Abstract
The great difficulty in eliciting broadly cross-reactive neutralizing antibodies (NAbs) against human immunodeficiency virus type 1 (HIV-1) isolates has been attributed to several intrinsic properties of their viral envelope glycoprotein, including its complex quaternary structure, extensive glycosylation, and marked genetic variability. Most previously evaluated vaccine candidates have utilized envelope glycoprotein from a single virus isolate. Here we compare the breadth of NAb and protective immune response following vaccination of pigtailed macaques with envelope protein(s) derived from either single or multiple viral isolates. Animals were challenged with Simian/human immunodeficiency virus strain DH12 (SHIVDH12) following priming with recombinant vaccinia virus(es) expressing gp160(s) and boosting with gp120 protein(s) from (i) LAI, RF, 89.6, AD8, and Bal (Polyvalent); (ii) LAI, RF, 89.6, AD8, Bal, and DH12 (Polyvalent-DH12); (iii) 89.6 (Monovalent-89.6); and (iv) DH12 (Monovalent-DH12). Animals in the two polyvalent vaccine groups developed NAbs against more HIV-1 isolates than those in the two monovalent vaccine groups (P = 0.0054). However, the increased breadth of response was directed almost entirely against the vaccine strains. Resistance to SHIVDH12 strongly correlated with the level of NAbs directed against the virus on the day of challenge (P = 0.0008). Accordingly, the animals in the Monovalent-DH12 and Polyvalent-DH12 vaccine groups were more resistant to the SHIVDH12 challenge than the macaques immunized with preparations lacking a DH12 component (viz. Polyvalent and Monovalent-89.6) (P = 0.039). Despite the absence of any detectable NAb, animals in the Polyvalent vaccine group, but not those immunized with Monovalent-89.6, exhibited markedly lower levels of plasma virus than those in the control group, suggesting a superior cell-mediated immune response induced by the polyvalent vaccine.
Neutralizing antibodies (NAbs) have been shown to be critical components of the immune response that controls a variety of viral infections. However, the protective role(s) of NAbs directed against human immunodeficiency virus type 1 (HIV-1) and other primate lentiviruses, which become detectable following acute infections, has been debated over the years and remains unresolved. For example, the clearance of HIV-1 from plasma during the primary infection occurs prior to the appearance of NAbs in newly infected individuals (37). Furthermore, in many studies, vaccinated macaques are able to efficiently control a virus challenge in the absence of detectable NAb, particularly those animals immunized with live, attenuated-virus vaccines (2, 16, 45, 51). Nonetheless, passive immunization experiments have demonstrated the protective effects of NAbs against subsequent challenges with primate lentiviruses (17, 20, 30, 32, 33, 42, 44, 47, 48). In some of these studies, sterilizing immunity could be achieved when high plasma concentrations of NAbs were present prior to virus inoculation. However, the design and/or development of immunogens capable of prospectively eliciting broadly reactive NAbs against multiple virus isolates has been frustratingly unproductive. None of the envelope glycoprotein-based vaccine candidates tested in primates thus far have been able to elicit broadly reactive NAbs, especially against primary isolates.
The HIV-1 envelope glycoprotein contains five highly variable regions, designated V1 through V5, the first four of which form loops through intramolecular disulfide linkage. These variable regions very likely cover significant portions of the exposed surface on the trimeric gp120 complex, as suggested from antigenic probing with monoclonal antibodies (38) and crystallographic data of the envelope core (28). The variable regions of HIV-1 and simian immunodeficiency virus (SIV) gp120 have long been known to be targeted by NAbs, possibly explaining the antigenic variation associated with these regions (9, 19, 22, 23, 27, 34, 43, 46, 55). In contrast, the conserved domains of gp120 are either extensively shielded by carbohydrate moieties, obscured beneath the variable regions, or hidden due to intermolecular protein-protein interactions and do not elicit antibodies that neutralize virions (38, 57).
Conserved neutralizing epitopes, present on the unmodified native gp120, have been nearly impossible to identify. To date, only two human anti-gp120 monoclonal NAbs (2G12 and b12), which exhibit relatively broad neutralizing activity, have been isolated (4, 6, 53, 54). Immunization with a variety of envelope glycoprotein preparations (e.g., monomeric gp120, soluble gp160, oligomeric gp140, and virions or virus-like particles) and the use of different vaccine strategies (e.g., whole inactivated virus, subunit, live vector, and DNA vector) usually result in extremely narrow and/or immunogen-specific NAb responses. Theoretically, it might be possible to elicit broadly reactive NAbs by two alternative vaccine strategies: (i) forcing the immune system to preferentially target a conserved gp120 neutralization epitope (assuming its existence) associated with the majority of HIV-1 isolates circulating in a given geographic region, or (ii) immunization with a cocktail of envelope proteins (if feasible) representing the majority of circulating virus isolates, thereby eliciting NAbs against the variable regions of all of the gp120s in the mixture.
Most lentivirus vaccine studies have utilized envelope glycoproteins from either one or, at most, two virus isolates; protective efficacy has usually been assessed using a homologous virus challenge (i.e., the virus isolate used to challenge animals contains the same gp120 as that used for immunization). In reality, however, vaccinated individuals would be expected to encounter an HIV-1 isolate or viral quasispecies containing a gp120 unrelated to the immunogen used for vaccination (heterologous challenge). In this study, we have evaluated a vaccine regimen, based solely on envelope glycoproteins, which utilizes individual or mixtures of both recombinant vaccinia viruses and gp120 boosts to address the following questions pertaining to protection against heterologous virus strains. (i) Is it possible to elicit a broader NAb response by immunization with a mixture (polyvalent) of envelope glycoproteins? (ii) If so, does the breadth of the NAb response extend beyond the virus isolates included in the immunization cocktail? (iii) How does the protective efficacy of the immune response elicited by a polyvalent envelope vaccine compare to that elicited by a monovalent (homologous or heterologous) envelope vaccine? (iv) Will there be antigenic competition between the different envelope glycoprotein components of the polyvalent envelope vaccine cocktail?
MATERIALS AND METHODS
Plasmids and recombinant vaccinia viruses.
Recombinant vaccinia viruses that express gp160 of HIV-1 isolates Bal, LAI, RF (vCB43, vCB41, and vCB36, respectively [3]), 89.6 (vBD3 [15]), DH12, and AD8 (vvDHenv and vvADenv, respectively [8]) have been previously described. Virus stocks were propagated in HeLa cells, purified on linear sucrose gradients (29, 31), and resuspended in phosphate-buffered saline (PBS).
To express histidine-tagged gp120 (gp120H), HeLa cells were infected with recombinant vaccinia virus vTF7-3 (18), which expresses T7 RNA polymerase, together with recombinant vaccinia viruses vTM-DHgp120H, vTM-ADgp120H, vTM-LAIgp120H, vTM-RFgp120H, vTM-BALgp120H, or vTM-89.6gp120H, each at a multiplicity of infection of 5 and maintained for 2 to 3 days in the absence of fetal bovine serum. Recombinant vaccinia viruses that express DH12, AD8, and LAI gp120Hs were generated using plasmids (29) that encode the corresponding gp120H as previously described (11). The plasmids for the other three HIV-1 isolates (BAL, RF, and 89.6) were generated using a similar cloning strategy. Briefly, the gp120 coding sequences were amplified by PCR from pCB43, pCB36, and pBD3 (3, 15). The forward primers used for the PCR amplification were 5′-GGGCCCCATGGGAGTGTTGGAGAAATATCAG-3′ (BAL), 5′-GGGCCCCATGGGAGTGATGGAGATGAGGAAG-3′ (RF), and 5′-GGGCCCCATGGGAGTGAAGGAGATCAGGAAG-3′ (89.6). The reverse primers 5′-GGGCCCCTCGAGTTAATGGTGATGATGGTGATGTCTTTTTTCTCTCTGCACCACTC-3′ (BAL and RF) and 5′-GGGCCCCTCGAGTTAATGGTGATGATGGTGATGTCTTTTTTCTCTTTGCACTGTTC-3′ (89.6) encoded six appended histidine residues (in bold print). The amplified PCR fragments were digested with NcoI and XhoI (introduced into the primers as indicated by underlines) and cloned into pTM-1 (39) for expression under the T7 promoter. The plasmid constructs were sequenced to confirm that no mutations were introduced inadvertently during PCR amplification. All of the recombinant vaccinia viruses employed in this study have been derived from the WR strain of the vaccinia viruses.
