ABSTRACT
Borrelia burgdorferi, the agent of Lyme disease, is estimated to cause >400,000 annual infections in the United States. Serology is the primary laboratory method to support the diagnosis of Lyme disease, but current methods have intrinsic limitations that require alternative approaches or targets. We used a high-density peptide array that contains >90,000 short overlapping peptides to catalog immunoreactive linear epitopes from >60 primary antigens of B. burgdorferi. We then pursued a machine learning approach to identify immunoreactive peptide panels that provide optimal Lyme disease serodiagnosis and can differentiate antibody responses at various stages of disease. We examined 226 serum samples from the Lyme Biobank and the National Institutes of Health, which included sera from 110 individuals diagnosed with Lyme disease, 31 probable cases from symptomatic individuals, and 85 healthy controls. Cases were grouped based on disease stage and presentation and included individuals with early localized, early disseminated, and late Lyme disease. We identified a peptide panel originating from 14 different epitopes that differentiated cases versus controls, whereas another peptide panel built from 12 unique epitopes differentiated subjects with various disease manifestations. Our method demonstrated an improvement in B. burgdorferi antibody detection over the current two-tiered testing approach and confirmed the key diagnostic role of VlsE and FlaB antigens at all stages of Lyme disease. We also uncovered epitopes that triggered a temporal antibody response that was useful for differentiation of early and late disease. Our findings can be used to streamline serologic targets and improve antibody-based diagnosis of Lyme disease.
IMPORTANCE
Serology is the primary method of Lyme disease diagnosis, but this approach has limitations, particularly early in disease. Currently employed antibody detection assays can be improved by the identification of alternative immunodominant epitopes and the selection of optimal diagnostic targets. We employed high-density peptide arrays that enabled precise epitope mapping for a wide range of B. burgdorferi antigens. In combination with machine learning, this approach facilitated the selection of serologic targets early in disease and the identification of serological indicators associated with different manifestations of Lyme disease. This study provides insights into differential antibody responses during infection and outlines a new approach for improved serologic diagnosis of Lyme disease.
KEYWORDS: Lyme disease, diagnostics, peptide arrays, VlsE, FlaB, serology
INTRODUCTION
Lyme disease, caused by infection with the spirochete Borrelia burgdorferi, is the most common tick-borne disease in the United States (1). An estimated >400,000 B. burgdorferi infections occur annually, with the severity ranging from mild to a systemic febrile illness (2). For clinical purposes, Lyme disease is divided into early localized, early disseminated, and late stages (3). The infection starts at the site of the tick bite, where the spirochetes are deposited in the dermis, multiply, and spread centrifugally through the dermis. The interaction with the host’s innate immune system results in an expanding erythema migrans rash, the typical primary sign of the infection, and is classified as early localized Lyme disease. If untreated, spirochetes can enter the bloodstream, disseminate, and establish infection at distant sites, causing diverse clinical manifestations (4). Early disseminated Lyme disease presentations include multiple erythema migrans lesions, early Lyme neuroborreliosis, and Lyme carditis. The hallmark of late Lyme disease in the United States is Lyme arthritis (5, 6).
Most laboratory tests used to support the diagnosis of Lyme disease are based on the detection of the antibody responses against B. burgdorferi in serum. Due to the time interval between infection and production of a detectable antibody response, patients with erythema migrans are usually negative at presentation. The United States Centers for Disease Control and Prevention (CDC)-recommended standard two-tier algorithm is positive in about 40% and the modified two-tier algorithms in about 50% of acute-phase samples from patients with erythema migrans (7). While patients with erythema migrans typically receive antibiotic therapy based on potential for exposure and the clinical presentation, improvements in laboratory testing that would shorten this window period would be helpful.
Differential expression of outer surface proteins (Osp) enables B. burgdorferi to adapt to the diverse environments that the spirochete encounters in vertebrate and arthropod hosts and plays a key role in facilitating dissemination during vertebrate infection (8, 9). Along with a high degree of genetic heterogeneity among strains, variable antigenic expression plays a key role in the challenges of serological diagnosis of Lyme disease. A better understanding of temporal antigenic expression of B. burgdorferi could result in greater insights into pathogenesis as well as serological targets. Although comprehensive in vivo omics analyses of B. burgdorferi antigenic expression have been hampered by low spirochetemia, the examination of antibody responses could prove useful to identify stage-specific serologic indicators. In this study, we used the TBD-Serochip, a linear peptide microarray, to analyze IgG and IgM antibody responses to linear B. burgdorferi epitopes from patients diagnosed with different stages and manifestations of Lyme disease. We identified peptides that can be used to improve early diagnosis, as well as peptides that could be used to differentiate among disease manifestations and have the potential to improve antibody-based diagnosis of Lyme disease.
MATERIALS AND METHODS
TBD-Serochip
The Tick-Borne Disease Serochip (TBD-Serochip) is a slide-based peptide array used to catalog antibody responses to tick-borne pathogens (10). For each antigen selected for inclusion on the array, all protein sequences available as of October 2016 were downloaded from the NCBI protein database, aligned, and used to design 12-mer peptides that tile each protein with an 11-amino acid (aa) overlap to the preceding peptide in a sliding window pattern. Our prototype version of the TBD-Serochip included approximately 170,000 12-mer peptides per subarray and contained 12-mer peptides designed from antigenic sequences of eight tick-borne pathogens present in North America. For B. burgdorferi, this included 62 different antigens (including all paralogs) that are known to elicit an antibody response in humans (Fig. S1) (11, 12). For each antigen, we included the sequence of every genetic variant in the database for the 12-mer design. This included 12-mer peptides for 20 distinct OspC types and a wide range of recombinant sequences for VlsE. This approach enables the identification of all reactive portions for every examined antigen and demonstrates the impact of amino acid variation within a given epitope on antibody binding. Conversely, it can also inflate the number of significant reactive peptides due to cross-reactivity between different variants of the same 12-mer fragment (Fig. S2). The B. burgdorferi peptide component of the TBD-Serochip consisted of 91,338 peptides. The arrays were manufactured by Nimble Therapeutics.
Sample descriptions
The Lyme Disease Biobank
The Lyme Disease Biobank (LDB) sample repository includes well-characterized samples collected from patients with Lyme disease and healthy controls living in areas endemic for tick-borne disease (the Northeast and Upper Midwest) (13). The samples used in this study included sera from 38 confirmed acute Lyme disease cases, as determined by positive two-tiered serology, two positive ELISAs with erythema migrans > 5 cm, quantitative PCR (qPCR) and/or culture followed by PCR of the culture fluid for B. burgdorferi of whole blood from the acute-phase blood draw, or IgG seroconversion, with most being confirmed by two-tier serology (Table 1). The presence of an erythema migrans of >5 cm was noted in 25 patients (designated SEM-A), with six patients having >1 lesion. Four samples had evidence of B. burgdorferi infection by PCR and/or culture (Table 1). The cohort also included 31 probable Lyme disease cases, consisting of individuals with an erythema migrans rash of >5 cm but no confirmatory laboratory evidence (Table 2). We also included sera from 38 healthy controls living in endemic areas without a history of Lyme disease, all with negative serology. Samples were collected under IRB-approved protocols, and all participants provided written informed consent. Males represented the majority of enrolled cases (25 vs 13).
TABLE 1.
