding domain; mBC, metastatic breast cancer; MUT, mutant; NTD, N-terminal domain; TMB, tumor mutational burden; WT, wild-type.
Finally, in the multivariable binary logistic regression model, older age (P < 0.001), presence of liver metastasis (P = 0.001), and IDC histology (P = 0.05) were all associated with the presence of ESR1MUT (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.103731).
ESR1 genomic alterations
A total of 136 ESR1MUT were observed among 131 cases. Of them, 97.79% (133 of 136) consisted of SNV, while indels accounted for the remaining 3 (2.21%) cases.
The most common ESR1MUT consisted of the missense point mutation p.D538G (41.91%, 57 of 136), followed by p.Y537S (22.06%, 30 of 136), and p.E380Q (10.29%, 14 of 136) (Figure 1B). Considering ESR1MUT mutations according to the functional protein domain, 85.29% (116 of 136) of ESR1MUT consisted of class I alterations located around helix 11-12 and within the LBD, followed by mutations affecting helix 5 (p. E380Q) (10.29%, 14 of 136), and class II mutations located in the ESR1 dimerizing domain (4.41%, 6 of 136), including in this latter group p.M421V, p.F461V, p.V422del, p.V418E, p.G442R, and p.S463P mutations.
Five patients (3.8%) displayed multiple ESR1MUT, which included p.D538G and p.Y537C; p.E380Q and p.Y537C; p.Y537N and p.V422del; p.D538G and p.G442R; p.Y537S and p.Y537N.
Genomic co-alterations with ESR1MUT
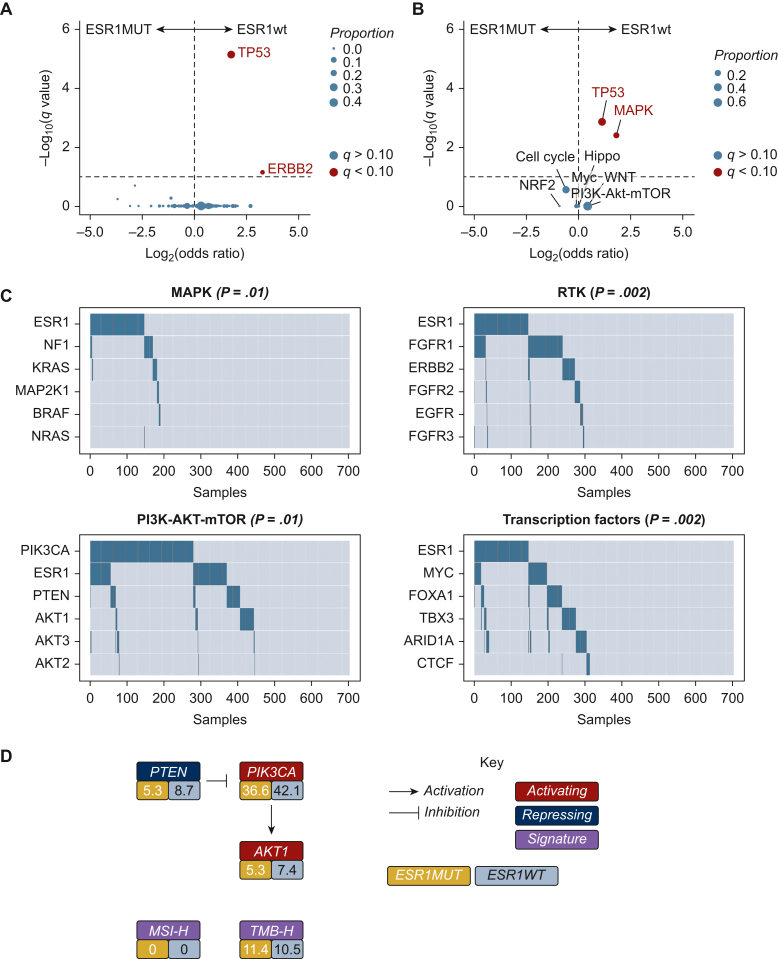
In both ESR1MUT and ESR1WT tumors, PIK3CA represented the most commonly altered gene (41% and 38%, respectively; Figure 1C, Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2024.103731). Compared with ESR1WT tumors, ESR1MUT tumors showed a lower rate of genomic alterations in TP53 {15% versus 36%, odds ratio (OR) 0.33 [95% confidence interval (CI) 0.19-0.56], q < 0.001} and ERBB2 [1% versus 7%, OR 0.10 (95% CI 0.01-0.62), q = 0.07] (Figure 2A). Considering signaling pathways, ESR1MUT showed a lower occurrence of MAPK [OR 0.34 (95% CI 0.14-0.73), q = 0.02] and TP53 pathway [OR 0.49 (95% CI 0.31-0.74), q = 0.003] alterations (Figure 2A and B, Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.103731).