Protein purification.
Recombinant gp120H was purified from the culture supernatant using a one-step metal-chelate affinity purification procedure (Ni-NTA; Qiagen). The culture supernatant was prepared by removing the cells by centrifugation at 1,000 × g for 10 min at 4°C. Then 1 M Na2HPO4 was added to the supernatant (50 mM final concentration) to raise the pH (to >8.0), and vaccinia virus was inactiviated with NP-40 (0.5%). The mixture was stored overnight at 4°C, the resulting CaPO4 precipitate was removed by centrifugation (10,000 × g for 30 min), and the supernatant was further clarified by filtration through a 0.2-μ ZapCap bottletop filter unit (Schleicher and Shuell). The Ni-NTA resin (10-ml bed volume per liter of supernatant) was equilibrated using three wash cycles of 50 mM sodium phosphate buffer (pH 8.0; 10 bed volumes per cycle) and was added to the filtrate while being continuously stirred for 4 to 16 h at 4°C. The resin was collected by pouring the mixture into a 25-ml EconoColumn (Bio-Rad) and washed using 10 bed volumes of the equilibration buffer containing 500 mM NaCl. The column was connected to an UV detector (Pharmacia), and the absorption at 280 nm was monitored during the purification procedure. Nonspecifically bound material was removed using approximately 10 to 20 bed volumes of 50 mM sodium phosphate buffer (pH 8.0) containing 500 mM NaCl and 20 mM imidazole. Recombinant gp120H was eluted using 50 mM sodium phosphate buffer (pH 8.0) containing 200 mM imidazole. The peak fractions were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, and dialyzed against PBS (100 volumes) at 4°C for 16 to 24 h. The purified gp120H was concentrated to approximately 1 mg/ml using a 10,000-dalton cutoff ultrafilter unit (Millipore). Typically, 5 mg of purified gp120H was produced from 1 liter of culture supernatant.
Animal experiments.
Pigtailed macaques (Macaca nemestrina) were maintained in accordance with American Association for Accreditation of Laboratory Animal Care standards and were housed in a biosafety level 2 facility; biosafety level 3 practices were followed. Animal anesthetization and bleeding were done as previously described (48). The macaques were immunized (see Fig. 1 for schedule) intradermally with the indicated recombinant vaccinia viruses on four separate sites on their backs, about 4 cm from each other. A total of 5 × 107 PFU in 0.5 ml (0.125 ml on each location) was injected per animal per immunization. Thus, in the two polyvalent vaccination groups, in which the macaques were immunized with mixtures containing either five or six different envelope glycoproteins, individual animals received either 107 or 0.83 × 107 PFU of each recombinant vaccinia virus, respectively. The monkeys were boosted intramuscularly with a total of 100 μg of recombinant gp120H combined with 100 μg of QS-21 adjuvant (Aquila Biopharmaceuticals) in 0.5 ml. The animals vaccinated with mixtures of gp120 were immunized with either 20 or 16.7 μg of each gp120H. The macaques were subsequently challenged intravenously with 100 50% tissue culture infectious doses (TCID50) of simian/human immunodeficiency virus strain DH12 (SHIVDH12) stock prepared in macaque peripheral blood mononuclear cells (PBMC) (49) (previously referred to as SHIVDH12MD14YE).
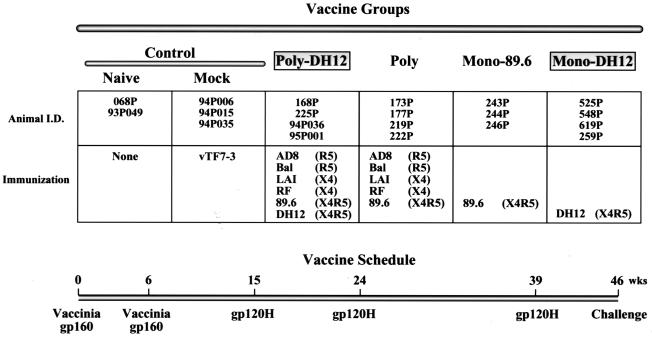
FIG. 1.
Vaccine strategy and immunization schedule. Twenty animals were divided into five vaccine groups: control (naïve and mock), Polyvalent-DH12, Polyvalent, Monovalent-89.6, and Monovalent-DH12. Animals were immunized twice (weeks 0 and 6) with recombinant vaccinia viruses expressing gp160 of the HIV-1 isolates indicated, followed by three immunizations (weeks 15, 24, and 39) with gp120s from the same isolates. The coreceptors used by the HIV-1 isolates are indicated. The animals in the mock immunization group were infected with recombinant vaccinia virus vTF7-3, which expresses T7 RNA polymerase. The animals were challenged with SHIVDH12 on week 46. I.D., identity.
Enzyme-linked immunosorbent assay (ELISA).
Nunc Maxisorp 96-well plates were coated with purified HIV-1DH12 gp120H (20 ng per well) in 50 μl of coating buffer (15 mM Na2CO3, 35 mM NaHCO3, 3 mM NaN3, pH 9.6) for 1 h at 37°C. The coating mixture was replaced with 200 μl of blocking buffer (2.5% skim milk, 25% fetal bovine serum, in PBS) and incubated for 1 h at 37°C. Plates were washed twice with PBS-T (PBS containing 0.1% Tween 20). Serially diluted antiserum in 200 μl of blocking buffer was added to each well and incubated for 1 h at 37°C. The plates were washed three times with PBS-T and 100 μl of horseradish peroxidase-conjugated goat anti-human antibodies were added (1:5,000 dilution; Pierce, Rockford, Ill.) for 1 h at 37°C. The plates were washed again three times with PBS-T, and 100 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added. The reaction was stopped after 10 min with 50 μl of 2 N H2SO4, and absorbency was measured at 450 nm using a 96-well plate spectrophotometer (Bio-Tek Instruments).
Western immunoblot.
SHIVDH12 particles were concentrated 100-fold from virus-infected MT-4 cell culture medium by ultracentrifugation as previously described (26). Concentrated virus particles (40 μl), supplemented with 300 ng of purified DH12 gp120H (see above), were resuspended in SDS-PAGE sample buffer and subjected to SDS-PAGE on a preparatory gel. Following electrotransfer onto a nitrocellulose membrane, immunoblotting was performed in a multichamber immunoblot apparatus (Bio-Rad) as previously described (29). Briefly, blots were incubated with macaque plasma samples (1:50 dilution) followed by goat anti-human immunoglobulin G conjugated with horseradish peroxidase (Pierce). Protein bands were visualized with SuperSignal chemiluminescent substrates (Pierce) using the manufacturer's protocol.
Neutralization assay.
Three different neutralization assays were employed in this study: complete virus neutralization in MT-4 cells, MT-2 cell killing reduction assay, and gag antigen reduction assay using PBMC. SHIVDH12, propagated in either chimpanzee or macaque PBMC, was used in the complete virus neutralization assay as previously described (48). In some cases a chimeric HIV-1, AD8-DHenv (HIV-1AD8 containing the gp160 coding region of HIV-1DH12 [10]), was also used. Serially diluted plasma samples, collected at the indicated times postimmunization, were incubated with 100 TCID50 for 1 h at room temperature. Triplicate- or quadruplicate-infected MT-4 cell cultures were maintained for 2 weeks. Virus replication was determined by measuring reverse transcriptase activity in culture supernatants as previously described (56). The titer represents the inverse of the serum dilution (before adding cells) that resulted in no detectable virus replication in all of the replicate wells.
