Abstract
Cell tropism of human and simian immunodeficiency viruses (HIV and SIV, respectively) is governed in part by interactions between the viral envelope protein and the cellular receptors. However, there is evidence that envelope-host cell interactions also affect postentry steps in viral replication. We used a helper-free replication-defective SIV macaque (SIVmac)-based retroviral vector carrying the enhanced jellyfish green fluorescent protein inserted into the nef region (V1EGFP) to examine SIV tropism in a single cycle of infection. Vector stocks containing envelope proteins from three different SIVmac clones, namely, SIVmac239 (T-lymphocyte tropic [T-tropic]), SIVmac316 (macrophage tropic [M-tropic]), and SIVmac1A11 (dualtropic), were tested. SIVmac239 replicates efficiently in many human T-cell lines, but it does not efficiently infect primary rhesus macrophages. Conversely, SIVmac316 efficiently infects primary macrophages, but it does not replicate in Molt4-Clone8 (M4C8) T cells. SIVmac1A11 replicates efficiently in both cell types. When primary macrophages were infected with V1EGFP pseudotyped by SIVmac316 or SIVmac1A11 envelopes, the infection was substantially (ca. 200- to 300-fold) more efficient than for the SIVmac239 pseudotype. Thus, in primary macrophages, a major component of M versus T tropism involves relatively early events in the infection cycle. Quantitative PCR studies indicated that synthesis and transport of vector DNA into the nucleus were similar for macrophages infected with the clone 239 and 316 pseudotypes, suggesting that the restriction for SIVmac239 infection is after reverse transcription and nuclear import of viral DNA. When the same vector pseudotypes were used to infect M4C8 cells, they all showed approximately equivalent infectivities, even though replication-competent SIVmac316 does not continue to replicate in these cells. Therefore, in M4C8 cells, restriction involves a late step in the infection cycle (after proviral integration and expression). Thus, depending on the cell type infected, envelope-dependent cell interactions that govern SIV M and T tropism may involve different steps in infection.
Simian immunodeficiency viruses (SIVs) are important model systems for studying human immunodeficiency virus (HIV), the etiologic agent of AIDS. The infection of rhesus macaques with SIV macaque (SIVmac) results in a clinical immunodeficiency that closely mimics AIDS in humans. As for HIV, the primary receptor for SIV on cells is the CD4 molecule; this molecule is present on the surface of T-helper lymphocytes, macrophages, and dendritic cells. During the course of infection by HIV, there is a shift in biological properties and cell tropism of the virus (4, 17, 31). In initially infected people, the predominant virus replicates well in macrophages and is considered macrophage tropic (M-tropic); as individuals progress to clinical AIDS, virus that replicates preferentially in T lymphocytes (T-tropic virus) appears. For HIV, the determinants of cell tropism have been localized to the V3 loop of envelope SU (gp120) protein (14, 15, 36, 46, 47). More recently, HIV cell tropism has been associated with differential use of cellular coreceptors. Typically, viruses that use CCR5 coreceptor in engineered cells are M-tropic, whereas viruses that use CXCR4 can infect T-cell lines and are T-tropic (replicating poorly in macrophages) (1, 2, 7, 9, 16, 18, 22, 25). Macrophages (and dendritic cells) express CCR5 on the cell surface, while activated T lymphocytes express high levels of CXCR4 (8, 13, 21, 28).
In SIV-infected animals, a similar shift from M tropism to T tropism has also been observed (5, 20, 33, 40, 45), and closely related clones of SIV differ in their cell tropism. Clones SIVmac239 (T-tropic) and SIVmac1A11 (dualtropic) have 98% sequence homology, but only SIVmac1A11 can replicate in macrophages (5, 30). Likewise, SIVmac316 (M-tropic), which was isolated from alveolar macrophages from a monkey inoculated with SIVmac239, replicates more than 100-fold better than SIVmac239 in primary alveolar macrophage cultures (33). The primary determinants of M tropism for SIVmac map to specific regions of the envelope protein (3, 5, 33, 37). However, in the case of SIVs, cell tropism cannot be attributed to the same coreceptor preferences observed for HIV. In particular, both M-tropic and T-tropic SIVs efficiently utilize CCR5, while neither class of viruses recognizes CXCR4 (23, 24). While other alternate coreceptors have been identified for SIV (e.g., GPR15 [BOB], STRL-33 [Bonzo], CCR8, ChemR23, and GPR-1 [10, 19, 26, 38, 42]), the cellular distribution of these coreceptors has not provided an explanation for SIV cell tropism.
In light of the fact that SIV cell tropism does not appear to be governed by coreceptor preference, the mechanisms by which M-tropic and T-tropic SIVs infect or are restricted in different cell types have been of considerable interest. Mori et al. (32) addressed this by comparing infection of primary alveolar macrophages by the T-tropic clone SIVmac239 and by SIVmac239/316, an M-tropic recombinant clone of SIVmac239 containing the envelope from M-tropic SIVmac316. They found that these two viruses generated quite similar (within fivefold) levels of viral DNA when infected into the macrophages, suggesting that the major restriction for replication of SIVmac239 in macrophages was at a step after reverse transcription. Similar conclusions were reached by other investigators (27, 49).
One of the limitations of the previous studies of SIV cell tropism was the fact that replication-competent viruses were used. This made it somewhat difficult to distinguish between different steps in the infection cycle, since infection by SIV in vitro can be somewhat asynchronous and multiple (undetermined) rounds of infection may take place during the course of an experiment. We therefore have examined the issue of SIV cell tropism by using replication-defective, helper-free SIVmac-based vectors pseudotyped with envelope proteins from SIVs with different cell tropisms. These experiments limited infection to a single cycle, because these SIV-based vectors are replication defective. Moreover, a vector expressing a readily detected reporter gene made it possible to obtain sensitive and precise quantification of vector infection and restriction. As described in the experiments reported here, this approach led to evidence for two distinct modes of envelope-dependent restriction of SIV replication in primary macrophages and T-lymphocyte lines.
MATERIALS AND METHODS
Vector plasmids.
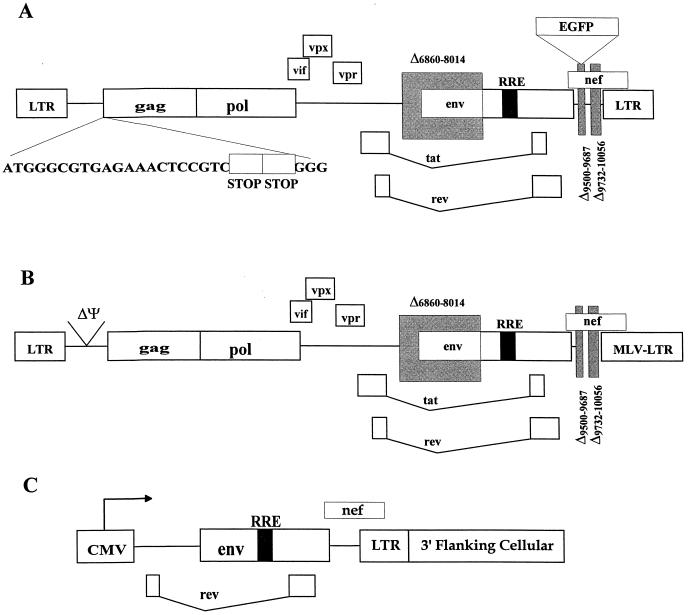
The plasmid pV1EGFP carries a SIVmac239-based vector expressing the enhanced jellyfish green fluorescent protein (EGFP) in place of the nef gene. Its construction has been described elsewhere (26a). The vector carried by pV1EGFP is replication defective due to two stop codons at the beginning of gag and deletions in vif and env. The organization of V1EGFP is shown in Fig. 1.
FIG. 1.
SIV vector and packaging plasmids. Schematic diagrams of the SIVmac239-based vector V1EGFP (A), the packaging plasmid pUpSVOΔΨ (B), and the envelope expression plasmids (C) are shown. (A) V1EGFP is replication defective due to a deletion in env and two consecutive stop codons at the beginning of gag. (B) pUpSVOΔΨ contains a deletion in env and the packaging sequence (Ψ) and contains a heterologous murine leukemia virus 3′ long terminal repeat (LTR). (C) The envelope expression plasmids all contain the cytomegalovirus immediate-early promoter driving the expression of env, rev, and nef; different plasmids contain env sequences from different SIVmac strains.
