Abstract
African green monkeys can maintain long-term persistent infection with simian immunodeficiency viruses (SIVagm) without developing AIDS and thus provide an important model for understanding mechanisms of natural host resistance to disease. This study assessed the levels and anatomic distribution of SIVagm in healthy, naturally infected monkeys. Quantitative competitive reverse transcriptase PCR assays developed to measure SIVagm from two African green monkey subspecies demonstrated high levels of SIV RNA in plasma (>6 × 106 RNA copies/ml) in sabaeus and vervet monkeys. Infectious virus was readily recovered from plasma and peripheral blood mononuclear cells and shown to be highly cytopathic in human cell lines and macrophages. SIVagm DNA levels were highest in the gastrointestinal tract, suggesting that the gut is a major site for SIVagm replication in vivo. Appreciable levels of virus were also found within the brain parenchyma and the cerebrospinal fluid (CSF), with lower levels detected in peripheral blood cells and lymph nodes. Virus isolates from the CSF and brain parenchyma readily infected macrophages in culture, whereas lymph node isolates were more restricted to growth in human T-cell lines. Comparison of env V2-C4 sequences showed extensive amino acid diversity between SIVagm recovered from the central nervous system and that recovered from lymphoid tissues. Homology between brain and CSF viruses, macrophage tropism, and active replication suggest compartmentalization in the central nervous system without associated neuropathology in naturally infected monkeys. These studies provide evidence that the nonpathogenic nature of SIVagm in the natural host can be attributed neither to more effective host control over viral replication nor to differences in the tissue and cell tropism from those for human immunodeficiency virus type 1-infected humans or SIV-infected macaques.
The family of CD4+ T-lymphotropic primate lentiviruses is comprised of two human viruses (human immunodeficiency virus type 1 [HIV-1] and HIV-2) and as many as 30 distinct simian immunodeficiency viruses (SIVs) found naturally in African nonhuman primates (44). Eight primate immunodeficiency viruses have been previously isolated and characterized and have been placed within five phylogenetic groups (3, 9, 17, 34, 40, 43, 46, 49, 76). It has been suggested that these distinct lentivirus groups arose as a consequence of the coevolution of the primate host and its resident virus (3, 31). The predominant HIV (HIV-1) appears to have resulted from cross-species transmission of SIVcpz from chimpanzees that evolved into the current AIDS pandemic (34, 76). Similarly, a second human AIDS virus (HIV-2) arose by cross-species transmission of SIVsm found naturally in sooty mangabeys (Cercocebus atys) (17, 40, 49). African green monkeys (AGMs) (Cercopithecus aethiops), due to their numbers and geographical distribution in sub-Saharan Africa, represent the largest single reservoir of SIV (SIVagm), as upwards of 50% of wild monkeys are infected with the virus (2, 54).
In host species that have not experienced long-term endemic infection by the primate immunodeficiency viruses, such as humans and Asian macaques, HIV or SIV infection causes an immunodeficiency syndrome characterized by inexorable CD4+ T-cell depletion. In addition, significant numbers of infected humans and macaques with AIDS also develop neurologic complications due to virus infection of the central nervous system (CNS) (35, 95). In these nonnatural host species, high steady-state levels of virus in the plasma are associated with rapid CD4 decline and disease progression (47, 66, 86, 91). High levels of HIV in the blood are maintained by continuous rounds of viral infection of activated CD4+ T cells and the destruction of the infected cells, as evidenced by the rapid decay of virus in the plasma following initiation of effective antiviral therapy (highly active antiretroviral therapy) (51, 77, 92). These virus and cell turnover studies have led to the formulation of a dynamic model of AIDS pathogenesis whose central assertion is that active HIV replication induces an accelerated rate of CD4+ T-cell destruction (51, 77). As a result of the envisioned extreme demand on CD4+ T-cell proliferation, lymphopoietic reserves are overwhelmed and collapse of the immune system ensues. Thus, this model places virus-induced CD4+ T-cell destruction as the central event in HIV-1 immunopathogenesis. In addition to the direct cytopathic effects of HIV-1 and SIV, pathogenic lentivirus infections are also associated with a pronounced tendency of uninfected T lymphocytes of both CD4+ and CD8+ lineages to undergo activation-induced programmed cell death or apoptosis, destruction of lymphoid tissue architecture, and suppression of bone marrow and thymic production of requisite hematolymphoid precursor cells (30, 38, 39). Thus, progression to AIDS in nonnatural hosts may result from the cumulative deleterious effects of both direct and indirect consequences of virus infection that lead to accelerated destruction and compromised host immune regenerative capacity.
The natural host reservoirs for primate lentiviruses in sub-Saharan Africa, including sooty mangabeys and AGMs, are generally considered to be resistant to AIDS-related diseases and thus represent important models for understanding the differential pathogenesis of SIV (44). The study of infected unnatural hosts such as Asian macaques has provided valuable information regarding AIDS pathogenesis and the development of vaccines; examples of disease-free equilibrium between the natural simian hosts and their viruses provide a unique opportunity to study the basis of nonpathogenic virus infection. Understanding this disease-free equilibrium is likely to provide important clues regarding mechanisms of AIDS pathogenesis. SIVagm-infected AGMs provide one such model of nonpathogenic SIV infection. Experimental transfer of SIVagm to Asian pigtailed macaques (Macaca nemestrina) yields high levels of virus replication and induction of simian AIDS characterized by immunodeficiency and neuropathology (45). In contrast, SIVagm does not replicate to high levels or induce disease in rhesus macaques (Macaca mulatta), perhaps as the result of a species-specific restriction in the function of viral accessory gene products such as vpr (45, 88). Whether immunopathology and neuropathology follow infection with SIV appears to depend on the levels of virus replication achieved, as well as additional, yet-to-be-defined host-specific variables. The observation that SIVagm, like SIVsm, has the potential to cause AIDS upon transfer to a new host species suggests that AIDS results from differential host responses to SIV infection rather than from fundamental properties of the virus alone. Understanding the basis of nonpathogenic SIV infection may help to define precisely the features of the host-virus interaction that are responsible for the irreversible CD4 decline seen with pathogenic HIV and SIV infections.
In naturally infected AGMs, SIVagm seropositivity parallels the onset of sexual activity and is nearly universal in females of reproductive age (78). In addition to intraspecies transmission of SIVagm, cross-species transmission to patas monkeys and yellow baboons has been reported elsewhere (11, 53). High seroprevalence rates for AGMs in the wild and in captivity suggest that a substantial level of viremia is likely to be present in naturally infected animals. This is supported by the ready isolation of infectious virus from the peripheral blood of infected AGMs (2, 21). Further, the extensive genetic diversity of SIVagm observed within and between animals is compatible with frequent cycles of virus infection in vivo accomplished via the error-prone processes of retroviral replication (65). In addition, we and others have elsewhere demonstrated chronic high levels of viremia in another natural reservoir host, SIVsm-infected sooty mangabeys (82; R. Grant, A. Kaur, S. I. Staprans, S. P. O'Neil, H. M. McClure, R. P. Johnson, and M. B. Feinberg, submitted for publication; G. Silvestri, D. Sodora, R. A. Koup, S. P. O'Neil, W. J. Kaiser, H. M. McClure, S. I. Staprans, and M. B. Feinberg, submitted for publication).
Earlier studies of SIVagm infection of AGMs have theorized that the lack of disease in AGMs results from restricted virus replication and reduced destruction of CD4+ T cells (8, 41). For example, in contrast to what is observed with pathogenic immunodeficiency virus infections, infected AGMs appear to express lower levels of virus in the lymph nodes (LNs) (8, 45). The low levels of virus observed in the LNs could reflect either effective host immune control of virus replication or simply the presence of fewer resident cells that are susceptible to virus infection. At any anatomic location, target cell availability will depend upon the number of total CD4+ T cells present, as well as their state of activation and expression of appropriate coreceptors for infection (12). The levels of viral load and sites of SIVagm production in naturally infected AGMs determined using sensitive quantitative gene amplification techniques have not been reported to date. Both SIVagm-infected green monkeys and SIVsm-infected mangabeys exhibit only a slight decline in peripheral CD4+ T-cell counts below those for uninfected animals (7, 16; Silvestri et al., submitted). Due to the relatively low numbers of CD4+ T cells present in the peripheral blood of both SIV-infected and uninfected AGMs and the difficulties of maintaining these cells in culture, studies addressing the cytopathic potential of SIVagm for AGM cells have been difficult to perform (71). Very little is known about the extent to which SIVagm-specific immune responses may control virus infection in vivo. Similar to those of humans, AGM CD8+ T cells are reported to secrete a factor that suppresses SIVagm replication in vitro, although it is not clear to what extent, if any, this factor modulates SIVagm replication in vivo (6, 28). Humoral immune responses are evident in naturally infected monkeys; however, neutralizing antibody levels are low (2, 4, 37, 73). In naturally infected sooty mangabeys, SIV-specific cytolytic CD8+ T-cell responses are limited, consistent with the observed high levels of SIVsm replication (56; Grant et al., submitted; Silvestri et al., submitted).
To better understand the biology of SIV infection in AGMs, we sought to define the levels and locations of virus replication in naturally infected animals. In doing so, we hoped to reconcile the paradox of their asymptomatic infection with their high prevalence of infection and the extensive genetic diversity observed in viral quasispecies in infected animals. Toward these ends, we developed sensitive quantitative competitive reverse transcriptase PCR (QC RT-PCR) methods based on conserved regions of the SIVagm genome to enumerate plasma SIV RNA levels in AGM subspecies naturally infected by diverse SIVagm isolates. The tissue distribution of SIVagm was documented using similarly tailored PCR-based methods for the enumeration of viral DNA copy number. Infectious virus was also isolated from the CNS and lymphoid tissues and characterized with respect to viral variation and cell tropism. Our findings, together with similar findings for the natural sooty mangabey host for SIVsm, suggest that the natural hosts for primate lentiviruses remain clinically healthy despite high-level virus replication, although this replication may be preferentially localized to those sites where antigen-driven activation is most pronounced, such as the gut. These studies suggest that the direct consequences of high-level HIV replication alone may not account for the progressive CD4+ T-cell depletion and AIDS in humans.
MATERIALS AND METHODS
Animals.