Neutralization of SHIV-HXB2, SHIV-89.6, and HIV-1 strains RF and MN was determined in MT-2 cells by a reduction in virus-induced cell killing, measured by neutral red uptake as previously described (36). All of the virus stocks were produced in H9 cells, except for SHIV-89.6, which was produced in human PBMC. Virus (500 TCID50) was incubated in triplicate with dilutions of serum for 1 h at 37°C. Cells were added and the incubation continued until approximately 80% of cells in virus control wells (cells plus virus but no serum sample) exhibited syncytium formation (usually 4 to 6 days). Neutralization titers are defined as the dilution of serum in the presence of virus (before the addition of cells) at which 50% of cells were protected from virus-induced killing. A 50% reduction in cell killing corresponds to an 85 to 90% reduction in viral gag antigen synthesis in this assay (5, 41).
Neutralization of SHIVDH12 and HIV-1 strains AD8 and BAL, all produced in human PBMC, was determined by a reduction in gag antigen synthesis, as previously described (14, 35). Serum samples were diluted in interleukin 2 (IL-2)-containing (4%) growth medium and mixed with virus (500 TCID50) in triplicate for 1 h at 37°C. Phytohemagglutinin-stimulated PBMC were subsequently added to each well. The virus inoculum and antibodies were removed 24 h later by 3 washes with 200 μl of growth medium, and the washed cells were maintained in 200 μl of IL-2-containing growth medium. Culture supernatants (25 μl) were collected on a daily basis thereafter and mixed with 225 μl of 0.5% Triton X-100 for quantification of Gag antigen. SHIV p27 and HIV-1 p24 were quantified by antigen ELISA as described by the supplier (SHIV p27 was from Organon-Teknika/Akzo, Durham, N.C.; HIV-1 p24 was from DuPont/NEN Life Sciences, Boston, Mass.). The 25-μl volume of culture fluids removed each day was replaced with 25 μl of fresh IL-2-containing growth medium. The percent reduction in Gag antigen synthesis is reported relative to the amount of the protein synthesized in the presence of preimmunization serum.
Virus load measurements.
Plasma samples were prepared from blood collected with Acid Citrate Dextrose (ACD)-A solution (Becton Dickinson) as the anticoagulant and stored at −70°C. Plasma viral RNA levels were determined by real-time PCR (ABI Prism 7700 sequence detection system; Perkin-Elmer) using reverse-transcribed viral RNA as templates. Viral RNA extraction, reverse transcription, and cDNA amplification was done as previously described (26). Viral p27 antigenemia was measured by using an SIV core antigen assay kit (Coulter) and by following the manufacturer's protocol. The amount of proviral DNA in axillary lymph node cells was determined by PCR as previously described (50).
Lymphocyte immunophenotyping.
EDTA-treated blood samples were stained with fluorochrome-conjugated monoclonal antibodies (anti-CD3, anti-CD4, anti-CD8, and anti-CD20) and analyzed by flow cytometry (FACSort; Becton Dickinson) as previously described (26).
Amino acid sequence analyses.
Amino acid sequences of the V1/V2 and V3 loops of gp120s from various HIV-1 isolates were compared by the BestFit alignment program from Genetics Computer Group.
RESULTS
Immunization.
To accomplish the multiple objectives of this study, animals were divided into the five different vaccine groups indicated in Fig. 1: control (including naïve and mock-immunized), Polyvalent-DH12, Polyvalent, Monovalent-89.6, and Monovalent-DH12. Since all of the macaques ultimately were to be challenged with a SHIV bearing the HIV-1DH12 envelope glycoprotein, animals in the Polyvalent-DH12- and Monovalent-DH12-vaccinated groups modeled potential resistance to a homologous virus challenge, whereas those in the Polyvalent and Monovalent-89.6 groups measured the response to heterologous virus.
Immunizations with recombinant vaccinia viruses followed by boosts with recombinant proteins have previously been shown to elicit superior immune responses compared to vaccination with either vaccinia virus or subunit proteins alone (12, 13, 21, 24, 25). We have employed such a live-vector prime followed by protein boost vaccination approach in this study. Animals were immunized twice (weeks 0 and 6) with recombinant vaccinia viruses (WR strain) that express full-length HIV-1 gp160(s) (Fig. 1, bottom). This was followed by three immunizations (weeks 15, 24, and 39) with purified recombinant gp120(s), which were tagged with six histidine residues (gp120H) to facilitate their purification. In the Monovalent-89.6 or Monovalent-DH12 groups, macaques were immunized with recombinant vaccinia viruses expressing gp160 from either HIV-189.6 or HIV-1DH12, respectively. In the Polyvalent-DH12 group, animals were vaccinated with a mixture of six different recombinant vaccinia viruses expressing gp160s from the HIV-1 isolates AD8, Bal, LAI, RF, 89.6 and DH12. Macaques in the Polyvalent group were immunized with a mixture of five envelope glycoprotein immunogens that did not include DH12. HIV-1 isolates AD8 and Bal have been classified into R5, LAI and RF have been classified into X4, and 89.6 and DH12 have been classified into X4R5 coreceptor usage groups. This combination of immunogens was chosen to ascertain whether a preferential response might be elicited against envelope glycoproteins with specific coreceptor requirements. For mock immunizations, animals were vaccinated with recombinant vaccinia virus vTF7-3, which expresses T7 RNA polymerase (18).
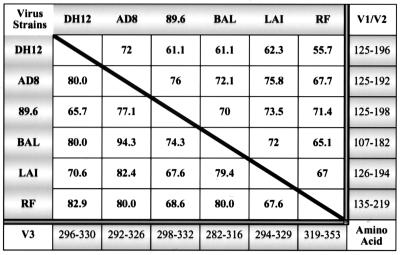
The amino acid (aa) sequences of the hypervariable V1/V2 and V3 loops of the gp120s from the six HIV-1 isolates used in this study are quite heterogeneous (Fig. 2). In general, the sequence of the V1/V2 loops was more diverse than that of the V3 loops. For example, the V1/V2 loop of the DH12 isolate (aa 125 to 196) showed a range of amino acid identity of 55.7% with RF and 72% with AD8. In contrast, the V3 loop of DH12 (aa 296 to 330) was 65.7 and 82% identical to 89.6 and RF, respectively.
FIG. 2.
Comparative amino acid sequence analyses of the gp120 variable loops V1/V2 and V3 of the HIV-1 isolates used for the immunization. The amino acid residue numbers in the V1/V2 and V3 loops included in the analyses are indicated on the right side and the bottom of the figure, respectively. The percent amino acid sequence identity for the V1/V2 and V3 loops are shown on the upper right and lower left side of the diagonal line, respectively.
Immune response.
