Abstract
Triple-negative breast cancer (TNBC), the most aggressive subtype, presents a critical challenge due to the absence of approved targeted therapies. Hence, there is an urgent need to identify effective therapeutic targets for this condition. While epidermal growth factor receptor (EGFR) is prominently expressed in TNBC and recognized as a therapeutic target, anti-EGFR therapies have yet to gain approval for breast cancer treatment due to their associated side effects and limited efficacy. Here, we discovered that intercellular adhesion molecule-1 (ICAM-1) exhibits elevated expression levels in metastatic breast cancer and serves as a pivotal binding adaptor for EGFR activation, playing a crucial role in malignant progression. The activation of EGFR by tumor-expressed ICAM-1 initiates biased signaling within the JAK1/STAT3 pathway, consequently driving epithelial-to-mesenchymal transition and facilitating heightened metastasis without influencing tumor growth. Remarkably, ICAM-1-neutralizing antibody treatment significantly suppressed cancer metastasis in a breast cancer orthotopic xenograft mouse model. In conclusion, our identification of ICAM-1 as a novel tumor intrinsic regulator of EGFR activation offers valuable insights for the development of TNBC-specific anti-EGFR therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02150-4.
Keywords: ICAM-1, Triple-negative breast cancer, EGFR, JAK1/STAT3 signaling, Targeted therapy
Introduction
Breast cancer, the leading form of cancer among women worldwide, also remains the leading cause of death [1, 2]. This high mortality rate is mainly due to metastatic spread to distant organs [3–5]. However, the factors driving metastasis, a major hallmark and cause of death in all cancer types, remain poorly understood [6]. Breast cancer can be categorized into several molecular subtypes: Luminal A and B, HER2 + , and triple-negative breast cancer (TNBC). TNBC is characterized by the absence of estrogen, progesterone, and human epidermal growth factor receptor 2 (HER2) receptors. This molecular subtype overlaps with basal-like breast cancer (BLBC), a subgroup that expresses cytokines and other non-luminal (basal) genes [7]. Most basal-like breast cancers (BLBCs) are triple-negative. However, approximately 5% of estrogen receptor-positive tumors and between 6 and 35% of HER2-positive tumors exhibit a basal-like gene expression profile [8, 9]. Patients diagnosed with TNBC endure a more aggressive and profoundly metastatic manifestation of this disease and have a worst prognosis than those with other breast cancer subtypes [10, 11].
Traditional chemotherapeutic agents, such as taxanes, anthracyclines, and platinum-based drugs, remain the cornerstone of first-line treatment for TNBC patients. Although responses to treatment are common, TNBC often relapses quickly, and resistance to chemotherapy eventually develops [12, 13]. Recent advances in molecular biology and immunology have paved the way for the development of highly targeted therapies for breast cancer. The primary goal of targeted therapy is to inhibit specific molecules that promote tumor progression. Key targets include AKT, cyclin-dependent kinases, poly (ADP-ribose) polymerase (PARP), and various growth factors, which have shown potential for treating specific breast cancer subtypes [14–16]. However, targeted drugs have not yet shown significant therapeutic promise against triple-negative breast cancer (TNBC). Currently, the only approved targeted drug for later-line treatment of metastatic TNBC is sacituzumab govitecan, an antibody–drug conjugate that targets Trop-2, which has demonstrated improved survival in a recent phase III trial [17].
Immunotherapy has also emerged as a promising treatment approach specifically for TNBC patients. Various immunotherapeutic modalities, including immune checkpoint blockade, vaccination, and adoptive cell transfer, have been extensively studied in clinical settings, particularly for TNBC [14, 18]. Immunotherapy combined with chemotherapy is available for patients with advanced TNBC that express PD-L1, showing a significant overall survival benefit in patients with TNBC [19–21]. However, the impact of checkpoint inhibitors (anti-PD1/PD-L1 antibodies) on the overall survival of TNBC patients is modest suggesting that additional treatment options are still needed for TNBC.
Abnormal activation or mutation of epidermal growth factor receptor (EGFR) is frequently reported in various types of cancers [22–24]. EGFR is highly expressed in TNBC and correlates with poor survival [25–27]. Accordingly, EGFR has been proposed as a biomarker and therapeutic target in TNBC [28]. However, anti-EGFR therapies, including monoclonal antibodies (mAbs) and tyrosine kinase inhibitors (TKIs), have not been approved for breast cancer treatment [29, 30]. This lack of success is likely because of EGFR expression in normal breast epithelial cells, which results in low response rates and high toxicities of anti-EGFR therapies in therapeutic applications. Therefore, development of novel strategies to target cancer-specific EGFR is imperative.
Intercellular adhesion molecule-1 (ICAM-1), a transmembrane glycoprotein receptor, belongs to the immunoglobulin superfamily and consists of a small intracellular domain and IgG-like extracellular domains [31–33]. ICAM-1 has been extensively studied for its role in the adhesion of macrophages and leukocytes to endothelial cells during their transmigration [34]. ICAM-1 has been implicated in promoting cancer in many cancer types such as leukemia, liver, pancreatic cancers [35–37]. In addition, ICAM-1 contributes to tumor removal through activating T cells through its roles in cell adhesion and co-stimulatory functions [38, 39]. Lymphocyte function-associated antigen-1 (LFA-1), expressed on T cells, acts as a receptor for ICAM-1 and is essential for this function through its interaction with ICAM-1 on antigen-presenting cells [38–40]. However, the tumor intrinsic role of ICAM-1 in cancer progression remains unclear.
Here, our comprehensive analysis unveiled a specific ICAM1 upregulation in TNBC, strongly associated with breast cancer metastasis. Consequently, we embarked on investigating the intricate molecular mechanisms governing the tumor-intrinsic interactions between ICAM-1 and EGFR, as well as their activation pathways in the context of breast cancer metastasis. Furthermore, we evaluated the therapeutic viability of targeting ICAM-1 as a potential treatment strategy for TNBC.
Results
ICAM-1 as a potential candidate for targeted metastatic breast cancer therapy
Despite the high incidence of breast cancer, metastatic breast cancer-specific treatment protocols have not been developed [3, 6, 41]. To investigate potential candidates for metastatic cancer-targeted therapy, we accessed previously published microarray datasets of patients with breast cancer (from the GEO database). We used gene ontology (GO) to identify cell surface molecules. We then selected targets with increased expression in breast cancer tissues relative to normal breast and with higher specific expression in TNBC relative to other breast cancer types (Fig. 1A, Supplementary Fig. 1A, Supplementary Table 1). Significantly, ICAM-1 and LY6D emerged as potential target genes notably upregulated in four different TNBC datasets. Building upon our prior investigation into the functional significance of soluble ICAM-1 (sICAM-1) within the glioblastoma tumor microenvironment, as well as our exploration of protein stability regulation in breast cancer, we directed our attention towards unraveling the clinical significance and underlying mechanisms of ICAM-1 in metastatic breast cancer.
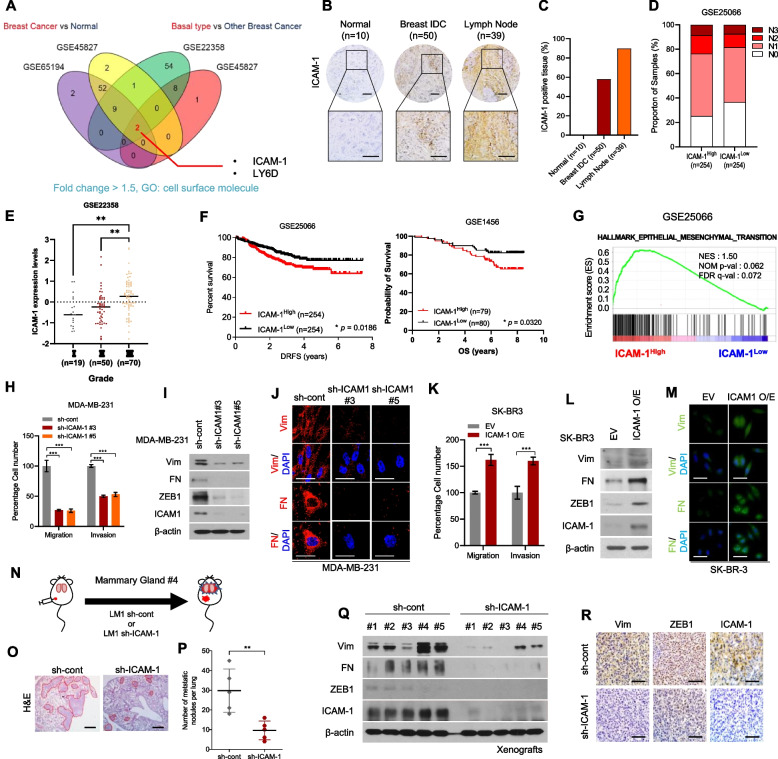
Fig. 1.