Description and testing data of the confirmed acute Lyme disease samples from the Lyme Disease Biobank cohorta,b
| Sample ID | Patient origin | EM > 5 cm at enrollment | Antibiotic therapy at enrollment (days) | B. burgdorferi culture | B. burgdorferi culture fluid PCR |
B. burgdorferi qPCR | Whole-cell lysate ELISA | C6 peptide ELISA | VlsE/ PepC10 |
Western blot IgM |
Western blot IgG |
Two-tier testing Result |
Initial discriminatory model prediction | Tester Set Prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LYM-997 | NY | YES | YES (1) | NA | NA | NEG | RE | POS | NA | POS | IT | POS | POS | NA |
| LYM-1237 | NY | YES | NO | NA | NA | NEG | RE | POS | NA | POS | IT | POS | POS | NA |
| LYM-1232 | WI | YES | YES (1) | NA | NA | NEG | NA | POS | POS | NEG | NEG | NEG | POS | NA |
| LYM-1227 | NY | YES | NO | NO | POS | NEG | NR | NEG | NA | IT | NEG | NEG | POS | NA |
| LYM-1214 | WI | YES | NO | NA | NA | NEG | NA | POS | NA | NEG | POS | POS | POS | NA |
| LYM-1206 | NY | YES | NO | NA | NA | NEG | BL | POS | NA | POS | IT | POS | POS | NA |
| LYM-1203 | NY | YES | NO | NO | POS | NEG | RE | POS | NA | IT | IT | NEG | POS | NA |
| LYM-1200 | NY | YES | YES (1) | NO | POS | NEG | RE | NEG | NA | POS | IT | POS | NEG | NA |
| LYM-1199 | NY | YES | NO | NO | NEG | NEG | NR | POS | NA | POS | IT | POS | NEG | NA |
| LYM-1191 | NY | YES | YES (1) | NA | NA | NEG | RE | POS | NA | POS | IT | POS | POS | NA |
| LYM-1160 | MA | YES | NO | NA | NA | NEG | RE | POS | NA | IT | IT | NEG | POS | POS |
| LYM-1131 | MA | YES | NO | POS | POS | NEG | NR | NEG | NA | IT | IT | NEG | NEG | NA |
| LYM-1114 | NY | YES | NO | NA | NA | POS | NR | POS | NA | IT | IT | NEG | NEG | NEG |
| LYM-1110 | NY | YES | NO | NA | NA | POS | NR | NEG | NA | NEG | IT | NEG | POS | NA |
| LYM-1107 | NY | YES | NO | NA | NA | NEG | RE | POS | NA | IT | POS | POS | POS | NA |
| LYM-1099 | WI | YES | NO | NA | NA | POS | NA | NEG | NEG | NEG | NEG | NEG | NEG | NEG |
| LYM-1097 | NY | YES | YES (10) | NA | NA | NEG | RE | POS | NA | POS | IT | POS | POS | POS |
| LYM-1093 | WI | YES | YES (1) | NA | NA | NEG | NA | POS | POS | POS | NEG | POS | POS | NA |
| LYM-1034 | WI | YES | YES (1) | NA | NA | NEG | NA | POS | POS | POS | NEG | POS | POS | NA |
| LYM-1031 | NY | YES | NO | NA | NA | NEG | BL | POS | NA | IT | IT | NEG | POS | NA |
| LYM-1016 | MA | YES | NO | NEG | NEG | NEG | RE | POS | NA | POS | POS | POS | POS | NA |
| LYM-1015 | MA | YES | YES (1) | NEG | NEG | NEG | NR | POS | NA | POS | IT | POS | POS | NA |
| LYM-1005 | NY | YES | NO | NA | NA | NEG | NR | POS | NA | POS | IT | POS | POS | POS |
| LYM-1002 | NY | YES | NO | NA | NA | NEG | NR | POS | NA | POS | IT | POS | POS | NA |
| LYM-1001 | NY | YES | NO | NA | NA | NEG | RE | POS | NA | IT | IT | NEG | POS | NA |
| LYM-998 | NY | NO | NO | NA | NA | NEG | RE | POS | NA | POS | IT | POS | POS | POS |
| LYM-1207 | WI | NO | NO | NA | NA | NEG | NA | POS | NA | NEG | NEG | NEG | POS | NA |
| LYM-1181 | NY | NO | NO | NA | NA | NEG | NR | POS | NA | POS | IT | POS | POS | NA |
| LYM-1127 | NY | NO | YES (1) | NEG | NEG | NEG | RE | POS | NA | IT | POS | POS | POS | NA |
| LYM-1055 | NY | NO | NO | NA | NA | NEG | BL | POS | NA | POS | IT | POS | POS | NA |
| LYM-1204 | NY | NO | NO | NA | NA | NEG | NR | POS | NA | POS | IT | POS | NEG | NA |
| LYM-1137 | MA | NO | NO | NEG | NEG | NEG | RE | POS | NA | POS | POS | POS | POS | NA |
| LYM-1123 | NY | NO | NO | NEG | NEG | NA | NR | POS | NA | POS | NEG | POS | POS | NA |
| LYM-1086 | WI | NO | YES (2) | NA | NA | NEG | NA | POS | POS | POS | NEG | POS | POS | NA |
| LYM-1080 | WI | NO | NO | NA | NA | NEG | NA | POS | POS | POS | POS | POS | POS | NA |
| LYM-1022 | WI | NO | YES (2) | NA | NA | NEG | NA | NEG | POS | POS | NEG | POS | POS | POS |
| LYM-1013 | NY | NO | NO | NA | NA | POS | BL | NEG | NA | NEG | IT | NEG | NEG | NA |
| LYM-1010 | NY | NO | NO | NA | NA | NEG | RE | NEG | NA | POS | IT | POS | POS | NA |
NA: data not available; RE: reactive; NR: not reactive; IT: indeterminate; BL: borderline; POS: positive; NEG: negative; NY; New York; WI: Wisconsin; MA: Massachussetts.
Samples with an infection identified by qPCR and/or culture are shown in bold. B. burgdorferi qPCR was performed in whole blood samples.
TABLE 2.
| Sample ID | Patient origin | EM > 5 cm at enrollment | Antibiotic therapy at enrollment (days) | B. burgdorferi qPCR | Whole-cell lysate ELISA |
C6 Peptide ELISA |
VlsE/ PepC10 |
Western blot IgM |
Western blot IgG |
Two-tier Testing result |
Discriminatory model prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LYM-1008 | NY | YES | NO | NEG | BL | NEG | NA | IT | IT | NEG | POS |
| LYM-1011 | NY | YES | NO | NEG | NR | NEG | NA | NEG | IT | NEG | POS |
| LYM-1048 | NY | YES | NO | NEG | NA | NEG | NEG | NEG | NEG | NEG | POS |
| LYM-1081 | NY | YES | NO | NEG | NR | POS | NA | IT | NEG | NEG | POS |
| LYM-1105 | NY | YES | NO | NEG | NA | NEG | POS | NEG | NEG | NEG | POS |
| LYM-1178 | NY | YES | NO | NEG | NR | NEG | NA | IT | IT | NEG | POS |
| LYM-1186 | NY | YES | YES (2) | NEG | NR | NEG | NA | IT | IT | NEG | POS |
| LYM-1210 | NY | YES | NO | NEG | NR | NEG | NA | IT | IT | NEG | POS |
| LYM-991 | NY | YES | NO | NEG | NR | POS | NA | IT | NEG | NEG | POS |
| LYM-1006 | WI | YES | NO | NEG | NR | NEG | NA | NEG | IT | NEG | NEG |
| LYM-1014 | NY | YES | NO | NEG | NR | NEG | NA | NEG | IT | NEG | NEG |
| LYM-1032 | NY | YES | NO | NEG | NR | NEG | NA | IT | IT | NEG | NEG |
| LYM-1039 | WI | YES | YES (1) | NEG | NA | IND | NA | NEG | NEG | NEG | NEG |
| LYM-1054 | WI | YES | NO | NEG | NA | NEG | NA | NEG | NEG | NEG | NEG |
| LYM-1082 | WI | YES | NO | NEG | NA | NEG | EQUIV | NEG | NEG | NEG | NEG |
| LYM-1083 | WI | YES | NO | NEG | NA | NEG | NEG | POS | NEG | NEG | NEG |
| LYM-1094 | NY | YES | NO | NEG | RE | NEG | NA | IT | IT | NEG | NEG |
| LYM-1096 | WI | YES | YES (1) | NEG | NA | NEG | NEG | NEG | NEG | NEG | NEG |
| LYM-1098 | WI | YES | YES (1) | NEG | NA | POS | NEG | NEG | NEG | NEG | NEG |
| LYM-1100 | WI | YES | YES (2) | NEG | NA | NEG | NEG | NEG | NEG | NEG | NEG |
| LYM-1109 | CA | YES | YES (1) | NEG | NR | NEG | NA | IT | NEG | NEG | NEG |
| LYM-1133 | UT | YES | NO | NEG | NR | NEG | NA | NEG | IT | NEG | NEG |
| LYM-1153 | WI | YES | NO | NEG | NA | NEG | NEG | NEG | NEG | NEG | NEG |
| LYM-1154 | NY | YES | YES (1) | NEG | NR | NEG | NA | POS | IT | NEG | NEG |
| LYM-1177 | NY | YES | NO | NEG | NR | POS | NA | IT | IT | NEG | NEG |
| LYM-1219 | NY | YES | NO | NEG | NR | NEG | NA | IT | NEG | NEG | NEG |
| LYM-1233 | NY | YES | NO | NEG | NR | NEG | NA | IT | NEG | NEG | NEG |
| LYM-1244 | NY | YES | NO | NEG | NA | NEG | NEG | NEG | NEG | NEG | NEG |
| LYM-1248 | NY | YES | NO | NEG | NA | NEG | NEG | NEG | NEG | NEG | NEG |
| LYM-989 | CA | YES | NO | NEG | RE | NEG | NA | IT | IT | NEG | NEG |
| LYM-993 | WI | YES | YES (1) | NEG | NA | NEG | NEG | NEG | NEG | NEG | NEG |
NA: data not available; RE: reactive; NR: not reactive; IT: indeterminate; BL: borderline; POS: positive; NEG: negative; EQUIV: equivocal; NY; New York; WI: Wisconsin; UT: Utah; CA: California.
B. burgdorferi qPCR was performed in whole blood samples.
The National Institutes of Health cohort
This cohort consisted of 82 patients diagnosed with Lyme disease and 47 healthy controls from an endemic area without a history of Lyme disease (Table 3). Serum samples were collected under clinical protocols approved by the National Institutes of Health (NIH) Institutional Review Board (ClinicalTrials.gov Identifier: NCT00028080 and NCT00001539), and written informed consent was obtained from all participants. Patients with Lyme disease acquired the infection in the mid-Atlantic region of the United States and fulfilled the 2017 CDC case definition of confirmed or probable Lyme disease (14). Patients were grouped according to their main clinical manifestations and disease stage. Most samples were collected after the start of antibiotic therapy (Table 3). The NIH cohort included 27 patients with single erythema migrans (designated SEM-C), 13 patients with multiple erythema migrans (MEM), 15 patients with acute Lyme neuroborreliosis (ALNB), and 27 patients with Lyme arthritis (LA). There was a male predominance among cases of neuroborreliosis and arthritis.