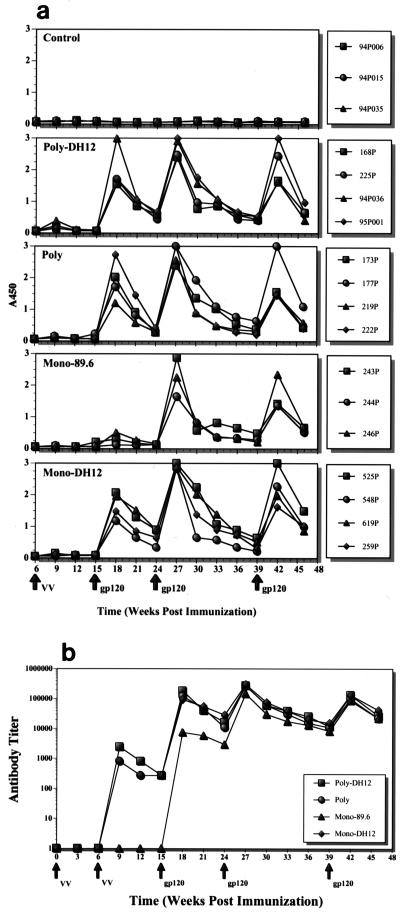
Following the first immunizations with vaccinia virus, small lesions of less than 1 cm in diameter, which rapidly healed during the next 2 weeks, were observed. After the second vaccinia virus immunization, extremely small skin lesions appeared in some of the animals. No other side effects were observed in the 18 monkeys vaccinated with the WR strain of vaccinia virus. The humoral immune response elicited by this vaccinia virus prime and protein boost regimen was monitored primarily by measuring the level of antibodies binding to HIV-1DH12 gp120 in an ELISA. ELISA antibody responses for individual animals are presented in Fig. 3a. No significant differences were observed between the animals in any particular vaccine group. The levels of anti-gp120 antibodies increased after each immunization and then declined until a subsequent boost was administered. Of note, however, was the very low antibody levels for the Monovalent-89.6 vaccine group on week 18 (3 weeks after the first gp120H boost). This is better seen in Fig. 3b, which shows the average endpoint antibody titers for each group of monkeys. While gp120-specific antibodies did not appear until after the first protein boost for the Monovalent-89.6 group, antibodies were detected immediately after the second vaccinia virus immunization in the other vaccine groups. In these latter animals, antibody levels increased about 1,000-fold after the second vaccinia virus immunization and reached titers over 1:100,000 following the first protein boost. At this time, the titer for the Monovalent-89.6 group was about 10-fold lower. However, after the second protein boost, the titers for all of the vaccine groups were virtually indistinguishable. The difference in antibody titers in the Monovalent-89.6 and the other vaccine groups may be due to two factors. First, analyses of gp160 expression by each of the recombinant vaccinia viruses in cultured HeLa cells indicated that the level of 89.6 gp160 expressed was approximately twofold lower than that of the other envelope proteins (data not shown). Second, when ELISAs were conducted using plates coated with gp120s from different HIV-1 isolates, preferential binding to homologous proteins was observed (i.e., antibodies generated against the DH12 gp120 bound more efficiently to DH12 gp120 than to 89.6 gp120, and vice versa [unpublished observation]).
FIG. 3.
Antibody response during the course of immunization. (a) The antibody levels in the plasma samples of individual animals were analyzed by ELISA. Twenty nanograms of HIV-1DH12 gp120 was coated in each well and results from one plasma sample dilution (1:2,430) are shown. (b) Average endpoint antibody titers for the vaccine groups immunized with envelope glycoproteins. The times at which the animals were immunized with either recombinant vaccinia virus (VV) or gp/120 are indicated by the arrows.
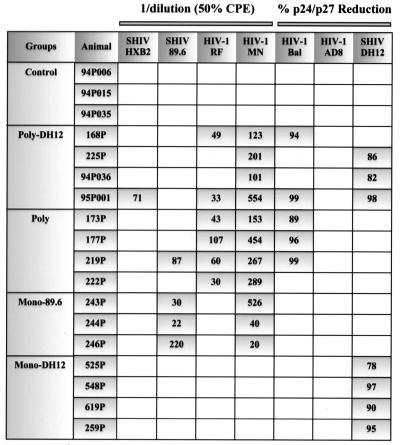
Protection of animals against SIV or HIV-1 infection correlates with the presence of NAbs, not gp120 binding activity as measured by ELISA, Western immunoblot, or immunoprecipitation assays (48). Since the immunized animals were to be challenged with SHIVDH12, assays monitoring NAbs directed against either SHIVDH12 or HIV-1AD8-DH12 (AD8-DHenv [10]), a chimeric HIV-1 containing the env gene from the DH12 isolate, were carried out. As shown in Table 1, anti-DH12 NAbs initially appeared after the first gp120H protein boost in the Monovalent-DH12-vaccinated monkeys. Thus, the monomeric gp120H used was able to elicit or recall an antibody response capable of neutralizing virus. None of the other groups, including the Polyvalent-DH12 group, produced any neutralizing at this time. Following the second gp120H boost, NAb directed against virus bearing the DH12 gp120 also became detectable in the Polyvalent-DH12 group. Animal 95P001, in particular, generated very high levels of neutralizing activity. On the day of virus challenge, only the animals in the vaccine groups immunized with the DH12 envelope glycoprotein (Polyvalent-DH12 and Monovalent-DH12) were producing antibodies that neutralized SHIVDH12. Two macaques, one in the Polyvalent-DH12 group (95P001) and the other in the Monovalent-DH12 group (259P), had extremely high titers of neutralizing activity (1:81). Two other animals in the Monovalent-DH12 group (548P and 619P) had intermediate levels of neutralizing activity (1:9), while the other four monkeys had relatively low titers of NAbs [<(1:9) or ≤(1:3)]. It would appear that the production of NAbs may very well be antigen dose-dependent, judging by the delayed appearance and generally lower titers elicited by polyvalent immunization (8.3 × 106 PFU of the recombinant vaccinia virus expressing DH12 gp160 and 16.7 μg of DH12 gp120) than by vaccination with monovalent DH12 Env (5 × 107 PFU of the recombinant vaccinia virus and 100 μg of gp120).
TABLE 1.
Neutralization antibody titers against DH12
| Time of neutralization assay | Vaccine group and antibody titer for each animala
|
||||
|---|---|---|---|---|---|
| Control | Polyvalent-DH12 | Polyvalent | Monovalent-89.6 | Monovalent-DH12 | |
| 3 wk after first gp120H boost | None | None | None | None | 525P (1:3) |
| 548P (1:9) | |||||
| 619P (1:27) | |||||
| 259P (1:3) | |||||
| 3 wk after second gp120H boost | None | 168P (noneb) | None | None | 525P (noneb) |
| 225P (1:10) | 548P (1:10) | ||||
| 94P036 (1:10) | 619P (1:50) | ||||
| 95P001 (1:50) | 259P (1:50) | ||||
| Day of challenge | None | 168P [<(1:3c)] | None | None | 525P [<(1:9c)] |
| 225P (1:3) | 548P (1:9) | ||||
| 94P036 [<(1:3c)] | 619P (1:9) | ||||
| 95P001 (1:81) | 259P (1:81) | ||||
The antibody titer (in parentheses) is the dilution of serum that results in complete neutralization of 100 TCID50 in MT-4 cells.
The lowest dilution tested was 1:10.
Only two of the quadruplicate samples showed complete neutralization.
Virus challenge.
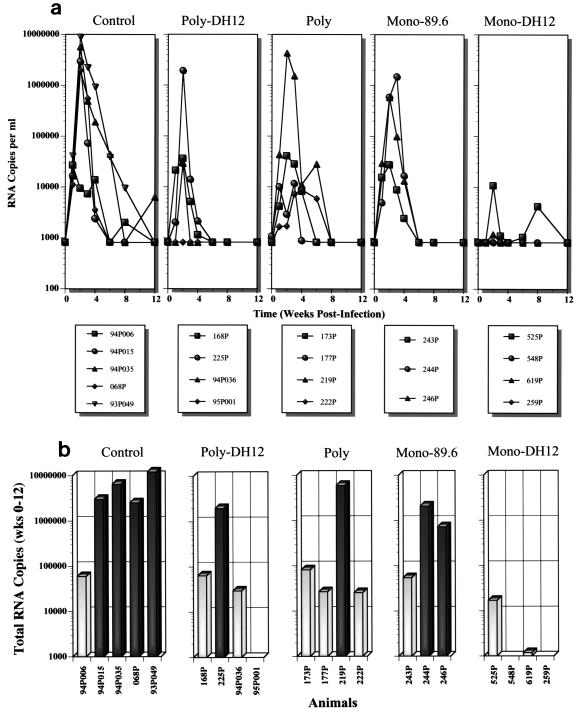
Seven weeks after the final gp120H boost, all 18 immunized animals plus 2 naïve macaques were challenged intravenously with 100 TCID50 of SHIVDH12, produced in macaque PBMC. In general, viral RNA in the plasma became detectable on week 1, peaked on week 2, and then declined (Fig. 4a). The majority of the viral burden was observed during the first 4 weeks after challenge. After week 12 (the monitoring continued up to 32 weeks postinfection), all of the animals had plasma virus loads below the level of detection. CD4+ T-lymphocyte numbers in the blood did not change significantly (data not shown), as the challenge virus used is not pathogenic.
