Abstract
During viral maturation, the cytoplasmic tail of the murine leukemia virus (MuLV) envelope (Env) protein undergoes proteolytic cleavage by the viral protease to release the 16-amino-acid R peptide, and this cleavage event activates the Env protein's fusion activity. We introduced Gly and/or Ser residues at different positions upstream of the R peptide in the cytoplasmic tail of the Friend MuLV Env protein and investigated their effects on fusion activity. Expression in HeLa T4 cells of a mutant Env protein with a single Gly insertion after I619, five amino acids upstream from the R peptide, induced syncytium formation with overlaid XC cells. Env proteins containing single or double Gly-Ser insertions after F614, 10 amino acids upstream from the R peptide, induced syncytium formation, and mutant proteins with multiple Gly insertions induced various levels of syncytium formation between HeLa T4 and XC cells. Immunoprecipitation and surface biotinylation assays showed that most of the mutants had surface expression levels comparable to those of the wild-type or R peptide-truncated Env proteins. Fluorescence dye redistribution assays also showed no hemifusion in the Env proteins which did not induce fusion. Our results indicate that insertion mutations in the cytoplasmic tail of the MuLV Env protein can suppress the inhibitory effect of the R peptide on membrane fusion and that there are differences in the effects of insertions in two regions in the cytoplasmic tail upstream of the R peptide.
Retroviruses enter the host cell through a membrane fusion process mediated by the viral envelope proteins in a pH-independent manner (25, 45). The retroviral envelope proteins are synthesized as precursor proteins, which are processed by a cellular protease into two subunits: the surface (SU) subunit, containing the receptor binding domain and the transmembrane (TM) subunit, which interacts with the SU protein and is involved in subsequent membrane fusion (21, 47, 48). The TM subunit contains three basic structural domains: an extracellular domain containing the highly hydrophobic N-terminal fusion peptide, which is thought to be directly involved in the fusion process, a membrane-spanning region of 19 to 27 amino acids for anchorage to the cell membrane, and a cytoplasmic tail (7, 11). Studies with the influenza virus hemagglutinin (HA) protein have shown that it undergoes a conformational change at low pH, which exposes the buried fusion peptide at the distal tip of the protein for insertion into the target cell surface and which induces fusion between viral and cell membranes, allowing the joining of formerly separated aqueous compartments (45, 46).
The envelope protein (Env) of Friend murine leukemia virus (F-MuLV) has structural features similar to those of other retroviral envelope proteins (9, 44). However, the Env protein of MuLV undergoes another processing event during virus assembly that removes its C-terminal 16-amino-acid segment, designated the R peptide (13). It has been shown that the cleavage of the R peptide from the Env protein of MuLV is important in activating the fusion activity of the Env protein and virus infectivity (16, 33, 34).
Although several studies have analyzed the inhibitory effect of the R peptide on membrane fusion (12, 16, 50), the molecular mechanism of the inhibition remains unclear. Previous studies with simian immunodeficiency virus (SIV) indicated that truncations in the cytoplasmic tail region altered the conformation of the external domain of the viral envelope protein (39). Therefore, it is possible that there is communication between the cytoplasmic domain and the extracellular domain of the viral envelope protein during the fusion process, and the R peptide may regulate membrane fusion by affecting the conformation of the Env protein extracellular domain. In this study, we introduced different numbers of Gly and/or Ser residues, commonly used helix breakers (8, 17, 37), into the cytoplasmic tail of the F-MuLV Env protein upstream of the R peptide coding sequence to interrupt possible α-helix formation in the cytoplasmic tail of the Env protein (36, 49, 52) and to potentially alter an effect of the R peptide on the external domain. We determined the expression levels and transport of the insertion mutants and analyzed the fusion activities of these mutant Env proteins. We also monitored hemifusion and fusion activity to investigate the steps of the fusion process which are affected by the R peptide and by alterations in the cytoplasmic domain.
MATERIALS AND METHODS
Cells and viruses.
HeLa T4 cells and XC cells were obtained from the American Type Culture Collection, Manassas, Va. They were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (GIBCO BRL). The recombinant vaccinia virus (vTF7-3) expressing the T7 polymerase was provided by Bernard Moss (National Institutes of Health, Bethesda, Md.). VTF7-3 was grown on HeLa T4 cells, and the titers of the virus were determined on CV-1 cells.
Plasmid construction and site-directed mutagenesis.
The insertion mutation was carried out using a Stratagene Quick Change site-directed mutagenesis kit. All other restriction endonucleases and DNA modification enzymes used for plasmid construction were purchased from Roche and Stratagene. Construction of the insertion mutants was done as follows. A pair of completely complementary primers was synthesized; these primers contained the desired mutation at the middle, with 10 to 15 bases of correct sequence on both sides, based on the cytoplasmic tail sequence of the wild-type F-MuLV Env protein (20). The primers for mutant M619GS1 (see Fig. 1) were as follows: forward primer, 5′-GTTAAAGACAGGATCGGATCAGTAGTCCAGG-3′; reverse primer. 5′ CCTGGACTACTGATCCGATCCTGTCTTTAAC-3′. All the other constructs at this site had the same flanking sequence except for the central insertion sequence (boldface) coding for the desired mutants. The primers for M619G were as follows: forward primer, 5′-CAATTTGTTAAAGACAGGGGATCAGTAGTCCAGGC-3′; reverse primer, 5′-GCCTGGACTACTGATCCCCTGTCTTTAACAAATTG-3′. Those for M619A were as follows: forward primer, 5′- CAATTTGTTAAAGACAGGGCTTCAGTAGTCCAGGC-3′; reverse primer, 5′- GCCTGGACTACTGAAGCCCTGTCTTTAACAAATTG-3′. The same strategy was used for the design of primers at the other site for insertion mutations. PCR amplification was carried out by using the pGEM3-Menv plasmid as the template, which contains the full-length MuLV Env protein coding sequence in the pGEM-3 vector. PCR consisted of 20 cycles of 94°C for 1 min, 50°C for 1 min 10 s, and 72°C for 10 min. The PCR products were incubated with DpnI at 37°C for 1 h to digest the parental supercoiled double-stranded DNA and then transformed into Escherichia coli DH5α competent cells to repair the nicks in the mutated plasmid. All constructs were sequenced to confirm the presence of the desired insertion mutations and the absence of additional mutations.
