Abstract
The N-terminal part of the mouse polyomavirus T antigens contains a highly conserved segment (-LLELLKL-), including amino acid residues 13 to 19. The sequence motif is predicted to form alpha helix I in the DnaJ domain of the T antigens. Four mutants with conservative substitutions of amino acid residues 13 and 14 were constructed. Of the four substitutions, L13M, L13I, L13V, and L14V, only L13V resulted in a phenotypic change. In transfected mouse cells, L13V large T antigen showed a more than 100-fold-reduced viral DNA synthesis. The viral replication could not be rescued by cotransfection of the cells with DNA expressing small t antigen or a large T antigen truncated at the C terminus that would compensate for a defect in host cell stimulation. In contrast to the effect on DNA replication, the L13V substitution in large T antigen did not prevent complex formation with Hsc70 and the Rb protein. Also, the activity of the protein in transactivation of transcription from the adenovirus E2 promoter was unimpaired, showing that the transcription factor E2F was released from pRb. The L13V substitution also caused a defect in small t antigen. However, this phenotypic change was due to protein instability. In contrast, middle T antigen with the L13V substitution remained stable and functional in cellular transformation. Together, the data show that the effect of the L13V substitution did not abrogate the Hsc70 interaction of the DnaJ domain. However, it is possible that the substitution of amino acid residue 13 affected specific DnaJ functions of large T antigen.
Polyomaviruses establish persistent infections in their hosts. The limited size of the viral genome makes the replication of viral DNA dependent on cellular enzymes. For this reason, viral early proteins, the T antigens, induce quiescent cells to enter the cell division cycle. Mouse polyomavirus encodes four early proteins: large, middle, small, and tiny T antigen (47, 51). These proteins are translated from mRNAs formed from a precursor by differential splicing. The four mRNAs have a common 5′-terminal sequence corresponding to 79 translated codons. Downstream of the splice points in mRNA, translation results in polypeptide segments unique to each T antigen.
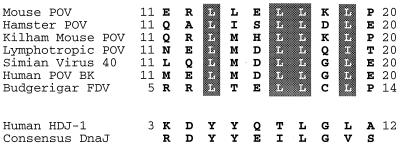
The N-terminal common region of the T antigens (crt) is homologous to the conserved domain of the DnaJ family of molecular chaperones (5, 28). The J domain, consisting of approximately 70 amino acid residues that form four alpha helices, binds to and stimulates the ATPase activity of proteins belonging to the Hsp70/DnaK family (9). In a loop between helices II and III there is a conserved amino acid motif, the J box (-HPD-), which contacts Hsp70 in the formation of a binary complex (21). The DnaJ-homologous structure of polyomavirus T antigen has DnaJ activity, since binding to Hsp70 protein and activation of its ATPase activity have been demonstrated (27, 47). Besides the -HPD- motif in the J domain, the T antigens of many polyomaviruses contain a second, highly conserved leucine-rich motif located at a position corresponding to alpha helix I. However, this leucine-rich motif is not particularly conserved in other DnaJ proteins (see Fig. 1), suggesting that it might confer a T-antigen-specific function. In mouse polyomavirus the motif at the putative alpha helix I has the sequence -LLELLKL-.
FIG. 1.
Conserved amino acid motif in the N-terminal part of polyomavirus (POV) T antigens. Deduced amino acid sequence of indicated T-antigen segments encoded by mouse polyomavirus (EMBL accession no. J02288), hamster polyomavirus (accession no. M26281), Kilham mouse polyomavirus (accession no. M55904), lymphotropic polyomavirus (accession no. K02562), simian virus 40 (accession no. V01380), BK virus (accession no. V01108), and budgerigar fledgeling disease virus (accession no. M20775) are shown. The homologous segment of the HDJ-1 protein (accession no. X62421) and the consensus amino acid sequence of DnaJ proteins (Prosite accession no. PD000231) are included as a reference. The highly conserved leucine residues in T antigens are highlighted by shading.
Large T antigen controls viral DNA synthesis by binding to the origin of replication and forming a homomultimeric complex that unwinds the two DNA strands (14). In this process, large T antigen also interacts with cellular replication proteins, directing them to the viral replicon. In simian virus 40 large T antigen, the J domain was shown to be involved in the initiation reaction, probably in the formation or dissociation of protein complexes (8). The J domain of large T antigen is also involved in the interaction with cellular proteins indirectly involved in replication. One such interaction is with the Rb family of proteins (48, 53). In mouse polyomavirus large T antigen, the segment -DLFCYE-, located on the C-terminal side of the J domain at amino acid residues 141 to 146, mediates binding to the pocket of these proteins (pRb, p107, and p130) (17, 31, 50). However, for dissociation of the complex with transcription factor E2, interaction of pRb with the J domain of large T antigen appears to be necessary, since mutant polypeptides with amino acid substitutions of the -HPD- motif are defective in this respect (48). The released transcription factors positively regulate the expression of a set of genes whose products participate in replication, including the synthesis of viral DNA. Binding of pRb to large T antigen is also necessary for its activity in immortalization of rodent embryonic cells (1, 10, 24, 25, 46).
Middle T antigen is the main transforming protein of mouse polyomavirus. It has no known enzyme activity but acts by binding to cellular polypeptides involved in transduction of growth signals (reviewed in references 12 and 26). The contribution of the J domain to the function of middle T antigen has not been systematically studied. However, deletions affecting this segment of the polypeptide do not damage its transforming activity or binding to protein phosphatase 2A (7, 19). Small t antigen is also able to bind to the A-subunit of protein phosphatase 2A (43). An apparently separate function of small t antigen is to stimulate the activity of large T antigen in viral DNA replication. Whether this effect of small t antigen is caused by direct activation of large T antigen or by influence on cellular replication factors is not known.
Functional studies of the J domain, using mutant proteins, have been focused on the highly conserved J box. Here we report on the effects of mutations altering alpha helix I of the J domain of polyomavirus T antigens.
MATERIALS AND METHODS
Cells, genomes, and transfection methods.