TABLE 3.
| Sample ID | Group | Gender | Age bracket | Direct microbiological evidence of Bb infection | Interval sample from start of antibiotic therapy (days) | Lyme EIA (C6 Peptide ELISA or VlsE/PepC10) | Western blot IgM | Western blot IgG | Two-tier testing | Cohort differential model | Match observed/predicted |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LA_01 | Lyme Arthritis | Female | >55 | POS | >45 | POS | NEG | POS | POS | LA | YES |
| LA_02 | Lyme Arthritis | Male | 41–55 | POS | pre-therapy | POS | NEG | POS | POS | LA | YES |
| LA_03 | Lyme Arthritis | Female | <20 | POS | >45 | POS | NEG | POS | POS | LA | YES |
| LA_04 | Lyme Arthritis | Male | >55 | POS | >45 | POS | POS | POS | POS | LA | YES |
| LA_05 | Lyme Arthritis | Male | 20–40 | POS | 3 to 8 | POS | POS | POS | POS | LA | YES |
| LA_06 | Lyme Arthritis | Male | <20 | POS | 22 to 45 | POS | NEG | POS | POS | LA | YES |
| LA_07 | Lyme Arthritis | Male | 41–55 | NA | >45 | POS | NEG | POS | POS | LA | YES |
| LA_08 | Lyme Arthritis | Male | <20 | POS | 22 to 45 | POS | NEG | POS | POS | LA | YES |
| LA_09 | Lyme Arthritis | Male | 20–40 | NA | >45 | POS | NEG | POS | POS | LA | YES |
| LA_10 | Lyme Arthritis | Male | 41–55 | NA | 22 to 45 | POS | NEG | POS | POS | LA | YES |
| LA_11 | Lyme Arthritis | Female | 41–55 | NA | 22 to 45 | POS | POS | POS | POS | LA | YES |
| LA_12 | Lyme Arthritis | Female | 41–55 | NA | >45 | POS | NEG | POS | POS | LA | YES |
| LA_13 | Lyme Arthritis | Female | >55 | NEG | >45 | POS | POS | POS | POS | LA | YES |
| LA_14 | Lyme Arthritis | Male | 41–55 | NEG | >45 | POS | NEG | POS | POS | LA | YES |
| LA_15 | Lyme Arthritis | Male | >55 | POS | 22 to 45 | POS | POS | POS | POS | LA | YES |
| LA_16 | Lyme Arthritis | Male | >55 | NA | >45 | POS | POS | POS | POS | LA | YES |
| LA_17 | Lyme Arthritis | Male | >55 | POS | >45 | POS | NEG | POS | POS | LA | YES |
| LA_18 | Lyme Arthritis | Female | <20 | POS | three to 8 | POS | NEG | POS | POS | LA | YES |
| LA_19 | Lyme Arthritis | Male | >55 | POS | >45 | POS | NEG | POS | POS | LA | YES |
| LA_20 | Lyme Arthritis | Male | >55 | NEG | >45 | POS | POS | POS | POS | LA | YES |
| LA_21 | Lyme Arthritis | Female | 41–55 | NA | >45 | POS | POS | POS | POS | LA | YES |
| LA_22 | Lyme Arthritis | Male | 20–40 | POS | 22 to 45 | POS | NEG | POS | POS | LA | YES |
| LA_23 | Lyme Arthritis | Male | 41–55 | NA | >45 | POS | NEG | POS | POS | LA | YES |
| LA_24 | Lyme Arthritis | Male | >55 | POS | >45 | POS | POS | POS | POS | LA | YES |
| LA_25 | Lyme Arthritis | Male | 41–55 | NA | 22 to 45 | POS | POS | POS | POS | LA | YES |
| LA_26 | Lyme Arthritis | Female | 41–55 | NEG | >45 | POS | NEG | POS | POS | LA | YES |
| LA_27 | Lyme Arthritis | Female | 20–40 | POS | 22 to 45 | POS | POS | POS | POS | LA | YES |
| MEM_01 | Multiple EM | Female | 41–55 | NA | 22 to 45 | POS | POS | POS | POS | MEM | YES |
| MEM_02 | Multiple EM | Male | >55 | NA | >45 | POS | POS | NEG | NEG | SEM-C | NO |
| MEM_03 | Multiple EM | Female | >55 | POS | 22 to 45 | POS | POS | POS | POS | SEM-C | NO |
| MEM_04 | Multiple EM | Female | >55 | NA | 22 to 45 | POS | POS | POS | POS | MEM | YES |
| MEM_05 | Multiple EM | Male | 41–55 | NA | 9 to 21 | POS | POS | POS | POS | LA | NO |
| MEM_06 | Multiple EM | Female | 41–55 | NA | 22 to 45 | POS | POS | POS | POS | MEM | YES |
| MEM_07 | Multiple EM | Female | 41–55 | NA | >45 | POS | POS | POS | POS | MEM | YES |
| MEM_08 | Multiple EM | Male | 41–55 | NA | 22 to 45 | POS | POS | NEG | POS | MEM | YES |
| MEM_09 | Multiple EM | Male | 20–40 | NA | 22 to 45 | POS | POS | NEG | NEG | MEM | YES |
| MEM_10 | Multiple EM | Female | 20–40 | NA | 22 to 45 | POS | POS | NEG | NEG | MEM | YES |
| MEM_11 | Multiple EM | Female | >55 | NEG | 22 to 45 | POS | POS | NEG | NEG | MEM | YES |
| MEM_12 | Multiple EM | Female | 41–55 | POS | 22 to 45 | POS | POS | NEG | NEG | SEM-C | NO |
| MEM_13 | Multiple EM | Female | 41–55 | NA | 22 to 45 | POS | POS | NEG | POS | ALNB | NO |
| ALNB_01 | Acute LNB | Male | >55 | NEG | 1 to 2 | POS | NEG | POS | POS | LA | NO |
| ALNB_02 | Acute LNB | Male | 41–55 | NEG | 1 to 2 | POS | POS | NEG | POS | Acute LNB | YES |
| ALNB_03 | Acute LNB | Male | 20–40 | NEG | 1 to 2 | POS | POS | POS | POS | Acute LNB | YES |
| ALNB_04 | Acute LNB | Female | 20–40 | NEG | 3 to 8 | POS | POS | NEG | POS | Acute LNB | YES |
| ALNB_05 | Acute LNB | Female | 41–55 | NEG | 9 to 21 | POS | POS | POS | POS | Acute LNB | YES |
| ALNB_06 | Acute LNB | Male | 20–40 | NEG | 9 to 21 | POS | POS | POS | POS | Acute LNB | YES |
| ALNB_07 | Acute LNB | Male | 20–40 | NEG | 3 to 8 | POS | POS | NEG | POS | Acute LNB | YES |
| ALNB_08 | Acute LNB | Male | 41–55 | NEG | 1 to 2 | POS | POS | NEG | POS | Acute LNB | YES |
| ALNB_09 | Acute LNB | Male | <20 | NEG | Pre-therapy | POS | POS | POS | POS | Acute LNB | YES |
| ALNB_10 | Acute LNB | Male | 20–40 | NEG | 9 to 21 | POS | POS | NEG | NEG | Acute LNB | YES |
| ALNB_11 | Acute LNB | Male | >55 | NEG | Pre-therapy | POS | POS | POS | POS | MEM | NO |
| ALNB_12 | Acute LNB | Male | 41–55 | NEG | 3 to 8 | POS | POS | NEG | POS | Acute LNB | YES |
| ALNB_13 | Acute LNB | Female | 41–55 | NEG | 9 to 21 | POS | POS | NEG | POS | Acute LNB | YES |
| ALNB_14 | Acute LNB | Male | 20–40 | NEG | 9 to 21 | POS | POS | POS | POS | Acute LNB | YES |
| ALNB_15 | Acute LNB | Male | <20 | NEG | Pre-therapy | POS | POS | POS | POS | Acute LNB | YES |
| SEM-C_01 | Single EM | Male | 20–40 | NA | >45 | POS | NEG | NEG | NEG | MEM | NO |
| SEM-C_02 | Single EM | Female | >55 | NA | 22 to 45 | POS | POS | NEG | POS | Single EM | YES |
| SEM-C_03 | Single EM | Female | >55 | NA | >45 | POS | NEG | NEG | NEG | Single EM | YES |
| SEM-C_04 | Single EM | Male | <20 | NA | 22 to 45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_05 | Single EM | Female | >55 | POS | 22 to 45 | POS | POS | NEG | POS | Single EM | YES |
| SEM-C_06 | Single EM | Female | 41–55 | NEG | 22 to 45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_07 | Single EM | Female | 20–40 | NEG | 22 to 45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_08 | Single EM | Male | >55 | NA | 9 to 21 | POS | POS | POS | POS | Single EM | YES |
| SEM-C_09 | Single EM | Female | <20 | POS | 3 to 8 | POS | NA | NA | NA | Single EM | YES |
| SEM-C_10 | Single EM | Female | 20–40 | POS | 22 to 45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_11 | Single EM | Female | 41–55 | POS | 22 to 45 | POS | NEG | POS | POS | Single EM | YES |
| SEM-C_12 | Single EM | Female | >55 | NA | 22 to 45 | POS | NEG | NEG | NEG | Single EM | YES |
| SEM-C_13 | Single EM | Male | 20–40 | NA | 22 to 45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_14 | Single EM | Male | >55 | NA | >45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_15 | Single EM | Female | >55 | POS | 22 to 45 | POS | NEG | POS | POS | Single EM | YES |
| SEM-C_16 | Single EM | Female | 41–55 | NA | 22 to 45 | NEG | NA | NA | NA | Single EM | YES |
| SEM-C_17 | Single EM | Female | >55 | POS | >45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_18 | Single EM | Male | >55 | NA | >45 | POS | POS | NEG | NEG | MEM | NO |
| SEM-C_19 | Single EM | Female | 41–55 | POS | 22 to 45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_20 | Single EM | Male | 20–40 | POS | 22 to 45 | NEG | NA | NA | NA | Single EM | YES |
| SEM-C_21 | Single EM | Female | 20–40 | NEG | 22 to 45 | POS | POS | POS | POS | LA | NO |
| SEM-C_22 | Single EM | Male | 41–55 | NEG | 22 to 45 | NEG | NA | NA | NA | Single EM | YES |
| SEM-C_23 | Single EM | Male | >55 | NA | >45 | POS | POS | NEG | NEG | Single EM | YES |
| SEM-C_24 | Single EM | Female | 20–40 | NA | 22 to 45 | POS | POS | NEG | POS | Single EM | YES |
| SEM-C_25 | Single EM | Female | 20–40 | NEG | Pre-therapy | NEG | NA | NA | NA | Single EM | YES |
| SEM-C_26 | Single EM | Male | 41–55 | NA | 22 to 45 | NEG | NA | NA | NA | SEM-A | NO |
| SEM-C_27 | Single EM | Male | >55 | NEG | 22 to 45 | POS | POS | POS | POS | SEM-A | NO |
Direct microbiological evidence of Borrelia burgdorferi infection by culture and/or PCR in blood, synovial fluid, skin biopsies, and/or cerebrospinal fluid.
NA: data not available; POS: positive; NEG: negative. LA: Lyme arthritis; MEM: multiple erythema migrans; ALNB: acute Lyme neuroborreliosis; SEM-C: single erythema migrans convalescent.
