Abstract
Introduction:
Oral squamous cell carcinoma (OSCC) includes about 90% of all oral malignant tumors, and most of them are diagnosed in advanced stages. This study investigated the expression changes of miR-24, miR-200, and miR-34 in saliva samples of patients with oral squamous cell carcinoma, for early diagnosis.
Methods:
In this study, 30 patients and 30 healthy individuals were selected. After RNA extraction and cDNA synthesis, the expression levels of miR-24, miR-200, and miR-34 in saliva samples were measured and evaluated using the Real-Time PCR technique.
Results:
Folding change calculation using 2^(-∆∆ Ct) refers to the relative difference in the expression of the markers of the two groups. The expression level of two biomarkers, miR-200 and miR-34, is decreased in patients compared to healthy people; and the expression level of miR-24 is increased in patients compared to healthy people.
Conclusion:
In general, considering the availability and convenience of saliva sample collection for early detection of the disease, this research result can be considered a diagnostic screening test. To further prove the research results, conducting more extensive studies with more samples is recommended.
Key Words: Oral Squamous Cell Carcinoma, Early Diagnosis, miR-34, miR-24, miR-200
Introduction
Oral squamous cell carcinoma is a relatively rare cancer that unfortunately its mortality rate is high [1]. Squamous Cell Carcinoma is the most common oral epithelial cancer observed in the oral cavity. So that it covers about 90% of oral malignancies [2-5]. Most oral cancers are diagnosed in advanced stages, and this leads to the unfavorable prognosis of oral cancer in most parts of the world [6, 7]. Local invasion and metastasis in this cancer are high and increase the mortality rate of patients with this cancer. Despite the recent therapeutic advances such as chemotherapy, radiotherapy, and surgery, unfortunately, the mortality rate of patients with this cancer has not yet decreased [8].
miRNAs usually have about 18 to 25 nucleotides. These molecules are completely specific in regulating gene expression [9]. miRNAs are expressed quite specifically in different tissues and diseases, which has led researchers to use these molecules as a diagnostic marker [10]. miRNA24 is a multifunctional biomarker with different biological processes. One of its important roles is to regulate apoptosis and increase inhibition through cyclin-dependent kinases [11]. Moreover, studies have reported that increased expression of miR-24 can lead to increased proliferation and decreased apoptosis in tongue squamous cell carcinoma cells. According to the results, miR-21, miR-24, and miR-29a in the serum of patients with oral squamous cell carcinoma, compared to the normal group, can be used as strong markers for cancer diagnosis [11].
In general, miRNA-24 affects many molecules. This miRNA-24 plays a significant role in the cell cycle and cellular differentiation [11, 12]. 200-miR plays an important role in tumor prognosis. The genes of this family are widely involved in tumor progression and metastasis. 200-miR in hepatocellular carcinoma cells increases cell proliferation and inhibits apoptosis. 200-miR plays a dual role in tumors, and the reason for these different roles in different types of cancers may be due to differences in target genes and cell type [13]. miR-34 is a known tumor suppressor in various types of cancers, including lung cancer, breast, prostate, and liver cancers, etc. however, the role of miR-34a in oral cancer is still unknown [14]. Abnormal expression of miR-34a has significant effects on cell growth, and cell migration, leading to reduced invasion. This suggests that miR-34a downregulation may play an important role in OSCC invasion and metastasis. Evidence supports that overexpression of miR-34a in OSCC cells inhibits cell migration, invasion, and lung colony formation [14].
This research aims to investigate the expression of miR-24, miR-200, and miR-34 in the saliva samples of patients with oral squamous cell carcinoma, compared to the saliva samples of healthy people.
Materials and Methods
This research has received the permission of the Ethics Committee with ID IR.SBMU.NRITLD.REC.1402.117. 30 patients, who were suspected of oral cancer based on physical examination and diagnosis by a specialist doctor, were selected before any treatment. Moreover, 30 healthy people, after being examined by a doctor, voluntarily and by filling out the consent form, were selected as the control group. The people of the two groups were considered in the same groups in terms of age factor, in the age group of 24 to 68 years. After selecting the subjects, saliva samples were taken, and the RNA extraction phase began immediately. RNA extraction steps were performed using RNeasy Mini Kit (Qiagen Cat no.75144). To evaluate the quality of the extracted RNA, NanoDrop was used, and in the next step, the ZIST ROYESH kit was used to make cDNA. This kit contains the necessary materials to perform Real-time PCR, including Forward and Reverse primers and SYBR Master Mix Green. U6 was used as housekeeping. Real-time PCR was performed in a final volume of 25 mL containing: 10mL Ampliqon master mix, 0.5 mL of each of F and R primers, 5 μL DNA, and distilled water. The test was done in 35 cycles using a rotor-gene cycler. Temperature and time conditions: initial denaturation of 95 °c, for 5 minutes, denaturation of 95 °c for 20 seconds, primers connection of 56 °C, for 40 seconds, and amplification of 72 °C, for 30 seconds (for 35 cycles), and the final amplification was 72 °C for 5 min.
