Abstract
Aims
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been linked to cardiovascular complications, notably cardiac arrhythmias. The open reading frame (ORF) 3a of the coronavirus genome encodes for a transmembrane protein that can function as an ion channel. The aim of this study was to investigate the role of the SARS-CoV-2 ORF 3a protein in COVID-19-associated arrhythmias and its potential as a pharmacological target.
Methods and results
Human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) and cultured human fibroblasts were infected with SARS-CoV-2. Subsequent immunoblotting assays revealed the expression of ORF 3a protein in hiPSC-CM but not in fibroblasts. After intracytoplasmic injection of RNA encoding ORF 3a proteins into Xenopus laevis oocytes, macroscopic outward currents could be measured. While class I, II, and IV antiarrhythmic drugs showed minor effects on ORF 3a-mediated currents, a robust inhibition was detected after application of class III antiarrhythmics. The strongest effects were observed with dofetilide and amiodarone. Finally, molecular docking simulations and mutagenesis studies identified key amino acid residues involved in drug binding.
Conclusion
Class III antiarrhythmic drugs are potential inhibitors of ORF 3a-mediated currents, offering new options for the treatment of COVID-19-related cardiac complications.
Keywords: 3a protein, Antiarrhythmic drugs, Arrhythmias, COVID
Graphical Abstract
Graphical Abstract.
Class III antiarrhythmic drugs are capable of inhibiting SARS-CoV-2 ORF 3a-mediated cation currents, thus offering novel therapeutic avenues for the management of cardiac arrhythmias associated with COVID-19. Created with the help of DALL-E 2, OpenAI.
Translational perspective.
This study aims at exploring the SARS-CoV-2 ORF 3a protein’s role in COVID-19-associated arrhythmias and its interaction with antiarrhythmic drugs. The findings reveal that hiPSC-derived cardiomyocytes express ORF 3a proteins upon infection with SARS-CoV-2. Further, class III antiarrhythmics were found to significantly inhibit ORF 3a-mediated currents. Molecular docking and mutagenesis studies identified key amino acid residues crucial for drug binding, enhancing our understanding of the mechanistic basis of ORF 3a’s role in arrhythmogenesis and its potential as a therapeutic target. This significant finding not only advances our understanding of COVID-19’s impact on cardiovascular health but also points towards novel pharmacological strategy for treating COVID-19-related cardiac complications.
What’s new?
When infected with SARS-CoV-2, hiPSC-derived cardiomyocytes express ORF 3a proteins.
Class III antiarrhythmics inhibit SARS-CoV-2 ORF 3a-mediated currents.
Key amino acid residues of the ORF 3a protein, identified to mediate interaction with class III antiarrhythmics, are highly conserved among different SARS-CoV-2 strains.
Taken together, the findings of this study point towards a novel pharmacological strategy for treating COVID-19-related cardiac arrhythmias.
Introduction
The medical field has faced numerous challenges due to the emergence of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1–4 Among these challenges, cardiovascular disorders have emerged as a significant extrapulmonary manifestation of SARS-CoV-2 infection.5–8 Case series from the early phase of the pandemic reported a frequent occurrence of cardiac arrhythmias in COVID-19 patients.9,10 It was described that SARS-CoV-2 infection may result in various cardiac arrhythmias, such as sinus bradycardia or high-grade atrioventricular block, as well as tachycardia, ranging from increased numbers of supraventricular or ventricular premature contractions to fatal ventricular arrhythmias. In particular, atrial fibrillation (AF) was reported to be frequently observed in COVID-19 patients.5,6 Arrhythmias can pose significant risks, as they not only complicate treatment of COVID-19 infection but also carry consequences of their own, such as stroke, heart failure, and even sudden cardiac death. The causal link between the documented increased incidence of arrhythmias and SARS-CoV-2 infection remains a subject of ongoing debate.11–13 Since the prevalence of arrhythmias like AF in elderly populations with cardiovascular risk factors and comorbidities is high, a direct causal relationship of COVID-19 and these arrhythmias has been questioned. It has been discussed whether infection may primarily act as a trigger for the induction of AF episodes and that increased medical surveillance during COVID-19 infection may lead to the clinical diagnosis of previously unrecognized arrhythmias.11 Recent studies indicate that the incidence of AF in COVID-19 may be comparable to that of influenza.11,13 The aetiology of these arrhythmias can be attributed to fever, autonomic aberration, hypoxia, systemic inflammation, hypercoagulopathy, myocardial ischaemia, mechanical ventilation strategies, electrolyte abnormalities, medication effects, or induction of the hepatic cytochrome P450 system, increasing the bioavailability of QT-prolonging agents.5 However, viral infections have historically been associated with cardiac complications. Cardiac magnetic resonance imaging in some trials has shown frequent abnormalities indicative of active myocardial inflammation or fibrosis after infection.13–15 Moreover, the onset of new arrhythmias has been shown to independently predict worse outcomes, including in-hospital mortality, the need for mechanical ventilation, and cardiovascular death.5,11,16–18 This finding warrants further investigation. Further, the incidence of arrhythmias after COVID-19 is observed in up to 10% of cases.19 A cohort study followed over 1700 discharged COVID-19 patients for 6 months and found that 9.3% experienced palpitations.20 The COVID-19 virus has been found to enter cells via the angiotensin-converting enzyme-2 (ACE2), which is present on cardiomyocytes.21 This has raised concerns about direct cardiac injury and arrhythmogenesis, as evidenced by the increase in cardiac biomarkers in a significant number of COVID-19 patients.5
In 2006, it was reported that the open reading frame (ORF) 3a protein of the coronavirus genome, a viroporin involved in regulation of autophagy, inflammasome activation, and apoptosis, can function as a non-selective cation channel.22 The ORF 3a protein of SARS-CoV-2 shares ∼72% homology with the SARS coronavirus studied in 2006.23 Thus, it was considered whether the COVID-19 3a protein could also conduct ion currents. If replication of the coronavirus in cardiomyocytes may result in the integration of SARS-CoV-2 ORF 3a proteins into the sarcolemma, ORF 3a-mediated currents may contribute to COVID-19-associated arrhythmogenesis by altering the action potential shape of cardiomyocytes.
Based on this hypothesis, this study was designed to further investigate the role of ORF 3a in the cardiac complications of COVID-19. By exploring the expression of ORF 3a proteins in cardiomyocytes upon SARS-CoV-2 infection, its ion channel activity, and its interaction with existing antiarrhythmic drugs, this study aims to elucidate the mechanisms by which SARS-CoV-2 may predispose to arrhythmias and to explore potential therapeutic strategies to counteract these effects.
Methods
Ethics statement
Animal experiments were conducted in compliance with guidelines set forth by the U.S. National Institutes of Health (NIH publication No. 86-23), the EU Directive 2010/63/EU, and German laws for animal protection. Approval was obtained from the local Animal Welfare Committee (Regierungspraesidium Karlsruhe, reference number G165/19). Additional experiments with human tissue samples were approved by the ethics committee of the University of Heidelberg (Medical Faculty Heidelberg, S-017/2013) and performed in accordance with the Declaration of Helsinki on patients who had given their written consent.
Cell culture, differentiation, and maintenance of human-induced pluripotent stem cell-derived cardiomyocytes
Human-induced pluripotent stem cells were derived from healthy donors by the Stanford Cardiovascular Institute Biobank, directed by Jospeh C Wu, via episomal reprogramming using Sendai-Virus and transferred to T.S. using a material transfer agreement. The cells were differentiated to a cardiomyocyte-like phenotype using established protocols.24,25 After differentiation, they were kept in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with B27 (Gibco, Thermo Fisher Scientific). On Days 25–35, they were seeded for viral infection on 24-culture plates (Sarstedt, Nürmbrecht, Germany).
Culture of human cardiac fibroblasts
Human atrial tissue samples were cut into small pieces (∼1 mm3) and placed in a 25 mm dish with 1 mL medium consisting of DMEM (Gibco, Thermo Fisher Scientific), 10% foetal calf serum (Gibco, Thermo Fisher Scientific), and 1% penicillin/streptomycin (Gibco, Thermo Fisher Scientific) and cultured at 37°C, 5% CO2. The medium was changed two times per week, and after 3 weeks of culture, cells were trypsinized, replated, and further cultivated for 4–6 weeks before infection.