Array data analyses
The method for microarray assays is demonstrated in Fig. S1. Sera were tested at a 1:50 dilution. After incubation with sera and fluorescently labeled secondary anti-IgG and anti IgM antibodies, arrays were scanned on a NimbleGen MS 200 Microarray Scanner (Roche) at 2 µm resolution, with an excitation wavelength of 532 nm for Cy3/IgM and 635 nm for Alexa Fluor/IgG. After scanning, a file was generated that included a relative fluorescent unit (RFU) signal for each 12-mer peptide on the array. Next, an aggregate file was generated by combining data files from all subarrays, including 129 samples from the NIH cohort and 107 from the LDB cohort. The final aggregate file included combined data from all Lyme disease cohorts and controls (n = 236), which included 182,676 data points for B. burgdorferi and 91,338 each for IgG and IgM. The analyses were conducted separately for IgG and IgM data sets. The DESeq2 package in R was used to identify peptides with different signal intensity comparing control and case groups (15). Slide-to-slide variation was considered in the differential analysis. The FDR-adjusted P-values ≤ 0.05 were applied to obtain significantly different signal intensity among peptides, and only the peptides with increased signal intensity in the cases were selected. To further narrow down the numbers of potential sero-reactive peptides for the differential analysis, peptides were retained only if its signal intensity was greater than three times of the median signal intensity for all peptides (intensity threshold = 3,000) in at least 30% of the Lyme case samples (signal intensity >3,000 and case prevalence >30%). The peptide-array differential analysis was performed in R version 4.2.2 within RStudio. Data munging was performed by reshape2, dplyR, and tidyR packages in R. The array data have been deposited and are available under the following link: https://datadryad.org/stash/share/Ws_tDf9_WNMl524GfeM6mgYliBSIbCwNByQKZKpsEMA.
Generation of a classification model for Lyme disease
Once both the IgG and IgM sero-reactive peptides were identified, we implemented random forest analyses using the random forest package in R to evaluate their classification performance with subsets of sero-reactive peptides (16). In a random forest model, the measure of importance of a peptide is based on its mean decrease in impurity (MDI) value. For the initial selection of peptides, we calculated the mean MDI and used it as a threshold. We followed an iterative model building approach where peptides with MDI values above the mean MDI threshold were selected to build another model with better accuracy. This process was continued until no further improvement in accuracy was obtained with the subsequent model. Once the minimal number of peptides needed for diagnostic accuracy was selected, we pursued further classification with random forest model using the R package caret (17). For each iteration, our primary data set was randomly split into a training set (80%) and a testing set (20%). In addition, the models were trained with tenfold cross-validation. The receiver operating characteristic (ROC) analysis was conducted to illustrate performances of classification models, using the R package pROC (18). To accurately assess the performance and select the best models with biomarker combinations, the random resampling process was repeated 20 times, and the model with the median AUC score (area under the curve) was obtained to represent the performance of the final model classification.
RESULTS
Peptide selection—Lyme disease diagnostic model
We pursued a machine learning approach to identify reactive 12-mer linear peptides of B. burgdorferi that could be used in a stepwise fashion to (i) identify serologic signatures unique to Lyme disease and (ii) distinguish cohorts with different stages and/or manifestations of Lyme disease. We first used a case/control data set to identify the minimum set of peptides that could differentiate sera of patients with early Lyme disease from healthy controls. The Lyme disease cases consisted of 38 sera samples from confirmed early Lyme disease patients presenting with erythema migrans (LDB cohort), collected at the time of the diagnosis (acute sera) (Table 1). For controls, we used a merged data set of 85 sera samples from LDB (N = 38) and NIH (N = 47) cohorts. The combined case and control data set consisted of 123 samples. The initial differential analysis identified 1,169 (12.8%) IgG or IgM-reactive peptides with a significantly higher expression in cases vs controls. We used the random forest method to downselect this peptide panel into the minimum number of peptides with the lowest degree of predictive error. The final panel consisted of 62 reactive peptides (31 IgG and 31 IgM) and generated an error rate of 7.3% (Table 4). By using this panel, a total of 31 out of 38 early acute Lyme disease samples were predicted as cases. Of the 85 healthy controls, two were also classified as cases.
TABLE 4.
List of peptides and their importance for the Lyme disease diagnostic model
| Mean decrease Gini | Peptide sequence | Antigen | Antibody class |
|---|---|---|---|
| 4.34 | QIAAAIALRGRA | VlsE C6 | IgG |
| 3.55 | NQIAAAIALRGM | VlsE C6 | IgG |
| 3.14 | QIAAAIALRGMA | VlsE C6 | IgG |
| 2.62 | HIAAAIALRGMA | VlsE C6 | IgG |
| 1.98 | DNQIAAAIALRG | VlsE C6 | IgG |
| 1.79 | PIAAAIALRGMA | VlsE C6 | IgG |
| 1.72 | NPIAAAIALRGM | VlsE C6 | IgG |
| 1.63 | DQIAAAIALRGM | VlsE C6 | IgG |
| 1.57 | DQIAAAIALRGR | VlsE C6 | IgG |
| 1.22 | PAQEGAQQEGVQ | FlaB | IgG |
| 1.09 | AAMNGNDKIAAA | VlsE C6 | IgM |
| 1.04 | VQQEGAQQPALA | FlaB | IgM |
| 0.9 | QSAPVQEGVQQE | FlaB | IgG |
| 0.81 | DDHIAAAIALRG | VlsE C6 | IgG |
| 0.81 | DAGKLFAKKNDA | VlsE C3 | IgG |
| 0.77 | DAGKLFAKKNDE | VlsE C3 | IgG |
| 0.76 | AGDGGEKAGVKA | VlsE | IgM |
| 0.74 | LFGKAGAGGDSE | VlsE | IgM |
| 0.73 | DAGKLFAKKNDD | VlsE C3 | IgG |
| 0.72 | QEGAQQPALATA | FlaB | IgM |
| 0.71 | KDGKFAVKSNDE | VlsE C6 | IgM |
| 0.71 | GKLFAKKNDDGD | VlsE C3 | IgM |
| 0.7 | DDQIAAAIALRG | VlsE C6 | IgG |
| 0.66 | GVQQEGAQQPAL | FlaB | IgM |
| 0.65 | AGMNGNDKIAAA | VlsE C6 | IgM |
| 0.64 | IGEGNGDAEFNQ | VlsE | IgM |
| 0.64 | QAAPVQEGAQQE | FlaB | IgG |
| 0.63 | GKLFAKKNDDGD | VlsE C3 | IgG |
| 0.6 | VQQEGAQQPAPV | FlaB | IgM |
| 0.6 | GCNLDDNSKMER | S2 | IgG |
| 0.57 | CNLDDNSKMERE | S2 | IgG |
| 0.55 | QEGVQQEGAQQQ | FlaB | IgM |
| 0.55 | VKLTISDDLNKT | OspA | IgM |
| 0.54 | GCNLDDNSKIER | S2 | IgG |
| 0.53 | GGMNGNDKIAAA | VlsE C6 | IgM |
| 0.53 | LKNSEELNKKIE | OspC | IgM |
| 0.52 | QEGAQQEGVQAA | FlaB | IgM |
| 0.51 | EGAQQEGAQQPT | FlaB | IgM |
| 0.51 | KDGKFAVKKDEE | VlsE C6 | IgM |
| 0.5 | IKAIVDAAGNGG | VlsE | IgM |
| 0.49 | KDKDGKYSLDAT | OspA | IgM |
| 0.49 | CNLDDNSKMERK | S2 | IgG |
| 0.47 | NEDAGKLFAAKN | VlsE C3 | IgG |
| 0.45 | KGLNAKIDSLDV | BdrK | IgM |
| 0.44 | IVDAAGGGEQDG | VlsE | IgM |
| 0.43 | QQEGAQQPALAT | FlaB | IgM |
| 0.42 | CNLDDNSKIERK | S2 | IgG |
| 0.41 | QEGVQQEGAQQS | FlaB | IgM |
| 0.41 | EKQFGIKFDNLI | BdrN | IgM |
| 0.41 | QSAPVQEGVQQE | FlaB | IgM |
| 0.4 | VQDGVQQEGAQQ | FlaB | IgM |
| 0.38 | KDGKFAVKSDGD | VlsE C6 | IgG |
| 0.36 | QEGVQQEGAQQP | FlaB | IgM |
| 0.35 | DAGKLFAAKNAN | VlsE C3 | IgG |
| 0.35 | TNPIAAAIALRG | VlsE C6 | IgG |
| 0.35 | EGVQQEGAQQPA | FlaB | IgM |
| 0.32 | QAAPVQEGVQQE | FlaB | IgM |
| 0.32 | QEGVQQEGARQP | FlaB | IgM |
| 0.3 | PVQEGVQQEGAR | FlaB | IgG |
| 0.29 | KDGKFAVKDERE | VlsE C6 | IgG |
| 0.23 | QVAPVQEGVQQE | FlaB | IgG |
| 0.22 | VQEGVQQEGAQQ | FlaB | IgG |
Characterization of peptides in the diagnostic model
The selected 62 peptides mapped to 14 different regions within the B. burgdorferi proteome and often included multiple versions of the same 12-mer fragment, with each version containing variations in the amino acid (aa) sequence associated with differences in strain origin (Fig. 1). The majority of the 62 peptides originated from VlsE and FlaB and included the key peptides driving the diagnostic model (Table 4). Most of the VlsE peptides mapped to two invariable (IR) domains. Six IgG-reactive peptides and one IgM-reactive peptide were mapped to a IR3 and partial variable (VR3) fragment corresponding to aa 197 to 212 of the B31 strain (Fig. 1A). Twelve IgG-reactive peptides clustered within a 14-aa portion that corresponds to aa 4–17 (shown in bold) of the B31 IR6 (C6) sequence MKKDDQIAAAIALRGMAKDGKFAVKD. All these IgG-reactive peptides contained a conserved internal IAAAIALRG motif (Fig. 1B). In addition, three IgM-reactive peptides mapped 5 aa upstream of the IgG peptides, and all included a MNGNDKIAAA motif. Two IgG and two IgM peptides mapped to the C-terminal part of the C6, and they all contained a KDGKFAVK motif. All IgG (N = 15) and IgM (N = 6) FlaB peptides mapped to a highly reactive 23-aa fragment located within residues 207 and 229 (Fig. 1C). Combined, 46 out of the 62 peptides in our model included peptides within these three fragments in VlsE and FlaB. The remaining 16 peptides included five peptides that clustered within a 13-aa portion of the N terminus of the S2 antigen, as well as peptides from within Borrelia direct repeat proteins K and N, OspA, OspC, and other regions within VlsE (Table 4).