Upregulation of ICAM-1 Correlates with Poor Prognosis and Metastasis of Breast Cancer. A Expression of cell surface molecules and comparison of breast cancer tissue versus normal tissue and basal-type breast cancer patients versus other subtype breast cancer patients (GSE65194, GSE45827, GSE22358). Fold change > 1.5 (B) IHC of ICAM-1 in human breast cancer tissue array (BR1008). C IHC scoring of human breast cancer tissue array using IHC profiler. Scale bar = 200 μm. D Lymph node stage analysis using GEO dataset (GSE25066); ICAM-1High vs. ICAM-1Low. E ICAM-1 expression levels in three different grades of breast cancer patients (GSE22358). F Kaplan–Meier survival analysis of breast cancer patients (GSE25066, GSE1456); ICAM-1High vs. ICAM-1Low. G GSEA of hallmark epithelial-mesenchymal transition signature in human breast cancer patients (GSE25066); ICAM-1High vs. ICAM-1Low. NES, normalized enrichment score; Nom p-val, normalized p-value; FDR q-val, false discovery rate q-value. H Transwell migration and invasion assay of MDA-MB-231 cells transduced with ICAM-1 shRNA. I Western blots of EMT markers and regulators in MDA-MB-231 cells transduced with ICAM-1 shRNA. J ICC of vimentin (Vim) and fibronectin (FN) in MDA-MB-231 cells transduced with ICAM-1 shRNA. Scale bar = 20 μm. K Transwell migration and invasion assay of SK-BR3 transfected with ICAM-1 or control empty vector. L Western blots of EMT markers and regulators in SK-BR3 transfected with ICAM-1 or control empty vector. M ICC of vimentin (Vim) and fibronectin (FN) in SK-BR3 transfected with ICAM-1 or control empty vector. Scale bar = 20 μm. N Schematic illustration of animal experiments. O, P H&E staining (O) and the number of metastatic nodules (P) of lung in LM-1sh−cont or LM-1sh−ICAM−1 orthotopic xenograft mice. Scale bar = 200 μm. Q, R Western blots (Q) and IHC (R) of EMT markers, regulators, and ICAM-1 in LM-1sh−cont or LM-1.sh−ICAM−1 orthotopic xenograft tumors. Scale bar = 100 μm. Data are presented as mean ± SD and analyzed using one way ANOVA or Student’s t-tests. n.s, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
Next, we analyzed ICAM-1 expression in two independent datasets from the TCGA breast cancer and GOBO databases and confirmed that ICAM-1 expression was substantially increased in TNBC relative to other breast cancer types and normal breast tissue (Supplementary Fig. 1B-C). Similarly, ICAM-1 was highly expressed in TNBC cell lines (Supplementary Fig. 1D-E). Through GSEA, we discovered a positive correlation between elevated ICAM-1 expression and the downregulation of gene sets within luminal-type breast cancer cells among patients with breast cancer (Supplementary Fig. 1F). However, no significant upregulation of other cell adhesion molecules was observed in TNBC (Supplementary Fig. 1G). To substantiate the clinical significance of ICAM-1 expression in TNBC, we examined breast cancer tissue microarrays (Fig. 1B-C). ICAM-1 was more highly expressed in breast cancer tissues than in normal tissues and was most highly expressed in lymph node metastases (Fig. 1C). In agreement with the tissue microarray results, patients with breast cancer with high ICAM-1 levels (GSE25066) showed an elevated lymph node stage (Fig. 1D). Patients with later stages of breast cancer had higher ICAM-1 expression levels (Fig. 1E). Furthermore, high ICAM-1 expression levels were significantly associated with poor overall survival in breast cancer patients (p = 0.0320; Fig. 1F). These findings suggest that ICAM-1 represents a promising therapeutic target in the context of TNBC and metastatic breast cancer.
ICAM-1-mediated epithelial-to-mesenchymal transition (EMT) promotes metastasis of breast cancer cells
The metastatic capacity of cancer cells has often been attributed to the phenomenon of EMT [42, 43]. We found that the EMT signature gene set was upregulated in patients with breast cancer with high ICAM-1 expression levels (Fig. 1G). Consistently, shRNA-mediated knockdown of ICAM-1 decreased the migration and invasion of MDA-MB-231 and BT20 cell lines (Fig. 1H, Supplementary Fig. 2A-B). Additionally, we observed that ICAM-1 knockdown led to a decrease in EMT markers and regulators in TNBC MDA-MB-231 cells (Fig. 1I-J).
In contrast, ICAM-1 overexpression enhanced the migration and invasion capacity and expression of EMT markers and regulators in HER2 + breast cancer subtype SK-BR3 cells (Fig. 1K-M). Next, we evaluated the role of ICAM-1 in vivo by orthotopically injecting LM1 cells, which is a lung metastatic MDA-MB-231 breast cancer cell line derived from lung lesions after colonization, with control shRNA or ICAM-1 shRNA (sh-ICAM-1) into mammary glands of NSG mice and analyzing the primary tumors and metastatic lung foci (Fig. 1N, Supplementary Fig. 2C-D). Tumor metastasis to lung tissue and EMT marker expression in primary tumors decreased in LM1 sh-ICAM-1 tumor-bearing mice (Fig. 1O-R); however, no effect on tumor growth was observed (Supplementary Fig. 2E-H). Overall, our findings strongly indicate that the ICAM-1-mediated EMT facilitates the metastasis of breast cancer cells.
ICAM-1 directly interacts with EGFR and promotes its activation in breast cancer
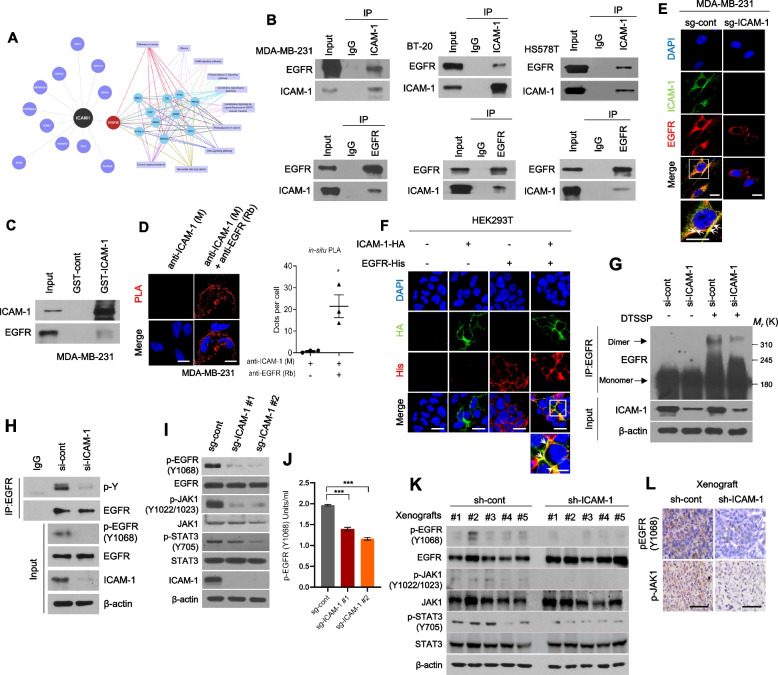
To understand how ICAM-1 contributes to breast cancer metastasis, we isolated and studied proteins that interact with ICAM-1. We analyzed mass spectrometry (MS) data from ICAM-1 IPs in MDA-MB-231 cells using bioinformatics tools which led us to identify EGFR as a likely interactor of ICAM-1 (Fig. 2A, Supplementary Fig. 3A-B). The interactions between ICAM-1 and EGFR were confirmed in TNBC cells using co-IP, GST pull-down assay, and in situ PLA (Fig. 2B-D). Immunocytochemistry (ICC) for ICAM-1 and EGFR revealed that these proteins were colocalized in the cell membrane (Fig. 2E-F). Given the critical correlation between EGFR activation and metastasis in various cancers, [24, 44, 45], we hypothesized that ICAM-1 promotes cancer progression by modulating EGFR activity. In TNBC cells, the use of siRNA or CRISPR to either knock down or knock out ICAM-1 resulted in the suppression of EGFR dimerization and phosphorylation at Y1068, as well as decreased phosphorylation of JAK1 and STAT3, which are downstream targets of EGFR activation, without changing total protein levels of EGFR (Fig. 2G-J, Supplementary Fig. 3C-D). In contrast, ICAM-1 overexpression enhanced EGFR activity, as indicated by increased phosphorylation of EGFR, JAK1, and STAT3 and EGFR dimerization (Supplementary Fig. 3E-H). Comparable outcomes were observed in mice bearing LM1 sh-ICAM-1 tumors. (Fig. 2K-L). Taken together, our data suggest that the direct interaction between ICAM-1 and EGFR increases EGFR activity and downstream signaling pathways, which likely promotes metastasis of breast cancer cells in vitro and in vivo.
Fig. 2.
ICAM-1 directly interacts with EGFR and promotes its activation in breast cancer. A Analysis of identified ICAM-1 interactors using Cytoscape. ICAM-1 interactors were detected using MS and cell surface molecules were selected. B Co-IP with ICAM-1 or EGFR antibody and western blot to evaluate the interaction between ICAM-1 and EGFR in basal-type breast cancer cell lines. C GST pull-down assay in MDA-MB-231 cells. D Representative images and quantification of in situ PLA showing the interaction between ICAM-1 and EGFR in MDA-MB-231 cells. Scale bar = 20 μm. E ICC of ICAM-1 and EGFR in MDA-MB-231 cells. Scale bar = 20 μm. F ICC of HA tag and His-tag in HEK293T cells transfected with either ICAM-1-HA or EGFR-His, or co-transfected with both. Scale bar = 20 μm. G Dimerization of EGFR in MDA-MB-231 cells transfected with siRNA targeting ICAM-1 or control siRNA. H Co-IP with EGFR antibody and western blotting to evaluate the total phospho tyrosine in MDA-MB-231 cells transfected with siRNA targeting ICAM-1 or control siRNA. I, J Western blots (I) and phospho EGFR ELISA (J) of MDA-MB-231 cells transfected with sgRNA targeting ICAM-1 or control as indicated. K, L Western blots (K) and IHC (L) of phospho EGFR, phospho JAK1, and phospho STAT3 in LM-1sh−cont or LM-1.sh−ICAM−1 orthotopic xenograft tumors. Scale bar = 100 μm. Data are presented as mean ± SD and analyzed using one-way ANOVA. n.s, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
ICAM-1 expression correlates with EGFR expression and poor prognosis of breast cancer
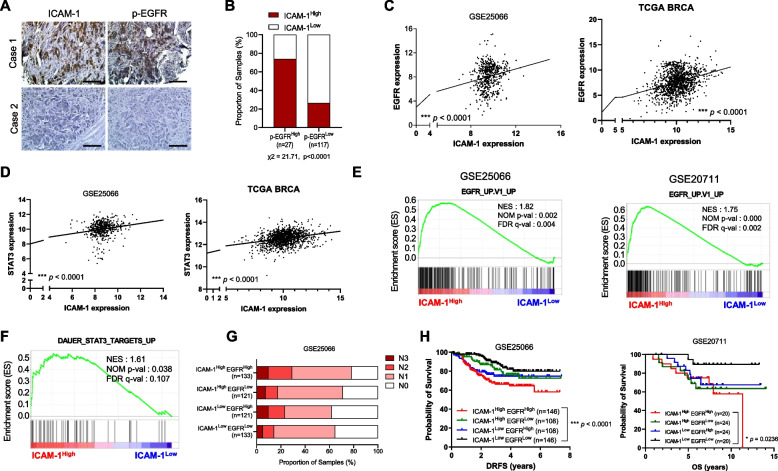
To explore the clinical significance of targeting ICAM-1 as a therapeutic approach in combating metastatic breast cancer, we investigated the correlation between ICAM-1 and EGFR activation in patients with breast cancer. We conducted a tissue array analysis of patients with breast cancer, presenting representative IHC results (Fig. 3A). Results indicated a positive correlation between ICAM-1 expression and phospho EGFR (Fig. 3B, χ2 = 21.71, p < 0.0001). Considering the induction of ICAM-1 expression by the EGFR/STAT3 axis, we conducted an analysis of ICAM-1, EGFR, and STAT3 expression using patient datasets retrieved from the GEO database. Surprisingly, ICAM-1 expression was positively correlated with EGFR and STAT3 expressions (Fig. 3C-D). The GSEA also revealed that patients with breast cancer with high ICAM-1 levels had higher expression of genes related to EGFR activity and STAT3 targets (Fig. 3E-F). Furthermore, we examined the lymph node stages and survival rates in response to ICAM-1 and EGFR status and found that patients with breast cancer with high ICAM-1 and EGFR expression represented those with the highest proportion of advanced lymph node stage and shortest survival rates (Fig. 3G-H). In contrast, patients with low ICAM-1 and EGFR levels had the longest survival. Moreover, in the subset of patients with triple-negative breast cancer (TNBC), the combination of high ICAM-1 and high EGFR expression levels approached statistical significance in predicting poor outcomes (p = 0.0538; Supplementary Fig. 3I). However, the specific phenotype of ICAM-1 and EGFR may be specific to breast cancer, as high ICAM1 and/or EGFR expression is not correlated with poor clinical outcomes in other cancer types such as lung cancer and melanoma (Supplementary Fig. 3 J-K). Collectively, our findings indicate a positive correlation between ICAM-1 and EGFR expression levels, which are closely associated with poor prognosis in patients with breast cancer.
