Abstract
Human herpesvirus 8 (HHV-8; Kaposi's sarcoma herpesvirus) encodes four open reading frames with homology to cellular proteins of interferon regulatory factor (IRF) family. Three of them, viral IRF-1 (vIRF-1), vIRF-2, and vIRF-3, have been cloned and found, when overexpressed, to down-regulate the transcriptional activity of interferon type I gene promoters in infected cells by interfering with the transactivating activity of cellular IRFs. In this study, we have further characterized vIRF-2 and shown that it is a nuclear protein which is constitutively expressed in HHV-8-positive pleural effusion lymphoma cell lines. Nuclear localization of vIRF-2 was confirmed by in situ detection of ectopically expressed enhanced green fluorescent protein/vIRF-2 fusion protein. We found that the expression of vIRF-2 in HEK293 cells inhibited the antiviral effect of interferon and rescued translation of vesicular stomatitis virus mRNA from interferon-induced translational block. To provide insight into the mechanism of this effect we have demonstrated that vIRF-2 physically interacts with PKR consequently inhibiting autophosphorylation of double-stranded RNA-activated protein kinase (PKR) and blocking phosphorylation of PKR substrates histone 2A and eukaryotic translation initiation factor 2α. These results suggest that the latently expressed vIRF-2 has a role in viral mimicry which targets the activity of interferon-induced PKR kinase. By inhibiting the kinase activity of PKR and consequent down-modulation of protein synthesis, HHV-8 has evolved a mechanism by which it can overcome the interferon-mediated antiviral effect. Thus, the anti-interferon functions of vIRF-2 may contribute to the establishment of a chronic or latent infection.
Human herpesvirus 8 (HHV-8; Kaposi's sarcoma herpesvirus) is consistently found in all clinical forms of Kaposi's sarcoma (2, 6), AIDS-associated body cavity-based lymphoma or pleural effusion lymphoma (5), and multicentric Castleman's disease (15, 42). HHV-8 belongs to family Rhadinovirinae; its closest relative is a recently discovered rhesus monkey rhadinovirus that contains high number of homologous open reading frames (ORFs) with similar genomic arrangements (1). The HHV-8 genome has been found to contain a unique set of nonstructural genes which may be part of viral mimicry and essential for viral replication in vivo and for pathogenicity (36). HHV-8 encodes four homologues of cellular interferon (IFN) regulatory factors (IRFs); of these, ORF K9-encoded viral IRF-1 (vIRF-1) (3, 16, 27, 47) and ORF k11.1-encoded vIRF-2 (4), have been cloned and characterized, and the cloning of vIRF-3 (ORF k10.5 and 10.6) has been described recently (29). The expression of vIRF-1 is very low in BCBL-1 cells but can be induced by tetradecanoyl phorbol acetate (TPA) treatment (31). Several groups (16, 29, 33, 47) have shown that vIRF-1 can function as a repressor of promoters containing an IFN-sensitive response element. This inhibition is at least partially due to the binding of vIRF-1 to the cellular IRFs as well as competitive binding to the transcriptional coactivator CBP/p300 (3). NIH 3T3 cells overexpressing vIRF-1 gained the ability to grow in soft agar and to form tumors in nude mice (18, 27). We have previously cloned and characterized a second HHV-8-encoded vIRF, vIRF-2, that encodes a short protein of 163 amino acids. It is constitutively transcribed, and vIRF-2 transcripts can be detected in HHV-8-positive B-lymphoma cell lines. We have shown that vIRF-2 is a DNA binding protein with a specificity distinct from that of the cellular IRF, since it binds to an oligonucleotide corresponding to the NF-κB site (4). In a transient transfection assay, vIRF-2 inhibits the virus-mediated induction of IFN type I gene transcription as well as RelA-stimulated activity of the human immunodeficiency virus long terminal repeat. vIRF-2 specifically binds to several transcription factors such as IRF-1, IRF-2, ICSBP (interferon concensus sequence binding protein), and RelA/p65, as well as to the transcription coactivator CBP/p300.