Fig 1.
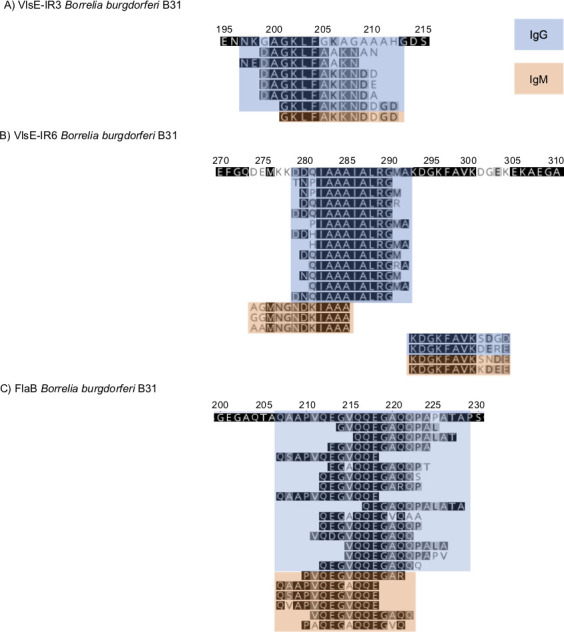
Mapping of the key VlsE (panels A and B) and FlaB (panel C) peptides identified by the diagnostic model, which differentiates between patients with Lyme disease and controls. The peptides were mapped to the B31 sequence. The numbers above the sequence correspond to the amino acid position in the protein. IgG peptides are indicated in blue and IgM in orange.
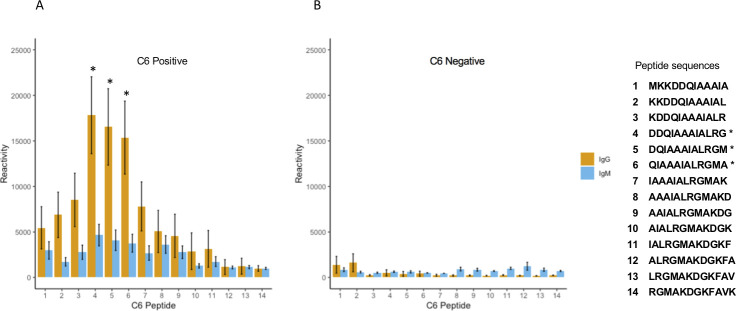
A closer examination of individual peptides within the C6 revealed predominantly IgG reactivity, which was mostly confined within the N-terminal half of the 26-aa sequence of the C6 (Fig. 2). Using the B31 C6 sequence as a reference, we noted that the fourth, fifth, and sixth peptides (DDQIAAAIALRG, DQIAAAIALRGM, and QIAAAIALRGMA) were the predominant reactive peptides in the samples from the LDB cohort. All three peptides were identified as key predictive drivers of our differential peptide panel.
Fig 2.
IgG and IgM reactivity of individual peptides that make up the C6 fragment. Shown is the reactivity of sera from SEM-A samples. Each number on the X axis corresponds to a unique 12-mer peptide sequence displayed on the right. Panel A shows the average from 30 C6 ELISA positive samples. Panel B demonstrates the average from eight C6 ELISA negative samples. The three dominant reactive peptides are indicated with *.
Two healthy control samples (LYM-518 and LYM-1211) were classified as Lyme disease by our model. We examined the array data to determine if these two samples yielded antibody signatures consistent with Lyme disease. Sample LYM-518, part of the NIH control cohort, generated elevated IgG reactivity against multiple peptides within the 207–229 FlaB fragment (Fig. S3). The misclassification of sample LYM-1211, from the LDB cohort, was less clear. Nonetheless, we did note slight (>3 fold) elevated reactivity to multiple VlsE and FlaB peptides in our model compared to controls that could account for the positive classification.
Prediction algorithm results
Using the same 123-sample data set, we trained our model on a set of randomly selected 99 samples (80% of the data set) and then used it on a tester set that consisted of the remaining 24 samples (20% of the data set). The tester set included seven Lyme disease cases and 17 healthy controls. The clinical status of five out of seven Lyme disease cases and all controls were correctly predicted (AUC = 0.96). Of the five predicted Lyme disease cases, four were positive by the standard two-tiered (STT) algorithm. The two Lyme disease samples classified as controls by our model (LYM-1099 and LYM-1114) were B. burgdorferi PCR-positive and STT-negative, respectively, although LYM-1114 was positive by a commercial C6 peptide ELISA. However, neither sample displayed any significant reactivity with any of the C6 peptides on the array.
We next employed our model on 31 sera of patients with probable Lyme disease from the LDB cohort. All samples were classified as STT-negative, although four samples had a positive C6-peptide ELISA, and several others had a single positive whole-cell ELISA or Western Blot IgM test (Table 2). Our model predicted nine samples (29%) as representing subjects with Lyme disease. These included two samples (LYM-991 and LYM-1081) that were positive on the C6-peptide ELISA.
We also determined if clinical features could be used as a predictor of positive serology in both confirmed and probable cases of early Lyme disease. There was no significant correlation between the size of the erythema migrans rash or the presence of multiple symptoms with positive results as determined by our model (Wilcoxon test, P = 0.65 and 0.52, respectively).
Next, we applied our model to predict the Lyme disease status of subjects in the NIH cohort comprising 82 cases and 47 healthy controls. Of the 82 cases, 77 were positive by STT or C6 alone with the five negative samples, all from the SEM-C group. Our model correctly identified all controls and 81 out of 82 Lyme disease samples. The lone misclassified sample, LYM-465, was an SEM-C sample, which, upon review, was non-reactive for all B. burgdorferi antigens on the array. This sample was also negative by a commercial C6 ELISA and STT.
FlaB and C6 include the major immunodominant linear epitopes of B. burgdorferi
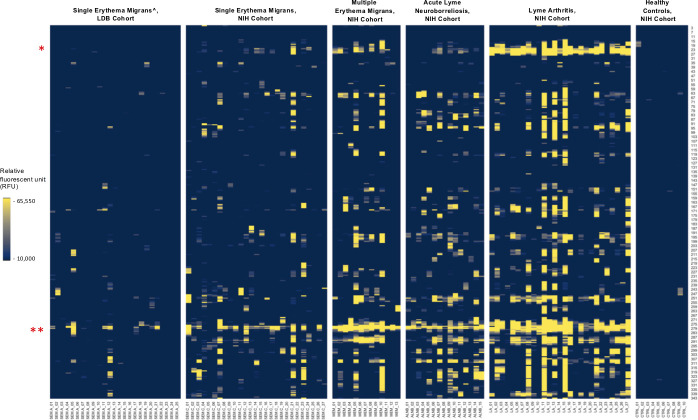
Because VlsE and FlaB peptides were the key peptides in our diagnostic model, we examined the array intensity to determine antibody abundance to these targets relative to other antigens. For each of the five Lyme disease groups, we identified peptides reactive in the cases vs all 85 controls and sorted them by intensity. As anticipated, we observed a wide range of redundancy among the reactive peptides. Nonetheless, intensity data revealed that the key peptides from the VlsE C6, VlsE IR3, and the 207–229 FlaB fragments that were driving diagnosis in our model were also among the most immunoreactive peptides on the array throughout all five groups (Fig. 3). The lone exception was in the ALNB group, where the FlaB 207–229 peptides displayed lower IgG reactivity, but instead were the highest reactive IgM targets (Fig. 4; Fig. S4).
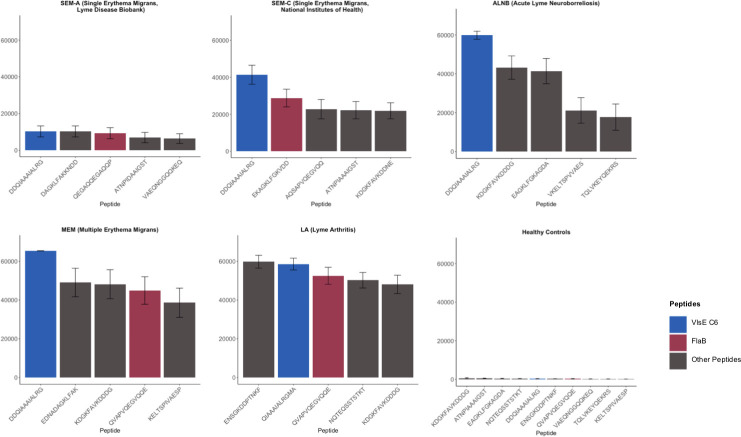
Fig 3.
List of top five reactive peptides from each Lyme disease group. A single representative peptide was selected for each reactive epitope. The C6 representative peptide are shown in blue, the FlaB peptides are shown in red; the rest of the peptides are shown in gray. The SEM-A cohort included only confirmed Lyme disease cases.
Fig 4.