Fig. 3.
High levels of ICAM-1 expression correlate with EGFR expression and poor prognosis in patients with breast cancer. A IHC of ICAM-1 and p-EGFR in human breast cancer tissue array (BR2082b). Scale bar = 200 μm. B Contingency table for statistical analysis of IHC showing the relationship between ICAM-1 and p-EGFR in human breast cancer tissue array using IHC profiler. C Pearson correlation analysis of ICAM-1 and EGFR in patient cohorts from GEO dataset (GSE25066) and TCGA. D Pearson correlation analysis of ICAM-1 and STAT3 in patient cohorts from GEO dataset (GSE25066) and TCGA. E GSEA plots for a significant correlation between high expression of ICAM-1 and EGFR signaling (GSE25066, GSE20711). F GSEA plots for a significant correlation between high expression of ICAM-1 and STAT3 signaling (GSE25066). G Lymph node stage analysis using GEO dataset (GSE25066); ICAM-1High EGFRHigh vs. ICAM-1High EGFRLow vs. ICAM-1Low EGFRHigh vs. ICAM-1Low EGFRLow. H Kaplan–Meier survival analysis of patients with breast cancer (GSE25066, GSE20711); ICAM-1High EGFRHigh vs. ICAM-1High EGFRLow vs. ICAM-1Low EGFRHigh vs. ICAM-1Low EGFR.Low. Data are presented as mean ± SD and analyzed using one-way ANOVA. n.s, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
ICAM-1 increases ligand-binding affinity for EGFR and promotes metastasis of breast cancer
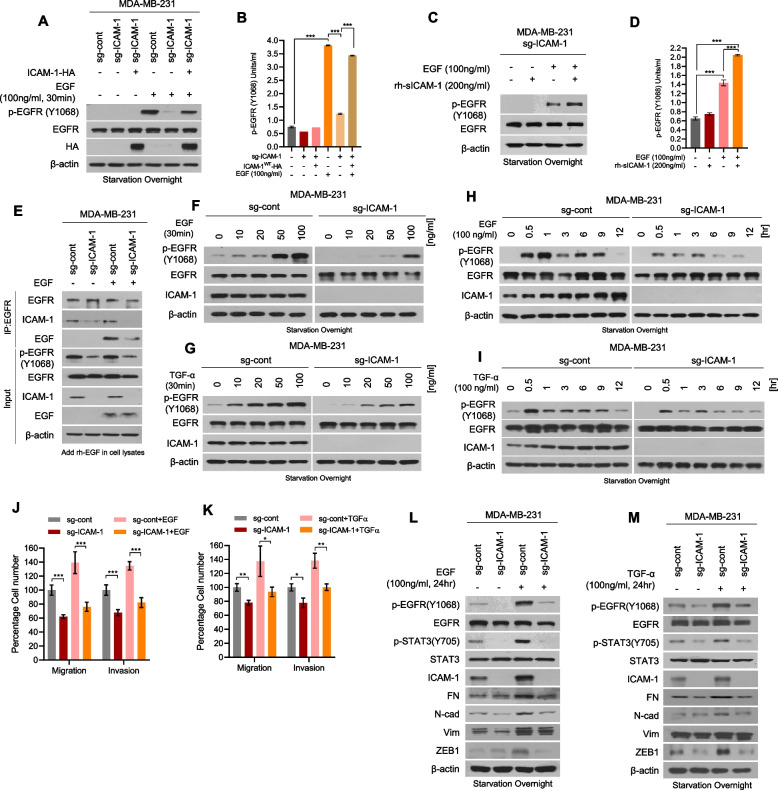
Our demonstration of the connection between ICAM-1 expression and EGFR activation in both in vitro and in vivo settings, along with its impact on clinical outcomes in patients with breast cancer, highlights its clinical significance. Therefore, we explored the underlying molecular mechanisms by which ICAM-1's direct interaction with EGFR influences specific cellular processes, including EMT, ultimately facilitating breast cancer metastasis. Beyond having the classical function of stabilizing cell–cell interaction, ICAM-1 can also be shed from the cell surface via proteolytic cleavage [46, 47]. Notably, sICAM-1 acts as an endocrine and paracrine factor [48, 49]. To examine whether the soluble and membrane-bound forms of ICAM-1 regulate EGFR activity differentially, we transfected ICAM-1-knockout MDA-MB-231 cells with ICAM-1WT, sICAM-1, or mutant ICAM-1P404E, in which 404 proline (an MMP9-mediated cleavage site) was substituted with glutamic acid (P404E) (Supplementary Fig. 4A). Significantly, both membrane-bound and soluble forms of ICAM-1 amplified EGFR phosphorylation, migration, and invasion in breast cancer cells (Supplementary Fig. 4B-D). In addition, treatment with recombinant human (rh)-sICAM-1 augmented EGFR phosphorylation (Supplementary Fig. 4E-F). Given the lack of documented kinase activity associated with ICAM-1, we postulated that ICAM-1 acts as an EGFR ligand. After starvation of breast cancer cells, ICAM-1 had no effect on EGFR activation (Fig. 4A-B, Supplementary Fig. 5A). Nevertheless, both ICAM-1 overexpression and EGF treatment elicited EGFR phosphorylation (Fig. 4A-B). Similar results were observed upon treatment of cells with rh-sICAM-1 and EGF (Fig. 4C-D, Supplementary Fig. 5B-C). Hence, we examined the ligand-binding affinity of EGFR towards ICAM-1. By employing CRISPR to knock out ICAM-1 in MDA-MB-231 cells, reduced binding affinity between EGFR and EGF was observed (Fig. 4E). Conversely, ICAM-1 overexpression augmented EGF and EGFR binding (Supplementary Fig. 5D). Therefore, we investigated dose- and time-dependent EGFR activation following treatment with EGF or TGF-α. ICAM-1 knockout resulted in a dose- and time-dependent suppression of EGFR phosphorylation following treatment with EGF or TGF-α (Fig. 4F-I). Furthermore, we found that knockout of ICAM-1 decreased EGF and TGF-a -induced cell migration and invasion and EMT-associated marker expression, which suggests that ICAM-1 inhibition may play a role in attenuating metastatic potential of breast cancer cells through decreasing EGFR activation (Fig. 4J-M, Supplementary Fig. 5E-H).
Fig. 4.
ICAM-1 increases ligand-binding affinity for EGFR and promotes metastasis of breast cancer. A, B Western blots (A), and phospho EGFR ELISA (B) of MDA-MB-231 sg-cont and MDA-MB-231 sg-ICAM-1 cells transfected with either ICAM-1 or control empty vector. Cells were starved overnight and treated with EGF or PBS. C, D Western blots (C), and phospho EGFR ELISA (D) of MDA-MB-231 sg-ICAM-1 cells. Cells were starved overnight and treated with either rh-sICAM-1 or EGF, or co-treated. E Co-IP with EGFR antibody and western blotting to evaluate the interaction between EGF and EGFR in MDA-MB-231 sg-cont and MDA-MB-231 sg-ICAM-1 cells. rh-EGF was added in cell lysates. F, G Western blots of EGFR phosphorylation in MDA-MB-231 sg-cont and MDA-MB-231 sg-ICAM-1 cells. Cells were starved overnight and treated with increasing doses of EGF (F) or TGF-α (G). H, I Western blots of EGFR phosphorylation in MDA-MB-231 sg-cont and MDA-MB-231 sg-ICAM-1 cells. Cells were starved overnight and treated with EGF (H) or TGF-α (I) for various time intervalsin a time-dependent manner. J, K Migration and invasion by MDA-MB-231 sg-cont and MDA-MB-231 sg-ICAM-1 cells. Cells were starved overnight and treated with EGF (J) or TGF-α (K) for 24 h. L, M Western blots of EMT markers and regulators in MDA-MB-231 sg-cont and MDA-MB-231 sg-ICAM-1 cells. Cells were starved overnight and treated with EGF (L) or TGF-α (M) for 24 h. Data are presented as mean ± SD and analyzed using one-way ANOVA or Student’s t-tests. n.s, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
EGFR reportedly promotes cancer progression, including cell growth and metastasis [24, 27, 44, 45]. In contrast to that of EGFR, suppression of ICAM-1 did not affect cell growth (Supplementary Fig. 6A-B and Supplementary Fig. 2A). Therefore, we confirmed that the phosphorylation of EGFR downstream signaling kinases were decreased following ICAM-1 knockout and found that the interaction of EGFR with EMT-related kinases, including JAK1, STAT3, and Src, was significantly suppressed in ICAM-1 knockout cells (Supplementary Fig. 6C-D). Similar outcomes were observed in the mouse xenograft model (Supplementary Fig. 6E). Previous research has detailed biased EGFR signaling, showing that its sustained activation promotes cell differentiation rather than proliferation [50–53]. Our results demonstrated the prolonged activation of EGFR by ICAM-1 (Fig. 4H-I), suggesting that ICAM-1 regulates the biased EGFR signaling in breast cancer, enhancing cell migration and invasion without promoting the growth of cancer cells.