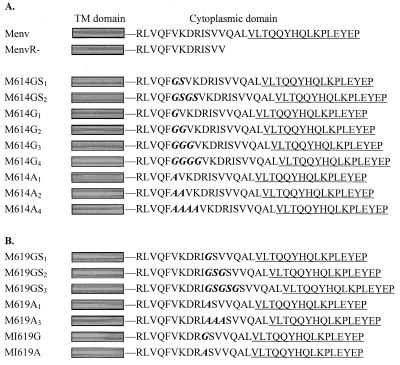
FIG. 1.
Schematic diagram of the wild-type F-MuLV Env protein and cytoplasmic tail insertion mutants. Construction of the plasmids containing the insertion mutations is described in Materials and Methods. The designation of each Env protein is given on the left. Shaded box, TM domain. The amino acid sequence of the cytoplasmic tail is shown, and the R peptide-coding region is underlined. The inserted and substituted amino acids are in boldface italics. (A) The full-length 32-amino-acid sequence of the F-MuLV Env protein (Menv) and the sequence of the R peptide-truncated Env protein (MenvR-) are shown. Mutants with insertions after residue F614 are shown below. (B) Mutants with insertions after I619 and the substitution at I619 are aligned at the transmembrane and cytoplasmic domains with other F-MuLV Env proteins.
Protein expression, radioactive labeling, and immunoprecipitation.
Protein expression was carried out using the recombinant vaccinia virus T7 transient-expression system (10). HeLa T4 cells were grown in 35-mm-diameter dishes to 70 to 80% confluence and then infected with vTF7-3, which expresses T7 polymerase, at a multiplicity of infection of 10 for 1 h. Lipofectin (GIBCO BRL) was used to transfect the infected HeLa T4 cells with 3 μg of plasmid DNA containing the MuLV Env protein coding sequences. At 12 h posttransfection, the cells were starved in Eagle's medium deficient in methionine and cysteine for 45 min, labeled with 100 μCi of [35S]Met-Cys (Du Pont, NEN) in 600 μl of Eagle's deficient medium for another 45 min, and then chased in DMEM with 10% fetal calf serum for 4 h. Cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate [pH 7.5]) plus protease inhibitor (Roche) and immunoprecipitated with goat anti-MuLV Env antibody and protein A-agarose beads (Pierce) at 4°C overnight. Samples were washed with lysis buffer three times and prepared in reducing gel-loading buffer (125 mM Tris-HCl [pH 7.5], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 10% β-mercaptoethanol). Samples were heated at 95°C for 5 min before they were loaded onto an SDS–10% polyacrylamide gel electrophoresis (PAGE) gel for subsequent autoradiography.
Biotinylation of cell surface proteins.
Cell surface proteins were detected by a surface biotinylation assay (22). At 12 h posttransfection, HeLa T4 cells were starved with methionine- and cysteine-deficient Eagle's medium for 45 min and then pulse-labeled with 100 μCi of [35S]methionine and [35S]cysteine for 45 min and chased for various periods with complete medium containing 10% fetal calf serum. At the end of labeling, cells were washed three times with ice-cold PBS-CM (phosphate-buffered saline [PBS] containing 0.1 mM CaCl2 and 1 mM MgCl2) and incubated with 0.5 mg of NHS-SS-biotin (Pierce) in 1 ml of PBS-CM at 4°C for 30 min. Unreacted biotin was quenched by adding fresh DMEM. Cells were lysed with lysis buffer and then immunoprecipitated with goat anti-MuLV Env antibodies and protein A-agarose beads at 4°C overnight. Samples were washed three times in lysis buffer and then divided into two equal aliquots. One aliquot was used for immunoprecipitation, and the other one was treated with 10 μl of 10% SDS and heated at 95°C for 5 min to release the Env proteins. The dissociated proteins were then dissolved in 1 ml of lysis buffer and incubated with 10 μl of streptavidin-agarose (Pierce) for 5 h at 4°C. Biotinylated samples were washed three times with lysis buffer and heated at 95°C for 5 min before analysis by SDS-PAGE.
Fusion assay of wild-type and mutant MuLV Env proteins.
The fusion activities of MuLV Env proteins were determined by the following procedure. HeLa T4 cells were infected with vTF7-3 and transfected with plasmids containing the wild-type or mutant MuLV env genes using Lipofectin (GIBCO BRL) as described above. At 12 h posttransfection, HeLa T4 cells were overlaid with XC cells, a transformed rat cell line which has the receptors for MuLV Env proteins (18). Syncytium formation was observed 1 h later under a phase-contrast microscope. Syncytia were defined as giant cells with more than four nuclei within one single membrane. Ten fields from each sample were randomly selected, and the fusion activity was calculated based on the average value of the ratio of the nuclei in syncytia to the total nuclei in the same field.
Fluorescence dye redistribution fusion assay.