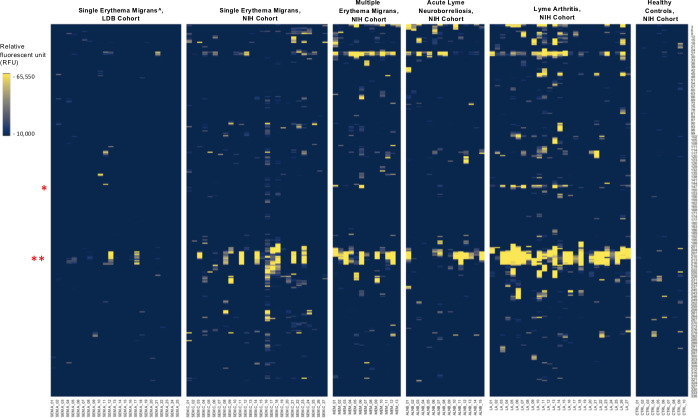
IgG reactivity to FlaB peptides in the five sample types. The Y axis indicates the relative amino acid coordinates of the peptides within the full-length FlaB protein. Reactivity is shown in yellow. For clarity, only peptides with the reactivity above 10,000 RFU are shown. Samples are indicated on the X axis. To illustrate baseline reactivity, ten random normal control samples were selected and shown on the right. The red asterisks indicate the position of the key regions identified by our models. * indicates the peptides within residues 147–158; ** - indicates the highly immunoreactive region encompassing residues 207–229; ^ - includes only confirmed Lyme disease cases.
Cohort differential model
Next, we used the random forest approach to identify a panel of peptides that could differentiate between different clinical manifestations of Lyme disease. Our combined data set consisted of 107 samples and included 25 SEM-A samples that had an erythema migrans >5 cm from the LDB cohort (Table 1) and the four Lyme disease types (SEM-C, MEM, ALNB, and LA) from the NIH cohort. By downselecting the number of differential peptides with a random forest model, we selected 20 peptides as the optimal combination with an OOB error of 12.15% (Table 5). The model provided 100% accuracy in predictions for SEM-A (25/25) and LA (27/27) samples. The predictive accuracy for SEM-C samples was 81.5% (22/27), with two samples classified as SEM-A, two samples as MEM, and one sample as LA. For patients with ALNB, the prediction was 87% (13/15), with one sample classified as LA and another as MEM. The lowest accuracy was observed for MEM samples, with six out of 13 samples misclassified. One sample was misclassified as ALNB, two as LA, and three as SEM-C. We generated a three-dimensional principal component analysis (PCA) plot using the IgG raw intensity values of the 20 peptides selected by our model to visualize the separation of the five groups (Fig. 5). We observed a clear separation for the LA group and an association of the selected peptides with late disease.
TABLE 5.
List of peptides and their importance in the differential model
| Mean decrease gini | Sequence | Antibody class | Antigen | Sequence origin - Annotation |
|---|---|---|---|---|
| 5.330794088 | MIINHNTSAINA | IgG | FlaB | NP_212281.1 flagellin [Borreliella burgdorferi B31] |
| 5.177076454 | GKDDPFSAYIKV | IgG | p66 | NP_212737.1 integral outer membrane protein p66 [Borreliella burgdorferi B31] |
| 4.857495488 | NNQTEQSSTSTK | IgG | p66 | NP_212737.1 integral outer membrane protein p66 [Borreliella burgdorferi B31] |
| 4.669973934 | DKDDPTNKFYQS | IgG | VlsE N | YP_004940414.1 outer surface protein VlsE1 (plasmid) [Borreliella burgdorferi B31] |
| 4.512979257 | TAEELGMQPAKT | IgG | FlaB | ABW79842.1 flagellin, partial [Borreliella burgdorferi] |
| 4.501736876 | SGKDDPTNKFYQ | IgG | VlsE N | ADQ30189.1 vlsE protein (truncated), partial (plasmid) [Borreliella burgdorferi JD1] |
| 4.446413545 | LGKDDPFSAYIK | IgG | p66 | NP_212737.1 integral outer membrane protein p66 [Borreliella burgdorferi B31] |
| 4.325870964 | ENSGKDDPTNKF | IgG | VlsE N | ADQ30189.1 vlsE protein (truncated), partial (plasmid) [Borreliella burgdorferi JD1] |
| 4.268780356 | ESIKNEFLNKGF | IgM | BdrK | ADQ44869.1 BdrK (plasmid) [Borreliella burgdorferi 297] |
| 4.124715706 | KDDPTNKFYQSV | IgG | VlsE N | YP_004940414.1 outer surface protein VlsE1 (plasmid) [Borreliella burgdorferi B31] |
| 4.025256035 | MAKDGKFAVKKG | IgG | VlsE C6 | ACD00653.1 VlsE, partial (plasmid) [Borreliella burgdorferi] |
| 3.971709872 | KDGKFAVKSGGG | IgG | VlsE C6 | ACD00984.1 VlsE, partial (plasmid) [Borreliella burgdorferi] |
| 3.968590071 | KDDDAKAFGKGK | IgG | VlsE | ACN55594.1 outer surface protein VlsE (plasmid) [Borreliella burgdorferi WI91-23] |
| 3.842963268 | GKKPADAKNPIA | IgM | VlsE V5 C5 | ACD00940.1 VlsE, partial (plasmid) [Borreliella burgdorferi] |
| 3.744512829 | ILKAIVEAAGVS | IgG | VlsE | ACN55594.1 outer surface protein VlsE (plasmid) [Borreliella burgdorferi WI91-23] |
| 3.738322943 | NAAAFGGNMKKK | IgG | VlsE V6-C6 | WP_002662199.1 variable large family protein [Borreliella burgdorferi] |
| 3.736929574 | ANGDAGHLFAAA | IgM | VlsE | ACO38545.1 borrelia lipoprotein (plasmid) [Borreliella burgdorferi 29805] |
| 3.339991462 | TAEELGMQPAKI | IgG | FlaB | NP_212281.1 flagellin [Borreliella burgdorferi B31] |
| 3.313878237 | DGAEFNKEGMKK | IgM | VlsE | ACC99986.1 VlsE, partial (plasmid) [Borreliella burgdorferi] |
| 3.168644553 | KKPGDAKNPIAA | IgM | VlsE V5-C5 | ACD01023.1 VlsE, partial (plasmid) [Borreliella burgdorferi] |
Fig 5.
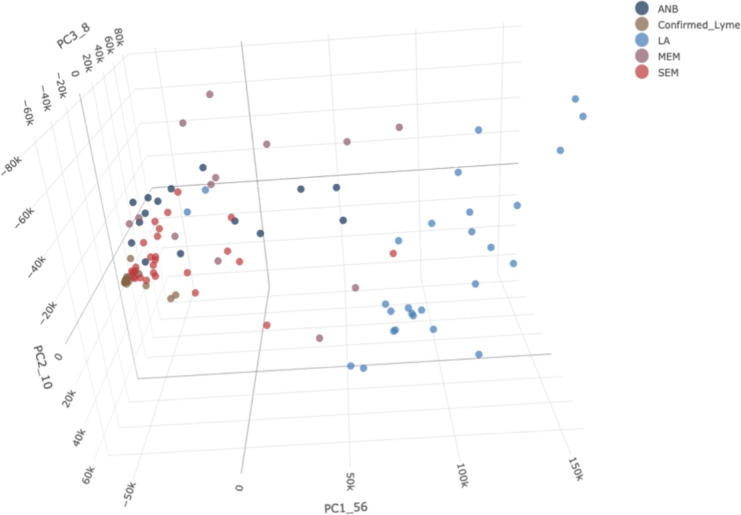
PCA plot for individual samples from the five groups. Each point on the plot represents an individual sample. The different colors represent samples from subjects with different types of disease. SEM-A: , SEM-C: The full 3D PCA plot can be accessed at https://magical-muffin-f665c4.netlify.app/.
Characterization of peptides in a differential model
The differential model was driven by IgG-reactive peptides from FlaB, p66, and VlsE. Similar to the diagnostic model, there was redundancy within the selected peptide panel, with 12 distinct regions represented within the 20 peptides. The three FlaB peptides mapped to two regions; the key peptide driving the model was the peptide MIINHNTSAINA encompassing the first 12 aa residues of FlaB (Fig. 5). This peptide was not reactive in SEM-A and SEM-C samples. Two redundant peptides mapped to coordinates 147–158 and were reactive primarily in LA samples (Fig. 5). The three p66 peptides originated from two dominant reactive regions (Fig. 6). Two peptides were mapped to aa 78–90 and consisted of the aa sequence LGKDDPFSAYIKV that was highly reactive in most of LA sera. Another p66 peptide mapped to aa 497–508 and consisted of the sequence NNQTEQSSTSTK that was highly reactive in the majority of LA samples and, overall, was among the highest reactive peptides in this cohort (Fig. 3). These p66 peptides were mostly nonreactive in SEM-A, SEM-C, and ALNB sera and reactive in only four of 13 MEM sera.
Fig 6.
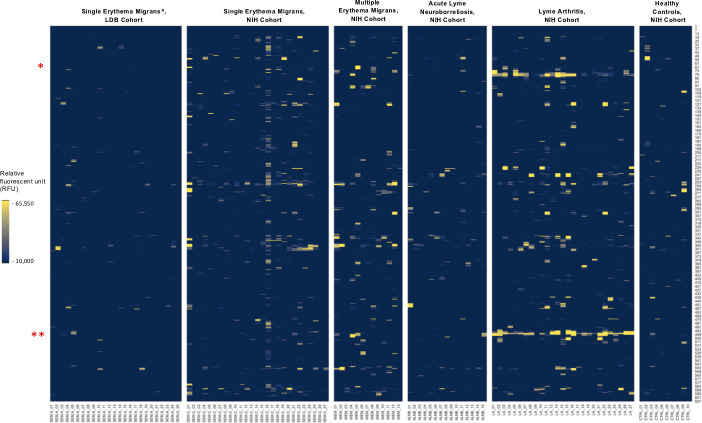
IgG reactivity to p66 peptides in the five sample types. The Y axis indicates the relative amino acid coordinates of the peptides within the full-length p66 protein. The reactivity is shown in yellow. For clarity, only peptides with reactivity above 10,000 RFU are shown. Samples are indicated on the X axis. To illustrate baseline reactivity, ten random control samples were selected and shown on the right. The red asterisks indicate the position of the key peptides identified by our differential model, at position 78–90 and 497–508. ^ - includes only confirmed Lyme disease cases.
