ABSTRACT
Clostridioides difficile, the major cause of antibiotic-associated diarrhea, is a strict anaerobic, sporulating Firmicutes. However, during its infectious cycle, this anaerobe is exposed to low oxygen (O2) tensions, with a longitudinal decreasing gradient along the gastrointestinal tract and a second lateral gradient with higher O2 tensions in the vicinity of the cells. A plethora of enzymes involved in oxidative stress detoxication has been identified in C. difficile, including four O2-reducing enzymes: two flavodiiron proteins (FdpA and FdpF) and two reverse rubrerythrins (revRbr1 and revRbr2). Here, we investigated the role of the four O2-reducing enzymes in the tolerance to increasing physiological O2 tensions and air. The four enzymes have different, yet overlapping, spectra of activity. revRbr2 is specific to low O2 tensions (<0.4%), FdpA to low and intermediate O2 tensions (0.4%–1%), revRbr1 has a wider spectrum of activity (0.1%–4%), and finally FdpF is more specific to tensions > 4% and air. These different O2 ranges of action partly arise from differences in regulation of expression of the genes encoding those enzymes. Indeed, we showed that revrbr2 is under the dual control of σA and σB. We also identified a regulator of the Spx family that plays a role in the induction of fdp and revrbr genes upon O2 exposure. Finally, fdpF is regulated by Rex, a regulator sensing the NADH/NAD+ ratio. Our results demonstrate that the multiplicity of O2-reducing enzymes of C. difficile is associated with different roles depending on the environmental conditions, stemming from a complex multi-leveled network of regulation.
IMPORTANCE
The gastrointestinal tract is a hypoxic environment, with the existence of two gradients of O2 along the gut, one longitudinal anteroposterior decreasing gradient and one proximodistal increasing from the lumen to the epithelial cells. O2 is a major source of stress for an obligate anaerobe such as the enteropathogen C. difficile. This bacterium possesses a plethora of enzymes capable of scavenging O2 and reducing it to H2O. In this work, we identified the role of the four O2-reducing enzymes in the tolerance to the physiological O2 tensions faced by C. difficile during its infectious cycle. These four enzymes have different spectra of action and protect the vegetative cells over a large range of O2 tensions. These differences are associated with a distinct regulation of each gene encoding those enzymes. The complex network of regulation is crucial for C. difficile to adapt to the various O2 tensions encountered during infection.
KEYWORDS: oxygen detoxication, oxygen tolerance, stress response, flavodiiron enzymes, rubrerythrin, Sigma B, Spx, Rex
INTRODUCTION
Clostridioides difficile, formerly known as Clostridium difficile, is a Gram-positive sporulating bacterium and a strict anaerobe. C. difficile is also an opportunistic pathogen, the first cause of antibiotic-associated diarrhea in developed countries, and the etiological agent of pseudomembranous colitis (1). Spores, which are the form of resistance and dissemination of C. difficile, are transmitted through the orofecal route. Following ingestion, spores can germinate in the gastrointestinal tract (GIT) (2). C. difficile colonization of the GIT is favored by a dysbiosis of the microbiota often associated with antibiotic treatments (3–5). Indeed, this dysbiosis modifies the pools of metabolites available in the gut including a diminution of the pool of secondary bile acids, which are toxic for C. difficile vegetative cells, and an increase in the concentration of primary bile acids, favoring germination (1, 4). Changes in the metabolite pools further allow the colonization of the GIT by the vegetative cells and the production of toxins, which modify the cytoskeleton of enterocytes and disrupt the intestinal barrier (6). Toxin production then triggers an important local inflammation associated with diarrheal symptoms (1, 2).
During its infectious cycle, C. difficile is exposed to inflammation-induced oxidative and nitrosative stresses, as well as to low O2 tensions, a major stress for this obligate anaerobe. Indeed, there is a decreasing longitudinal gradient of O2 along the GIT ranging from 4% to 5% in the small intestine, where spores germinate, to 0.1% to 0.4% in the lumen of the colon (7). In the colon, there is a second lateral O2 gradient increasing from the lumen toward the mucus (1%–2%) and the tissues (5%). The O2 tensions in the GIT increase during microbiota dysbiosis, which are conditions favoring C. difficile infection (8). Indeed, dysbiosis is often associated with a decrease in butyrate-producing commensal bacteria. In physiological conditions, the oxidation of butyrate by the colonocytes consumes O2, maintaining hypoxia (9, 10), whereas, in a dysbiotic gut, the depletion of butyrate leads to a switch of colonocytes metabolism to glucose fermentation, decreasing O2 consumption (11). This rerouting of epithelial cell metabolism thus favors an increase in O2 tensions in the colon (8, 12). C. difficile, which encounters O2 during the infection, possesses a variety of protection mechanisms and especially detoxication enzymes. In anaerobic bacteria, “Flavodiiron proteins” (Fdp) and rubrerythrins/reverse-rubrerythrins (Rbr/revRbr) play a major role in O2 and reactive oxygen species (ROS), namely hydrogen peroxide (H2O2), detoxication (13). Rbr and revRbr contain both a “diiron” in a four-helix scaffold and a rubredoxin-like (Rd) domain, with an inverted organization between the two families. These proteins act mainly as peroxidases, converting H2O2 to H2O, but they were already reported to also harbor an O2-reductase activity, although lower than hydrogen peroxide (14, 15). The Fdp proteins, which act either as NO- or O2-reductases (16–18), are a modular family of proteins that share a minimal dimeric form in which each monomer is built by a metallo-β-lactamase-like domain, harboring the catalytic diiron center, and a flavodoxin-like domain containing an FMN (16, 17). Class A Fdps only have this common core, whereas more complex classes have extra domains. For example, class F enzymes have a Rd and a NADH:Rd oxidoreductase (NROR) domain allowing the direct transfer of electrons from NADH to its substrates. Although C. difficile is an obligate anaerobe, some strains can grow in the presence of up to 2% O2 (19). This rather high O2 tolerance can be explained, at least partly, by the presence of four O2-reducing enzymes: two Fdp proteins, a Class A (FdpA, CD1157) and a Class F (FdpF, CD1623), and two revRbr, revRbr1 (CD1474) and revRbr2 (CD1524) (15, 18). All four purified enzymes have O2-reductase activity in vitro (15, 18), and for this reason, will be called O2-reductases thereon. While FdpF is a standalone enzyme, receiving electrons directly from NADH, FdpA, and both revRbrs cannot directly take electrons from NADH and therefore need partners that remain to be identified in C. difficile (13, 16, 17). The role of these O2-reductases in the tolerance to low O2 tensions (<0.4%) has been studied (15). At these tensions, a C. difficile fdpF mutant is not affected for growth and a fdpA mutant has a reduced growth at 0.4% O2. A double revrbr mutant is almost unable to grow at 0.1 or 0.4% O2 but neither single mutant has a phenotype at these O2 tensions. A functional redundancy is thus observed for revRbr1 and revRbr2, which share 95% amino acid identity at the protein level (15). Moreover, the genes encoding the four O2-reductases are controlled by the alternative sigma factor σB, which is the sigma factor of the general stress response in C. difficile (15, 20, 21). The σB regulon of C. difficile also includes many other genes involved in responses to various stress stimuli. Accordingly, a sigB mutant is more sensitive to many stresses including low O2 tensions but also oxidative and nitrosative stresses (20). In the absence of stress, σB is sequestered by the anti-sigma factor RsbW, preventing the transcription of its regulon. However, upon stress exposure, the phosphatase RsbZ dephosphorylates the anti-anti-sigma factor RsbV, which then competes with σB for binding to RsbW, thus relieving the σB protein (22). The signal sensed and its receptor still remain to be identified.
Identifying a global expression signature of C. difficile upon O2 exposure is challenging due to the different strains and growth conditions used (19, 23–26). The transcriptomic studies have shown various and sometimes conflicting results suggesting the existence of a rather complex adaptive response of C. difficile to O2 exposure (19, 24–26). Many genes found differentially regulated upon O2 exposure are involved in C. difficile metabolism, suggesting the existence of various complex O2-dependent rerouting of C. difficile metabolism. Some regulators, such as CodY, are known to link metabolic control with stress. Indeed, CodY is inactivated upon metabolic starvation and will thus allow the expression of genes involved both in metabolic rerouting in the stationary phase and toxin production and sporulation. It is worth noting that neither the genes encoding the four O2-reductases nor other members of the σB regulon were found to be differentially regulated upon exposure to hypoxia (8 h at 1.5% O2, 4 h at 2% O2 or 15 and 60 min at 5% O2) (19, 24, 26). However, after a short air exposure to strain 630, the expression of fdp and revrbr genes is upregulated along with about 100 other genes, including several σB targets (25). This suggests that those enzymes might be more important for short-term exposure to air than for adaptation to hypoxia.
In this work, we aim to further characterize the physiological role of the O2-reductases in the presence of intermediate to high O2 tensions, as well as during air exposure. We were able to show that the four O2-reductases have different yet overlapping spectra of activity. To better understand the origin of those different spectra of activity, we tested the induction of fdp and revrbr gene expression in the presence of various O2 tensions and identified the different regulators controlling their expression. Each gene encoding an O2-reductase seems to have a specific regulation. Indeed, while all genes are transcribed by σB, revrbr2 and fdpA are also transcribed by σA. Moreover, all those genes are controlled by a newly identified oxygen-sensing regulator, while fdpF is specifically regulated by Rex.
RESULTS
C. difficile tolerance to intermediate O2 tensions
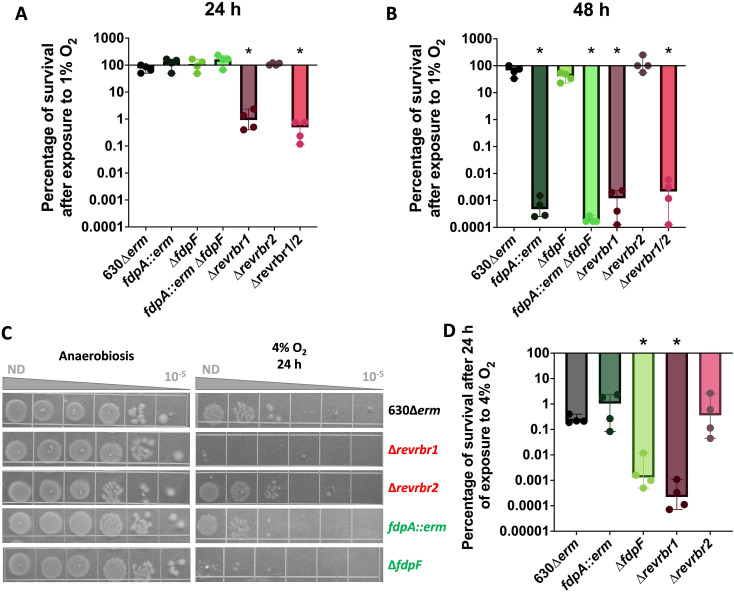
The physiological role of the four O2-reductases of C. difficile in the presence of 0.1%– 0.4% O2 tensions has already been studied (15). However, their contribution to the tolerance of C. difficile to 1% O2, a tension found near the mucus layer and the epithelial cells in the colon that vegetative cells reached during infection (27, 28), remains to be determined. As both triple and quadruple mutants are unable to grow at 0.4% O2 (15), we focused our analysis on single and double mutants inactivated for the fdp or the revrbr genes. We incubated these mutants on agar plates for 24 or 48 h in the presence of 1% O2 or anaerobiosis (Fig. 1A and B; Fig. S1A and B). No significant loss of survival was observed for the 630∆erm strain after 48 h of growth at 1% O2 compared to anaerobiosis (Fig. 1B; Fig. S1A). By contrast, we observed an almost complete loss of survival of the fdpA::erm and the double fdp mutant after 48 h of exposure to 1% O2 (Fig. 1B; Fig. S1A). However, these strains were not affected after a 24-h exposure (Fig. 1A; Fig. S1A). In addition, the phenotype of the ∆fdpF mutant remained rather similar to that of the wild-type (WT) strain even after 48 h (Fig. 1A and B; Fig. S1A).