West African green monkeys (Cercopithecus aethiops sabaeus) were imported from Senegal (animals 9307 to 9312); four of them were determined to be naturally infected (3). Vervet monkeys (Cercopithecus aethiops pygerythrus) were either imported from Tanzania (AGMs 9648 and 9649), purchased from a commercial facility (AGMs 6242 and 6243), or housed at another facility (AGMs 684, 687, and 692). Grivets (Cercopithecus aethiops aethiops) were also housed at another facility, and blood samples were sent by overnight delivery (AGMs 654 and 677). AGMs and humans were deemed SIVagm negative by Western blot analysis before their lymphocytes were used for in vitro assays. AGM 9309 died of congestive heart failure, and AGM 9315 was euthanatized due to an incidental partial obstruction of the cecum. Tissues were collected from these animals at the time of necropsy, snap frozen in liquid nitrogen, and stored at −80°C. Rhesus macaques, used as controls, were inoculated with SIVmac251 (MAC 8357 and MAC 8358). For inoculations, animals were sedated with ketamine by intramuscular injection, and 1 ml of virus stock was inoculated into the femoral vein. The monkeys were housed in biosafety level 2 conditions and fed a commercial monkey diet supplemented with fresh vegetables. Housing and handling of animals were in accordance with the Guide for the Care and Use of Laboratory Animals (U.S. Public Health Service) and the Animal Welfare Act. The protocols and procedures were approved by the Institutional Animal Care and Use Committee. The Southwest Foundation for Biomedical Research is accredited by the American Association for Accreditation of Laboratory Animal Care.
Cell lines, PBMC, macrophages, and viruses.
Molt4(cl8) cells were grown in RPMI 1640 supplemented with 15% fetal bovine serum, 2 mM l-glutamine, 50 U of penicillin/ml, and 50 μg of streptomycin/ml (complete medium) (58). AGM and human peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood by Ficoll-Hypaque density gradient centrifugation (Histopaque; Sigma Chemical Co., St. Louis, Mo.), grown in complete medium (20% fetal bovine serum) with 10 μg of phytohemagglutinin (PHA)/ml for 24 h, and then grown in complete medium containing 10% interleukin-2. Human monocytes were isolated from PBMC (Leukopak; Southwest Texas Blood Bank, San Antonio) by adding 106 PBMC to 12-well plates and then removing the nonadherent population the next day. The adherent cells were maintained in Dulbecco modified Eagle medium supplemented with 10% human serum (heat inactivated, 56°C, 30 min) and 10% giant cell tumor (GCT)-conditioned medium (Origen, Inc., Rockville, Md.) for 5 days until the monocytes differentiated into macrophages. The cells were demonstrated to be greater than 95% macrophages on the basis of morphology, adherence to plastic, and nonspecific esterase activity. The viral isolates SIVagm(tyo-1), SIVagm(gri-1), SIVagm(MJ8), and SIVmac239 have been described previously (31, 32, 52, 72). SIVagm(gri-1)-infected CEM cells were derived by transfection with a molecular clone of gri-1, and SIVagm(MJ8)-infected Molt4/8 cells were derived from a molecular clone (MJ8) of SIVagm(sab-1) (31, 52). HIV-1/Ba-L was propagated on human macrophages after receipt from the AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (reagent 510) (36). The concentrations of virus stocks were determined by endpoint titration with an antigen-capture enzyme-linked immunosorbent assay (p27/p24; Coulter Immunologics) and are as follows: SIVagm(sab-4CSF), 433 ng/ml; SIVagm(sab-4ln), 1,190 ng/ml; SIVagm(sab-4br), 1,720 ng/ml; SIVagm(gri-1), 565 ng/ml; SIVagm(MJ8), 153 ng/ml; and HIV-1/Ba-L, 95 ng/ml.
Virus isolation.
Virus was isolated from PBMC by cocultivation of 2 × 106 Molt4(cl8) cells with 2 × 106 stimulated AGM PBMC. To isolate virus from the plasma, 2 × 106 Molt4(cl8) cells were centrifuged at 1,600 × g, resuspended in 250 μl of plasma, and incubated at 37°C for 2 h. For both isolation protocols, the volume was then adjusted with medium to 106 cells/ml. The cultures were monitored for 28 days, with growth medium being replaced every 3 to 4 days, and the culture fluid was stored at −80°C until analysis for virus by SIV p27 antigen capture (Coulter Immunologics). Cell-free virus was isolated from cerebrospinal fluid (CSF) and homogenized brain and LN tissues of AGM 9315 at the time of necropsy. Tissue specimens (∼1 g) were homogenized with a Tissumizer (Tekmar, Cincinnati, Ohio) equipped with a dispersing tool (probe 10G). Samples were subjected to homogenization on average for 30 s on ice at 11,500 rpm followed by centrifugation at 13,500 × g to remove cells and debris. The supernatants were then filtered through a 0.45-μm-pore-size nylon filter, and virus isolation was attempted by incubation with Molt4(cl8) cells.
Western blot analysis.
Western blotting was performed as previously described (3). Virus in culture supernatants taken from infected cell lines at 48 to 72 h was purified through a 20% sucrose cushion, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then blotted onto nitrocellulose sheets. The nitrocellulose paper was blocked with 3% bovine serum albumin and subsequently incubated with a 1:50 dilution of serum from an experimentally infected rhesus macaque (mac7695). Detection of viral proteins was accomplished by the streptavidin-biotin system (Amersham Inc.) with diaminobenzidine as the substrate for color development.
Virus infection.
Macrophage cultures or Molt4(cl8) cells (2 × 106 cells) were incubated with 250 μl of virus stock for 2 h, and the volume was adjusted to 500 μl for overnight incubation in Dulbecco modified Eagle medium supplemented with 10% human serum and GCT-conditioned medium or complete RPMI 1640, respectively. Cells were washed five times and resuspended in complete medium, and the culture fluids were harvested every 3 to 4 days and stored at −80°C. Viral antigen levels (p27/p24) were determined by endpoint titration with the Coulter antigen-capture enzyme-linked immunosorbent assay.
Quantitative and semiquantitative PCR analysis.
QC RT-PCR was used to measure SIV plasma viremia and was modified as previously described (86, 87). Blood was collected in acid citrate dextrose tubes, and the plasma was separated and frozen at −80°C. Viral RNA was isolated from 1.0 ml of plasma. Virions were concentrated from the plasma by centrifugation at 39,500 × g and 4°C for 1 h. The virus pellet was suspended in TRIZOL (Gibco/BRL, Grand Island, N.Y.) and extracted with chloroform, after which the RNA was precipitated in isopropanol.
For quantitating viral RNA copy equivalents per milliliter from vervets, the QC RT-PCR protocol was modified from our previously published protocol to make the DNA templates for RNA transcription with the following primers and templates: forward primer gag519, 5′-CAAITIAAACAITTAATATGGGCAGG-3′, nucleotides 519 to 544 (26-mer), and reverse primer gag734, 5′-TACIGCTTCITCTGTGTCTTTCACTT-3′, nucleotides 734 to 709 (26-mer) (86). For making the internally deleted control RNA template, the forward primer gag594 (5′-CAAITIAAACATTTAATATGGGCAGGGAGGGGTGTAAAAGAATC AT-3′, nucleotides 519 to 544 and 594 to 613 [46-mer]), which resulted in a 49-bp deletion, was used. The full-length and deleted templates were cloned into the pGEM-T vector (Promega, Madison, Wis.), resulting in 298- and 249-nucleotide in vitro transcripts for use as positive control and competitive templates, respectively, and for generating a standard curve. Primers gag519 and gag734 were used to amplify the cDNA products at 94°C for 50 s, 61°C for 40 s, and 72°C for 40 s through 45 cycles. Nucleotide numbering was based on the published sequence of SIVagm(tyo-1) (32).
For generating a QC RT-PCR standard curve for SIVagm sabaeus viruses, a 176-bp region of the SIVagm(sab-1) gag gene was generated by PCR amplification from the full-length MJ8 clone and then cloned into the pCR2.1 expression vector (Invitrogen Corp., Carlsbad, Calif.). The primers used to generate the nondeleted control template were as follows: forward primer gag1212, 5′-CCCCTAGTTCCTACAGGGTCAG-3′, nucleotides 1212 to 1233, and reverse primer gag1386, 5′-TTTGGCCACTAGATGTCGCTG-3′, nucleotides 1386 to 1366. The internally deleted competitive template was synthesized by overlap primer extension PCR using the forward primer gag1282, 5′-CCCCTAGTTCCTACAGGGTCAGGATCCACGCAGAAATAAAAGTGAAAG-3′, nucleotides 1282 (1212) to 1302 (49-mer), which was used in conjunction with the reverse primer (gag1386). A BamHI restriction site was also inserted to allow for orientation of the insert in the pCR2.1 vector. RNA was synthesized from the plasmid by in vitro runoff transcription according to the Promega RIBOMAX protocol, and RNA concentration and copy number were determined. The RNA samples and competitive template were diluted, added to a 96-well plate, and reverse transcribed using random primers, and the cDNAs were coamplified. Forward primer gag1208, 5′-CCTTCCCCTAGTTCCTACAGGGT-3′, nucleotides 1208 to 1230, and reverse primer gag1378, 5′-CTAGATGCGCTGTGGCACTTTC-3′, nucleotides 1378 to 1356, were used for PCR through 45 cycles of 94°C for 50 s, 60°C for 40 s, and 72°C for 40 s, resulting in a 168-bp control fragment and a 125-bp internally deleted fragment. This protocol was modified as previously described by Staprans et al. and incorporates [32P]dCTP during PCR amplification (86, 87). The radiolabeled PCR products were then separated by polyacrylamide gel electrophoresis, and the gel was dried and exposed to a Molecular Dynamics (Sunnyvale, Calif.) imaging plate. A PhosphorImager 445 SI (Molecular Dynamics) was used for analysis of gels in conjunction with IPLab Gel software (Signal Analysis Corporation). The RNA copy number was normalized for milliliters of plasma. Less than threefold interassay variation was observed, and the sensitivity was determined to be 500 copies based on the standard curve.
Levels of viral DNA in lymphocytes and tissues were assessed by semiquantitative PCR coupled with Southern blot analysis. Total cellular DNA was isolated from PBMC or homogenized tissues as previously described (3). For quantitation of PBMC load in sabaeus monkeys, serial 10-fold dilutions of DNA were subjected to PCR using SIVagm sabaeus-specific gag primers: forward primer gag1668, 5′-ATCAATGAAGAAGCTGCAGATTGG-3′, and reverse primer gag2037, 5′-TGGCATTCTGGATCCACAGAGACTG-3′. For SIVmac PCR, forward primer gag1658, 5′-TATAAATGAGGAGGCTGCAGATTGGGACTT-3′, and reverse primer gag2571, 5′-TGAGCAGTGACTACTGGTCTCCTCCAAAGA-3′, were used. Reaction conditions for both sets of primers were 94°C for 1 min, 55°C for 45 s, and 72°C for 1 min through 30 cycles.