The gag-pol helper plasmid pUpSVOΔΨ was also described previously (26a). This plasmid carries the gag and pol genes of SIVmac239; the putative packaging sequence (ψ) was deleted, and a simian virus 40 (SV40) origin of replication was added for amplification in T antigen-containing cells. The env expression plasmid pCDSenv, which expresses SIVmac239 env (as well as tat and rev), was also described previously (44). The sequences downstream of SphI at position 6450 of pCDSenv were replaced with the corresponding sequences from SIVmac1A11 (30), kindly provided by Marta Marthas, to produce p1A11env. p316env was produced by replacing the 3′ sequences downstream of SphI (position 6450) of pCDSenv with the corresponding sequences from SIVmac316 (33), kindly provided by R. C. Desrosiers. Each envelope expression plasmid was sequenced using an ABI prism automated sequencer and verified with corresponding sequences in GenBank.
Cell culture.
293T cells (a human embryonic kidney cell line that expresses the adenovirus early proteins and SV40 large T antigen) were obtained from the American Type Tissue Culture Collection (Rockville, Md.). These cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 300 μg of l-glutamine/ml. CMMT-CD4 cells (a rhesus macaque mammary tumor cell line that expresses human CD4 [11]) were maintained in DMEM supplemented with 10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 300 μg of glutamine/ml, and 0.2 mg of gentamicin (G418, at an active concentration of 700 μg/mg)/ml. Molt4-Clone8 cells (a human T-cell line; AIDS Research and Reference Reagent Program) were maintained in RPMI complete (10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 300 μg of l-glutamine/ml). Macrophage cultures were established from rhesus macaque (Macaca mulatta) whole blood. Briefly, 20 ml of heparinized rhesus macaque whole blood was harvested by centrifugation at 12,000 × g for 10 min, resuspended in 9.0 ml of RPMI complete without serum, and then separated by centrifugation over 8 ml of lymphocyte separation medium (Cappel, Aurora, Ohio). Cells were washed twice in 50 ml of RPMI complete without serum by low-speed centrifugation, resuspended in 20.5 ml of macrophage adherence medium (20% FBS–10% human AB serum in RPMI complete), and then plated onto 12-well plates and maintained in culture with macrophage growth medium (RPMI complete supplemented with 20% FBS, 200 U of human recombinant granulocyte-macrophage colony-stimulating factor [Genzyme, Cambridge, Mass.]). Cells were washed rigorously three times to remove nonadherent cells with RPMI complete without serum at 48 and 72 h. Cells were refed with macrophage growth medium every 3 days thereafter.
Vector production.
Vector stocks were made by transient transfections of 293T cells. Briefly, 2 × 105 293T cells were plated 2 days prior to transfection in 6-cm-diameter dishes. The plates were refed 2 h prior to transfection with 5 ml of fresh DMEM with 10% FBS. Transfections were performed by the calcium phosphate method using the Calphos Maximizer transfection kit (Clontech, Palo Alto, Calif.). Fifteen micrograms of plasmid DNA (5 μg each of V1EGFPSVO, pUpSVOΔΨ, and an envelope expression plasmid) was used, and transfection reaction mixtures were incubated at 37°C under 5% CO2. Plates were refed with a half volume (2.5 ml) of medium at 12 h posttransfection and incubated at 32°C under 5% CO2. At 24 h after refeeding, vector supernatants were collected, filter clarified through a 0.45-μm-pore-size filter, and stored frozen at —140°C.
Titration of vector stocks.
A total of 5 × 104 CMMT-CD4 cells were plated in 2 ml of medium in 12-well plates 24 h prior to infection. Infections were carried out by aspirating the wells and then adding 295 μl of fresh medium, 15 μg of DEAE-dextran/ml, and 5 μl of diluted vector stock. After 2 h, 2 ml of fresh medium was added to each well. Four days later, the cells were microscopically scanned for EGFP expression using a fluorescent microscope with a fluorescein isothiocyanate filter designed for optimal EGFP detection (Chroma Technology, Brattleboro, Vt.). The number of green fluorescent cells reached a plateau at 4 days postinfection. Titers of each vector stock (green-fluorescence units [GFU] per milliliter) were calculated by multiplying the total number of EGFP-positive colonies by 200 (to correct for volume of supernatant used) and then multiplying by the dilution factor. In some cases, vector titrations were carried out by flow cytometry (see below).
Characterization of vectors.
Reverse transcriptase inhibition assays were performed by preincubating CMMT-CD4 cells, Molt4-Clone8 cells, and macrophages with 25 μM 9-(2-phosphonylmethoxypropyl) adenine (PMPA; Gilead Sciences, Hayward, Calif.) (48) 1 h prior to infection. Fifty microliters of a 1.3 × 106-GFU/ml concentration of the SIVmac239 (pCDSenv) pseudotype, a 3.2 × 105-GFU/ml concentration of the SIVmac1A11 pseudotype, or a 1.4 × 106-GFU/ml concentration of the SIVmac316 pseudotype was used for infection of these cells. Ninety-six hours after initiation of infection, the cells were screened by fluorescent microscopy or flow cytometry (see below) for EGFP expression. Vector stocks were tested for replication-competent recombinants by long-term infection of CEMX174 cells. Briefly, 106 CEMX174 cells in 500 μl of medium (RPMI 1640 plus 10% FBS) were infected with 500 μl of serially diluted and undiluted vector stocks in triplicate. Twice weekly for 4 weeks, a half volume of medium (500 μl) was removed and the cultures were refed with an equal volume of fresh medium. The medium removed from the cells was clarified by low-speed centrifugation and then precipitated with polyethylene glycol. Virion pellets were resuspended, and the level of reverse transcriptase activity was assayed using a standard reverse transcriptase assay as previously described (41).
Vector infections.
In infections of rhesus monocyte-derived macrophages, 5 × 104 macrophages were refed with 500 μl of macrophage growth medium and infected with 500 μl of viral stocks diluted so that infection would be in the linear range (1.3 × 105 GFU of pCDSenv pseudotype/ml, 3.2 × 103 GFU of p1A11env pseudotype/ml, or 3.4 × 103 GFU of p316env pseudotype/ml) and incubated at 37°C under 5% CO2 for 48 h. The macrophages were refed with 2 ml of fresh macrophage growth medium, and EGFP expression was evaluated 96 h after the initiation of infection. In infection of Molt4-Clone8 cells, 106 cells in 200 μl of RPMI complete were plated into six-well plates 2 h prior to the start of infection. Each envelope pseudotype was added to the cells at 105 GFU in a final volume of 500 μl and then incubated for 24 h in 37°C under 5% CO2. Two milliliters of fresh RPMI complete was added 24 h after initiation of the infections, and 1/20 volume of cells was evaluated for EGFP expression at 96 h by fluorescent microscopy.
Flow cytometry.
For flow cytometry, cultures of infected primary macrophages, Molt4-Clone8 cells, or CMMT-CD4 cells were aspirated and washed one time with phosphate-buffered saline (PBS). The cells were then removed from the culture dish by trypsinization (0.05% trypsin–1 mM EDTA–0.5 ml per 10-cm-diameter dish) for 2 to 5 min at 37°C. One milliliter of PBS was then added, and the cell suspension was harvested by low-speed centrifugation. The cell pellet was suspended in 0.5 ml of PBS, and 0.5 ml of 4% paraformaldehyde in PBS was added. After 30 min at room temperature, cell suspensions were analyzed by flow cytometry with a Becton-Dickinson FACScalibur in the analytical mode. EGFP-positive cells were detected in the green fluorescence channel. Events of 5,000 to 10,000 cells were recorded.
Flow cytometry was also used to measure titers of SIV vectors. In this case, CMMT-CD4 cells were infected with different dilutions of vector stocks, and then 10,000 cells from the infected cultures were analyzed for EGFP-positive cells. The numbers of total EGFP-positive cells in the cultures were then calculated from the total number of cells in the cultures and the percentage of EGFP-positive cells. In addition, the total numbers of EGFP-positive cells were divided by a factor of 2.8, to correct for the average number of cell divisions between the start of infection and time of assay. This factor was determined by comparing the numbers of EGFP-positive colonies determined by fluorescence microscopy on parallel cultures with those analyzed by flow cytometry.
PMPA timed infections.