Fig 1.
Role of O2-reductases in the tolerance to 1% and 4% O2. Serial dilutions of the 630∆erm strain and single or double mutants inactivated for revrbr or fdp genes were spotted on TY Tau plates. Plates were incubated either in anaerobiosis for 24 h, in the presence of 1% O2 for 24 h or 48 h (Fig. S1A and B) or in the presence of 4% O2 for 24 h. Following 24 h incubation in the presence of O2, plates were subsequently incubated in anaerobiosis for 24 h. CFU were determined for each experiment. The percentage of survival after exposure to 1% O2 relative to the survival in anaerobiosis was plotted at 24 h (A) and 48 h (B). Pictures are representative of four independent experiments at 4% O2 (C). The percentage of survival after exposure to 4% O2 relative to survival in anaerobiosis was plotted (D). Four biological replicates were performed per experiment. For all plots, a median with 95% CI is shown. Mann-Whitney statistical tests were performed. All comparisons were made with the corresponding 630∆erm strain. *: P-value < 0.05.
For the double mutant inactivated for both revrbrs, we observed a 2-log reduction after 24 h of exposure to 1% O2 and a more drastic loss of survival (6-logs) after 48 h (Fig. 1A and B; Fig. S1B). This is in agreement with the drastic phenotype observed at 0.4% O2 for this mutant (15). We then tested the phenotype of the single revrbr mutants in the same conditions. Upon exposure to 1% O2, the ∆revrbr1 mutant had a phenotype similar to that of the double revrbr mutant, both at 24 and 48 h. Surprisingly, the survival of the ∆revrbr2 mutant was not affected even at 48 h (Fig. 1A and B; Fig. S1B). The difference between both single revrbr mutants is striking as the two proteins share 95% amino acid identity and have similar O2 reduction activities in vitro (15). The revRbr1 seems to play a more important role in the tolerance to 1% O2 than revRbr2. After 24 h of growth, the phenotype of the single ∆revrbr1 and double mutants were, respectively, fully and partially complemented with a revrbr1 copy carried by a plasmid (Fig. S1C). The revrbr2 gene expressed under the control of its promoter on a plasmid also fully or partially restored the survival at 24 h of the single ∆revrbr1 and double revrbr mutants (Fig. S1C). The high similarity of both enzymes allows cross-complementation for compensations, even though the role of revRbr2 seems to be minor at 1% O2 and is visible only when revRbr1 is absent and when revrbr2 is overexpressed. Finally, it is interesting to note that the survival of the ∆revrbr1 mutant is more affected than that of the fdpA::erm mutant upon exposure to 1% O2 for 24 h (Fig. 1A and B; Fig. S1A and B), suggesting that revRbr1 is more important than FdpA in the tolerance to 1% O2. All these results suggest that the tolerance to 1% O2 is mainly due to revRbr1 and to a lesser extent to FdpA, whereas FdpF and revRbr2 do not seem to be involved.
C. difficile tolerance to high O2 tensions
We then studied the role of the four O2-reductases at 4% O2, which is physiologically encountered in the small intestine where spores germinate (29). After 24 h of incubation at 4% O2, the 630∆erm strain had lost around 3 logs of survival, as also observed for the ∆revrbr2 and the fdpA::erm mutants (Fig. 1C and D). However, a significantly more important loss of survival was obtained in the ∆revrbr1 mutant compared to the WT strain (Fig. 1C and D). This phenotype was fully complemented by expressing the revrbr1 gene on a plasmid under the control of its promoter (Fig. S1D), whereas the complementation of the ∆revrbr1 mutant by the revrbr2 gene was only partial (Fig. S1D). It is worth noting that we observed for the first time a more important loss of survival of the ∆fdpF mutant compared with the parental strain (Fig. 1C and D). However, the phenotype of the mutant is not complemented by the introduction of a plasmid-borne copy of the fdpF gene with its promoter (Fig. S1D). Altogether, these results suggest that revRbr1 and FdpF contribute to the tolerance of C. difficile to high O2 tensions, whereas FdpA and revRbr2 seem to play more marginal roles.
C. difficile tolerance to air
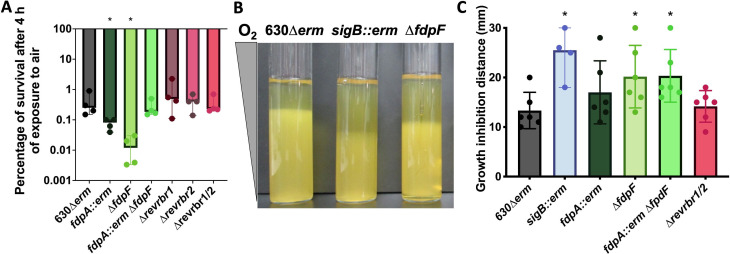
We also investigated C. difficile tolerance to air, a high non-physiological O2 tension (21%), to test the ability of C. difficile to survive harsher conditions at least transiently. Since C. difficile is not able to grow in air, we thus exposed transiently vegetative cells to air on plates (4 h) before incubating the plates back for 24 h in anaerobiosis. Only the cells that survived were then able to form colonies. We observed a loss of survival around 3 logs for the 630∆erm after 4 h of air exposure (Fig. 2A; Fig. S2A). This result confirmed that air exposure is toxic for C. difficile, but that strain 630∆erm can still survive a longer air exposure than expected, as previously published (30, 31). After 4 h under air exposure, the survival of the two single revrbr mutants and the double mutant inactivated for both revrbr was not affected compared to the 630∆erm strain (Fig. 2A; Fig. S2A). For the single fdpA::erm mutant, we observed a slight but significant decrease in survival compared to the parental strain (1 log) (Fig. 2A; Fig. S2A). Interestingly, the ∆fdpF mutant also showed a more drastic survival defect compared to the parental 630∆erm strain after 4 h in air (Fig. 2A; Fig. S2A). This phenotype was complemented when we expressed, in this mutant, the fdpF gene under the control of its promoter (Fig. S2B). We confirmed the phenotype of the ∆fdpF mutant following inoculation of soft agar tubes. After incubation in air, the growth inhibition length reflects the air sensitivity of the strain. As observed for the sigB::erm mutant used as a control (20), the zone of growth inhibition for the ∆fdpF mutant or for the double fdp mutant was significantly longer than that of the parental strain (Fig. 2B and C). This phenotype was complemented by the reintroduction of a copy of fdpF on a plasmid (Fig. S2C). By contrast, we did not observe any differential phenotype between the 630∆erm and either the fdpA::erm mutant or the double revrbr mutant (Fig. 2B and C). These results suggest that FdpF is the main O2-reductase in air.
Fig 2.
Role of O2-reductases in the tolerance to air. Serial dilutions of the 630∆erm and the mutants were spotted on TY Tau plates. Plates were incubated either in anaerobiosis for 24 h, or in the presence of air for 4 h (Fig. S2A). Plates incubated in air were subsequently incubated in anaerobiosis for 24 h. CFUs were determined for each experiment and the percentage of survival after exposure to air relative to the survival in anaerobiosis was plotted (A). Soft agar tubes were inoculated with various strains and incubated in air (B). The growth inhibition distance for each replicate was determined and plotted (C). At least four biological replicates were performed per experiment. For all plots, a median with 95% CI is shown. Mann-Whitney statistical tests were performed. All comparisons were made with the 630∆erm strain. *: P-value < 0.05.
Contribution of the Fdps and revRbrs to O2 reduction in crude extracts
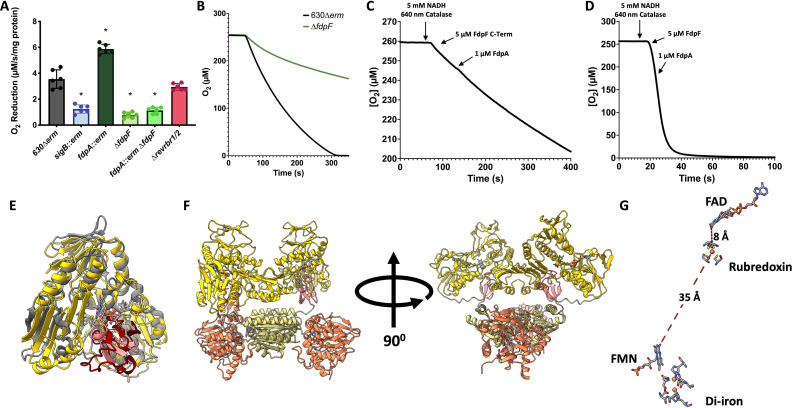
To evaluate the contribution of each of these enzymes to O2 elimination, we measured in vitro in a buffer equilibrated in air the O2-reducing activities of crude extracts prepared from anaerobically grown 630∆erm and the mutant strains inactivated for the genes encoding the different O2-reductases. First, an important loss of O2-reductase activity was observed in the sigB::erm mutant compared to the 630∆erm strain (Fig. 3A), in agreement with the key role of σB in the control of the expression of the genes involved in O2 reduction (15). For the double revrbr mutant, the O2-reductase activity was similar to that of the parental strain (Fig. 3A). For the single ∆fdpF mutant and the double fdp mutant, we detected a significant decrease of O2-reductase activity comparable to that observed for the sigB::erm mutant (Fig. 3A and B). This result indicates that FdpF has the highest ability to reduce O2 and cements the importance of FdpF for an efficient O2 reduction. In a cellular context, the O2-reduction activity of FdpF is more significant than the one from the other enzymes in agreement with the previously reported in vitro assays using the purified enzymes (15, 18). It is intriguing to observe a significant increase of the O2-reductase activity (+ 60%) in the fdpA::erm mutant compared to the WT strain (Fig. 3A). This could be explained by the existence of some compensation at the genetic level between the genes encoding the other O2-reductases. To test this hypothesis, we performed RT-qPCR on RNA extracted from the fdpA::erm mutant grown in the conditions used for crude extract preparation, that is, after 16 h of growth in anaerobiosis. We showed that neither the revrbr genes nor the fdpF gene were differentially expressed in the fdpA::erm mutant compared to the 630∆erm strain (Fig. S3A) suggesting another mechanism involved in this increased activity. Another explanation could be that, in the WT strain, FdpA would hijack electrons from FdpF, that is, electrons would transfer directly from the NROR and Rd domains of FdpF to FdpA (Fig. S3B). We thus tested whether electron transfer can occur between FdpA and either the C-terminal domains of FdpF (NROR and Rd) or the full FdpF protein. We observed no differences in the O2 consumption after the addition of FdpA to a medium containing either the C-terminal domain of FdpF or the full FdpF protein (Fig. 3C and D), strongly suggesting that this electron transfer is not efficient or possible. The Alphafold3 model structure of the FdpF dimer is presented in Fig. 3 (Fig. 3E and F) and corroborates this observation (see below). This model predicts a relative position of the Rd and NROR domains on its C-terminal part similar (r.m.s.d. of ~1.2 Å) to what was observed in the complex between the Pseudomonas aeruginosa Rd and its NROR (PDB 2V3B) (32). A very short distance of ~8.2 Å is also predicted between the FAD of NROR and the iron from Rd, indicating a direct electron transfer between these two domains and then to the FMN domain (Fig. 3G). These points were demonstrated in vitro for both FdpF and FdpH (class H FDP) where mutations in the Rd domain broke the intramolecular electron transfer pathway, leading to a considerable or even total abolishment of their activity (33). In addition, the relative position of the C-terminal and the N-terminal (diiron and FMN domains) parts, with a distance of ~35 Å between the Rd iron and the FMN (Fig. 3F and G), should impair the presence of FdpA in a proper orientation to receive the electrons from the Rd domain of FdpF in agreement with our in vitro data (Fig. 3C and D). Finally, we tested the reduction of FdpA with soluble extracts of either the 630∆erm strain or the sigB::erm mutant. This experiment was done under anaerobic conditions to avoid reoxidation of the enzyme. FdpA can be completely reduced in the presence of both soluble crude extracts and NADH, albeit slowly (Fig. S3D and E). These results thus indicate that the gene encoding the FdpA electron donor is not expressed under σB control, also supporting ruling out FdpF. This electron donor is, most probably, a so far unidentified NADH oxidoreductase.