IFNs, as major mediators of innate antiviral defense mechanisms, stimulate the expression of a wide range of cellular genes. Some of these genes encode proteins that can modulate viral replication, with consequent generation of an antiviral state in IFN-treated cells. Among these, the IFN-induced, double-stranded RNA (dsRNA)-activated serine-threonine protein kinase (PKR) is a key mediator of the antiviral and antiproliferative effects of IFN (10, 17, 32, 40). PKR is present in an inactive form in cytosol, and its expression can be further enhanced by treatment with IFN. PKR is activated by number of agents; however, the activation by dsRNA has been studied most extensively (45). Many viruses produce highly structured viral transcripts which can also activate PKR by a mechanism similar to that used by dsRNA (43). The binding of dsRNA to PKR was shown to induce conformational changes and expose the catalytic site for autophosphorylation (13). Furthermore, the dimerization of PKR was shown to be an important part of the autophosphorylation process, suggesting that autophosphorylation is intermolecular (22, 32). Activated PKR was reported to target a number of proteins, but the α subunit of eukaryotic initiation factor 2 (eIF-2α) (37) is the only physiological substrate characterized in detail. The phosphorylation of this factor results in inhibition of protein synthesis, consequently contributing to the antiviral, antiapoptotic, and tumor-suppressing functions of PKR (45, 46). However, in addition to the role in protein synthesis, PKR can regulate gene expression at the transcriptional level. It was demonstrated that PKR regulates NF-κB signaling by phosphorylation of the inhibitory subunit IκB (23), and PKR-mediated phosphorylation of histones (13) and of a 90-kDa protein with unknown function (35) was also demonstrated.
A number of studies have shown that overexpression of wild-type PKR results in the inhibition of cellular growth in mammalian and insect cells as well as in yeast (9, 21, 30, 44). Although the mechanism of growth suppression in eukaryotic cells remains to be clarified, the repression of translation through inhibition of the eIF-2α pathway is assumed to play a role in the toxic phenotype.
The aim of this study was to analyze the expression of vIRF-2 on the protein level and to determine the function of vIRF-2 in virus-host interactions. We have shown that vIRF-2 is expressed constitutively in HHV-8-positive pleural effusion lymphoma lines, and the majority of vIRF-2 protein can be detected in the nucleus. We further show that ectopically overexpressed vIRF-2 counteracts the IFN-induced block of viral protein synthesis. The anti-IFN activity is a result of direct interaction between vIRF-2 and PKR, with consequent inhibition of PKR activity. Thus, by interfering with PKR function, vIRF-2 can facilitate virus invasion by down-regulation of the early inflammatory response; as a constitutively expressed protein, it may also contribute to establishment and maintenance of viral persistence.
MATERIALS AND METHODS
Cell culture and transfections.
HEK293 and HeLa cells were grown in Dulbecco's modified Eagle medium with 10% fetal bovine serum BCBL-1, HBL-6, and JSC-1 cells were grown in RPMI medium supplemented with 10% fetal bovine serum. Subconfluent HeLa cells (4.5 × 106 cells/90-mm-diameter plate) were transfected with 30 μg of empty or vIRF-2-expressing plasmid, using the Superfect (Qiagen) reagent. Where indicated, cells were mock infected or infected with vesicular stomatitis virus (VSV) at a multiplicity of infection (MOI) of 10 in serum-free medium for 45 min and then incubated in complete medium for 5 h. Alternatively, cells were transfected using the Superfect (Qiagen) reagent with poly(I-C) (10 μg/ml; Sigma) for 2 h.
Plasmids.
The cloning and construction of plasmids pcDNA3.1/vIRF-2, pGEXT4/vIRF-2, and pGEXT4/IRF-3 were described previously (3, 39). FLAG-tagged vIRF-2 fusion protein was expressed from the same plasmid with a FLAG epitope coding region inserted at the 5′ end of the vIRF-2 cDNA. This modification was done by PCR using Pfu polymerase (Stratagene) with primers 5′-CTTAAGCTTGCCGCCATGGATTACAAGGATGACGACGATAAGGGATCCATGCCTCGC TACACGGAGTCGG and 3′-GGGAATTCTACATCAACCATCCTACCTCTGG.