NIH 3T3 and Swiss 3T6 mouse fibroblast cells were obtained from ATCC (Manassas, Va.), and Fischer rat FR3T3 cells were a gift from F. Cuzin (Nice University). Primary cell cultures of rat embryo fibroblasts (REF) were established from 15- to 16-day-old whole rat embryos (50). Cell cultures in 6-cm-diameter petri dishes were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with serum as indicated. Transfection experiments were carried out with growing cells that had been plated 20 h earlier at a density of 3 × 105 cells per petri dish. In various DNA transfection experiments we used DEAE-dextran-chloroquine (36), Lipofectamine as recommended by the manufacturer (Life Technologies), or coprecipitation with calcium phosphate (56). Polyomavirus genomes were propagated as recombinants of plasmid pBR322 or pML (35), joined at the single BamHI sites. Mutants dl1061 (40), MT-1, and ST-1 (58) have deletions that restrict early viral gene expression to large, middle, or small T antigen, respectively. Mutant mu1355/dl1061 produces an N-terminal fragment of large T antigen which is inactive in viral DNA synthesis but retains the ability to bind the cellular pRb (50), and mutant dl1384/dl1061 has a deletion at nt 986 to 997, encoding a large T antigen that is deficient in pRb binding (50). The reporter gene constructs pPYLcat and pE2cat have been described elsewhere (34, 55). In these plasmids the chloramphenicol acetyltransferase gene is located downstream of the polyomavirus late promoter or the adenovirus E2 promoter, respectively. The previously published genotypes and expression properties of polyomavirus genomes are summarized in Table 1. For analysis of viral DNA replication a reporter plasmid, PyOrirep/pUC18, was used. It contained the polyomavirus origin of DNA replication (nt 4634 to 5293 and 1 to 174) inserted into the BamHI site of pUC18 DNA.
TABLE 1.
Polyomavirus genomes and T-antigen expression
| DNA type | Early regiona | Expressed T antigensb |
|---|---|---|
| dl1061 | Δ in intron (nt 713–765) | LT |
| dl1384/dl1061 | Δ(nt 986–997)c | LT (Δ143–146) |
| mu1355/dl1061 | Δ(nt 1412–2524)c | LT(Δ288–785) |
| MT-1 | Δ of complete intron (nt 749–810) | MT |
| ST-1 | Δ of complete intron (nt 749–796) | ST |
| PYΔE1 | Δ of early region (nt 174–3612) | None |
Nucleotide numbers are nominal and refer to the standard sequence with EMBL accession number J02288. Deleted nucleotides (nt) are indicated by Δ.
LT, MT, and ST designate large, middle, and small T antigen, respectively. Deleted amino acid residues are indicated by Δ.
The mutant also has the dl1061 deletion.
Wild-type and mutant large T antigen were expressed using the pcDNA3 vector (InVitrogen). The large-T-antigen-coding sequence of polyomavirus mutant LT-1 DNA and its derivatives, crtL13V/E15D, crtL14V/E15D, and crtP43S, were inserted into the EcoRV site of pcDNA3. Wild-type and mutant crtL13V/E15D small t antigen were expressed from the early coding sequences of ST-1 DNA inserted into pcDNA3. The Rb protein p105 was expressed from plasmid pSGRb (kindly provided by W. G. Kaelin), containing cDNA cloned in pSG5 (Stratagene).
Analysis of protein expression.
For radioactive labeling of polypeptides, the cells were first incubated for 30 min in methionine-free DMEM buffered with 20 mM HEPES. [35S]methionine was added (150 μCi per culture), and the incubation of the cells was continued for 4 h. The cell were lysed in a buffer consisting of 10 mM Tris-HCl (pH 8.0), 137 mM NaCl, 1.0 mM dithiothreitol, 1.0 mM MgCl2, 1.0 mM CaCl2, 1.0 mM EDTA, 10% (vol/vol) glycerol, 1.0% (vol/vol) Nonidet P-40, 50 μg of phenylmethylsulfonyl fluoride, and 1.0 μg of aprotinin per ml. After 30 min at 0°C, nuclei and cell debris were removed by centrifugation. For immunoprecipitation and immunoblot analysis of T antigen, the monoclonal antibodies (MAb) LT1 (13), F4, and F5 (42) were used. For immunoprecipitation of pRb and Hsc70 we used the MAb Ab1 (Oncogene Sciences) and MAb SP822 (Stressgene), respectively. In immunoprecipitation, antibodies were captured using protein G-Sepharose (Pharmacia Biotech).
Analysis of viral DNA replication.
Viral DNA was excised from the recombinant plasmids by digestion with BamHI, recircularized by treatment with T4 DNA ligase at a concentration of 5 μg of DNA per ml, and used for transfection of 3T6 cells. Low-molecular-weight DNA was selectively extracted from cells, and viral DNA was partially purified (40). It was cleaved with DpnI and BamHI, resolved by agarose gel electrophoresis, and transferred to a hybridization membrane (52). DNA on the membrane was annealed with 32P-labeled polyomavirus DNA (15), and bound radioactivity was quantified using a Molecular Imager G-450 (Bio-Rad). In analyses of viral DNA replication with large T antigen expressed at a high level, the origin of viral DNA replication was present on a separate plasmid, PyOrirep/pUC18. Here, newly replicated DNA was isolated after digestion with DpnI and PstI and was detected using a 32P-labeled PstI fragment excised from the same plasmid.
Analysis of transcriptional transactivation.
REF cultures were transfected with a mixture of pPYLcat or pE2cat DNA and a second plasmid expressing large T antigen. Protein extracts were prepared at 40 h posttransfection by addition of 0.10 ml of 10 mM Tris-HCl (pH 7.9), 150 mM NaCl, 1.5 mM MgCl2, and 0.5% (vol/vol) Nonidet P-40. Chloramphenicol acetyltransferase (CAT) activity in 0.04-ml portions was assayed by the method of Gorman et al. (20), as modified by Herbomel et al. (23). Incubations were done at 37°C for 3 h. The substrate and the products were separated by thin-layer chromatography. For quantitation of the reaction, the thin-layer chromatograms were analyzed in a Molecular Imager.
RESULTS
Mutant construction.
The conserved amino acid motif -LLELLKL- in the common region of mouse polyomavirus T antigens (Fig. 1) is predicted to form the alpha helix I (5) of the J domain. To construct mutants with base-pair substitutions in the codons of this conserved motif, a unique BglII cleavage site was introduced by changing bp 219 of polyomavirus DNA from A-T to T-A. This transversion resulted in the conservative amino acid replacement E15D. The mutation did not detectably alter the activity of mouse polyomavirus large T antigen in the initiation of viral DNA replication (data not shown). In further mutagenesis we replaced the highly conserved residues L13 and L14 of polyomavirus T antigens. Mutations leading to the amino acid substitutions L13M, L13V, and L13I were introduced. In addition, a mutation leading to the P43S substitution was introduced to provide a reference for defective DnaJ activity (8, 49). The mutants were denoted crt (common region T antigen). None of these amino acid substitutions at residue 13 led to disruption of the predicted alpha-helical structure. All these mutations were made in the genetic background of dl1061, having a deletion that restricts early gene expression to large T antigen. To investigate the effect of the crt mutations on middle and small t antigen, an AvaI restriction fragment (nucleotides 658 to 1018) was substituted for the homologous fragment of MT-1 and ST-1 DNA. In these viral genomes deletions corresponding to intervening sequences in RNA splicing restrict early gene expression to middle T antigen and small t antigen, respectively.