We also determined fusion activity based on the redistribution of fluorescence dyes between the effector and target cells upon fusion, using the following procedures.
(i) Labeling with the cytoplasmic probe calcein-acetoxymethyl(AM)ester (AM).
XC cells were incubated with 5 μM of calcein-AM (Molecular Probes) for 45 min at 37°C in the dark, washed once, and then incubated in fresh medium for another 30 min at 37°C. Cells were detached with trypsin and then resuspended in DMEM with 10% bovine serum and washed twice with PBS.
(ii) Labeling with the lipophilic probes.
XC cells were incubated with 2 μM octadecyl rhodamine B (R-18; Molecular Probes) in 10 ml of PBS for 30 min at room temperature in the dark. Unbound probes were absorbed by adding 10 ml of DMEM with 10% serum and shaken for 20 min in the dark. Cells were then washed five times in PBS (26, 28).
(iii) Fusion assay.
Doubly labeled XC cells were resuspended in DMEM and overlaid on HeLa T4 cells, which were transfected with wild-type or mutant MuLV Env proteins, and then incubated at 4°C for at least 30 min to allow cells to bind to each other. Cells were then transferred to 37°C to allow fusion to occur. Dye redistribution was monitored 5 min after cells were transferred to 37°C using a Nikon fluorescence microscope. Fluorescein isothiocyanate and rhodamine optical filter cubes were used for observing green and red fluorescent-dye transfer, respectively. The percentages of the fused cells to the total number of labeled cells were determined at different times and defined as the relative fusion activities.
RESULTS
Construction and expression of MuLV Env protein cytoplasmic tail mutants.
Previous studies showed that the R peptide has a potent inhibitory effect on the membrane fusion activity of the MuLV Env protein (33, 34). To further investigate the mechanism of the inhibitory effect of the R peptide, we introduced different numbers of Gly and/or Ser residues upstream from the R peptide coding sequence in the cytoplasmic tail of the F-MuLV Env protein. The amino acid sequences of the insertion mutants are shown in Fig. 1. The cytoplasmic tail of the MuLV Env protein has 32 amino acids, and the C-terminal 16-amino-acid segment is designated the R peptide. Two sites were selected for insertion mutations. One site was located at the membrane-distal region, five amino acids upstream from the R peptide, after residue I619. We inserted a single Gly residue or Gly-Ser-Gly or Gly-Ser-Gly-Ser-Gly residues, which formed single, double, and triple Gly-Ser pairs, respectively, because of the already-existing Ser residue (S620) adjacent to I619. We also inserted different numbers of Ala residues at the same position for comparison. In order to avoid possible effects of the cytoplasmic tail elongation caused by the insertion mutations, we also constructed a mutant Env gene encoding a glycine residue in place of Ile-619, which generated a Gly-Ser pair without altering the length of the cytoplasmic tail. The other site that we chose for insertion mutation was located at the membrane-proximal region, which was 10 amino acids upstream from the R peptide, after the F614 residue. We introduced a single or double Gly-Ser or multiple Gly residues after F614 and also inserted a corresponding number of Ala residues for comparison. Sequence analysis confirmed that there were no additional mutations which occurred during the PCR amplification or plasmid construction.
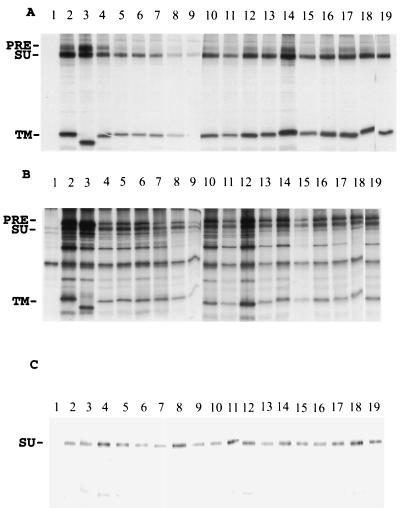
To compare the synthesis and processing of wild-type and mutant Env proteins, we used the vaccinia virus T7 transient expression system to express the MuLV Env proteins in HeLa T4 cells. Immunoprecipitation and surface biotinylation assays were employed to detect the intracellular and cell surface expression and secretion of the Env proteins. As shown in Fig. 2, except for M614G4, which had a very low surface expression, all the mutant envelope proteins were effectively expressed and transported to the cell surface at levels comparable to that for the wild-type or the R peptide-truncated Env proteins, indicating that the mutations did not significantly affect the expression or transport of the MuLV Env proteins.
FIG. 2.
Expression of wild-type and mutant MuLV Env proteins. HeLa T4 cells were infected with vTF7-3 at a multiplicity of infection of 10 for 1 h at 37°C and then transfected with plasmids containing genes encoding wild-type or insertion mutant Env proteins. At 12 h posttransfection, cells were labeled with 100 μCi of [35S]methionine and [35S]cysteine as described in Materials and Methods. After the labeling, cells were biotinylated and immunoprecipitated with antibodies against the F-MuLV Env protein plus protein A-agarose beads at 4°C overnight. The samples were prepared with reducing sample buffer and analyzed by SDS–10% PAGE. (A) Cell surface. (B) Cell lysate. (C) Culture medium. Lanes: 1, pGEM-3 vector control; 2, full-length MuLV Env (Menv); 3, MenvR-; 4, M614GS1; 5, M614GS2; 6, M614G1; 7, M614G2; 8, M614G3; 9, M614G4; 10, M614A1; 11, M614A2; 12, M614A4; 13, M619G; 14, M619A; 15, M619GS1; 16, M619GS2; 17, M619GS3; 18, M619A1; 19, M619A3. PRE, precursor protein.