To construct the green fluorescent protein (GFP)/vIRF-2 fusion protein, the vIRF-2 coding sequence was amplified by PCR using primers 5′-GAAGAATTCTGCTCGCTACACGGAGTCGG and 3′-TGGGGATCCTACATCAACCATCCTACCTCTGG and subcloned into pEGFP-C vector (Invitrogen). All constructs were tested for possible mutations by direct DNA sequencing.
RT-PCR analysis.
Total RNA was isolated by the Trizol reagent (Life Technologies) and treated with RNase-free DNase I (Boehringer Mannheim) for 20 min at 37°C. Four micrograms of total RNA was used for cDNA synthesis using SuperScript II reverse transcriptase (RT; Life Technologies) and oligo(dT)20 primer. Two microliters of cDNA-containing reaction mixture was used as the template in 30 cycles of PCR amplification using sequence-specific primers as follows: 5- GAAGAATTCATGGCTCGCTACACGGAGTCGG and 3′-TGGGGATCCTACATCAACCATCCTACCTCTGG for the vIRF-2 transcript, 5′-AGCGGATCCCACAGTTTGTTTTTTGAAGAGC and 3′-GCTGAATTCCTAGTCTCTGTGGTAAAATGGG for the K11 transcript, and 5′-GAAGAATTCATGGCTCGCTACACGGAGTCGG and 3′-GCTGAATTCCTAGTCTCTGTGGTAAAATGGG for the vIRF-2 and K11.1 region. PCR products (10 μl) were resolved by 1.5% Tris-borate-EDTA agarose electrophoresis.
Preparation of recombinant vIRF-2 protein and antibodies.
Preparation of glutathione S-transferase (GST) and His6 recombinant fusion proteins was described previously (3). The cleared lysates from isopropyl-β-d-thiogalactopyranoside (Sigma)-stimulated bacteria were mixed with glutathione-agarose beads (200 μl of a 1:1 slurry in phosphate-buffered saline) (Pharmacia) at 4°C for 2 h, and the beads were washed three times with ice-cold sonication buffer (3). Bound proteins were eluted with elution buffer containing 50 mM Tris (pH 8.5), 150 mM NaCl, and 20 mM reduced glutathione. The purity and quantity of fusion proteins were examined by Tricine-sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) followed by Coomassie blue staining.
The SDS-PAGE-purified His6/vIRF-2 protein was used to generate a polyclonal antibody in New Zealand White rabbits according to the standard 7-week immunization protocol. The vIRF-2 antiserum was further purified on a Ni2+/chelate affinity column containing His6/vIRF-2 protein, and the antigen-specific antibodies were eluted with high-pH solution (0.1 M triethylamine [pH 11.5]). Using purified antibody, we were able to detect as little as 50 pg of vIRF-2/GST protein by serial dilution analysis, with virtually no cross-reactions to any other recombinant or endogenous proteins. FLAG-tagged proteins were detected by mouse monoclonal antibody M2 (Stratagene).
GST pull-down assay.
The 35S-labeled proteins were synthesized in vitro using the coupled TNT T7 transcription/translation system (Promega) according to the manufacturer's instructions. Each reaction mixture contained 1 μg of nonlinearized expression plasmid and was incubated (90 min, 30°C) in the presence of 4 μl of Translabel (DuPont) amino acid mixture. GST fusion proteins (0.5 μg) bound to glutathione-agarose beads were incubated with 10-μl reaction mixture aliquots of 35S-labeled proteins in 250 μl of binding buffer (10 mM Tris [pH 7.6], 100 mM NaCl, 0.1 mM EDTA [pH 8.0], 1 mM dithiothreitol, 5 mM MgCl2, 0.05% NP-40, 8% glycerol, mammalian protease inhibitor cocktail [Sigma]) at 4°C for 90 min. After three washes (10 min at room temperature) with binding buffer, the proteins bound to the beads were solubilized in sample buffer and resolved by Tricine-SDS-PAGE (8% gel). The gel was dried and exposed to film. Where indicated, HeLa whole-cell lysates were used instead of rabbit reticulocyte lysates for detection of GST protein interactions. Bound proteins were electrophoresed, transferred onto polyvinylidene difluoride membranes (Bio-Rad), and immunoblotted with anti-PKR antiserum (kindly provided by G. N. Barber). Detection was done with an enhanced chemiluminescence kit (Pharmacia/Amersham).