Activity of large T antigens produced by crt mutants in DNA replication.
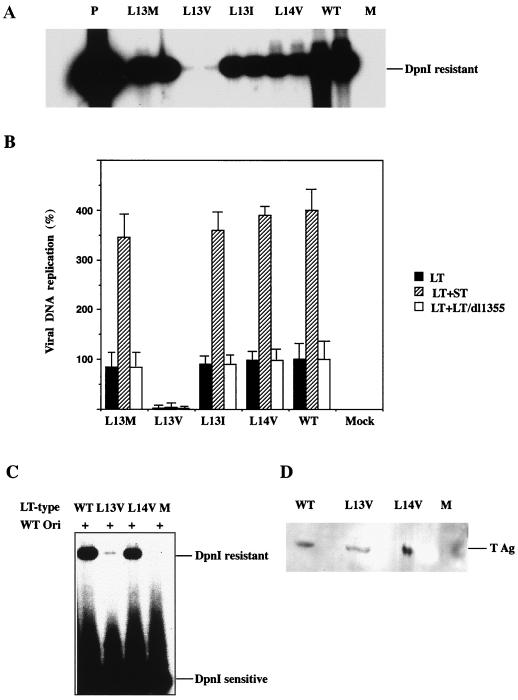
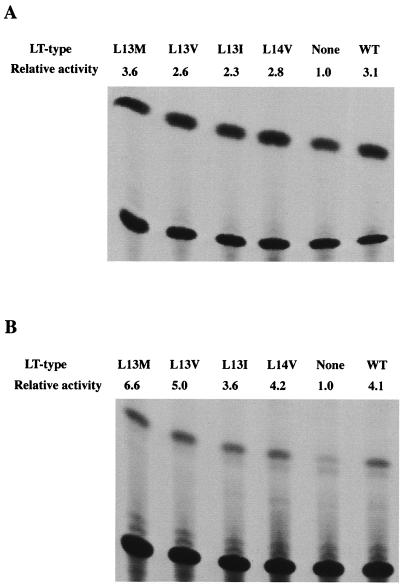
To test the activity of the mutant large T antigens in viral DNA replication, mouse 3T6 cells were transfected with the mutant genomes prepared by excision from recombinant plasmids. At 40 h posttransfection, viral DNA was selectively extracted from the cells and partially purified. After digestion with the methylation-dependent restriction endonucleases DpnI and BamHI that linearized the DNA molecules, they were resolved by agarose gel electrophoresis and blotted onto a hybridization membrane. Detection of polyomavirus DNA was done by annealing with a 32P-labeled probe. The autoradiogram is shown in Fig. 2A, and quantitative data from a similar experiment (Fig. 2B) showed that large T antigens with the L13I, L13M, and L14V substitutions supported viral DNA replication at 75 to 90% of the level reached with the wild-type protein. In contrast, the mutant expressing large T antigen with the L13V substitution replicated very poorly. The amount of viral DNA produced during 40 h was only 0.1% of that for the dl1061 control.
FIG. 2.
Activity of wild-type (WT) and mutant large T antigen in viral DNA replication. (A and B) Cultures of mouse 3T6 cells (5 × 105 cells) grown in DMEM containing 10% horse serum were transfected with 0.1 μg of viral DNA using the DEAE-dextran method. The cells were transfected either with a viral genome expressing wild-type or mutant large T antigen (LT) alone (A) (black columns in panel B) or together with a second viral genome expressing small t antigen (ST-1) or truncated large T antigen (dl1355). Low-molecular-weight DNA was selectively extracted at 42 h posttransfection, partially purified, cleaved with DpnI and BamHI, and subjected to agarose gel electrophoresis. DNA was transferred from the gel to a hybridization membrane and was annealed with 32P-labeled polyomavirus DNA. Radioactivity retained on the filters was then determined by autoradiography (A) or quantified in a Molecular Imager. The columns represent the average values of determinations of samples from duplicate cultures, and the error bars show the variation. (C and D) Cultures of NIH 3T3 cells (5 × 105 cells) grown in DMEM containing 10% fetal bovine serum were transfected, using Lipofectamine, with 1.0 μg of PyOrirep/pUC18 and 1.0 μg of pcDNA3LT-wt, -L13V/E15D, or -L14V/E15D, encoding large T antigen. One culture was transfected with only PyOrirep/pUC18 DNA (lanes M). (C) Newly replicated DNA was analyzed as described above. (D) Cell extracts from parallel transfected cultures were analyzed by immunoblotting, using MAb F5. T Ag shows the position of large T antigen.
To rule out that the L13V substitution made large T antigen unstable, DNA replication was analyzed with large T antigen expressed at a high level, allowing parallel determination of protein. NIH 3T3 cells were transfected with pcDNA3/LT-wt, pcDNA3/LT-L13V/E15D, pcDNA3/LT-L14V/E15D or, as a negative control, pcDNA3 mixed with PyOrirep/pUC18. Analysis of newly replicated DNA isolated at 40 h posttransfection showed (Fig. 2C) that the L13V substitution in large T antigen severely decreased its activity in the initiation of viral DNA replication also in this experiment. The synthesis of a reporter plasmid, containing an origin of DNA replication, in cells transfected with pcDNA3/LT-L13V/E15D was only 1% of the amount obtained in cells expressing wild-type or L14V/E15D large T antigen. To determine the quantity of large T antigen in the cells, protein was extracted from parallel, transfected cultures and was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Analysis of large T antigen by immunoblotting showed (Fig. 2D) that the amount of immunoreactive protein in cells expressing the L13V mutant protein was slightly lower than the corresponding amounts of wild-type and L14V mutant protein. However, this difference was too small to explain the large effect of the L13V substitution on viral DNA replication. Immunofluorescence analysis using MAb LT1 showed that both L13V and L14V large T antigens were located in the nucleus (data not shown). Hence, large T antigen with the L13V substitution appeared to have lost most of its activity in the initiation of viral DNA synthesis in both 3T6 and NIH 3T3 cells.
Attempts to rescue the replication function of mutant large T antigens.
Besides controlling the initiation of each round of viral DNA synthesis, large T antigen participates in the induction of cellular DNA synthesis that is a prerequisite for the viral replication. If the substitution of amino acid residue 13 or 14 inhibited the binding of large T antigen to the origin of viral DNA replication, or any ensuing cis activity in viral DNA synthesis, then other T-antigen proteins would probably be unable to rescue that function. If, on the other hand, a mutant large T antigen was primarily deficient in the perturbation of cell cycle control, rescue of function might be possible.