The effect of insertion mutations after I619 on the fusion activity of the MuLV Env protein.
To determine whether the insertion mutations could interfere with the inhibitory effect of the R peptide on fusion activity, we analyzed the fusion activities of these mutant proteins. As shown in Table 1, cells expressing the full-length MuLV Env (Menv) protein on the surface did not show any syncytium formation, whereas cells expressing the R peptide-truncated Env protein (MenvR-) showed significant syncytium formation after overlay with XC cells. Expression of the Env protein with a single Gly insertion after I619, which formed a single Gly-Ser pair because of the Ser (S620) residue downstream of I619, was also found to induce syncytium formation between HeLa T4 cells and XC cells. In contrast, Env proteins having insertions of double or triple Gly-Ser pairs or multiple Gly residues after I619 (data not shown) did not induce any syncytium formation. When the effect of replacement with Ala residues at this region was examined, a mutant with insertion of only a single Ala residue was found to induce syncytium formation which was comparable to that of the I619GS1 mutant. Mutants with other numbers of Ala insertions did not induce syncytium formation. The mutant with a replacement of I619 with G619, resulting in an internal Gly-Ser pair without altering the length of the cytoplasmic tail, or the Ala substitution (MI619A) did not induce syncytium formation either. As shown in Fig. 2, those mutants which didn't exhibit fusion activities still had high expression levels on cell surfaces, indicating that the defect in syncytium formation is not due to a low expression level on cell surfaces. We also observed that the fusion process induced by the M619GS1 and M619A1 Env proteins was delayed compared with that induced by the MenvR- Env protein. The cells expressing the MenvR- protein began to fuse about 1 to 2 h after overlay with XC cells, while the syncytium formation induced by M619GS1 and M619A1 was first observed at about 5 h after the overlay with XC cells. The syncytia were also smaller than those induced by MenvR-. These results showed that a Gly-Ser insertion at the membrane-distal region of the cytoplasmic tail upstream of the R peptide suppressed the inhibitory effect of the R peptide and that the suppressive effect of the insertion at this region was sensitive to the lengths of the inserted sequences, though it was not amino acid specific.
TABLE 1.
Fusion activity of MuLV Env proteins with insertion mutations after I619
| Construct | Fusion activitya at the indicated time (h)
|
||||||
|---|---|---|---|---|---|---|---|
| 2 | 5 | 6 | 7 | 8 | 10 | 24 | |
| Menv | − | − | − | − | − | − | − |
| MenvR- | ++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| M619GS1 | − | + | + | + | ++ | ++ | +++ |
| M619GS2 | − | − | − | − | − | − | − |
| M619GS3 | − | − | − | − | − | − | − |
| M619A1 | − | + | + | + | + | ++ | +++ |
| M619A3 | − | − | − | − | − | − | − |
| MI619G | − | − | − | − | − | − | − |
| MI619A | − | − | − | − | − | − | − |
Fusion activity was designated as follows: +++++, more than 50% of nuclei are in syncytia; ++++, 30 to 50% of nuclei are in syncytia; +++, 20 to 30% of nuclei are in syncytia; ++, 10 to 20% of nuclei are in syncytia; +, less than 10% of nuclei are in syncytia; −, no syncytia were observed.
The effect of insertion mutations after F614 on the fusion activity of the MuLV Env protein.
When the mutant Env proteins with insertions at the membrane-proximal region were analyzed for fusogenic activity, it was found that cells expressing mutants with a double Gly-Ser insertion after F614 (M614GS2) or insertion of four Gly residues (M614G4) showed the most-extensive syncytium formation (Table 2). Expression of Env mutants with a single Gly-Ser insertion or insertion of three Gly residues caused syncytium formation at a reduced level, and expression of single- or double-Gly insertion mutants also induced a low level of syncytium formation after 10 h of incubation with XC cells, suggesting that single- and multiple-amino-acid insertions have similar effects on the Env protein at this region. In contrast to what was found for the membrane-distal region, none of the mutants with Ala substitutions in this region showed any syncytium formation, indicating that the effect of the insertion mutations in this region was amino acid specific. The time course of the fusion process caused by these mutants was similar to that for M619GS1 and M619A1. HeLa T4 cells expressing Gly-Ser insertion mutant Env proteins began to fuse about 5 to 6 h after being overlayed with the XC cells, and the syncytia caused by these mutants were also smaller than those induced by the MenvR-protein.
TABLE 2.
Fusion activity of MuLV Env proteins with insertion mutations after F614
| Construct | Fusion activitya at the indicated time (h)
|
||||||
|---|---|---|---|---|---|---|---|
| 2 | 5 | 6 | 7 | 8 | 10 | 24 | |
| Menv | − | − | − | − | − | − | − |
| MenvR- | ++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| M614GS1 | − | + | + | + | + | ++ | ++ |
| M614GS2 | − | + | + | + | ++ | ++ | +++ |
| M614G1 | − | − | − | − | − | + | + |
| M614G2 | − | − | − | − | − | + | + |
| M614G3 | − | − | − | + | + | ++ | ++ |
| M614G4 | − | + | + | ++ | ++ | +++ | +++ |
| M614A1 | − | − | − | − | − | − | − |
| M614A2 | − | − | − | − | − | − | − |
| M614A4 | − | − | − | − | − | − | − |
Fusion activities were determined by counting the nuclei in syncytia and comparing this number with the total number of nuclei. +++++, more than 50% of nuclei are in syncytia; ++++, 30 to 50% of nuclei are in syncytia; +++, 20 to 30% of nuclei are in syncytia; ++, 10 to 20% of nuclei are in syncytia; +, less than 10% of nuclei are in syncytia; −, no syncytia were observed.