In vitro kinase reaction.
Human wild-type PKR was immunoprecipitated from 107 HeLa cells as follows. High-salt (500 mM NaCl) cell extracts (600 μl) were incubated with 3 μl of either rabbit polyclonal or mouse monoclonal antibody against PKR (25) overnight in the presence of protease and phosphatase inhibitors (inhibitor cocktail; Sigma). Beads with bound PKR were washed twice with high-salt lysis buffer and twice with kinase buffer (10 mM Tris [pH 7.6], 50 mM KCl, 2 mM magnesium acetate, 20% glycerol, 7 mM β-mercaptoethanol, 1 mM MnCl2, 1 μM ATP, 5 μCi of [γ-32P]ATP, protease and phosphatase inhibitors). Where indicated, immunocomplexes were preincubated for 10 min on ice with poly (I-C) (100 μg/ml; Sigma), 2 μg of histone 2A or 400 ng of purified eIF-2α (kindly provided by J. Jefferson) and the indicated amount of soluble GST fusion protein. ATF2/GST protein was purchased from New England Biolabs. Reaction mixtures were incubated 25 min at 30°C, and reactions were stopped by boiling in sample buffer. Labeled proteins were resolved by Tricine-SDS-PAGE (8% gel), dried, and exposed to film.
Metabolic labeling of VSV-infected cells.
HEK293 cells transfected with either empty or vIRF-2-expressing pcDNA3.1 vector were mock infected or infected with VSV (MOI of 10) 24 h after transfection as described above. Cells were pulse-labeled 5 h after infection with Pro-mix l-35S cell labeling mix (50 μCi/ml; Amersham/Pharmacia) in Met- and Cys-depleted medium for 1 h. Cells were than washed with phosphate-buffered saline and lysed in cell lysis buffer. Labeled proteins were resolved by Tricine-SDS-PAGE (8% gel), dried, and then exposed to film or a PhosphorImager screen for quantification.
RESULTS
Distinct transcription regulation of vIRF-2 and K11 ORFs in BCBL-1 cells.
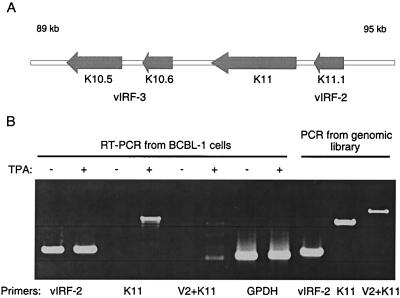
We have previously shown by Northern blotting and RT-PCR analysis that the vIRF-2 ORF (designated K11.1) is constitutively expressed in HHV-8-positive BCBL-1 cells (4). To analyze vIRF-2 expression in more detail and examine the possibility that vIRF-2 (K11.1) is transcribed together with the proximal K11 ORF (Fig. 1A) as one transcript, we amplified vIRF-2 transcripts in BCBL-1 cells by RT-PCR using primers specific for vIRF-2 and K11, and set of primers that correspond to 5′ vIRF-2 and 3′ K11 regions. These primers should amplify the putative common K11 and K11.1 transcript. As seen in Fig. 1B, RT-PCR amplification of poly(dT)-primed cDNA with primers specific either for vIRF-2 or K11 amplified only a single fragment which correspond in size to the K11 ORF and vIRF-2 and has the same size as fragments amplified from the genomic DNA. However, no specific product could be amplified with the combined vIRF-2 and K11 primers, while these two primers were able to amplify a 2.2-kb fragment from genomic DNA of BCBL-1 cells. Furthermore, while a K11-specific product was detected by RT-PCR only in samples from TPA-stimulated cells, the vIRF-2-specific fragment was amplified from both TPA-stimulated and unstimulated cells. These data suggest that both vIRF-2 and K11 ORFs are expressed in BCBL-1 cells as distinct full-length polyadenylated transcripts, and in neither induced nor uninduced cells could we detect a 2.2-kb transcript that would indicate that the transcription proceeds from vIRF-2 to K11. Furthermore, these data indicate that vIRF-2 and K11 ORFs are expressed during different phases of the viral replication cycle.