For infection of macrophages with timed addition of PMPA, 5 × 104 macrophages were plated onto 12-well plates and infected with the different vector pseudotypes as described above. A 100 μM concentration of PMPA was added to one well at 0, 1, 3, 6, 12, 18, and 24 h. In one well, infection was performed without the addition of PMPA. At the 36-h time point, all wells were fed with 2 ml of macrophage growth medium containing 100 μM PMPA. EGFP-positive macrophages were counted 4 days postinfection by fluorescent microscopy or flow cytometry, and the percentage of infected (EGFP-positive) cells relative to the number in infected macrophages in the absence of PMPA was calculated. For timed addition of PMPA in Molt4-Clone8 cells, 106 Molt4-Clone8 cells were plated into six-well plates 2 h prior to the start of infection. Each envelope pseudotype was added to the cells at 105 GFU in a final volume of 500 μl. A 100 μM concentration of PMPA was added to wells at —1, 0, 3, 9, 15, 24, and 30 h. At the 36-h time point, all wells were refed with 2 ml of RPMI complete containing 100 μM PMPA. The percentage of EGFP-positive cells, relative to the number of EGFP-positive cells in infected Molt4-Clone8 cells in the absence of PMPA, was calculated.
Quantitative real-time PCR for detection of newly synthesized SIV DNA.
Prior to infections, V1EGFP vector stocks pseudotyped with either SIVmac239 or SIVmac316 envelopes were digested with RNase-free pancreatic DNase (250 μg/ml in PBS and 10 mM MgCl2; 30 min at 37°C). Infections were carried out on 10-cm-diameter tissue culture dishes containing 2 × 106 peripheral blood mononuclear cell (PBMC)-derived macrophages. One milliliter of diluted vector stock (5 × 104 GFU/ml on CMMT-CD4 cells) was adsorbed to the macrophage cultures (multiplicity of infection = 0.025) for 2 h; the virus was then aspirated, and the cells were washed once with PBS and then refed with 10 ml of macrophage growth medium. As a control, macrophages were pretreated for 75 min with 200 μg of PMPA/ml, and the infections were carried out in the presence of PMPA as well. Twenty-four hours after infection, the macrophages were harvested by trypsinization (0.05% trypsin–1 mM EDTA) and washed once with PBS. The cells were then suspended in hypotonic buffer (0.01 M NaCl, 0.01 M MgCl2 [pH 7.4]) for 10 min on ice and then lysed by the addition of NP-40 to 1% followed by vortex mixing for 15 s. Nuclei were recovered by centrifugation (12,000 × g for 2 min), and nuclear DNA was extracted with the Qiagen tissue kit and suspended in 100 μl of TE (0.01 mM Tris [pH 7.4], 1 mM EDTA) buffer containing 0.2 mg of yeast tRNA per ml.
Quantification of vector DNA from the infected macrophages was performed by real-time PCR using the TaqMan amplification system. PCR amplification for the SIV gag region (present in V1EGFP vector DNA) was carried out. Forward and reverse PCR primers were SIVgag1120F (AGTACGGCTGAGTGAAGGCAGTA) and SIVgag1192R (GACCCGCGCCTTTATAGGA), respectively, and the fluorescent gag probe was SIVgagprobe1147 (6-carboxyfluorescein-CGGCAGGAACCAACCACGACG-NNN′N′-tetramethyl-6-carboxy rhodamine). Nuclear DNA samples corresponding to equal numbers of cells infected by the different vector pseudotypes were analyzed in parallel; fluorescence was recorded as a function of PCR amplification cycle.
Infection with replication-competent SIVmac.
Infection of T-cell lines with replication-competent SIVmac239, SIVmac316, and SIVmac1A11 was performed as described previously (41).
RESULTS
Generation of helper-free SIV vector stocks.
For these experiments, we used a helper-free vector based on SIVmac239, described elsewhere (26a). This vector, V1EGFP, expresses EGFP; the EGFP gene was inserted into the SIV genome in place of the nef gene (Fig. 1). In addition, the vector was rendered replication defective by two consecutive stop codons at the beginning of the gag gene and by deletion of coding sequences from the env gene. To generate vector stocks, a plasmid containing the V1EGFP vector organization along with an SV40 origin of replication (V1EGFPSVO) was cotransfected into human 293T cells along with two helper plasmids. One helper plasmid (pUpSVOΔΨ) expressed the SIVmac239 gag, pol, vpx, vpr, vif, tat, and rev genes from a deleted form of the provirus; the other plasmid (pCDSenv) expressed the env, nef, tat, and rev genes under control of the cytomegalovirus immediate-early promoter. Both of the helper plasmids lacked the SIV RNA packaging signals (39) so that the mRNAs encoded by them should not be packaged into virus particles, and they both contained SV40 origins of replication for efficient expression in 293T cells. Vector stocks were harvested from the cotransfected 293T cells at 48 and 72 h posttransfection. It was possible to change the Env protein on the vectors by changing the env helper plasmid; for these experiments, we generated versions of this plasmid containing genes from SIVmac239 (T-tropic), SIVmac1A11 (dualtropic), and SIVmac316 (M-tropic). The resulting vector particles contained all of the SIV structural proteins, including accessory proteins such as Vpr and Vpx.
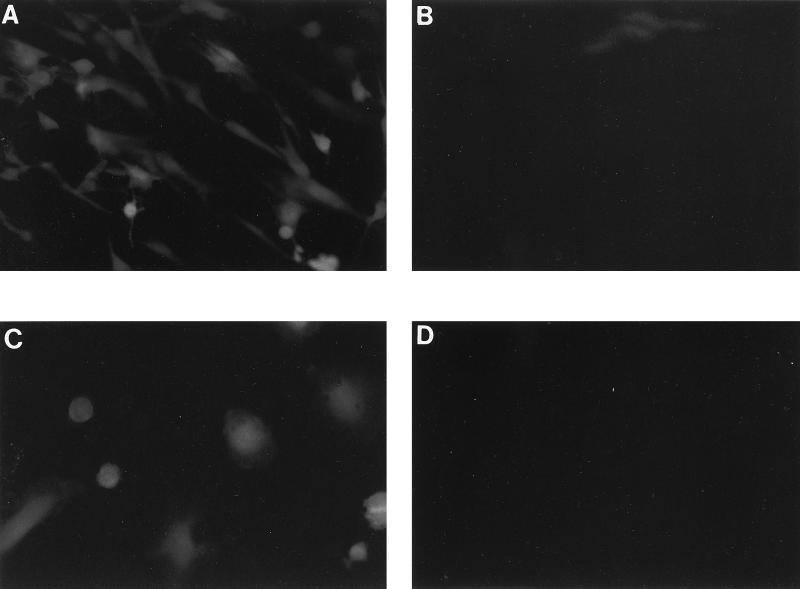
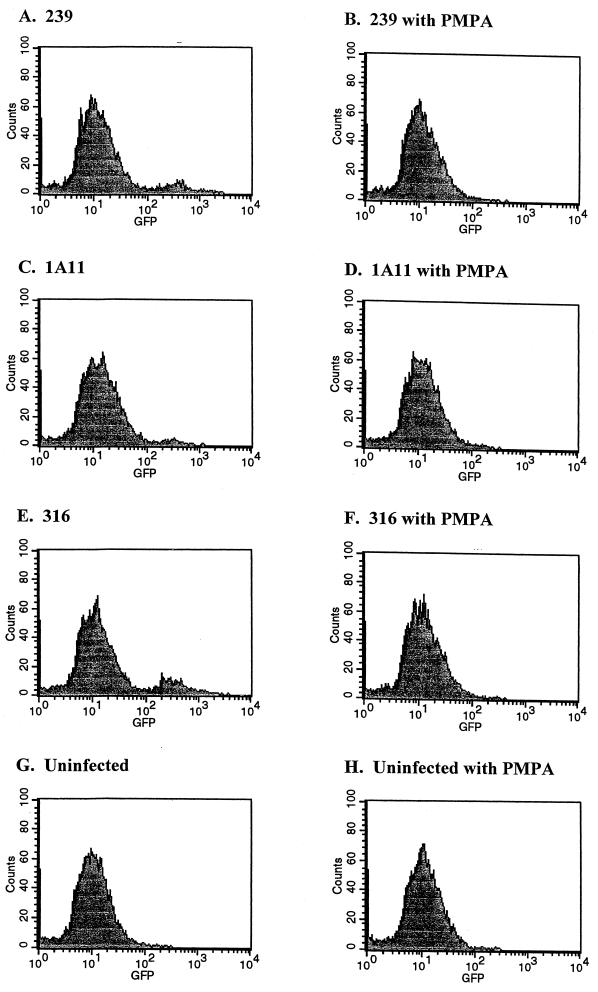
Infection of CMMT-CD4 cells by the V1EGFP vectors was carried out to assess the efficiency of vector expression. CMMT-CD4 cells are macaque mammary tumor cells that express the human CD4 protein (11). The cells are infectible by most strains of SIV, and they show similar efficiencies of infection for M- and T-tropic SIV strains (11). Infection of the cells with the different V1EGFP pseudotypes resulted in readily detectable green fluorescence 4 days postinfection (Fig. 2a). As an alternate detection technique, infected cells were trypsinized and single-cell suspensions were screened by flow cytometry (fluorescence-activated cell sorting [FACS]). Fluorescent cells could be readily detected by the FACS analysis, and there were no differences in the fluorescent intensities of cells infected with the different envelope pseudotypes (Fig. 3). Uninfected CMMT-CD4 cells did not show detectable fluorescence either by fluorescence microscopy or FACS analysis (Fig. 2B and 3G). The vector stocks efficiently infected other cells that are susceptible to SIV infection, including CEMX174 cells, Molt4-Clone8 cells, rhesus PBMCs, and primary rhesus macrophages. To further test whether the fluorescent signals were the result of retroviral infection, infections were carried out in the presence of the reverse transcriptase inhibitor PMPA (25 μM). Pretreatment of the cells 1 h prior to infection effectively eliminated green fluorescent cells (Fig. 2D and 3).