Thirteen peptides from seven different fragments mapped to VlsE. Four of them were from the N terminal region of the protein (VlsE 18–38), and all included a conserved KDDPTNKF motif (Fig. 7). The immunoreactivity to this region was strongly associated with late disease (Fig. 8). Along with C6, peptides within this region were the most reactive of all Borrelia peptides in the LA samples (Fig. 3). Other VlsE peptides consisted of peptides within the IR5 region, VR5-IR6, and IR6 (Fig. 7).
Fig 7.
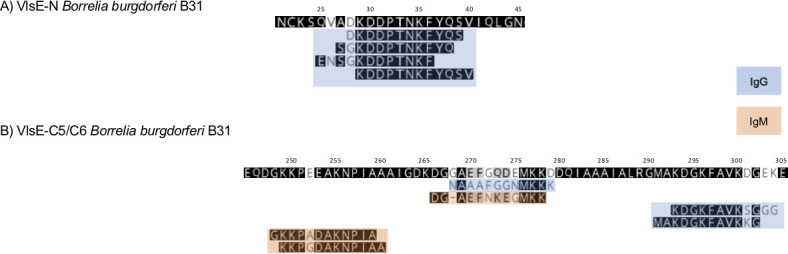
Mapping of the main VlsE peptides identified by the differential model. Panel A shows the peptides that mapped to a fragment within the N terminus, and panel B indicates peptides within the C5/VR5 and C6/VR6 region. The numbers above the sequence correspond to the amino acid positions in the protein. IgG peptides are indicated in blue. IgM peptides are indicated in orange.
Fig 8.
IgG reactivity to VlsE peptides in the five sample types. The Y axis indicates the relative amino acid coordinates of the peptides within the full-length VlsE protein. Reactivity is shown in yellow. For clarity, only peptides with reactivity above 10,000 RFU are shown. Samples are indicated on the X axis. To illustrate baseline reactivity, ten random control samples were selected and shown on the right. The asterisks indicate the regions encompassing the peptides within aa 18–38 (*) and the C6 (**). ^ - includes only confirmed Lyme disease cases.
DISCUSSION
Our aim in this study was to identify diagnostic immune signatures for progressive stages of Lyme disease. We used a combination of high-density peptide arrays and machine learning in a two-step approach. In the first step, we used a diagnostic model for selection of diagnostic Lyme disease antibody-reactive peptides. In the second step, we used a differential model to select reactive peptides associated with disease stage.
FlaB and VlsE were the major antibody targets throughout all stages of disease and contained peptides with the foremost predictive value in both of our models. The diagnostic model was driven primarily by peptides originating from a FlaB fragment located within residues 191–231 and two invariable regions (IR) of VlsE. The sequence encompassing the FlaB 191–231 fragment is a major immunodominant region of FlaB in B. burgdorferi and other Borreliae (19). IgM immunoreactivity was detected to peptides from throughout the length of this fragment, whereas IgG reactivity was confined to approximately 33 aa within residues 207–229. The FlaB 191–231 fragment included some of the most immunoreactive peptides for both IgG and IgM in all Lyme disease stages, although IgM reactivity waned in LA patients. We also detected intermittent, mostly IgM reactivity of peptides within this fragment to control sera. FlaB is a key component of both IgM and IgG Western blots used for Lyme disease serodiagnosis, and cross-reactivity to FlaB on both blots is not uncommon in patients without a documented history of Lyme disease (20, 21). The reactivity we observed in our control samples may explain the source of the false-positive Western blot results. Therefore, despite clear diagnostic utility of this large immunoreactive fragment, only a focus on select smaller peptides like the ones identified by our model is likely to provide the required specificity.
The majority of the approximate 350-aa sequence of VlsE is divided into alternating fragments of genetically heterogenous (or variable, VR) and invariable regions (IR) (22, 23). The 26-aa-long IR6, or C6, region is a well-known target of specific B. burgdorferi antibodies and has been exploited in Lyme disease serodiagnostic assays (24, 25). Peptides within the C6 were typically among the first and most reactive B. burgdorferi peptides in patients with early disease, and peptides located at both the N and C termini of this region were selected in our diagnostic model. However, in agreement with other studies, we observed that the N terminal portion constitutes the primary immunoreactive linear antigenic portion within the C6 (26). We also found that the 9-aa fragment IAAAIALRG serves as the key antigenic motif in C6, and 12-mer peptides that included this sequence were the primary peptides driving the diagnostic model. Additional VlsE peptides, particularly within the IR3 and containing a GKLF motif, were also included in the diagnostic model. However, the diagnostic utility of the IR3 peptides may be partially compromised by their higher degree of sequence divergence relative to C6 in different strains of B. burgdorferi. In addition, we surprisingly found substantial IgM reactivity to VlsE peptides, including within the C6 region. Other peptides in the diagnostic model included peptides within the S2 (BB_RS05130, old designation as BBA04), Bdr, and OspA antigens. Both S2 and Bdr are plasmid-encoded antigens that are expressed at higher levels in B. burgdorferi during vertebrate infection. The selection of the two OspA IgM-reactive peptides in the model was surprising, as OspA is a tick-associated lipoprotein that is not expressed by B. burgdorferi during vertebrate infection (27). Nonetheless, the presence of anti-OspA antibodies and their potential utility for diagnosis have previously been demonstrated (28, 29). We propose the reactivity to these peptides could stem from the immune interaction with a limited number of spirochetes that did not clear OspA from their surface during early infection.
Our differential model was utilized to determine if temporal antigen expression and the subsequent development of the antibody response could be associated with a particular disease presentation. Despite examining a wide range of antigens, the optimal predictive model was built with peptides only from VlsE, FlaB, and p66. The predictive accuracy of the differential model was most pronounced for SEM-A (early disease) and LA (late disease) samples, mostly because the primary drivers of the model were peptides from epitopes reactive predominately in LA sera and nonreactive in SEM-A.
The p66 is one of the 10 antigens recognized on the Lyme disease serodiagnostic IgG Western blot (20). Previous epitope mapping efforts by Arnaboldi et al. revealed a lack of specific regions within p66 that were useful for serodiagnosis of early Lyme disease (30). We also did not identify consistently reactive p66 epitopes in samples from early disease. Although we did identify reactive peptides within several p66 fragments, including regions located at aa 223–271 and 331–361, they were reactive in <50% of sera from each group. The p66 peptides selected in our differential model originated from two distinct reactive portions of p66, located at aa 78–90 and 497–508, and were reactive almost exclusively in the LA samples. Thus, our results indicate that antibodies to p66 78–90 and 497–508 arise late in disease and represent IgG fragments of p66 that can be useful for serologic differentiation between early and late disease.
Similarly, the 18–38 N terminal region of VlsE is also a major target of antibodies late in disease. Along with C6, peptides from the 18–38 fragment included the most immunoreactive peptides in sera from LA patients. However, unlike the C6, this region was largely nonreactive in non-LA sera. A strong antibody response to this region was uncovered in patients with posttreatment Lyme disease syndrome (31). Our data suggest that the 18–38 region and the C6 represent two major sequence-conserved VlsE targets of antibodies during disease, with the C6 antibodies arising first, and the antibodies to the N terminal region only arising during the latter progressive stages of infection.
In agreement with prior studies, our findings indicate that epitopes within VlsE and FlaB are key targets for Lyme disease antibody detection assays. Accordingly, both of these antigens have been utilized in the majority of Lyme disease serologic tests. Similar to our work here, other studies that employed epitope mapping have identified peptides within the IR6 of VlsE and the FlaB 191–231 fragment as targets with high utility for serologic diagnosis (26, 32) Consequently, these shorter peptide fragments, either separately or combined, have been included in several peptide-based serologic assays. The utility of a concatemer using both the partial IR6 and a FlaB-13 mer for all Lyme disease stages has been demonstrated (33) (34). Our finding that these peptides represent the optimum serologic targets throughout the course of disease adds further validity to these earlier studies.
Of the 31 samples listed as “probable Lyme disease,” only nine were predicted as serologically positive by our model. In the absence of conclusive laboratory molecular or serologic findings, the primary rationale for diagnosis of this cohort as probable Lyme disease was the presence of an EM >5 cm. However, EM rashes can be heterogenous in appearance, and skin lesions originating from other, often noninfectious causes can be erroneously characterized as erythema migrans (35). One potential cause of misdiagnosis is the skin lesion associated with the bites of the Lone Star ticks, called Southern Tick-Associated Rash Illness (STARI), a condition of unknown etiology (36). Since Lone Star ticks are increasingly found in Lyme disease endemic areas, there is a growing likelihood of STARI rashes being misdiagnosed as EM (37, 38). It is possible that some of these probable Lyme disease cases may not be caused by B. burgdorferi infection.
A limitation of our study was that we used a partial B. burgdorferi proteome. Although we included the major antigens known to elicit an antibody response, we cannot exclude that other proteins could also improve predictive diagnosis. In addition, our approach only applies to non-conformational epitopes. Nonetheless, our study provides insights into antibody responses at different stages of disease and identified peptides with diagnostic utility.
ACKNOWLEDGMENTS
We would like to thank Shreyas Joshi and Teresa Tagliafierro for their contributions.
This study was funded by grants from the Global Lyme Alliance, The Steven & Alexandra Cohen Foundation, and the R01AI182237 (Tokarz). It was also supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (A.M., S.P.T., and A.E.).
Footnotes
This article is a direct contribution from W. Ian Lipkin, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Steven Schutzer, New Jersey Medical School Department of Medicine, and Maria Gomes-Solecki, University of Tennessee Health Science Center.
Contributor Information
Rafal Tokarz, Email: rt2249@cumc.columbia.edu.
Adriana Marques, Email: amarques@niaid.nih.gov.