Fig 3.
Contribution of FdpA and FdpF to O2 reduction in crude extracts and the impossibility of electron transfer between FdpA and FdpF. O2-reductase activity was measured in crude extracts of various mutants. The O2-reduction capacity is expressed in µM/s/mg protein. Six assays per strain from three independent biological replicates of each strain were performed. Mean and standard deviation are represented. ANOVA tests were performed with a confidence interval of 95% between the mutant strains and the 630∆erm strain. *: P-value < 0.05 (A). O2 consumption of crude extracts of the 630∆erm strain and the ∆fdpF mutant was measured over time. A representative replicate for each strain is shown (B). O2-reduction assays were performed to assess whether the C-terminal (C-Term) domain of FdpF (C) or the full FdpF protein (D) could transfer electrons to FdpA. Assays were performed in 50 mM Tris-HCl, pH 7.5 containing 18% glycerol, at 25°C in a buffer equilibrated in air (≈250 µM of O2), in the presence of catalase (640 nM, from bovine liver). Assays were initiated by the addition of 5 µM of either FdpF C-Term or FdpF and then 1 µM of FdpA. Assays were performed at least in triplicate and a representative assay is shown. Superposition of the Alphafold3 model of Rd (pink) and NROR (yellow) domains of FdpF (residues 409–843) with the crystallographic structure of P. aeruginosa NADH:Rd oxidoreductase complex (PDB 2V3B, gray and dark red) (32) (E). AlphaFold3 model structure of FdpF dimer, showing the four domains of each monomer (β-lactamase-like in orange, flavodoxin-like in light yellow, Rd in pink and NROR in yellow) (F). Distance between the FAD from the NROR domain, iron from the Rd domain, and FMN from the flavodoxin-like domain predictor from the FdpF AlphaFold3 model (G).
Role of PerR in the control of genes encoding O2-reductases
In Clostridium acetobutylicum, a perR mutant is aerotolerant and PerR strongly represses the expression of the genes encoding the Fdps and the revRbrs, all being induced upon air exposure (34, 35). In C. difficile, PerR negatively controls the expression of the rbr1 operon involved in oxidative stress response (31). PerR senses H2O2 but might also detect O2 as proposed for the PerR regulators of Staphylococcus aureus and Bacillus subtilis (36, 37). The 630∆erm strain harbors a point mutation (T41A) in the perR gene making it non-functional and leading to an increased air tolerance (31). So, we wanted to determine whether PerR could still be involved in the regulation of the O2-reductase genes in C. difficile. To do so, we performed RT-qPCR both on the 630∆erm strain and a 630∆erm strain harboring a chromosomic perRWT allele. We did not observe a significant differential regulation of the O2-reductase encoding genes in these two strains (Fig. S4A). Moreover, the perRWT strain shows a similar phenotype to the perRT41A strain in the presence of 1% O2 (Fig. S4B). These results suggest that PerR is not involved in the regulation of O2-reductase genes in C. difficile.
C. difficile O2-reductase gene expression at various O2 tensions
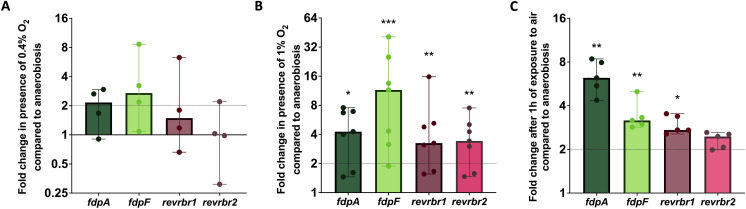
The diversity of the role of the O2-reductases in the presence of various O2 tensions could, at least partly, stem from differences in the expression of the genes encoding these enzymes. We first tested the expression of the four O2-reductase genes in the presence of increasing O2 tensions. RNAs were sampled in the conditions of our phenotypical assays, that is, a long-term exposure on agar plates. At 0.4% O2, we did not observe any differences in expression compared to anaerobiosis (Fig. 4A). By contrast, a significant induction of expression of the four genes (revrbr1, revrbr2, fdpA, and fdpF) was shown at 1% O2 (Fig. 4B). Testing the exposure to higher O2 tensions in similar conditions was more complex, as cells are only able to survive but not to grow at 4% O2 and in air. Thus, we performed a transient air exposure in six-well plates. After 1 h of air exposure, we also observed a significant induction of the expression of the genes encoding the O2-reductases, except revrbr2, which only showed a slight non-significant increase (Fig. 4C). These results show that intermediate O2 tensions and air are sensed by C. difficile and trigger an O2 detoxication response.
Fig 4.
Expression of the genes encoding the O2-reductases in the presence of various O2 tensions. The expression of the genes encoding the four O2-reductases was evaluated by RT-qPCR in the 630∆erm strain exposed to different O2 tensions [0.4% O2 (A), 1% O2 (B) or air (C)] and compared with their expression in anaerobiosis. For 0.4% and 1% O2, cells were grown for 48 h on agar plates. For air, mid-exponential growing cells were exposed for 1 h to air in six-well plates at 37°C. For all experiments, the control condition was cells grown in anaerobiosis. gyrA was used as a reference gene. For all plots, a median with 95% CI is shown. Mann-Whitney statistical tests were performed between the ∆Ct of both conditions compared. *: P-value < 0.05; **<0.01; ***<0.001.
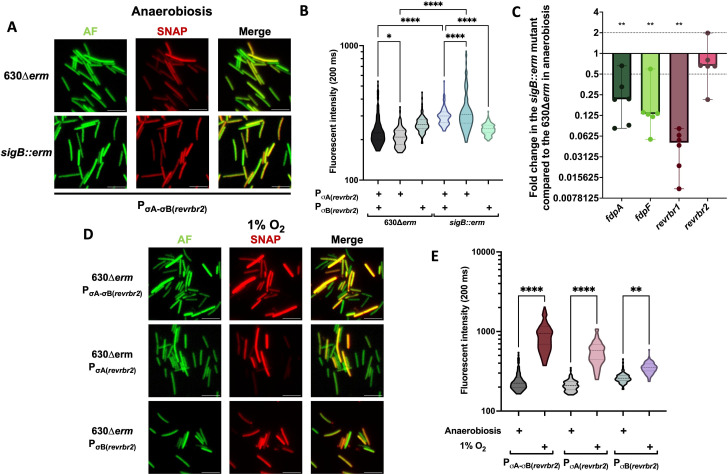
The revrbr2 gene is expressed under the dual control of a σA- and a σB-dependent promoter
Even if the two revRbr enzymes share 95% of identity at the protein level, different phenotypes for the single revrbr mutants were observed. Differences in the expression of the revrbr genes might contribute to the distinct role of revRbr1 in the tolerance to intermediate and high O2 tensions. Both genes have been previously shown to be controlled by σB (15, 20). However, the genome-wide transcriptional start site mapping indicated the presence of a second putative σA-dependent promoter upstream of the revrbr2 gene (Fig. S5A), whereas a second promoter seems to be absent upstream of the revrbr1 gene (38). This could explain a differential regulation. To confirm the existence of dual regulation of revrbr2, we grew the strain carrying a PσA-σB(revrbr2)-SNAPCd fusion on agar plates for 48 h in anaerobiosis. We were still able to detect fluorescence in the sigB::erm mutant (Fig. 5A) and the intensity of fluorescence even slightly increased in this mutant compared to the 630∆erm strain (Fig. 5B). In addition, in RT-qPCR experiments, the expression of the revrbr2 gene was not significantly downregulated in the sigB::erm mutant after growth on plates in anaerobiosis (Fig. 5C). These results confirm that the expression of revrbr2 is not strictly σB-dependent. To analyze more precisely the dual control of the revrbr2 gene, we removed either the σA- or the σB-dependent promoter in the PσA-σB(revrbr2)-SNAPCd fusion (Fig. S5A). A fluorescence was detected for all the constructions in the WT and sigB::erm backgrounds (Fig. 5B; Fig. S5B). These results indicate that the σA-dependent promoter also plays a role in the expression of the revrbr2 gene on plates.
Fig 5.
σA- and σB-dependent control of the revrbr2 gene expression in anaerobiosis and in the presence of O2. The expression of the revrbr2 gene was evaluated using a transcriptional fusion between the promoter region PσA-σB(revrbr2) and the SNAPCd fluorescent reporter gene. Constructions containing either only the PσA(revrbr2) or the PσB(revrbr2) were also used. All fusions were transferred to the 630∆erm and the sigB::erm mutant. Bacteria were cultured for 48 h on agar plates in anaerobiosis. Autofluorescence (AF), SNAP fluorescence, and merge are shown (A). Quantification of the fluorescence intensity of the SNAP reporter of images was performed (B). 600 cells out of 2 independent experiments were quantified. RT-qPCR experiments were performed using RNA extracted from the 630∆erm and the sigB::erm strains grown for 48 h on agar plates in anaerobiosis (C). Fold changes of expression in the sigB::erm mutant compared to the 630∆erm strain were plotted. Medians with 95% CI are shown. Six biological replicates were performed per experiment. The different fusions in the 630∆erm background were also exposed for 48 h on agar plates to 1% O2. Autofluorescence (AF), SNAP fluorescence, and merge are shown (D). Quantification of the fluorescence intensity of the SNAP reporter of images was performed (E). 600 cells out of 2 independent experiments were quantified. For RT-qPCR, Mann-Whitney statistical tests were performed, while one-way ANOVA were performed for the fluorescence intensity. *: P-value < 0.05; **<0.01, ***<0.001, ****<0.0001.
We also compared the expression of the revrbr1, fdpF and fdpA genes by RT-qPCR in the WT strain and the sigB::erm mutant grown on plates. We showed that the expression of the revrbr1, fdpA, and fdpF genes decreased in the sigB::erm compared to the WT strain (Fig. 5C). When using transcriptional fusions, we also showed that the fluorescence of the P(revrbr1)-SNAPCd and the P(fdpF)-SNAPCd fusion was undetectable in a sigB::erm mutant during growth on plates (Fig. S5C and D). This shows that the transcription of the revrbr1 gene is strictly σB-dependent, while the regulation of the revrbr2 gene is more complex, as the gene is expressed under the dual control of σA and σB. The differential physiological role of each revRbr could, at least partly, be explained by these differences of expression.
We then wanted to determine the possible role of σB, the sigma factor of the general stress response (20, 21), in the induction upon long-term exposure to 1% O2 of the genes encoding the O2-reductases. To test this hypothesis, we used the PσA-σB(revrbr2)-SNAPCd, the PσA(revrbr2)-SNAPCd, and the PσB(revrbr2)-SNAPCd fusions. In the 630∆erm strain, we first observed a significant increase in the fluorescence intensity of the PσA-σB(revrbr2)-SNAPCd fusion when cells were grown on plates in the presence of 1% O2 (Fig. 5A, D, and E), confirming the induction of revrbr2 expression at this O2 tension. Using the construction containing only the σB or the σA promoter, we still observed an increase in the fluorescence intensity in hypoxia (Fig. 5D and E). These results indicate that the induction by O2 of the revrbr2 expression is not exclusively σB-dependent.