Viral infection of human-induced pluripotent stem cell-derived cardiomyocytes and human cardiac fibroblasts
SARS-CoV-2 (strain BavPat1) was obtained from Prof. Christian Drosten at the Charité in Berlin, Germany, and provided via the European Virology Archive. The virus was amplified in Vero E6 cells and used at passage 2. All SARS-CoV-2 infections were performed at the multiplicity of infection (MOI) indicated in the text. Medium was removed from cells, and virus was added to cells for 1 h at 37°C. Virus was removed, cells were washed 1× with PBS, and the respective media were added back to the cells.
Protein isolation and immunoblot analysis
Protein immunodetection was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting as described.26,27 Human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) and cultured human cardiac fibroblasts (hCFs) were lysed in radioimmunoprecipitation lysis buffer containing 50 mM Tris–HCl (pH 7.4), 0.5% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and protease inhibitors (cOmplete mini protease inhibitor cocktail, Roche Diagnostics, Mannheim, Germany). The protein concentration was determined using the Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific). Equal amounts of protein were separated using SDS-PAGE. Nitrocellulose membranes were developed by sequential exposure to blocking reagent containing 5% dry milk and 3% bovine serum albumin, primary antibodies directed against the SARS ORF 3a protein (1:500; ORF 3a polyclonal antibody, NCP0019, Bioworld Technology, St. Louis Park, MN, USA) and appropriate HRP-conjugated secondary antibodies (1:3000, Anti-Rabbit Na934, Cytiva, Marlborough, MA, USA). Signals were recorded using the enhanced chemiluminescence assay (WesternBright ECL HRP substrate, Advansta, San Jose, CA, USA) and quantified with ImageJ (National Institutes of Health, Bethesda, MD, USA). Protein content was normalized to β-actin using monoclonal mouse anti-β-actin antibodies (1:1000; sc-47778; Santa Cruz Biotechnology, Heidelberg, Germany) and corresponding secondary antibodies (1:3000; sc-2005; Santa Cruz Biotechnology) for quantification of optical density. The complete set of original image data for the immunoblot membranes is presented in the Supplementary material.
Molecular biology and RNA preparation
Plasmid DNA encoding for the SARS-CoV-2 ORF 3a protein (GenBank accession number UXU20787.1) was constructed by gene synthesis (Eurofins Genomics, Ebersberg, Germany) and subcloned to the Xenopus laevis expression vector pMax+. For generation of the ORF 3a pore mutants, site-directed mutagenesis was performed as described previously,28 and the sequences of the mutant constructs were confirmed by Sanger sequencing. Copy RNA was prepared as described previously.26,29 In short, after plasmid linearization and in vitro transcription with mMessage mMachine T7 Transcription Kit (Thermo Fisher Scientific), the integrity of the RNA transcript was confirmed by agarose gel electrophoresis. RNA concentrations were measured via spectrophotometry (ND–2000, peqLab Biotechnology GmbH, Erlangen, Germany).
Oocyte preparation
Oocytes were prepared as described.26,29 In short, ovarian lobes from X. laevis (Xenopus Express, Vernassal, France) were extracted under tricaine (Pharmaq, Fordingbridge, UK) anaesthesia (1 g/L, pH 7.5). After manual dissection and collagenase treatment (Collagenase D, Roche Diagnostics), collagenase free oocytes were selected and transferred to standard oocyte solution, containing 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 2.5 mM pyruvic acid, and 50 mg/L gentamicin sulfate, adjusted to pH 7.7 with NaOH. Oocytes were injected by a Nanoject II system (46 nL per oocyte; Drummond Scientific Company, Broomall, PA, USA) with cRNA of ORF 3a protein (30 ng/oocyte).
Electrophysiology
Two-electrode voltage clamp (TEVC) recordings from X. laevis oocytes were performed 24–72 h after injection as described earlier.26,29 Whole cell currents were measured with an OC-725C amplifier (Warner Instruments, Hamden, CT, USA) using pCLAMP10 software (Axon Instruments, Foster City, CA, USA) for data acquisition and analysis. The standard bath solution contained 100 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5 mM HEPES, adjusted to pH 7.4 with KOH. The microelectrodes were manufactured by a Flaming/Brown P-87 micropipette puller (Sutter Instruments, Novato, CA, USA) from glass pipettes (GB 100F-10, Science Products, Hofheim, Germany). The pipettes were backfilled with a 3 M KCl and had a resistance between 1.5 and 3.0 MΩ. Holding potentials were −80 mV, and the experiments were performed at room temperature (20–22°C). Leak currents were not subtracted. Unless otherwise stated, steady-state currents were quantified at the end of the test pulse.
Pharmacological compounds
The test substances were purchased from Bristol Myers Squibb GmbH (Regensburg, Germany), Cardiome Pharma (Vancouver, Canada), MedChemExpress (Monmouth Junction, NJ, USA), Sigma-Aldrich (Steinheim, Germany), and Selleck Chemicals (Cologne, Germany) as listed in Table 1. Stock solutions of the indicated concentrations were prepared with the indicated solvents and stored at −20°C.
Table 1.
Pharmacological compounds—manufacturers, concentrations, and solvents
| Substance | Manufacturer | Concentration of stock solution in mmol/L | Solvent |
|---|---|---|---|
| Ajmaline | MedChemExpress | 100 | DMSO |
| Amiodarone | Sigma-Aldrich | 10 | Ethanol absolute |
| Bisoprolol | Selleck Chemicals | 100 | DMSO |
| Carvedilol | Selleck Chemicals | 100 | DMSO |
| Dofetilide | Sigma-Aldrich | 5 | DMSO |
| Dronedarone | MedChemExpress | 50 | DMSO |
| D-Sotalol | Bristol Myers Squibb | 10 | Sterile water |
| Etripamil | MedChemExpress | 100 | DMSO |
| Flecainide | MedChemExpress | 100 | DMSO |
| Ibutilide | Sigma-Aldrich | 100 | DMSO |
| Lidocaine | Sigma-Aldrich | 10 | Bath solution |
| Metoprolol | Selleck Chemicals | 25 | DMSO |
| Nebivolol | Sigma-Aldrich | 100 | DMSO |
| Procaine | Sigma-Aldrich | 100 | DMSO |
| Propafenone | Sigma-Aldrich | 100 | DMSO |
| Propranolol | Selleck Chemicals | 100 | DMSO |
| Quinidine | Sigma-Aldrich | 100 | DMSO |
| Ranolazine | Selleck Chemicals | 100 | DMSO |
| Verapamil | Sigma-Aldrich | 100 | DMSO |
| Vernakalant | Cardiome Pharma | 51.8 | Sterile water |
DMSO, dimethyl sulfoxide.
Computational modelling
For the molecular docking of the active compounds into the channel, we chose the homodimeric model of the protein structure with a nominal resolution of 2.1 Å, which is deposited in the Protein database under the PDB-code 7KJR.22 This structure got prepared by deleting all water molecules and the two DOPE lipids. Next, the hydrogens were added, charges assigned, and the best hydrogen bonding arrangement for sidechains containing amide groups selected. Then, the cavities inside this protein structure were determined and shown with Molcad, as implemented in the software SYBYL-X1.3 package (Tripos, St. Louis, MO, USA). The big cavity served as input for the docking algorithm SurflexDock v.2.51 from BioPharmics IT (Sonoma County, CA, USA) as implemented in the software package SYBYL-X1.3 to place the ligands.
Statistical analysis and data visualization
The data acquisition and analysis were performed using the pCLAMP10 software (Axon Instruments), and statistical analysis and visualization were done using Prism 10 (GraphPad Software, La Jolla, CA, USA). Visualization of the SARS-CoV-2 ORF 3a 3D cryo-electron microscopy structure recently revealed by Kern et al.22 (PDB-ID:6XDC) was performed using the PyMOL Molecular Graphics System, Version 1.8 (Schrödinger LLC, Cambridge, MA, USA). Data are presented as the mean ± standard error of the mean (SEM). Statistical comparisons were made using paired and unpaired Student’s t-tests, with a significance level of P < 0.05. The Bonferroni method was used to correct for multiple comparisons.