Viral DNA from vervet monkeys was amplified using forward primer tyo25, 5′-TTGAGCCTGGGTGTTCGCTG-3′, nucleotides 25 to 44, and reverse primer tyo548, 5′-CTTGCCTGCCCATATTAAATG-3′, nucleotides 548 to 528, which encompasses the 5′ long terminal repeat (LTR) region and gag gene. Reaction conditions were 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min through 40 cycles. The amplified samples were separated on agarose gels and blotted onto positively charged nylon membranes. The SIVagm sabaeus and SIVmac samples were then hybridized with 32P-labeled SIVagm(sab-1) clone MJ8 or SIVmac239 corresponding to the amplified gag gene sequence. The SIVagm vervet samples were hybridized with a 32P-labeled internal probe generated from SIVagm(tyo-1) using forward primer tyo194, 5′-GACCCAGGCGAGAGAAACTCC-3′, nucleotides 194 to 214, and reverse primer tyo418, 5′-CCTGCCCTACTTGCCCAGAGTA-3′, nucleotides 418 to 397.
For SIVagm-infected vervets, individual LTR/gag genes were amplified from AGM6242, AGM9648, and AGM9649 using primers tyo25 and tyo744 (5′-TTACTGTTGCTACTGCTTCCTCTGTG-3′) (nucleotides 744 to 719), and assay sensitivity was determined with the tyo-1 probe. Reaction conditions for this set of primers were 94°C for 30 s, 48°C for 1 min, and 72°C for 1 min through 40 cycles. PCR products were gel purified and cloned into the pCRII-TOPO vector (Invitrogen). Plasmid DNA from characterized molecular clones was diluted to the appropriate copy number and used as template for the standard curves. The detection limit of the PCR-Southern blot protocol was determined to be five copies for all primer pairs using serial dilutions of the probe represented by plasmids containing either MJ8, SIVmac239, or the SIVagm(tyo-1) LTR/gag gene in 100 ng of total cellular DNA.
Molecular cloning and DNA sequencing.
Forward primer env7164 (5′-TGCTCCAGCAGGTTATGCTTTG-3′, nucleotides 7164 to 7185) and reverse primer env7881 (5′-GGCTGTCGAGTTACACTCCAAGTG-3′, nucleotides 7881 to 7858) were used to amplify a 672-bp env fragment from total cellular DNA of SIVagm-infected human T-cell lines. Numbering for the nucleotide sequence was based on the published sequence of SIVagm(sab-1) (52). Thermocycler conditions for amplification were as follows: step 1, one cycle at 95°C for 5 min; step 2, 25 cycles at 94°C for 1 min, 57°C for 45 s, and 72°C for 1 min; and step 3, one cycle at 72°C for 7 min. PCR products were cloned using the TA cloning kit from Invitrogen (San Diego, Calif.). Two clones from each virus were randomly selected and then purified using the S.N.A.P. miniprep kit (Invitrogen Corp.). One microgram of plasmid was sequenced using an Applied Biosystems automated sequencer, model 373A, version 1.2.0. Sequences were aligned and analyzed using the GCG Package, version 7.0 (Genetics Computer Group, Inc.), and corrected by hand.
Nucleotide sequence accession number.
DNA sequences from the env genes have been submitted to GenBank and assigned accession no. AF332984 to AF332995.
RESULTS
High levels of SIV RNA in plasma in AGMs.
African host species of the simian lentiviruses experience long-term natural infection without associated disease. Elucidation of the dynamic process of host-virus equilibrium leading to natural disease resistance is key to understanding AIDS pathogenesis. One explanation for the lack of SIV-associated disease is host-specific restriction in virus replication in the natural host. In infected humans and nonhuman primates, the levels of virion-associated HIV or SIV RNA in the plasma are indicative of the overall level of ongoing virus replication extant in the host. In nonnatural or new hosts for CD4+ T-cell-tropic lentiviruses, such as humans and Asian macaques, measurement of plasma HIV RNA levels provides a powerful tool to predict rates of progression to AIDS and death (66, 79, 87). High levels of plasma viral RNA are associated with rapid progression to AIDS, whereas slowly progressive or nonprogressive infections are characterized by very low levels of plasma viral RNA. To assess the level of SIVagm replication in naturally infected AGMs, we developed a QC RT-PCR assay to measure plasma SIV RNA levels. This assay is similar in principle to the SIVmac QC RT-PCR assay that we previously developed and validated for SIVmac viruses by comparison with the SIV branched DNA signal amplification assay (64, 87, 91). We initially screened seven AGMs (four sabaeus and three vervet monkeys) from 14 September 1994 with primer pairs designed to amplify conserved gag-related sequences. These primer sets were optimized to amplify gag gene sequences from SIVagm of vervets, grivets, and tantalus monkeys (gag519 and gag734; see Materials and Methods), and inosine bases were substituted where variation was common between strains. Internally deleted in vitro RNA transcripts from the SIVagm gag gene were used as internal standards for calibration as previously described (see Materials and Methods and reference 86). All five SIVagm-infected AGMs had high viral RNA levels in their plasma, ranging from 3.5 × 105 to more than 9.5 × 106 RNA copies/ml, while no viral RNA was detected in the plasma from two seronegative AGMs (Table 1). The viral RNA levels from the two SIVagm-infected sabaeus monkeys were generally lower than those for the vervets in this assay; however, this difference might have reflected greater variation in the gag gene between SIVagm subtypes.
TABLE 1.
Quantitation of SIVagm RNA from plasma and CSF of naturally infected AGMs
| Animal subspecies and no. | Bleed datea (mo-day-yr) | RNA copy eq/ml of:
|
|
|---|---|---|---|
| Plasma | CSF | ||
| Sabaeus | |||
| AGM 9307 | 9-14-94 | 0 | |
| AGM 9312 | 9-14-94 | 0 | |
| 1-14-97 | 0 | 0 | |
| AGM 9308 | 9-14-94 | 6.4 × 105 | |
| 8-10-96 | 5.1 × 106 | 4.8 × 105 | |
| AGM 9310 | 9-14-94 | 6.1 × 105 | |
| 8-10-96 | 3.9 × 106 | 1.2 × 105 | |
| Vervet | |||
| AGM 6242 | 9-14-94 | 1.2 × 106 | |
| AGM 9648 | 9-14-94 | >9.5 × 106 | |
| 1-14-97 | 6.3 × 106 | ||
| AGM 9649 | 9-14-94 | 3.5 × 105 | |
| 1-14-97 | 3.5 × 106 | ||
All plasma samples from 14 September 1994 were assayed by QC RT-PCR with the gag519-gag734 primer set based on the SIVagm(tyo-1) vervet gag gene sequence (see Materials and Methods). Samples from 10 August 1996 and 14 January 1997 were assessed for viral RNA with either the gag519-gag734 primer set (vervet) or gag1208-gag1378 primers (sabaeus).
The overall genetic similarity between the gag genes of SIVagm(sab) and SIVagm(ver) strains is less than 65%, and we found 8 to 10 mismatches between the vervet-derived forward primer gag519 and the SIVagm(sab) sequences, indicative of this diversity (data not shown). To enable a more accurate measurement of these genetically diverse SIVagm strains, we therefore designed primer pairs and an internally deleted standard based on the published sequence from a clone derived from AGM 9308 (MJ8) to enhance the detection of SIVagm from sabaeus monkeys. Plasma samples were taken 2 to 3 years after the initial sample from four of five SIVagm-positive AGMs and one seronegative AGM and assessed for viral RNA load using either vervet-specific or sabaeus-specific primer pairs for each species (Table 1). High levels of viral RNA, ranging from 3.5 × 106 to 6.3 × 106 (AGM 9648) RNA copies/ml of plasma, were detected in all four seropositive monkeys, with an average of 4.0 × 106 copies/ml. SIV RNA levels did not vary by more than threefold in duplicate samples. The RNA levels in the sabaeus monkeys were approximately 1 log higher than those for samples from the earlier bleed and may have been due to greater sensitivity of the primer pairs. No viral RNA was detected in plasma from the seronegative animal.
Infectious virus is readily isolated from the peripheral blood of naturally infected AGMs.
In order to determine whether the high level of plasma viral RNA reflects the presence of infectious virus, PBMC and plasma from SIV-infected AGMs were cocultivated with the human CD4+ T-cell line Molt4(cl8). None of the 10 wild-caught AGMs included in this analysis had any clinical disease associated with immunodeficiency during more than 5 years in captivity. Virus could be recovered from both plasma (7 of 10) and PBMC (11 of 11) of naturally infected animals (Table 2), indicating the presence of significant amounts of infectious virus in the peripheral blood. With HIV-infected humans, the ability to culture virus from the plasma is associated with high virus loads. Given that most virions in the plasma appear to be replication defective, it is estimated that RNA copy number measures of viremia can exceed infectious virus titers by approximately 50,000-fold (79). The subspecies of green monkey (sabaeus, vervet, and grivet) studied did not influence the success rate of virus isolation. Thus, the frequent isolation of SIVagm from the PBMC and plasma of infected animals is consistent with the observed high plasma SIV RNA levels.
TABLE 2.
Virus isolation and proviral burden in the peripheral blood of naturally infected monkeys
| Animal no. | AGM subspecies | SIVagm serostatusa | Virus isolation fromb:
|
PBMC DNA (no. of copies/1.5 × 105 cells) | |
|---|---|---|---|---|---|
| Plasma | PBMC | ||||
| 9308 | Sabaeus | + | 1/1 | 1/1 | 50 |
| 9310 | Sabaeus | + | NDc | 1/1 | 5 |
| 9312 | Sabaeus | − | ND | ND | 0 |
| 6242 | Vervet | + | 0/1 | 1/1 | 5 |
| 9648 | Vervet | + | 1/2 | 1/2 | 5 |
| 9649 | Vervet | + | 2/2 | 2/2 | 50 |
| 9309 | Sabaeus | + | 1/1 | 1/1 | ND |
| 684 | Vervet | + | 0/1 | 1/2 | ND |
| 687 | Vervet | + | 1/1 | 1/2 | ND |
| 692 | Vervet | + | 0/1 | 1/2 | ND |
| 654 | Grivet | + | 1/1 | 2/2 | ND |
| 677 | Grivet | + | 1/1 | 2/2 | ND |
+, positive; −, negative
Number of successful isolations/number of attempted isolations.
ND, not determined.
SIV DNA copy number in peripheral blood lymphocytes of AGMs.