FIG. 4.
Plasma viral RNA loads in animals subsequent to SHIVDH12 challenge. (a) Viral RNA copies in the plasma of infected animals, determined by quantitative real time reverse transcription-PCR, during the first 12 weeks after the virus infection. (b) Total plasma viral load (arithmetic sum) during the first 12 weeks of virus infection.
The total plasma virus load measured during the first 12 weeks of infection is compiled in Fig. 4b. In the control group, four out of five animals (94P015, 94P035, 068P, and 93P049) produced large amounts of viral RNA. As shown in Fig. 4a, these same animals had peak virus loads of 2 × 106 to 9 × 106 RNA copies per ml of plasma on week 2. Unexpectedly, one of the control monkeys (94P006) had a lower total virus load even though it had viral RNA levels similar to those of the other control macaques on week 1 (Fig. 4a). In contrast to the control group, plasma viremia was undetectable in two of the four animals in the Monovalent-DH12 group throughout the 32-week observation period (macaques 548P and 259P). One additional monkey in this group (619P) exhibited a plasma viremia barely detectable over background on week 2 (Fig. 4a). The fourth animal in the Monovalent-DH12 group (525P) produced relatively small amounts of viral RNA during the first 12 weeks of infection (approximately 280-fold less than that measured in control monkeys [∼4.8 × 106 copies/ml]). The difference in plasma viremia between the control and Monovalent-DH12 groups was statistically significant (P = 0.016 by the Wilcoxon rank sum test). In the Polyvalent-DH12 group, one macaque (95P001) had no detectable viremia, two animals (168P and 94P036) produced small amounts of virus, and a single monkey (225P) experienced a viremia indistinguishable from the control group. In the Polyvalent group, three animals produced relatively small quantities of virus, and only one animal (219P) had a viremia similar to that measured in the control group. Although the macaques in the Polyvalent group generated no detectable NAb against SHIVDH12, significant protection against the virus challenge occurred, suggesting that a nonhumoral immune response induced by the vaccine (e.g., envelope-specific cytotoxic T-lymphocyte [CTL] activity) might be responsible for the low plasma virus loads observed. This response is to be contrasted with the animals in the Monovalent-89.6 group, two of which developed high virus loads and one which had a relatively low viremia.
The protection against the SHIVDH12 challenge observed in the eight animals immunized with HIV-1DH12 envelope glycoprotein (Polyvalent-DH12 and Monovalent-DH12 groups) was significantly better than in animals that did not (Polyvalent and Monovalent-89.6; P = 0.039). It should be noted that the virus loads in the individual Polyvalent-DH12-, Polyvalent-, and Monovalent-89.6-vaccinated groups were not statistically different from that of the control group (P > 0.15 after Bonferroni correction for multiple comparisons) despite the marked reduction of virus loads in several of the animals in the Polyvalent-DH12 and Polyvalent groups. This undoubtedly reflects the small number of animals in each group, since the viral load in the three groups as a whole (11 animals) was significantly lower than that of the control group (P = 0.027). Viral p27 antigenemia was detected only on week 2 in all of the animals with high (>105 RNA copies/ml) plasma viral RNA loads (data not shown).
To determine, in fact, whether or not a virus infection had occurred in animals with undetectable or barely detectable plasma viremia (monkeys 95P001, 548P, 619P, and 259P), lymph node biopsies were collected at week 2 postinfection and analyzed for proviral DNA by PCR. Viral DNA was readily amplified from samples of 100,000 or 4,000 lymphocytes, prepared from lymph node specimens collected from two control group animals (94P015 and 94P035) (data not shown). In contrast, no proviral DNA was detected in specimens from macaques 95P001, 548P, and 259P. Proviral DNA could be amplified from the 100,000 cells but not from the 4,000 cells from the lymph node of animal 619P, suggesting that a low level of virus replication had occurred. Despite limited viral replication in lymph node cells, this animal was able to control its plasma virus load to barely detectable levels.
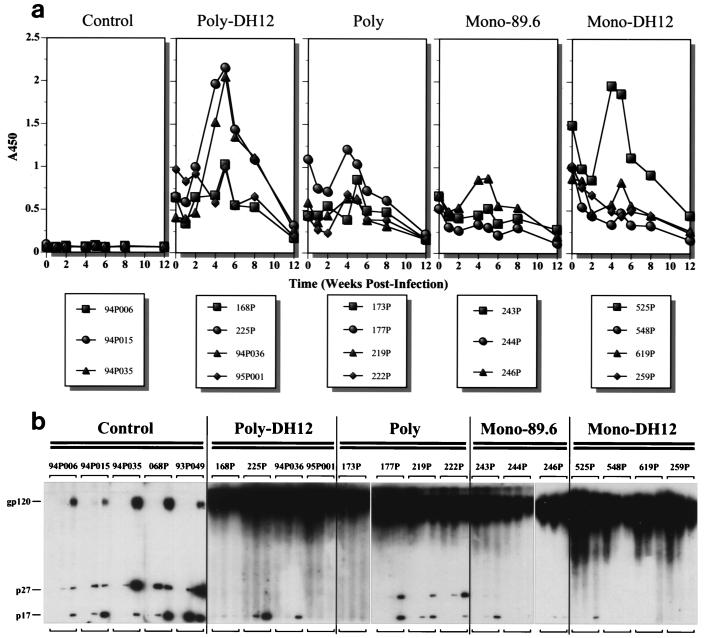
The strength of the postvirus challenge anamnestic antibody responses against DH12 gp120, measured by gp120 ELISA, did not necessarily correlate with the virus load measurements (Fig. 5a). Among four vaccinated animals with high plasma viremia (225P, 219P, 244P, and 246P), only one exhibited a significant anamnestic response (225P). Strong anamnestic responses were observed in only two other monkeys (94P036 and 525P), both of which had low plasma virus loads. All three animals with strong anamnestic responses were members of either the Polyvalent-DH12 or Monovalent-DH12 group. This suggests that the anamnestic responses were primarily directed against the epitopes specific to DH12 envelope glycoprotein (i.e., the variable regions of DH12 gp120). In contrast, negligible or no anamnestic responses occurred in the three macaques (95P001, 548P, and 259P) in which no plasma viral RNA or lymph node proviral DNA was detected. Relatively weak anamnestic responses were observed in animals 168P and 619P. For animal 619P, this weak response suggested that virus did replicate to some degree and was consistent with the low but detectable levels of proviral DNA and plasma viremia described earlier. The anamnestic responses in all of the Polyvalent vaccine group animals were also quite low and only one of three animals in the Monovalent-89.6 group (246P) exhibited a significant anamnestic response, although all three macaques in this vaccine group had a substantial plasma viremia. Anti-gp120 antibodies were first detected 5 weeks postinfection in control group animals, but only at much lower plasma dilutions (1:90) (data not shown).
FIG. 5.
Antibody responses after SHIVDH12 challenge. (a) The antibody levels in the plasma samples of individual animals were analyzed by ELISA as described for Fig. 3. A450, absorbance at 450 nm. (b) Western immunoblot detection of antibodies against HIV-1 gp120 and SIV Gag proteins. The antiserum collected at times 0, 8, and 24 weeks postinfection from each animal were analyzed. Bands corresponding to HIV-1 gp120 and SIV p27 and p17 are indicated on the left.