Results
ORF 3a protein expression in SARS-CoV-2-infected human-induced pluripotent stem cell-derived cardiomyocytes
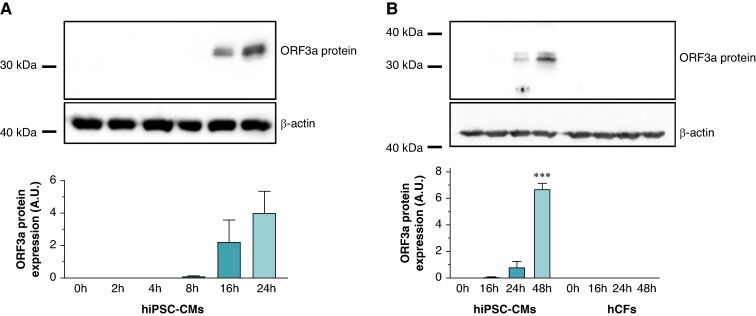
To determine whether ORF 3a proteins are expressed in cardiomyocytes after infection with coronavirus, in vitro cultures of hiPSC-CMs were infected with a SARS-CoV-2 viral strain from early 2020. Following infection at a MOI of 1.0, hiPSC-CMs were harvested at 0–24 h. Protein lysates were then subjected to immunoblotting to quantify ORF 3a expression, demonstrating protein bands 16- and 24-h post-infection, as illustrated in Figure 1A. In contrast to hiPSC-CM, no ORF 3a protein was detected in hCFs even with an extended 48-h incubation period, as depicted in Figure 1B.
Figure 1.
Time course of ORF 3a protein expression in SARS-CoV-2-infected human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) and human cardiac fibroblasts (hCFs). (A) Human-induced pluripotent stem cell-derived cardiomyocytes were subjected to SARS-CoV-2 infection [multiplicity of infection (MOI) of 1.0], and cells were harvested at various time points post-infection as specified. The upper section shows representative immunoblot signals for ORF 3a protein and β-actin as loading control. The lower section depicts the average ORF 3a protein signals, obtained from n = 3 independent experiments, normalized to β-actin and presented in arbitrary units (A.U.). (B) Both hiPSC-CMs and hCFs were exposed to SARS-CoV-2, and cells were harvested at designated time intervals. The top section illustrates the immunoblot signals for ORF 3a protein and β-actin from the respective cell lysates. The bottom section presents the average ORF 3a protein signals from n = 3 experiments, adjusted relative to β-actin expression and measured in A.U. Statistical significance is indicated by ***, representing a P < 0.001 from a two-tailed unpaired Student’s t-test. Data are expressed as mean ± standard error of the mean.
Functional expression of SARS-CoV-2 ORF 3a protein in X. laevis oocytes
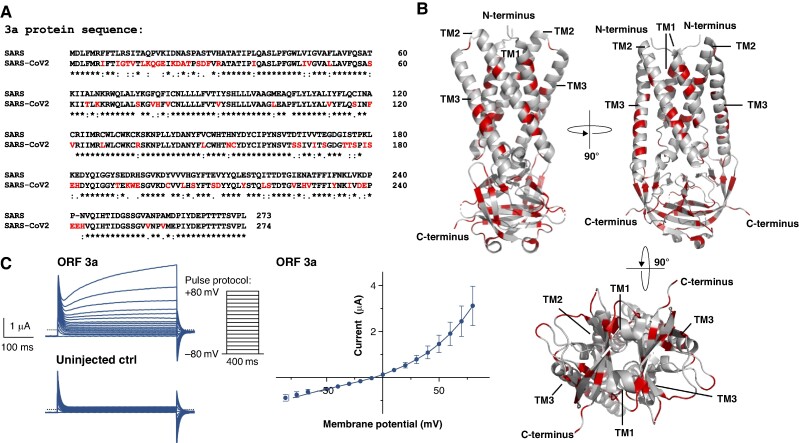
For the ORF 3a protein of the conventional SARS-CoV-1 virus, previous studies have demonstrated its functionality as an ion channel when expressed heterologously in X. laevis oocytes, resulting in the generation of macroscopic outward currents.23 Nevertheless, the amino acid sequence of SARS-CoV-2’s ORF 3a protein reveals only an about 72% similarity with that of SARS-CoV-1’s ORF 3a protein (Figure 2A). The differences in amino acid sequence are not just limited to the peripheral domains, but also affect the central pore-forming regions (Figure 2B). To investigate the SARS-CoV-2 ORF 3a protein’s ion channel capabilities, the gene encoding the ORF 3a protein of the early 2020 SARS-CoV-2 virus was generated by gene synthesis, subcloned, and used to synthesize copy RNA (cRNA) by in vitro transcription. Macroscopic cation currents, as shown in Figure 2C, can be derived by conducting TEVC measurements on X. laevis oocytes 48–72 h after intracytoplasmic cRNA injection, which differ significantly from background currents, recorded from uninjected control cells. To determine the current–voltage relationship, 400 ms depolarizing pulses were applied in 10 mV increments from a resting membrane potential of −80 mV to values of +80 mV according to the voltage protocol shown. The current–voltage relationship of the macroscopic cation currents, mediated by ORF 3a, is presented in the right part of Figure 2C (mean value of n = 5 measurements).
Figure 2.
The SARS-CoV-2 ORF 3a protein gives rise to outward potassium currents upon heterologous expression in X. laevis oocytes. (A) Protein sequence alignment of the ORF 3a protein sequences derived from SARS-CoV-2 and the classic SARS-CoV-1 variant (SARS2018). Differences in the amino acid sequence are highlighted in red *, conserved amino acid residue; :, conservative amino acid residue exchange; ., semiconservative amino acid residue exchange. (B) 3D visualization of the SARS-CoV-2 ORF 3a dimer based on the cryo-EM structure recently revealed by Kern et al.22 (PDB-ID: 6XDC); again the differences to the SARS-CoV-1 variant are highlighted in red. TM1–3, transmembrane domain 1–3. (C) Representative family of current trances recorded from X. laevis oocytes, heterologously expressing SARS-CoV-2 ORF 3a proteins by application of the pulse protocol depicted and respective uninjected control (ctrl) oocytes. Current–voltage relationship of SARS-CoV-2 ORF 3a protein-mediated current. Current amplitudes of ORF 3a-expressing X. laevis oocytes were quantified at the end of each 400 ms voltage pulse. Data are expressed as mean ± standard error of the mean from n = 5 cells. Dotted line represents zero current level, and scale bar is depicted as inset.
Modulation of SARS-CoV-2 ORF 3a protein-related currents by clinically employed antiarrhythmic drugs
Based on the observation that SARS-CoV-2 infects hiPSC-CM in vitro and induces the expression of the ORF 3a protein, and that the SARS-CoV-2 ORF 3a protein induces macroscopic outward cation currents upon heterologous expression, it was hypothesized that ORF 3a protein-mediated currents may affect the electrophysiological properties of cardiomyocytes in patients with COVID-19 infection and thereby contribute to arrhythmogenesis. To address the question of which clinically used antiarrhythmic drugs can inhibit ORF 3a protein-mediated currents, SARS-CoV-2 ORF 3a proteins heterologously expressed in X. laevis oocytes were subjected to a broad screening involving several clinically employed antiarrhythmic drugs.
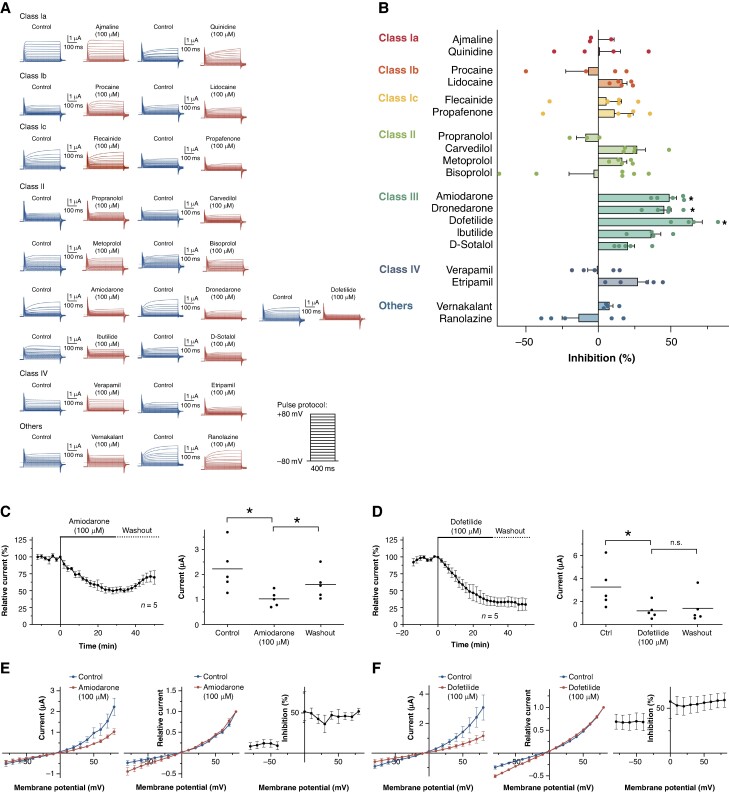
According to the described method, SARS-CoV-2 ORF 3a protein-expressing X. laevis oocytes were subjected to TEVC measurements. Macroscopic outward currents were quantified at the end of the +80 mV test pulse. Cells were stabilized by applying the depicted voltage protocol at 2-min intervals until current variance within a 10-min period was <10%. The appropriate antiarrhythmic drugs were then added to the extracellular bath solution at a concentration of 100 µM, and measurements were continued for 30 min. The results of the screening are shown in Figure 3A and B.