ICAM-1 is induced via regulation of EGFR/STAT3 positive feedback loop
Our current data suggests that ICAM-1 plays a pivotal role in EGFR activation, with evidence showing ICAM-1 protein levels increase over time with EGFR ligand treatment (Fig. 4H-I). Next, we examined ICAM-1 expression levels after treatment with AG1478 (an EGFR inhibitor) or transfection with EGFR siRNA (si-EGFR) and found that ICAM-1 expression decreased in response to EGFR suppression (Supplementary Fig. 7A-D). In contrast, EGFR overexpression augmented ICAM-1 expression (Supplementary Fig. 7E-F). Previous studies have demonstrated STAT3's role in promoting ICAM-1 expression [54–56], and also STAT3 is recognized as a key downstream regulator of EGFR [57]. Therefore, we investigated whether STAT3 could regulate ICAM-1 expression in breast cancer cells. Blocking STAT3 using a STAT3 inhibitor or si-STAT3 transfection attenuated ICAM-1 expression (Supplementary Fig. 7G-J). This blockade also reduced EGF-induced migration, invasion, and EMT of breast cancer cells (Supplementary Fig. 7 K-L). Consistent with what we have shown previously, ICAM1 knockout reduced STAT3 phosphorylation and nuclear translocation, demonstrating a reduction in STAT3 activity (Supplementary Fig. 7 M-N). To further identify whether ICAM-1 expression is directly regulated by STAT3, we predicted STAT3-binding sites within the ICAM-1 promoter using the JASPAR online tool (http://jaspar.genereg.net) and investigated each site using a chromatin immunoprecipitation (ChIP) assay (Supplementary Fig. 7O). STAT3 chromatin immunoprecipitation indicated that STAT3 directly binds to the F2, 3, and 4 fragments of the ICAM-1 promoter, and its binding was suppressed by transfection with si-ICAM-1 (Supplementary Fig. 7P). Overall, these findings suggest that ICAM-1 expression is induced through positive feedback regulation involving ICAM-1, EGFR, and STAT3.
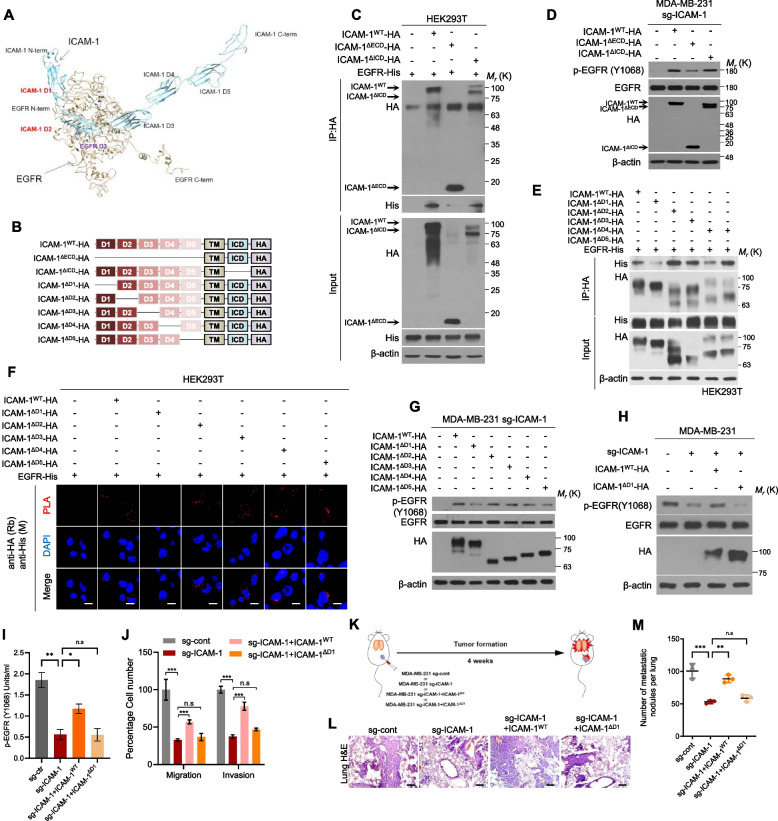
The ICAM-1 D1 domain is necessary for EGFR activation and its downstream signaling
Next, we investigated the interactions between ICAM-1 and EGFR using protein–protein docking simulations. Interestingly, the anticipated protein structure suggests potential interaction between the D1 and D2 domains of ICAM-1 and the D3 domain of EGFR (Fig. 5A). Accordingly, we generated a series of deletion constructs for ICAM-1, in which each domain was removed, and a HA tag was fused at the C-terminus (Fig. 5B). The expression constructs encoding ICAM-1ΔECD or ICAM-1ΔICD, in which the extracellular domain (ECD) or intracellular domain (ICD) were deleted, were co-transfected with the EGFR vector in HEK293T cells. Co-IP results revealed that the interactions between EGFR and ICAM1 were retarded only when ECD was deleted (Fig. 5C). EGFR phosphorylation was also not enhanced by overexpression of the ICAM-1ΔECD construct (Fig. 5D). Five IgG-like ECD deletion mutants of ICAM-1 were co-transfected with the EGFR vector in HEK293T cells, and co-IP and in situ PLA were performed. Almost all ICAM-1 deletion mutants, except ΔD1, were able to bind to EGFR (Fig. 5E-F). Moreover, EGFR phosphorylation, migration, and invasion were not rescued by the expression of the ICAM-1ΔD1 construct in ICAM-1 knockout cells (Fig. 5G-J). Subsequently, we generated a set of constructs encoding EGFR deletion mutants and investigated their interactions with ICAM-1 (Supplementary Fig. 8A). The D3 domain deletion construct of EGFR demonstrated reduced binding affinity for ICAM-1 (Supplementary Fig. 8B-D). Here, our investigation into the interaction between ICAM-1 and EGFR through protein–protein docking simulations and deletion construct experiments reveals a potential binding mechanism involving the D1 domain of ICAM-1 with the D3 domain of EGFR.
Fig. 5.
The ICAM-1 D1 domain is necessary for EGFR activation and its downstream signaling. A Protein–protein docking simulation to evaluate the interaction domain between ICAM-1 and EGFR. B Schematic diagram of expression constructs encoding ICAM-1WT, ICAM-1ΔECD, ICAM-1ΔICD, ICAM-1ΔD1, ICAM-1ΔD2, ICAM-1ΔD3, ICAM-1ΔD4, and ICAM-1ΔD5. C Co-IP with HA antibody and western blot to evaluate the interaction domain between ICAM-1 and EGFR in HEK293T cells co-transfected with EGFR and ICAM-1WT, ICAM-1ΔECD, or ICAM-1ΔICD vector. D Western blots of EGFR phosphorylation in MDA-MB-231 sg-ICAM-1 cells transfected with ICAM-1WT, or ICAM-1ΔECD, or ICAM-1ΔICD, or control empty vector. E, F Co-IP (E) and in situ PLA (F) to evaluate the interaction domain between ICAM-1 and EGFR in HEK293T cells co-transfected with EGFR and either ICAM-1WT, ICAM-1ΔD1, ICAM-1ΔD2, ICAM-1ΔD3, ICAM-1ΔD4, or ICAM-1ΔD5. Scale bar = 20 μm. G Western blots of EGFR phosphorylation in MDA-MB-231 sg-ICAM-1 cells transfected with either control empty vector or ICAM-1WT, ICAM-1ΔD1, ICAM-1ΔD2, ICAM-1ΔD3, ICAM-1ΔD4, or ICAM-1ΔD5. H-J Western blotting (H), phospho EGFR ELISA (i), migration and invasion assay (J) in MDA-MB-231 sg-ICAM-1 cells transfected with either control empty vector, ICAM-1WT, or ICAM-1ΔD1. K Schematic illustration of animal experiments. L, M H&E staining (L) and the number of metastatic nodules (M) of lung in MDA-MB-231 sg-cont or MDA-MB-231 sg-ICAM-1 transfected with control empty vector, ICAM-1WT, or ICAM-1.ΔD1 orthotopic xenograft mice. Scale bar = 200 μm. Data are presented as mean ± SD and analyzed using one-way ANOVA. n.s, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
To validate our findings in vivo, we assessed breast cancer cell metastasis using an orthotopic xenograft mouse model (Fig. 5K). Tumor metastasis to lung tissue and EMT of primary tumors decreased in the mouse group injected with ICAM-1 knock out MDA-MB-231 compared with that in the control. Similar to the in vitro results, ICAM-1WT overexpression in ICAM-1 knockout cells rescued breast cancer cell lung metastasis but ICAM-1ΔD1 expression had no effect (Fig. 5L-M). Similar to our previous xenograft experiment, primary tumor growth was unaffected by the deletion or expression of the ICAM-1 constructs (Supplementary Fig. 8E-F). In brief, these data indicated that the D1 domain of ICAM-1 is required for EGFR-mediated metastasis of breast cancer cells.
Blockade of ICAM-1 using a neutralizing antibody suppresses breast cancer metastasis
To assess the therapeutic potential of blocking ICAM-1 in breast cancer metastasis, we treated MDA-MB-231 cells with a neutralizing antibody against ICAM-1 and this treatment notably suppressed EGFR activity, indicated by decreased EGFR and STAT3 phosphorylation, in a dose-dependent manner (Fig. 6A-B). Blockade of ICAM-1 by a neutralizing antibody effectively suppressed cancer cell migration, invasion, and EMT marker levels (Fig. 6C-D). The interactions among ICAM-1, EGFR, and EGF also decreased following antibody treatment (Fig. 6E-F).
Fig. 6.