Figure 2.
Enrichment gene alteration analysis among ESR1-mutated and ESR1 wild-type tumors. (A) Different distribution of gene-level genomic alterations among ESR1MUT and ESR1WT tumors. (B) Different distribution of pathway-level genetic alterations among ESR1MUT and ESR1WT tumors. (C) Groupwise comparisons of mutual exclusivity of selected list of genes from included in the MAPK, RTK, PI3K-AKT-mTOR, and transcription factors pathways. (D) Proportion of detected genetic biomarkers with approved biomarker-matched targeted therapy available among ESR1MUT (in yellow) and ESR1WT (in gray) tumors. NTRK1/2/3 fusions, RET fusions and MSI-H not displayed as no case was observed. MUT, mutant; WT, wild-type.
In the co-occurrence and mutual exclusivity analysis, ESR1MUT showed mutual exclusivity with genomic variants in TP53 (q < 0.001), CDH1 (q < 0.001), and ERBB2 (q < 0.001) (Figure 2C; Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2024.103731). Conversely, ESR1MUT did not demonstrate any significant co-occurrence with other genetic alterations (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2024.103731).
Finally, considering the availability of different genomically-driven therapies in ER+/HER2− advanced BC, we further evaluated the different distribution of genomic biomarkers for which at least one therapy is clinically approved among ESR1MUT and ESR1WT tumors. While we did not observe any case showcasing microsatellite instability, genomic fusion involving NTRK1/2/3 or RET and class I BRAF alterations, between ESR1MUT and ESR1WT BC we did not observe any statistically different distribution of gene alterations in PIK3CA (38.6% versus 44.1%, P = 0.29), PTEN (5.3% versus 8.7%, P = 0.26), AKT1 (7.6% versus 5.3%, P = 0.50), and in the proportion of TMB-high tumors (11.4% versus 10.5%, P = 0.77) (Figure 2D).
Genomic signatures
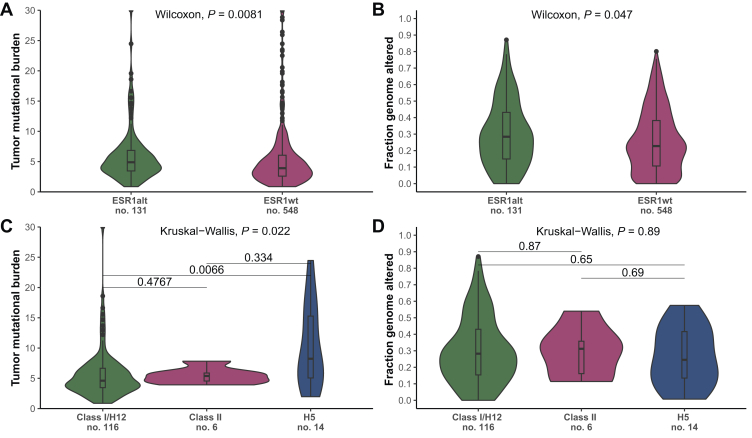
Compared with ESR1WT tumors, tumors showing ESR1MUT exhibited a higher median TMB [4.89 (IQR 3.46-6.85) versus 3.92 (IQR 2.59-6.05), respectively; P < 0.001] and FGA [0.28 (IQR 0.15-0.43) versus 0.22 (IQR 0.10-0.38), respectively; P = 0.047] (Figure 3A and B). Considering the class of ESR1MUT, tumors carrying class H5 mutations showed higher median TMB [8.24 (IQR 5.06-15.3)] compared with class I/H11-12 ESR1MUT [4.32 (IQR 3.46-6.66), P = 0.01] and class II ESR1MUT [4.66 (IQR 4.31-5.14), P = 0.39] (Figure 3C). Of note, 5 of 14 p.E380Q ESR1MUT (class H5) showed a TMB above 10 mutations per megabase (mut/Mb). Conversely, no difference in FGA was observed according to the class of ESR1MUT [0.28 (IQR 0.15-0.43) Class I/H11-12; 0.24 (0.13-0.41) H5; 0.24 (0.11-0.41) Class II; P = 0.88] (Figure 3D).
Figure 3.