Yasuko Rikihisa, The Ohio State University, Columbus, Ohio, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02360-24.
Figures S1 to S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Rosenberg R, Lindsey NP, Fischer M, Gregory CJ, Hinckley AF, Mead PS, Paz-Bailey G, Waterman SH, Drexler NA, Kersh GJ, Hooks H, Partridge SK, Visser SN, Beard CB, Petersen LR. 2018. Vital signs: trends in reported vectorborne disease cases - United States and territories, 2004-2016. MMWR Morb Mortal Wkly Rep 67:496–501. doi: 10.15585/mmwr.mm6717e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kugeler KJ, Schwartz AM, Delorey MJ, Mead PS, Hinckley AF. 2021. Estimating the frequency of lyme disease diagnoses, United States, 2010-2018. Emerg Infect Dis 27:616–619. doi: 10.3201/eid2702.202731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steere AC, Strle F, Wormser GP, Hu LT, Branda JA, Hovius JWR, Li X, Mead PS. 2016. Lyme borreliosis. Nat Rev Dis Primers 2:16090. doi: 10.1038/nrdp.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wormser GP, Dattwyler RJ, Shapiro ED, Halperin JJ, Steere AC, Klempner MS, Krause PJ, Bakken JS, Strle F, Stanek G, Bockenstedt L, Fish D, Dumler JS, Nadelman RB. 2006. The clinical assessment, treatment, and prevention of lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the infectious diseases society of America. Clin Infect Dis 43:1089–1134. doi: 10.1086/508667 [DOI] [PubMed] [Google Scholar]
- 5. Wormser GP, Nadelman RB, Schwartz I. 2012. The amber theory of Lyme arthritis: initial description and clinical implications. Clin Rheumatol 31:989–994. doi: 10.1007/s10067-012-1964-x [DOI] [PubMed] [Google Scholar]
- 6. Nardelli DT, Callister SM, Schell RF. 2008. Lyme arthritis: current concepts and a change in paradigm. Clin Vaccine Immunol 15:21–34. doi: 10.1128/CVI.00330-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marques AR. 2018. Revisiting the Lyme disease serodiagnostic algorithm: the momentum gathers. J Clin Microbiol 56:e00749-18. doi: 10.1128/JCM.00749-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kenedy MR, Lenhart TR, Akins DR. 2012. The role of Borrelia burgdorferi outer surface proteins. FEMS Immunol Med Microbiol 66:1–19. doi: 10.1111/j.1574-695X.2012.00980.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caine JA, Coburn J. 2016. Multifunctional and redundant roles of Borrelia burgdorferi outer surface proteins in tissue adhesion, colonization, and complement evasion. Front Immunol 7:442. doi: 10.3389/fimmu.2016.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tokarz R, Mishra N, Tagliafierro T, Sameroff S, Caciula A, Chauhan L, Patel J, Sullivan E, Gucwa A, Fallon B, Golightly M, Molins C, Schriefer M, Marques A, Briese T, Lipkin WI. 2018. A multiplex serologic platform for diagnosis of tick-borne diseases. Sci Rep 8:3158. doi: 10.1038/s41598-018-21349-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. 2008. A genome-wide proteome array reveals A limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun 76:3374–3389. doi: 10.1128/IAI.00048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y, Bruno JF, Luft BJ. 2008. Profiling the humoral immune response to Borrelia burgdorferi infection with protein microarrays. Microb Pathog 45:403–407. doi: 10.1016/j.micpath.2008.09.006 [DOI] [PubMed] [Google Scholar]
- 13. Horn EJ, Dempsey G, Schotthoefer AM, Prisco UL, McArdle M, Gervasi SS, Golightly M, De Luca C, Evans M, Pritt BS, Theel ES, Iyer R, Liveris D, Wang G, Goldstein D, Schwartz I. 2020. The Lyme disease biobank: characterization of 550 patient and control samples from the east coast and upper midwest of the United States. J Clin Microbiol 58:e00032-20. doi: 10.1128/JCM.00032-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention . 2017. Lyme disease (Borrelia burgdorferi) 2017 case definition. National notifiable disease surveillance system (NNDSS). Available from: https://ndc.services.cdc.gov/case-definitions/lyme-disease-2017. Retrieved Mar 2023.
- 15. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liaw A, Wiener M. 2002. Classification and regression by randomforest. R News 3:18–22. [Google Scholar]
- 17. Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Softw 28:1–26. doi: 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- 18. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. doi: 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tokarz R, Tagliafierro T, Caciula A, Mishra N, Thakkar R, Chauhan LV, Sameroff S, Delaney S, Wormser GP, Marques A, Lipkin WI. 2020. Identification of immunoreactive linear epitopes of Borrelia miyamotoi. Ticks Tick Borne Dis 11:101314. doi: 10.1016/j.ttbdis.2019.101314 [DOI] [PubMed] [Google Scholar]
- 20. Centers for Disease Control and Prevention . 1995. Recommendations for test performance and interpretation from the second national conference on serologic diagnosis of Lyme disease. MMWR MMWR 44:590–591. [PubMed] [Google Scholar]
- 21. Seriburi V, Ndukwe N, Chang Z, Cox ME, Wormser GP. 2012. High frequency of false positive IgM immunoblots for Borrelia burgdorferi in clinical practice. Clin Microbiol Infect 18:1236–1240. doi: 10.1111/j.1469-0691.2011.03749.x [DOI] [PubMed] [Google Scholar]
- 22. Zhang J-R, Hardham JM, Barbour AG, Norris SJ. 1997. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275–285. doi: 10.1016/S0092-8674(00)80206-8 [DOI] [PubMed] [Google Scholar]
- 23. Zhang JR, Norris SJ. 1998. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun 66:3698–3704. doi: 10.1128/IAI.66.8.3698-3704.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang FT, Alvarez AL, Gu Y, Nowling JM, Ramamoorthy R, Philipp MT. 1999. An immunodominant conserved region within the variable domain of VlsE, the variable surface antigen of Borrelia burgdorferi . J Immunol 163:5566–5573. doi: 10.4049/jimmunol.163.10.5566 [DOI] [PubMed] [Google Scholar]
- 25. Liang FT, Steere AC, Marques AR, Johnson BJB, Miller JN, Philipp MT. 1999. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi vlsE. J Clin Microbiol 37:3990–3996. doi: 10.1128/JCM.37.12.3990-3996.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gomes-Solecki MJC, Meirelles L, Glass J, Dattwyler RJ. 2007. Epitope length, genospecies dependency, and serum panel effect in the IR6 enzyme-linked immunosorbent assay for detection of antibodies to Borrelia burgdorferi . Clin Vaccine Immunol 14:875–879. doi: 10.1128/CVI.00122-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwan TG, Piesman J. 2000. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol 38:382–388. doi: 10.1128/JCM.38.1.382-388.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Magni R, Espina BH, Shah K, Lepene B, Mayuga C, Douglas TA, Espina V, Rucker S, Dunlap R, Petricoin EFI, Kilavos MF, Poretz DM, Irwin GR, Shor SM, Liotta LA, Luchini A. 2015. Application of nanotrap technology for high sensitivity measurement of urinary outer surface protein a carboxyl-terminus domain in early stage Lyme borreliosis. J Transl Med 13:346. doi: 10.1186/s12967-015-0701-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schutzer SE, Coyle PK, Dunn JJ, Luft BJ, Brunner M. 1994. Early and specific antibody response to OspA in Lyme disease. J Clin Invest 94:454–457. doi: 10.1172/JCI117346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arnaboldi PM, Dattwyler RJ. 2015. Cross-reactive epitopes in Borrelia burgdorferi p66. Clin Vaccine Immunol 22:840–843. doi: 10.1128/CVI.00217-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chandra A, Latov N, Wormser GP, Marques AR, Alaedini A. 2011. Epitope mapping of antibodies to VlsE protein of Borrelia burgdorferi in post-Lyme disease syndrome. Clin Immunol 141:103–110. doi: 10.1016/j.clim.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnaboldi PM, Katseff AS, Sambir M, Dattwyler RJ. 2022. Linear peptide epitopes derived from ErpP, p35, and FlaB in the serodiagnosis of Lyme disease. Pathogens 11:944. doi: 10.3390/pathogens11080944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nayak S, Sridhara A, Melo R, Richer L, Chee NH, Kim J, Linder V, Steinmiller D, Sia SK, Gomes-Solecki M. 2016. Microfluidics-based point-of-care test for serodiagnosis of Lyme disease. Sci Rep 6:35069. doi: 10.1038/srep35069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arumugam S, Nayak S, Williams T, di Santa Maria FS, Guedes MS, Chaves RC, Linder V, Marques AR, Horn EJ, Wong SJ, Sia SK, Gomes-Solecki M. 2019. A multiplexed serologic test for diagnosis of Lyme disease for point-of-care use. J Clin Microbiol 57. doi: 10.1128/JCM.01142-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schutzer SE, Berger BW, Krueger JG, Eshoo MW, Ecker DJ, Aucott JN. 2013. Atypical erythema migrans in patients with PCR-positive Lyme disease. Emerg Infect Dis 19:815–817. doi: 10.3201/eid1905.120796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masters EJ, Grigery CN, Masters RW. 2008. STARI, or masters disease: lone star tick-vectored lyme-like illness. Infect Dis Clin North Am 22:361–376, doi: 10.1016/j.idc.2007.12.010 [DOI] [PubMed] [Google Scholar]
- 37. Feder HM, Hoss DM, Zemel L, Telford SR, Dias F, Wormser GP. 2011. Southern tick-associated rash illness (STARI) in the north: STARI following a tick bite in Long Island, New York. Clin Infect Dis 53:e142–6. doi: 10.1093/cid/cir553 [DOI] [PubMed] [Google Scholar]
- 38. Monzón JD, Atkinson EG, Henn BM, Benach JL. 2016. Population and evolutionary genomics of Amblyomma americanum, an expanding arthropod disease vector. Genome Biol Evol 8:1351–1360. doi: 10.1093/gbe/evw080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S4.