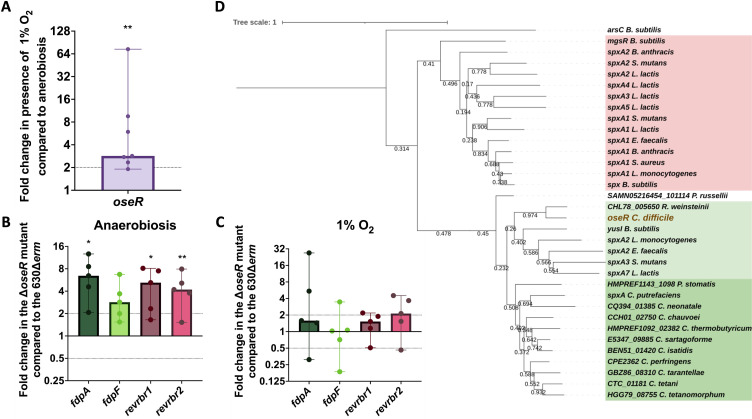
OseR, a new O2-sensing repressor, regulates all genes encoding O2-reductases
Since σB does not seem to be the main O2-sensing factor controlling revrbr and fdp genes, we thus looked for another regulator likely involved in the adaptation to O2, which would sense either directly O2 or the redox status of the cells. In Streptococcus mutans, the nox gene encoding the NADH oxidase, which reduces O2 to form H2O and protects the bacterium from ROS, is induced in the presence of O2 via a regulator of the Spx family, a family of redox sensing transcriptional factors (39–42). Thanks to a BLAST analysis, we proposed the uncharacterized CD1777 protein (renamed OseR for “Oxygen-sensitive Regulator”), which shares a high identity with regulators of the Spx family, as a good candidate to control the expression of the genes encoding the O2-reductases in C. difficile. Using 5′-RACE, we mapped a σB-dependent promoter upstream of the oseR gene (Fig. S6A) that is in agreement with the 10-fold downregulation of oseR expression in the transcriptome of the sigB::erm mutant (20, 21). The expression of the oseR gene is thus controlled by σB. We also showed that the expression of oseR was increased upon long-term exposure to 1% O2 (Fig. 6A). Then, we investigated whether OseR could be the regulator involved in the O2-dependent induction of fdp and revrbr genes. We compared by RT-qPCR the impact of oseR inactivation on the expression of the genes encoding the O2-reductases after growth of the 630∆erm or ∆oseR mutant in anaerobiosis or the presence of 1% O2. In anaerobiosis, we observed a derepression of fdpA, fdpF, revrbr1, and revrbr2 genes in the ∆oseR mutant compared to the 630∆erm strain (Fig. 6B), whereas this upregulation disappeared in the presence of 1% O2 (Fig. 6C). These results confirm that OseR is involved in the O2-dependent control of the expression of the revrbr and fdp genes. OseR most likely acts as a repressor of these genes in anaerobiosis, whereas O2 exposure releases the repression. In accordance, the ∆oseR mutant did not show any loss of survival compared to 630∆erm when exposed to 1% O2 for 48 h (Fig. S6C).
Fig 6.
The Spx-like regulator, OseR, regulates the genes encoding the O2-reductase in response to O2. The expression of the oseR gene in the 630∆erm strain exposed for 48 h to 1% O2 and in anaerobiosis was compared by RT-qPCR (A). The expression of the genes encoding the four O2-reductases was measured by RT-qPCR in the ∆oseR and the 630∆erm strain grown for 48 h on agar plates in anaerobiosis or at 1% O2. Fold changes correspond to the ratio between the RNA extracted from cells of the ∆oseR mutant and the 630∆erm strain in anaerobiosis (B) or at 1% O2 (C). Medians with 95% CI are shown. Five biological replicates were performed for each experiment. The sequence of known Spx and Spx-type proteins of Firmicutes was aligned with the sequence of OseR of C. difficile and a dendogram was constructed (D). Two sub-families are visible: highlighted in red, the canonical Spx and highlighted in green are Spx-like proteins of the YusI subfamily, including OseR (in brown) with two sub-families: in light green, the YusI-like subfamily and in dark green the other proteins of the YusI subfamily found in Clostridia. Mann-Whitney statistical tests were performed between the ∆Ct of the two conditions compared. *: P-value < 0.05; ** <0.01.
As many Firmicutes encode several proteins of the Spx family (40), we wanted to know whether OseR was a canonical Spx or a Spx-like protein. We thus compared the sequences of OseR to those of proteins of the Spx family in diverse Firmicutes and constructed the corresponding dendogram (Fig. 6D; Fig. S6B). The first striking feature is that there are two major branches in the dendogram, one with all the canonical Spx and some Spx-like proteins, like MgsR of B. subtilis (Fig. 6D, red), and a second branch containing more distant proteins and less well characterized Spx-like proteins, including YusI from B. subtilis (Fig. 6D, green). Using AlphaFold, we also performed a superposition of the potential structure of the OseR protein with that of YusI and Spx of B. subtilis. This suggested that the structures of OseR and YusI seem to be very similar (Fig. S6F) while an additional α-helix is present in Spx (Fig. S6G). OseR clearly belongs to the YusI subfamily of Spx-type proteins (40).
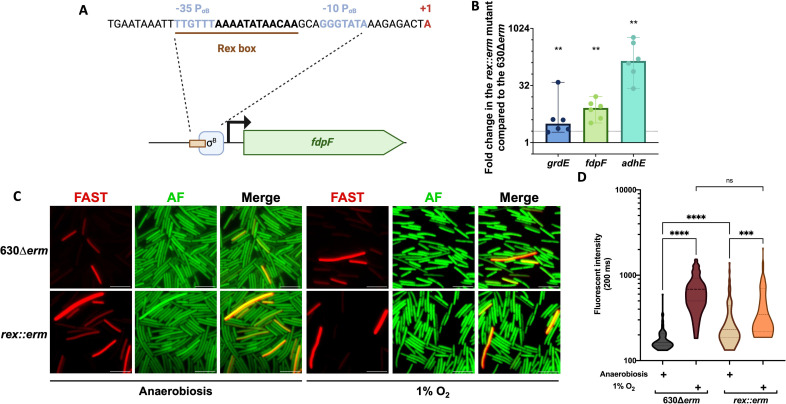
fdpF is also regulated by Rex
Since FdpF seems to participate more actively under high O2 tensions and air, we postulated whether this could be associated with a specific regulation. A putative Rex-binding site overlapping the −35 region of the σB-dependent promoter of fdpF (Fig. 7A) has previously been predicted (43), suggesting that Rex might control fdpF expression. To test this hypothesis, we compared the expression of fdpF in a rex::erm mutant and the 630∆erm strain. As a control, we also tested known Rex targets such as adhE encoding the alcohol dehydrogenase (24, 44, 45) and grdE encoding a protein involved in glycine reduction (44, 46, 47). After 48 h of growth on plates, the expression of adhE and grdE was upregulated as expected (Fig. 7B). The expression of fdpF was also significantly upregulated under similar conditions (Fig. 7B). To confirm the regulation of fdpF by Rex, we introduced a fusion between the fdpF promoter and the SNAPCd fluorescent reporter in the 630∆erm strain or the rex::erm mutant. In anaerobiosis, we were able to show a significant increase in the fluorescence intensity in the mutant compared to the parental strain (Fig. 7C and D), confirming that Rex represses fdpF expression in the absence of O2. Upon long-term exposure to 1% O2, we still observed induction of fdpF expression in the rex::erm mutant (Fig. 7C and D). In accordance, the rex::erm mutant has no differences in survival compared to the 630∆erm in the presence of 4% O2 and air, conditions where FdpF plays a major role (Fig. S6D and E).
Fig 7.
The control of fdpF gene expression by Rex. The identified Rex binding box in the promoter region of fdpF and the σB-dependent promoter are indicated (A). To confirm the regulation of fdpF by Rex, RT-qPCR experiments were performed on RNA extracted from the rex::erm mutant and the 630∆erm strain grown for 48 h on agar plates in anaerobiosis (B). Six biological replicates were performed. Medians with 95% CI are shown. Expression of fdpF was also tested using a transcriptional fusion between the promoter P(fdpF) and the SNAPCd fluorescent reporter gene introduced into the 630∆erm strain and the rex::erm mutant. Bacteria were grown for 48 h on agar plates in anaerobiosis or at 1% O2. Autofluorescence (AF), SNAP fluorescence, and merge are shown (C). Quantification of the fluorescence intensity of the SNAP reporter of images in panel C was performed (D). 600 bacteria out of 2 independent experiments were quantified. Mann-Whitney statistical tests were performed for qPCR and one-way ANOVA for fluorescence intensity. *: P-value < 0.05; **<0.01; ***<0.001; ****<0.0001.
DISCUSSION
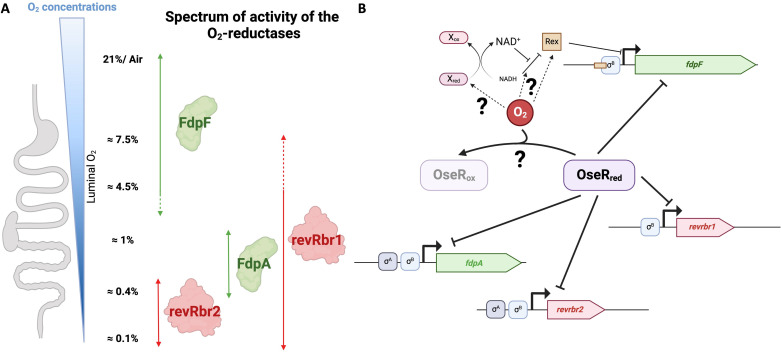
By combining our data with previous results (15), we showed that the four O2-reductases of C. difficile have different spectra of activity (Fig. 8A). revRbr2 is indeed more specific to the tolerance to very low O2 tensions (<0.4%), whereas FdpA is involved in the tolerance to low and intermediate tensions (0.4%–1% O2) and FdpF is important to resist high O2 tensions and air (4%–21% O2). The spectrum of action of revRbr1 is wider with a role ranging from 0.1% to 4% O2. It is worth noting that the two revRbr, sharing more than 95% of homology at the protein level (15), have distinct physiological roles with a more crucial role for the revRbr1 at intermediate to high O2 tensions. This is the first study showing a diversity of spectra of activity for O2 detoxication enzymes in an anaerobe. In C. difficile, the multiplicity of O2-reductases with different, yet overlapping spectra of activity, might allow complementary physiological roles covering the large range of the O2 tensions encountered during C. difficile infection cycle. Tensions from 0.1% to 0.4% O2 are found in the lumen of the colon during dysbiosis (48). The role of revRbr and FdpA is thus crucial for the initiation of colonization of the colon. 1% O2 is found at the mucus layer and near the epithelial cells that vegetative cells of C. difficile reach during infection (27, 28), and O2 tensions of 4% are found in the upper GIT, where spores germinate (29). revRbr1 and FdpF, which are important for the tolerance of vegetative cells to 4% O2, might play a role in the survival of cells before they attain the colon. It thus seems important for the bacterium to have an arsenal of active protection systems at such O2 tensions.
Fig 8.