Another measure of lentivirus replication is the number of viral DNA-positive CD4+ T cells or PBMC. The levels of viral DNA present in PBMC generally correlate with the plasma viremia levels of HIV-1-infected individuals (79). In untreated HIV-infected humans, the most prevalent form of HIV DNA is full-length, linear unintegrated DNA that, although not replication competent, is thought to be a good marker of the number of recent infection events and thus the level of ongoing viral replication (18). To quantify the number of SIV DNA-positive PBMC in the peripheral blood of AGMs, a semiquantitative DNA PCR assay that detects total (unintegrated and integrated) SIV DNA was used. Two sabaeus subspecies (C. aethiops sabaeus) and three vervet subspecies (C. aethiops pygerythrus) of AGMs were analyzed.
Primers and probes were individually designed to detect viral DNA from each subspecies because of a high degree of genetic diversity among different AGM types. Strain-specific LTR/gag gene molecular clones were constructed by RT-PCR amplification of the LTR/gag region of RNA isolated from the plasma of each of the infected vervets (AGM 6242, AGM 9648, and AGM 9649) and were used to determine the sensitivity of amplification by the PCR primers. Regardless of subspecies and interanimal SIVagm variability, the sensitivity of this quantitative assay was five copies with each of the plasmids (pgag9648, pgag9649, and pgag6242) (data not shown).
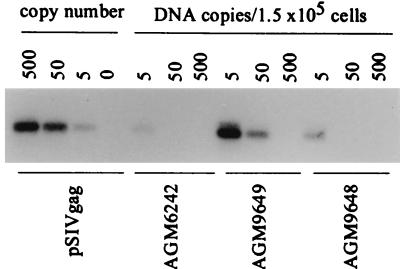
DNA purified from PBMC of each of five seropositive monkeys was subjected to DNA PCR followed by Southern blotting. An example of the viral DNA present in the PBMC samples from these three vervets is shown (Fig. 1). Overall, SIVagm DNA was detected for all five seropositive monkeys with endpoint titers ranging from 5 to 50 copies of viral DNA/1.5 × 105 PBMC (Table 2). Four of five monkeys had only five DNA copies per 1.5 × 105 cells. For comparison, the levels of HIV DNA that are typically found in HIV-infected humans range between 6 and 50,000 copies/105 PBMC, with an average of about 300 copies (20). The CD4+ T-cell population in the peripheral blood of AGMs is typically two to five times less than that in humans or macaques; however, CD4 T-cell counts from these bleeds were not available, and thus DNA copies per CD4+ T cell could not be calculated. These data suggest that the target cells for active SIVagm production in AGMs are relatively poorly represented in the peripheral blood, which likely results from low numbers of circulating CD4+ T cells in AGMs (71).
FIG. 1.
Levels of viral DNA present in peripheral blood cells from naturally infected green monkeys. Semiquantitative PCR followed by Southern blotting was performed with DNA from the PBMC of three vervets (6242, 9648, and 9649). The number designation for each animal represents the amount of total cellular DNA amplified (in nanograms). To determine the sensitivity of the assay, 10-fold dilutions of pSIVgag were amplified in the presence of 100 ng of cellular DNA. The numbers represent copy number based on the control pSIVgag plasmid.
Lymphoid tissues harbor higher viral burdens than does peripheral blood in AGMs.
Representative tissue compartments from naturally infected AGMs were examined to determine if there might be sites other than PBMC that are responsible for production of the high levels of plasma viremia. Lymphoid tissues, such as those found in the gut, LNs, and spleen, are considered to be the primary reservoirs for lentiviruses in both humans and monkeys (50, 75). Necropsied tissues from two naturally infected AGMs were studied to assess SIVagm viral DNA distribution in both lymphoid and nonlymphoid tissues. Two SIVmac251-infected rhesus macaques (MAC 8357 and MAC 8358) from a separate study were analyzed in order to compare the relative levels of virus in AGMs to those found for SIVmac-infected rhesus macaques which developed simian AIDS over a 2-year period. It is important to note that SIVmac251-infected macaques are known to harbor extraordinarily high levels of SIV RNA in the plasma, with levels sometimes exceeding 108 SIV RNA copies/ml in rapidly progressing animals (87).
By semiquantitative PCR, between 10- and 100-fold-more viral DNA was detected in LNs and spleens from both AGMs (AGM 9309 and AGM 9315) than in their peripheral blood lymphocytes, ranging from 50 to 500 viral DNA copies/1.5 × 105 cells in these tissues (Table 3). All three lymphoid samples taken from AGM 9309 had 500 copies/105 cells. These values are similar to those reported for HIV DNA copy number in LN cells obtained from asymptomatic humans (94). As might be expected from the high-level growth of SIVmac251, the two SIVmac-infected rhesus monkeys had higher viral DNA levels in both PBMC and lymphoid tissues, with ∼10,000 viral DNA copies/105 PBMC and ∼100,000 viral DNA copies/105 LN cells at the end stage of the disease (Table 3). The highest levels were observed for the spleens of the SIVmac251-infected animals (5 × 104 to 5 × 105 copies/105 cells), which is in agreement with the high levels reported by Hirsch and others for SIVmac-infected macaques (50, 57).
TABLE 3.
Tissue distribution of viral DNA in SIVagm-infected AGMs and SIVmac-infected rhesus macaques
| Tissue type | No. of viral DNA copies/1.5 × 105 cellsa
|
|||
|---|---|---|---|---|
| AGM 9315 | AGM 9309 | MAC 8358 | MAC 8357 | |
| Lymphoid | ||||
| Axillary LN | 50 | 500 | 5 × 104 | |
| Mesenteric LN | 500 | 500 | ||
| Spleen | 500 | 500 | 5 × 105 | 5 × 104 |
| Nonlymphoid | ||||
| Cerebellum | 500 | 500 | 500 | 500 |
| Cerebrum | 50 | 500 | 500 | |
| Liver | 500 | 500 | 5 × 103 | |
| Kidney | 50 | 500 | 5 × 103 | |
| Duodenum | 5 × 103–5 × 104 | 5 × 104 | ||
| Jejunum | 5 × 103 | 5 × 104 | ||
| Colon | 5 × 103–5 × 104 | 5 × 104 | ||
The values represent data from two experiments. Results given as two values (duodenum and colon, AGM 9315) represent 10-fold differences between experiments.
Viral DNA from the gastrointestinal tract, liver, and kidney of SIVagm-infected AGMs was also analyzed (Table 3). DNA was isolated directly from tissues, and the DNA copy number was calculated based on 1 μg of DNA representing 1.5 × 105 cells. The highest levels of virus were found in tissues taken from the gastrointestinal tract of AGM 9315, ranging from 50 to 5,000 copies of viral DNA/1.5 × 105 cells in the duodenum, jejunum, and colon, corresponding to approximately 1 in 10,000 to 1 in 100 infected cells, assuming that only one copy is present per cell. SIVmac251-infected rhesus monkeys harbored higher levels of viral DNA in these tissues, including the gastrointestinal tract (5 × 104 copies/1.5 × 105 cells), kidney (5 × 103 copies/1.5 × 105 cells), and liver. It is interesting to note that while the AGMs had a clear dichotomy between levels of virus in the gut and those in PBMC, this was not the case for end-stage disease in the SIVmac251-infected macaques (Table 3). This pattern might be due to the higher levels of immune activation in peripheral lymphoid tissues in the macaques and the possibly lower levels of immune activation in non-gut-associated lymphoid tissues in the natural host. Indeed, LNs from AGMs and sooty mangabeys naturally infected with SIV do not display the same levels of lymphocyte activation and follicular hyperplasia that characterize pathogenic HIV and SIV infections (8; Silvestri et al., submitted).
SIVagm replication in the CNS of naturally infected monkeys.
One major tissue compartment for viral replication and associated pathology in HIV-infected humans and SIV-infected macaques is the CNS (33, 95). High levels of CSF RNA may correspond with significant clinical and pathological findings for humans, especially in end-stage disease (27). In naturally infected AGMs, high levels of viral RNA were found in the CSF (>105 SIV RNA copies/ml [Table 1]), suggesting that active virus replication might be taking place within the CNS. As determined by DNA PCR, the cerebellum from AGM 9315 had greater numbers of infected cells than PBMC from infected monkeys (Table 3), suggesting that virus in the CSF reflects viral replication within the brain parenchyma rather than the product of blood cells trafficking to the CNS. To determine whether these tissues represent sites of active viral replication in vivo, virus isolation was performed using cell-free filtrates of tissue homogenates from cerebrum and CSF and an LN of AGM 9315 obtained at necropsy. Virus was readily recovered from all three samples, as evidenced by both syncytium formation in Molt4(cl8) indicator cells and increasing levels of p27 antigen detected in culture supernatants. Histopathologic examination of each of these tissues was unremarkable, demonstrating that SIVagm is expressed within the CNS and lymphoid tissues of naturally infected monkeys without causing significant pathological changes (data not shown). The virus isolates from AGM 9315 brain, CSF, and LN were designated SIVagm(sab-4br), SIVagm(sab-4CSF), and SIVagm(sab-4ln), respectively.
We have previously shown that each of the SIVagm subspecies varies considerably in the relative sizes of its gp120 and p24 proteins (2, 3). Differences in the apparent molecular weights between SIV gp120s typically result from differences in glycosylation patterns, which contribute to the overall mass of the proteins. To observe whether there were differences in the structural genes which might correspond with tissue-specific viral replication, the SIVagm tissue isolates were first characterized by Western blotting using viral antigens from each of the SIVagm strains. As seen in Fig. 2, protein species with apparent molecular masses in the range of 120 kDa were recognized by sera from SIVagm-infected monkeys and are consistent with the exterior gp120 glycoproteins. Presumptive gp120 bands from the brain and CSF isolates of SIVagm(sab-4) were significantly smaller than those of gp120 from the corresponding LN isolate SIVagm(sab-4ln). No significant size differences were noted for the other major viral proteins. These data suggest that the brain and CSF isolates are more closely related to each other than they are to the virus found in the LNs.
FIG. 2.
Immunoblot analysis of SIVagm virus isolates recovered from selected tissues of a naturally infected monkey (AGM 9315). Viruses were purified from culture supernatants of infected cell lines and reacted with serum from an SIVagm-infected rhesus macaque (MM7695). Lane B, SIVagm(tyo-1); lane C, SIVagm(sab-4br); lane D, SIVagm(sab-4CSF); lane E, SIVagm(sab-4ln). Lane A represents molecular mass markers. Viral proteins were identified as gp120(env), p34(pol), p24(gag), and p17(gag).