Statistical Analysis
The results were analyzed using the statistical software SPSS22, and the mean and standard deviation were calculated. Paired t-test was used to analyze the difference between gene expression and clinicopathological characteristics, or the relationship between them. The difference was considered significant at P≤0.05.
Results
As mentioned before, the studied population consists of 30 healthy people and 30 people with oral squamous cell cancer. These two groups corresponded to each other in terms of age variables.The average age in the patients was 46.25 and in the control group was 47.84. The groups were compared in terms of mean age using the t-test, and there was no significant difference between them in terms of mean age, so, it can be concluded that the age factor in the studied groups does not lead to problems. Real-Time RT-PCR reaction was performed. Thus, 2 vials of cDNA made from patients and healthy people were tested for the expression of the reference genes and studied biomarkers. The results were interpreted according to the melting curve .
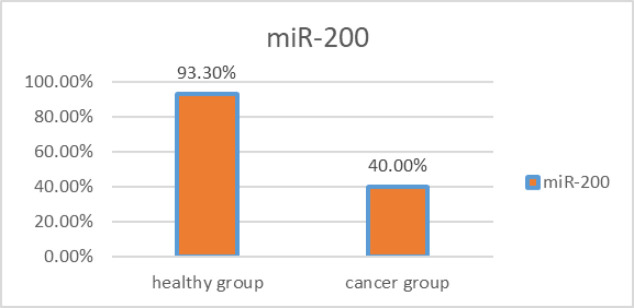
In the group of patients, the miR-200 biomarker was positive in 12 out of 30 people. This biomarker in the group of healthy people was 28 out of 30 people. The statistical comparison of the positivity rate of this biomarker in the group of patients and the group of healthy people was done using the two-sample binomial test, which indicates a statistically significant difference between these two groups (P-value < 0.001) (Figure 1).
Figure 1.
The Rate of miR-200 Positivity in the Saliva of Patients with OSCC and Healthy People
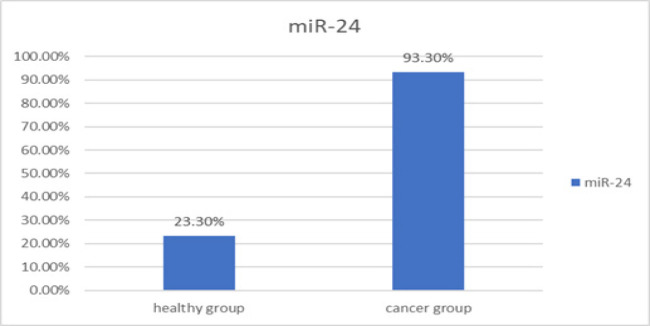
In the patient group, the miR-24 biomarker was positive in 28 out of 30 people. This biomarker in the group of healthy people was 7 out of 30 people. The statistical comparison of the positivity of this biomarker in the group of patients and the group of healthy people was done using the two-sample binomial test, which shows a statistically significant difference between these two groups (P-value < 0.001) (Figure 2).
Figure 2.
The Rate of miR-24 Positivity in the Saliva of Patients with OSCC and Healthy People
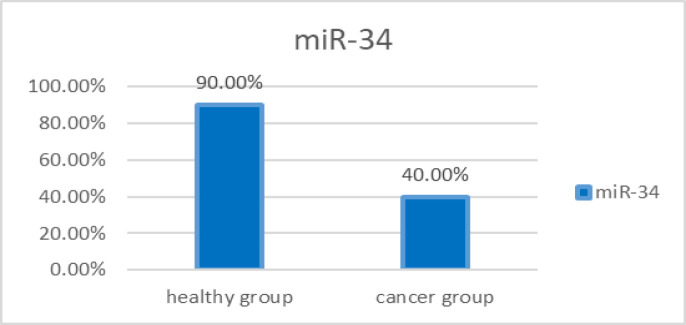
In the patient group, miR-34 biomarker was positive in 12 out of 30 people. This biomarker in the group of healthy people was 27 out of 30 people. The statistical comparison of the positivity of this biomarker in the group of patients and the group of healthy people was done using the two-sample binomial test, which shows a statistically significant difference between these two groups (P-value < 0.001) (Figure 3).