Genomic signatures among ESR1-mutated and ESR1 wild-type tumors, and across classes of ESR1 mutations. (A) Comparison of tumor mutational burden and (B) fraction genome altered among ESR1MUT and ESR1WT tumors. Comparison of tumor mutational burden (C) and fraction genome altered (D) across different class of ESR1 mutations. MUT, mutant; WT, wild-type.
Discussion
In the present work, we outlined the clinical and genomic landscape of ET-resistant ER+/HER2− mBC showing ESR1MUT, and aimed to define clinical variables to better select diagnostic strategies and genomic correlates that could potentially inform and tailor therapeutic decision-making.
Distinctive genomic mechanisms may account for ET resistance in ER+/HER2− mBC.
Consistent with previous reports,7,33 in our cohort tumors showcasing ESR1MUT represented about 20% of the cases. In the multivariable analysis, ESR1MUT tumors demonstrated enrichment in liver metastasis, as previously shown34,35 and occurred with lower frequency in younger patients and among ILC. The genomic landscape of ER+/HER2− BC differs according to age,36 with tumors arising in younger women demonstrating lower endocrine dependence and reduced expression of ESR1 and ER-related genes.37, 38, 39 Therefore, it is possible different mechanisms of ET resistance may emerge in tumors affecting younger women, which may not be directly related to the ET pathway, but rather occur in non-overlapping signaling pathways. Similarly, ILCs have been described to carry recurrent genomic alterations in oncogenes including MYC, PIK3CA, FGFR1, AKT1, ERBB2, and ERBB3.40, 41, 42 In our analysis, we observed ILC to carry a lower frequency of ESR1MUT compared with IDC (11.9% versus 20.1%, respectively; P = 0.052). Moreover, in contrast to previous studies showing recurrent ESR1 copy number gains occurring in ILC,41 in our analysis, we observed only one case of ILC exhibiting ESR1 amplification, not suggesting ESR1 amplifications to be enriched among ILC. Accordingly, our result supports the notion that ILC may leverage with higher frequency on non-estrogen-related genomic pathways to acquire ET, with our hypothesis further reinforced by the mutual exclusivity we observed between ESR1MUT and genetic alterations occurring in the CDH1 gene (q < 0.001).
Alterations involving oncogenes included within the MAPK pathway carry a critical role in determining ET resistance. Copy number losses and truncating mutations in NF1, along with hotspot mutations in KRAS, BRAF, and MAP2K1 (encoding MEK1) promote secondary resistance to ET and hence are found to be enriched among pre-treated ER+ mBC.43,44 Importantly, in our analysis, of 51 oncogenic alterations in the MAPK pathway among 46 cases, 42 (91.3%) of them occurred in ESR1WT tumors, the association of which with the ESR1WT status was further confirmed by the significant inferior rate of MAPK alterations at the pathway level among ESR1MUT tumors (q = 0.02). Accordingly, our results support the concept that mutations in ESR1 and MAPK represent two non-overlapping mechanisms of ET resistance, with agents targeting the MAPK pathway potentially representing a ground of research for overcoming ET resistance particularly among ESR1WT tumors.
Besides MAPK oncogenes, alterations in tyrosine kinase receptors (TKR) play a pivotal role in determining estrogen-independent tumor growth in ER+/HER2− BC. While ERBB2 copy number gains and hotspot mutations have been reported to be shared by primary tumors and thus contribute to primary ET resistance,40,43,45,46 alterations involving EGFR and FGFR are positively selected by acquired mechanisms of resistance to ET.40,43,47, 48, 49, 50 Similarly to MAPK alterations, we observed a superior rate of TKR alterations occurring among ESR1WT tumors (27.3% versus 19.0%, respectively, P = 0.051). Of note, ESR1WT tumors exhibited 38 of 39 (97.4%) and 10 of 11 (90.9%) ERBB2 and EGFR alterations, respectively. Conversely, we did not observe a different distribution of FGFR1 alterations among ESR1MUT and ESR1WT tumors (15.2% and 16.9%, respectively), which may be related to the reported co-occurrence of FGFR1MUT and ESR1MUT in tumors showing resistance to cyclin-dependent kinase (CDK) 4 and 6 inhibitors (CDK4/6i) plus ET.50 Notably, most ERBB2 alterations (89.1%) consisted of SNV and indels, supporting the notion that ERBB2 alterations other than amplifications represent a relevant and potentially actionable mechanism of ET resistance, which should be further investigated along with agents targeting other TRK, such as EGFR, particularly for tumors not showing ESR1MUT and for tumors showcasing pre-treatment TRK genomic alterations.