Effects of mutations on lipid mixing versus content mixing.
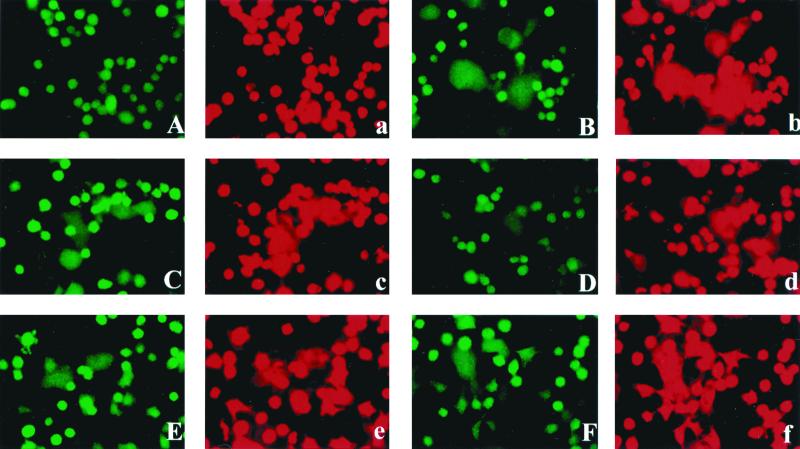
During the first step in the fusion process, the outer membrane leaflets of the two closely contacted cell membranes fuse to each other without mixture of the cellular contents. The molecules in the outer membrane leaflet can be transferred between cells, and this step has been called hemifusion (53). Following the formation of hemifusion, fusion pores between the two inner membrane leaflets can be established, resulting in the mixture of the cellular contents and complete fusion of cells (45, 46). With some viral glycoproteins, including the influenza virus HA protein, changes in the cytoplasmic tail, such as substitution of a glycosylphosphatidylinositol anchor or elongation of the C terminus, caused an alteration of the fusion process and abolished the ability of the HA protein to induce complete fusion, but hemifusion was still observed (19, 27, 30), indicating that, with some HA constructs, the fusion process can be arrested at an intermediate state. It was therefore of interest to determine if hemifusion could still be detected upon expression of Env constructs containing the R peptide or insertion mutations. We used fluorescence microscopy to monitor the dye redistribution between effector cells expressing MuLV Env proteins and the target cells with MuLV Env protein receptors on the cell surface. We loaded XC cells with two different fluorescent dyes, R-18 (red fluorescence), a membrane-impermeant dye which specifically labels the outer leaflet of the cell membrane (14, 24), and calcein-AM (green fluorescence), a cytoplasmic dye which can freely diffuse through the cell membrane and label the inside of the cell (5, 23, 32). We overlaid the doubly labeled XC cells on HeLa T4 cells which were transfected with plasmids encoding MuLV Env proteins and allowed binding by incubation at 4°C for at least 30 min. We then transferred the cells to 37°C to induce fusion. As shown in Fig. 3, cells expressing the full-length MuLV Env protein did not show any dye redistribution, while cells expressing MenvR- showed significant membrane and cytoplasmic dye transfer as soon as 15 min after overlay with XC cells. All the other constructs which had observable fusion activities, as indicated above, were found to exhibit both membrane and cytoplasmic dye redistribution. Compared with those for MenvR-, the membrane dye redistribution and cytoplasmic dye transfer for the other constructs were found to be delayed. Those constructs that did not show any cell fusion activities did not show any dye redistribution, indicating that they were unable to induce even hemifusion. Taken together, these results indicate that none of the MuLV Env protein constructs induced hemifusion unless they also induced cell fusion. Therefore, none of the molecules which are inactive in fusion activity are able to initiate the step of lipid mixing.
FIG. 3.
Fusion activities of Env proteins monitored by fluorescent-dye transfer. HeLa T4 cells expressing the MuLV Env constructs were used as effector cells, and target XC cells were labeled with both calcein-AM (green fluorescence) and R-18 (red fluorescence). Doubly labeled XC cells were overlaid on the HeLa T4 cells and incubated at 4°C for 30 min to allow binding. Then cells were transferred to 37°C for fusion assays. Fluorescent-dye redistribution was monitored 5 min after the temperature increased to 37°C. Green, calcein label; red, R-18 label. A and a, Menv; B and b, MenvR-; C and c, M619GS1; D and d, M619A1; E and e, M614GS1; F and f, M614GS2.
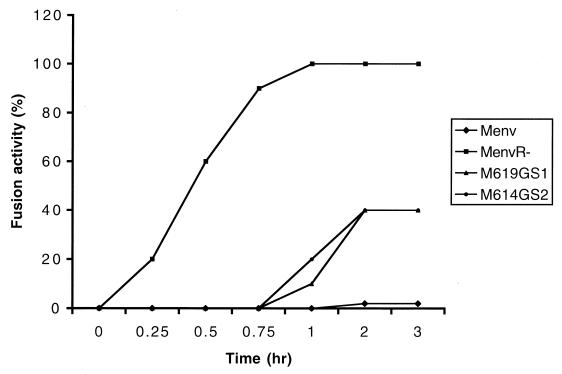
To investigate the kinetics of membrane fusion, we overlaid the doubly labeled XC cells on HeLa T4 cells which were transfected with MuLV Env proteins and incubated at 4°C for at least 30 min to allow the cells sufficient contact with each other. Then we transferred the cells from 4°C immediately to 37°C to induce fusion. Dye redistribution was monitored 5 min after cells were transferred to 37°C. As shown in Fig. 4, no dye redistribution was observed in the cells expressing the full-length MuLV Env protein. Cells expressing the MenvR- protein were first found to show dye redistribution at about 15 to 20 min after the temperature increased and reached maximal level after 40 min. The dye redistribution found with the insertion mutants started at about 1 h and reached peak levels after 2 h. Unlike the results of the fusion assay described above, dye redistribution was found to occur much faster than syncytium formation. Cells expressing the MenvR- protein showed visible syncytium formation at about 1 to 2 h, while the dye redistribution occurred shortly after the temperature was increased. Similar results were obtained with the insertion mutants. The rates of dye transfer were about five times faster than that of syncytium formation, indicating that the fluorescence dye transfer assay is a faster and more sensitive assay for fusion activity than the syncytium formation assay. Also, dye transfer could detect the fusion between as few as two cells, while the fusion assay primarily detects more cells fused together to generate visible syncytia (4).