Figure 3.
Effects of clinically employed antiarrhythmic drugs on SARS-CoV-2 ORF 3a protein-mediated currents—class III antiarrhythmic drugs amiodarone and dofetilide block SARS-CoV-2 ORF 3a protein-mediated currents. (A) Representative macroscopic currents recorded under control conditions and after application of the respective antiarrhythmic drug as indicated (100 µM, 30 min) (B) Effects of the respective antiarrhythmics on SARS-CoV-2 ORF 3a protein-mediated currents, quantified at the end of the +80 mV test pulse (n = 3–6 cells). The drugs are arranged according to Vaughan Williams classes I–IV. Data are expressed as mean ± standard error of the mean from. (C and D) Time course of SARS-CoV-2 ORF 3a current inhibition by amiodarone (C) or dofetilide (D) and the partial reversibility of inhibition upon washout. In the right part of the panel, mean current levels are depicted: under control conditions, subsequent to a 30-min superfusion with the respective drug, and after a 20-min washout period. (E and F) Left: activation curves (total current amplitudes, quantified at the end of the 400 ms test pulse), investigated under isochronal recording conditions for control conditions and after administration of the respective drug. Center: normalized activation curves, investigated under isochronal recording conditions (current amplitudes normalized to the maximum current), investigated under isochronal recording conditions. Right: fraction of blocked step currents, plotted as function of the test pulse potential. Data are presented as mean ± standard error of the mean from n = 5 cells. Dotted lines represent zero current level, and scale bars are depicted as insets. Statistical significance is indicated by *, representing a P < 0.05 from two-tailed paired Student’s t-tests followed by Bonferroni correction.
No significant inhibitory effects were observed with the Vaughan Williams class Ia (ajmaline and quinidine; n = 3–4), Ib (procaine and lidocaine; n = 4–5), or Ic (flecainide and propafenone; n = 5) sodium channel inhibitors. Among the Vaughan Williams class II beta-blockers, propranolol, carvedilol, metoprolol, and bisoprolol were investigated. While propranolol and bisoprolol had no clear effect on ORF 3a-mediated currents, application of metoprolol and carvedilol resulted in a trend towards inhibition of ORF 3a currents that did not reach statistical significance. The Vaughan Williams class III antiarrhythmic agents, which act as potassium channel blockers, exhibited the most significant inhibitory effects on the SARS-CoV-2 ORF 3a protein. Dronedarone, amiodarone, and dofetilide caused a 45.4 ± 4.7% (P = 0.004), 49.1 ± 4.6% (P = 0.003), and 65.1 ± 6.7% (P = 0.01) inhibition of ORF 3a currents, respectively. Slightly lower inhibitory effects were seen after administering ibutilide (36.4 ± 6.6%, P = 0.04) and D-sotalol (20.6 ± 4.5%, P = 0.06). As Vaughan Williams class IV antiarrhythmic agents, the calcium channel blockers verapamil and etripamil were investigated. Verapamil had no significant effect on ORF 3a-mediated currents. Perfusion with 100 µM etripamil resulted in a non-statistically significant trend towards inhibition of ORF 3a-mediated currents. Finally, vernakalant and ranolazine, two antiarrhythmic drugs not traditionally classified in the Vaughan Williams classification system, were tested to evaluate their interaction with ORF 3a. Both drugs did not alter ORF 3a-mediated currents significantly.
SARS-CoV-2 ORF 3a current inhibition by amiodarone and dofetilide
Electrophysiological implications of SARS-CoV-2 ORF 3a current inhibition by clinically used antiarrhythmic drugs were assessed in detail, focusing on the class III antiarrhythmic drugs amiodarone and dofetilide, which had shown the strongest effects in the previous screening. The time course of SARS-CoV-2 ORF 3a current inhibition by amiodarone and dofetilide is shown in Figure 3C and D. Following a control period with no significant amplitude changes, the SARS-CoV-2 ORF 3a protein currents decreased rapidly upon administration of 100 μM amiodarone or dofetilide (Figure 3C and D). Interestingly, effects of 100 μM amiodarone were partially reversible and current amplitudes reached 69.7 ± 9.9% (P = 0.04) of the control values following washout. The block by dofetilide persisted upon washout.
Drug effects on SARS-CoV-2 ORF 3a protein current–voltage (I–V) relationship were investigated under isochronal recording conditions. Representative families of SARS-CoV-2 ORF 3a protein current traces are shown for control conditions, after application of 100 μM amiodarone or dofetilide for 30 min, and after a 20-min washout period (Figure 3E and F). The current–voltage relationship was not affected by 100 μM amiodarone or dofetilide (Figure 3E and F). Relative current inhibition was plotted as function of the test pulse potential in Figure 3E and F. Reduction of SARS-CoV-2 ORF 3a current was not significantly different between +20 and +60 mV or between −80 and −40 mV (Figure 3E and F). However, the current inhibition was significantly reduced at potentials below the reversal potential compared positive potentials.
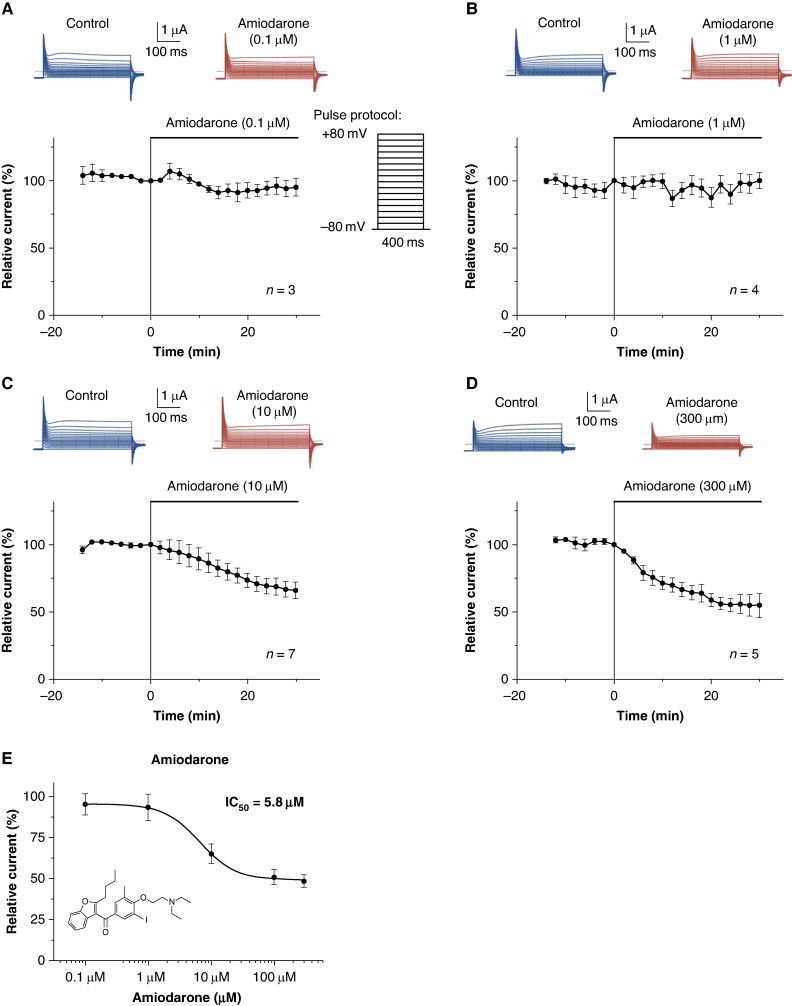
Amiodarone inhibits SARS-CoV-2 ORF 3a currents in a concentration-dependent manner
To study the concentration-dependent effect of amiodarone on SARS-CoV-2 ORF 3a currents, different concentrations of amiodarone were examined in the experimental setting described above (Figure 4A–D). In Figure 4E, currents in the presence of the drug were normalized to their respective control values and plotted as relative current amplitudes. The IC50 value for amiodarone inhibition of SARS-CoV-2 ORF 3a protein currents yielded 5.8 μM (n = 3–7 cells per concentration). It is noteworthy that amiodarone’s inhibition of SARS-CoV-2 ORF 3a protein currents in oocytes remained incomplete, with a maximum current reduction of 51.5 ± 3.8%.
