Abstract
Background
NPM1‐mutated acute myeloid leukemia (AML) is the most frequent AML subtype. As wild‐type NPM1 is known to orchestrate ribosome biogenesis, it has been hypothesized that altered translation may contribute to leukemogenesis and leukemia maintenance in NPM1‐mutated AML. However, this hypothesis has never been investigated. We reasoned that if mutant NPM1 (NPM1c) directly impacts translation in leukemic cells, loss of NPM1c would result in acute changes in the ribosome footprint.
Methods
Here, we performed ribosome footprint profiling (Ribo‐seq) and bulk messenger RNA (mRNA) sequencing in two NPM1‐mutated cell lines engineered to express endogenous NPM1c fused to the FKBP (F36V) degron tag (degron cells).
Results and discussion
Incubation of degron cells with the small compound dTAG‐13 enables highly specific degradation of NPM1c within 4 hours. As expected, RNA‐sequencing data showed early loss of homeobox gene expression following NPM1c degradation, confirming the reliability of our model. In contrast, Ribo‐seq data showed negligible changes in the ribosome footprint in both cell lines, implying that the presence of NPM1c does not influence ribosome abundance and positioning on mRNA. While it is predictable that NPM1c exerts its leukemogenic activity at multiple levels, ribosome footprint does not seem influenced by the presence of mutant NPM1.
Keywords: acute leukemia, cell biology, transcription
1. BACKGROUND
NPM1‐mutated AML represents approximately 30–35% of all adult AML [1]. The 5th edition of the World Health Organization [2] and the International Consensus Conference (ICC) [3] classifications of myeloid neoplasms recognize NPM1‐mutated AML as an entity with distinctive clinical, pathological, and genetic features [4]. NPM1 is a nucleolar protein exhibiting multiple functions, including ribosome biogenesis and maintenance of genome stability [4]. Mutations at the C‐terminus of the NPM1 protein abrogate its ability to localize within the nucleolus and result in the generation of a nuclear export signal (NES) motif, leading to enhanced interaction with the nuclear exporter XPO1 and accumulation of mutant NPM1 (NPM1c) in the cytoplasm of AML cells [5].
NPM1c likely exerts its oncogenic functions in the cytoplasm and in the nucleus [6]. While recent work has shed light on how NPM1c facilitates the expression of homeobox (HOX) genes directly at the chromatin level, less is known about possible oncogenic activities of NPM1c in the cytoplasm [6]. In this regard, a gain‐of‐function in the cytoplasm, leading to the inhibition of caspase‐6 and ‐8 with consequent deregulation of cell death and myeloid differentiation has been reported [7]. Moreover, NPM1c has been also demonstrated to hamper the formation of promyelocytic leukemia nuclear bodies, which are regulators of mitochondrial fitness [8].
Translation, the process by which genetic information in messenger RNA (mRNA) is converted into a functional protein by ribosomes and transfer RNA, is one of the critical processes of life that occurs in the cytoplasm. Many cancer‐promoting factors, including cyclins, antiapoptotic factors, regulators of cell metabolism, immune modulators, and proteins involved in DNA repair are translationally regulated and altered translation has been involved in cancer development [9].
One of the putative physiologic functions of NPM1 is the regulation of ribosome biogenesis. Specifically, NPM1 binds to nascent rRNA and several ribosomal proteins [10]. It has been suggested that in NPM1‐mutated AML, ribosome biogenesis and translation may be impacted either by the loss of one copy of wild‐type NPM1 or the presence of NPM1c in the cytoplasm or both [11]. A possible impact of NPM1 on translation is also consistent with the expression pattern of nucleophosmin, that is, diffuse cytoplasmic positivity, in NPM1‐mutated AML cells. However, there is no formal demonstration that NPM1c interacts with ribosomes in the cytoplasm of living cells, and whether NPM1c alters translation in NPM1‐mutated AML remains unclear.
2. METHODS AND RESULTS
It has recently become possible to comprehensively study translation using ribosome footprint profiling [12]. This technique allows us to determine the mRNAs that are bound to ribosomes, hence actively translated. Combining standard bulk RNA‐sequencing (RNA‐seq) and ribosome footprint profiling (Ribo‐seq), the rate of protein synthesis is then inferred by evaluating the ribosome density at each mRNA [13] (Figure 1A).
FIGURE 1.