Model of the role and regulation of O2-reductases of C. difficile. Spectrum of activity of C. difficile O2-reductases (A). The model is proposed based on the data from the phenotypical assays performed in this work and reference (15). The arrows indicate the activity limits of each O2-reductase. The dashed arrows indicate that the spectra of activity might be slightly underestimated since we tested only a limited number of O2 tensions. Model of the transcriptional regulation of the genes encoding the four O2-reductases (B). The genes fdpA and revrbr2 encoding the two enzymes more specific at the lowest O2 tensions are expressed under the control of both σB- and σA-dependent promoters, whereas fdpF and revrbr1 are transcribed only by σB. O2 induces the expression of the genes encoding the four enzymes in an OseR-dependent manner. OseR is a repressor under anaerobic conditions but does not repress the genes in the presence of O2. Rex represses the expression of the fdpF gene in response to the NADH/NAD+ ratio.
A striking result is the increased O2-reduction activity in crude extracts of the fdpA::erm mutant compared to those of the 630∆erm strain. We showed that this increase does not arise from transcriptional compensation, either through an overexpression of the fdpF gene or of the revrbr genes in this mutant or from a diminished electron transfer between the C-terminal domain of FdpF and FdpA, that would result in a larger availability of electrons to be used by FdpF. This later hypothesis was not observed in vitro, where no electron transfer was detected, and is supported by the model structures predicted for FdpF, that showed an arrangement that should impair a proper docking and electron transfer with FdpA. Electron transfer partners of FdpA and of revRbrs still remain to be identified even if we showed that the genes encoding the partners are not under the control of σB. Several proteins could correspond to NRORs in C. difficile, but we can already exclude those controlled by σB like CD0176. Only two proteins, FdpF and CD0828 (a putative oxidative stress glutamate synthase), contain a Rd domain suggesting that another type of small proteins involved in electron transfer such as flavodoxins or ferredoxins might be involved. Flavodoxins (eight present in C. difficile) are of a peculiar interest (49). Indeed, they are less sensitive to oxidative stress, as they rely on an FMN co-factor rather than O2-sensitive iron-sulfur clusters found in ferredoxins and would thus be a better electron transfer shuttle upon exposure to O2 (50, 51). Interestingly, some genes encoding flavodoxins (fldX and CD2825) are also induced upon O2 exposure in C. difficile (26, 49).
All genes encoding O2-reductases in C. difficile are controlled by the sigma factor of the general stress response, σB (20, 21). In liquid culture, the inactivation of the sigB gene abolished the expression of revrbr2, revrbr1, and fdpF but not of fdpA, which is expressed under the dual control of a σA- and a σB-dependent promoter (15). When grown for 48 h on plates, we confirmed that the expression of fdpF and revrbr1 is strictly σB-dependent, but this is not the case for the revrbr2 gene. Indeed, a revrbr2-SNAP fusion remains fluorescent in the sigB::erm mutant, due to the existence of a second σA-dependent promoter upstream of revrbr2 (38). During the late stationary phase on plates, the expression of revrbr2 seems to rely more on PσA than on PσB, while the opposite is observed in liquid culture. Even though σB plays a role in transcriptional control during the stationary phase (20, 52), a change in the regulation of revrbr2 can occur during the late stationary phase or on plates. The revrbr2 gene seems to be regulated by distinct regulatory programs (53, 54). This gene is mainly controlled by σB during planktonic growth and also by σA on plates, conditions closer to a microcolony/biofilm-like lifestyle. The differences could arise from a quorum-sensing-dependent control by an uncharacterized regulator or one of the already characterized systems in C. difficile (LuxS, RstA, or Agr) (54, 55). We also showed that revrbr2 expression is induced by O2 whether both or just one of the two promoters are present. More importantly, we demonstrated that the induction upon long-term exposure to O2 is not exclusively σB-dependent. This has also been previously shown with part of the genes encoding the thioredoxin systems of C. difficile under similar conditions (56).
Contrary to C. acetobutylicum, the PerR regulator does not seem to be involved in the control of the transcription of genes encoding the O2-reductases in C. difficile. We identified OseR, which belongs to the Spx family, as a transcriptional factor involved in the regulatory response to O2. In other Firmicutes, Spx can act both as a repressor and an activator (39, 57, 58). Spx-dependent activation is mediated via an interaction with the α subunit of the RNA polymerase and with promoter regions (59). Spx interacts with a DNA motif (AGCAW12AGCG) found upstream of some upregulated genes such as trxB of B. subtilis and nox of S. mutans (42, 60, 61). Moreover, upon oxidative stress, the oxidation of the CXXC motif leads to the formation of a disulfide bond, which increases Spx-dependent activation of transcription, by facilitating interactions between the RNA polymerase/Spx complex and the promoter region (59, 62, 63). However, this oxidation is not strictly required for Spx activation (64). OseR, like other clostridial regulators, belongs to the YusI subfamily of proteins rather than to canonical Spx proteins (Fig. 6; Fig. S6). Interestingly, a Spx or YusI homolog could not be identified in C. acetobutylicum. Few things are known of B. subtilis YusI or its closest relatives in other Firmicutes (40). The YusI subfamily seems to be divided into a group containing most Spx-type regulators found in Clostridia and a group containing YusI-type proteins from non-Clostridial Firmicutes and OseR from C. difficile. It is also interesting to note that YusI-type and Spx-type proteins share common motifs: the redox-sensing CXXC and a RPI motif (40, 57). However, all proteins of the YusI subfamily lack the Gly52 residue involved in the interaction of Spx with the α C-terminal domain of the RNA polymerase (57, 65, 66) and nothing is known on the mode of action of those proteins. We showed that OseR seems to act as a repressor of the expression of the fdp and revrbr genes in anaerobiosis, while this repression is released upon long-term exposure to 1% O2. OseR could directly sense O2, other sources of oxidative stress, like ROS, or more generally the redox state. Interestingly, in the OseR protein, the CXXC motif (Fig. S6H) is replaced by a CXXV motif (Fig. S6I). An intramolecular disulfide bond, increasing Spx activity in other Firmicutes, cannot be formed in OseR. However, the cysteine residue is exposed at the surface (Fig. S6I) suggesting a possible formation of intermolecular disulfide bonds (65, 67, 68). Residues involved in the interaction of Spx with either σA or the α C-terminal domain of the RNA polymerase (60) are poorly conserved in OseR, for instance, an important hydrophobic cluster in Spx of B. subtilis formed by L46, G52, V71, M74, L76, and L79 is replaced by hydrophilic residues in OseR (Fig. S6J) (60). In addition, neither the DNA motif recognized by Spx (59, 61) nor any other common DNA motif was found upstream of the revrbr and fdp genes of C. difficile, strongly suggesting a different uncharacterized mode of action for OseR. In B. subtilis, Spx negatively controls gene expression by a proposed anti-alpha mechanism (57), which could also be a possibility for OseR. Finally, in other Firmicutes, the Spx regulon is quite large and includes many genes involved in stress response (58). In this study, we only focused on the regulation by OseR of the genes encoding O2-reductases, but OseR may control other genes involved in oxidative stress response, like systems involved in thiol homeostasis, ROS detoxication, or DNA repair.
We showed that fdpF expression is repressed by Rex, a redox regulator that senses the NADH/NAD+ ratio (44, 69). However, we did not observe a significant Rex-dependent regulation of fdpF after long-term exposure to 1% O2 or any differences in survival of a rex::erm mutant exposed to increasing O2 tensions. In the late stationary phase, the metabolism is slowed down (70) and the NADH/NAD+ ratio can be stabilized. C. difficile metabolism relies on fermentation for ATP production. Fermentations and Stickland reductive pathways consume NADH leading to NAD+ formation. Glycolysis, the incomplete Krebs cycle, transamination reactions, and Rnf regenerate NADH from NAD+. As growth is halted during the stationary phase, fermentation pathways are probably slowed leading to a stabilization of the NADH/NAD+ ratio. In facultative anaerobes, Rex often controls the hierarchization of the metabolic pathways used by the bacteria. In Staphylococcus aureus, Rex acts as a repressor of fermentation pathways (71, 72) in aerobic environments and its target genes are derepressed upon growth in anaerobiosis, where fermentation will be favored (71). Rex also controls genes involved in oxidative stress response. In S. mutans, Rex directly regulates the nox gene encoding an O2 scavenging enzyme (42), whereas in C. acetobutylicum Rex regulates indirectly many genes encoding oxidative stress detoxication enzymes, including the revrbr and the fdp genes (73). In C. acetobutylicum, exposure to H2O2 reduces the NADH/NAD+ ratio leading to a repression of the Rex regulon. This control might increase the availability of NADH for detoxication enzymes (73). In C. difficile, it is interesting to note that fdpF is the sole O2-reductase gene directly controlled by Rex (43). In C. difficile, Rex represses alternative NAD+ regeneration pathways (44), suggesting that FdpF might act as one. Indeed, the standalone enzyme, FdpF, might be a more reliable and faster pathway for NAD+ regeneration than the other O2-reductases. This would be coherent with the data obtained in crude extracts, suggesting that FdpF is the fastest and most efficient O2-reducing enzyme of C. difficile and the combination of an increased NAD+ regeneration with an efficient O2 detoxication could be an advantage for cells exposed to O2. Finally, many stress-response-associated enzymes, including the O2-reductases, consume NADH for their activity (13). When FdpA and both revRbrs are active upon O2 exposure, a decrease in the NADH/NAD+ ratio might occur and thus leading to a repression of the Rex regulon, fdpF included.
Finally, a model of regulation of the four genes encoding O2-reductases in C. difficile can be proposed (Fig. 8B). These genes are all expressed under the control of σB, even if fdpA and revrbr2 are also transcribed by σA. It is interesting to note that the expression of fdpF and revrbr1, encoding the O2-reductases involved in the tolerance to higher O2 tensions, are strictly dependent on σB, the sigma factor of the general stress response. FdpF and revRbr1 are likely mainly produced under stressful conditions triggering σB activity (22). By contrast, fdpA and revrbr2 are also transcribed by σA, that ensure a basal level of O2 reduction and protection even at very low O2 tension. The production of FdpA and revRbr2 might be further enhanced under the control of σB when the stress increases. Moreover, we showed that all four genes are repressed in anaerobiosis by OseR, a regulator from the Spx/YusI family and that this repression is released upon exposure to O2, probably through oxidation of OseR. Finally, fdpF is also regulated by Rex, and thus is repressed upon increase of the NADH/NAD+ ratio, even though a direct or indirect link with exposure to O2 remains to be established. Multiple levels of controls exist. For instance, the fdpF gene is dually regulated by OseR and by Rex in addition to its control by σB. Other uncharacterized cross-regulations for fdpA and revrbr genes yet to be characterized might complete the proposed model in the future (Fig. 8B). A rather more complex regulatory network exists as the oseR gene itself is transcribed under the control of σB and its expression is induced by O2 (20, 21). In this paper, we focused on the transcriptional regulation of the O2-reductase genes, but we cannot exclude additional levels of control. First, as FdpA and both revRbr require electron transfer partners for their activity, the expression of the genes encoding those partners could also be regulated. Indeed, in C. acetobutylicum, the genes encoding the Rd and the NROR are also controlled by Rex (73). Control at post-translational levels could also modulate the activity or the stability of O2-reductases. Indeed, even though FdpA and both revRbr seem to have similar O2-reductase activity in vitro (15), their in vivo activity might also depend on the O2 tensions. In addition, the O2-reductases seem to be phosphorylated, with even FdpF being phosphorylated at multiple sites (74). The effect of phosphorylation on the activity of the Fdp or revRbr enzymes will deserve further investigations. As a conclusion, we started to decipher a complex multi-level network of control of the O2-reductases of C. difficile. The regulatory network identified strengthen the spectra of activity of the various enzymes and contributes to the different physiological roles of these multiple O2-reductases in an obligate anaerobe, exposed to many varying O2 tensions during its infectious cycle.