Postinfection humoral immune responses were also examined by Western blot analysis (Fig. 5b). All animals in the control group, including the macaque with low virus loads (94P006), developed antibodies against Gag proteins by 8 weeks postinfection. Antibodies against both p27 and p17 were detected in these animals. By 24 weeks postinfection, antibodies against gp120 were also detected, although the reactivity was considerably weaker than the reactivity measured in vaccinated monkeys. In contrast to the control animals, only one of four macaques in the Monovalent-DH12 group (525P) developed antibodies against p17. This was the only animal with clearly demonstrable plasma viremia. Although monkey 619P had proviral DNA in the lymph node, no antibodies against Gag proteins were generated. In the Polyvalent-DH12 group, the three animals with plasma viremia all developed antibodies to p17, although the response was barely demonstrable in animal 168P. Interestingly, no antibodies were detected against p27. As expected, monkey 95P001, which experienced no plasma viremia, failed to mount an anti-Gag antibody response. In the Polyvalent vaccine group, three of the four animals generated antibodies against both p17 and p27. The fourth macaque, 173P, made no antibody to either p27 or p17, although it sustained a robust plasma viremia. In the Monovalent-89.6 group, weak antibody responses against p17 were detected in animals 243P and 246P, whereas monkey 244P developed no anti-Gag antibodies despite having relatively high virus loads. At present, the factors determining whether or not animals immunized with HIV-1 envelope glycoproteins generate antibodies against Gag proteins during a subsequent virus infection are not known. Although a general correlation between plasma viremia and an antibody response to Gag proteins existed, several exceptions (e.g., 244P and 173P) were observed.
Breadth of NAb response.
Since one of the major aims of this study was to determine whether broader NAb response might be elicited by immunizing animals with a mixture of HIV-1 envelope glycoproteins, the neutralization sensitivity of viruses other than HIV-1DH12 was evaluated (Fig. 6). Assays were performed with plasma samples collected 3 weeks after the final gp120H boost, using either human PBMC or the MT-2 continuous T-cell line. No neutralizing activity was detected in the plasma collected from control group macaques. Animals in the Monovalent-DH12 group developed NAbs that were highly specific for HIV-1DH12; their antisera failed to neutralize any of the other test viruses, including relatively neutralization-sensitive HIV-1MN. Antisera from the macaques immunized with the 89.6 envelope glycoprotein exhibited a slightly broader NAb response than those vaccinated with DH12, being able to neutralize virus strains bearing both the homologous 89.6 and heterologous MN gp120s.
FIG. 6.
Neutralization activity against various HIV-1 and SHIV strains. The antisera collected 3 weeks after the final gp120 boost (4 weeks prior to the challenge) were analyzed for neutralizing activity. NAb against SHIV-HXB2, SHIV-89.6, HIV-1RF, and HIV-1MN were tested using cell-killing reduction assays in MT-2 cells while HIV-1Bal, HIV-1AD8, and SHIVDH12 were tested using p24 reduction assays with human PBMC as target cells. In the cell killing assays, the number indicates 1 divided by the dilution of the serum samples that yield 50% reduction in cytopathic effects. In the p24 reduction assays, the number indicates the percentage of reduction in Gag protein production at the dilution of serum of 1:4.
In general, animals in the Polyvalent-DH12 and Polyvalent groups exhibited significantly broader NAb responses than those in the two monovalent vaccine groups (P = 0.0054 by the Wilcoxon rank sum test). There was, however, significant animal-to-animal variation as well as differences in the ability to elicit NAbs with different envelope glycoproteins. While macaques 95P001 and 219P generated NAbs that could neutralize up to five and four different isolates, respectively, some of the other animals were able to neutralize only two isolates, which invariably included the very sensitive HIV-1MN (monkeys 225P, 94P036, and 222P). None of the vaccinated macaques could neutralize HIV-1AD8, possibly because it may be an intrinsically neutralization-resistant primary isolate. Alternatively, the HIV-1AD8 envelope glycoprotein may be relatively nonimmunogenic and may be unable to elicit detectable levels of NAb. Another possibility is that antigenic competition from other envelope glycoproteins included in the mixture may have muted an immune response. We are not presently able to distinguish between these alternative explanations. In this regard, only one macaque (219P) of the eight animals in the Polyvalent-DH12 and Polyvalent vaccine groups was able to neutralize SHIV89.6. This result is in contrast to that obtained with animals in the Monovalent-89.6 group, where plasma from all three macaques neutralized SHIV89.6. This would indicate that the 89.6 envelope glycoprotein is immunogenic, SHIV89.6 is neutralizable, and antigenic competition may have muted the immune response against the 89.6 envelope glycoprotein. Alternatively, the lower amounts of 89.6 antigen administered to the monkeys in the Polyvalent-DH12 and Polyvalent vaccine groups (i.e., either 1/6 or 1/5 of the dose given to the Monovalent-89.6 group, respectively) may have been insufficient to elicit detectable anti-SHIV89.6 NAbs.
Because antisera from monkeys 95P001 and 219P exhibited the broadest cross-reactive neutralizing activity, their ability to neutralize primary HIV-1 isolates not included in the vaccine mixture was examined. Six isolates from clade B (P15, P27, 1168, 1196, Pvo, and Tro) (5) and two isolates from clade C (DU151 and DU123) were selected for analysis. All were R5 isolates obtained during early seroconversion, and the neutralization assay was performed in human PBMC with a 1:4 dilution of the antisera. No significant neutralizing activity was detected with either antiserum (data not shown).
DISCUSSION
In this study, the recombinant vaccinia virus prime and subunit protein boost approach was used to demonstrate that mixtures of HIV-1 envelope glycoproteins could elicit broader immune responses than vaccinations with individual Env immunogens. Animals in the two monovalent vaccine groups developed NAbs against only one or two HIV-1 isolates, whereas five of eight animals in the polyvalent vaccine groups made NAbs against three or more viral strains (P = 0.0054). Unfortunately, however, this increased breadth of neutralization was limited almost entirely to the virus strains used for vaccination. This was best illustrated with the antisera from two macaques (95P001 and 219P) which possessed neutralizing activity against five and four different viruses, respectively, but were unable to neutralize any of eight heterologous primary HIV-1 isolates tested. As far as protective immunity was concerned, resistance to SHIVDH12 strongly correlated with the levels of NAbs specifically directed against SHIVDH12 at the time of challenge. Disappointingly, the most potent protective humoral responses against the SHIVDH12 challenge were elicited only in monkeys vaccinated with preparations containing a DH12 Env component. Nonetheless, it might still be possible to elicit more broadly protective virus neutralizing responses by immunization with a pool of HIV-1 glycoproteins representing a larger cross section of the neutralization subtypes in circulation.
Because polyvalent vaccination regimens have not been extensively used to elicit protective immune responses against primate lentiviruses, some of the results obtained were unexpected. For example, the immunogenicity of the envelope glycoprotein mixture was quite variable in stimulating NAbs in the pigtailed macaques under study. In the case of DH12, four of four recipients of monovalent immunogen and three of four vaccinees given the mixture of Env proteins (including DH12) developed NAbs against SHIVDH12, as measured in p27 reduction assays (Fig. 6). This is in contrast to the 89.6 Env protein which elicited NAbs against SHIV89.6 in all three recipients of monovalent 89.6 Env but in only one of eight animals when 89.6 Env was administered in a mixture of other envelope glycoproteins. This also appeared to be the case for the AD8 Env, which failed to elicit NAbs in any of eight polyvalent vaccinees, although a comparable monovalent AD8 control group was not included in this study. At present, it is not clear whether these variable responses reflect the innate poor immunogenicity of HIV-1 envelope proteins, possible antigenic competition among the various components of the polyvalent vaccine, or simply the effect of antigen dilution resulting from the administration of the same amount of total Env immunogen as a polyvalent or monovalent vaccine.