Considering the expanding genomically-driven therapeutic option for pre-treated ER+/HER2− BC, and particularly for agents targeting the PI3K-AKT-mTOR cascade, we investigated whether ESR1MUT could account for a different prevalence of alterations in this signaling pathway. While previous studies reported a higher prevalence of PIK3CAMUT among ESR1MUT BC,9 in our cohort, PIK3CA represented the most commonly altered gene in both ESR1MUT and ESR1WT tumors, with no association between the ESR1 status and PI3K-AKT-mTOR alterations in our pathway-level analysis. As such, our results demonstrate that concomitant alterations in the PI3K-AKT-mTOR pathway occur with a relevant frequency also among ESR1MUT tumors, which may be particularly true for tumors showing resistance to CDK4/6i, as previously shown,50 which emphasizes the necessity to gather additional biomarkers to further personalize second-line treatment options for ESR1MUT tumors showing PIK3CA/AKT1/PTEN alterations.
In addition to targeting ESR1MUT and the PI3K-AKT-mTOR pathway, several trials assessed the continuation of CDK4/6i beyond progression, with conflicting clinical results that underscore the importance of biomarker selection. Different mechanisms of resistance may account for CDK4/6i resistance, including alterations in RB1, AURKA, CCNE1, ERBB2, and FGFR1.51, 52, 53 Of note, in ESR1MUT compared with ESR1WT tumors, we observed a relevant lower occurrence of alterations in RB1 (2 of 131, versus 23 of 548, P = 0.19) and AURKA (2 of 131 versus 26 of 548, P = 0.13), suggesting a different mechanism of resistance to CDK4/6i may emerge according to the ESR1 status. To support our observation, in the phase II PACE study, continuing palbociclib while switching to ET showed superior outcomes among ESR1MUT (n = 78),54 whose finding was yet not replicated in the phase II MAINTAIN trial, in which ribociclib in combination with fulvestrant following progression to ET plus CDK4-6i (mostly receiving palbociclib) showed inferior benefits among ESR1MUT tumors (n = 36).55 It must be noted that in the MAINTAIN study, 50% of ESR1MUT tumors showed co-occurrent FGFR1 alterations,55 which along with the small population size, does not discredit our hypothesis for which resistance mechanisms to CDK4/6i may occur with a lower frequency among ESR1MUT tumors, warranting further investigation as to whether ESR1MUT may act as a surrogate biomarker for which CDK4/6i continuation may portend clinical benefits.
Consistent with previous reports,56,57 we observed a mutual exclusivity between ESR1MUT and TP53 genomic alterations, the relationship of which remained significant at the pathway-level analysis. A reciprocal antagonism between ER and TP53 signaling has been reported, with TP53 aberrations negatively affecting the downstream ER activation, and conversely by ER blocking the transactivation of TP53 by means of activation of TP53 co-repressors.35,50,57,58 Consequently, our results further confirm the concept that ER+ mBC can leverage distinctively on either ESR1MUT or TP53 pathway under the selective pressure of ET. Moreover, tumors carrying co-occurrent TP53MUT-ESR1MUT have been described to be enriched in liver metastasis and to carry an immune-enriched imprinting with a higher prevalence of CD8+ T cells and programmed death ligand 1 (PD-L1) expression.35 In our study cohort, of 19 patients affected by tumors showing co-occurrent TP53MUT-ESR1MUT, 15 of them (78.9%) showed liver involvement. Furthermore, tumors showing TP53MUT-ESR1MUT exhibited higher TMB compared with both TP53WT-ESR1MUT and TP53MUT-ESR1WT tumors (5.87 versus 4.89 mut/Mb, P = 0.28; and 5.87 versus 4.89 mut/Mb, P = 0.39, respectively), as well as higher FGA (0.36 versus 0.24, P = 0.002; and 0.36 versus 0.30, P = 0.02, respectively). As such, our findings expand the limited evidence suggesting tumors carrying TP53MUT-ESR1MUT exhibit higher genomic complexity, potentially rendering them susceptible to immunotherapy, and to display aggressive clinical behavior, warranting further characterization of their genomic landscape and investigation for tailored treatment regimens.