FIG. 1.
Expression of vIRF-2 and K11 ORFs in BCBL-1 cells. (A) Scheme of the 89- to 95-kbp region of the HHV-8 genome showing the cluster of vIRF-2 (K11.1), K11, and vIRF-3 (K10.5 and K10.6) homologues of cellular IRFs. (B) RT-PCR analysis from control and TPA (50 ng/ml)-induced BCBL-1 cells for 24 h. Constitutive expression of vIRF-2 (V2) is in contrast with inducible expression of K11. No specific product could be detected using a combination of 5′ vIRF-2 and 3′ K11 primers (V2 + K11), detecting, a theoretical common transcript. Control PCR products amplified from the BCBL-1 genomic library are shown. RT-PCR amplification of GPDH mRNA is shown as a control.
vIRF-2 protein is constitutively expressed and localized in nucleis of HHV-8-positive pleural effusion lymphoma cell lines.
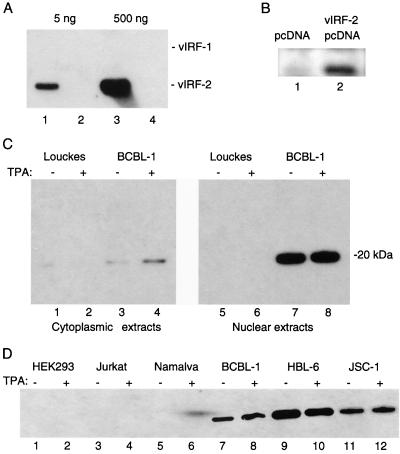
To detect the expression of vIRF-2 in HHV-8-positive B-cell lymphoma lines, we generated a rabbit polyclonal antiserum against recombinant full-length His6/vIRF-2 fusion protein. The antiserum was purified by immunoaffinity chromatography as described in Materials and Methods and reference 27. Figure 2A shows the specificity of the purified vIRF-2 antiserum as analyzed by Western blot hybridization. While the antiserum easily detected 5 ng of vIRF-2/GST protein, no cross-reaction was observed with as much as 500 ng of vIRF-1/GST protein. The antiserum also detected vIRF-2 protein expressed ectopically in transiently transfected HEK293 cells (Fig. 2B). We also detected in cell lysates from HHV-8-positive BCBL-1 cells immunoblotted with vIRF-2 antiserum a specific protein with an apparent molecular mass of 20 kDa (Fig. 2C). No signal was detected in lysates of HHV-8-negative Louckes cells. The mobility of vIRF-2 as determined on SDS-PAGE was about 20 kDa, which corresponds well to the predicted molecular mass of 18.5 kDa. The slight difference in mobility could be due to posttranslational modification of vIRF-2 or to its high overall basic nature (pI 9.8). Fractionation of BCBL-1 cells showed that the majority of vIRF-2 localized in the nuclear fraction; the cytoplasmic fraction showed only low levels of the protein. These data indicate that vIRF-2 is a predominantly nuclear protein. In agreement with analyzes of vIRF-2 mRNA, the relative levels of vIRF-2 protein detected in unstimulated and TPA-stimulated BCBL-1 cells were the same. Thus, vIRF-2, like HHV-8-encoded latency-associated nuclear antigen (LANA) and v-cyclin, is a constitutively expressed nuclear protein (38).
FIG. 2.
Detection of vIRF-2 protein in cell lysates from HHV-8-positive lymphomas. (A) Specificity of the His6/vIRF-2-immunopurified antibody is demonstrated by detection of 5 and 500 ng of vIRF-2/GST fusion protein (lanes 1 and 3). No cross-reaction with the same amount of vIRF-1/GST protein was observed (lanes 2 and 4). (B) Detection of ectopically expressed vIRF-2 in NIH 3T3 cells. Cells were transfected with 4 μg of empty or vIRF-2-expressing pcDNA vector. Whole-cell lysates (20 μg) were prepared 48 h later and immunoblotted with vIRF-2 antibody. (C) Detection of vIRF-2 in nuclear and cytoplasmic fractions from BCBL-1 cells. BCBL-1 and control HHV-8-negative Louckes cells were treated with TPA (50 ng/ml) for 24 h. Cytoplasmic and nuclear fractions (20 μg) were prepared from control or TPA-stimulated cells by differential centrifugation and immunoblotted with vIRF-2 antibody. The only immunoreactive protein, with an apparent mobility of 20 kDa, is marked. (D) Immunoblot analysis of nuclear extracts (20 μg) from different cell lines using immunopurified anti-vIRF-2 antibody. The region around 20 kDa is shown.