FIG. 4.
Kinetics of MuLV Env protein-induced membrane fusion. Fusion activity by fluorescence assay was defined as the percentage of the fused cells to the total labeled cells. The percentage was determined at different periods of time after cells were transferred to 37°C.
In order to slow down the fusion process and to attempt to detect an intermediate state by decreasing the temperature, we incubated the cells at different temperatures after preincubation at 4°C. Compared with cells incubated at 37°C, the cells at 4 or 25°C (room temperature) did not show any dye redistribution, except for the cells expressing the MenvR- protein, which showed both outer membrane and cytoplasmic dyes transferred after a relatively long time (about 1 to 2 h), compared with the fast dye transfer rate (transfer began as early as 15 min) when cells were incubated at 37°C. The observed lack of any detectable intermediate fusion state induced by the MuLV Env proteins suggests that the transition from the hemifusion to the fusion process occurs quickly. Once the hemifusion diaphragm forms, the fusion pore is opened and steadily enlarges (26).
DISCUSSION
In this study, we investigated the effects of insertion mutations in the cytoplasmic tail of the Friend MuLV Env protein on membrane fusion activity. We introduced different numbers of Gly and/or Ser residues in the cytoplasmic tail of the MuLV Env protein upstream of the R peptide by site-directed mutagenesis. We found that, at different insertion sites, there were different effects on the fusion activity. At the membrane-distal region upstream of the R peptide, we found that cells expressing a mutant protein with a single Gly insertion caused syncytium formation but that cells expressing proteins with insertions of more than one Gly or Gly-Ser residue did not show any syncytia. Also, cells expressing an Env protein containing only a single Ala insertion caused syncytium formation. In contrast, at the membrane-proximal region upstream of the R peptide, cells expressing mutant proteins with single or double Gly-Ser insertions caused syncytium formation and mutant proteins with insertion of multiple Gly residues showed various levels of syncytium formation. Also at this region, no Env proteins with Ala insertions were found to cause syncytia at all when expressed on the cell surface. These results indicate that there are differences in the effects of insertion in the two regions in the cytoplasmic tail of the MuLV Env protein upstream of the R peptide. We have also observed that, when the R peptide is truncated from the mutant Env proteins, full fusion activity is recovered in all the mutant Env proteins (data not shown), indicating that inability to induce fusion in some mutant proteins is due to the presence of the R peptide. We hypothesize that insertion mutations which result in recovery of fusion activity interfere with an interaction between the R peptide and the external domain and thus suppress the inhibitory effect of the R peptide. The hemifusion assay also showed that the Env mutants which fail to induce fusion could not induce hemifusion either, indicating that they are unable to initiate lipid mixing. Melikyan et al. (26) reported that cells expressing the Moloney MuLV full-length Env protein could induce both hemifusion and fusion when expressed in 293 T cells overlaid with XC cells but not overlaid with NIH 3T3 cells. They also reported that cells expressing Moloney MuLV Env proteins lacking the entire cytoplasmic tail still induced both hemifusion and fusion with XC cells but that mutants with a further deletion of part of the transmembrane domain could not induce even hemifusion.
Studies on the mechanism of viral envelope protein-induced fusion have been largely focused on the extracellular domain and the transmembrane anchor (6, 29, 41). The extracellular domains of several viral envelope proteins have been crystallized, and their structures have been elucidated; these include the influenza virus HA protein, human immunodeficiency virus gp41, and Ebola virus GP2 (2, 3, 15, 43). The cytoplasmic tail was not expected to play a role in membrane fusion; however previous studies have shown that the cytoplasmic tails of some viral envelope proteins can modulate fusion activity. Truncation of the cytoplasmic tail of parainfluenza virus type 3 F protein and simian virus type 5 F protein abolished fusion activity (1, 51), and elongation of the cytoplasmic tail of the influenza virus HA protein by as little as one amino acid reduced fusion activity significantly, whereas addition of five amino acids abolished fusion activity completely (30). In other circumstances, the cytoplasmic tail seems to be dispensable; removal of the cytoplasmic domain of the parainfluenza virus type 2 F protein did not affect its fusion activity (51). In mammalian C-type retroviruses, the cytoplasmic tail is thought to function as a regulatory factor for membrane fusion activity as well as virus infectivity (16, 34, 42). In SIV and human immunodeficiency virus type 1, truncation of the cytoplasmic tail caused increased syncytium formation, indicating a regulatory effect by the cytoplasmic tail on fusion activity (35). MuLV is different from most other enveloped viruses in that its fusion activity is regulated by proteolytic cleavage of the cytoplasmic tail of its Env protein. The C-terminal 16-amino acid-segment, the R peptide, of the MuLV Env cytoplasmic tail is cleaved by the viral protease to render the Env protein fusogenic during virus budding (33, 38). This cleavage is controlled during virus maturation to ensure appropriate propagation of the virus. It seems that the R peptide serves as a safety gate to prevent extensive fusion events from occurring at the surfaces of infected cells.