Lastly, we investigated whether different ESR1MUT account for different genomic signatures. In line with previous reports, we observed most ESR1MUT (85.29%) occur in the loop bridging helix 11 (H11) and H12, which are known to promote conformational changes in ESR1 to drive ligand-independent downstream activation. Conversely, a second hotspot alteration involves the aminoacidic position 380, affecting H5, which represented 10.29% of our observed cases and lies topographically near the dimer interface in which class II ESR1MUT occur, which is believed to promote ESR1-ligand independent heterodimerization, as these latter. While we observed superior TMB and FGA for ESR1MUT compared with ESR1WT tumors, we observed a relevant higher TMB particularly for H5 mutations (8.24 mut/Mb) compared with class I ESR1MUT (4.32 mut/Mb, P = 0.01) and class II ESR1MUT (4.66 mut/Mb, P = 0.39), with 35.7% (5 of 14) showing a TMB above 10 mut/Mb. To our knowledge, no previous reported data revealed a higher TMB specifically in the context of E380Q ESR1MUT, which in light of the encouraging progression-free survival observed in the PACE trial with avelumab added to fulvestrant and continuation of palbociclib in ET-resistant ER+ mBC,54 should be further investigated to possibly identify subgroups of ER+ MBC which may potentially benefit from immunotherapy. Of note, E380Q ESR1MUT has been described as a context-mutation frequently occurring in mBC carrying an APOBEC3 dominant signature, which in turn has been shown to associate with higher TMB and to account for most ER+/HER2− BC showing ≥10 mut/Mb,59 Accordingly, our data suggest the tumor detection of E380Q ESR1MUT could act as a surrogate biomarker of HR+/HER2− BC with potential high neoantigen load, for which the use of immunotherapy should be further explored.
It must be noted our work presents limitations. First, our analysis refers to a retrospective, single-institution case series, for which findings should gather external replication and validation. Second, regardless we selected post-treatment, metastatic ER+ BC, we did not have access to specific treatment data and to the line of therapy on which ESR1MUT was detected, and accordingly to whether the genomic landscape could differ according to the previous exposure to distinct ET regimens; nevertheless, it is reasonable most of the patients were exposed to AI and CDK4/6i, according to international guidelines, and corroborated by the relevant frequency of ESR1MUT we observed. Third, for the same reason, we could not perform correlations between genomic biomarkers and clinical outcomes to ET as well as analyze different genomic mechanisms possibly governing different resistance to AI and fulvestrant.
Despite these limitations, our study provides a comprehensive analysis of the genomic landscape differentiating ESR1MUT from ESRWT tumors, revealing distinct mechanisms of ET resistance and further potential and distinct therapeutic targets among the two groups of ER+/HER2− mBC.
Conclusion
Our genomic analysis revealed divergent acquired mechanisms driving clinical resistance to ET, with non-overlapping signaling cascades possibly separating resistance trajectories according to the ESR1 status. While optimal second-line targeted treatments should be guided in a biomarker-driven fashion, further research is needed to elucidate the best treatment options for tumors carrying multiple actionable biomarkers. Moreover, novel therapeutic options, such as agents targeting MAPK and TRK, should be further explored to revert ET resistance, and particularly among ESR1WT tumors. Lastly, our findings showcased specific mutations in ESR1 associate with predictive biomarkers of immunotherapy efficacy, warranting further research to address their potential actionability by using immuno-oncology agents.
Acknowledgments
Funding
None declared.
Disclosure
EGR has received honoraria and/or advisory fees and/or research funding from AstraZeneca, Exact Sciences, Novartis, Roche, and Thermo Fisher. CB received travel expenses reimbursement from Gilead, and has received honoraria for an advisory role from Roche. EM received consulting or advisory fees from Eisai, Exact Sciences, MSD Oncology, Daiichi-Sankyo/AstraZeneca, Pfizer, and Seagen outside the submitted work. NF reports financial interests, personal, invited speaker: Merck Sharp & Dohme (MSD), AstraZeneca, Daiichi Sankyo, GlaxoSmithKline, Gilead, Novartis, Roche, Menarini, Lilly, Sysmex; financial interests, personal, advisory board: Merck Sharp & Dohme (MSD), AstraZeneca, Menarini; financial interests, personal and institutional, research grant: Novartis, Sysmex, Veracyte. CC reports personal fees for consulting, advisory role, and speakers’ bureau from Lilly, Roche, Novartis, MSD, Seagen, Gilead and Pfizer. GC received honoraria for speaker’s engagement: Roche, Seattle Genetics, Novartis, Lilly, Pfizer, Foundation Medicine, NanoString, Samsung, Celltrion, BMS, MSD; honoraria for providing consultancy: Roche, Seattle Genetics, NanoString; honoraria for participating in advisory board: Roche, Lilly, Pfizer, Foundation Medicine, Samsung, Celltrion, Mylan; honoraria for writing engagement: Novartis, BMS; honoraria for participation in Ellipsis Scientific Affairs Group; institutional research funding for conducting phase I and II clinical trials: Pfizer, Roche, Novartis, Sanofi, Celgene, Servier, Orion, AstraZeneca, Seattle Genetics, AbbVie, Tesaro, BMS, Merck Serono, Merck Sharp Dome, Janssen-Cilag, Philogen, Bayer, Medivation, Medimmune. All other authors have declared no conflicts of interest.