FIG. 2.
Infection with V1EGFP vector. Cultures of CMMT-CD4 cells or primary rhesus macrophages were infected with undiluted stocks of V1EGFP pseudotyped with SIVmac1A11 envelope. The cultures were then examined by fluorescence microscopy with a green filter. (A) Infected CMMT-CD4 cells 4 days postinfection. (B) Uninfected CMMT-CD4 cells. (C) Infected rhesus macrophages 4 days postinfection. (D) CMMT-CD4 cells infected in the presence of the reverse transcriptase inhibitor PMPA.
FIG. 3.
Fluorescence intensity of V1EGFP-infected cells. Cultures of CMMT-CD4 cells (5 × 104) were infected with V1EGFP pseudotyped with different SIVmac envelopes, in the presence or absence of PMPA. The cultures were then harvested at 4 days postinfection, fixed, and analyzed for green fluorescence by flow cytometry. The x axis shows log fluorescence intensity and the y axis shows cell number; 10,000 cells were analyzed in each case. (A and B) Cells infected with a SIVmac239 pseudotype (1.6 × 104 GFU); (C and D) cells infected with a SIVmac1A11 pseudotype (4 × 103 GFU); (E and F) cells infected with a SIVmac316 pseudotype (8 × 103 GFU); (G and H), uninfected cells. (B, D, F, and H), cultures infected in the presence of 100 μM PMPA. The vector-infected cells are evident as the peak with a mean fluorescence of ca. 200 to 300. Note that the fluorescence values for the cultures infected with the different pseudotypes were approximately the same.
The titers of the vector stocks were determined by infecting CMMT-CD4 cells at different dilutions, followed by counting fluorescent cells (by microscopy or by FACS), and the results are shown in Table 1. Vector titers ranged from 3 × 105 to 1.3 × 106.
TABLE 1.
Titers of V1EGFP vectors pseudotyped with different SIVmac envelopes
| Vector plasmid | Packaging plasmid | Envelope plasmid | Titer (GFU/ml)a |
|---|---|---|---|
| pV1EGFPSVO | pUpΔΨSVO | pCDSenv | 1.26 × 106 |
| pV1EGFPSVO | pUpΔΨSVO | p1A11env | 3.20 × 105 |
| pV1EGFPSVO | pUpΔΨSVO | p316env | 1.36 × 106 |
Titers were determined by standardized infection of CMMT-CD4+ cells.
Because our experiments were designed to study a single round of infection, it was important for the vector stocks to be free of replication-competent SIV. In principle, they should have lacked infectious SIV, since the helper plasmids encoding the SIV structural proteins lacked the RNA packaging signals. As described elsewhere (26a), the V1EGFP stocks prepared as described here lacked detectable replication-competent SIV as measured by serial passage on CEMX174 cells followed by assays for reverse transcriptase activity.
Linearity of infection.
It was important to establish the linear range of infection for the vectors in each of the cell lines or primary cell types, since our goal was to quantify the efficiency of infection in these different cells. Moreover, as mentioned in Discussion, linearity of infection proved essential for obtaining results that reflected actual efficiencies of infection. Table 2 shows data from infection of primary macrophages with different concentrations of V1EGFP vectors pseudotyped with different SIVmac envelopes. In macrophages from animal 30440, linearity was achieved when the vector stocks were diluted at least 20-fold. Similar results were obtained for vectors pseudotyped with the SIVmac239 and SIVmac316 envelopes used to infect primary macrophages or Molt4-Clone8 cells, but the range of dilutions over which linearity was achieved differed for various vector-cell combinations. For instance, for macrophages from animal 25980, when V1EGFP vector pseudotyped with the SIVmac239 envelope was used to infect primary macrophages, linearity occurred even with a vector stock diluted 1:5. In contrast, vectors pseudotyped with clone 316 or 1A11 envelopes required dilutions of 1:320 to achieve linearity for this animal. All of the studies described below were carried out with diluted vector stocks that were in the linear range. In practice, linearity was achieved when no more than 5% of the cells were infected.
TABLE 2.
Linearity of macrophage infectionsa
| Animal no. | Vector envelope | Dilution | No. of green cells | Calculated titer |
|---|---|---|---|---|
| 30440 | 1A11 | Undiluted | 9,668 | 1.9 × 104 |
| 1:5 | 6,376 | 6.4 × 104 | ||
| 1:20 | 3,080 | 1.23 × 105 | ||
| 1:80 | 736 | 1.17 × 105 | ||
| 1:320 | 168 | 1.07 × 105 | ||
| 25980 | 239 | Undiluted | 296b | 23b |
| 1:5 | 4,876 | 1.9 × 103 | ||
| 1:10 | 2,928 | 2.3 × 103 | ||
| 1:20 | 976 | 1.5 × 103 | ||
| 1:80 | 244 | 1.5 × 103 | ||
| 1:320 | 92 | 2.3 × 103 | ||
| 1:1,280 | 16 | 2.1 × 103 | ||
| 1A11 | Undiluted | 29,160 | 9.1 × 103 | |
| 1:5 | 17,292 | 2.7 × 104 | ||
| 1:10 | 14,672 | 4.6 × 104 | ||
| 1:20 | 10,712 | 6.7 × 104 | ||
| 1:80 | 3,716 | 9.3 × 104 | ||
| 1:320 | 1,232 | 1.23 × 105 | ||
| 1:1,280 | 332 | 1.32 × 105 | ||
| 316 | Undiluted | 19,440 | 1.4 × 103 | |
| 1:5 | 18,552 | 6.8 × 103 | ||
| 1:10 | 17,423 | 1.3 × 104 | ||
| 1:20 | 15,496 | 2.3 × 104 | ||
| 1:80 | 12,664 | 7.4 × 104 | ||
| 1:320 | 4,472 | 1.05 × 105 | ||
| 1:1,280 | 1,288 | 1.21 × 105 |
Cultures of 4 × 105 rhesus macrophages growing in 12-well plates were infected with 500 μl of various dilutions of V1EGFP stocks pseudotyped with SIVmac1A11 (undiluted = 1.6 × 105 GFU/ml titered on CMMT-CD4 cells), SIVmac239 (undiluted = 6.3 × 105 GFU/ml), or SIVmac316 (undiluted = 6.8 × 105 GFU/ml) envelopes. Green fluorescent cells were counted after 4 days by fluorescent microscopy. Macrophages from two different animals were tested.
In other infections of this vector stock in primary macrophages, the number of green cells for the undiluted sample was slightly higher than that for the sample diluted 1:5.
Infection in primary rhesus macrophages.
Previous experiments by other investigators have shown differences in the ability of various SIVmac strains to infect primary alveolar or blood-derived rhesus macrophages over multiple rounds of infection (5, 27, 30, 32, 33, 35, 49). In particular, SIVmac239 does not replicate (31) (or replicates poorly [26, 47]) in primary rhesus macrophages, while SIVmac316 efficiently replicates in the cells. We wished to reexamine this issue by using the replication-defective V1EGFP vector pseudotypes, since these vectors would limit infection to only one round. Moreover, these vectors will carry out early steps in the infection cycle, including entry, reverse transcription, integration, and transcription. However, detection of vector infection does not require late events such as virion protein expression, virus particle assembly, or virion maturation. Thus, these vectors would also allow discrimination between early and late blocks in viral replication; if the block for SIVmac239 replication in macrophages is at a late step, then V1EGFP pseudotypes with either SIVmac239 or SIVmac316 envelopes would be expected to infect and express in primary macrophages with equal efficiency. On the other hand, if the block is at an early step, then the SIVmac316 pseudotype would be expected to efficiently infect the macrophages, while the SIVmac239 pseudotype would not.