SIVagm isolates from the CNS are macrophage tropic.
The CNS tropism of primate lentiviruses is thought to be a reflection of the ability of these viruses to readily infect brain macrophages and/or microglial cells (33, 36). To assess the cellular tropism of SIV isolates obtained from different anatomic sites, five SIVagm isolates were compared for infection of human T cells and macrophages. Human cells were used because of the lack of AGM CD4+ T-cell lines, the low numbers of CD4+ T-cell targets in the PBMC of AGMs, and difficulty in culturing AGM macrophages. Two molecularly cloned SIVagm viruses from sabaeus and grivet monkeys (MJ8 and gri-1) and three tissue-derived isolates were grown for comparison. All of the SIVagm 9315 tissue isolates replicated to higher titers in the Molt4(cl8) CD4+ T-cell line, with the highest virus levels detected in SIVagm strains derived from the brain and LN, followed closely by the CSF isolate (Fig. 3A). Only the brain and CSF viruses replicated well in macrophage cultures, although the overall levels were severalfold less than that seen for human T-cell lines. Lower virus titers in the macrophage cultures are likely due to the lower total number of cells present in macrophage cultures and the lack of active cell division by macrophages. In contrast to the CNS SIVagm isolates, the LN-derived SIVagm(sab-4ln) gave approximately 1,000-fold-lower virus titers in the macrophage cultures than in the Molt4(cl8) cells.
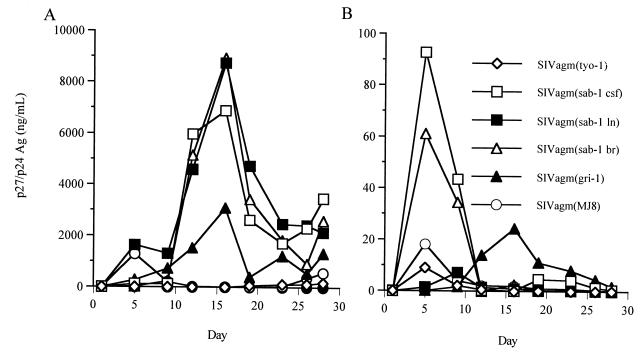
FIG. 3.
In vitro growth characteristics of SIVagm variants in a human T-cell line and macrophages. SIVagm viruses recovered from brain (sab-4br), CSF (sab-4CSF), and LNs (sab-4ln) were normalized for p27 (nanograms per milliliter) and used to infect Molt4 clone 8 cells (A) and human macrophages (B). Well-characterized isolates [SIVagm(tyo-1), SIVagm(MJ8), and SIVagm(gri-1)] were run for comparison. HIV-1/Ba-L is not shown in panel B due to the very high virus peak at day 6 (1,200 ng/ml). Ag, antigen.
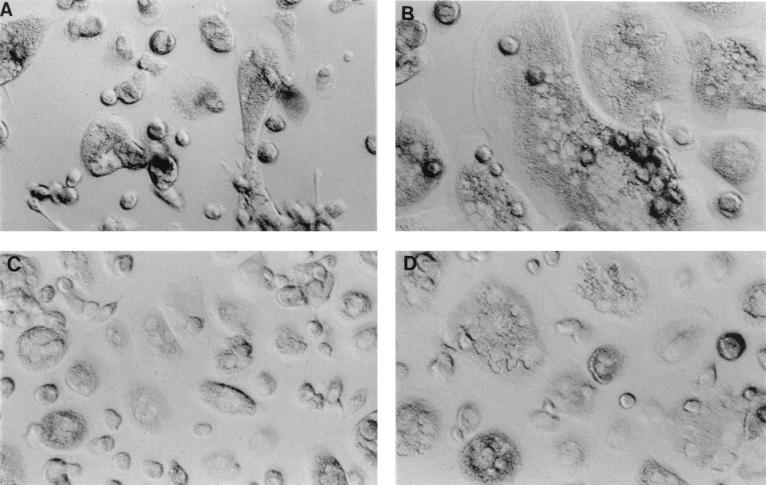
SIVagm strains from the CNS (sab-4br and sab-4CSF) readily infected both T cells and macrophages and displayed similar replication kinetics in both cell types despite having been initially isolated on a human T-cell line. Of the SIVagm molecularly cloned viruses, gri-1 replicated in both Molt4(cl8) cells and macrophages. All SIVagm strains were highly cytopathic for Molt4(cl8) cells, causing massive syncytia, suggesting that these viruses, like HIV, are highly cytopathic for CD4+ T cells. This cytopathicity likely influenced the amount of cell-free virus production in Molt4 cultures. The low level of virus expression in the MJ8 culture in particular is likely due to pronounced cytopathic effects that result in an overall decrease in the number of surviving Molt4(cl8) cells. Consistent with this notion, when the culture was recovering from cytopathic crisis at day 26, p27 antigen levels again began to rise. A similar rebound effect was seen for the other virus cultures (Fig. 3A). HIV-1/Ba-L, a highly macrophage-tropic isolate, was used as a control for lentivirus growth in macrophages (36). HIV-1/Ba-L replicated to the highest titers in the macrophages (data not shown) and induced massive syncytia (Fig. 4B). The SIVagm isolates were also cytopathic for human macrophages, with more multinucleated cells present in the SIVagm(sab-4br) (Fig. 4D) and SIVagm(sab-4CSF) cultures than were observed for the SIVagm(sab-4ln) isolate (Fig. 4C). SIVagm MJ8 and gri-1 infection of macrophages also induced multinucleated cells, although this induction was not as pronounced as the effect of the SIVagm(sab-4) isolates (data not shown); these molecularly cloned viruses still appeared to be primarily T cell tropic. It is interesting to note the rapid kinetics of virus replication (peak virus production by day 6) and the high virus titers observed in these macrophage studies.
FIG. 4.
Cytopathic effects of SIVagm and HIV-1 on human macrophages. Macrophages prepared from human PBMC were infected with medium alone (A), HIV-1/Ba-L (B), SIVagm(sab-4ln) (C), or SIVagm(sab-4br) (D). Cultures are shown at day 14 postinfection. Magnification, ×40.
Sequence analysis of env genes from SIVagm tissue variants.
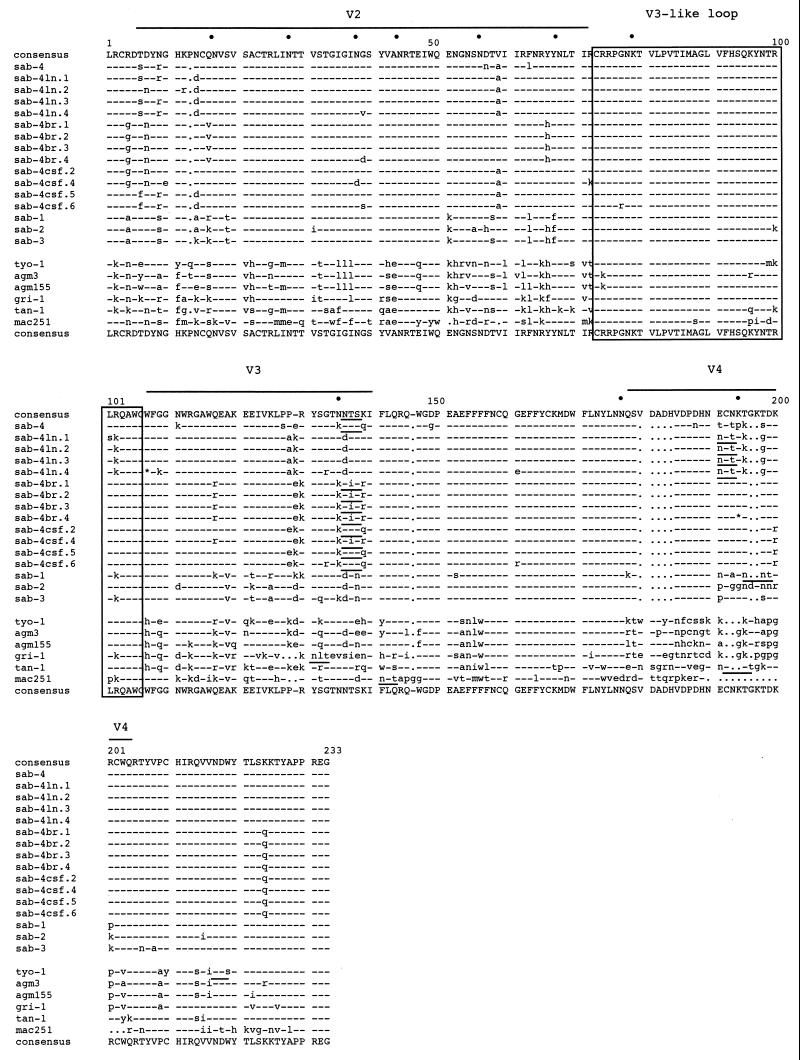
To define genetic differences between tissue variants which might correlate with cell tropism or variations in the size of gp120 species, we amplified a 672-bp fragment representing the V2-C4 env region of SIVagm gp120 and sequenced four clones each from the brain (br), CSF, and LN (ln) isolates of SIVagm(sab-4). The variable regions of SIVagm have been described previously and represent regions that are hypervariable between SIVagm subtypes from grivet, vervet, tantalus, and sabaeus monkeys (31, 69). In HIV-1, the hypervariable V3 region defined by a cysteine-cysteine loop is an important neutralizing domain and also functions in determining coreceptor binding and viral entry (19, 68). For SIVagm, this region is highly conserved with only rare mutations and is designated “V3-like loop” (1). For these sequence comparisons, an infectious clone isolated 5 years previously from the PBMC of the same monkey is presented (sab-4). No large deletions or additions which could have accounted for the difference in gp120 species seen by Western blotting were observed between the viruses derived from the CNS (br and CSF) and the LN variant (Fig. 5). Two clones had defective envelopes (br.4 and ln.4). The number of predicted glycosylation sites was highly conserved among the SIVagm strains, suggesting that gp120 size differences were not explained by differential glycosylation of the V2-C4 region. The overall heterogeneity between the sab-4 CNS and LN viruses was relatively large, ranging from 9 to 14% at the amino acid level, mostly occurring within the small stretches in V2, V3, and V4. Clones from SIVagm(sab-4br) were more homogeneous than those from the CSF or LN, consistent with the results of studies of HIV-1-infected humans (24). In addition, the viruses from the brain and CSF were more closely related to each other than to the LN-derived virus. In HIV-1, the env C4 region functions as part of the CD4 binding domain (63). In the SIVagm viruses analyzed here, a nonconservative mutation of Lys to Gln was seen in all clones derived from the CNS, possibly affecting CD4 or CCR5 binding (61, 83).