MATERIALS AND METHODS
Strains and growth conditions
Both Escherichia coli and C. difficile strains used in this study are listed in Table S1. E. coli strains were grown in LB (tryptone 10 g.L−1, yeast extract 5 g.L−1, and NaCl 5 g.L−1). C. difficile strains were grown under anaerobiosis (5% H2, 5% CO2, 90% N2) in TY medium (bacto tryptone 30 g.L−1, yeast extract 20 g.L−1, pH 7.4), BHI (Brain Heart Infusion 37 g.L−1) or a peptone-containing medium Pep-M [proteose peptone no. 2 at 40 g.L−1 (BD Diagnostics, USA), Na2HPO4 5 g.L−1, KH2PO4 1 g.L−1, NaCl g.L−1, MgSO4 0.1 g.L−1] (75). All media were solidified by the addition of 15 g.L−1 (BHI and TY) or 20 g.L−1 (LB) of agar. When necessary, antibiotics were added at the following concentrations: ampicillin (Amp) 100 µg.mL−1, chloramphenicol (Cm) 15 µg.mL−1, thiamphenicol (Tm) 15 µg.mL−1, erythromycin (Erm) 2.5 µg.mL−1, cefoxitin (Cfx) 25 µg.mL−1, and cycloserine 250 µg.mL−1. When needed, Taurocholate (Tau) at 0.05% wt/v was added to favor spore germination.
Plasmid and strain construction
The clostron gene knockout system (76, 77) was used to inactivate the rex gene (CD0171), yielding the insertional mutant strain 630Δerm rex::erm. We designed primers to retarget the group II intron of pMTL007 to insert it into the rex gene in sense orientation after nucleotide 193 in the coding sequence. The PCR product generated by overlap extension was cloned between the HindIII and BsrG1 sites of pMTL007 to obtain pDIA5908. C. difficile transconjugants obtained with E. coli HB101(RP4) containing pDIA5908 were selected on BHI agar containing Tm and Cfx and then plated on BHI agar containing Erm. We performed PCRs on the chromosomal DNA of transconjugants to verify the integration of the intron into the rex gene. The ∆oseR mutant was obtained using the CD25717.1-mediated allelic chromosomic exchange method. 1 kb fragments located upstream and downstream of this gene were PCR amplified from 630Δerm genomic DNA using the primer pair CM1-CM2 and CM3-CM4. Purified PCR fragments were then introduced into the pDIA6753 plasmid using a Gibson Assembly master mix (Biolabs). The plasmid obtained, pDIA6864, introduced in HB101(RP4) E. coli strain was transferred by conjugation into the C. difficile 630Δerm strain. Transconjugants were selected on BHI plates supplemented with Tm and C. difficile selective supplement (SR0096; Oxoid). Isolation of faster growing single crossing-over integrants was performed by serial restreaking on BHI plates containing Cfx and Tm. Single-crossover integrants were then restreaked on BHI plates supplemented with anhydrotetracycline (200 ng.mL−1), 1% glucose, and 0.1% cysteine, allowing the isolation of double-crossover events. After confirmation of plasmid loss (Tm-sensitive clones), the presence of the expected deletion in clones was checked by PCR using oligonucleotides CM5 and CM6.
Survival assays in the presence of intermediate/high O2 tensions or in air
A culture of C. difficile in TY Tau was inoculated at 1:50 from an overnight culture in the same medium. After 3 h of growth (OD600nm between 0.35 and 0.8), we prepared an inoculum at an OD600nm of 0.5 in TY Tau. For each strain, serial dilutions by 10 up to 10−5 were prepared. 5 µL of each dilution and the non-diluted inoculum were plated on calibrated square plates containing 28 mL of TY Tau agar to avoid the presence of spores. For all types of assays, a control plate was kept at 37°C in anaerobiosis for 24 h. The other plates were incubated at 37°C either in hypoxia (5% CO2, × % O2, 95 × % N2) or in air (21% O2) for various durations: 24 h and 48 h in the presence of 1% O2, 24 h in the presence of 4% O2 and 4 h in air. Plates exposed to those conditions were subsequently incubated again for 24 h at 37°C in anaerobiosis. Plates incubated for 48 h under hypoxia were not further incubated in anaerobiosis. CFU were numerated after incubations.
The inhibition of growth by air was tested in soft agar tubes (78). Briefly, 20 µL of an overnight culture of C. difficile grown anaerobically in the Pep-M medium was mixed with 10 mL of the Pep-M medium containing 0.4% agar at 45°C in a screw cap tube. The tubes were then incubated aerobically at 30°C for 40 h allowing the establishment of an O2 gradient. We measured the distance from the top of the agar to the edge of visible bacterial growth, that is, the zone of growth inhibition by air.
Determination of O2-reducing activities in C. difficile crude extracts
To prepare crude extracts, an overnight culture of C. difficile strains in TY was used to inoculate 10 mL of the same medium (1:50). After 8 h, this culture was diluted (1:100) in 200 mL of TY medium. After 16 h of growth, the OD600nm of the culture was measured and the cells were harvested by centrifugation at 6,000 rpm for 15 min at 4°C. Cells were resuspended in 25 mL of ice-cold sterile PBS, transferred to 50 mL tubes, and centrifugated. The pellets were resuspended in 10 mL of ice-cold sterile PBS and pelleted again by centrifugation for 10 min at 5,000 rpm and 4°C. Pellets were then stored at −20°C. Cells were resuspended in 50 mM Tris-HCl pH 7.5 containing 18% glycerol and disrupted by three cycles in a French-Press apparatus at 16,000 psi (Thermo) in the presence of 1 mg.mL−1 DNAse (Applichem). The crude extracts were centrifuged at 25,000 g for 30 min and at 138,000 g for 1.5 h at 4°C to remove cell debris and membrane fraction, respectively. The soluble fractions divided in aliquots were stored at −20°C.
The O2-reducing activity of the soluble fractions was measured amperometrically with a Clark-type electrode selective for O2 (Oxygraph-2K, Oroboros Instruments, Innsbruck, Austria). The assays were performed in 50 mM Tris–HCl, pH 7.5 containing 18% glycerol. The O2-reducing activity was evaluated at 25°C in an air-equilibrated buffer (≈250 µM of O2), in the presence of catalase (640 nM, from the bovine liver) and superoxide dismutase (240 nM, from bovine erythrocytes) to avoid the influence of ROS (superoxide and H2O2) formed by incomplete O2 reduction. The soluble fraction was added to the reaction mixture and the O2 concentration was allowed to stabilize (avoiding the variability originated by the different endogenous reductant concentrations). Then, the reaction was initiated by the addition of 5 mM of NADH. The turnover rates (s−1) were calculated by subtracting the experimental slope (µM.s−1) before and after the addition of NADH and dividing by the total protein amount used in each assay. The total protein concentration of the soluble fractions was determined by the Bradford method using bovine serum albumin as standard (79).
Electron transfer assays to FdpA
The anaerobic reduction of 30 µM of FdpA was performed in the presence of 60 µg.mL−1 and 50 µg.mL−1 of the soluble extracts of either 630∆erm or the sigB::erm mutant and 200 µM of NADH, and monitored by UV-Visible spectroscopy in a Shimadzu UV-1800 spectrophotometer, inside an anaerobic chamber (Coy Lab Products, Grass Lake, MI, USA).
The FdpA O2 reduction assays using FdpF C-Term or FdpF as electron donors were performed in 50 mM Tris-HCl, pH 7.5 containing 18% glycerol, at 25°C in an air-equilibrated buffer (≈250 µM of O2), in the presence of catalase (640 nM, from the bovine liver). Assays were initiated by the addition of 5 µM of either FdpF C-Term or FpdF and then 1 µM of FdpA.
Alphafold3 model structure predictions
The FdpF dimer model structure was predicted by Alphafold3 (80), using FdpF amino acid sequence and considering three iron atoms and 1 FAD molecule per protein monomer. The FMN molecules were added by running the PDB file generated by Alfaphold3 with Alphafill algorithm [https://alphafill.eu/ (81)]. Structure superimposition with the Rd and NADH:Rd oxidoreductase complex of Pseudomonas aeruginosa (PDB 2V3B), distance measurements, and figures were generated with ChimeraX v1.7.1 (82–84). Model structures of OseR from C. difficile and of YusI and Spx of B. subtilis were predicted by Alphafold3, using their respective amino acid sequences. Structure superposition was generated with ChimeraX v1.7.1.
RNA extraction, RT-qPCR, and 5′RACE
To check for compensation at the genetic level and the role of PerR, cells from the 630∆erm strain (both the perRT41A and perRWT) and the fdpA::erm mutant were harvested after 16 h of growth at 37°C in liquid TY medium. For hypoxia exposition, cells were harvested after 48 h of growth on a TY-calibrated agar plate incubated either in anaerobiosis or in hypoxia (0.4% or 1% O2) at 37°C. Cells from agar plates were resuspended in 3 mL of killing buffer (20 mM Tris HCl 7,5; 5 mM MgCl2; 20 mM NaN3) prior a rapid centrifugation. For air exposition, cells from an exponential phase culture were exposed for 1 h in air at 37°C in six-well plates containing 2 mL of medium per well to maximize the surface/volume ratio and optimize the gas exchange.
The culture pellets were resuspended in RNApro solution (MP Biomedicals) and RNA was extracted using the FastRNA Pro Blue Kit (MP Biomedicals). RNA was then purified using the Direct-Zol RNA Miniprep kit (Zymo). cDNA synthesis and quantitative PCR were performed as previously described (85, 86). Primers used are listed in Table S2. As both revRbrs share more than 95% of identity at the protein level, primers for RT-qPCR were carefully designed in regions that were different in revrbr1 and revrbr2 to distinguish each gene. The specificity of the primers was confirmed by RT-qPCR on RNA extracted from either a ∆revrbr1 or a ∆revrbr2 single mutant. The gyrA gene was used as a reference gene. The relative change in gene expression was determined using the ∆∆Ct approach (87).
To determine the transcription initiation site of oseR, the 5′ RACE (Rapid Amplification of cDNA Ends, Invitrogen kit) technique was used. mRNAs were extracted from the 630∆erm strain after 5 h of growth in TY. Using SuperScript II reverse transcriptase in the buffer provided [20 mM Tris-HCl (pH 8.4), 50 mM KCl, MgCl2 2.5 mM, dNTP 0.4 mM, DTT 0.01 mM], a single-strand cDNA was synthesized from a site internal to the unknown 5′ end of the oseR mRNA using the oligonucleotide IMV694. The matrix mRNA was eliminated by treatment with H and T1 RNAses. A polyC tail was then added to the 3′ ends of the cDNA via the terminal deoxynucleotidyl transferase. A PCR was then performed using a primer complementary to the polyC tail (containing an arbitrary sequence in 5′) and IMV753, a primer specific of oseR. A second amplification of the obtained PCR product was performed with oligonucleotides specific for the start of oseR (IMV1211) and a primer hybridizing with the arbitrary sequence located upstream of the polyC (Abridged Anchor Primer APP). The PCR products were cloned into the plasmid pGEMT easy (Promega). 4 to 8 plasmids corresponding to white colony-forming transformants were extracted and sequenced to determine the start of transcription.