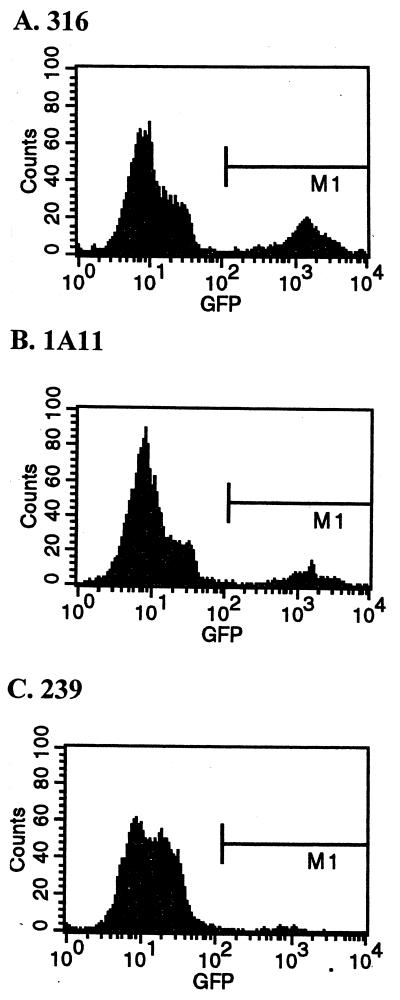
We used the V1EGFP vector stocks described in Table 1 to infect PBMC-derived macrophages from four rhesus macaques under conditions of linear infection, as shown in Table 3. There was a striking difference between the efficiencies of infection for the different V1EGFP pseudotypes. In particular, vector pseudotyped with the SIVmac239 envelope was substantially less efficient at infecting the macrophages than the same vector pseudotyped with either the SIVmac316 or SIVmac1A11 envelope. On average, vector pseudotyped with the SIVmac316 envelope was 295-fold more infectious on the primary macrophages than was the same vector pseudotyped with the SIVmac239 envelope. Likewise, vector pseudotyped with the SIVmac1A11 envelope was on average 167-fold more infectious than vectors pseudotyped with the SIVmac239 envelope. Further, comparison of the mean intensity of intracellular GFP signal from infection of macrophages with the different envelope pseudotypes did not show significant differences in fluorescence intensity, suggesting similar levels of EGFP expression in the cells (Fig. 4). These results indicate that a substantial portion of the replication block for virus carrying SIVmac239 envelope in primary macrophages can be explained by a defect or defects in relatively early steps in the infection cycle.
TABLE 3.
Infection of macrophages by different vector pseudotypes
| Macrophages from monkey no. | Titer (GFU/ml) for pseudotypea
|
Ratio of corrected titer for pseudotype
|
|||
|---|---|---|---|---|---|
| 239 | 1A11 | 316 | 1A11/239b | 316/239c | |
| 25980d | 492 | 53,250 | 103,764 | 108 | 210 |
| 30440 | 127 | 50,875 | 75,824 | 400 | 597 |
| 25629 | 130 | 10,200 | 35,059 | 78 | 270 |
| 21887 | 1,079 | 90,250 | 109,882 | 84 | 102 |
Macrophage cultures were infected with dilutions of V1EGFP vectors pseudotyped with the different SIVmac envelopes. For each pseudotype, infections were carried out at dilutions that were in the linear range for infection (see Table 2). The resulting macrophage titers for each pseudotype are shown, corrected to an initial input titer of 105 GFU/ml on CMMT-CD4 cells.
The ratio of the corrected titer for the 1A11 pseudotype to the 239 pseudotype titer is shown for macrophages from each animal. The average of the ratios is 167.
The ratio of the corrected titer for the 316 pseudotype to the 239 pseudotype is shown. The average of the ratios is 295.
PBMC-derived macrophages were established from four different rhesus macaques and used for the infections.
FIG. 4.
Fluorescence intensity of V1EGFP-infected macrophages. Cultures of PBMC-derived macrophages were infected with V1EGFP pseudotyped with different SIVmac envelopes (4 × 104 GFU on CMMT-CD4 cells per 105 cells). The cultures were then harvested at 4 days postinfection, fixed, and analyzed for green fluorescence by flow cytometry. The x axis shows log fluorescence intensity, and the y axis shows cell number; 5,000 cells were analyzed in each case. (A) Cells infected with a SIVmac316 pseudotype; (B) cells infected with a SIVmac1A11 pseudotype; (C) cells infected with a SIVmac239 pseudotype. The fluorescence values for the cultures infected with the different pseudotypes were approximately the same. The geometric mean intensities (in arbitrary fluorescence units) in the M1 region were as follows: 601 for the SIVmac239 pseudotype, 1,212 for the SIVmac1A11 pseudotype, and 1,265 for the SIVmac316 pseudotype.
Quantification of newly synthesized viral DNA in macrophages.
To further define restriction to replication of T-tropic SIV in macrophages, we compared the levels of vector DNA in nuclear fractions of macrophages infected by SIVmac239 and SIVmac316 pseudotypes of V1EGFP. Vector stocks were first treated with DNase to remove contaminating plasmid DNA and then used to infect primary PBMC-derived macrophages. Twenty-four hours after infection, nuclei were prepared and nuclear DNA was extracted. Nuclear DNA from equal numbers of cells infected by the two pseudotypes was then assayed for the level of reverse-transcribed vector DNA by quantitative real-time PCR (Fig. 5A). The results indicated that there was approximately threefold more vector DNA in the nuclei of macrophages infected with the M-tropic pseudotype than in those infected with the T-tropic pseudotype. Similar results were obtained on repeated assays of four independent macrophage infections. Thus, despite a ca. 300-fold-higher efficiency of macrophage infection for the M-tropic V1EGFP pseudotype (as measured by fluorescent cells), there was only a minor difference in nuclear DNA levels. Thus, a major block for vector infection of virus with SIVmac239 envelope appears to be located after transport of nuclear DNA into the nucleus and prior to viral DNA expression.
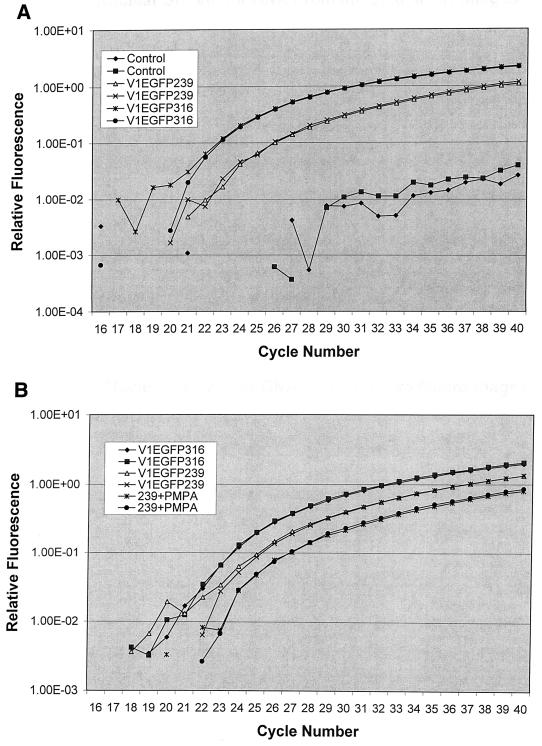
FIG. 5.
Quantification of vector DNA in infected macrophages. (A) PBMC-derived rhesus macrophages (105) were infected with DNase-treated V1EGFP pseudotyped with either SIVmac239 or SIVmac316 Env protein, at a multiplicity of 0.025 GFU (titered on CMMT-CD4 cells) per cell. At 24 h after infection, nuclei were prepared and DNA was extracted. Equal samples of nuclear DNA were tested for the presence of vector DNA by real-time PCR in a TaqMan thermal cycler, using the SIV-specific PCR primers and probe described in Materials and Methods. Quantification (A260) of the total nuclear DNAs prior to real-time PCR indicated equivalent efficiencies of recovery. The relative fluorescence signal for each PCR cycle is shown for each DNA sample. Duplicate amplifications were performed for each DNA. As a control, nuclear DNA from uninfected macrophages was analyzed in parallel. Fluorescence values below 10−2 were not significant. (B) In a second experiment, rhesus macrophages were infected with SIVmac239 and SIVmac316 pseudotypes of V1EGFP, and nuclear DNA was quantified as described for panel A. In addition, macrophages were infected with the SIVmac239 pseudotype of V1EGFP in the presence of 200 μM PMPA and analyzed in parallel. Results from duplicate real-time PCR assays are shown.