Figure 4.
Concentration–response relationship for the effect of amiodarone on SARS-CoV-2 ORF 3a proteins heterologously expressed in X. laevis oocytes. (A–E) Time course of SARS-CoV-2 ORF 3a current inhibition by amiodarone, administered in a concentration of (A) 0.1 µM, (B) 1 µM, (C) 10 µM, and (D) 300 µM. Representative macroscopic current traces elicited before (Control) and after administration of amiodarone by application of the voltage pulse protocol depicted in (A) are shown at the top of the respective panels. (E) Concentration–response relationships for the effects of amiodarone on SARS-CoV-2 ORF 3a protein-mediated currents. The mean inhibitory concentration (IC50) is provided as inset. Data are expressed as mean ± standard error of the mean from n = 3–7 cells as indicated. Dotted lines represent zero current level, and scale bars are given as insets.
Structural determinants of SARS-CoV-2 ORF 3a current inhibition by class III antiarrhythmic drugs
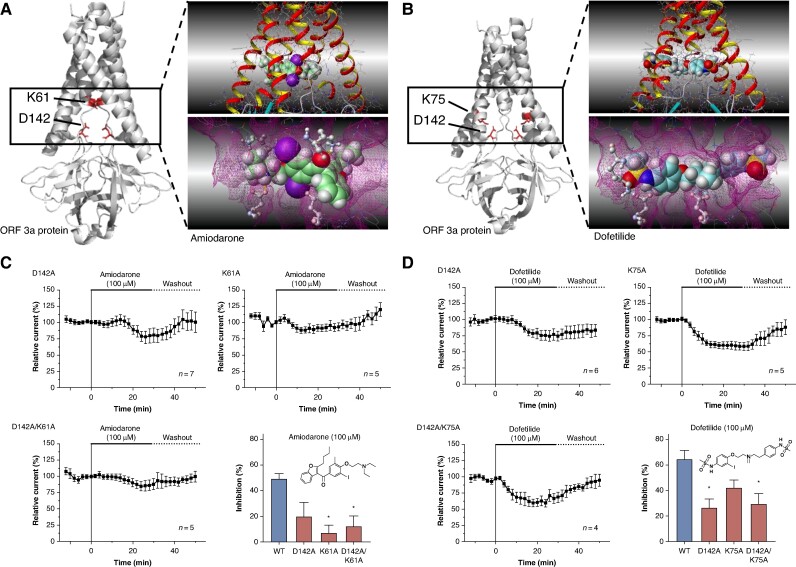
To gain a better understanding of the molecular interaction between the SARS-CoV-2 ORF 3a protein and the antiarrhythmic drugs amiodarone and dofetilide, the cryo-EM structure (PDB-ID: 7KJR) of the SARS-CoV-2 ORF 3a protein recently revealed by Kern et al.22 was used for computational docking simulations. The simulations indicated that amiodarone and dofetilide bind to the intracellular region of the channel pore, thereby inhibiting ion flux (Figure 5A and B). Almost all poses of amiodarone showed the charge enhanced hydrogen bond between the protonated tertiary amine and aspartic acid 142. Also, a strong interaction between the carbonyl oxygen to lysine 61 was detected. Both of these key interactions are with the same monomer. Dofetilide sits in the cavity and interacts via hydrogen bonds with both aspartic acid 142, i.e. with both monomers of the channel. Here, the sulfonamide-NH and the protonated tertiary amine of dofetilide are the respective interaction partners. Another but weaker hydrogen bond is seen between the sulfonamide oxygens to lysine 75 of one monomer. Of note, all three amino acids are highly conserved among different SARS-CoV-2 strains (see Supplementary material online, Figure S1).
Figure 5.
Structural determinants of SARS-CoV-2 ORF 3a protein interaction with amiodarone and dofetilide. (A and B) Computational docking simulation revealing the interaction of amiodarone (A) and dofetilide (B) with the intracellular channel pore of the SARS-CoV-2 ORF 3a protein dimer. The simulation is based on the cryo-EM structure delineated by Kern et al.22 (PDB-ID: 7KJR). It highlights the critical roles of amino acids lysine 61 (K61) and aspartic acid 142 (D142) in forming the molecular drug-binding site for amiodarone (A) and the involvement of lysine 75 (K75) along with aspartic acid 142 (D142) in the interaction site for dofetilide (B). (C) To confirm the contribution of the said amino acids to the drug-binding site, mutant proteins SARS-CoV-2 ORF 3a K61A, D142A, and the double-mutant K61A/D142A were expressed in X. laevis oocytes and exposed to amiodarone (100 µM; 30 min as described above). (D) In a similar fashion, SARS-CoV-2 ORF 3a-K75A, D142A, and the double-mutant K75A/D142A were superfused with dofetilide (100 µM; 30 min as described above) upon heterologous expression in X. laevis oocytes. Data are presented as mean ± standard error of the mean. Chemical structures of amiodarone and dofetilide are depicted as insets. Statistical significance is indicated by *, representing a P < 0.05 from a two-tailed unpaired Student’s t-test vs. WT channels followed by Bonferroni correction.
The effects of dofetilide inhibition were evaluated on mutant SARS-CoV-2 ORF 3a-D142A, SARS-CoV-2 ORF 3a-K75A, and double-mutant SARS-CoV-2 ORF 3a-D142A/K75A, along with wild-type (WT) SARS-CoV-2 ORF 3a proteins (Figure 5D). The degree of inhibition was determined at the end of the +80 mV test pulse following 30-min perfusion of 100 µM dofetilide. Dofetilide inhibited WT ORF 3a-mediated currents by 65.1 ± 6.7% (P = 0.01), while its inhibitory effects on ORF 3a-D142A, ORF 3a-K75A, and double-mutant ORF 3a-D142A/K75A were 26.6 ± 7.1% (P = 0.02), 42.7 ± 5.9% (P = 0.09), and 12.6 ± 18.2% (P = 0.007), respectively. In a similar fashion, amiodarone inhibited WT SARS-CoV-2 ORF 3a protein-mediated currents by 49.1 ± 4.6% (P = 0. 003), while inhibitory effects on SARS-CoV-2 ORF 3a protein D142A, ORF 3a-K61A, and SARS-CoV-2 ORF 3a-D142A/K61A protein were 19.4 ± 10.9% (P = 0.07), 6.6 ± 6.2% (P = 0.004), and 11.7 ± 8.4% (P = 0.01), respectively (Figure 5C).
Discussion
Cardiac arrhythmias are considered to be among the most important extrapulmonary manifestations of SARS-CoV-2. Therefore, understanding the mechanisms underlying arrhythmogenesis in COVID-19 is critical to reduce morbidity and mortality associated with COVID-19.5,6 This study aimed to explore the role of the SARS-CoV-2 viroporin ORF 3a in COVID-19-associated arrhythmias and its potential as a pharmacological target. To accomplish this goal, a two-fold approach was adopted. First, this study investigated whether ORF 3a is expressed in hiPSC-derived cardiomyocytes upon infection with SARS-CoV-2. Second, this study evaluated the ion channel activity of the SARS-CoV-2 ORF 3a protein and characterized its pharmacological fingerprint by assessing its interaction with various clinically used antiarrhythmic drugs.