Small t antigen enhances viral DNA replication in 3T6 mouse fibroblasts. The mechanism is unknown but may relate to site-specific phosphorylation of large T antigen required for its activity in viral DNA replication. To investigate the effect on the activity of the four crt mutant large T antigens, 3T6 cells were cotransfected with ST-1, expressing wild-type small t antigen, and each of the plasmids encoding wild-type or crt mutant large T antigen. The amounts of viral DNA formed during 40 h following transfection are shown in Fig. 2B. Small t antigen increased viral DNA synthesis in all cases. The synthesis of crtL13I/E15D/dl1061, crtL13M/E15D/dl1061, and crtL14V/E15D/dl1061 reached almost the same level as that of dl1061 DNA. In contrast, mutant crtL13V/E15D/dl1061 remained at 0.1% of the control level. However, the activity of this mutant large T antigen also was stimulated by small t antigen. We showed earlier (50) that mutant large-T-antigen polypeptides with substitutions in the pRb binding site (-DLFCYE-) at amino acid residues 141 to 146 were partially defective in viral DNA synthesis. In these cases cotransfection with a genome expressing a 287-amino-acid-residue N-terminal fragment of large T antigen but with a normal pRb binding site overcame the deficiency. Here we investigated whether this truncated large T antigen (mu1355/dl1061) had a stimulatory effect on the viral DNA synthesis of the four crt mutant forms of the protein. Mouse 3T6 cells were transfected with the two types of DNA, and the amount of newly synthesized viral DNA was determined. The result showed (Fig. 2B) that the activity in viral DNA replication of the crt mutant large T antigens was not increased by the coexpression of the mu1355 large T antigen. Hence the L13V substitution in large T antigen appeared to have a direct effect on the activity of the protein in viral DNA replication. However, this experiment does not exclude additional effects of the mutation on large-T-antigen activities.
Binding of crt mutant large T antigen to Hsc70 and pRb.
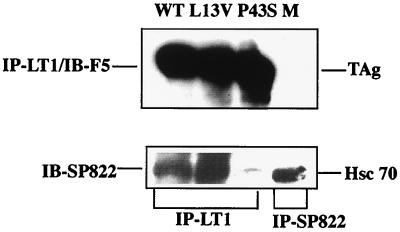
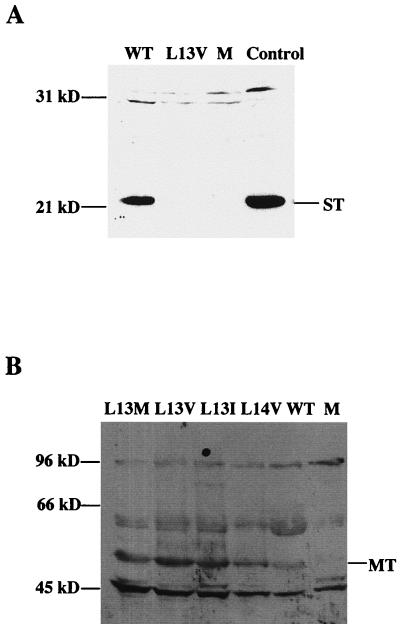
To investigate whether the L13V substitution affected the DnaJ activity of large T antigen, binding to the Hsc70 protein was tested. NIH 3T3 cells that have a high constitutive expression of this polypeptide were used for the analysis. The cells were transfected with plasmids expressing wild-type or mutant crtL13V/E15D large T antigen. As a negative control, a plasmid, pcDNA3/LT-P43S, was used that encoded a mutant large T antigen with a P43S substitution in the universally conserved -HPD motif. This mutant large T antigen does not form a stable complex with Hsc70 (8, 49). At 42 h posttransfection, protein was extracted and the cleared lysates were immunoprecipitated with either MAb LT1 directed against large T antigen or MAb SP822 directed against Hsc70. After SDS-PAGE, protein was transferred to nitrocellulose membranes by blotting. The membranes were then developed with MAb F5 directed against large T antigen or MAb SP822. Chemiluminograms show (Fig. 3) that cells transfected with plasmids encoding wild-type, mutant crtL13V/E15D, and crtP43S large T antigens expressed easily detectable amounts of the large-T-antigen protein. Moreover, there was no apparent difference in the association of wild-type and mutant crtL13V/E15D large T antigen with Hsc70. In contrast, in extracts of cells transfected with pcDNA3/LT-P43S there was no detectable Hsc70 protein coimmunoprecipitated with large T antigen.
FIG. 3.
Complex formation of mutant and wild-type large T antigen with Hsc70. Cultures of NIH 3T3 cells (5 × 105 cells) were transfected, using Lipofectamine, with plasmid pcDNA3/LT-wt (WT), -L13V/E15D (L13V), or P43S encoding large T antigen. As a negative control, cells were transfected with pcDNA3 without an insert (M). At 42 h posttransfection, cell extracts were prepared and incubated with either MAb LT1 (anti-large T antigen) or MAb SP822 (anti-Hsc70). Immunoprecipitated (IP) material was resolved by SDS-PAGE followed by immunoblotting, using either MAb F5 (anti-LT) or MAb SP822. The positions in the gel of large T antigen (TAg) and Hsc70 relative to markers with known size are indicated.
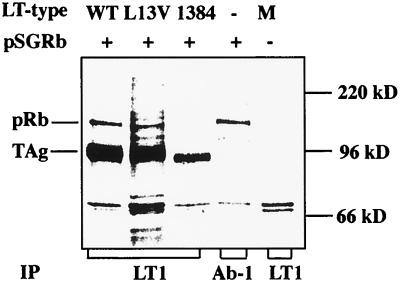
The -DLFCYE- segment of large T antigen (amino acid residues 142 to 146) binds to the Rb pocket family of proteins. However, for some functional interactions between large T antigen and the pRbs, a functional J domain of the former protein is also required (48, 49, 57). To test whether large T antigen with the L13V substitution bound to pRb, we cotransfected cells with one plasmid encoding pRb (pSGRb) and a second plasmid encoding wild-type or mutant crtL13V/E15D large T antigen. As a negative control, the mu1384 large T antigen, with amino acid residues 143 to 146 deleted, was used (50). At 42 h posttransfection, [35S]methionine was added to the cultures and protein was labeled for 4 h. After lysis of the cells, large T antigen was immunoprecipitated with MAb LT1, and pRb was immunoprecipitated with MAb Ab1 from the extract of cells transfected with the pRb-encoding plasmid alone. Polypeptides were resolved by SDS-PAGE and then analyzed by autoradiography (Fig. 4). Extracts of cells transfected with plasmids encoding wild-type or mutant large T antigen contained a 90-kDa polypeptide reacting with MAb LT1. Its electrophoretic mobility was somewhat heterogenous, consistent with the known modifications of large T antigen in mammalian cells (4, 22). A ca. 105-kDa polypeptide coimmunoprecipitated with wild-type and mutant crtL13V/E15D large T antigen but not with the mu1384 protein. A similar ca. 105-kDa polypeptide, reacting with MAb Ab1, was observed in cells transfected with only pSGRb, but was below the detection level in untransfected cells. Together, the data indicated that wild-type and mutant crtL13V/E15D large T antigen but not mu1384 large T antigen coimmunoprecipitated with pRb. Thus, the L13V substitution did not impair the formation of large T antigen-pRb complexes.