One potential artifact in the experiments illustrated in Fig. 5 could have been the uptake of contaminating plasmid DNA in the vector stocks, even though the stocks were treated with DNase prior to infection and nuclei were isolated from trypsinized infected macrophages prior to DNA extraction. This was of greater concern for the SIVmac239 pseudotypes, since they showed the lower levels of nuclear vector DNA. To address this concern, macrophages were infected in parallel with V1EGFP pseudotyped with SIVmac239 in presence of PMPA and analyzed, as shown in Fig. 5B and Table 4. The results indicated that while some vector DNA was detected in the nuclei of PMPA-treated cells, more vector DNA was present in the nuclei of cells infected without PMPA. As shown in Table 4, when only PMPA-sensitive DNA was considered, the levels of nuclear vector DNA in macrophages infected with the clone 316 or 239 pseudotype were still quite similar (within threefold).
TABLE 4.
Quantification of nuclear vector DNA by real-time PCRa
| PCR cycle no. | Fluorescence intensity
|
Corrected ratio for 316/239c | |||
|---|---|---|---|---|---|
| 316 | 239 | 239 + PMPA | 239 (PMPA sensitive)b | ||
| 24 | 12.6 | 5.8 | 2.9 | 2.9 | 3.3 |
| 25 | 19.8 | 9.1 | 4.8 | 4.3 | 3.5 |
| 26 | 28.7 | 14.1 | 7.7 | 6.4 | 3.3 |
The fluorescence values for the TaqMan real-time PCR studies shown in Fig. 5B (nuclear DNA from primary PBMC-derived macrophages infected by 316 and 239 pseudotypes of V1EGFP) are shown for the first PCR cycles where positive signals above background were obtained. The values shown are in TaqMan fluorescence units (10−2).
Fluorescence signal for V1EGFP pseudotype 239 corresponding to PMPA-sensitive vector DNA, obtained by subtracting the signal for cells infected by V1EGFP pseudotype 239 in the presence of PMPA from the signal from cells infected by the same vector without PMPA. TaqMan amplifications on diluted pV1EGFP plasmid DNA were carried out in parallel: the PMPA-sensitive vector DNA corresponded to ca. 023 molecule per cell (data not shown). These macrophage cultures were infected with 0.025 GFU (measured on CMMT-CD4) per cell.
The corrected ratio of PMPA-sensitive vector DNA in nuclei from cells infected by V1EGFP pseudotype 316 versus V1EGFP pseudotype 239. For these calculations, the assumption was made that there were equivalent amounts of PMPA-resistant vector DNA in the nuclei of cells infected with the 316 and 239 pseudotypes.
As mentioned, the most likely source for the PMPA-resistant vector DNA detected in Fig. 5B was uptake of contaminating plasmid DNA by the macrophages. However, this DNA was not integrated and expressed, since vector infection in the presence of PMPA eliminated the appearance of EGFP-positive cells (see above and below).
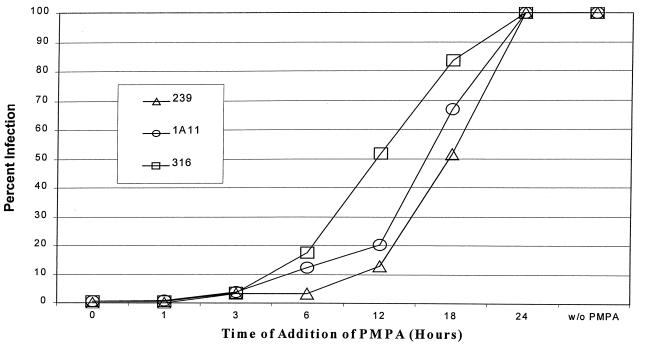
Kinetics of reverse transcription in macrophages.
While the results shown in Fig. 5 indicated a major block for SIVmac239 in macrophages at a step between nuclear import and gene expression, it was possible that an earlier block was also present. This would be consistent with the ca. threefold-less nuclear vector DNA in macrophages infected with the T-tropic V1EGFP pseudotype. To examine an earlier step in infection, we investigated the rate at which reverse transcription took place for the different vector pseudotypes. This was accomplished by adding the reverse transcriptase inhibitor PMPA to the infected cultures at different times after the initiation of infection. If PMPA is added after reverse transcription of the vector has taken place in a given infected cell, then it will not inhibit expression of EGFP. Figure 6 shows the results of adding the PMPA to primary macrophages at different times postinfection with the different V1EGFP pseudotypes. The results indicated that the kinetics of events up to and including reverse transcription occurred more rapidly when the vector was pseudotyped with the clone 316 envelope than when it was pseudotyped with the clone 239 envelope. As measured from the time points at which 50% of the vector infection was resistant to PMPA treatment, reverse transcription was completed on average by 11.5 h for the 316 pseudotype while this did not occur until 17.5 h postinfection for the 239 pseudotype. V1EGFP vector pseudotyped with the clone 1A11 envelope showed an intermediate time for completion of reverse transcription. The same rank order for the vector pseudotypes was observed in repeated experiments, some of which utilized macrophages from different rhesus macaques.
FIG. 6.
Timing of reverse transcription in macrophages infected with vector pseudotypes (monkey 25980). A total of 4 × 105 macrophages in 12-well plates were infected with 500 μl of diluted V1EGFP stocks pseudotyped with different SIVmac envelopes as described in Materials and Methods. Each well infected with the SIVmac239 pseudotype received 6.3 × 103 GFU, each well infected with the SIVmac1A11 pseudotype received 1.6 × 103 GFU, and each well infected with the SIVmac316 pseudotype received 1.7 × 103 GFU. A 100 μM concentration of PMPA was added at the time points indicated. All cultures were scored for infected cells at 4 days postinfection. The levels of infection are plotted as percent infection without PMPA.
Infection of T lymphocytes.
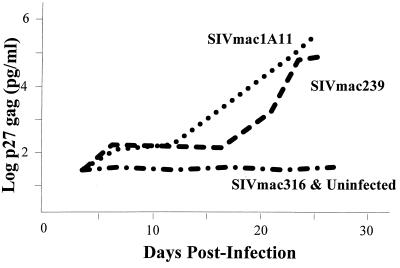
Since the experiments with the primary macrophages indicated that a major block for T-tropic SIVmac239 is at an intermediate step in the infection cycle, we wished to test whether restriction of M-tropic virus in T lymphocytes is also determined at early steps in infection. One challenge was that there are relatively few SIVmac isolates that do not replicate in the standardly used T-lymphocyte lines. Indeed, most M-tropic SIVmac isolates would be better considered dualtropic since they can replicate in both primary macrophages and CD4-positive T-lymphocyte lines. To identify a truly M-tropic SIVmac isolate, we first tested several human T-lymphocyte lines for infection by replication-competent SIVmac316. While this virus replicated in T-lymphocyte lines such as SupT1 (data not shown), it did not replicate in Molt4-Clone8 cells on multiple rounds of infection, as shown in Fig. 7. Thus, on the basis of Molt4-Clone8 cell infections, SIVmac316 can indeed be considered M-tropic. As expected, SIVmac239 efficiently replicated in Molt4-Clone8 cells, as did the dual-tropic SIVmac1A11.
FIG. 7.
Replication of SIVmac viruses in Molt4-Clone8 cells. A total of 5 × 105 Molt4-Clone8 cells were infected at a multiplicity of infection of 0.002 with different SIVmac viruses as described in Materials and Methods. p27 SIV gag antigen in culture supernatants was measured at different times and is plotted versus days postinfection.
Molt4-Clone8 cells were infected with equal concentrations of the V1EGFP pseudotypes, and the results are shown in Table 5. The results indicate that all three pseudotypes infected the Molt4-Clone8 cells with similar efficiencies. Thus, in contrast to the situation in primary macrophages where the SIVmac239 pseudotype was restricted at a relatively early event in infection, the SIVmac316 pseudotype was not affected in its ability to carry out early steps in the infection cycle. Further, comparison of the GFP signal intensity in infection of Molt4-Clone8 cells showed no significant difference in the fluorescence intensities between cells infected with the SIVmac239 and -316 envelope pseudotypes, as expected. Together, these indicate that the block results for SIVmac316 infection of Molt4-Clone8 cells is at a late step in infection.
TABLE 5.
Vector infection of Molt4-Clone8 cellsa
| Vector pseudotype | No. of GFP-positive cellsb |
|---|---|
| SIVmac239 | 8,687 |
| SIVmac1A11 | 11,956 |
| SIVmac316 | 9,449 |
Infection of 106 Molt4-Clone8 cells with 105 GFU (titered on CMMT-CD4 cells) of each vector pseudotype. Infections were scored after 4 days of fluorescence microscopy. Numbers of GFP-positive cells were corrected for cell division as described in Materials and Methods.