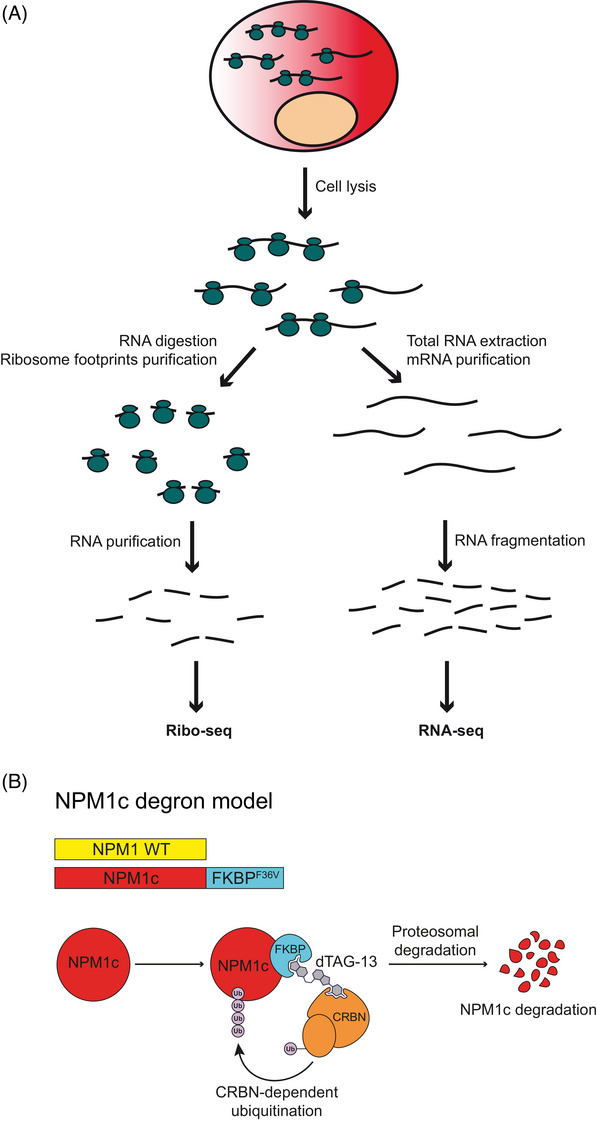
(A) Cartoon depicting the simultaneous processing of RNA for ribosome footprint sequencing (Ribo‐seq, left) and messenger RNA (mRNA) sequencing (RNA‐seq, right). (B) Schematic representation of the NPM1c degron system. The addition of the small compound dTAG‐13 enables CRBN‐dependent ubiquitination and proteasome‐mediated degradation of the target protein.
Here, we combined ribosome footprint profiling with selective NPM1c degradation to explore whether NPM1c influences translation in NPM1‐mutated AML cells. We hypothesized that if NPM1c directly impacts the positioning of ribosomes in leukemic cells, loss of NPM1c would result in acute changes in the ribosome footprint. Hence, we performed ribosome footprint profiling and bulk RNA sequencing in two NPM1‐mutated cell lines (i.e., OCI‐AML3 and IMS‐M2), engineered to express endogenous NPM1c fused to the FKBP (F36V) degron domain (hereafter referred as to degron cells) (Figure 1B). Incubation of degron cells with the small compound dTAG‐13 enables highly specific degradation of > 85% of NPM1c within 4 h [14].
The experiments were performed comparing cells treated with either dimethyl sulfoxide (DMSO) or dTAG for 6 h. We chose this timepoint as it guarantees maximal NPM1c degradation (achieved within 3–4 h [14]) and minimal differentiation, which is known to happen shortly after NPM1c loss [14, 15], influencing translation. Collected cells from each condition were split into aliquots and simultaneously processed for bulk RNA‐sequencing and for ribosome footprint profiling as previously reported [12]. Differential expression (DE) analysis enabled exploring the impact of NPM1c loss on transcriptional regulation (RNA‐seq data), ribosome occupancy (Ribo‐seq data), and translational efficiency (ribosomal mRNA occupancy accounting for mRNA quantity, that is, Ribo‐seq/RNA‐seq). We performed one replicate for OCI‐AML3 and two replicates for IMS‐M2 cells. For DE, when two replicates where used, genes that passed a false discovery rate (FDR) of 5% and with a z‐score of ≥ 4 and ≤ −4 were considered regulated, while when one replicate was used, genes with a z‐score of ≥ 4 and ≤ −4 were considered regulated. Further details are provided in the Supplementary Appendix. Efficient NPM1c degradation was confirmed by western blot in all replicates (Figure S1).