FIG. 4.
Complex formation of wild-type and mutant large T antigen with pRb. Cultures of NIH 3T3 cells were transfected, using Lipofectamine, with plasmid pSGRb (2.0 μg) alone or mixed with 1.0 μg of pcDNA3/LT-wt (WT), -L13V/E15D (L13V), or dl1384 (1384), encoding large T antigen. As a control (M), cells were transfected with pcDNA3 without an insert. At 42 h posttransfection, cells were labeled for 4 h with [35S]methionine. Protein extracts were prepared, and immunoprecipitation (IP) was done with MAb LT1 directed against large T antigen or with MAb Ab-1 directed against pRb. Immunoprecipitated material was resolved by SDS-PAGE in an 8% gel, and radioactivity was detected by autoradiography. The positions in the gel of large T antigen (TAg at 90 kDa) and pRb (105 kDa) relative to markers with known size are indicated.
Transcriptional transactivation by crt mutant large T antigens.
Polyomavirus large T antigen is able to transactivate transcription by more than one mechanism. The transcription factor E2F is activated by release from the Rb family of proteins after binding of large T antigen (39). Mutant large T antigens with amino acid substitutions in the conserved loop of the J domain were shown to bind to pRb but were defective in the induction of E2F release (48, 57). A second, for the polyomavirus protein as yet uncharacterized, transactivation mechanism operates on various transcription units, including the viral late genes (6, 29, 34). This mechanism may involve binding of the cellular p300 to the C-terminal part of large T antigen (33, 38). To test the ability of our crt mutant large T antigens in transcription transactivation, they were expressed together with reporter plasmids containing the adenovirus E2 (pE2cat) or polyomavirus late promoter (pPYLcat) upstream of the cat gene. To ensure that wild-type pRb was expressed, secondary REF cells were used in this experiment. They were transfected with a recombinant plasmid encoding wild-type or mutant large T antigen mixed with the plasmid carrying the reporter gene. As a negative control for large T antigen expression, we used the plasmid pPYΔE1, which has most of the T-antigen-coding sequences deleted but retains the regulatory elements of the viral genome. Under the conditions of the experiment, CAT expression from the polyomavirus late promoter (Fig. 5A) was relatively high even in the absence of large T antigen. Coexpression of wild-type large T antigen or any of the four mutant forms stimulated the activity of the viral late promoter two- to fourfold. The small differences in the activity of the wild-type and various mutant large T antigens are probably not significant. The level of CAT expression from the adenovirus E2 promoter was low in REF cells (Fig. 5B). However, in the presence of large T antigen it was stimulated four- to sevenfold. The wild-type and all four mutant forms of large T antigen had similar effects on the activity of the E2 promoter. Hence the substitutions of amino acid residues 13 and 14 of large T antigen did not impair the DnaJ function of the protein that is required for a functional interaction with pRb to release the transcription factor E2F (48).
FIG. 5.
Effect of wild-type and mutant large T antigen on the polyomavirus late promoter and the adenovirus E2 promoter. Growing REF cultures (5 × 105 cells) were transfected, using Lipofectamine, with 1.0 μg of pPYLcat (A) or pE2cat (B) DNA mixed with 1.0 μg of polyomavirus DNA cloned in plasmid pML. As a control to expression of wild-type and mutant large T antigen, the mutant PYΔE1 with a deletion of the early region was used. Cell extracts were prepared at 40 h posttransfection, and CAT activity was assayed. The 14C-labeled chloramphenicol substrate and the acetylated products were separated by thin layer chromatography. The radioactive spots were identified by autoradiography and quantified in a phosphorimager. The relative enzyme activity in the absence of large-T-antigen expression was set to 1. Wild-type and mutant large T antigen (LT) are denoted by WT and the amino acid substitutions, respectively.
Stability of mutant crtL13V small and middle T antigen.
An analysis of the small-t-antigen activity on large-T-antigen-dependent viral DNA synthesis showed that the L13M, L13I, and L14 substitutions did not impair this function (the activity of wild-type small t antigen is shown in Fig. 2B). However, the crtL13V/E15D small t antigen was completely inactive (data not shown). To test whether this result was due to instability of the mutant small t antigen, the expression plasmid pcDNA3/ST-L13V/E15D was constructed. NIH 3T3 cells were transfected with this plasmid or pcDNA3/ST-wt. As a negative control, pcDNA3 vector DNA without an insert was used. At 42 h posttransfection, cells were lysed and extracted protein was analyzed by immunoblotting using MAb F4. The result shows (Fig. 6A) that the cells did not contain detectable amounts of the mutant crtL13V/E15D small t antigen, although the wild-type protein was easily detectable. Together with the activity determination of mutant crtL13V/E15D/ST-1, the data indicated that a substitution at position 13 of leucine for valine, but not for isoleucine or methionine, made small t antigen labile.
FIG. 6.
Stability of mutant small and middle T antigens. (A) Cultures of NIH 3T3 cells were transfected as described in the legend to Fig. 5. At 42 h posttransfection, cell extracts were prepared and subjected to SDS-PAGE in a 12% gel, followed by immunoblotting using MAb F4 reacting with small t antigen. Lane M, material from cells transfected with pcDNA3 without insert; control lane, small t antigen produced in insect cells. (B) FR3T3 cells were transfected, using the calcium phosphate coprecipitation method with MT-1 DNA, encoding wild-type protein or mutant protein with the indicated substitutions at residues 13 and 14, respectively. Clones of transformed cells were isolated, and extracts were prepared from cultures of these cloned cells. Proteins were resolved by SDS-PAGE in an 8% gel, and T antigen was identified by immunoblotting using MAb F5. Lane M, extract of untransformed FR3T3 cells. The positions in gels of middle T antigen (MT), small T antigen (ST), and of markers with known sizes are indicated.