To further test the specificity of the vIRF-2 antibody, we analyzed nuclear fractions of several cell lines for the presence of the 20-kDa immunoreactive protein. The 20-kDa protein was also detected in two other HHV-8-positive pleural effusion lymphoma cell lines, HBL-6 and JSC-1 (Fig. 2D), while no signal was detected in HEK293, Jurkat, Namalwa, and NIH 3T3 cells (data not shown).
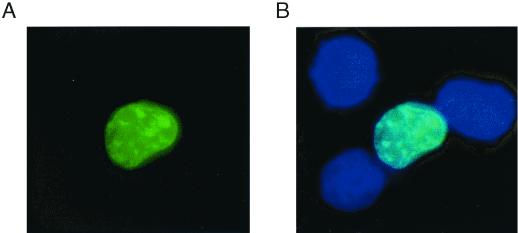
To confirm the nuclear localization of vIRF-2, HeLa cells were transiently transfected with a plasmid expressing the enhanced GFP (EGFP)/vIRF-2 fusion protein. Figure 3A shows the image captured by a GFP-specific filter, Fig. 3B shows a computer-generated overlay combining images captured by GFP and Hoechst stain-specific filters. EGFP/vIRF-2 protein shows evident co localization with Hoechst nuclear staining, as indicated by the bright blue color. In cells transfected with empty EGFP vector the GFP-specific signal was distributed throughout the cells without any pattern (data not shown). We were also interested in determining whether subcellular localization of vIRF-2 is modulated by an extracellular signal. However, neither TPA nor virus infection had any effect on nuclear localization of vIRF-2 (data not shown). These data suggest that vIRF-2 is transported into nucleus independently of virus infection or extracellular signals leading to activation of protein kinase C.
FIG. 3.
Nuclear localization of EGFP/vIRF-2 fusion protein. HeLa cells were transfected with 4 μg of EGFP/vIRF-2 expression vector. Cells were fixed 36 h later and counterstained with DNA stain (Hoechst). (A) Visualization of EGFP/vIRF-2 using a GFP-specific filter. (B) Computer-generated overlay of sequentially captured images from the same field, using GPF- and Hoechst-specific filters. Dark blue shows nuclei; bright blue indicates colocalization of nuclear and EGFP signals.
vIRF-2 inhibits the antiviral effect of IFN-α: rescue of VSV mRNA translation from IFN-induced block.