As the impact of NPM1c loss on transcriptome has been already studied [14, 15, 16], we first analyzed RNA‐seq data to confirm the fidelity of our experimental system. In OCI‐AML3 13 genes were upregulated and 12 downregulated (Figure 2A and Table S1) upon NPM1c loss. In line with NPM1c directly promoting the expression of HOX genes, 11 of the 12 downregulated genes were part of the HOX family, including members of the HOXA and HOXB clusters (Figure 2A and Table S1). In IMS‐M2 cells only one gene was upregulated, while none of the downregulated genes had an average z‐score of ≤ −4. However, among the 8 genes with an average z‐score of ≤ −3, 4 were members of the HOXA and HOXB clusters (Figure 2B and Table S1), indicating consistent downregulation of HOX genes across both cell lines. Altogether, these results confirmed that NPM1c drives HOX expression and that our model was reliable.
FIGURE 2.
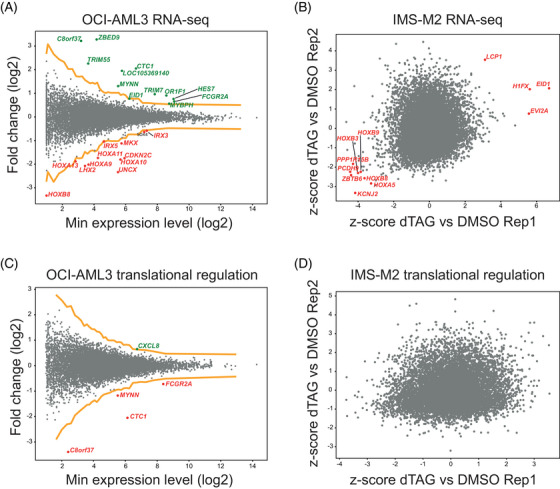
(A) Messenger RNA (mRNA) fold change of OCI‐AML3 degron cells treated for 6 h with dTAG‐13 versus dimethyl sulfoxide (DMSO). The yellow lines represent a z‐score of +4 and −4, respectively. Genes with a z‐score of ≤ ‐4 are highlighted in red, while genes with a z‐score of ≥ 4 are highlighted in green. Single replicate. (B) mRNA fold change of IMS‐M2 degron cells treated for 6 h with dTAG versus DMSO. Genes with a significant average z‐score with a false discovery rate (FDR) of 5% are highlighted in red. Two replicates. (C) Translational efficiency (TE) fold change of OCI‐AML3 degron cells treated for 6 h with dTAG‐13 versus DMSO. The yellow lines represent a z‐score of +4 and −4, respectively. Genes with a z‐score of ≤ ‐4 are highlighted in red, while genes with a z‐score of ≥ 4 are highlighted in green. Single replicate. (D) TE fold change of IMS‐M2 degron cells treated for 6 h with dTAG‐13 versus DMSO. Genes with a significant average z‐score with an FDR of 5% are highlighted in red. Two replicates.
Then, we combined RNA‐seq and Ribo‐seq data to infer translational efficiency (TE) upon loss of NPM1c. TE data revealed that only one gene (CXCL8) was translationally upregulated and 4 genes (CTC1, C8orf37, FCGR2A, and MYNN) were downregulated in OCI‐AML3 (Figure 2C), while none was found to be translationally regulated (any z‐score) in IMS‐M2 using 5% FDR (Figure 2D and Table S1). Altogether, these results indicate that the impact of NPM1c loss on the transcript footprint of functional ribosomes and TE is marginal or likely absent.
3. DISCUSSION
Combining RNA‐seq, Ribo‐seq, and selective NPM1c degradation through dTAG‐13, we explored for the first time the impact of NPM1c on ribosome footprint in the two available NPM1‐mutated cell lines. Analysis of translatome (referring to all mRNAs recruited to ribosomes for protein synthesis) showed that NPM1c only marginally impacts ribosome abundance and positioning on mRNA in NPM1‐mutated cells. Although it is possible that NPM1 mutations may result in deranged translation either by the presence of NPM1c or by the reduction of wild‐type NPM1 levels, there is still no evidence that this could be of biological relevance in AML cells. Furthermore, as each NPM1‐mutated cell harbors one wild‐type and one mutant NPM1 copy, it is possible that residual wild‐type NPM1 is still able to control ribosome biogenesis, ribosome positioning, and translational efficiency. As we did not perform comparative proteomic studies, we cannot exclude that the stability of specific proteins and post‐translational modifications may be still impacted by the presence of NPM1c in the cytoplasm.