FIG. 5.
Deduced amino acid sequence alignment of the V2-C4 env gene region from SIVagm viruses isolated from the CSF, brain, and LNs of a naturally infected green monkey (AGM 9315). The dots above the sequence indicate potential N-linked glycosylation sites, and underlined sequences represent positional changes in these sites. The outlined regions indicate relative variable regions (V) for SIVagm strains. The dashes represent amino acid homology, and lowercase letters are nonsynonymous mutations.
DISCUSSION
This study was performed to elucidate the virologic nature of nonpathogenic, naturally acquired SIV infection of AGMs. The observed high rates of SIVagm seroprevalence for AGMs in the wild and in captivity, the apparent ease of SIVagm transmission between AGMs, and the high levels of genetic diversity delineated among SIVagm isolates all suggest that a substantial level of virus replication takes place in vivo in infected animals (2, 70, 78). Yet, asymptomatic CD4+ T-cell-tropic lentivirus infections have commonly been assumed to reflect low virus loads and effective host immunologic control of virus replication. However, there are no reports to date that employ contemporary methods of virus quantitation to characterize levels of ongoing virus replication in vivo in AGMs naturally infected with SIV. We and others have developed and applied sensitive assays to quantitatively detect variable SIVsm species and demonstrated that high steady-state levels of virus are present in sooty mangabeys naturally infected with SIVsm. In addition, we have observed high rates of plasma virus decay following the initiation of antiviral therapy in healthy SIV-infected sooty mangabeys that are indistinguishable from those calculated for pathogenic SIV and HIV infections (Grant et al., submitted; Silvestri et al., submitted). The high steady-state level of plasma viremia in infected mangabeys is therefore a reflection of high ongoing rates of de novo infection and turnover of infected cells. These data clearly indicate that high levels of viremia and high rates of virus replication do not necessarily lead to disease and that specific host factors or the particular nature of the host response plays a critical role in determining whether disease arises following infection.
We therefore were interested in elucidating whether virus-infected sooty mangabeys and AGMs manifest similar or distinct host-virus equilibria underlying their preserved states of health. In order to precisely quantitate SIVagm RNA levels in AGMs, two different QC RNA PCR assays for the SIVagm(sab) and SIVagm(ver) variants were developed and used to measure SIVagm levels in naturally infected AGMs. High steady-state levels of viral RNA, ranging from 3.5 × 106 to 6.3 × 106 RNA copies/ml of plasma, were observed. In infected humans, similar plasma HIV RNA levels are usually associated with rapid progression to AIDS (66). A recent report describing experimental infection of AGMs has demonstrated high-level plasma viremia and substantial replication in LN cells and PBMC at early times after experimental infection with a decline thereafter (25). In the present study, we demonstrated high levels of plasma viremia for naturally infected animals that had been infected for more than 10 years.
In order to determine whether the high level of plasma viral RNA reflected the presence of infectious virus, PBMC and plasma from SIV-infected AGMs were cocultivated with the human CD4+ T-cell line Molt4. Virus was readily recovered from both plasma and PBMC of naturally infected animals (Table 2), indicating the presence of significant amounts of infectious virus in the peripheral blood. Frequent isolation of infectious virus is consistent with the observed high plasma SIVagm RNA levels and the high frequency of SIVagm transmission (78). The SIVagm isolates obtained in this study were shown to be highly cytopathic for both human macrophages and T cells; however, we were unable to ascertain their cytopathic properties on AGM cells because of difficulties with in vitro growth of CD4+ T cells (J. Allan, unpublished data). Studies of virus and cell turnover rates, such as those performed with HIV-infected humans, SIV-infected macaques, and SIV-infected sooty mangabeys, will be necessary to determine whether SIVagm-infected cells also turn over rapidly in vivo (51, 74, 92; Grant et al., submitted).
To gain insight into the anatomic sites of SIVagm replication and production, we measured SIVagm DNA levels in several different body tissues using a quantitative DNA PCR assay designed to detect diverse SIVagm variants. In HIV-infected humans, the inaccessibility of lymphoid tissues such as those associated with the gut or spleen has made it difficult to unequivocally determine their contribution to overall levels of HIV production in vivo. However, these tissues may contribute significantly to systemic viremia due to the large number of activated CD4+ T cells present within them (5, 50, 89). The immunologically activated milieu of the gastrointestinal tract has been reported to provide fertile substrates for active SIV replication in macaques, likely reflecting the known high frequency of resident activated CD4+ T lymphocytes that are responding to the many environmental and food antigens encountered in the gut (89). Similarly, the highest levels of SIVagm DNA were observed in the gut tissues of naturally infected AGMs. Given that the gut-associated lymphoid tissue represents the largest mass of body lymphoid tissue (>80%), it is likely for AGMs, as has been reported previously for rhesus macaques, that the gut-associated lymphoid tissue represents a major site of virus production in vivo (89).
Despite having persistently high levels of plasma viremia, SIV-infected AGMs display much lower levels of viral DNA in peripheral blood cells than are commonly observed for SIV-infected macaques or HIV-1-infected humans (15, 50, 79). The intrinsically much lower numbers of CD4+ T cells in the peripheral blood of AGMs than in that of humans or mangabeys might help to explain the overall lower numbers of infected cells (71). In addition, the relatively low levels of SIVagm DNA in the PBMC compared to what is observed for pathogenic infections may reflect lower numbers of circulating activated CD4+ T-cell targets for viral infection. In support of this hypothesis, SIV-infected AGMs do not manifest the elevated lymphocyte apoptosis seen in association with the states of generalized immune system activation characteristic of pathogenic HIV or SIV infections (29). Similarly, naturally infected sooty mangabeys do not exhibit the excessive activation and apoptosis of circulating lymphocytes observed for nonnatural hosts of SIV (Silvestri et al., submitted).
Levels of SIVagm DNA in peripheral lymphoid tissues of AGMs, ranging from 50 to 500 viral DNA copies/1.5 × 105 cells (Table 3), were also lower than are observed for pathogenic SIV and HIV infections (8, 41, 50, 64, 79). Relatively low levels of productively infected cells have also been observed by in situ hybridization methods in tissues of experimentally infected AGMs, whereas greater numbers have been detected in naturally and experimentally infected sooty mangabeys (8, 45, 82). The source of high levels of cell-free virus in the blood of the natural host may therefore be in other lymphoid tissues such as the gut, where potentially greater activation is occurring in response to antigens. Activated and proliferating cells are known to provide better targets for virus infection and expression (93). In addition, the activation state of T cells is also known to influence the expression of cell surface coreceptors necessary for viral entry (12). As most SIVagm viruses isolated to date are primarily CCR5 tropic, virus infection may be limited to a small pool of susceptible cells ultimately determined by which tissues represent the sites of greatest immune activation (23, 26).
SIVagm infection, like other lentivirus infections, is marked by extensive viral genetic diversity seen both within and between infected hosts (10). This viral diversity reflects the error-prone nature of the process of reverse transcription, the number of viral replication cycles that lead to introduction of new mutations, and the evolutionary selection of different quasispecies to fill distinct host niches (55). To study the diversity of SIVagm populations within a naturally infected animal, viruses were isolated from the brain, CSF, and LNs of an infected AGM by cocultivation with Molt4 cells. Virus was readily isolated from LNs, brain, and CSF, suggesting considerable virus replication in these tissues in vivo. Because the selective pressures of cell culture may select for only a subset of the viral quasispecies present in vivo, the tissue-specific SIVagm variants described in this study may underrepresent the extent of viral diversity present in the infected animal (22).
The CNS- and LN-derived SIVagm isolates were subjected to nucleotide sequence analysis of the env V2-C4 region and compared to each other and to previously described SIVagm sequences. These studies demonstrated significant predicted amino acid diversity between CNS and lymphoid tissue viruses, on the order of 9 to 14% at the amino acid level; however, only 3 to 8% nucleotide sequence diversity was found. While limited in scope, a greater nonsynonymous/synonymous ratio between viruses isolated from distinct tissue compartments would suggest an adaptive evolution of viral variants to specific anatomic sites. In contrast, interanimal SIVagm diversity has revealed a high ratio of synonymous to nonsynonymous mutation frequencies, indicating that SIVagm is a more ancient virus lineage that has reached some equilibrium outweighing the positive selection for change that has been reported previously for pathogenic HIV-1 and SIV infections (59, 70, 84).
Similar to previous reports of brain HIV env V3 sequences, SIVagm isolated from the brain was shown to have the most homogeneous sequence (three of four clones were identical) (60, 67). Such homogeneity may reflect biological constraints imposed by the available target cells for infection, likely brain microglial cells (90). env V2-C4 sequence analysis also demonstrated the presence of two notable differences between CNS- and lymphoid tissue-derived viruses. A conservative change in the V3 region could conceivably affect viral tropism, while a nonconservative mutation in the CD4 binding region (C4) could affect CD4 binding and possibly coreceptor usage (63, 83). Future mutagenesis studies will be required to determine the functional significance of these mutations. Overall, the observed intra-animal diversity of SIVagm sequences is significant and may reflect both active replication and compartmentalization of viral variants.
Virus isolates were subjected to Western blot analysis to investigate potential differences in SIVagm structural proteins that correspond to tissue-specific compartmentalization of virus replication. Our previous studies demonstrated that each SIVagm subspecies differs considerably with respect to the relative sizes of gp120 and p24 proteins (2, 3). Differences in the apparent molecular weights between SIV gp120s typically result from differences in glycosylation patterns, which contribute to the overall mass of the proteins. The brain and CSF isolates of SIVagm expressed putative gp120 species that were significantly smaller than gp120 from the corresponding LN isolate SIVagm(sab-4ln). These data suggest that the brain and CSF isolates may be less extensively glycosylated and that the CNS-derived isolates are more closely related to each other than they are to the LN virus. The env V2-C4 region sequence analyses described above did not localize the changes responsible for the fewer glycosylation sites underlying the observed variability in gp120 size. However, variability in the number of N-linked sites could exist in other regions of env for which sequence information was not obtained, such as the V1 region. Analyses of HIV-1 isolates from the brain have suggested that similar differences are due to a reduction in the number of N-linked glycosylations (67). Similarly, isolates from the brains of SIVmac/sm-infected monkeys have also been reported previously to manifest differentially glycosylated gp120 species, especially in the V1 region, which are linked to neutralization sensitivities (14, 81).