Blockade of ICAM-1 using a neutralizing antibody can suppress breast cancer metastasis. A, B Western blotting (A) and phospho EGFR ELISA (B) of MDA-MB-231 cells treated with various doses of ICAM-1 neutralizing antibody for 24 h. C, D Transwell migration, invasion assay (C) and (D) western blotting of MDA-MB-231 cells treated with ICAM-1 neutralizing antibody (500 ng/ml, 24 h). E Co-IP with EGFR antibody and western blotting to evaluate the interaction between ICAM-1 and EGFR in MDA-MB-231 cells treated with ICAM-1 neutralizing antibody. F Co-IP with EGFR antibody and western blotting to evaluate the interaction between EGFR and EGF in MDA-MB-231 cells treated with ICAM-1 neutralizing antibody. rh-EGF was added in cell lysates. G Schematic illustration of animal experiments. H, I H&E staining (H) and the number of metastatic nodules (I) of lung in MDA-MB-231 orthotopic xenograft mice treated with ICAM-1 neutralizing antibody. Scale bar = 200 μm. J-M Phospho EGFR ELISA (J), IHC (K), western blotting (L), and in situ PLA (M) of in MDA-MB-231 orthotopic xenograft tumors mice treated with ICAM-1 neutralizing antibody. Scale bar = 100 μm. N Schematic illustration of the mechanism underlying ICAM-1-mediated EGFR activation, leading to cancer metastasis in breast cancer. Highlighted arrows indicate the pathways suggested by our study. Data are presented as mean ± SD and analyzed using one-way ANOVA or Student’s t-tests. n.s, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
To explore the therapeutic potential of targeting ICAM-1, we injected LM-1 cells orthotopically into the mammary fat pads of NSG mice and then treated these animals with an ICAM-1 neutralizing antibody introduced intratumorally two weeks after the initial treatment (Fig. 6G). Importantly, lung metastasis of breast cancer cells decreased in the mice treated with ICAM-1 neutralizing antibody (Fig. 6H-I). Aligned with the in vitro findings, inhibiting ICAM-1 demonstrated a suppression of EGFR phosphorylation, EMT marker expression, and the interaction between ICAM-1 and EGFR within primary tumor tissues, suggesting that ICAM-1-targeted therapy significantly inhibited EGFR activity and breast cancer cell metastasis in a mouse xenograft model (Fig. 6J-M). Overall, our findings here reveal that ICAM-1 is overexpressed in metastatic breast cancer, acting as a crucial mediator of EGFR activation and driving malignant progression through the JAK1/STAT3 signaling pathway, ultimately promoting metastasis without affecting tumor growth. Our data positions ICAM-1 as a promising target for developing TNBC-specific anti-EGFR therapies, offering valuable insights into therapeutic strategies for combating metastatic breast cancer (Fig. 6N).
Discussion
Numerous targeted drugs are currently in clinical trials, and some have already received FDA approval for use as monotherapies or in combination with other treatments for different subtypes of breast cancer. Also, the FDA has approved some immune checkpoint blockers in combination with chemotherapeutic drugs for the treatment of TNBC, and several trials are ongoing [16, 18]. However, we identified the cell surface protein ICAM-1 as a potential candidate for targeted metastatic breast cancer therapy. Our research uncovered ICAM-1's critical role in promoting the metastatic potential of TNBC cells through prolonged activation of EGFR. Specifically, we found that metastatic breast cancer cells exhibit elevated ICAM-1 expression. ICAM-1 directly interacts with EGFR, activating the EGFR-mediated STAT3 pathway and inducing epithelial-mesenchymal transition (EMT). Importantly, blocking ICAM-1 significantly reduced metastasis by inhibiting the EGFR/JAK1/STAT3 signaling pathway. These findings highlight ICAM-1's potential as both a prognostic marker and a therapeutic target in TNBC.
ICAM-1 has been reported as a key molecule in the adhesion of immune cells to endothelial cells [34]. Recently, studies have indicated a correlation between elevated ICAM-1 expression and inflammatory disorders, including multiple sclerosis [48, 58, 59]. While ICAM-1 upregulation has been noted in several cancer types including pancreatic cancer, hepatocellular carcinoma, and breast cancer, the precise mechanism underlying its influence on cancer progression remains unclear, making it an attractive target for further investigation [36, 60, 61]. ICAM-1 is also well-known for its anti-tumor effects, as it plays a role in T cell priming through the trans-endothelial trafficking of effector T cells and facilitates immune cell adhesion to tumors [62]. Therefore, increased ICAM-1 levels positively correlate with immune cell infiltration, suggesting the potential of ICAM-1 as a novel immunotherapy. sICAM-1 selectively co-stimulates tumor-specific T cells and promotes their interaction, leading to an increase in effector T cells and their differentiation into memory T cells [63, 64]. Due to side effects to patients based on drug safety, many clinical trials have also been conducted on T cell activation by other costimulatory molecules as a combination strategy with anti-PD-1, as in the ICAM-1/LFA-1 axis [65]. Recently, patients with NSCLC indicates that high ICAM-1 expression, rather than other checkpoints, is associated with higher survival probability and serves as a predictive marker for response to anti-PD-1 therapy [66].
Here, we examined the expression patterns of cell surface molecules on cancer cells that are primed for therapeutic intervention, identifying ICAM-1 as a promising target for combating metastatic TNBC. Elevated ICAM-1 expression aligns with dismal patient survival and increased metastatic potential in breast cancer cases. We prioritized the significant discovery that ICAM-1 directly interacts with EGFR in cancer cells, thereby modulating EGFR’s kinase activity. Given that ICAM-1 can undergo shedding from the cell surface, generating a soluble form, it emerges as a recognized paracrine factor [67]. Here, we examined the function of ICAM-1 as an EGFR ligand. Although ICAM-1 alone is not sufficient to induce EGFR activation, it enhances the ligand-binding affinity of EGFR. It is crucial to understand how ICAM-1-mediated EGFR activation contributes to breast cancer metastasis while having no impact on cell growth. Growth factor receptors and G protein-coupled receptors (GPCRs) exhibit functional selectivity for particular downstream signaling pathways in response to various types of ligands or specific microenvironments [50, 51, 68]. Further, transient EGFR activation induces proliferation, and sustained EGFR activation causes breast cancer cell differentiation [52]. Our findings reveal that the enhanced ligand-binding affinity of EGFR, facilitated by the interaction between ICAM-1 and EGFR, results in amplified and prolonged EGFR activation. This corresponds with previous studies indicating that persistent EGFR activation induces EMT in breast cancer cells by influencing EMT-associated kinases, including those in the JAK1, STAT3, and Src signaling pathways [24, 27, 44]. Moreover, STAT3 stimulates EMT signature gene expression and establishes a positive feedback loop with ICAM-1 through the EGFR/STAT3 axis. This feedback loop indicates that blocking ICAM-1 not only suppresses EGFR activity but also reduces ICAM-1 expression, highlighting ICAM-1 as a clinically significant therapeutic target.
Additionally, we delineated the interaction domains of ICAM-1 and EGFR through protein–protein docking simulations, co-immunoprecipitation (co-IP), and in situ PLA. Our investigation revealed that the IgG-like D1 domain of ICAM-1 interacts with domain III of EGFR, a crucial site for ligand binding. This finding strongly suggests that the EGFR-ICAM-1 interaction triggers a conformational shift, enhancing the affinity for ligand binding. Detailed biochemical analyses at the molecular level are crucial for the effective production of mAbs, particularly in determining the optimal therapeutic target within the D1 region of ICAM1. Given that our study provides key evidence for the relationship between EGFR and ICAM-1, it is worth considering whether there might also be an important role for the interaction between EGFRvIII, a truncated form of EGFR that acts as a tumor-specific antigen, and ICAM-1. Our findings revealed that the EGFR construct lacking the D3 domain exhibited reduced binding affinity for ICAM-1. Notably, the EGFRvIII variant retains the complete D3 domain, suggesting that it could theoretically maintain its ability to bind ICAM-1. This hypothesis opens up the possibility of EGFRvIII serving as a more tumor-specific target, which could be valuable for developing ICAM-1 targeted therapeutic interventions.
Elevated EGFR expression is a common feature in TNBC cohorts, correlating with poorer survival rates [27, 28]. Nevertheless, clinical trials targeting EGFR have encountered limited success due to low response rates and a multitude of adverse effects [30]. The ineffectiveness of anti-EGFR therapies in TNBC clinical trials can be attributed to several underlying factors. One primary reason is that TNBC tumors are not solely driven by EGFR, due to considerable heterogeneity within this subtype of breast cancer. Moreover, the intricate web of signaling networks and feedback loops often results in resistance to anti-EGFR treatments. Preclinical research has demonstrated that inhibiting EGFR can trigger the activation of alternative signaling pathways, such as the JAK/STAT pathway, thereby promoting tumor growth and metastasis.
We provided strong evidence to confirm that the blockade of ICAM-1 with neutralizing antibodies exerted a robust inhibitory effect on the lung metastasis of breast cancer cells in an orthotopic xenograft model. Our current study focused on lung metastasis, future research could explore additional cancer models to broaden our understanding of ICAM-1's role in organotropic metastasis, which is known to be regulated by breast cancer subtypes, the host organ microenvironment, and cancer cell-organ interactions [69]. These additional studies could include investigating ICAM-1's potential involvement in metastasis to other common sites for breast cancer spread, such as the liver and brain. Such studies would provide a more comprehensive view of ICAM-1's function across different metastatic sites and breast cancer subtypes.
Our data underscores particular considerations for developing anti-EGFR therapy for TNBC. ICAM-1-mediated EGFR activation promotes tumor metastasis through biased signaling via the JAK/STAT pathway, which may not necessarily correlate with resistance to anti-EGFR therapies. ICAM-1 is known to play a crucial role in forming the immunological synapse, and its inhibition can impair T cell cytotoxicity. While our study did not specifically investigate ICAM-1's potential role in immune regulation or its broader effects on the anti-cancer immune response, recent research has shed light on this aspect [70]. Disruption of IFN-gamma signaling mediates partial resistance of solid tumors to T-cell bispecific antibody- and CAR T-cell-mediated killing [70–72]. The resistance is in part caused by disruption of IFN-gamma leading to downregulation of ICAM-1, which reduces adhesion of CAR T cells to tumor cells [70]. Future studies should aim to unravel the intricate interplay between ICAM-1, tumor progression, and immune response in the context of cancer immunotherapy, as these findings suggest a more complex role for ICAM-1 than previously understood. Additionally, while EGFR-targeted CAR T cell therapy seems promising, it might unintentionally promote more aggressive cancer behavior in breast cancer due to the complex signaling interactions between IFN-gamma, ICAM-1, and EGFR. This complexity suggests that caution is needed when considering EGFR as a target for immunotherapy in breast cancer.