Expression of the SARS-CoV-2 ORF 3a protein in human-induced pluripotent stem cell-derived cardiomyocytes
Immunoblot analysis showed that ORF 3a is expressed in hiPSC-derived cardiomyocytes as quickly as 16 h after SARS-CoV-2 exposure. This expression was cardiomyocyte specific, as hCF did not express ORF 3a post-infection. This selective cellular tropism correlates with expression of ACE2, known to facilitate SARS-CoV-2 entry, which, while not detectable in fibroblasts, has been shown to be preferentially expressed in cardiomyocytes.21 However, further research is needed to confirm these findings in primary human cardiomyocytes and to investigate the broader implications of ORF 3a expression in the heart in patients suffering from COVID-19. To date, the question of whether coronaviruses replicate in cardiomyocytes has not been conclusively answered.5 In a rabbit model published more than three decades ago, it was shown that infection with coronaviruses led to viral myocarditis and heart failure.30 Similarly, a more recent model of engineered heart tissue containing hiPSC-CM, fibroblasts, and macrophages showed direct cardiomyocyte cell damage under COVID-19 infection.31 In autopsy studies of COVID-19 patients, however, no direct viral presence in cardiomyocytes was found in most cases, with interstitial macrophage infiltration without cardiomyocyte degeneration being observed in over 80% of cases.32,33 In contrast, studies that have conducted cardiovascular magnetic resonance imaging on patients who have recovered from COVID-19 but still exhibit cardiac symptoms show a different result. These studies were able to detect signs of cardiac involvement (myocardial oedema, fibrosis, and impaired right ventricular function) in 58–78% of patients.14,34 It is noteworthy that other studies have found an incidence of cardiac arrhythmias around 10% in patients post COVID-19.19
Ion channel activity of SARS-CoV-2 ORF 3a proteins
This study demonstrated that ORF 3a forms a cation-permeable ion channel, displaying mild outwardly rectifying characteristics. This confirms previous findings about SARS-CoV-2 ORF 3a and its homologue in SARS-CoV-1, suggesting a common ion channel activity due to sequence similarity. Together with the evidence of ORF 3a protein expression in hiPSC-CM after SARS-CoV-2 infection, this strengthens the hypothesis that ion channel activity of ORF 3a contributes to dysregulation of cardiac action potential formation upon viral replication within cardiomyocytes. Since different regions of the heart can be affected by the infection, SARS-CoV-2 infection may lead to the occurrence of various arrhythmias in the atria or ventricles. If the conduction system is affected, bradycardias may also occur. Simultaneous work performed by others could demonstrate that HEK293T cells and H9c2 cardiac myoblasts, upon transfection with recombinant DNA encoding the SARS-CoV-2 ORF 3a, give rise to cation currents that exhibit electrophysiological properties, similar to the endogenous transient outward K+ current (Ito). Computational modelling showed that integration of the aforementioned current component into existing mathematical cardiomyocyte models increases the risk of early afterdepolarization in response to increased heart rate and dysregulated cardiac Ca2+ handling as well as action potential formation.35,36 Over a decade and a half ago, it was reported that SARS-CoV ORF 3a forms a Ba2+-sensitive cation channel when expressed in X. laevis oocytes.23 In the meantime, other groups have demonstrated this in X. laevis oocytes,37–40 HEK293,41 and lipid membranes.22,42 Recently, Miller et al.43 published a critical re-evaluation of these findings, questioning whether the membrane-spanning pore in the cryo-EM structure is wide enough to allow for the movement of dehydrated cations. They also noted the presence of a positively charged aqueous vestibule, which would not favour cation permeation. After Miller et al.43 were unable to measure ion currents upon heterologous expression in HEK293, X. laevis oocytes, and vesicle-reconstituted ORF 3a protein, they questioned the role of the ORF 3a protein as a viroporin. In fact, the pharmacological profile and single channel conduction properties of the ORF 3a protein in bilayer experiments were reported to be strikingly different and varying.44 In the present study, the SARS-CoV-2 ORF 3a protein expressed in X. laevis oocytes consistently produced macroscopic currents. Further studies are needed to determine whether differences in the ORF 3a sequence or variations in the measurement conditions are responsible for these discrepancies, or if they are due to interactions with endogenous proteins of the respective expression systems.
Interaction with antiarrhythmic drugs
Investigation into ORF 3a’s interaction with antiarrhythmic drugs revealed significant inhibition of ORF 3a currents by certain class III antiarrhythmics. Dofetilide and amiodarone, in particular, showed strong inhibition, highlighting their potential in modulating ORF 3a-mediated ion currents. Notably, amiodarone’s inhibition was more pronounced at depolarized potentials, indicating voltage-dependent blocking. The inhibition was not fully reversible upon washout, especially for dofetilide, possibly due to its entrapment within ORF 3a’s inner cavity. Using a combined approach of in silico simulations and in vitro experiments on alanine pore mutants, it was possible to identify the three amino acid residues K61, K75, and D142 as potential components of the drug-binding site in the SARS-CoV-2 ORF 3a protein. Substitution of these key amino acid residues with alanine resulted in decreased inhibitory effects of the substances. All three key amino acid residues are located in the inner cavity of the transmembrane region of ORF 3a. The recently published cryo-EM structure of SARS-CoV-2 ORF 3a shows that the inner cavity resides in the lower half of the transmembrane region and is continuous with the surrounding lipid bilayer through the upper tunnels.22 It can thus be hypothesized that lipophilic molecules such as dofetilide and amiodarone may diffuse through the cell membrane directly into the inner cavity of ORF 3a and subsequently interact with the key amino acid residues. In particular, the negatively charged pore-lining amino acid residue aspartic acid 142, located at the apex of the short alpha helix connecting the transmembrane and cytoplasmic domain, appears to play an important role in mediating interaction with class III antiarrhythmic drugs. The three identified amino acids and their surrounding region appear to be highly conserved among different common strains of SARS-CoV-2 virus (see Supplementary material online, Figure S1). A previous study identified mutations at positions K61 and K75 in only three and two of 2782 substrains analysed, and no mutations in position D142 were reported in this study.45 This suggests that the binding site of class III antiarrhythmics is highly conserved, and that the described effect could be expected across strains.
Clinical relevance of SARS-CoV-2 ORF 3a protein inhibition by class III antiarrhythmic drugs
The present study suggests that inhibition of ORF 3a by class III antiarrhythmics, particularly dofetilide and amiodarone, exhibits inhibitory effects on SARS-CoV-2 ORF 3a protein-mediated currents. During chronic amiodarone therapy, plasma levels ranging from 0.15 to 18.4 µM have been described, with a mean therapeutic range of 1–3 µM.46–48 While the markedly high plasma protein binding of ∼99.8% significantly reduces the free plasma levels of amiodarone, the pronounced lipophilicity of the compound leads to drug accumulation in cardiac tissue.48,49 Regarding the IC50 value of ORF 3a protein inhibition by amiodarone of 5.8 µM, the inhibitory effect of amiodarone on ORF 3a proteins could be expected to be present at therapeutic plasma levels, especially considering that the IC50 values in mammalian cell models are up to 30-fold lower than in X. laevis oocytes.50 Further experiments on concentration dependence, performed in mammalian cells, are warranted to comprehensively clarify this question.
Recently, the polycationic dye ruthenium red and the polyamine spermidine were likewise identified as blockers of the SARS-CoV-2 ORF 3a protein.22 The affinity of amiodarone is, however, about 10-fold higher than that of ruthenium red (IC50 = 90 ± 10 μM; patch clamp experiments performed on proteoliposomes) and 1000-fold higher than spermidine (where the block occurs at concentrations between 1 and 10 mM).22 Notably, class III antiarrhythmic drugs were also identified as off-target blockers of other cation channels such as the K2P channels, of which TREK-1 (K2P2.1) is also inhibited by spermidine and TASK-3 (K2P9.1) is also inhibited by ruthenium red.51–53 In summary, the pharmacological profile of SARS-CoV-2 ORF 3a proteins nonetheless shows a unique fingerprint.