As noted earlier, a strong inverse correlation was observed between the levels of vaccine-induced NAbs on the day of virus challenge and the subsequent plasma viremia (P = 0.0008 by the Jonckheere-Terpstra test for trend). Specifically, the four animals with either no demonstrable (548P, 259P, and 95P001) or barely detectable (619P) levels of plasma viremia all had NAb titers of 1:9 or greater (Table 1). This result is consistent with a previously published passive immunization study, which reported that NAb titers of approximately 1:8 (based on 100% neutralization assay in MT-4 cells), but not 1:4, conferred complete protection against a 100-TCID50 challenge with SHIVDH12 (48). Conversely, there was no statistical correlation of the postchallenge viral RNA levels in animals with anti-virus neutralization titers of less than 1:9 and with no detectable NAbs (viz. the monkeys in the Polyvalent and Monovalent-89.6 vaccine groups; P = 0.53 by the Wilcoxon rank sum test). These and previously published results strongly suggest the existence of a NAb threshold for complete or near-complete neutralization of the virus inoculum (and/or progeny virions produced during the first replication cycle). If the NAb titer is below the critical threshold, some fraction of the input virus will escape and will be able to establish a productive infection that may be refractory to subsequent cell-mediated immune responses.
Immunization with live virus vectors (e.g., vaccinia virus) or DNA vaccines, which allow de novo synthesis of antigens, are known to elicit CTL responses (7, 52). Although none of the macaques in either the Monovalent-89.6 or Polyvalent (lacking DH12) vaccine groups produced NAbs against SHIVDH12, only the virus loads in the Polyvalent group seemed to be markedly lower following the SHIVDH12 challenge (Fig. 4). This result suggests the possibility that the monkeys immunized with a mixture of HIV-1 envelope glycoproteins mounted a more effective nonhumoral immune response (possibly of CTL origin) than the Monovalent-89.6 group. We were unable to monitor possible CTL responses, because priming animals with recombinant vaccinia viruses precluded these vectors from being used to express viral proteins in autologous target cells for subsequent CTL assays. The tetramer binding assay (1, 40) could not be used because the macaques under study had not been previously classified for their major histocompatibility complex class I genotype. If CTLs were indeed responsible for the relatively low plasma viremia in the Polyvalent vaccine group animals, the protective effect observed might reflect the immunologic response of multiple cross-reactive CTL epitopes to the mixture of gp120 variable-loop peptides. Alternatively, such a hypothetical CTL response could be directed against the more highly conserved Env domains within the gp160 cocktail administered to animals in the Polyvalent group. CTL responses to epitopes mapping to lentiviral Gag proteins, but not to envelope glycoproteins, have received primary attention in the context of controlling the primary infection or developing a protective HIV-1 vaccine. It would therefore seem prudent to include mixtures of Gag and possibly other more conserved viral proteins in polyvalent vaccine formulations to elicit more effective immune responses against heterologous virus isolates.
ACKNOWLEDGMENTS
We are grateful to David Venzon for statistical analyses, to Ed Berger, Bernard Moss, and Bob Doms for recombinant envelope vaccinia viruses and plasmids, and to Lynn Morris, Carolyn Williamson, and Susan Fiscus for the clade C isolates. We also thank Charlotte Kensil for QS-21 and Ron Willey for valuable scientific discussions.
This study was supported in part by NIH grant AI-85343 to D. C. Montefiori and an IPA grant from NIAID to K. C. Gupta.
REFERENCES
- 1.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 2.Bogers W M, Niphuis H, ten Haaft P, Laman J D, Koornstra W, Heeney J L. Protection from HIV-1 envelope-bearing chimeric simian immunodeficiency virus (SHIV) in rhesus macaques infected with attenuated SIV: consequences of challenge. AIDS. 1995;9:F13–F18. [PubMed] [Google Scholar]
- 3.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retrovir. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 5.Bures R, Gaitan A, Zhu T, Graziosi C, McGrath K M, Tartaglia J, et al. Immunization with recombinant canarypox vectors expressing membrane-anchored gp120 followed by soluble gp160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retrovir. 2000;16:2015–2031. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 6.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W, Sawyer L S, Hendry R M, Dunlop N, Nara P L, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 7.Cho M W. Assessment of HIV vaccine development: past, present, and future. In: Jeang K-T, editor. Advances in pharmacology. Vol. 49. New York, N.Y: Academic Press; 2000. pp. 263–313. [DOI] [PubMed] [Google Scholar]
- 8.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho M W, Lee M K, Chen C H, Matthews T, Martin M A. Identification of gp120 regions targeted by a highly potent neutralizing antiserum elicited in a chimpanzee inoculated with a primary human immunodeficiency virus type 1 isolate. J Virol. 2000;74:9749–9754. doi: 10.1128/jvi.74.20.9749-9754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho M W, Shibata R, Martin M A. Infection of chimpanzee peripheral blood mononuclear cells by human immunodeficiency virus type 1 requires cooperative interaction between multiple variable regions of gp120. J Virol. 1996;70:7318–7321. doi: 10.1128/jvi.70.10.7318-7321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 12.Clements-Mann M L, Weinhold K, Matthews T J, Graham B S, Gorse G J, Keefer M C, McElrath M J, Hsieh R-H, Mestecky J, Zolla-Pazner S, Mascola J, Schwartz D, Siliciano R, Corey L, Wright P F, Belshe R, Dolin R, Jackson S, Xu S, Fast P, Walker M C, Stablein D, Excler J-L, Tartaglia J, Duliege A-M, Sinangil F, Paoletti E the NIAID AIDS Vaccine Evaluation Group. Immune responses to human immunodeficiency virus (HIV) type 1 induced by canarypox expressing HIV-1MN gp120, HIV-1SF2 recombinant gp120, or both vaccines in seronegative adults. J Infect Dis. 1998;177:1230–1246. doi: 10.1086/515288. [DOI] [PubMed] [Google Scholar]
- 13.Cooney E L, McElrath M J, Corey L, Hu S-L, Collier A C, Arditti D, Hoffman M, Coombs R W, Smith G E, Greenberg P D. Enhanced immunity to human immunodeficiency virus (HIV) envelope elicited by a combined vaccine regimen consisting of priming with a vaccinia recombinant expressing HIV envelope and boosting with gp160 protein. Proc Natl Acad Sci USA. 1993;90:1882–1886. doi: 10.1073/pnas.90.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford J M, Earl P L, Moss B, Reimann K A, Wyand M S, Manson K H, Bilska M, Zhou J T, Pauza C D, Parren P W, Burton D R, Sodroski J G, Letvin N L, Montefiori D C. Characterization of primary isolate-like variants of simian-human immunodeficiency virus. J Virol. 1999;73:10199–10207. doi: 10.1128/jvi.73.12.10199-10207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 16.Dunn C S, Hurtrel B, Beyer C, Gloeckler L, Ledger T N, Moog C, Kieny M P, Mehtali M, Schmitt D, Gut J P, Kirn A, Aubertin A M. Protection of SIVmac-infected macaque monkeys against superinfection by a simian immunodeficiency virus expressing envelope glycoproteins of HIV type 1. AIDS Res Hum Retrovir. 1997;13:913–922. doi: 10.1089/aid.1997.13.913. [DOI] [PubMed] [Google Scholar]