Transcriptional SNAPCd fusions
The transcriptional fusions of the promoter region of revrbr1, revrbr2, fdpA, and fdpF with the SNAPCd reporter gene using the pFT47 vector have been already published (88, 89). The deletion of the σA and σB promoter (Fig. S3A) has been performed by inverse PCR using pDIA6459 (pFT47-PσA-σB(revrbr2)-SNAPCd) as template and oligonucleotides IMV1588 and IMV1576 or IMV1577 and IMV1578, respectively. This allowed to construct plasmids pDIA7285 (pFT47-PσA(revrbr2)-SNAPCd) and pDIA7290 (pFT47-PσB(revrbr2)-SNAPCd). The plasmids as well as the other SNAP fusions were introduced into E. coli HB101(RP4) and then transferred by conjugation into the C. difficile 630Δerm strain, the sigB::erm or the rex::erm mutant.
Fluorescence microscopy and image analysis
To monitor the expression of the different transcriptional fusions, the strains were grown for 48 h at 37°C on calibrated TY plates either in anaerobiosis or in the presence of 1% O2. The plates were initially inoculated using a 4 h culture of the different strains. SNAP labeling and fluorescence microscopy were performed as previously described (89). The images were taken with exposure times of 200 ms for autofluorescence and 300 ms for SNAP. Cells were observed on a Nikon Eclipse TI-E microscope 60× objective and captured with a CoolSNAP HQ2 Camera. For quantification of the SNAP-TMR Star signal resulting from transcriptional fusions, the mean fluorescence intensity of each bacterium was determined using ImageJ. 600 bacteria from two biological replicates were analyzed per condition.
Bioinformatic analysis
OseR was identified as a Spx family protein using the BLAST tool from the Uniprot website (www.uniprot.org) (90, 91) to BLAST Spx from B. subtilis against the database restricted to C. difficile. Spx-like regulators in Clostridia were also identified by using the BLAST tool from the Uniprot website to BLAST Spx from B. subtilis against the database restricted to Clostridia. Spx and Spx-like regulator sequences from Firmicutes (File S1) were aligned using the MEGA software (version 11.0.13) based on the MUSCLE algorithm (92). A distance tree was then constructed using neighbor-joining method with a JTT substitution model with 500 bootstraps. ArsC from B. subtilis (93) was used as an outgroup to root the tree. The final dendrogram was produced using iTOL (94).
Statistical analysis
For survival assays on a plate or in soft agar tubes, Mann-Whitney tests were performed. For qPCR, Mann-Whitney tests were performed between the ∆Ct of both conditions compared. The results obtained for the different mutant strains were compared with the wild type (630Δerm) ones (at least five assays per strain from three independent biological replicates of each strain) using ANOVA test with a confidence interval of 95%. All data are presented as the mean value ±standard deviation (SD). Finally, for microscopy quantification, one-way ANOVA followed by Tukey’s multiple comparison test was performed. Graphs and statistical analysis were performed using GraphPad Prism (version 10.2.1). Schematics were drawn using Biorender (www.biorender.com).
ACKNOWLEDGMENTS
We thank Bruno Dupuy for the helpful discussions and Olga Soutourina for the construction of the rex::erm mutant.
This work was supported by the ANR Difox (ANR-22-CE15-0026-01), by the Fondation pour la Recherche Médicale (grant numbers ECO202006011710 and FDT202304016494) and by the Institut Pasteur for the funding of the PhD contract of Cyril Anjou and by the Institut Universitaire de France for Isabelle Martin-Verstraete. This work was financially supported by the Portuguese Fundação para a Ciência e Tecnologia (FCT), PTDC/BIA-BQM/0562/2020 project, MOSTMICRO-ITQB R&D Unit (references UIDB/04612/2020 and UIDP/04612/2020), and LS4FUTURE Associated Laboratory (LA/P/0087/2020). MCM is a recipient of FCT PhD grant SFRH/BD/143651/2019.
Contributor Information
Isabelle Martin-Verstraete, Email: isabelle.martin-verstraete@pasteur.fr.
Eleftherios T. Papoutsakis, University of Delaware, Newark, Delaware, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01591-24.
Figures S1 to S6.
Tables S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. doi: 10.1038/nrdp.2016.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. doi: 10.1128/mSphere.00045-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seekatz AM, Safdar N, Khanna S. 2022. The role of the gut microbiome in colonization resistance and recurrent Clostridioides difficile infection. Therap Adv Gastroenterol 15:17562848221134396. doi: 10.1177/17562848221134396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kordus SL, Thomas AK, Lacy DB. 2022. Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat Rev Microbiol 20:285–298. doi: 10.1038/s41579-021-00660-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keeley TP, Mann GE. 2019. Defining physiological normoxia for improved translation of cell physiology to animal models and humans. Physiol Rev 99:161–234. doi: 10.1152/physrev.00041.2017 [DOI] [PubMed] [Google Scholar]
- 8. Rivera-Chávez F, Lopez CA, Bäumler AJ. 2017. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med 105:93–101. doi: 10.1016/j.freeradbiomed.2016.09.022 [DOI] [PubMed] [Google Scholar]
- 9. Velázquez OC, Lederer HM, Rombeau JL. 1997. Butyrate and the colonocyte, p 123–134. In Kritchevsky D, Bonfield C (ed), Dietary fiber in health and disease. Springer, Boston, MA, USA. [Google Scholar]
- 10. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17:662–671. doi: 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donohoe DR, Wali A, Brylawski BP, Bultman SJ. 2012. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One 7:e46589. doi: 10.1371/journal.pone.0046589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Litvak Y, Byndloss MX, Tsolis RM, Bäumler AJ. 2017. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol 39:1–6. doi: 10.1016/j.mib.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 13. Morvan C, Folgosa F, Kint N, Teixeira M, Martin-Verstraete I. 2021. Responses of Clostridia to oxygen: from detoxification to adaptive strategies. Environ Microbiol 23:4112–4125. doi: 10.1111/1462-2920.15665 [DOI] [PubMed] [Google Scholar]
- 14. Riebe O, Fischer R-J, Wampler DA, Kurtz DM, Bahl H. 2009. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology (Reading) 155:16–24. doi: 10.1099/mic.0.022756-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kint N, Alves Feliciano C, Martins MC, Morvan C, Fernandes SF, Folgosa F, Dupuy B, Texeira M, Martin-Verstraete I. 2020. How the anaerobic enteropathogen Clostridioides difficile tolerates low O2 tensions. mBio 11:e01559-20. doi: 10.1128/mBio.02678-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martins MC, Romão CV, Folgosa F, Borges PT, Frazão C, Teixeira M. 2019. How superoxide reductases and flavodiiron proteins combat oxidative stress in anaerobes. Free Radic Biol Med 140:36–60. doi: 10.1016/j.freeradbiomed.2019.01.051 [DOI] [PubMed] [Google Scholar]
- 17. Folgosa F, Martins MC, Teixeira M. 2018. Diversity and complexity of flavodiiron NO/O2 reductases. FEMS Microbiol Lett 365. doi: 10.1093/femsle/fnx267 [DOI] [PubMed] [Google Scholar]
- 18. Folgosa F, Martins MC, Teixeira M. 2018. The multidomain flavodiiron protein from Clostridium difficile 630 is an NADH:oxygen oxidoreductase. Sci Rep 8:10164. doi: 10.1038/s41598-018-28453-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giordano N, Hastie JL, Carlson PE. 2018. Transcriptomic profiling of Clostridium difficile grown under microaerophillic conditions. Pathog Dis 76. doi: 10.1093/femspd/fty010 [DOI] [PubMed] [Google Scholar]
- 20. Kint N, Janoir C, Monot M, Hoys S, Soutourina O, Dupuy B, Martin-Verstraete I. 2017. The alternative sigma factor σB plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environ Microbiol 19:1933–1958. doi: 10.1111/1462-2920.13696 [DOI] [PubMed] [Google Scholar]
- 21. Boekhoud IM, Michel A-M, Corver J, Jahn D, Smits WK. 2020. Redefining the Clostridioides difficile σB regulon: σB activates genes involved in detoxifying radicals that can result from the exposure to antimicrobials and hydrogen peroxide. mSphere 5:e00728-20. doi: 10.1128/mSphere.00728-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kint N, Alves Feliciano C, Hamiot A, Denic M, Dupuy B, Martin-Verstraete I. 2019. The σB signalling activation pathway in the enteropathogen Clostridioides difficile. Environ Microbiol 21:2852–2870. doi: 10.1111/1462-2920.14642 [DOI] [PubMed] [Google Scholar]
- 23. Kint N, Morvan C, Martin-Verstraete I. 2022. Oxygen response and tolerance mechanisms in Clostridioides difficile. Curr Opin Microbiol 65:175–182. doi: 10.1016/j.mib.2021.11.009 [DOI] [PubMed] [Google Scholar]
- 24. Neumann-Schaal M, Metzendorf NG, Troitzsch D, Nuss AM, Hofmann JD, Beckstette M, Dersch P, Otto A, Sievers S. 2018. Tracking gene expression and oxidative damage of O2-stressed Clostridioides difficile by a multi-omics approach. Anaerobe 53:94–107. doi: 10.1016/j.anaerobe.2018.05.018 [DOI] [PubMed] [Google Scholar]
- 25. Emerson JE, Stabler RA, Wren BW, Fairweather NFY. 2008. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J Med Microbiol 57:757–764. doi: 10.1099/jmm.0.47657-0 [DOI] [PubMed] [Google Scholar]
- 26. Weiss A, Lopez CA, Beavers WN, Rodriguez J, Skaar EP. 2021. Clostridioides difficile strain-dependent and strain-independent adaptations to a microaerobic environment. Microb Genom 7:000738. doi: 10.1099/mgen.0.000738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soavelomandroso AP, Gaudin F, Hoys S, Nicolas V, Vedantam G, Janoir C, Bouttier S. 2017. Biofilm structures in a mono-associated mouse model of Clostridium difficile infection. Front Microbiol 8:2086. doi: 10.3389/fmicb.2017.02086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Engevik MA, Engevik AC, Engevik KA, Auchtung JM, Chang-Graham AL, Ruan W, Luna RA, Hyser JM, Spinler JK, Versalovic J. 2021. Mucin-degrading microbes release monosaccharides that chemoattract Clostridioides difficile and facilitate colonization of the human intestinal mucus layer. ACS Infect Dis 7:1126–1142. doi: 10.1021/acsinfecdis.0c00634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawler AJ, Lambert PA, Worthington T. 2020. A revised understanding of Clostridioides difficile spore germination. Trends Microbiol 28:744–752. doi: 10.1016/j.tim.2020.03.004 [DOI] [PubMed] [Google Scholar]
- 30. Edwards AN, Karim ST, Pascual RA, Jowhar LM, Anderson SE, McBride SM. 2016. Chemical and stress resistances of Clostridium difficile spores and vegetative cells. Front Microbiol 7:1698. doi: 10.3389/fmicb.2016.01698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Troitzsch D, Zhang H, Dittmann S, Düsterhöft D, Möller TA, Michel A-M, Jänsch L, Riedel K, Borrero-de Acuña JM, Jahn D, Sievers S. 2021. A point mutation in the transcriptional repressor PerR results in a constitutive oxidative stress response in Clostridioides difficile 630Δerm. mSphere 6:e00091-21. doi: 10.1128/mSphere.00091-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hagelueken G, Wiehlmann L, Adams TM, Kolmar H, Heinz DW, Tümmler B, Schubert W-D. 2007. Crystal structure of the electron transfer complex rubredoxin–rubredoxin reductase of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104:12276–12281. doi: 10.1073/pnas.0702919104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martins MC, Alves CM, Teixeira M, Folgosa F. 2023. The flavodiiron protein from Syntrophomonas wolfei has five domains and acts both as an NADH:O2 or an NADH:H2 O2 oxidoreductase. FEBS J 291:1275–1294. doi: 10.1111/febs.17040 [DOI] [PubMed] [Google Scholar]
- 34. Hillmann F, Fischer R-J, Saint-Prix F, Girbal L, Bahl H. 2008. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol Microbiol 68:848–860. doi: 10.1111/j.1365-2958.2008.06192.x [DOI] [PubMed] [Google Scholar]
- 35. Hillmann F, Döring C, Riebe O, Ehrenreich A, Fischer R-J, Bahl H. 2009. The role of PerR in O2-affected gene expression of Clostridium acetobutylicum. J Bacteriol 191:6082–6093. doi: 10.1128/JB.00351-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sethu R, Gouré E, Signor L, Caux-Thang C, Clémancey M, Duarte V, Latour J-M. 2016. Reaction of PerR with molecular oxygen may assist H2O2 sensing in anaerobes. ACS Chem Biol 11:1438–1444. doi: 10.1021/acschembio.5b01054 [DOI] [PubMed] [Google Scholar]