Our study has several limitations that should be addressed in future research. First, our human clinical data focused only on breast cancer patients, particularly TNBC. To gain a comprehensive understanding of the tumor intrinsic mechanism of the ICAM-1 and EGFR interaction role, it is crucial to extend this study to other types of human cancers. Second, the efficacy of ICAM-1 treatment was only confirmed in mouse models. Consequently, the potential side effects of ICAM-1 targeting in the human body have not been verified. A clinical trial involving cancer patients is necessary to assess both efficacy and safety in humans. Finally, while we demonstrated the tumor-intrinsic role of the ICAM-1 and EGFR interaction in driving metastatic progression, we did not explore the broader immune system implications. Targeting ICAM-1 through a blocking antibody in an immunocompetent setting could potentially impair T cell-mediated killing, leading to increased tumor growth and metastasis. Further studies on the immune regulatory aspects of ICAM-1 are needed to fully evaluate its potential as a target for cancer therapy.
Recent advancements in targeted therapy have introduced a more precise and effective treatment option for managing breast cancer. Treatment options for TNBC remain limited, and long-term remissions are rare. Therefore, there is a significant need for novel and improved therapeutic strategies for this patient group. The primary goal of targeted therapy is to inhibit specific molecules that promote tumor growth and survival. Building on this, we propose that targeting the tumor intrinsic role of the ICAM1-EGFR interaction offers an effective strategy and mechanism to combat malignancies. Our study provided strong evidence that targeting ICAM-1 may be a promising therapeutic approach for TNBC, as inhibiting ICAM-1 effectively suppressed tumor metastasis without impacting tumor growth. These findings suggest that ICAM-1 could play a role in the resistance mechanisms observed in some TNBC cases, potentially including resistance to anti-EGFR therapies. Consequently, the development of TNBC treatments that modulate ICAM-1 activity, possibly in combination with existing therapies, presents a potential strategy to improve outcomes in resistant cases. However, further investigations are necessary to fully elucidate the involvement of ICAM-1 in therapy resistance and to assess the efficacy of ICAM-1-targeted treatments for TNBC. Such research could contribute to a clearer understanding of breast cancer subtypes and help refine therapeutic approaches for patients with TNBC.
Conclusion
Our study elucidated the crucial role of ICAM-1 as a regulator of breast cancer metastasis, revealing its molecular mechanism. We observed elevated ICAM-1 expression in metastatic breast cancer cells, where it directly interacted with EGFR. This interaction triggered EGFR-mediated STAT3 activation, leading to ICAM-1 upregulation and induction of EMT in breast cancer cells. These findings shed light on the functional significance of ICAM-1 in targeting metastatic breast cancer. Furthermore, blocking ICAM-1 significantly attenuated breast cancer metastasis by inhibiting the EGFR/JAK1/STAT3 signaling pathway. Overall, our results uncover ICAM-1 as a critical binding adaptor of EGFR and indicate its potential utility as both a prognostic marker and therapeutic target in TNBC.
Materials and methods
Cell culture
HEK-293 T cells and the human breast cancer cell lines MDA-MB-231, BT-20, SK-BR-3, MCF-7, and MCF-10A were purchased from American Type Culture Collection (Manassas, VA, USA). Cell cultures were maintained in a 5% CO2 incubator at 37 °C. MDA-MB-231 and HEK-293 T cells were maintained in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 g/ml). BT-20 and SK-BR-3 cells were cultured in Roswell Park Memorial Institute Medium (RPMI) provided with 10% FBS, penicillin (100 units/mL), and streptomycin (100 g/mL). MCF-7 cells were grown in minimum essential medium (MEM) supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 g/mL). DMEM, RPMI, MEM, FBS, penicillin, streptomycin and trypsin were purchased from GIBCO (Seoul, Korea).
Chemical reagents and antibodies
Antibodies against ICAM1, vimentin (VIM), SLUG, TWIST, STAT3, JAK1, phospho-JAK1, EGFR (anti-mouse and anti-rabbit), phospho EGFR (Y1068), MYC (anti-mouse and anti-rabbit), and HA were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Polyclonal antibodies against SNAIL, phospho-STAT3 (Y705) and FLAG (M2) were purchased from Cell Signaling Technology (Danvers, MA, USA). Polyclonal antibodies against N-cadherin were purchased from BD Biosciences (Franklin Lakes, NJ, USA). Polyclonal antibody to ZEB1, monoclonal antibody to β-actin, and 4,6-diamidino- 2-phenylindole (DAPI) were purchased from Sigma (St. Louis, MO, USA). Anti-rabbit IgG Alexa 488 and anti-mouse IgG Alexa Fluor 488 antibodies were purchased from Invitrogen (Carlsbad, CA, USA). Control anti-mouse and anti-rabbit IgG antibodies were purchased from Santa Cruz Biotechnology.
Invasion and migration assays
For invasion and migration assays, MDA-MB-231, BT-20, SK-BR-3, and MCF-7 cells were seeded on the upper well of Transwell chambers (8 µm pore size; Corning Glass, Corning, NY, USA). For the invasion assay, Transwell chambers were pre-coated with 10 mg/mL growth factor-reduced Matrigel (BD Biosciences) on the upper side, whereas for the migration assay, the wells were left uncoated. The lower chambers were filled with 800 µL of growth medium. Non-invading cells were cleared using a cotton swab. Invading cells were stained with the Diff-Quick kit (Fisher) and photographed using an inverted microscope. The number of cells in three microscopic fields from each well was counted. The percentages of invading and migrating cells were determined relative to those of control cells. Cells were imaged using a phase-contrast microscope (Leica Microsystems, Bannockburn, IL, USA).
Transfection
For target molecule knockdown, cells were transfected with small interfering RNA (siRNA) targeting the respective molecule using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were incubated for 48 h after transfection for further experiments. The siRNAs against ICAM1, ICAM2, ICAM3, ICAM4, ICAM5, and EGFR were purchased from Genolution Pharmaceuticals Inc. (Seoul, Korea). For ICAM1, EGFR, and JAK1 over expression, the cells were transfected with vectors using Lipofectamine and Plus Reagent according to the manufacturer’s protocol.
Plasmid Construction
Human wild-type pCDM8-ICAM1 was gifted by Timothy Springer (Addgene Plasmid # 8632) [32]. To construct deletion mutants of ICAM1, pCDM8-ICAM1 was used as a template, and ICAM1 was amplified using Forward Primer 1 (containing the restriction enzyme site EcoRI) and Reverse Primer 2 (containing the restriction enzyme site NotI and HA tag sequence). Overlapping PCR was performed to generate deletion mutants. For the first PCR, two DNA fragments were amplified and the sequence of the intended domain that contained a common overlapping sequence was removed. The two amplified products were then mixed, and the final PCR was performed using Forward Primer 1 and Reverse Primer 2 to add the EcoRI and NotI restriction enzyme sites. Amplified products were subcloned into the MSCV vector via ligation. The primer sequences of the deletion mutants are shown in Supplementary Table 2.
Construction of knock-out cell line
MDA-MB-231 cells were seeded in a 24-well plate (120,000 cells/well) one day before transfection and transfected with 750 ng of Cas9-expressing plasmid and 250 ng of sgRNA-expressing plasmid using Lipofectamine 2000 according to the manufacturer’s instructions. The bulk CRISPR-Cas9-treated cells were then seeded into 96-well plates (1 cell/well) to grow single clones. Following expansion, the clones were subjected to high-throughput sequencing to select knockout cell lines. The target Cas sequences were designed using Cas-designer (http://www.rgenome.net/cas-designer/). To construct sgRNA-expressing plasmids, two complementary oligos for each target were annealed and cloned into pRG2 (#104,174; Addgene). The target sequence with PAM is as follows: 5’-CCGAGCAGGACCAGGAGTGCGGG-3’.
Western blot analysis
Cell lysates were prepared by incubating the cell pellets with lysis buffer (40 mM Tris–HCl [pH 8.0], 120 mM NaCl, and 0.1% Nonidet-P40) supplemented with protease inhibitors. Proteins in whole-cell lysates were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Amersham, Arlington Heights, IL). The membrane was blocked with 5% non-fat dry milk in phosphate-buffered saline (PBS) with Tween 20 (PBST) and incubated with primary antibodies overnight at 4 °C. Blots were incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies, and proteins were visualized using enhanced chemiluminescence (ECL) (Amersham, Arlington Heights, IL, USA) according to the manufacturer’s protocol. Secondary antibodies anti-mouse IgG-HRP, anti-goat IgG-HRP, and anti-rabbit IgG-HRP were purchased from Santa Cruz Biotechnology.
Co-immunoprecipitation assay
Cell lysates were prepared with lysis buffer as described for western blot analysis. Lysates were precleared using protein A-agarose beads (Santa Cruz Biotechnology) and then incubated with primary antibodies suitable for immunoprecipitation overnight at 4 °C. After 2 h of incubation with protein A-agarose beads, the supernatant was collected through centrifugation. Protein A-agarose beads were washed thrice with ice-cold PBS, and the immunoprecipitates were dissolved in SDS and analyzed using western blotting. For the determination of binding proteins, co-immunoprecipitates were visualized by Coomassie staining, sliced, and analyzed by LC–MS/MS (PROTIA, Seoul, Korea).
Immunocytochemistry
Cells cultured on coverslips were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton X-100 and 10% FBS in PBS. Following fixation, cells were incubated at 4 °C overnight with vimentin, fibronectin, Zeb1 E-cadherin and N-cadherin primary antibodies in PBS containing 10% FBS and 0.1% Triton X-100. Protein immunostaining was visualized using Alexa Fluor 488-conjugated anti-rabbit and anti-mouse secondary antibodies (Molecular Probes, Seoul, Korea). Nuclei were counterstained with DAPI (Sigma). Immunostaining was performed using an Olympus IX71 fluorescence microscope (Olympus, Seoul, Korea).
Immunohistochemistry
Mouse tissues (lungs and xenografts) were fixed in 4% formalin to prepare paraffin sections. Paraffin-embedded mouse and human breast microarray tissue sections (BR1008 and BR2082b; US Biomax, MD, USA) were deparaffinized in xylene and 100%, 95, 70%, and 50% ethanol, followed by PBS. Antigen retrieval was performed using 20 mg/mL proteinase K in PBS containing 0.1% Triton X-100. Sections were stained with hematoxylin and eosin (H&E) or immunostained overnight at 4 °C with primary antibody. After washing in PBS, biotinylated goat anti-rabbit IgG or anti-mouse IgG antibody was then applied to the sections for 30 min. After washing with PBS, ABC reagent (Vector laboratories Inc., Burlingame, CA, USA) was applied to the sections for 30 min. Color reaction was performed with 3, 3′-diaminobenzidine (DAB) (Vector laboratories). After counterstaining with hematoxylin and clearing with a graded ethanol series and xylene, the sections were mounted using Canada balsam. Images were captured using a DP71 digital imaging system and an IX71 microscope (Olympus).