The values are averages from five sets of infections.
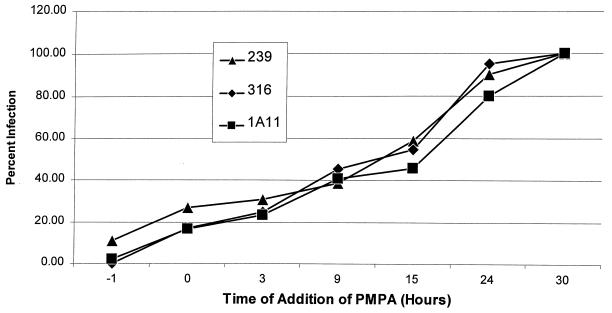
We also investigated the kinetics of reverse transcription for the different V1EGFP pseudotypes in Molt4-Clone8 cells, as shown in Fig. 8. Consistent with the vector infection results, there was no systematic difference in the kinetics of reverse transcription for the different pseudotypes.
FIG. 8.
Timing of reverse transcription in Molt4-Clone8 cells infected with vector pseudotypes. A total of 106 Molt4-Clone8 cells in six-well plates were infected with 105 GFU of V1EGFP pseudotyped with different SIVmac envelopes. A 100 μM concentration of PMPA was added at the times indicated, and the numbers of GFP-positive cells were measured at 4 days postinfection by flow cytometry. Infection levels are shown as percentage of GFP-positive cells in the absence of PMPA.
DISCUSSION
Previous studies of SIV tropism have employed analysis of multiple rounds of infection, making it difficult to determine the steps in replication where the virus is restricted. In the studies reported here, we used helper-free SIV-based vectors to study the mechanisms of SIV cell tropism. These vectors carry out only a single cycle of infection since they are replication defective; in fact, they can only carry out steps in infection from binding and entry through reverse transcription, DNA integration, and gene expression. V1EGFP vectors pseudotyped with either M-tropic or T-tropic envelope proteins allowed us to determine which steps were blocked for SIVs under the restrictive conditions. Two interesting results were obtained. In the case of infection in primary macrophages, the block for T-tropic SIVmac239 appeared to be at a relatively early step(s), since V1EGFP pseudotyped with the clone 239 envelope was substantially less infectious (ca. 200- to 300-fold) than the same vector pseudotyped with the clone 316 or 1A11 envelope. Furthermore, there were only minor differences (ca. threefold) in the level of nuclear SIV DNA between macrophages infected with the 239 and 316 pseudotypes (Fig. 5). This suggests that a major block to replication of T-tropic SIVs (SIVmac239) lies beyond nuclear transport but before early gene expression. On the other hand, the block for infection of M-tropic SIVmac316 in Molt4-Clone8 T cells was at a different step in the infection cycle, since V1EGFP pseudotyped with the SIVmac316 envelope infected these cells with the same efficiency as the same vector pseudotyped with the SIVmac239 envelope. Thus, M-tropism and T-tropism restrictions for SIV may involve steps for primary macrophages different from those for T lymphocytes.
The finding of a block in replication for SIVmac239 in primary macrophages at a postreverse transcription step in infection cycle confirms and extends work by other investigators. As mentioned above, in a study with replication-competent virus, Mori et al. suggested that the block for T-tropic SIVmac239 infection of primary rhesus alveolar macrophages was at a step after reverse transcription, since the amount of reverse-transcribed viral DNA was within ca. fivefold of the levels detected in SIVmac316-infected cells (32). Kirchhoff et al. also concluded that the restriction of T-tropic SIVmac239 in PBMC-derived macrophages is not at an early step of the viral infection cycle (27). However, Stephens et al. (49) reported restriction of T-tropic SIVmac in PBMC-derived macrophages at extremely late steps of infection, namely, virion assembly, release, and/or polyprotein processing, which would differ from the conclusions reached in the present study. Thus, our results agree with those of Mori et al. and Kirchhoff et al., and they further suggest a major block of the SIVmac239 pseudotype of V1EGFP in macrophages at some step beyond nuclear translocation of viral DNA and before expression of the integrated provirus. Another laboratory has independently used T-tropic pseudotypes of a replication-defective SIV vector and found a substantial reduction in infection efficiency in primary macrophages compared to an M-tropic pseudotype (N. Bannert, D. Schenten, and J. Sodroski, personal communication).
In these experiments, we found it necessary to ensure that infections were carried out at appropriate multiplicities. As described in Results, it was necessary to verify that the vector infections were carried out under conditions of linearity; in some cases, this was achieved only after the vector stocks were diluted. Indeed, when we carried out macrophage infections with undiluted vector pseudotypes, we observed only a fivefold difference between the efficiencies of infection for the SIVmac316 and SIVmac239 pseudotypes, because the amount of SIVmac316 pseudotype was saturating.
The results shown in Fig. 6 indicate that reverse transcription for V1EGFP pseudotyped by SIVmac316 may be completed more rapidly than that for the SIVmac239 pseudotype. This might suggest an impairment of SIVmac239 in macrophages at a quite early step in infection, before reverse transcription. At first glance, this might seem to be inconsistent with the major block at integration or gene expression described above. However, Fig. 6 allowed us to measure the rate of reverse transcription only for the small fraction of SIVmac239 pseudotypes that successfully completed infection to the point where EGFP was expressed. Thus, it is possible that there may be two blocks for SIVmac239 infection in macrophages: a minor block at a quite early step (e.g., binding, entry, or reverse transcription) and the later major block discussed above. It is noteworthy that Mori et al. found a ca. fivefold reduction in reverse-transcribed viral DNA for SIVmac239 compared to that for SIVmac316 when infecting macrophages, consistent with a minor early block (32). Recently, Bannert et al. (personal communication) found that the low efficiency of SIVmac239 infection in primary PBMC-derived macrophages can be enhanced by vectored overexpression of CD4 protein.
Evidence for an intracellular blockage of HIV infection has also been reported. Prior to the attribution of M versus T tropism of HIV to coreceptor usage, Schmidtmayerova et al. suggested that intracellular events may be involved (43). Our previous studies (12) showed that SIVs encoding the HIV envelope are unable to replicate in CD4+ macaque cells, unless those cells also express the appropriate human coreceptors. When the level of restriction was examined, it was found that in the nonpermissive cells, DNA from SIVs encoding the HIV envelope was synthesized, but there appeared to be a block at the level of nuclear import. Thus, blocks for T-tropic SIV in macrophages might reflect similar processes for HIV cell tropism; in the latter case, this may be superimposed on coreceptor binding. More recently, it has been shown that human macrophages do express CXCR4 coreceptor and that this coreceptor can function on dualtropic HIV-1 isolates but not on T-tropic isolates (29, 51, 52). Moreover, the restriction for T-tropic virus in macrophages is after reverse transcription (43), very similar to the major block for T-tropic SIV in macrophages described here.
It is interesting to consider possible mechanisms by which T-tropic SIV envelope protein could lead to restriction at postentry steps in macrophages. One possible mechanism is preferential activation of macrophages. Weissman et al. showed that recombinant gp120 from the M-tropic SIV clone PBj1.9 induces an intracellular calcium flux in the CCR5-positive B10 lymphocyte cell line, while gp120 from SIVmac239 does not (50). If gp120-mediated activation is necessary for productive infection of macrophages, a differential ability of M-tropic and T-tropic envelope proteins to trigger signals through CCR5 interactions might be responsible for cell tropism. However, incubation of macrophages with SIVmac316 envelope protein (or vectored expression in these cells) did not increase their ability to be infected by vectors pseudotyped with SIVmac239 envelope (X. J. You and H. Fan, unpublished data) (6). Thus, preferential activation of macrophages by M- versus T-tropic SIV envelope protein does not appear to be the mechanism involved.
Another possible mechanism could be that when T-tropic SIV is incubated with macrophages, it is taken up, but the viral particles enter a dead-end intracellular pathway due to an inappropriate interaction of envelope protein with CD4 or coreceptor. Thus, some postentry events may take place (e.g., reverse transcription or nuclear import), but the products will not ultimately result in virus expression and/or production. Such a mechanism would be consistent with the multiple reports of postentry blocks associated with cell tropism described above. Recent experiments have indicated that PBMC-derived macrophages express suboptimal levels of CD4 for infection by SIVmac239 (but not SIVmac316) and that this can be corrected by overexpression of CD4 in those cells (6). Alveolar macrophages have also been found to express low levels of CD4 (34). Taken together with the results reported here, this might suggest that in the absence of sufficient CD4, standard fusion-mediated viral entry does not occur, but virus may still enter cells by another (nonproductive) process such as endocytosis.