A hallmark of pathogenic lentivirus infections is the ability to infect CNS microglial cells and the development of neurologic complications in a significant fraction of AIDS patients (33, 42). Viral infection of the CNS likely serves to facilitate viral persistence in an anatomically sequestered site. This study demonstrated substantial levels of cell-free SIVagm in CNS tissues. SIVagm RNA levels in the CSF were similar to the HIV levels found in the CSF of HIV-1-infected humans with AIDS-associated dementia (13, 85). Infectious SIVagm was recovered directly from cell-free homogenates of the cerebral cortex and grew to high titers, demonstrating active viral replication in the CNS. Viral DNA copy numbers in the cerebrum exceeded those found in the PBMC. Despite these high virus titers in the CNS, the results of histopathologic examination of AGM brain tissues were unremarkable, demonstrating that substantial levels of SIVagm antigens can be expressed in the CNS without causing pathological changes. In contrast, high viral loads in the CSF have also been reported for SIVsm-infected macaques, correlating with severe neuropathologic changes (96). The CNS viruses also differed from lymphoid tissue-derived SIVagm strains by their ability to infect human macrophages in addition to human CD4+ T-cell lines. Macrophage tropism is a common property ascribed to HIV-1 strains of CNS origin and has been linked primarily to coreceptor usage (CCR5 and CCR3) (42). Further studies are required to determine if CNS-derived SIVagm strains will also replicate with similar characteristics, i.e., cytopathic effect, in AGM CD4+ T cells and macrophages.
Similarity of SIVagm in the CSF to that found in the brain was demonstrated by similarities in env gp120 size, env sequence, and macrophage tropism. This suggests that SIV detected in the CSF of natural hosts is reflective of active viral replication in the brain parenchyma and not a result of the inflammation-associated trafficking of systemically infected cells to the CSF space (27, 80). Together with the observation of significant viremia in the CSF of naturally SIV-infected mangabeys, these studies suggest that the CNS is an important viral niche in natural hosts of SIV (Grant et al., submitted). The expression of SIVagm gene products in the absence of CNS lesions suggests that the neurologic complications observed for HIV-infected humans may not be due to the toxic effects of viral gene products alone (e.g., tat or gp120) (62). Theoretically, the innate or adaptive host responses to SIV or HIV infection may significantly contribute to neuropathogenesis in nonnatural hosts.
In summary, persistent high-level SIV replication in naturally infected African nonhuman primates as described in this report and elsewhere suggests that the variation between host species in the levels of viral replication alone cannot account for the progressive CD4+ T-cell depletion or neurologic complications of AIDS. Rather, AGM species appear to exist in a nonpathogenic equilibrium with their virus. For sooty mangabeys, immunologic studies suggest that infected animals fail to mount significant antiviral cellular immune responses and show far lower levels of generalized immune system activation and associated lymphocyte apoptosis than are seen for pathogenic SIV and HIV infections (Silvestri et al., submitted). It is presently unknown if SIVagm is directly cytopathic for CD4+ T cells in the natural host, with previous studies having used unfractionated PBMC populations which have a low percentage of CD4+ targets for replication. Natural host species may plausibly still experience the direct cytopathic consequences of virus infection of CD4+ T cells but far less of the bystander damage characteristic of pathogenic primate lentivirus infections. Detailed analysis of the cytopathic potential of SIV for CD4+ T cells derived from the natural hosts is necessary to determine if SIVagm is indeed cytopathic on AGM cells. A lack of cytopathic effect of SIVagm on target cells from the natural host species would also presumably lead to lower overall levels of CD4+ T-cell destruction and little, if any, compromise of their immune regenerative capacity (56; Silvestri et al., submitted). Natural reservoir hosts are in a nonpathogenic equilibrium with their virus, wherein active virus replication is tolerated without development of disease. The extent to which the nonpathogenic SIV infections of AGMs and sooty mangabeys reflect similar or distinct host-virus interactions remains to be determined. Future comparisons of such equilibrium and nonequilibrium hosts promise to help define the pathogenic determinants of T-cell immunodeficiency and other sentinel complications of AIDS. The development of new strategies to prevent and treat HIV infection that are based on modulation of host responses in addition to chemotherapeutic suppression of virus replication may be envisioned.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants RO1 AI28273 and RO1 AI41396 (J.S.A.), R21 AI44763 (M.B.F.), and RR00165–40 (S.I.S.). M.B.F. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
We thank M. Hayami for the generous gift of SIVagm(tyo-1) and Molt4 clone 8 cells, Gene Hubbard for necropsies and pathological evaluations, Kathleen Brasky for veterinary assistance, Brian Corliss for technical assistance, and the monkeys for providing blood and tissue samples. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: HIV-1/Ba-L from Suzanne Gartner, Mikulas Popovic, and Robert Gallo.
REFERENCES
- 1.Allan J S. Receptor-mediated activation of the viral envelope and viral entry. AIDS. 1993;7(Suppl. 1):S43–S50. [PubMed] [Google Scholar]
- 2.Allan J S, Kanda P, Kennedy R C, Cobb E K, Anthony M, Eichberg J W. Isolation and characterization of simian immunodeficiency viruses from two subspecies of African green monkeys. AIDS Res Hum Retrovir. 1990;6:275–285. doi: 10.1089/aid.1990.6.275. [DOI] [PubMed] [Google Scholar]
- 3.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allan J S, Strauss J, Buck D W. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 5.Bagasra O, Steiner R M, Ballas S K, Castro O, Dornadula G, Embury S, Jungkind D, Bobroski L, Kutlar A, Burchott S. Viral burden and disease progression in HIV-1-infected patients with sickle cell anemia. Am J Hematol. 1998;59:199–207. doi: 10.1002/(sici)1096-8652(199811)59:3<199::aid-ajh4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 6.Baier M, Werner A, Bannert N, Metzner K, Kurth R. HIV suppression by interleukin-16. Nature. 1995;378:563. doi: 10.1038/378563a0. [DOI] [PubMed] [Google Scholar]
- 7.Beer B, Denner J, Brown C R, Norley S, zur Megede J, Coulibaly C, Plesker R, Holzammer S, Baier M, Hirsch V M, Kurth R. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:210–220. doi: 10.1097/00042560-199807010-00003. [DOI] [PubMed] [Google Scholar]
- 8.Beer B, Scherer J, zur Megede J, Norley S, Baier M, Kurth R. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long-term simian immunodeficiency virus infection of African green monkeys. Virology. 1996;219:367–375. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 9.Beer B E, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. doi: 10.1128/jvi.73.9.7734-7744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibollet-Ruche F, Brengues C, Galat-Luong A, Galat G, Pourrut X, Vidal N, Veas F, Durand J P, Cuny G. Genetic diversity of simian immunodeficiency viruses from West African green monkeys: evidence of multiple genotypes within populations from the same geographical locale. J Virol. 1997;71:307–313. doi: 10.1128/jvi.71.1.307-313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibollet-Ruche F, Galat-Luong A, Cuny G, Sarni-Manchado P, Galat G, Durand J P, Pourrut X, Veas F. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J Gen Virol. 1996;77:773–781. doi: 10.1099/0022-1317-77-4-773. [DOI] [PubMed] [Google Scholar]
- 12.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brew B J, Pemberton L, Cunningham P, Law M G. Levels of human immunodeficiency virus type 1 RNA in cerebrospinal fluid correlate with AIDS dementia stage. J Infect Dis. 1997;175:963–966. doi: 10.1086/514001. [DOI] [PubMed] [Google Scholar]
- 14.Campbell B J, Hirsch V M. Extensive envelope heterogeneity of simian immunodeficiency virus in tissues from infected macaques. J Virol. 1994;68:3129–3137. doi: 10.1128/jvi.68.5.3129-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y, Qin L, Zhang L, Safrit J, Ho D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 16.Chakrabarti L A, Lewin S R, Zhang L, Gettie A, Luckay A, Martin L N, Skulsky E, Ho D D, Cheng-Mayer C, Marx P A. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J Virol. 2000;74:1209–1223. doi: 10.1128/jvi.74.3.1209-1223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chun T W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 19.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 20.Cone R W, Gowland P, Opravil M, Grob P, Ledergerber B. Levels of HIV-infected peripheral blood cells remain stable throughout the natural history of HIV-1 infection. Swiss HIV Cohort Study. AIDS. 1998;12:2253–2260. doi: 10.1097/00002030-199817000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Daniel D M, Li Y, Naidu M, Durda P J, Schmidt D K, Troup C D, Silva D P, MacKey J J, Kestler H W, Sehgal P K, King N W, Ohta Y, Hayami M, Desrosiers R C. Simian immunodeficiency virus from African green monkeys. J Virol. 1988;62:4123–4128. doi: 10.1128/jvi.62.11.4123-4128.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 24.Diop O M, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Muller-Trutwin M C. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J Virol. 2000;74:7538–7547. doi: 10.1128/jvi.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Stefano M, Gray F, Leitner T, Chiodi F. Analysis of ENV V3 sequences from HIV-1-infected brain indicates restrained virus expression throughout the disease. J Med Virol. 1996;49:41–48. doi: 10.1002/(SICI)1096-9071(199605)49:1<41::AID-JMV7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Edinger A L, Hoffman T L, Sharron M, Lee B, O'Dowd B, Doms R W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 27.Ellis R J, Gamst A C, Capparelli E, Spector S A, Hsia K, Wolfson T, Abramson I, Grant I, McCutchan J A. Cerebrospinal fluid HIV RNA originates from both local CNS and systemic sources. Neurology. 2000;54:927–936. doi: 10.1212/wnl.54.4.927. [DOI] [PubMed] [Google Scholar]
- 28.Ennen J, Findeklee H, Dittmar M T, Norley S, Ernst M, Kurth R. CD8+ T lymphocytes of African green monkeys secrete an immunodeficiency virus-suppressing lymphokine. Proc Natl Acad Sci USA. 1994;91:7207–7211. doi: 10.1073/pnas.91.15.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estaquier J, Idziorek T, de Bels F, Barre-Sinoussi F, Hurtrel B, Aubertin A M, Venet A, Mehtali M, Muchmore E, Michel P, et al. Proc. Natl. Acad. Sci. USA 91:9431–9435. 1994. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkel T H, Tudor-Williams G, Banda N K, Cotton M F, Curiel T, Monks C, Baba T W, Ruprecht R M, Kupfer A. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat Med. 1995;1:129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 31.Fomsgaard A, Hirsch V M, Allan J S, Johnson P R. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991;182:397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- 32.Fukasawa M, Miura T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333:457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 33.Gabuzda D H, Ho D D, de la Monte S M, Hirsch M S, Rota T R, Sobel R A. Ann. Neurol. 20:289–295. 1986. Immunohistochemical identification of HTLV-III antigen in brains of patients with AIDS. [DOI] [PubMed] [Google Scholar]