In conclusion, while it is predictable that NPM1c likely exerts its oncogenic functions at multiple levels [6], our work shows that the ribosome footprint is not directly impacted by mutant NPM1.
AUTHOR CONTRIBUTIONS
Lorenzo Brunetti, Michael C. Gundry, and Giulia Pianigiani performed the experiments; Margaret A. Goodell provided supplies and contributed to the discussion; Brunangelo Falini coordinated the study. All authors contributed to the manuscript.
CONFLICT OF INTEREST STATEMENT
The corresponding author holds a patent on NPM1 mutants (number 102004901256449). The remaining authors declare no conflict of interest.
ETHICS STATEMENT
The authors have confirmed ethical approval statement is not needed for this submission.
PATIENT CONSENT STATEMENT
The authors have confirmed patient consent statement is not needed for this submission.
CLINICAL TRIAL REGISTRATION
The authors have confirmed clinical trial registration is not needed for this submission.
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC IG 2019 23604 and AIRC Start‐Up 2019 22895), the European Research Council (Advanced Grant 2016 740230), and the National Institutes of Health R01‐CA183252.
Brunetti L, Pianigiani G, Gundry MC, Goodell MA, Falini B. Mutant NPM1 marginally impacts ribosome footprint in acute myeloid leukemia cells. eJHaem. 2024;5:1028–1032. 10.1002/jha2.996
DATA AVAILABILITY STATEMENT
Sequencing data are available in the GEO repository GSE251919.
REFERENCES
- 1. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. [DOI] [PubMed] [Google Scholar]
- 2. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic /dendritic neoplasms. Leukemia. 2022;36(7):1703–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falini B, Dillon R. Criteria for diagnosis and molecular monitoring of NPM1‐mutated AML. Blood Cancer Discov. 2024;5(1):8–20. 10.1158/2643-3230.BCD-23-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23(10):1731–1743. [DOI] [PubMed] [Google Scholar]
- 6. Falini B, Martelli MP, Brunetti L. Mutant NPM1: nuclear export and the mechanism of leukemogenesis. Am J Hematol. 2023;98(4):550–552. [DOI] [PubMed] [Google Scholar]
- 7. Leong SM, Tan BX, Bte Ahmad B, Yan T, Chee LY, Ang ST, et al. Mutant nucleophosmin deregulates cell death and myeloid differentiation through excessive caspase‐6 and ‐8 inhibition. Blood. 2010;116(17):3286–3296. [DOI] [PubMed] [Google Scholar]
- 8. Wu HC, Rerolle D, Berthier C, Hleihel R, Sakamoto T, Quentin S, et al. Actinomycin D targets NPM1c‐primed mitochondria to restore PML‐driven senescence in AML therapy. Cancer Discov. 2021;11(12):3198–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kovalski JR, Kuzuoglu‐Ozturk D, Ruggero D. Protein synthesis control in cancer: selectivity and therapeutic targeting. EMBO J. 2022;41(8):e109823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, et al. Nucleophosmin integrates within the nucleolus via multi‐modal interactions with proteins displaying R‐rich linear motifs and rRNA. Elife. 2016;5:e13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ranieri R, Pianigiani G, Sciabolacci S, Perriello VM, Marra A, Cardinali V, et al. Current status and future perspectives in targeted therapy of NPM1‐mutated AML. Leukemia. 2022;36(10):2351–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165(1):22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015;16(11):651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34(3):499–512 e499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uckelmann HJ, Haarer EL, Takeda R, Wong EM, Hatton C, Marinaccio C, et al. Mutant NPM1 directly regulates oncogenic transcription in acute myeloid leukemia. Cancer Discov. 2023;13(3):746–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang XQ, Fan D, Han Q, Liu Y, Miao H, Wang X, et al. Mutant NPM1 hijacks transcriptional hubs to maintain pathogenic gene programs in acute myeloid leukemia. Cancer Discov. 2023;13(3):725–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Data Availability Statement
Sequencing data are available in the GEO repository GSE251919.


