The number of therapeutic options continues to increase for men with high-risk localized prostate cancer [1]. Risk stratification within a given National Comprehensive Cancer Network (NCCN) risk group remains challenging [2]. Arter-aAI Prostate, a multimodal artificial intelligence (MMAI) prognostic biomarker, was developed and validated using data from randomized trials in localized prostate cancer for prognostication of multiple clinically relevant endpoints [3]. Here we report the performance of this MMAI biomarker for men with high-risk prostate cancer from six phase 3 randomized trials.
After approval from NRG Oncology, histopathology slides from six phase 3 randomized trials that included men with prostate cancer with at least one NCCN high/very high (H/VH) risk factor were collected from the NRG/RTOG biobank and digitized. The six trials included the four randomized trials used for the original MMAI validation cohort (NRG/RTOG 9202, 9408, 9413, and 9910; n = 426) and two independent trials that were not part of the initial MMAI development and validation process (NRG/RTOG 0521, n = 344; NRG/RTOG 9902, n = 318).
Digitized images were combined with clinical information to generate an MMAI score from the locked model for each patient (Supplementary material). Fine-Gray and cumulative incidence analyses were performed to evaluate time to distant metastasis (DM), prostate cancer-specific mortality (PCSM), and death with DM (DDM) stratified by the MMAI model score, both as a continuous score (per increment in standard deviation) and categorically by quartile. Multivariable analyses (MVAs) were conducted to demonstrate the independent effect of the MMAI model and the number of NCCN H/VH risk factors (defined as cT3–4, Gleason 8–10, prostate-specific antigen [PSA] >20 ng/ml, and primary Gleason pattern 5). Other clinical variables (age; PSA; primary, secondary, and Gleason sum score; T stage) were not included in the primary MVA as they are included in the MMAI model, but were included in supplementary MVAs to further characterize the effect of the MMAI. We calculated the absolute difference in estimated event rates, and the associated 95% confidence intervals (CIs) were determined via bootstrapping 10 000 times. Death from other causes was treated as a competing risk. The study adhered to the TRIPOD reporting requirements for prognostic studies [4].
The high-risk validation cohort (n = 1088) had median follow-up of 10.4 yr (interquartile range [IQR] 9.1–11.8). The median PSA was 21.4 ng/ml (IQR 9.5–39.3), 60% had Gleason 8–10 disease, 37% had cT3–4 disease, and 20% were Black (Supplementary Table 1). On MVA, after adjusting for the number of H/VH risk features (n = 1034), the MMAI model remained significantly associated with DM (subdistribution hazard ratio [sHR] 1.90, 95% CI 1.57–2.31; p < 0.001), PCSM (sHR 2.06, 95% CI 1.67–2.54; p < 0.001), and DDM (sHR 2.12, 95% CI 1.72–2.62; p < 0.001; Fig. 1A). The MMAI model also remained significantly associated with DM after adjusting for individual clinical variables (Supplementary Table 3).