In situ proximity ligation assay (PLA)
PFA-fixed cells were permeabilized with 0.1% Triton X-100 and 10% FBS in PBS. In the case of xenografts, antigen-retrieved tumor tissues were blocked with a blocking solution as per the immunohistochemistry (IHC) protocol. After blocking, cells and tissues were incubated with primary antibodies (1:500) at 4 °C overnight. In situ PLA was performed according to the manufacturer’s protocol, using a Duolink detection kit with a pair of nucleotide-labeled secondary antibodies. After ligation and amplification of PLA probes, signals were analyzed using confocal microscopy and quantified using ImageJ software.
qRT-PCR
Total RNA was isolated manually using Trizol (Invitrogen). qRT-PCR was performed using the KAPA SYBR FAST qPCR kit (KAPA Biosystems, Wilmington, MA, USA) according to the manufacturer's instructions. Reactions were carried out in Rotor Gene Q (Qiagen, Seoul, Korea), and the results are expressed as fold change calculated using the ΔCt method relative to the control sample. β-actin was used as an internal normalization control.
ChIP assay
Prior to the ChIP assay, cells were cross-linked using a final concentration of 1% formaldehyde. ChIP assay was performed using EZ-ChIP (Merck, Darmstadt, Germany) according to the manufacturer's instructions. MDA-MB-231 cells were transfected with si-control or si-ICAM-1 prior to IP. IP was performed using anti-STAT3 and anti-IgG antibodies. The STAT3 binding site in the promoter region was predicted using the JASPER software (http://jaspar.genereg.net/).
Animal studies
All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Hanyang University. All experiments using 5–6-week-old female NOD SCID gamma (NSG) mice were performed by Orient Bio. For these experiments, 40 μL of metastatic MDA-MB-231-LM1 (sh-cont-LM1 and sh-ICAM-1-LM1) and MDA-MB-231 (sg-cont, sg-ICAM-1, sg-ICAM-1 + ICAM-1WTO/E, and sg-ICAM-1 + ICAM-1ΔD1O/E) cells were orthotopically injected into the fourth mammary fat pad of female mice. ICAM-1 neutralizing antibody was used to confirm the therapeutic effects of ICAM-1 blockade. Two weeks after the orthotopic injection of MDA-MB-231-LM1, the ICAM-1 neutralizing antibody was intratumorally injected 3 times (2 mg/kg). Tumor size was measured using a digital caliper, and tumor volumes were assessed using width (W) and length (I) and calculated using the following formula: (longest diameter/2) × (shortest diameter2). To detect lung metastases, lungs were removed and fixed in 9% PFA. Detectable tumor nodules on the entire lung surface were counted to calculate the metastatic index. Tumor tissues were homogenized, and the expression of target genes and proteins was analyzed using qRT-PCR and western blotting.
Statistical analysis
All experimental data were reported as mean ± standard deviation (SD, represented by error bars), and all experiments were repeated three times. Statistical comparisons between two groups were conducted using Student's t-test, while comparisons among multiple groups were performed using one-way ANOVA. Significance was determined by ordinary one-way ANOVA with Tukey pairwise multiple comparison testing. Significance was determined by T-test with Unpaired t-testing. Correlation analyses were evaluated using simple linear regression. Overall survival analyses were assessed using the Log-rank (Mantel-Cox) test. All analyses were carried out using GraphPad Prism software (Version 9.0). P values ≤ 0.05 were considered significant. Sample size for each statistical analysis was indicated.
Data and software availability
The datasets used in this study are publicly available in the UCSC Xena (http://xena.ucsc.edu; Cancer Genome Atlas Program [TCGA] breast cancer), Gene Expression-Based Outcome (GOBO) (http://co.bmc.lu.se/gobo/gsa.pl), and Gene Expression Omnibus (GEO) databases (GSE45827, GSE65194, GSE22358, GSE25066, GSE20711, GSE41313). ImageJ and gene set enrichment analysis (GSEA) software are currently available on their respective websites (https://imagej.nih.gov/ij; https://www.gsea-msigdb.org/gsea/index.jsp). The GraphPad Prism software (version 9.0) was purchased from https://www.graphpad.com/scientific-software/prism/. Cytoscape was used to draw the ICAM-1-protein interaction network [73].
Supplementary Information
Authors’ contributions
J.H.K., N.U., J.M.Y., and S.J.L. conceptualized the study and designed the experiments. J.H.K., J.M.Y, and S.M.K. prepared the manuscript. N.U., J.H.K., Y.Z., and K.C.Y. performed in vitro and in vivo experiments. I.C.S., J.Y.L., Y.W.J., and M.J.K. performed the clinicopathological characterization of the experimental animal tumor models. S.S.B. and S.A.H. generated ICAM-1 knockout cells. J.M.Y., S.J.L., and H.M.O reviewed and edited the manuscript. All the authors have read and approved the final version of the manuscript.
Funding
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MSIT) (NRF-2019M3E5D1A01069361 to S.J.L. and NRF-2023R1A2C1003916 to J.M.Y.) and also supported by the Ministry of Science and ICT (MSIT) (RS-2023–00256076, to S.J.L, Y.W.J), and Ministry of SMEs and Startups (S3306756, to S.J.L, Y.W.J).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Hanyang University.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jae-Hyeok Kang, Nizam Uddin and Seungmo Kim contributed equally to this work.
Contributor Information
Joo Mi Yi, Email: jmyi76@inje.ac.kr, Email: jooyi@iu.edu.
Su-Jae Lee, Email: sjlee@fnctbiotech.com.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2.Fahad Ullah M. Breast Cancer: Current Perspectives on the Disease Status. Adv Exp Med Biol. 2019;1152:51–64. [DOI] [PubMed] [Google Scholar]
- 3.Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9:311–20. [PubMed] [Google Scholar]
- 4.Kozłowski J, Kozłowska A, Kocki J. Breast cancer metastasis - insight into selected molecular mechanisms of the phenomenon. Postepy Hig Med Dosw (Online). 2015;69:447–51. [DOI] [PubMed] [Google Scholar]
- 5.Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganesh K, Massagué J. Targeting metastatic cancer. Nat Med. 2021;27:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 8.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol. 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Fan Q, Zhang Z, Li X, Yu H, Meng F. Basal-HER2 phenotype shows poorer survival than basal-like phenotype in hormone receptor-negative invasive breast cancers. Hum Pathol. 2008;39:167–74. [DOI] [PubMed] [Google Scholar]
- 10.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F. Breast cancer Nat Rev Dis Primers. 2019;5:66. [DOI] [PubMed] [Google Scholar]
- 11.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–67. [DOI] [PubMed] [Google Scholar]
- 12.Foulkes William D, Smith Ian E, Reis-Filho Jorge S: Triple-negative breast Cancer. New Engl J Med. 2010;363:1938–48. [DOI] [PubMed]
- 13.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-Negative Breast Cancer: Clinical Features and Patterns of Recurrence. Clin Cancer Res. 2007;13:4429–34. [DOI] [PubMed] [Google Scholar]
- 14.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–92. [DOI] [PubMed] [Google Scholar]
- 15.Heeke AL, Tan AR. Checkpoint inhibitor therapy for metastatic triple-negative breast cancer. Cancer Metastasis Rev. 2021;40:537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye F, Dewanjee S, Li Y, Jha NK, Chen Z-S, Kumar A. Vishakha, Behl T, Jha SK, Tang H: Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol Cancer. 2023;22:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardia A, Hurvitz Sara A, Tolaney Sara M, Loirat D, Punie K, Oliveira M, Brufsky A, Sardesai Sagar D, Kalinsky K, Zelnak Amelia B, et al. Sacituzumab Govitecan in Metastatic Triple-Negative Breast Cancer. N Engl J Med. 2021;384:1529–41. [DOI] [PubMed] [Google Scholar]
- 18.Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer — expanded options, evolving needs. Nat Rev Clin Oncol. 2022;19:91–113. [DOI] [PubMed] [Google Scholar]
- 19.Cortes J, Rugo Hope S, Cescon David W, Im S-A, Yusof Mastura M, Gallardo C, Lipatov O, Barrios Carlos H, Perez-Garcia J, Iwata H, et al. Pembrolizumab plus Chemotherapy in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2022;387:217–26. [DOI] [PubMed] [Google Scholar]
- 20.Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im S-A, Yusof MM, Gallardo C, Lipatov O, Barrios CH, Holgado E, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. The Lancet. 2020;396:1817–28. [DOI] [PubMed] [Google Scholar]
- 21.Schmid P, Adams S, Rugo Hope S, Schneeweiss A, Barrios Carlos H, Iwata H, Diéras V, Hegg R, Im S-A, Shaw Wright G, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379:2108–21. [DOI] [PubMed] [Google Scholar]
- 22.da Cunha SG, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z. ErbB Receptors and Cancer. Methods Mol Biol. 2017;1652:3–35. [DOI] [PubMed] [Google Scholar]
- 24.Rajaram P, Chandra P, Ticku S, Pallavi BK, Rudresh KB, Mansabdar P. Epidermal growth factor receptor: Role in human cancer. Indian J Dent Res. 2017;28:687–94. [DOI] [PubMed] [Google Scholar]
- 25.Levva S, Kotoula V, Kostopoulos I, Manousou K, Papadimitriou C, Papadopoulou K, Lakis S, Koukoulias K, Karavasilis V, Pentheroudakis G, et al. Prognostic Evaluation of Epidermal Growth Factor Receptor (EGFR) Genotype and Phenotype Parameters in Triple-negative Breast Cancers. Cancer Genomics Proteomics. 2017;14:181–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, Kim JH, Kang E, Kim SW, Kim IA, Park SY. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol. 2014;27:1212–22. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima H, Ishikawa Y, Furuya M, Sano T, Ohno Y, Horiguchi J, Oyama T. Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer. 2014;21:66–74. [DOI] [PubMed] [Google Scholar]