- 34.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 35.Gartner S. HIV infection and dementia. Science. 2000;287:602–604. doi: 10.1126/science.287.5453.602. [DOI] [PubMed] [Google Scholar]
- 36.Gartner S, Markovits P, Markovitz D, Kaplan M, Gallo R, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 37.Gicheru M M, Otsyula M, Spearman P, Graham B S, Miller C J, Robinson H L, Haigwood N L, Montefiori D C. Neutralizing antibody responses in Africa green monkeys naturally infected with simian immunodeficiency virus (SIVagm) J Med Primatol. 1999;28:97–104. doi: 10.1111/j.1600-0684.1999.tb00257.x. [DOI] [PubMed] [Google Scholar]
- 38.Gougeon L M, Lecoeur H, Dulioust A, Enouf M G, Crouvoiser M, Goujard C, Debord T, Montagnier L. Programmed cell death in peripheral lymphocytes from HIV-infected persons: increased susceptibility to apoptosis of CD4 and CD8 T cells correlates with lymphocyte activation and with disease progression. J Immunol. 1996;156:3509–3520. [PubMed] [Google Scholar]
- 39.Haase A T. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 40.Hahn B H, Shaw G M, De Cock K M, Sharp P M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 41.Hartung S, Boller K, Cichutek K, Norley S G, Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch V M, Campbell B J, Bailes E, Goeken R, Brown C, Elkins W R, Axthelm M, Murphey-Corb M, Sharp P M. Characterization of a novel simian immunodeficiency virus (SIV) from l'hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J Virol. 1999;73:1036–1045. doi: 10.1128/jvi.73.2.1036-1045.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirsch V M, Dapolito G, Goeken R, Campbell B J. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr Opin Genet Dev. 1995;5:798–806. doi: 10.1016/0959-437x(95)80014-v. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirsch V M, Dapolito G A, Goldstein S, McClure H, Emau P, Fultz P N, Isahakia M, Lenroot R, Myers G, Johnson P R. A distinct African lentivirus from Sykes' monkeys. J Virol. 1993;67:1517–1528. doi: 10.1128/jvi.67.3.1517-1528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirsch V M, McGann C, Dapolito G, Goldstein S, Ogen-Odoi A, Biryawaho B, Lakwo T, Johnson P R. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993;197:426–430. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- 49.Hirsch V M, Olmsted R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 50.Hirsch V M, Zack P M, Vogel A P, Johnson P R. Simian immunodeficiency virus infection of macaques: end-stage disease is characterized by widespread distribution of proviral DNA in tissues. J Infect Dis. 1991;163:976–988. doi: 10.1093/infdis/163.5.976. [DOI] [PubMed] [Google Scholar]
- 51.Ho D, Neumann A, Perelson A, Chen W, Leonard J, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 52.Jin J M, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin J M, Rogers J, Phillips-Conroy J E, Allan J S, Desrosiers R C, Shaw G M, Sharp P M, Hahn B H. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J Virol. 1994;68:8454–8460. doi: 10.1128/jvi.68.12.8454-8460.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolly C, Phillips-Conroy J E, Turner T R, Broussard S, Allan J S. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops) J Med Primatol. 1996;25:78–83. doi: 10.1111/j.1600-0684.1996.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 55.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 56.Kaur A, Grant R M, Means R E, McClure H, Feinberg M, Johnson R P. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J Virol. 1998;72:9597–9611. doi: 10.1128/jvi.72.12.9597-9611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kestler H W, Ringler D J, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for the maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 58.Kikukawa R, Koyanagi Y, Harada S, Kobayashi N, Hatanaka M, Yamamoto N. Differential susceptibility to the acquired immunodeficiency syndrome retrovirus in cloned cells of human leukemic T-cell line MOLT-4. J Virol. 1986;57:1159–1162. doi: 10.1128/jvi.57.3.1159-1162.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korber B T, Allen E E, Farmer A D, Myers G L. Heterogeneity of HIV-1 and HIV-2. AIDS. 1995;9(Suppl. A):S5–S18. [PubMed] [Google Scholar]
- 60.Korber B T, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lannuzel A, Barnier J V, Hery C, Huynh V T, Guibert B, Gray F, Vincent J D, Tardieu M. Human immunodeficiency virus type 1 and its coat protein gp120 induce apoptosis and activate JNK and ERK mitogen-activated protein kinases in human neurons. Ann Neurol. 1997;42:847–586. doi: 10.1002/ana.410420605. [DOI] [PubMed] [Google Scholar]
- 63.Lasky L A, Nakamura G, Smith D H, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, Capon D J. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–985. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- 64.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. . (Erratum, 275:14, 1997.) [DOI] [PubMed] [Google Scholar]
- 67.Monken C E, Wu B, Srinivasan A. High resolution analysis of HIV-1 quasispecies in the brain. AIDS. 1995;9:345–349. [PubMed] [Google Scholar]
- 68.Moore J P, Nara P L. The role of the V3 loop of gp120 in HIV infection. AIDS. 1991;5:S21–S33. doi: 10.1097/00002030-199101001-00004. [DOI] [PubMed] [Google Scholar]
- 69.Muller M C, Saksena N K, Nerrienet E, Chappey C, Herve V M A, Durand J-P, Legal-Campodonico P, Lang M-C, Digoutte J-P, Georges A J, Georges-Courbot M C, Sonigo P, Barré-Sinoussi F. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller-Trutwin M C, Corbet S, Tavares M D, Herve V M, Nerrienet E, Georges-Courbot M C, Saurin W, Sonigo P, Barre-Sinoussi F. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology. 1996;223:89–102. doi: 10.1006/viro.1996.0458. [DOI] [PubMed] [Google Scholar]
- 71.Murayama Y, Amano A, Mukai R, Shibata H, Matsunaga S, Takahashi H, Yoshikawa Y, Hayami M, Noguchi A. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int Immunol. 1997;9:843–851. doi: 10.1093/intimm/9.6.843. [DOI] [PubMed] [Google Scholar]
- 72.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Norley S G, Kraus G, Ennen J, Bonilla J, Konig H, Kurth R. Immunological studies of the basis for the apathogenicity of simian immunodeficiency virus from African green monkeys. Proc Natl Acad Sci USA. 1990;87:9067–9071. doi: 10.1073/pnas.87.22.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nowak A M, Lloyd A L, Vasquez G M, Wiltrout T A, Wahl L M, Bischofberger N, Williams J, Kinter A, Fauci A S, Hirsch V M, Lifson J D. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pantaleo G, Graziosi C, Demarest J, Butini L, Montroni M, Fox C, Orenstein J, Kotler D, Fauci A. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 76.Peeters M, Honore C, Huet T, Bedjaba L, Ossari S, Bussi P, Cooper R W, Delaporte E. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS. 1989;3:625–630. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 77.Perelson A, Neumann A, Markowitz M, Leonard J, Ho D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 78.Phillips-Conroy J E, Jolly C J, Petros B, Allan J S, Desrosiers R C. Sexual transmission of SIVagm in wild grivet monkeys. J Med Primatol. 1994;23:1–7. doi: 10.1111/j.1600-0684.1994.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 79.Piatak M, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K-C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 80.Price R W, Staprans S. Measuring the “viral load” in cerebrospinal fluid in human immunodeficiency virus infection: window into brain infection? Ann Neurol. 1997;42:675–678. doi: 10.1002/ana.410420502. [DOI] [PubMed] [Google Scholar]
- 81.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 82.Rey-Cuille M-A, Berthier J-L, Bomsel-Demontoy M-C, Chaduc Y, Montagnier L, Hovanessian A G, Chakrabarti L A. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol. 1998;72:3872–3886. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retrovir. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 84.Shpaer E G, Mullins J I. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J Mol Evol. 1993;37:57–65. doi: 10.1007/BF00170462. [DOI] [PubMed] [Google Scholar]
- 85.Staprans S, Marlowe N, Glidden D, Novakovic-Agopian T, Grant R M, Heyes M, Aweeka F, Deeks S, Price R W. Time course of cerebrospinal fluid responses to antiretroviral therapy: evidence for variable compartmentalization of infection. AIDS. 1999;13:1051–1061. doi: 10.1097/00002030-199906180-00008. [DOI] [PubMed] [Google Scholar]
- 86.Staprans S I, Corliss B C, Guthrie J L, Feinberg M B. Quantitative methods to monitor viral load in simian immunodeficiency virus infections. In: Adolph K W, editor. Viral genome methods. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 171–189. [Google Scholar]
- 87.Staprans S I, Dailey P J, Rosenthal A, Horton C, Grant R M, Lerche N, Feinberg M B. Simian immunodeficiency virus disease course is predicted by the extent of virus replication during primary infection. J Virol. 1999;73:4829–4839. doi: 10.1128/jvi.73.6.4829-4839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Veazey R S, DeMaria M, Chalifoux L V, Shvetz D E, Pauley D R, Knight H L, Rosenzweig M, Johnson R P, Desrosiers R C, Lackner A A. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 90.Watkins B A, Dorn H H, Kelly W B, Armstrong R C, Potts B J, Michaels F, Kufta C V, Dubois-Dalcq M. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science. 1990;249:549–553. doi: 10.1126/science.2200125. [DOI] [PubMed] [Google Scholar]
- 91.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 93.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yerly S, Rutschmann O T, Opravil M, Marchal F, Hirschel B, Perrin L. Cell-associated HIV-1 RNA in blood as indicator of virus load in lymph nodes. The Swiss HIV Cohort Study. J Infect Dis. 1999;180:850–853. doi: 10.1086/314932. [DOI] [PubMed] [Google Scholar]
- 95.Zink C M, Amedee A M, Mankowski J L, Craig L, Didier P, Carter D L, Munoz A, Murphey-Corb M, Clements J E. Pathogenesis of SIV encephalitis. Selection and replication of neurovirulent SIV. Am J Pathol. 1997;151:793–803. [PMC free article] [PubMed] [Google Scholar]
- 96.Zink C M, Suryanarayana K, Mankowski J L, Shen A, Piatak M, Jr, Spelman J P, Carter D L, Adams R J, Lifson J D, Clements J E. High viral load in the cerebrospinal fluid and brain correlates with severity of simian immunodeficiency virus encephalitis. J Virol. 1999;73:10480–10488. doi: 10.1128/jvi.73.12.10480-10488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]