Fig. 1 –
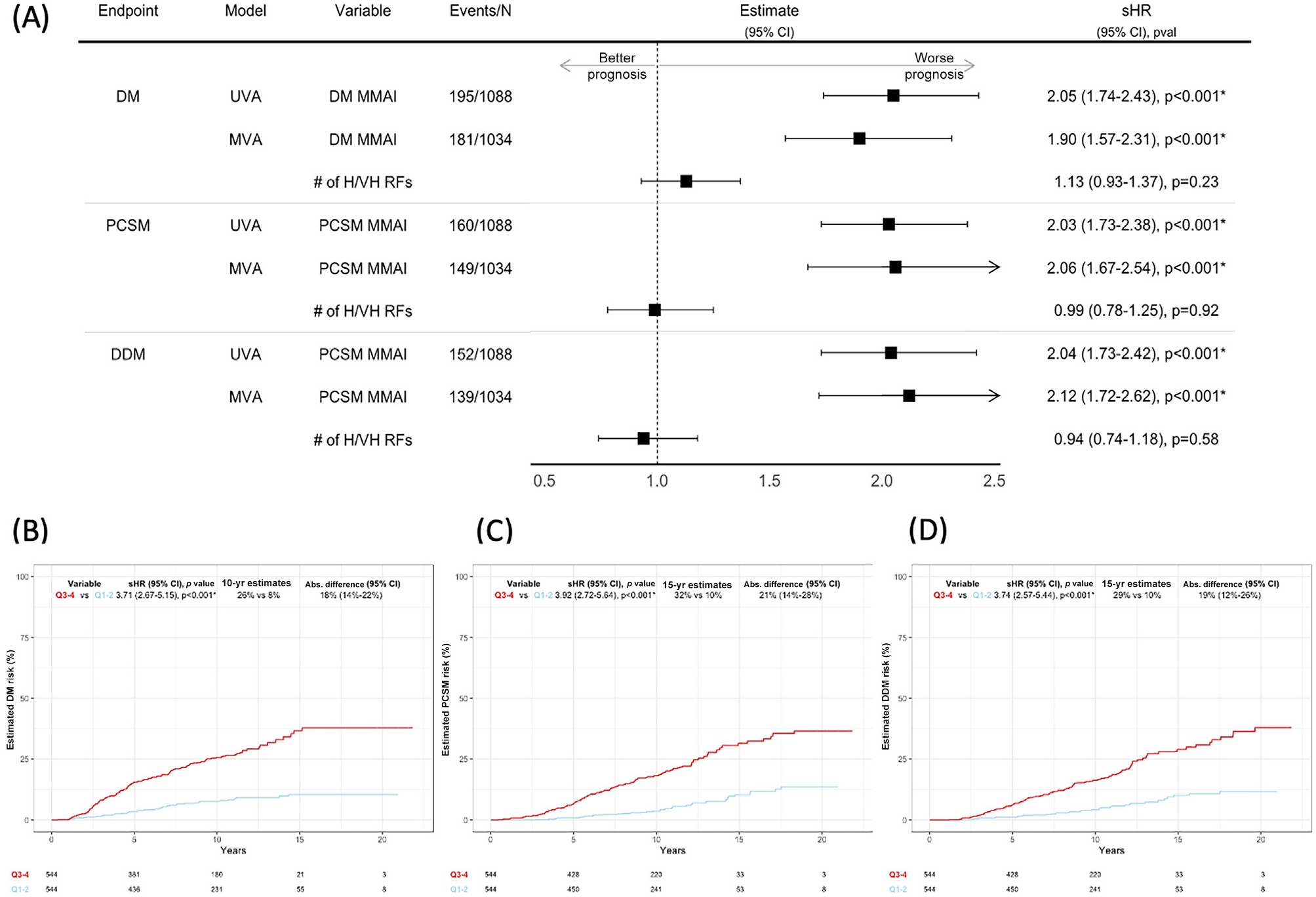
(A) UVA of the association between the MMAI score and DM and MVA of the association between the MMAI score and DM after adjustment for the number of H/VH risk factors. (B) Estimated 10-yr DM rates for MMAI Q1–2 versus Q3–4. (B) Estimated 15-yr PCSM rates for MMAI Q1–2 versus Q3–4. (D) Estimated 15-yr DDM rates for MMAI Q1–2 versus Q3–4. Abs. = absolute; CI = confidence interval; DDM = death with DM; DM = distant metastasis; MMAI = H/VH = high/very high; MMAI = multimodal artificial intelligence; MVA = multivariable analysis; PCSM = prostate cancer–specific mortality; Q = quartile; RF = risk factor; sHR = subdistribution hazard ratio; UVA = univariable analysis.
The estimated 10-year DM risk was 8% for MMAI quartiles Q1–2 versus 26% for MMAI Q3–4, with a total absolute difference of 18% (95% CI 14–22%; Fig. 1B). There were similar findings for PCSM and DDM (Fig. 1C,D).