Statement of ethical approval
Data used for the reported analyses were retrieved from a publicly available database (cBioPortal), with cases anonymized and assigned to a unique code for de-identification, for which no ethical approval nor specific informed consent for the present study was required.
Contributor Information
A. Marra, Email: antonio.marra@ieo.it.
G. Curigliano, Email: giuseppe.curigliano@ieo.it.
Supplementary data
References
- 1.Burstein H.J. Systemic therapy for estrogen receptor–positive, HER2-negative breast cancer. N Engl J Med. 2020;383(26):2557–2570. doi: 10.1056/NEJMra1307118. [DOI] [PubMed] [Google Scholar]
- 2.Yu F., Bender W. The mechanism of tamoxifen in breast cancer prevention. Breast Cancer Res. 2001;3(suppl 1):A74. doi: 10.1016/s0361-090x(02)00124-1. [DOI] [PubMed] [Google Scholar]
- 3.Osborne C.K., Wakeling A., Nicholson R.I. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. Br J Cancer. 2004;90(suppl 1):S2–S6. doi: 10.1038/sj.bjc.6601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emons G., Schally A.V. The use of luteinizing hormone releasing hormone agonists and antagonists in gynaecological cancers. Hum Reprod. 1994;9(7):1364–1379. doi: 10.1093/oxfordjournals.humrep.a138714. [DOI] [PubMed] [Google Scholar]
- 5.Miller W. Aromatase inhibitors: mechanism of action and role in the treatment of breast cancer. Semin Oncol. 2003;30:3–11. doi: 10.1016/s0093-7754(03)00302-6. [DOI] [PubMed] [Google Scholar]
- 6.Jeselsohn R., Yelensky R., Buchwalter G., et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor–positive breast cancer. Clin Cancer Res. 2014;20(7):1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toy W., Shen Y., Won H., et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzenellenbogen J.A., Mayne C.G., Katzenellenbogen B.S., Greene G.L., Chandarlapaty S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer. 2018;18(6):377–388. doi: 10.1038/s41568-018-0001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spoerke J.M., Gendreau S., Walter K., et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7(1) doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toy W., Weir H., Razavi P., et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7(3):277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeselsohn R., Buchwalter G., De Angelis C., Brown M., Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12(10):573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner N.C., Swift C., Kilburn L., et al. ESR1 mutations and overall survival on fulvestrant versus exemestane in advanced hormone receptor–positive breast cancer: a combined analysis of the phase III SoFEA and EFECT trials. Clin Cancer Res. 2020;26(19):5172–5177. doi: 10.1158/1078-0432.CCR-20-0224. [DOI] [PubMed] [Google Scholar]
- 13.van Kruchten M., de Vries E.G., Glaudemans A.W., et al. Measuring residual estrogen receptor availability during fulvestrant therapy in patients with metastatic breast cancer. Cancer Discov. 2015;5(1):72–81. doi: 10.1158/2159-8290.CD-14-0697. [DOI] [PubMed] [Google Scholar]
- 14.Bidard F.-C., Kaklamani V.G., Neven P., et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: results from the randomized phase III EMERALD trial. J Clin Oncol. 2022;40(28):3246–3256. doi: 10.1200/JCO.22.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martín M., Lim E., Chavez-MacGregor M., et al. Giredestrant for estrogen receptor–positive, HER2-negative, previously treated advanced breast cancer: results from the randomized, phase II acelERA breast cancer study. J Clin Oncol. 2024;42(18):2149–2160. doi: 10.1200/JCO.23.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burstein H.J., DeMichele A., Somerfield M.R., Henry N.L. Testing for ESR1 mutations to guide therapy for hormone receptor–positive, human epidermal growth factor receptor 2–negative metastatic breast cancer: ASCO guideline rapid recommendation update. J Clin Oncol. 2023;41(18):3423–3425. doi: 10.1200/JCO.23.00638. [DOI] [PubMed] [Google Scholar]
- 17.Venetis K., Pepe F., Pescia C., et al. ESR1 mutations in HR+/HER2-metastatic breast cancer: enhancing the accuracy of ctDNA testing. Cancer Treat Rev. 2023;121 doi: 10.1016/j.ctrv.2023.102642. [DOI] [PubMed] [Google Scholar]
- 18.André F., Ciruelos E., Rubovszky G., et al. Alpelisib for PIK3CA -mutated, hormone receptor–positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 19.Turner N.C., Oliveira M., Howell S.J., et al. Capivasertib in hormone receptor–positive advanced breast cancer. N Engl J Med. 2023;388(22):2058–2070. doi: 10.1056/NEJMoa2214131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karissawhiting. cbioportalR: Browse and Query Clinical and Genomic Data from cBioPortal. R package version 110. 2024. https://www.karissawhiting.com/cbioportalR/ Available at.