In contrast to the relatively early blocks found for infection of T-tropic SIVmac239 in primary macrophages, the block for M-tropic SIVmac316 in Molt4-Clone8 T-lymphoid cells was clearly at a late step in infection, since V1EGFP pseudotyped with SIVmac316 envelope infected these cells as efficiently as the SIVmac239 pseudotype did. This is reminiscent of the late block in infection described by Stephens et al. (49), although in that report the block was described for a T-tropic SIVmac in PBMC-derived macrophages. It will be interesting to determine how the envelope glycoprotein can affect virus replication at such a late step. One possible mechanism is aberrant viral assembly or maturation of virions containing SIVmac316 envelope protein in Molt4-Clone8 cells.
In summary, the use of helper-free replication-defective SIV-based vectors has allowed us to gain new insights into the mechanisms of SIV cell tropism and to more closely focus on the exact location of the restriction to replication of T-tropic SIV in macrophages. These vector particles were identical except for the envelope proteins. While the tropism was determined by the envelope protein, envelope-host cell interactions affect virus replication at several steps after entry. Moreover, different steps in the infection cycle appeared to be critical in primary macrophages and a T-lymphocyte cell line.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01AI39435. S.S.K. was supported by NIH training grant T32 AI07319 and by a fellowship from the American Heart Association. M.-E.H was supported by NIH training grant T32 CA09229. Institutional support of the UCI Cancer Research Institute and the Chao Family Comprehensive Cancer Center is acknowledged.
We thank R. Desrosiers, T. Kodama, M. Marthas, C. Miller, K. Uberla, and Gilead Sciences for providing materials and W. E. Robinson for advice.
REFERENCES
- 1.Alkhatib G, Broder C C, Berger E A. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Anderson G M, Hauer D, Sharma D P, Joag S V, Narayan O, Zink M C, Clements J E. Analysis of envelope changes acquired by SIVmac239 during neuroadaption in rhesus macaques. Virology. 1993;195:616–626. doi: 10.1006/viro.1993.1413. [DOI] [PubMed] [Google Scholar]
- 4.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 5.Banapour B, Marthas M L, Ramos R A, Lohman B L, Unger R E, Gardner M B, Pedersen N C, Luciw P A. Identification of viral determinants of macrophage tropism for simian immunodeficiency virus SIVmac. J Virol. 1991;65:5798–5805. doi: 10.1128/jvi.65.11.5798-5805.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophage-tropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brussel A, Pretet J L, Girard M, Butor C. Sequences and predicted structures of chimpanzee STRL33 (Bonzo) and gpr15 (BOB) AIDS Res Hum Retrovir. 1999;15:1315–1319. doi: 10.1089/088922299310214. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian B, Haigwood N L, Overbaugh J. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology. 1995;213:386–394. doi: 10.1006/viro.1995.0011. [DOI] [PubMed] [Google Scholar]
- 12.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chelucci C, Casella I, Federico M, Testa U, Macioce G, Pelosi E, Guerriero R, Mariani G, Giampaolo A, Hassan H J, Peschle C. Lineage-specific expression of human immunodeficiency virus (HIV) receptor/coreceptors in differentiating hematopoietic precursors: correlation with susceptibility to T- and M-tropic HIV and chemokine-mediated HIV resistance. Blood. 1999;94:1590–1600. [PubMed] [Google Scholar]
- 14.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng-Mayer C, Shioda T, Levy J A. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in tat and gp120. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 17.Connor R I, Ho D D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 20.Desrosiers R C, Hansen-Moosa A, Mori K, Bouvier D P, King N W, Daniel M D, Ringler D J. Macrophage-tropic variants of SIV are associated with specific AIDS-related lesions but are not essential for the development of AIDS. Am J Pathol. 1991;139:29–35. [PMC free article] [PubMed] [Google Scholar]
- 21.Di Marzio P, Tse J, Landau N R. Chemokine receptor regulation and HIV type 1 tropism in monocyte- macrophages. AIDS Res Hum Retrovir. 1998;14:129–138. doi: 10.1089/aid.1998.14.129. [DOI] [PubMed] [Google Scholar]
- 22.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC- CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 23.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edinger A L, Clements J E, Doms R W. Chemokine and orphan receptors in HIV-2 and SIV tropism and pathogenesis. Virology. 1999;260:211–221. doi: 10.1006/viro.1999.9819. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 26.Jinno A, Shimizu N, Soda Y, Haraguchi Y, Kitamura T, Hoshino H. Identification of the chemokine receptor TER1/CCR8 expressed in brain-derived cells and T cells as a new coreceptor for HIV-1 infection. Biochem Biophys Res Commun. 1998;243:497–502. doi: 10.1006/bbrc.1998.8130. [DOI] [PubMed] [Google Scholar]
- 26a.Kim, S., N. Kothari, X. J. You, W. E. Robinson, Jr., T. Schnell, K. Uberla, and H. Fan. Generation of replication-defective helper-free vectors based on simian immunodeficiency virus. Virology, in press. [DOI] [PubMed]
- 27.Kirchhoff F, Pohlmann S, Hamacher M, Means R E, Kraus T, Uberla K, Di Marzio P. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMX174 cells for efficient entry. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B, Ratajczak J, Doms R W, Gewirtz A M, Ratajczak M Z. Coreceptor/chemokine receptor expression on human hematopoietic cells: biological implications for human immunodeficiency virus-type 1 infection. Blood. 1999;93:1145–1156. [PubMed] [Google Scholar]
- 29.Liu Q H, Williams D A, McManus C, Baribaud F, Doms R W, Schols D, De Clercq E, Kotlikoff M I, Collman R G, Freedman B D. HIV-1 gp120 and chemokines activate ion channels in primary macrophages through CCR5 and CXCR4 stimulation. Proc Natl Acad Sci USA. 2000;97:4832–4837. doi: 10.1073/pnas.090521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luciw P A, Shaw K E, Unger R E, Planelles V, Stout M W, Lackner J E, Pratt-Lowe E, Leung N J, Banapour B, Marthas M L. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac) AIDS Res Hum Retrovir. 1992;8:395–402. doi: 10.1089/aid.1992.8.395. [DOI] [PubMed] [Google Scholar]
- 31.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori K, Ringler D J, Kodama T, Desrosiers R C. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori K, Rosenzweig M, Desrosiers R C. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 37.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohlmann S, Krumbiegel M, Kirchhoff F. Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. J Gen Virol. 1999;80:1241–1251. doi: 10.1099/0022-1317-80-5-1241. [DOI] [PubMed] [Google Scholar]
- 39.Rizvi T A, Panganiban A T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 41.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samson M, Edinger A L, Stordeur P, Rucker J, Verhasselt V, Sharron M, Govaerts C, Mollereau C, Vassart G, Doms R W, Parmentier M. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur J Immunol. 1998;28:1689–1700. doi: 10.1002/(SICI)1521-4141(199805)28:05<1689::AID-IMMU1689>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 43.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. Human immunodeficiency virus type 1 T-lymphotropic strains enter macrophages via a CD4- and CXCR4-mediated pathway: replication is restricted at a postentry level. J Virol. 1998;72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnell T, Foley P, Wirth M, Munch J, Uberla K. Development of a self-inactivating, minimal lentivirus vector based on simian immunodeficiency virus. Hum Gene Ther. 2000;11:439–447. doi: 10.1089/10430340050015905. [DOI] [PubMed] [Google Scholar]
- 45.Sharma D P, Zink M C, Anderson M, Adams R, Clements J E, Joag S V, Narayan O. Derivation of neurotropic simian immunodeficiency virus from exclusively lymphocytotropic parental virus: pathogenesis of infection in macaques. J Virol. 1992;66:3550–3556. doi: 10.1128/jvi.66.6.3550-3556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 47.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivas R V, Robbins B L, Connelly M C, Gong Y F, Bischofberger N, Fridland A. Metabolism and in vitro antiretroviral activities of bis(pivaloyloxymethyl) prodrugs of acyclic nucleoside phosphonates. Antimicrob Agents Chemother. 1993;37:2247–2250. doi: 10.1128/aac.37.10.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephens E B, McClure H M, Narayan O. The proteins of lymphocyte- and macrophage-tropic strains of simian immunodeficiency virus are processed differently in macrophages. Virology. 1995;206:535–544. doi: 10.1016/s0042-6822(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 50.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 51.Yi Y, Isaacs S N, Williams D A, Frank I, Schols D, De Clercq E, Kolson D L, Collman R G. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J Virol. 1999;73:7117–7125. doi: 10.1128/jvi.73.9.7117-7125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]