Previous studies in our laboratory showed that the R peptide has profound fusion-inhibitory effects not only in MuLV Env but also in SIV Env, a distantly related retroviral envelope protein that utilizes different receptors and fuses different cell types (49), and that the region upstream of the R peptide cleavage site in the cytoplasmic tail was also involved in fusion inhibition (16, 50). In other retroviruses the cytoplasmic tail appears to affect the conformation of the extracellular domain (40), indicating that the R peptide may be able to modulate the conformation of the external domain of the MuLV Env protein. In studies of the Moloney MuLV Env protein, it was reported that the palmitoylation of the intracytoplasmic R peptide tilts the TM molecule in the membrane, thus affecting the conformation of the external domain of the TM protein and regulating membrane fusion activity (31). It was suggested that the R peptide cleavage removes a conformational constraint on the cytoplasmic tail and causes a conformational rearrangement of the extracellular domain of the Env protein, promoting membrane fusion (52). Crystal structure studies of the MuLV Env protein have shown that several α-helical regions are expected to exist in the MuLV Env protein TM subunit, such as the heptad repeat sequence which forms trimeric coiled coils similar to those of the influenza virus HA protein (9, 36, 52). We hypothesized that the connecting region in the cytoplasmic tail between the R peptide and the external domain, which has been predicted to form an amphipathic α-helical region (16, 49), is involved in the fusion modulation function of the R peptide. We found that, by introducing Gly-Ser insertions in this connecting region, which would be expected to alter the rigidity of the helix, the inhibitory effect of the R peptide was suppressed. These results indicate the importance of this region in mediating the inhibitory effect of the R peptide. We also found that the two sites we examined for insertions had different responses to the mutations. The membrane-distal region was found to be sensitive to the length of the inserted sequence, and only a mutant with a single Gly insertion caused syncytium formation. Comparing the helical wheel structures of the cytoplasmic tails of the Env proteins, we found that the single Gly insertion changed the predicted amphipathic helix of the MuLV Env protein cytoplasmic tail and that the hydrophobic amino acid alignment was interrupted. The results obtained at the membrane-proximal region for insertion mutants were different from those found at the membrane-distal region. Mutants with insertions of as long as four Gly residues had significant syncytium-forming activity. Previous studies with the Moloney MuLV Env protein also have shown that the membrane-proximal and membrane-distal regions of the cytoplasmic tail have different tolerances to internal deletions and point mutations, which caused different membrane fusion phenotypes (16). It has also been suggested that the R peptide could either interact with a cellular factor which is fusion inhibitory or prevent the Env protein from interacting with a cellular factor which is fusion promoting (50, 52). It is possible that mutations in these two regions have different effects on interaction with a cellular factor. Taken together, the results indicate that, in addition to the R peptide, the size and conformation of the rest of the cytoplasmic tail are important in regulating fusion activity.
In conclusion, we found that insertion mutations in the cytoplasmic tail of the MuLV Env protein can suppress the inhibitory effect of the R peptide. We suggest that this occurs by breaking the rigidity of an α-helical region in the cytoplasmic tail of the MuLV Env protein and interfering with the communication between the R peptide and the external domain of the Env protein. The finding of different effects of insertion mutations at different regions supports the view that, although the cytoplasmic tail is not an absolute requirement for fusion activity, its structure is important for fusion modulation.
ACKNOWLEDGMENTS
This study was supported by grant CA 18611 from the National Institutes of Health.
We thank Tanya Cassingham for assistance in preparing the manuscript and Lawrence Melsen for help with the photography.
REFERENCES
- 1.Bagai S, Lamb R A. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J Cell Biol. 1996;135:73–84. doi: 10.1083/jcb.135.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 3.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 4.Cohen F S, Melikyan G B. Methodologies in the study of cell-cell fusion. Methods. 1998;16:215–226. doi: 10.1006/meth.1998.0670. [DOI] [PubMed] [Google Scholar]
- 5.De Clerck L S, Bridts C H, Mertens A M, Moens M M, Stevens W J. Use of fluorescent dyes in the determination of adherence of human leukocytes to endothelial cells and the effect of fluorochromes on cellular function. J Immunol Methods. 1994;172:115–124. doi: 10.1016/0022-1759(94)90384-0. [DOI] [PubMed] [Google Scholar]
- 6.Denesvre C, Sonigo P, Corbin A, Ellerbrok H, Sitbon M. Influence of transmembrane domains on the fusogenic abilities of human and murine leukemia retrovirus envelopes. J Virol. 1995;69:4149–4157. doi: 10.1128/jvi.69.7.4149-4157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durell S R, Martin I, Ruysschaert J M, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 8.Falcon C M, Matthews K S. Glycine insertion in the hinge region of lactose repressor protein alters DNA binding. J Biol Chem. 1999;274:30849–30857. doi: 10.1074/jbc.274.43.30849. [DOI] [PubMed] [Google Scholar]