- 28.Ueno NT, Zhang D. Targeting EGFR in Triple Negative Breast Cancer. J Cancer. 2011;2:324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016;6:1609–23. [PMC free article] [PubMed] [Google Scholar]
- 30.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30:2615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothlein R, Dustin ML, Marlin SD, Springer TA. A human intercellular adhesion molecule (ICAM-1) distinct from LFA-1. J Immunol. 1986;137:1270–4. [PubMed] [Google Scholar]
- 32.Staunton DE, Marlin SD, Stratowa C, Dustin ML, Springer TA. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families. Cell. 1988;52:925–33. [DOI] [PubMed] [Google Scholar]
- 33.Bella J, Kolatkar PR, Marlor CW, Greve JM, Rossmann MG. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci U S A. 1998;95:4140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman A, Fazal F. Hug tightly and say goodbye: role of endothelial ICAM-1 in leukocyte transmigration. Antioxid Redox Signal. 2009;11:823–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo W, Liu S, Cheng Y, Lu L, Shi J, Xu G, Li N, Cheng K, Wu M, Cheng S, Liu S. ICAM-1–Related Noncoding RNA in Cancer Stem Cells Maintains ICAM-1 Expression in Hepatocellular Carcinoma. Clin Cancer Res. 2016;22:2041–50. [DOI] [PubMed] [Google Scholar]
- 36.Liou GY, Döppler H, Necela B, Edenfield B, Zhang L, Dawson DW, Storz P. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015;5:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reina M, Espel E. Role of LFA-1 and ICAM-1 in cancer. Cancers (Basel). 2017;9(11):153. [DOI] [PMC free article] [PubMed]
- 38.Blank C, Brown I, Kacha AK, Markiewicz MA, Gajewski TF. ICAM-1 Contributes to but Is Not Essential for Tumor Antigen Cross-Priming and CD8+ T Cell-Mediated Tumor Rejection In Vivo1. J Immunol. 2005;174:3416–20. [DOI] [PubMed] [Google Scholar]
- 39.Anderson ME, Siahaan TJ. Targeting ICAM-1/LFA-1 interaction for controlling autoimmune diseases: designing peptide and small molecule inhibitors. Peptides. 2003;24:487–501. [DOI] [PubMed] [Google Scholar]
- 40.Van Seventer GA, Shimizu Y, Horgan KJ, Shaw S. The LFA-1 ligand ICAM-1 provides an important costimulatory signal for T cell receptor-mediated activation of resting T cells. J Immunol. 1990;144:4579–86. [PubMed] [Google Scholar]
- 41.Waks AG, Winer EP. Breast Cancer Treatment A Review. Jama. 2019;321:288–300. [DOI] [PubMed] [Google Scholar]
- 42.Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol. 2018;13:395–412. [DOI] [PubMed] [Google Scholar]
- 43.Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29:212–26. [DOI] [PubMed] [Google Scholar]
- 44.Shin DY, Na II, Kim CH, Park S, Baek H, Yang SH. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J Thorac Oncol. 2014;9:195–9. [DOI] [PubMed] [Google Scholar]
- 45.Day KC, Lorenzatti Hiles G, Kozminsky M, Dawsey SJ, Paul A, Broses LJ, Shah R, Kunja LP, Hall C, Palanisamy N, et al. HER2 and EGFR Overexpression Support Metastatic Progression of Prostate Cancer to Bone. Cancer Res. 2017;77:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarín C, Gomez M, Calvo E, López JA, Zaragoza C. Endothelial nitric oxide deficiency reduces MMP-13-mediated cleavage of ICAM-1 in vascular endothelium: a role in atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:27–32. [DOI] [PubMed] [Google Scholar]
- 47.Ramos TN, Bullard DC, Barnum SR. ICAM-1: isoforms and phenotypes. J Immunol. 2014;192:4469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Den Engel NK, Heidenthal E, Vinke A, Kolb H, Martin S. Circulating forms of intercellular adhesion molecule (ICAM)-1 in mice lacking membranous ICAM-1. Blood. 2000;95:1350–5. [PubMed] [Google Scholar]
- 49.Kim JY, Kim DH, Kim JH, Lee D, Jeon HB, Kwon SJ, Kim SM, Yoo YJ, Lee EH, Choi SJ, et al. Soluble intracellular adhesion molecule-1 secreted by human umbilical cord blood-derived mesenchymal stem cell reduces amyloid-β plaques. Cell Death Differ. 2012;19:680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ali R, Brown W, Purdy SC, Davisson VJ, Wendt MK. Biased signaling downstream of epidermal growth factor receptor regulates proliferative versus apoptotic response to ligand. Cell Death Dis. 2018;9:976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith JS, Lefkowitz RJ, Rajagopal S. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov. 2018;17:243–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, Lemmon MA. EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell. 2017;171:683-695.e618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindsey S, Langhans SA. Epidermal growth factor signaling in transformed cells. Int Rev Cell Mol Biol. 2015;314:1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han X, Wang Y, Chen H, Zhang J, Xu C, Li J, Li M. Enhancement of ICAM-1 via the JAK2/STAT3 signaling pathway in a rat model of severe acute pancreatitis-associated lung injury. Exp Ther Med. 2016;11:788–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kesanakurti D, Chetty C, Rajasekhar Maddirela D, Gujrati M, Rao JS. Essential role of cooperative NF-κB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene. 2013;32:5144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wung BS, Ni CW, Wang DL. ICAM-1 induction by TNFα and IL-6 is mediated by distinct pathways via Rac in endothelial cells. J Biomed Sci. 2005;12:91–101. [DOI] [PubMed] [Google Scholar]
- 57.Sabbah DA, Hajjo R, Sweidan K. Review on Epidermal Growth Factor Receptor (EGFR) Structure, Signaling Pathways, Interactions, and Recent Updates of EGFR Inhibitors. Curr Top Med Chem. 2020;20:815–34. [DOI] [PubMed] [Google Scholar]
- 58.Müller N. The Role of Intercellular Adhesion Molecule-1 in the Pathogenesis of Psychiatric Disorders. Front Pharmacol. 2019;10:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mycko MP, Kwinkowski M, Tronczynska E, Szymanska B, Selmaj KW. Multiple sclerosis: the increased frequency of the ICAM-1 exon 6 gene point mutation genetic type K469. Ann Neurol. 1998;44:70–5. [DOI] [PubMed] [Google Scholar]
- 60.Benedicto A, Romayor I, Arteta B. Role of liver ICAM-1 in metastasis. Oncol Lett. 2017;14:3883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosette C, Roth RB, Oeth P, Braun A, Kammerer S, Ekblom J, Denissenko MF. Role of ICAM1 in invasion of human breast cancer cells. Carcinogenesis. 2005;26:943–50. [DOI] [PubMed] [Google Scholar]
- 62.Figenschau SL, Knutsen E, Urbarova I, Fenton C, Elston B, Perander M, Mortensen ES, Fenton KA. ICAM1 expression is induced by proinflammatory cytokines and associated with TLS formation in aggressive breast cancer subtypes. Sci Rep. 2018;8:11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox MA, Barnum SR, Bullard DC, Zajac AJ. ICAM-1–dependent tuning of memory CD8 T-cell responses following acute infection. Proc Natl Acad Sci. 2013;110:1416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zumwalde NA, Domae E, Mescher MF, Shimizu Y. ICAM-1–Dependent Homotypic Aggregates Regulate CD8 T Cell Effector Function and Differentiation during T Cell Activation. J Immunol. 2013;191:3681–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi M, Zheng X, Niu M, Zhu S, Ge H, Wu K. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer. 2022;21:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SH, Kim Y, Jeon BN, Kim G, Sohn J, Yoon Y, Kim S, Kim Y, Kim H, Cha H, et al. Intracellular Adhesion Molecule-1 Improves Responsiveness to Immune Checkpoint Inhibitor by Activating CD8(+) T Cells. Adv Sci (Weinh). 2023;10: e2204378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seth R, Raymond FD, Makgoba MW. Circulating ICAM-1 isoforms: diagnostic prospects for inflammatory and immune disorders. Lancet. 1991;338:83–4. [DOI] [PubMed] [Google Scholar]
- 68.Seyedabadi M, Ghahremani MH, Albert PR. Biased signaling of G protein coupled receptors (GPCRs): Molecular determinants of GPCR/transducer selectivity and therapeutic potential. Pharmacol Ther. 2019;200:148–78. [DOI] [PubMed] [Google Scholar]
- 69.Chen W, Hoffmann AD, Liu H, Liu X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis Oncol. 2018;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larson RC, Kann MC, Bailey SR, Haradhvala NJ, Llopis PM, Bouffard AA, Scarfó I, Leick MB, Grauwet K, Berger TR, et al. CAR T cell killing requires the IFNγR pathway in solid but not liquid tumours. Nature. 2022;604:563–70. [DOI] [PubMed] [Google Scholar]
- 71.Arenas EJ, Martínez-Sabadell A, Rius Ruiz I, Román Alonso M, Escorihuela M, Luque A, Fajardo CA, Gros A, Klein C, Arribas J. Acquired cancer cell resistance to T cell bispecific antibodies and CAR T targeting HER2 through JAK2 down-modulation. Nat Commun. 2021;12:1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martínez-Sabadell A, Morancho B, Rius Ruiz I, Román Alonso M, Ovejero Romero P, Escorihuela M, Chicote I, Palmer HG, Nonell L, Alemany-Chavarria M, et al. The target antigen determines the mechanism of acquired resistance to T cell-based therapies. Cell Rep. 2022;41:111430. [DOI] [PubMed] [Google Scholar]
- 73.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study are publicly available in the UCSC Xena (http://xena.ucsc.edu; Cancer Genome Atlas Program [TCGA] breast cancer), Gene Expression-Based Outcome (GOBO) (http://co.bmc.lu.se/gobo/gsa.pl), and Gene Expression Omnibus (GEO) databases (GSE45827, GSE65194, GSE22358, GSE25066, GSE20711, GSE41313). ImageJ and gene set enrichment analysis (GSEA) software are currently available on their respective websites (https://imagej.nih.gov/ij; https://www.gsea-msigdb.org/gsea/index.jsp). The GraphPad Prism software (version 9.0) was purchased from https://www.graphpad.com/scientific-software/prism/. Cytoscape was used to draw the ICAM-1-protein interaction network [73].
No datasets were generated or analysed during the current study.