The MMAI prognostic biomarker was validated using data for men with high-risk prostate cancer within six phase 3 randomized trials and was independently prognostic over standard clinical and pathologic variables. Although patients had at least one NCCN H/VH risk factor, the MMAI biomarker identified those with highly variable risks for DM, PCSM, and DDM. Future efforts could extend this work beyond prognostication by evaluating how the MMAI biomarker interacts with specific treatments. This scalable tool can provide physicians and patients with more personalized information for shared decision-making on treatment [5].
The trials from which data were used are registered on ClinicalTrials.gov as NCT00004054, NCT00288080, NCT00767286, NCT00002597, NCT00769548, and NCT00005044.
This study was presented at the 2023 American Society of Clinical Oncology Genitourinary Cancers Symposium 2023.
Supplementary Material
Funding/Support and role of the sponsor:
This project was supported by grants UG1CA189867 (NCI Community Oncology Research Program), U10CA180868 (NRG Oncology Operations), U24CA196067 (NRG Specimen Bank), U10CA180822 (NRG Oncology SDMC) from the National Cancer Institute, Pfizer, Bristol-Myers Squibb, Sanofi, Takeda Pharmaceutical. Daniel E. Spratt is supported by grants U01 CA257638 and R21 CA274500 from the National Institutes of Health, the Vincent K. Smith Endowed Chair through the University Hospitals Seidman Cancer Center, and the Prostate Cancer Foundation.
Conflicts of interest:
Stephanie L. Pugh has nothing to disclose. Felix Y. Feng reports a consultancy role for Janssen Oncology, Astellas Pharma, Serimmune, Foundation Medicine, Exact Sciences, Bristol-Myers Squibb, Varian Medical Systems, Novartis, Roivant, Myovant Sciences, Bayer, BlueStar Genomics, Artera, Tempus, and Genentech; stock options from Artera for a medical advisor role; and stock options for advisory board participation from BlueStar Genomics and SerImmune. Angela Y. Jia reports payments/honoraria from Myovant for advisory board membership and meeting/travel support from DAVA Oncology. Vinnie Y. T. Liu and Trevor J. Royce report employment and stock/stock options with Artera Inc. Howard M. Sandler reports support for a role as chair of the GU Cancer Committee for ACR/NRG Oncology; consulting fees for clinical trials steering committee participation for Janssen; and a leadership role for ASTRO (President-elect and member of the board of directors). Daniel E. Spratt reports grants or contracts from Janssen; consulting fees from Boston Scientific; and payments/honoraria from Astellas, AstraZeneca, Bayer, Elekta, Gamma Tile, Varian, Janssen, Novartis/AAA, Pfizer, and Myovant for lectures or presentations. Phuoc T. Tran reports grants from the National Cancer Institute (U01CA212007, U01CA231776, R01CA271540, and U54CA273956), the US Department of Defense (W81XWH-21-1-0296), the Prostate Cancer Foundation, the Distinguished Gentlemen’s Ride, and the Movember Foundation; consulting fees from RefleXion Medical, Natsar Pharm, Bayer Healthcare, Regeneron, and J&J; meeting/travel support from RefleXion Medical, Bayer Healthcare, Regeneron, and J&J; intellectual interest in patent 9114158 held by Natsar Pharm; and leadership/fiduciary roles for the University of Maryland (Vice Chair of Research), NRG Oncology GU Translational Science (Chair), Cancer Research (Senior Editor), and the ASTRO Research Funding Development Committee (Chair of the Science Council).
Footnotes
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eururo.2024.06.019.
References
- [1].National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology. Prostate cancer v4. 2023. [DOI] [PubMed] [Google Scholar]
- [2].Dess RT, Suresh K, Zelefsky MJ, et al. Development and validation of a clinical prognostic stage group system for nonmetastatic prostate cancer using disease-specific mortality results from the International Staging Collaboration for Cancer of the Prostate. JAMA Oncol 2020;6:1912–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Esteva A, Feng J, van der Wal D, et al. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digit Med 2022;5:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Circulation 2015;131:211–9. [DOI] [PubMed] [Google Scholar]
- [5].Olleik G, Kassouf W, Aprikian A, et al. Evaluation of new tests and interventions for prostate cancer management: a systematic review. J Natl Compr Cancer Netw 2018;16:1340–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