- 37. Ji C-J, Kim J-H, Won Y-B, Lee Y-E, Choi T-W, Ju S-Y, Youn H, Helmann JD, Lee J-W. 2015. Staphylococcus aureus PerR is a hypersensitive hydrogen peroxide sensor using iron-mediated histidine oxidation. J Biol Chem 290:20374–20386. doi: 10.1074/jbc.M115.664961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soutourina O, Dubois T, Monot M, Shelyakin PV, Saujet L, Boudry P, Gelfand MS, Dupuy B, Martin-Verstraete I. 2020. Genome-wide transcription start site mapping and promoter assignments to a sigma factor in the human enteropathogen Clostridioides difficile. Front Microbiol 11:1939. doi: 10.3389/fmicb.2020.01939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schäfer H, Turgay K. 2019. Spx, a versatile regulator of the Bacillus subtilis stress response. Curr Genet 65:871–876. doi: 10.1007/s00294-019-00950-6 [DOI] [PubMed] [Google Scholar]
- 40. Rojas-Tapias DF, Helmann JD. 2019. Roles and regulation of Spx family transcription factors in Bacillus subtilis and related species, p 279–323. In Advances in microbial physiology. Vol. 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Antelmann H, Helmann JD. 2011. Thiol-based redox switches and gene regulation. Antioxid Redox Signal 14:1049–1063. doi: 10.1089/ars.2010.3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baker JL, Derr AM, Karuppaiah K, MacGilvray ME, Kajfasz JK, Faustoferri RC, Rivera-Ramos I, Bitoun JP, Lemos JA, Wen ZT, Quivey RG. 2014. Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol 196:2166–2177. doi: 10.1128/JB.01542-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ravcheev DA, Li X, Latif H, Zengler K, Leyn SA, Korostelev YD, Kazakov AE, Novichkov PS, Osterman AL, Rodionov DA. 2012. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J Bacteriol 194:1145–1157. doi: 10.1128/JB.06412-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bouillaut L, Dubois T, Francis MB, Daou N, Monot M, Sorg JA, Sonenshein AL, Dupuy B. 2019. Role of the global regulator Rex in control of NAD+-regeneration in Clostridioides (Clostridium) difficile. Mol Microbiol 111:1671–1688. doi: 10.1111/mmi.14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gencic S, Grahame DA. 2020. Diverse energy-conserving pathways in Clostridium difficile: growth in the absence of amino acid stickland acceptors and the role of the Wood-Ljungdahl pathway. J Bacteriol 202:e00233-20. doi: 10.1128/JB.00233-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andreesen JR. 2004. Glycine reductase mechanism. Curr Opin Chem Biol 8:454–461. doi: 10.1016/j.cbpa.2004.08.002 [DOI] [PubMed] [Google Scholar]
- 47. Neumann-Schaal M, Jahn D, Schmidt-Hohagen K. 2019. Metabolism the difficile way: the key to the success of the pathogen Clostridioides difficile. Front Microbiol 10:219. doi: 10.3389/fmicb.2019.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schnizlein MK, Young VB. 2022. Capturing the environment of the Clostridioides difficile infection cycle. Nat Rev Gastroenterol Hepatol 19:508–520. doi: 10.1038/s41575-022-00610-0 [DOI] [PubMed] [Google Scholar]
- 49. Troitzsch D, Knop R, Dittmann S, Bartel J, Zühlke D, Möller TA, Trän L, Echelmeyer T, Sievers S. 2024. Characterizing the flavodoxin landscape in Clostridioides difficile. Microbiol Spectr 12:e0189523. doi: 10.1128/spectrum.01895-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Campbell IJ, Bennett GN, Silberg JJ. 2019. Evolutionary relationships between low potential ferredoxin and flavodoxin electron carriers. Front Energy Res 7:79. doi: 10.3389/fenrg.2019.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zurbriggen MD, Tognetti VB, Fillat MF, Hajirezaei M-R, Valle EM, Carrillo N. 2008. Combating stress with flavodoxin: a promising route for crop improvement. Trends Biotechnol 26:531–537. doi: 10.1016/j.tibtech.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 52. Hecker M, Pané-Farré J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 61:215–236. doi: 10.1146/annurev.micro.61.080706.093445 [DOI] [PubMed] [Google Scholar]
- 53. Brauer M, Lassek C, Hinze C, Hoyer J, Becher D, Jahn D, Sievers S, Riedel K. 2021. What’s a biofilm?—How the choice of the biofilm model impacts the protein inventory of Clostridioides difficile. Front Microbiol 12:682111. doi: 10.3389/fmicb.2021.682111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meza-Torres J, Auria E, Dupuy B, Tremblay YDN. 2021. Wolf in sheep’s clothing: Clostridioides difficile biofilm as a reservoir for recurrent infections. Microorganisms 9:1922. doi: 10.3390/microorganisms9091922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rubio-Mendoza D, Martínez-Meléndez A, Maldonado-Garza HJ, Córdova-Fletes C, Garza-González E. 2023. Review of the impact of biofilm formation on recurrent Clostridioides difficile infection. Microorganisms 11:2525. doi: 10.3390/microorganisms11102525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anjou C, Lotoux A, Zhukova A, Royer M, Caulat LC, Capuzzo E, Morvan C, Martin-Verstraete I. 2024. The multiplicity of thioredoxin systems meets the specific lifestyles of Clostridia. PLoS Pathog 20:e1012001. doi: 10.1371/journal.ppat.1012001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol 186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakano S, Küster-Schöck E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A 100:13603–13608. doi: 10.1073/pnas.2235180100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakano MM, Lin A, Zuber CS, Newberry KJ, Brennan RG, Zuber P. 2010. Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One 5:e8664. doi: 10.1371/journal.pone.0008664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shi J, Li F, Wen A, Yu L, Wang L, Wang F, Jin Y, Jin S, Feng Y, Lin W. 2021. Structural basis of transcription activation by the global regulator Spx. Nucleic Acids Res 49:10756–10769. doi: 10.1093/nar/gkab790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reyes DY, Zuber P. 2008. Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation. Mol Microbiol 69:765–779. doi: 10.1111/j.1365-2958.2008.06330.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nakano S, Erwin KN, Ralle M, Zuber P. 2004. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol 55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x [DOI] [PubMed] [Google Scholar]
- 63. Lin AA, Walthers D, Zuber P. 2013. Residue substitutions near the redox center of Bacillus subtilis Spx affect RNA polymerase interaction, redox control, and Spx-DNA contact at a conserved cis-acting element. J Bacteriol 195:3967–3978. doi: 10.1128/JB.00645-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rojas-Tapias DF, Helmann JD. 2018. Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol Microbiol 107:659–674. doi: 10.1111/mmi.13906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Newberry KJ, Nakano S, Zuber P, Brennan RG. 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the α C-terminal domain of RNA polymerase. Proc Natl Acad Sci U S A 102:15839–15844. doi: 10.1073/pnas.0506592102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Birch CA, Davis MJ, Mbengi L, Zuber P. 2017. Exploring the amino acid residue requirements of the RNA polymerase (RNAP) α subunit C-terminal domain for productive interaction between Spx and RNAP of Bacillus subtilis. J Bacteriol 199:e00124-17. doi: 10.1128/JB.00124-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lamour V, Westblade LF, Campbell EA, Darst SA. 2009. Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase α subunit C-terminal domain complex. J Struct Biol 168:352–356. doi: 10.1016/j.jsb.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zuber P. 2009. Management of oxidative stress in Bacillus. Annu Rev Microbiol 63:575–597. doi: 10.1146/annurev.micro.091208.073241 [DOI] [PubMed] [Google Scholar]
- 69. McLaughlin KJ, Strain-Damerell CM, Xie K, Brekasis D, Soares AS, Paget MSB, Kielkopf CL. 2010. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol Cell 38:563–575. doi: 10.1016/j.molcel.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 70. Jaishankar J, Srivastava P. 2017. Molecular basis of stationary phase survival and applications. Front Microbiol 8:2000. doi: 10.3389/fmicb.2017.02000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pagels M, Fuchs S, Pané-Farré J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, von Wachenfeldt C, Liebeke M, Lalk M, Sander G, von Eiff C, Proctor RA, Engelmann S. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol 76:1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vesić D, Kristich CJ. 2013. A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J Bacteriol 195:1815–1824. doi: 10.1128/JB.02135-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zheng Y, Ko T-P, Sun H, Huang C-H, Pei J, Qiu R, Wang AH-J, Wiegel J, Shao W, Guo R-T. 2014. Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes. J Struct Biol 188:195–204. doi: 10.1016/j.jsb.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 74. Garcia-Garcia T, Douché T, Giai Gianetto Q, Poncet S, El Omrani N, Smits WK, Cuenot E, Matondo M, Martin-Verstraete I. 2022. In-depth characterization of the Clostridioides difficile phosphoproteome to identify Ser/Thr kinase substrates. Mol Cell Proteomics 21:100428. doi: 10.1016/j.mcpro.2022.100428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. 2013. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8:e56051. doi: 10.1371/journal.pone.0056051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. doi: 10.1016/j.mimet.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 77. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70:452–464. doi: 10.1016/j.mimet.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 78. Rocha ER, Tzianabos AO, Smith CJ. 2007. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. J Bacteriol 189:8015–8023. doi: 10.1128/JB.00714-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- 80. Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, et al. 2024. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630:493–500. doi: 10.1038/s41586-024-07487-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hekkelman ML, de Vries I, Joosten RP, Perrakis A. 2023. AlphaFill: enriching AlphaFold models with ligands and cofactors. Nat Methods 20:205–213. doi: 10.1038/s41592-022-01685-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27:14–25. doi: 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. doi: 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Meng EC, Goddard TD, Pettersen EF, Couch GS, Pearson ZJ, Morris JH, Ferrin TE. 2023. UCSF ChimeraX: tools for structure building and analysis. Protein Sci 32:e4792. doi: 10.1002/pro.4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193:3186–3196. doi: 10.1128/JB.00272-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée J-Y, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 88. Pereira FC, Saujet L, Tomé AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B, Henriques AO. 2013. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet 9:e1003782. doi: 10.1371/journal.pgen.1003782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cassona CP, Pereira F, Serrano M, Henriques AO. 2016. A fluorescent reporter for single cell analysis of gene expression in Clostridium difficile, p 69–90. In Roberts AP, Mullany P (ed), Clostridium difficile. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 90. Bateman A, Martin M-J, Orchard S, Magrane M, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bye-A-Jee H, Cukura A. 2023. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 51:D523–D531. doi: 10.1093/nar/gkac1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zaru R, Orchard S, UniProt Consortium . 2023. UniProt tools: BLAST, align, peptide search, and ID mapping. Curr Protoc 3:e697. doi: 10.1002/cpz1.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. doi: 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sato T, Kobayashi Y. 1998. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J Bacteriol 180:1655–1661. doi: 10.1128/JB.180.7.1655-1661.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S6.
Tables S1 and S2.