In vitro experiments have shown that transfection of ORF 3a in eukaryotic cells induces apoptosis. Apoptosis markers were significantly reduced in the presence of pharmacological inhibitors such as 4-AP or Ba2+ or upon induction of the loss of function mutations C133A/Y160A.41 This suggests that the cationic currents of the ORF 3a proteins, at least by part, directly influence the virulence of SARS-CoV-2. Furthermore, the attenuation of viral titre and morbidity in murine disease models was demonstrated by the genomic deletion of the SARS-CoV-1 or 2 ORF 3a.22,54,55
Ion channels are attractive drug targets, and the mechanism of ion channel inhibition underlies around 15% of clinically employed drugs.56 The development of vaccines and antiviral therapies has mainly focused on the essential virus-encoded spike protein, RNA-dependent RNA polymerase proteins, and the major protease. The results of this and other studies indicate that the ORF 3a protein may represent a novel promising target for the treatment of complications of COVID-19 infection, such as arrhythmias, as well as for the suppression of viral release.22 The identification and evaluation of such targets are essential to counteract the possible emergence of resistance and to protect against new viruses that may emerge in the future. Assessing the implications of the recently described expression of ORF 3a proteins in mitochondrial membranes and its consequences for myocardial damage and arrhythmogenesis also requires further studies.35 This study highlights the potential of SARS-CoV-2 ORF 3a as a pharmacological target for managing COVID-19-associated arrhythmias. The identification of ORF 3a’s potassium channel activity and its inhibition by class III antiarrhythmic drugs opens new avenues for therapeutic intervention. The mechanistic and biophysical insights provided here could facilitate the development of targeted ORF 3a inhibitors, offering novel strategies in the treatment of arrhythmias in COVID-19 patients. Further research in larger animal models and clinical studies are warranted to validate these findings and to explore the full therapeutic potential of ORF 3a inhibition.
Limitations
The research encountered limitations due to the immature phenotype of hiPSC-CMs, which may not fully replicate the properties of adult cardiomyocytes. Future studies should concentrate on verifying ORF 3a expression and its effects in primary human cardiomyocytes. Additionally, the study relied on the TEVC technique in X. laevis oocytes, which may underestimate drug potency compared to mammalian cells. Furthermore, the dual-electrode voltage clamp experiments were performed in a 100 K+ solution that did not contain sodium. This may have compromised the investigation of the effects of class I antiarrhythmic drugs. Future investigations using mammalian cell models, pulse protocols that are more physiological than square pulses like action potential clamping, or current clamp measurements of action potentials are needed to more accurately assess the role of the SARS-CoV-2 ORF 3a protein in cardiac electrophysiology and its pharmacological modulation.
Conclusions
In conclusion, this study has established the expression of the ORF 3a protein in hiPSC-CMs upon SARS-CoV-2 infection. Further studies are warranted to assess whether this mechanism actually contributes to the arrhythmogenesis described in COVID-19 patients. The fact that SARS-CoV-2 ORF 3a is inhibited by clinically employed class III antiarrhythmic drugs may be relevant to the future treatment of COVID-19-associated arrhythmias. Since the ORF 3a protein is further implicated in viral release, this observation could even have implications for the causal therapy of COVID patients.
Supplementary Material
Acknowledgements
We thank Anne Grube, Sabine Höllriegel, Björn Rogatzki, Lisa Kuenstler, and Katrin Kupser for excellent technical support. We further thank Christian Drosten at the Charité, Berlin, and the European Virus Archive (EVAg) for the provision of the SARS-CoV-2 strain BavPat1. The use of AI tools ChatGPT 4.o (OpenAI, San Francisco, CA, USA) and DeepL (DeepL GmbH Cologne, Germany) to improve the readability of the manuscript and for image generation (central illustration) is acknowledged, with the clarification that they were not employed for text generation and data evaluation/interpretation.
Contributor Information
Felix Wiedmann, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany; DZHK (German Center for Cardiovascular Research), partner site Heidelberg/Mannheim, University of Heidelberg, Heidelberg, Germany; HCR, Heidelberg Center for Heart Rhythm Disorders, University Hospital Heidelberg, Heidelberg, Germany.
Emika Boondej, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany.
Megan Stanifer, Department of Molecular Genetics and Microbiology, University of Florida, College of Medicine, Gainesville, FL, USA.
Amelie Paasche, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany; DZHK (German Center for Cardiovascular Research), partner site Heidelberg/Mannheim, University of Heidelberg, Heidelberg, Germany.
Manuel Kraft, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany; DZHK (German Center for Cardiovascular Research), partner site Heidelberg/Mannheim, University of Heidelberg, Heidelberg, Germany; HCR, Heidelberg Center for Heart Rhythm Disorders, University Hospital Heidelberg, Heidelberg, Germany.
Merten Prüser, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany; DZHK (German Center for Cardiovascular Research), partner site Heidelberg/Mannheim, University of Heidelberg, Heidelberg, Germany.
Timon Seeger, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany; DZHK (German Center for Cardiovascular Research), partner site Heidelberg/Mannheim, University of Heidelberg, Heidelberg, Germany.
Ulrike Uhrig, Chemical Biology Core Facility, EMBL, Heidelberg, Germany.
Steeve Boulant, Department of Molecular Genetics and Microbiology, University of Florida, College of Medicine, Gainesville, FL, USA.
Constanze Schmidt, Department of Cardiology, University Hospital Heidelberg, Im Neuenheimer Feld 410, D-69120 Heidelberg, Germany; DZHK (German Center for Cardiovascular Research), partner site Heidelberg/Mannheim, University of Heidelberg, Heidelberg, Germany; HCR, Heidelberg Center for Heart Rhythm Disorders, University Hospital Heidelberg, Heidelberg, Germany.
Supplementary material
Supplementary material is available at Europace online.
Funding
This work was supported by research grants from the German Cardiac Society (Research Clinician-Scientist programme to F.W.); German Heart Foundation (atrial fibrillation research funding to F.W. and C.S., F/03/19 to C.S., and Kaltenbach scholarship to A.P.); German Center for Cardiovascular Research (DZHK; TRP starter grant, Shared expertise, Innovation cluster); Federal Ministry of Education and Research Germany (BMBF); German Research Foundation (SCHM 3358/1-1 to C.S.); and Else-Kröner Fresenius Foundation (EKFS Fellowship and EKFS Clinician-Scientist professorship to C.S.). C.S., F.W., M.K., and A.P. are members of the CRC1425 and CRC1550, funded by the German Research Foundation (#422681845 and #464424253). M.S. and S.B. were funded by their start-up funds from the University of Florida.