The lability of small t antigen with the L13V substitution raised the question of whether it had a similar destabilizing effect on middle T antigen, since the sequences of the N-terminal 192 amino acid residues of the two proteins are colinear. The activity of middle T antigen in cellular transformation reflects its overall activity. Therefore, the mutant derivatives of polyomavirus MT-1 DNA were used for transfection of FR3T3 cells. Two days after transfection, the cells were replated at a fivefold-lower density, and after another 10 days, foci of cells were isolated by trypsinization in glass cylinders. Clones of transformed cells were obtained at similar yields with wild-type and all four crt mutant DNAs. Protein was extracted from cells of individual clones and was analyzed by immunoblotting using MAb F5, which recognizes middle T antigen. The experiment showed (Fig. 6B) that all the clones of transformed cells, but not the negative control, contained immunoreactive material with an electrophoretic mobility corresponding to 55 kDa. This result indicated that the L13V substitution did not have any negative effect on the stability or activity of middle T antigen.
DISCUSSION
Processing of mouse polyomavirus early RNA by differential splicing leads to four types of mRNA which share a 5′-terminal exon segment. Therefore, the four translation products, the T antigens, have identical N-terminal parts consisting of 79 amino acid residues. Most of this segment forms a structure homologous to the DnaJ domain (9, 28). Within this part of the T antigens, two short amino acid sequences are highly conserved in the polypeptides encoded by all known polyomaviruses. One is the -HPDKGG- motif containing the J box. The other is highly conserved in T antigens but not in other DnaJ proteins. Amino acid residues 11 to 20 of polyomavirus T antigens are predicted (30) to form an alpha helix, corresponding to helix I of the DnaJ protein. This putative helix is longer (residues 10 to 19) than the homologous structure in the human Hsp40 protein HDJ-1 (residues 5 to 10). In the alpha helix I of HDJ-1, the predicted structure was confirmed by nuclear magnetic resonance analysis (45). In large T antigen, leucine residue 13, which is sensitive to substitution, would be located in alpha helix I or just outside this structure if it has the same size as in the HJD-1 protein. The high sequence conservation of this T-antigen segment suggests that it has a strongly selected function. However, this function is not necessarily related to DnaJ activity. Interestingly, the conserved leucine-rich motif is related to the conserved region 1 of the adenovirus E1A protein [(E/D)X3LX(E/D)LX2(L/I)], which is known to participate in binding to several cellular proteins, such as pRb, Cdk2, and p300, also called CREB-binding protein. (16, 54). To study the function of the leucine-rich segment in polyomavirus T antigens that corresponds to alpha helix I, we introduced minimal amino acid changes that were unlikely to disrupt its potential helical structure. In mutagenesis of codons for amino acid residues 13 and 14 in the T antigens, we selected conservative shifts to residues which were not represented in the corresponding polypeptides of any known polyomavirus (Fig. 1). L13 was completely conserved in T antigens. Thus, we made the crt mutants L13I, L13M, and L13V. Position L14 is less conserved, but a valine residue has not been observed at this position in any of the known T antigens. Therefore, we made the mutant L14V. All these mutants were constructed with DNA containing the crtE15D mutation that results in a unique restriction endonuclease cleavage site. Since an aspartic acid residue is present at this position in the T antigens of the human and simian polyomaviruses, we did not anticipate any phenotypic effect from the replacement of the glutamic acid residue. This assumption was supported by several experiments that showed normal T-antigen functions of the crtE15D mutant (data not shown). To test the effect of the other mutations on individual polyomavirus T antigens, the crt mutations were combined with deletion mutants that restricted splicing of the viral early RNA. Effects on viral DNA synthesis transcriptional transactivation, binding to Hsc70 and pRb, and cellular immortalization were then analyzed in transfection experiments. We also analyzed the effect of the mutations on the function of middle T antigen. However, in keeping with earlier studies of deletion mutants (7, 19), the J domain of middle T antigen appeared to have little influence on the known functions of the protein. Previous analyses of the DnaJ domain of T antigens have been focused on the interaction with DnaK homologs, such as Hsp70 and Hsc70, and the specificity of interactions with cellular targets (8, 11). The J domain of T antigens can functionally substitute for DnaJ in Escherichia coli cells (27). It binds to Hsc70 in mammalian cells (8, 48) and interacts with this protein by stimulating its ATPase activity (47). The cooperation with Hsc70 or its homologs is required for functional interactions with other cellular polypeptides, such as pRb (48, 53, 57).
Mutation of the highly conserved J box (-HPD-) impairs the function of simian virus 40 large T antigen in initiation of viral DNA synthesis (8). A nonnuclear localization might explain this phenotype, since DnaK proteins have been reported to mediate nuclear translocation (41). However, Sheng et al. (48) showed that J-box-defective large T antigen had a nuclear localization. Large T antigens with substitutions of amino acid residue 13 or 14 also accumulated in the cell nucleus, as shown by immunofluorescence (data not shown). The defect of the protein with an alteration of the HPD box is probably in one or several of the numerous protein-protein interactions that are involved in the initiation process of DNA replication. The crt mutants analyzed in the present study produce T antigens with a normal J box. However, even one of the conservative amino acid substitutions in alpha helix I of the J domain, L13V, caused a distinctive phenotype in viral DNA synthesis. The impairment of viral DNA replication was traced to defects of both large (Fig. 2) and small (Fig. 6A) T antigen. In large T antigen the L13V substitution had a profound effect on viral DNA synthesis that was not rescuable by coexpression of a truncated form of the polypeptide with an intact J domain. This N-terminal fragment of large T antigen has been shown to stimulate cellular and viral DNA synthesis by advancing the cell cycle (50). The activity of crtL13V/E15D large T antigen was stimulated by coexpression of small t antigen (Fig. 2B). However, in relation to the activity of the wild-type protein, the defect of crtL13V/E15D large T antigen remained. Hence it is probable that large T antigen with the L13V substitution was impaired in its interaction with viral DNA or cellular replication factors. However, purified crtL13V/E15D large T antigen had normal DNA binding and unwinding activities (unpublished data). The polyomavirus crtL13V large T antigen was recently reported to be unstable when expressed in both mouse and rat cells (32). In our investigation, the L13V substitution had little effect on the stability of large T antigen in NIH 3T3 cells (Fig. 2C and D). Moreover, analysis of stability in BHK cells, following a pulse-chase protocol, showed that both wild-type and L13V/E15D large T antigens had half-lives of approximately 8 h (data not shown). It is possible that the combination of L13V and E15D substitutions resulted in a more stable protein than the isolated L13V substitution. Paradoxically, a C-terminal fragment of large T antigen, devoid of the DnaJ domain, is able to support viral DNA. However, it is probable that the full-length and truncated versions of the protein behave differently in the assembly of DNA replication complexes, providing an explanation for the involvement of the DnaJ structure in viral DNA synthesis.