Figure 3.
The Rate of miR-34 Positivity in the Saliva of Patients with OSCC and Healthy People
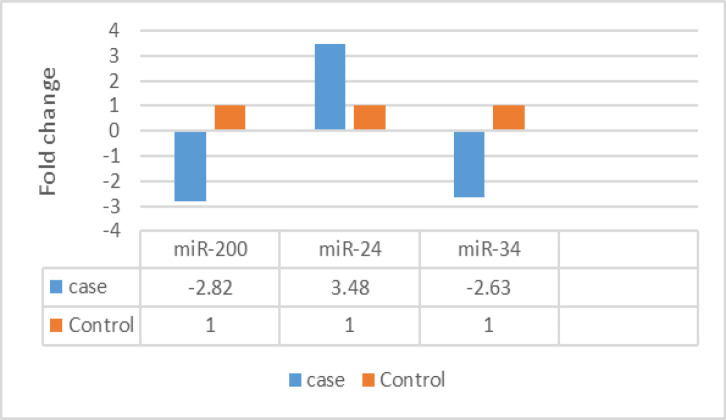
To calculate folding change, first of all, the Ct of each sample was determined. The relative difference in the markers expression of the two groups was calculated using 2(-∆∆ Ct). Therefore:
The expression of miR-200 in patients is 2.82 times lower than in healthy people.
The expression of miR-24 in patients is 3.48 times higher than in healthy people.
The expression of miR-34 in patients is 2.63 times lower than in healthy people (Figure 4).
Figure 4.
The Difference in miR-34, miR-200, and miR-24 Genes Expression of Control and Treatment Groups
Discussion
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous group of malignancies, including the oral cavity, nasal cavity, paranasal sinuses, pharynx, larynx, and salivary glands. Oral squamous cell carcinoma (OSCC) accounts for about 90% of all oral malignant tumors [15]. The diagnosis of OSCC usually depends on the clinical examination of the oral cavity, followed by a biopsy for histological analysis. However, despite easy access to visual examination, OSCC is often diagnosed in advanced stages, which leads to reduced patient survival [16, 17].
According to studies, microRNAs play an important role in the initiation and progression of cancer. MicroRNAs, depending on the type of mRNAs they inhibit, can be tumor suppressors or oncogenes. Therefore, microRNAs are useful genes for the diagnosis and prognosis of diseases including cancer, and they can be used for early diagnosis [18-23]. In the study conducted by Jain et al. (2014), it was found that the expression of miR-34 in the tissues of Adenocarcinoma of the gallbladder, in comparison with normal tissue, is significantly reduced in patients, and it is related to the poor prognosis of patients with Adenocarcinoma [24]. Gallardo et al. (2009), in their study, identified miR-34 as a new prognostic marker in patients with non-small-cell lung cancer (NSCLC), which provides a potential method to estimate the risk of recurrence of the disease and also is a useful tool to help decide on treatment [25].
These studies, from the point of view, that the reduction of miR-34 expression causes cancer progression and cancer formation, are consistent with the recent study.
miRNAs effectively exist in human body fluids such as blood, saliva, urine, and breathing air, so they can be made available by non-invasive methods. Wong et al. showed the existence of miR-184 in the plasma of 80% of patients with tongue squamous cell carcinoma [26]. In addition, Liu et al. showed a high level of miR-31 in the saliva of patients with oral cancer, while the level of miR-31 in plasma was increased. Furthermore, in the conducted study, an increase in miR-24 expression was seen in patients with OSCC compared to healthy people, and there was a significant statistical difference between them [27, 28].