- 23.Nguyen B., Fong C., Luthra A., et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022;185(3):563–575.e11. doi: 10.1016/j.cell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng D.T., Mitchell T.N., Zehir A., et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarty D., Gao J., Phillips S., et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;(1):1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holst F., Moelans C.B., Filipits M., Singer C.F., Simon R., van Diest P.J. On the evidence for ESR1 amplification in breast cancer. Nat Rev Cancer. 2012;12(2):149. doi: 10.1038/nrc3093-c3. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Vega F., Mina M., Armenia J., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irani S., Tan W., Li Q., et al. Somatic estrogen receptor α mutations that induce dimerization promote receptor activity and breast cancer proliferation. J Clin Invest. 2024;134(1) doi: 10.1172/JCI163242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middha S., Zhang L., Nafa K., et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Core Team R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. 2022. https://www.R-project.org Available at.
- 31.Canisius S., Martens J.W.M., Wessels L.F.A. A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol. 2016;17(1):261. doi: 10.1186/s13059-016-1114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrer-Bonsoms J.A., Jareno L., Rubio A. Rediscover: an R package to identify mutually exclusive mutations. Bioinformatics. 2022;38(3):844–845. doi: 10.1093/bioinformatics/btab709. [DOI] [PubMed] [Google Scholar]
- 33.Schiavon G., Hrebien S., Garcia-Murillas I., et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313) doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razavi P., Chang M.T., Xu G., et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Spoelstra N.S., Sikora M.J., et al. Mutual exclusivity of ESR1 and TP53 mutations in endocrine resistant metastatic breast cancer. NPJ Breast Cancer. 2022;8(1):62. doi: 10.1038/s41523-022-00426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luen S.J., Viale G., Nik-Zainal S., et al. Genomic characterisation of hormone receptor-positive breast cancer arising in very young women. Ann Oncol. 2023;34(4):397–409. doi: 10.1016/j.annonc.2023.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark G.M., Osborne C.K., McGuire W.L. Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J Clin Oncol. 1984;2(10):1102–1109. doi: 10.1200/JCO.1984.2.10.1102. [DOI] [PubMed] [Google Scholar]
- 38.Liedtke C., Rody A., Gluz O., et al. The prognostic impact of age in different molecular subtypes of breast cancer. Breast Cancer Res Treat. 2015;152(3):667–673. doi: 10.1007/s10549-015-3491-3. [DOI] [PubMed] [Google Scholar]
- 39.Qing T., Karn T., Rozenblit M., et al. Molecular differences between younger versus older ER-positive and HER2-negative breast cancers. NPJ Breast Cancer. 2022;8(1):119. doi: 10.1038/s41523-022-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanker A.B., Sudhan D.R., Arteaga C.L. Overcoming endocrine resistance in breast cancer. Cancer Cell. 2020;37(4):496–513. doi: 10.1016/j.ccell.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desmedt C., Zoppoli G., Gundem G., et al. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol. 2016;34(16):1872–1881. doi: 10.1200/JCO.2015.64.0334. [DOI] [PubMed] [Google Scholar]
- 42.Thomas A., Shatsky R., Kalinsky K. Moving precision forward: extending next generation sequencing to operable disease in less common breast cancer subtypes. Ann Oncol. 2024;35(1):7–9. doi: 10.1016/j.annonc.2023.10.127. [DOI] [PubMed] [Google Scholar]
- 43.Pearson A., Proszek P., Pascual J., et al. Inactivating NF1 mutations are enriched in advanced breast cancer and contribute to endocrine therapy resistance. Clin Cancer Res. 2020;26(3):608–622. doi: 10.1158/1078-0432.CCR-18-4044. [DOI] [PubMed] [Google Scholar]