Small t antigen stimulates viral DNA synthesis, at least in some types of mouse cells (2, 37). The simian virus 40 small t antigen has been shown to transactivate the cyclin A gene, and mutation of the J box disturbs this effect (44). Of the four mutants producing T antigens with amino acid substitutions of 13L and 14L, only the crtL13V/E15D small t antigen was defective in stimulating viral DNA synthesis. However, this phenotype was linked to instability of the protein (Fig. 6A). Since neither large nor middle T antigen was destabilized by the L13V substitution, the unique part of the proteins probably influences the structure of the DnaJ domain and its stability.
The effect of the L13V amino acid substitution on the function of large T antigen in viral DNA replication raised the question of whether the observed defect was in DnaJ activity on DnaK-like proteins. Since mutation of the J box results in loss of binding to Hsc70 and transcription factor E2F activation, we investigated whether the L13V and P43S substitutions in large T antigen caused similar defects. The experiment was done with NIH 3T3 cells, which have a high constitutive level of Hsc70. In a coimmunoprecipitation experiment there was no difference in binding to Hsc70 of the wild-type large T antigen and the protein with the L13V substitution. In contrast, large T antigen with the P43S substitution in the J box did not form a stable complex with Hsc70 (Fig. 3). This result suggests that conservative substitutions in alpha helix I of large T antigen were not critical for the interaction with Hsc70.
Binding of the pRb to large T antigen does not occur unless the -DLFCYE- segment (residues 141 to 146) is present (17, 31, 50). However, for displacement from pRb of the transcription factor E2F, the DnaJ domain of large T antigen has to be functional. In contrast, transactivation by large T antigen of the polyomavirus late promoter is independent of pRb binding, since the -DLFCY- motif in the protein is not required for this function (50). The cellular pRb formed a complex with crtL13V/E15D large T antigen but not with the truncated dl1384 protein lacking residues 143 to 146 (Fig. 4). The large T antigen with the L13V substitution also induced activation of the transcription factor E2F, since all the crt mutants (L13M, L13I, L13V, and L14V) were positive in transactivation of both the E2F-regulated E2 promoter from adenovirus and the polyomavirus late promoter (Fig. 5). Analysis of cellular immortalization corroborated the conclusion that E2F was activated, since lines of secondary REFs were established after transfection with genomes expressing each of the four mutations (data not shown).
Our data show that the four tested mutants have a DnaJ function, as analyzed by interactions of large T antigen with Hsc70 and pRb. However, the L13V substitution in large T antigen caused a distinct defect of the protein in the initiation of viral DNA replication. Since the amino acid replacement mapped in the core of the DnaJ domain, the functional defect must by definition reflect a DnaJ activity. Thus, we propose that the DnaJ domain of polyomavirus T antigens has more than one specificity for the interaction with other proteins. Support for this idea is provided by the work of Pipas (5), which shows that the L-to-F substitution of conserved residue 19 in simian virus 40 large T antigen (Fig. 1), which corresponds to residue 19 in the polyomavirus protein, did not inhibit viral DNA synthesis but instead abolished the activity of the protein in cellular transformation and in assembly of virus particles.
After completion of this paper, a paper by Berjanskii et al. was made public (3). These investigators demonstrated that residues 13 and 14 of polyomavirus T antigens form part of the J-domain alpha helix I. It was also shown that the L13V substitution in an N-terminal fragment of T antigen inactivated its DnaJ function in a complementation assay performed with E. coli cells, possibly due to disruption of alpha helix I. In the reported structure, the side chain of residue 13 is buried in the helix.
ACKNOWLEDGMENT
The experiments described in this report were supported financially by the Swedish Cancer Society.
REFERENCES
- 1.Asselin C, Bastin M. Sequences from polyomavirus and simian virus 40 large T genes capable of immortalizing primary rat embryo fibroblasts. J Virol. 1985;56:958–968. doi: 10.1128/jvi.56.3.958-968.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger H, Wintersberger E. Polyomavirus small T antigen enhances replication of viral genomes in 3T6 mouse fibroblasts. J Virol. 1986;60:768–770. doi: 10.1128/jvi.60.2.768-770.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berjanskii M V, Riley M I, Xie A, Semenchenko V, Folk W R, Van Doren S R. NMR structure of the N-terminal J domain of murine polyomavirus T antigens: implications for DnaJ-like domains and for mutations of T antigens. J Biol Chem, 2000;275:36094–36103. doi: 10.1074/jbc.M006572200. [DOI] [PubMed] [Google Scholar]
- 4.Bockus B J, Schaffhausen B. Localization of the phosphorylations of polyomavirus large T antigen. J Virol. 1987;61:1155–1163. doi: 10.1128/jvi.61.4.1155-1163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky J L, Pipas J M. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahill K B, Roome A J, Carmichael G G. Replication-dependent transactivation of the polyomavirus late promoter. J Virol. 1990;64:992–1001. doi: 10.1128/jvi.64.3.992-1001.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell K S, Auger K R, Hemmings B A, Roberts T M, Pallas D C. Identification of regions in polyomavirus middle T and small t antigens important for association with protein phosphatase 2A. J Virol. 1995;69:3721–3728. doi: 10.1128/jvi.69.6.3721-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham M E, Caplan A J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowie A, de Villiers J, Kamen R. Immortalization of rat embryo fibroblasts by mutant polyomavirus large T antigens deficient in DNA binding. Mol Cell Biol. 1986;6:4344–4352. doi: 10.1128/mcb.6.12.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCaprio J A. The role of the J domain of SV40 large T in cellular transformation. Biologicals. 1999;27:23–28. doi: 10.1006/biol.1998.0173. [DOI] [PubMed] [Google Scholar]
- 12.Dilworth S M. Polyoma virus middle T antigen: meddler or mimic? Trends Microbiol. 1995;3:31–35. doi: 10.1016/s0966-842x(00)88866-6. [DOI] [PubMed] [Google Scholar]