Lin et al showed that the level of miR-24 in the plasma of patients with OSCC is significantly higher than that of the control group. Likewise, it has been suggested that the high level of miR-21 and miR-146a in plasma is a diagnostic sign for OSCC [29]. The expression level of miR-31 in the saliva of patients with OSCC is significantly increased in all clinical stages, compared to the control group [30]. In a recent study, there is an increase in the expression of miR-24. Giulietti et al. (2002), who examined the expression level of microRNAs in the serum of 100 patients with PDAC and 150 healthy people, observed that miRNA-200 is also one of these microRNAs, and finally, the importance of using these microRNAs as a disease prognosis was confirmed using data analysis [31].
The research of B. J. Zhang et al. (2002) also mentioned the oncogenic role of mir-200, and also this research has suggested that mir-200 leads to cell stimulation, proliferation, and invasion, and inhibits programmed cell death, which is consistent with the present study [32].
In conclusion, in general, the results of our research, consistent with previous studies, showed that the expression of miR-24, miR-200 and miR-34 markers in patients with oral squamous cell carcinoma was different compared to healthy people, and there is an increase or decrease in their expression. Therefore, analyzing the expression level of these markers in the saliva of patients, which is a good, available, and non-invasive sample, can be considered as potential biomarkers in the diagnosis of OSCC, however, additional research is recommended.
Acknowledgements
None.
Author Contribution Statement
All authors contributed equally in this study.
References
- 1.Collins FS, Barker AD. Mapping the cancer genome. Sci Am. 2007;296(3):50–7. [PubMed] [Google Scholar]
- 2.Altekruse SF, Huang L, Cucinelli JE, McNeel TS, Wells KM, Oliver MN. Spatial patterns of localized-stage prostate cancer incidence among white and black men in the southeastern united states, 1999-2001. Cancer Epidemiol Biomarkers Prev. 2010;19(6):1460–7. doi: 10.1158/1055-9965.EPI-09-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolandparva F, Nasab MSH, Mohamadnia A, Garajei A, Nasab AF, Bahrami N. Early diagnosis of oral squamous cell carcinoma (oscc) by mir-138 and mir-424-5p expression as a cancer marker. Asian Pac J Cancer Prev. 2021;22(7):2185. doi: 10.31557/APJCP.2021.22.7.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Najafi Z, Mohamadnia A, Ahmadi R, Mahmoudi M, Bahrami N, Khosravi A, et al. Proteomic and genomic biomarkers for non‐small cell lung cancer: Peroxiredoxin, haptoglobin, and alpha‐1 antitrypsin. Cancer Med. 2020;9(11):3974–82. doi: 10.1002/cam4.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamaati H, Bahrami N, Tabarsi P, Khosravi A, Kiani A, Abedini A, et al. Multi-gene expression in anthracosis of the lungs as one of the risk factors for non-small cell lung cancer. Asian Pac J Cancer Prev. 2017;18(11):3129. doi: 10.22034/APJCP.2017.18.11.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wicha MS, Liu S, Dontu G. Cancer stem cells: An old idea—a paradigm shift. Cancer Res. 2006;66(4):1883–90. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 7.Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13(5):555–62. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 8.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao M, Zheng L, Liu J, Dobleman T, Hu S, Go VLW, et al. Micrornas as effective surrogate biomarkers for early diagnosis of oral cancer. Clin Oral Investig. 2018;22:571–81. doi: 10.1007/s00784-017-2317-6. [DOI] [PubMed] [Google Scholar]
- 10.Lodish H, Berk A, Kaiser CA, Kaiser C, Krieger M, Scott MP, et al. Molecular cell biology. Macmillan. 2008 [Google Scholar]
- 11.Mukherjee S, Shelar B, Krishna S. Versatile role of miR-24/24-1*/24-2* expression in cancer and other human diseases. Am J Transl Res. 2022;14(1):20. [PMC free article] [PubMed] [Google Scholar]
- 12.Amelio I, Lena AM, Viticchiè G, Shalom-Feuerstein R, Terrinoni A, Dinsdale D, et al. Mir-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J Cell Biol. 2012;199(2):347–63. doi: 10.1083/jcb.201203134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scully C, Bagan J. Oral squamous cell carcinoma: Overview of current understanding of aetiopathogenesis and clinical implications. Oral Dis. 2009;15(6):388–99. doi: 10.1111/j.1601-0825.2009.01563.x. [DOI] [PubMed] [Google Scholar]