- 44.Fribbens C., Garcia Murillas I., Beaney M., et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann Oncol. 2018;29(1):145–153. doi: 10.1093/annonc/mdx483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massarweh S., Osborne C.K., Jiang S., et al. Mechanisms of tumor regression and resistance to estrogen deprivation and fulvestrant in a model of estrogen receptor–positive, HER-2/neu-positive breast cancer. Cancer Res. 2006;66(16):8266–8273. doi: 10.1158/0008-5472.CAN-05-4045. [DOI] [PubMed] [Google Scholar]
- 46.Nayar U., Cohen O., Kapstad C., et al. Acquired HER2 mutations in ER+ metastatic breast cancer confer resistance to estrogen receptor–directed therapies. Nat Genet. 2019;51(2):207–216. doi: 10.1038/s41588-018-0287-5. [DOI] [PubMed] [Google Scholar]
- 47.Giltnane J.M., Hutchinson K.E., Stricker T.P., et al. Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci Transl Med. 2017;9(402) doi: 10.1126/scitranslmed.aai7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao P., Cohen O., Kowalski K.J., et al. Acquired FGFR and FGF alterations confer resistance to estrogen receptor (ER) targeted therapy in ER+ metastatic breast cancer. Clin Cancer Res. 2020;26(22):5974–5989. doi: 10.1158/1078-0432.CCR-19-3958. [DOI] [PubMed] [Google Scholar]
- 49.Formisano L., Stauffer K.M., Young C.D., et al. Association of FGFR1 with ERα maintains ligand-independent ER transcription and mediates resistance to estrogen deprivation in ER+ breast cancer. Clin Cancer Res. 2017;23(20):6138–6150. doi: 10.1158/1078-0432.CCR-17-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaudhary N., Chibly A.M., Collier A., et al. CDK4/6i-treated HR+/HER2− breast cancer tumors show higher ESR1 mutation prevalence and more altered genomic landscape. NPJ Breast Cancer. 2024;10(1):15. doi: 10.1038/s41523-024-00617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wander S.A., Cohen O., Gong X., et al. The genomic landscape of intrinsic and acquired resistance to cyclin-dependent kinase 4/6 inhibitors in patients with hormone receptor–positive metastatic breast cancer. Cancer Discov. 2020;10(8):1174–1193. doi: 10.1158/2159-8290.CD-19-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrera-Abreu M.T., Palafox M., Asghar U., et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor–positive breast cancer. Cancer Res. 2016;76(8):2301–2313. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Condorelli R., Spring L., O’Shaughnessy J., et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29(3):640–645. doi: 10.1093/annonc/mdx784. [DOI] [PubMed] [Google Scholar]
- 54.Mayer E.L., Ren Y., Wagle N., et al. PACE: a randomized phase II study of fulvestrant, palbociclib, and avelumab after progression on cyclin-dependent kinase 4/6 inhibitor and aromatase inhibitor for hormone receptor–positive/human epidermal growth factor receptor–negative metastatic breast cancer. J Clin Oncol. 2024;42(17):2050–2060. doi: 10.1200/JCO.23.01940. [DOI] [PubMed] [Google Scholar]
- 55.Kalinsky K., Accordino M.K., Chiuzan C., et al. Randomized phase II trial of endocrine therapy with or without ribociclib after progression on cyclin-dependent kinase 4/6 inhibition in hormone receptor–positive, human epidermal growth factor receptor 2–negative metastatic breast cancer: MAINTAIN trial. J Clin Oncol. 2023;41(24):4004–4013. doi: 10.1200/JCO.22.02392. [DOI] [PubMed] [Google Scholar]
- 56.Rao X., Chen Y., Beyrer J., et al. Clinical and genomic characteristics of patients with hormone receptor–positive, human epidermal growth factor receptor 2–negative metastatic breast cancer following progression on cyclin-dependent kinase 4 and 6 inhibitors. Clin Cancer Res. 2023;29(17):3372–3383. doi: 10.1158/1078-0432.CCR-22-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konduri S.D., Medisetty R., Liu W., et al. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci. 2010;107(34):15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu W., Konduri S.D., Bansal S., et al. Estrogen receptor-α binds p53 tumor suppressor protein directly and represses its function. J Biol Chem. 2006;281(15):9837–9840. doi: 10.1074/jbc.C600001200. [DOI] [PubMed] [Google Scholar]
- 59.Gupta A, Gazzo A, Selenica P, et al. APOBEC3 mutagenesis drives therapy resistance in breast cancer. [Preprint.] Advance Access published on bioRxiv May 1, 2024. 10.1101/2024.04.29.591453 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.