- 17.Emini E A, Schleif W A, Nunberg J H, Conley A J, Eda Y, Tokiyoshi S, Putney S D, Matsushita S, Cobb K E, Jett C M, et al. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature. 1992;355:728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- 18.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung M S, Sun C R, Gordon W L, Liou R S, Chang T W, Sun W N, Daar E S, Ho D D. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J Virol. 1992;66:848–856. doi: 10.1128/jvi.66.2.848-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauduin M C, Parren P W, Weir R, Barbas C F, Burton D R, Koup R A. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 21.Giavedoni L D, Planelles V, Haigwood N L, Ahmad S, Kluge J D, Marthas M L, Gardner M B, Luciw P A, Yilma T D. Immune response of rhesus macaques to recombinant simian immunodeficiency virus gp130 does not protect from challenge infection. J Virol. 1993;67:577–583. doi: 10.1128/jvi.67.1.577-583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorny M K, Moore J P, Conley A J, Karwowska S, Sodroski J, Williams C, Burda S, Boots L J, Zolla-Pazner S. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goudsmit J, Debouck C, Meloen R H, Smit L, Bakker M, Asher D M, Wolff A V, Gibbs C J, Jr, Gajdusek D C. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc Natl Acad Sci USA. 1988;85:4478–4482. doi: 10.1073/pnas.85.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham B S, Matthews T J, Belshe R B, Clements M L, Dolin R, Wright P F, Gorse G J, Schwartz D H, Keefer M C, Bolognesi D P the NIAID AIDS Vaccine Clinical Trials Network. Augmentation of human immunodeficiency virus type 1 neutralizing antibody by priming with gp160 recombinant vaccinia and boosting with rgp160 in vaccinia-naive adults. J Infect Dis. 1993;167:533–537. doi: 10.1093/infdis/167.3.533. [DOI] [PubMed] [Google Scholar]
- 25.Hu S L, Klaniecki J, Dykers T, Sridhar P, Travis B M. Neutralizing antibodies against HIV-1 BRU and SF2 isolates generated in mice immunized with recombinant vaccinia virus expressing HIV-1 (BRU) envelope glycoproteins and boosted with homologous gp160. AIDS Res Hum Retrovir. 1991;7:615–620. doi: 10.1089/aid.1991.7.615. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi T, Endo Y, Englund G, Sadjadpour R, Matano T, Buckler C, Buckler-White A, Plishka R, Theodore T, Shibata R, Martin M. Emergence of a highly pathogenic simian/human immunodeficiency virus in a rhesus macaque treated with anti-CD8 mAb during a primary infection with a nonpathogenic virus. Proc Natl Acad Sci USA. 1999;96:14049–14054. doi: 10.1073/pnas.96.24.14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurkiewicz E, Hunsmann G, Schaffner J, Nisslein T, Luke W, Petry H. Identification of the V1 region as a linear neutralizing epitope of the simian immunodeficiency virus SIVmac envelope glycoprotein. J Virol. 1997;71:9475–9481. doi: 10.1128/jvi.71.12.9475-9481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong P D, Wyatt R, Sattentau Q J, Sodroski J, Hendrickson W A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M K, Martin M A, Cho M W. Higher Western blot immunoreactivity of glycoprotein 120 from R5 HIV type 1 isolates compared with X4 and X4R5 isolates. AIDS Res Hum Retrovir. 2000;16:765–775. doi: 10.1089/088922200308765. [DOI] [PubMed] [Google Scholar]
- 30.Lewis M G, Elkins W R, McCutchan F E, Benveniste R E, Lai C Y, Montefiori D C, Burke D S, Eddy G A, Shafferman A. Passively transferred antibodies directed against conserved regions of SIV envelope protect macaques from SIV infection. Vaccine. 1993;11:1347–1355. doi: 10.1016/0264-410x(93)90106-8. [DOI] [PubMed] [Google Scholar]
- 31.Mackett M, Smith G Y, Moss B. The construction and characterization of vaccinia virus recombinants expressing foreign genes. In: Rickwood D, Hames B D, editors. DNA cloning. 2nd ed. Washington, D.C.: IRL Press; 1985. pp. 191–211. [Google Scholar]
- 32.Mascola J R, Lewis M G, Stiegler G, Harris D, VanCott T C, Hayes D, Louder M K, Brown C R, Sapan C V, Frankel S S, Lu Y, Robb M L, Katinger H, Birx D L. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascola J R, Stiegler G, VanCott T C, Katinger H, Carpenter C B, Hanson C E, Beary H, Hayes D, Frankel S S, Birx D L, Lewis M G. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 34.Matsushita S, Robert-Guroff M, Rusche J, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–2114. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montefiori D C, Pantaleo G, Fink L M, Zhou J T, Zhou J Y, Bilska M, Miralles G D, Fauci A S. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J Infect Dis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- 36.Montefiori D C, Robinson W E, Jr, Schuffman S S, Mitchell W M. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26:231–235. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J P, Cao Y, Ho D D, Koup R A. Development of the anti-gp120 antibody response during seroconversion to human immunodeficiency virus type 1. J Virol. 1994;68:5142–5155. doi: 10.1128/jvi.68.8.5142-5155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 40.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 41.Ourmanov L, Bilska M, Hirsch V M, Montefiori D C. Recombinant modified vaccinia virus ankara expressing the surface gp120 of simian immunodeficiency virus (SIV) primes for a rapid neutralizing antibody response to SIV infection in macaques. J Virol. 2000;74:2960–2965. doi: 10.1128/jvi.74.6.2960-2965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parren P W, Ditzel H J, Gulizia R J, Binley J M, Barbas III C F, Burton D R, Mosier D E. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9:F1–F6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 43.Pinter A, Honnen W J, Kayman S C, Trochev O, Wu Z. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine. 1998;16:1803–1811. doi: 10.1016/s0264-410x(98)00182-0. [DOI] [PubMed] [Google Scholar]
- 44.Putkonen P, Thorstensson R, Ghavamzadeh L, Albert J, Hild K, Biberfeld G, Norrby E. Prevention of HIV-2 and SIVsm infection by passive immunization in cynomolgus monkeys. Nature. 1991;352:436–438. doi: 10.1038/352436a0. [DOI] [PubMed] [Google Scholar]
- 45.Quesada-Rolander M, Makitalo B, Thorstensson R, Zhang Y J, Castanos-Velez E, Biberfeld G, Putkonen P. Protection against mucosal SIVsm challenge in macaques infected with a chimeric SIV that expresses HIV type 1 envelope. AIDS Res Hum Retrovir. 1996;12:993–999. doi: 10.1089/aid.1996.12.993. [DOI] [PubMed] [Google Scholar]
- 46.Rusche J R, Javaherian K, McDanal C, Petro J, Lynn D L, Grimaila R, Langlois A, Gallo R C, Arthur L O, Fischinger P J, et al. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci USA. 1988;85:3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safrit J T, Fung M S, Andrews C A, Braun D G, Sun W N, Chang T W, Koup R A. hu-PBL-SCID mice can be protected from HIV-1 infection by passive transfer of monoclonal antibody to the principal neutralizing determinant of envelope gp120. AIDS. 1993;7:15–21. doi: 10.1097/00002030-199301000-00002. [DOI] [PubMed] [Google Scholar]
- 48.Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho M W, Martin M A. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 49.Shibata R, Maldarelli F, Siemon C, Matano T, Parta M, Miller G, Fredrickson T, Martin M A. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J Infect Dis. 1997;176:362–373. doi: 10.1086/514053. [DOI] [PubMed] [Google Scholar]
- 50.Shibata R, Siemon C, Cho M W, Arthur L O, Nigida S M, Jr, Matthews T, Sawyer L A, Schultz A, Murthy K K, Israel Z, Javadian A, Frost P, Kennedy R C, Lane H C, Martin M A. Resistance of previously infected chimpanzees to successive challenges with a heterologous intraclade B strain of human immunodeficiency virus type 1. J Virol. 1996;70:4361–4369. doi: 10.1128/jvi.70.7.4361-4369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata R, Siemon C, Czajak S C, Desrosiers R C, Martin M A. Live, attenuated simian immunodeficiency virus vaccines elicit potent resistance against a challenge with a human immunodeficiency virus type 1 chimeric virus. J Virol. 1997;71:8141–8148. doi: 10.1128/jvi.71.11.8141-8148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinman R M, Germain R N. Antigen presentation and related immunological aspects of HIV-1 vaccines. AIDS. 1998;12:S97–S112. [PubMed] [Google Scholar]
- 53.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas III C F, Burton D R, Ho D D, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warrier S V, Pinter A, Honnen W J, Girard M, Muchmore E, Tilley S A. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J Virol. 1994;68:4636–4642. doi: 10.1128/jvi.68.7.4636-4642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]