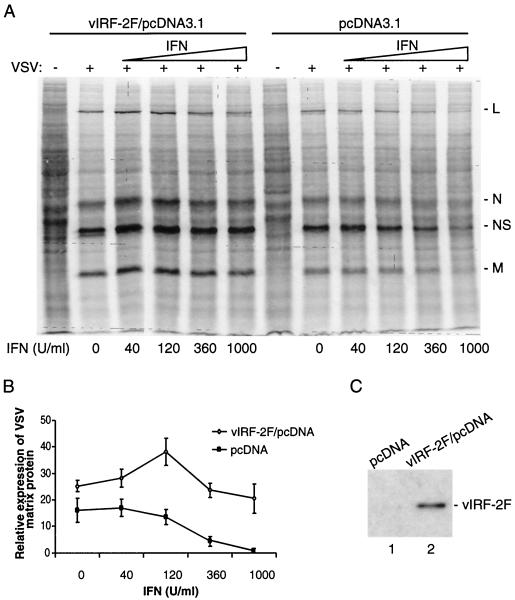
We have demonstrated previously that vIRF-2 protein is potent inhibitor of IFN-α and IFN-β promoters in transient transfection assays. The inhibitory mechanism involves (i) binding of vIRF-2 to transcription factors IRF-1 and RelA and to transcription coactivator CBP/p300 and (ii) inhibition of their transactivating potentials (4). To evaluate whether vIRF-2 can directly inhibit the antiviral effect of IFN, we tested the effect of vIRF-2 on the IFN-mediated inhibition of VSV protein synthesis. We selected VSV because of its high sensitivity to the antiviral effect of IFN. It also has a short replication cycle, allowing us to use cells transiently transfected with vIRF-2 (14). As shown in Fig. 4A, HEK293 cells transfected with empty vector were responsive to the antiviral effect of IFN, and synthesis of VSV early proteins was significantly reduced by IFN-α2 at concentrations higher than 360 U/ml. However, in cells transfected with a vIRF-2-expressing vector, viral protein synthesis was not significantly inhibited even when a high concentration (1,000 U/ml) of IFN-α2 was used (Fig. 4A). Quantitation of relative levels of VSV matrix protein detected in IFN-treated cells in the presence and absence of vIRF-2 is shown in Fig. 4B. The relative levels of VSV matrix protein were not affected by IFN treatment, even at a high IFN-α2 concentration (1,000 U/ml) in vIRF-2-expressing cells. The small increase in amount of labeled matrix protein at an IFN concentration of 120 U/ml is on the edge of statistical significance, and it could be due to a variance in cell samples. In contrast, the relative levels of matrix protein were proportionaly decreased in parental cells treated with increasing concentrations of IFN-α2. The level of ectopically expressed FLAG-tagged vIRF-2 protein in transfected cells is shown in Fig. 4C. These data provide evidence that vIRF-2 can mediate viral resistance to IFN and rescue the IFN-induced block of viral mRNA translation.
FIG. 4.
Expression of vIRF-2 rescues IFN-induced block of viral protein synthesis. (A) Autoradiography analysis of metabolically labeled cells transfected with empty (pcDNA3.1) or FLAG/vIRF-2-expressing (vIRF-2F/pcDNA3.1) vector. Cells were mock infected (−) or infected with VSV (+) and 5 h later pulse-labeled with 35S-labeled amino acid mixture as described in Materials and Methods. Positions of early VSV proteins are marked on the right. (B) Relative levels of 29-kDa VSV matrix protein (M) in pcDNA- or vIRF-2F/pcDNA-transfected cells and its dependency on increased concentrations of IFN. Amounts of viral proteins in cells pretreated with increased concentrations of IFN were quantified from autoradiograms by using a PhosphorImager densitometer and data analysis software. Error bars represent standard errors from three independent experiments. (C) Immunoblot detection of FLAG-tagged vIRF-2 in representative samples of pcDNA- or vIRF-2F/pcDNA-transfected cells by using anti-FLAG antibody.
vIRF-2 physically interacts with PKR in vitro.
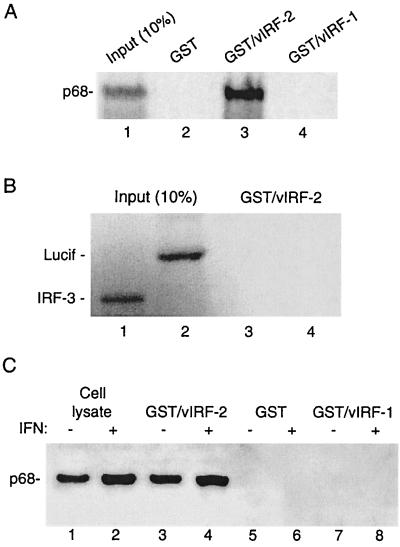
It has been shown that the IFN-mediated inhibition of VSV replication occurs at the level of viral protein synthesis and that PKR plays a major role in this inhibition (14, 26, 41). We therefore examined the possibility that the vIRF-2-mediated inhibition of IFN action observed in previous experiments could be related to modulation of PKR activity. As PKR is a well-known key regulator of protein synthesis that is very often targeted by viral factors, we examined whether vIRF-2 directly interacts with PKR. vIRF-2/GST-containing beads incubated with in vitro-translated radiolabeled PKR retained a significant amount (50% of the input) of the input PKR (Fig. 5A, lane 3), while no PKR was associated with GST-containing beads (lane 2). This high level of binding suggests that the interaction between vIRF-2 and PKR is very strong. The interaction was specific for vIRF-2, since PKR did not associate with the same amount of vIRF-1/GST fusion protein (lane 4). Also, in vitro-translated IRF-3 and luciferase showed no binding to vIRF-2/GST (Fig. 5B, lanes 3 and 4).