- 9.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallaher W R. Detection of a fusion peptide sequence in the transmembrane protein of human immunodeficiency virus. Cell. 1987;50:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- 12.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoekstra D, de Boer T, Klappe K, Wilschut J. Fluorescence method for measuring the kinetics of fusion between biological membranes. Biochemistry. 1984;23:5675–5681. doi: 10.1021/bi00319a002. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman L R, Kuntz I D, White J M. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J Virol. 1997;71:8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Javadpour M M, Eilers M, Groesbeek M, Smith S O. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemble G W, Danieli T, White J M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 20.Koch W, Hunsmann G, Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983;45:1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozarsky K, Penman M, Basiripour L, Haseltine W, Sodroski J, Krieger M. Glycosylation and processing of the human immunodeficiency virus type 1 envelope protein. J Acquir Immune Defic Syndr. 1989;2:163–169. [PubMed] [Google Scholar]
- 22.Le Bivic A, Quaroni A, Nichols B, Rodriguez-Boulan E. Biogenetic pathways of plasma membrane proteins in Caco-2, a human intestinal epithelial cell line. J Cell Biol. 1990;111:1351–1361. doi: 10.1083/jcb.111.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenfels R, Biddison W E, Schulz H, Vogt A B, Martin R. CARE-LASS (calcein-release-assay), an improved fluorescence-based test system to measure cytotoxic T lymphocyte activity. J Immunol Methods. 1994;172:227–239. doi: 10.1016/0022-1759(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald R I. Characteristics of self-quenching of the fluorescence of lipid-conjugated rhodamine in membranes. J Biol Chem. 1990;265:13533–13539. [PubMed] [Google Scholar]
- 25.McClure M O, Sommerfelt M A, Marsh M, Weiss R A. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 26.Melikyan G B, Markosyan R M, Brener S A, Rozenberg Y, Cohen F S. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J Virol. 2000;74:447–455. doi: 10.1128/jvi.74.1.447-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melikyan G B, White J M, Cohen F S. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol. 1999;73:6089–6092. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohuchi M, Fischer C, Ohuchi R, Herwig A, Klenk H D. Elongation of the cytoplasmic tail interferes with the fusion activity of influenza virus hemagglutinin. J Virol. 1998;72:3554–3559. doi: 10.1128/jvi.72.5.3554-3559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen K E, Andersen K B. Palmitoylation of the intracytoplasmic R peptide of the transmembrane envelope protein in Moloney murine leukemia virus. J Virol. 1999;73:8975–8981. doi: 10.1128/jvi.73.11.8975-8981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadopoulos N G, Dedoussis G V, Spanakos G, Gritzapis A D, Baxevanis C N, Papamichail M. An improved fluorescence assay for the determination of lymphocyte-mediated cytotoxicity using flow cytometry. J Immunol Methods. 1994;177:101–111. doi: 10.1016/0022-1759(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 33.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter G D, Jr, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 36.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 37.Saier M H, Jr, Yamada M, Suda K, Erni B, Rak B, Lengeler J, Stewart G C, Waygood E B, Rapoport G. Bacterial proteins with N-terminal leader sequences resembling mitochondrial targeting sequences of eukaryotes. Biochimie. 1988;70:1743–1748. doi: 10.1016/0300-9084(88)90033-8. [DOI] [PubMed] [Google Scholar]
- 38.Schultz A, Rein A. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- 39.Spies C P, Compans R W. Effects of cytoplasmic domain length on cell surface expression and syncytium-forming capacity of the simian immunodeficiency virus envelope glycoprotein. Virology. 1994;203:8–19. doi: 10.1006/viro.1994.1449. [DOI] [PubMed] [Google Scholar]
- 40.Spies C P, Ritter G D, Jr, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor G M, Sanders D A. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol Biol Cell. 1999;10:2803–2815. doi: 10.1091/mbc.10.9.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas A, Gray K D, Roth M J. Analysis of mutations within the cytoplasmic domain of the Moloney murine leukemia virus transmembrane protein. Virology. 1997;227:305–313. doi: 10.1006/viro.1996.8333. [DOI] [PubMed] [Google Scholar]
- 43.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 44.Weissenhorn W, Dessen A, Calder L J, Harrison S C, Skehel J J, Wiley D C. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 45.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 46.White J M. Membrane fusion: the influenza paradigm. Cold Spring Harbor Symp Quant Biol. 1995;60:581–588. doi: 10.1101/sqb.1995.060.01.062. [DOI] [PubMed] [Google Scholar]
- 47.White J M, Doms R W, Gething M-J, Kielian M, Helenius A. Viral membrane fusion proteins. In: Crowell K L R L, editor. Virus attachment and entry into cells. Washington, D.C.: American Society for Microbiology; 1986. pp. 54–59. [Google Scholar]
- 48.Witte O N, Wirth D F. Structure of the murine leukemia virus envelope glycoprotein precursor. J Virol. 1979;29:735–743. doi: 10.1128/jvi.29.2.735-743.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, Compans R W. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol. 1996;70:248–254. doi: 10.1128/jvi.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C, Compans R W. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 1997;71:8490–8496. doi: 10.1128/jvi.71.11.8490-8496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Q, Compans R W. Differences in the role of the cytoplasmic domain of human parainfluenza virus fusion proteins. J Virol. 1995;69:7045–7053. doi: 10.1128/jvi.69.11.7045-7053.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y, Zhu L, Benedict C A, Chen D, Anderson W F, Cannon P M. Functional domains in the retroviral transmembrane protein. J Virol. 1998;72:5392–5398. doi: 10.1128/jvi.72.7.5392-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zimmerberg J, Vogel S S, Whalley T, Plonsky I, Sokoloff A, Chanturia A, Chernomordik L V. Intermediates in membrane fusion. Cold Spring Harbor Symp Quant Biol. 1995;60:589–599. doi: 10.1101/sqb.1995.060.01.063. [DOI] [PubMed] [Google Scholar]