- 15.Avruch J, Khokhlatchev A, Kyriakis JM, Luo Z, Tzivion G, Vavvas D, et al. Ras activation of the raf kinase: Tyrosine kinase recruitment of the map kinase cascade. Recent Prog Horm Res. 2001;56:127–55. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 16.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, et al. S6k1−/−/s6k2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mrna translation and reveal a mitogen-activated protein kinase-dependent s6 kinase pathway. Mol Cell Biol. 2004;24(8):3112–24. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebolt-Leopold JS. Advances in the development of cancer therapeutics directed against the ras-mitogen-activated protein kinase pathway. Clin Cancer Res. 2008;14(12):3651–6. doi: 10.1158/1078-0432.CCR-08-0333. [DOI] [PubMed] [Google Scholar]
- 18.Herceg Z. Epigenetics and cancer: Towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 19.Ghadimi K, Bahrami N, Fathi M, Farzanegan B, Naji T, Emami M, et al. Diagnostic value of lunx mrna and cea mrna expression in pleural fluid of patients with non-small cell lung cancer. Minerva Pneumol. 2017;56(2):90–5. [Google Scholar]
- 20.Karimi S, Bahrami N, Sharifi K, Daustany M, Baghbani-Arani F, Kazempour M, et al. Investigating gene expression level of muc1 and cea in pleural fluid of nsclc lung cancer patients with real-time rt-pcr method. Minerva Pneumol. 2017;56(1):18–24. [Google Scholar]
- 21.Bahrami N, Gholami M, Jamaati HR, Mohamadnia A, Dargahi H, Kazempour Dizaji M, et al. Expression of two essential mrna biomarker in the peripheral blood as possible biomarkers for diagnosis of non-small cell lung carcinoma. Minerva Pneumol. 2016;55(3):31–6. [Google Scholar]
- 22.Jamaati H, Bahrami N, Abniki M, Tabarsi P, Farzanegan B, Doroudinia A, et al. Real-time rt-pcr detection of hcn4 and adam8 genes in ventilator-associated pneumonia patients hospitalized in intensive care unit. JCMA. 2016;1(4):163–7. [Google Scholar]
- 23.Azimi SA, Nia HRS, Jarrahi AM, Jamaati HR, Dizaji MK, Dargahi H, et al. Ectopic expression of mirna-21 and mirna-205 in non-small cell lung cancer. Int J Cancer Manag. 2019;12:1. [Google Scholar]
- 24.Marocchio LS, Lima J, Sperandio FF, Corrêa L, de Sousa SO. Oral squamous cell carcinoma: An analysis of 1,564 cases showing advances in early detection. J Oral Sci. 2010;52(2):267–73. doi: 10.2334/josnusd.52.267. [DOI] [PubMed] [Google Scholar]
- 25.La Vecchia C. Mouthwash and oral cancer risk: An update. Oral Oncol. 2009;45(3):198–200. doi: 10.1016/j.oraloncology.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Fu Y, Tu YY, Liu Y, Tan YR, Ju WT, et al. Mutation allele frequency threshold does not affect prognostic analysis using next-generation sequencing in oral squamous cell carcinoma. BMC Cancer. 2018;18:1–10. doi: 10.1186/s12885-018-4481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coutinho‐Camillo CM, Lourenço SV, Nishimoto IN, Kowalski LP, Soares FA. Caspase expression in oral squamous cell carcinoma. Head Neck. 2011;33(8):1191–8. doi: 10.1002/hed.21602. [DOI] [PubMed] [Google Scholar]
- 28.Liu PF, Hu YC, Kang BH, Tseng YK, Wu PC, Liang CC, et al. Expression levels of cleaved caspase-3 and caspase-3 in tumorigenesis and prognosis of oral tongue squamous cell carcinoma. PLoS One. 2017;12(7):e0180620. doi: 10.1371/journal.pone.0180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanojia D, Vaidya MM. 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol. 2006;42(7):655–67. doi: 10.1016/j.oraloncology.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Amôr NG, Buzo RF, Ortiz RC, Lopes NM, Saito LM, Mackenzie IC, et al. In vitro and in vivo characterization of cancer stem cell subpopulations in oral squamous cell carcinoma. J Oral Pathol Med. 2021;50(1):52–9. doi: 10.1111/jop.13101. [DOI] [PubMed] [Google Scholar]
- 31.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by erk, jnk, and p38 protein kinases. Science. 2002;298(5600):1911–2. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 32.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable rb–e2f switch underlies the restriction point. Nat Cell Biol. 2008;10(4):476–82. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]