- 13.Dilworth S M, Griffin B E. Monoclonal antibodies against polyoma virus tumor antigens. Proc Natl Acad Sci USA. 1982;79:1059–1063. doi: 10.1073/pnas.79.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 17.Freund R, Bauer P H, Crissman H A, Bradbury E M, Benjamin T L. Host range and cell cycle activation properties of polyomavirus large T-antigen mutants defective in pRB binding. J Virol. 1994;68:7227–7234. doi: 10.1128/jvi.68.11.7227-7234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gjørup O V, Rose P E, Holman P S, Bockus B J, Schaffhausen B S. Protein domains connect cell cycle stimulation directly to initiation of DNA replication. Proc Natl Acad Sci USA. 1994;91:12125–12129. doi: 10.1073/pnas.91.25.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glenn G M, Eckhart W. Amino-terminal regions of polyomavirus middle T antigen are required for interactions with protein phosphatase 2A. J Virol. 1995;69:3729–3736. doi: 10.1128/jvi.69.6.3729-3736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene M K, Maskos K, Landry S J. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassauer M, Scheidtmann K H, Walter G. Mapping of phosphorylation sites in polyomavirus large T antigen. J Virol. 1986;58:805–816. doi: 10.1128/jvi.58.3.805-816.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbomel P, Bourachot B, Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984;39:653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- 24.Jat P S, Sharp P A. Cell lines established by a temperature-sensitive simian virus 40 large-T-antigen gene are growth restricted at the nonpermissive temperature. Mol Cell Biol. 1989;9:1672–1681. doi: 10.1128/mcb.9.4.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jat P S, Sharp P A. Large T antigens of simian virus 40 and polyomavirus efficiently establish primary fibroblasts. J Virol. 1986;59:746–750. doi: 10.1128/jvi.59.3.746-750.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan D R, Pallas D C, Morgan W, Schaffhausen B, Roberts T M. Mechanisms of transformation by polyoma virus middle T antigen. Biochim Biophys Acta. 1989;948:345–364. doi: 10.1016/0304-419x(89)90006-1. [DOI] [PubMed] [Google Scholar]
- 27.Kelley W L, Georgopoulos C. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley W L, Landry S J. Chaperone power in a virus? Trends Biochem Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 29.Kern F G, Pellegrini S, Cowie A, Basilico C. Regulation of polyomavirus late promoter activity by viral early proteins. J Virol. 1986;60:275–285. doi: 10.1128/jvi.60.1.275-285.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kneller D G, Cohen F E, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 31.Larose A, Dyson N, Sullivan M, Harlow E, Bastin M. Polyomavirus large T mutants affected in retinoblastoma protein binding are defective in immortalization. J Virol. 1991;65:2308–2313. doi: 10.1128/jvi.65.5.2308-2313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemieux B, Bastin M. Polyomavirus large T antigen mutants affected in viral DNA replication. Virology. 2000;269:370–376. doi: 10.1006/viro.2000.0231. [DOI] [PubMed] [Google Scholar]
- 33.Lill N L, Tevethia M J, Eckner R, Livingston D M, Modjtahedi N. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J Virol. 1997;71:129–137. doi: 10.1128/jvi.71.1.129-137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linder S, Nilsson M, Martens I, Magnusson G. A viable mouse polyomavirus mutant without immortalizing or transforming activities. Virology. 1990;179:78–86. doi: 10.1016/0042-6822(90)90276-w. [DOI] [PubMed] [Google Scholar]
- 35.Lusky M, Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature (London) 1981;293:79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- 36.Luthman H, Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983;11:1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martens I, Nilsson S A, Linder S, Magnusson G. Mutational analysis of polyomavirus small-T-antigen functions in productive infection and in transformation. J Virol. 1989;63:2126–2133. doi: 10.1128/jvi.63.5.2126-2133.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nemethova M, Wintersberger E. Polyomavirus large T antigen binds the transcriptional coactivator protein p300. J Virol. 1999;73:1734–1739. doi: 10.1128/jvi.73.2.1734-1739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson S V, Magnusson G. T-antigen expression by polyoma mutants with modified RNA splicing. EMBO J. 1983;2:2095–2101. doi: 10.1002/j.1460-2075.1983.tb01708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuno Y, Imamoto N, Yoneda Y. 70-kDa heat-shock cognate protein colocalizes with karyophilic proteins into the nucleus during their transport in vitro. Exp Cell Res. 1993;206:134–142. doi: 10.1006/excr.1993.1129. [DOI] [PubMed] [Google Scholar]
- 42.Pallas D C, Schley C, Mahoney M, Harlow E, Schaffhausen B S, Roberts T M. Polyomavirus small t antigen: overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J Virol. 1986;60:1075–1084. doi: 10.1128/jvi.60.3.1075-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallas D C, Shahrik L K, Martin B L, Jaspers S, Miller T B, Brautigan D L, Roberts T M. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 44.Porras A, Bennett J, Howe A, Tokos K, Bouck N, Henglein B, Sathyamangalam S, Thimmapaya B, Rundell K. A novel simian virus 40 early-region domain mediates transactivation of the cyclin A promoter by small-t antigen and is required for transformation in small-t antigen-dependent assays. J Virol. 1996;70:6902–6908. doi: 10.1128/jvi.70.10.6902-6908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian Y Q, Patel D, Hartl F U, McColl D J. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol. 1996;260:224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- 46.Rassoulzadegan M, Naghashfar Z, Cowie A, Carr A, Grisoni M, Kamen R, Cuzin F. Expression of the large T protein of polyoma virus promotes the establishment in culture of “normal” rodent fibroblast cell lines. Proc Natl Acad Sci USA. 1983;80:4354–4358. doi: 10.1073/pnas.80.14.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riley M I, Yoo W, Mda N Y, Folk W R. Tiny T antigen: an autonomous polyomavirus T antigen amino-terminal domain. J Virol. 1997;71:6068–6074. doi: 10.1128/jvi.71.8.6068-6074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng Q, Love T M, Schaffhausen B. J domain-independent regulation of the Rb family by polyomavirus large T antigen. J Virol. 2000;74:5280–5290. doi: 10.1128/jvi.74.11.5280-5290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soderbarg K, Magnusson G. Lytic functions of mutant polyomavirus large T-antigen with deletion of retinoblastoma protein-binding motif. Virology. 1993;193:281–288. doi: 10.1006/viro.1993.1123. [DOI] [PubMed] [Google Scholar]
- 51.Soeda E, Arrand J R, Smolar N, Walsh J E, Griffin B E. Coding potential and regulatory signals of the polyoma virus genome. Nature (London) 1980;283:445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- 52.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 53.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H G, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weeks D L, Jones N C. E1A control of gene expression is mediated by sequences 5′ to the transcriptional starts of the early viral genes. Mol Cell Biol. 1983;3:1222–1234. doi: 10.1128/mcb.3.7.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Z Y, Veldman G M, Cowie A, Carr A, Schaffhausen B, Kamen R. Construction and functional characterization of polyomavirus genomes that separately encode the three early proteins. J Virol. 1984;51:170–180. doi: 10.1128/jvi.51.1.170-180.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]