Data availability
All relevant data are within the manuscript and its supporting information files. Any further data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miller IF, Becker AD, Grenfell BT, Metcalf CJE. Disease and healthcare burden of COVID-19 in the United States. Nat Med 2020;26:1212–7. [DOI] [PubMed] [Google Scholar]
- 3. Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep 2020;32:107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Triana S, Metz-Zumaran C, Ramirez C, Kee C, Doldan P, Shahraz M et al. Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut. Mol Syst Biol 2021;17:e10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huseynov A, Akin I, Duerschmied D, Scharf RE. Cardiac arrhythmias in post-COVID syndrome: prevalence, pathology, diagnosis, and treatment. Viruses 2023;15:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS et al. Extrapulmonary manifestations of COVID-19. Nat Med 2020;26:1017–32. [DOI] [PubMed] [Google Scholar]
- 7. Boulos PK, Freeman SV, Henry TD, Mahmud E, Messenger JC. Interaction of COVID-19 with common cardiovascular disorders. Circ Res 2023;132:1259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xie Y, Xu E, Bowe B, Al-Aly Z. Long-term cardiovascular outcomes of COVID-19. Nat Med 2022;28:583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gopinathannair R, Merchant FM, Lakkireddy DR, Etheridge SP, Feigofsky S, Han JK et al. COVID-19 and cardiac arrhythmias: a global perspective on arrhythmia characteristics and management strategies. J Interv Card Electrophysiol 2020;59:329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020;323:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Musikantow DR, Turagam MK, Sartori S, Chu E, Kawamura I, Shivamurthy P et al. Atrial fibrillation in patients hospitalized with COVID-19: incidence, predictors, outcomes, and comparison to influenza. JACC Clin Electrophysiol 2021;7:1120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wollborn J, Karamnov S, Fields KG, Yeh T, Muehlschlegel JD. COVID-19 increases the risk for the onset of atrial fibrillation in hospitalized patients. Sci Rep 2022;12:12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dewland TA, Marcus GM. SARS-CoV-2 infection and cardiac arrhythmias. Nat Cardiovasc Res 2022;1:1109–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Puntmann VO, Carerj ML, Wieters I, Fahim M, Arendt C, Hoffmann J et al. Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from coronavirus disease 2019 (COVID-19). JAMA Cardiol 2020;5:1265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Puntmann VO, Martin S, Shchendrygina A, Hoffmann J, Ka MM, Giokoglu E et al. Long-term cardiac pathology in individuals with mild initial COVID-19 illness. Nat Med 2022;28:2117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen MY, Xiao FP, Kuai L, Zhou HB, Jia ZQ, Liu M et al. Outcomes of atrial fibrillation in patients with COVID-19 pneumonia: a systematic review and meta-analysis. Am J Emerg Med 2021;50:661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spinoni EG, Mennuni M, Rognoni A, Grisafi L, Colombo C, Lio V et al. Contribution of atrial fibrillation to in-hospital mortality in patients with COVID-19. Circ Arrhythm Electrophysiol 2021;14:e009375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mountantonakis SE, Saleh M, Fishbein J, Gandomi A, Lesser M, Chelico J et al. Atrial fibrillation is an independent predictor for in-hospital mortality in patients admitted with SARS-CoV-2 infection. Heart Rhythm 2021;18:501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sudre CH, Murray B, Varsavsky T, Graham MS, Penfold RS, Bowyer RC et al. Attributes and predictors of long COVID. Nat Med 2021;27:626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang C, Huang L, Wang Y, Li X, Ren L, Gu X et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet 2021;397:220–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kern DM, Sorum B, Mali SS, Hoel CM, Sridharan S, Remis JP et al. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat Struct Mol Biol 2021;28:573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu W, Zheng BJ, Xu K, Schwarz W, Du L, Wong CK et al. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci U S A 2006;103:12540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD et al. Chemically defined generation of human cardiomyocytes. Nat Methods 2014;11:855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 2013;8:162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmidt C, Wiedmann F, Voigt N, Zhou XB, Heijman J, Lang S et al. Upregulation of K2P3.1K+ current causes action potential shortening in patients with chronic atrial fibrillation. Circulation 2015;132:82–92. [DOI] [PubMed] [Google Scholar]
- 27. Schmidt C, Wiedmann F, Zhou XB, Heijman J, Voigt N, Ratte A et al. Inverse remodelling of K2P3.1K+ channel expression and action potential duration in left ventricular dysfunction and atrial fibrillation: implications for patient-specific antiarrhythmic drug therapy. Eur Heart J 2017;38:1764–74. [DOI] [PubMed] [Google Scholar]
- 28. Wiedmann F, Kiper AK, Bedoya M, Ratte A, Rinné S, Kraft M et al. Identification of the A293 (AVE1231) binding site in the cardiac two-pore-domain potassium channel TASK-1: a common low affinity antiarrhythmic drug binding site. Cell Physiol Biochem 2019;52:1223–35. [DOI] [PubMed] [Google Scholar]
- 29. Wiedmann F, Beyersdorf C, Zhou XB, Kraft M, Paasche A, Jávorszky N et al. Treatment of atrial fibrillation with doxapram: TASK-1 potassium channel inhibition as a novel pharmacological strategy. Cardiovasc Res 2021;117:1790–801. [DOI] [PubMed] [Google Scholar]
- 30. Edwards S, Small JD, Geratz JD, Alexander LK, Baric RS. An experimental model for myocarditis and congestive heart failure after rabbit coronavirus infection. J Infect Dis 1992;165:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bailey AL, Dmytrenko O, Greenberg L, Bredemeyer AL, Ma P, Liu J et al. SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis. JACC Basic Transl Sci 2021;6:331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Basso C, Leone O, Rizzo S, De Gaspari M, van der Wal AC, Aubry MC et al. Pathological features of COVID-19-associated myocardial injury: a multicentre cardiovascular pathology study. Eur Heart J 2020;41:3827–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chung MK, Zidar DA, Bristow MR, Cameron SJ, Chan T, Harding CV 3rd et al. COVID-19 and cardiovascular disease: from bench to bedside. Circ Res 2021;128:1214–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang L, Zhao P, Tang D, Zhu T, Han R, Zhan C et al. Cardiac involvement in patients recovered from COVID-2019 identified using magnetic resonance imaging. JACC Cardiovasc Imaging 2020;13:2330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Polina I, Guo Y, Jhun BS, Tolkacheva E, O-Uchi J. Expression of SARS-CoV-2 viroporins triggers cardiac arrhythmia. FASEB J 2021;35. doi: 10.1096/fasebj.2021.35.S1.04486 [DOI] [Google Scholar]
- 36. Polina IA, Guo Y, Cypress MW, Tolkacheva EG, Sook Jhun B, O-Uchi J. Expression of SARS-CoV-2-ORF3a protein induces cardiomyocyte damage. Biophys J 2022;121:237a.34951981 [Google Scholar]
- 37. Schwarz S, Sauter D, Lu W, Wang K, Sun B, Efferth T et al. Coronaviral ion channels as target for Chinese herbal medicine. Onco Therapeutics 2012;3:1–13. [Google Scholar]
- 38. Schwarz S, Sauter D, Wang K, Zhang R, Sun B, Karioti A et al. Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med 2014;80:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwarz S, Wang K, Yu W, Sun B, Schwarz W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Res 2011;90:64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Toft-Bertelsen TL, Jeppesen MG, Tzortzini E, Xue K, Giller K, Becker S et al. Amantadine has potential for the treatment of COVID-19 because it inhibits known and novel ion channels encoded by SARS-CoV-2. Commun Biol 2021;4:1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chan CM, Tsoi H, Chan WM, Zhai S, Wong CO, Yao X et al. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int J Biochem Cell Biol 2009;41:2232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chien T-H, Chiang Y-L, Chen C-P, Henklein P, Hänel K, Hwang I-S et al. Assembling an ion channel: ORF 3a from SARS-CoV. Biopolymers 2013;99:628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B et al. The SARS-CoV-2 accessory protein ORF3a is not an ion channel, but does interact with trafficking proteins. Elife 2023;12:e84477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McClenaghan C, Hanson A, Lee SJ, Nichols CG. Coronavirus proteins as ion channels: current and potential research. Front Immunol 2020;11:573339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems 2020;5:e00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haffajee CI, Love JC, Alpert JS, Asdourian GK, Sloan KC. Efficacy and safety of long-term amiodarone in treatment of cardiac arrhythmias: dosage experience. Am Heart J 1983;106:935–43. [DOI] [PubMed] [Google Scholar]
- 47. Latini R, Tognoni G, Kates RE. Clinical pharmacokinetics of amiodarone. Clin Pharmacokinet 1984;9:136–56. [DOI] [PubMed] [Google Scholar]
- 48. Gierten J, Ficker E, Bloehs R, Schweizer PA, Zitron E, Scholz E et al. The human cardiac K2P3.1 (TASK-1) potassium leak channel is a molecular target for the class III antiarrhythmic drug amiodarone. Naunyn Schmiedebergs Arch Pharmacol 2010;381:261–70. [DOI] [PubMed] [Google Scholar]
- 49. Veronese ME, McLean S, Hendriks R. Plasma protein binding of amiodarone in a patient population: measurement by erythrocyte partitioning and a novel glass-binding method. Br J Clin Pharmacol 1988;26:721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Redfern WS, Carlsson L, Davis AS, Lynch WG, MacKenzie I, Palethorpe S et al. Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development. Cardiovasc Res 2003;58:32–45. [DOI] [PubMed] [Google Scholar]
- 51. Schmidt C, Wiedmann F, Schweizer PA, Becker R, Katus HA, Thomas D. Novel electrophysiological properties of dronedarone: inhibition of human cardiac two-pore-domain potassium (K2P) channels. Naunyn Schmiedebergs Arch Pharmacol 2012;385:1003–16. [DOI] [PubMed] [Google Scholar]
- 52. Schmidt C, Wiedmann F, Schweizer PA, Becker R, Katus HA, Thomas D. Class I antiarrhythmic drugs inhibit human cardiac two-pore-domain K+ (K2P) channels. Eur J Pharmacol 2013;721:237–48. [DOI] [PubMed] [Google Scholar]
- 53. Wiedmann F, Frey N, Schmidt C. Two-pore-domain potassium (K2P-) channels: cardiac expression patterns and disease-specific remodelling processes. Cells 2021;10:2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio 2018;9:e02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Silvas JA, Vasquez DM, Park JG, Chiem K, Allué-Guardia A, Garcia-Vilanova A et al. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J Virol 2021;95:e0040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McManus OB. HTS assays for developing the molecular pharmacology of ion channels. Curr Opin Pharmacol 2014;15:91–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its supporting information files. Any further data that support the findings of this study are available from the corresponding author upon reasonable request.