FIG. 5.
vIRF-2 binds to PKR in vitro. (A) In vitro-translated, 35S-labeled PKR was incubated with GST-tagged vIRF-2 or vIRF-1 coupled to glutathione-agarose beads as described in Materials and Methods. Lane 1 represents 10% of the input volume of in vitro-translated PKR (p68) used for the binding reactions. (B) Absence of binding of in vitro-translated, 35S-labeled luciferase (Lucif) or IRF-3 to GST/vIRF-2 beads. (C) Whole-cell lysates from IFN (500 U/ml, 16 h)-treated or untreated HeLa cells were incubated with GST-tagged vIRF-2 or vIRF-1 coupled to glutathione-agarose beads. Bound proteins were separated by SDS-PAGE and subjected to immunoblot analysis with anti-PKR antiserum. Lanes 1 and 2 show the presence of p68 in 10% of the input volume of cell lysates used for the binding reactions.
To test the ability of vIRF-2 to bind endogenously expressed PKR, whole-cell lysates from control cells and cells pretreated with IFN-α for 16 h were incubated with vIRF-2/GST-containing beads (Fig. 5B). In agreement with previous data, a high amount of PKR was bound to vIRF-2/GST-containing beads (Fig. 5B, lane 3), and binding was about threefold higher with the cell lysates from IFN-treated than untreated cells (lane 4). No binding of PKR to beads containing GST only or vIRF-1/GST fusion proteins was detected (lanes 5 to 8).
vIRF-2 inhibits PKR autophosphorylation and phosphorylation of histone 2A.
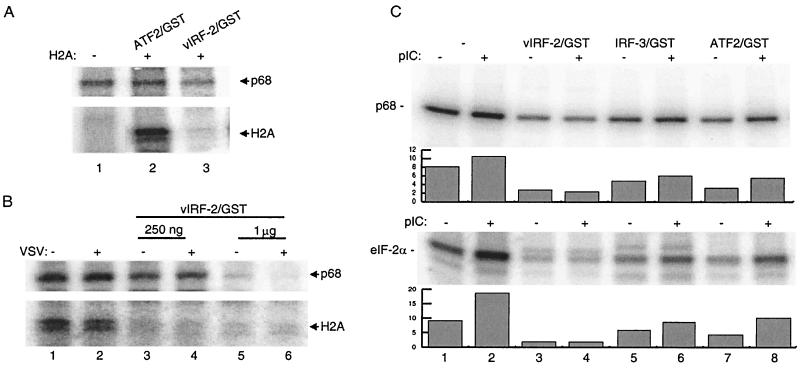
Next, we tested whether the vIRF-2–PKR interaction has any effect on the intrinsic kinase activity of PKR. To this end, we immunoprecipitated PKR from HeLa cells infected with VSV or stimulated by dsRNA and measured its activity by immunocomplex kinase assay in vitro. The autophosphorylation level of PKR in unstimulated cells in the absence of histone 2A is shown in Fig. 6A, lane 1. A control ATF2/GST fusion protein (2 μg) did not modulate the autophosphorylation of PKR activity (lane 2), but addition of vIRF-2/GST protein significantly inhibited PKR phosphorylation (lane 3). As shown in Fig. 6B, PKR was autophosphorylated in both control and VSV-infected cells and specifically phosphorylated the purified histone 2A protein (lanes 1 and 2). However, when vIRF-2/GST was included in kinase reaction, we observed a vIRF-2/GST concentration-dependent decrease in PKR autophosphorylation as well as in phosphorylation of histone 2A (lanes 3 to 6). Thus, 250 ng of purified vIRF-2/GST protein effectively inhibited autophosphorylation of PKR and completely inhibited the phosphorylation of histone 2A (lanes 3-4), while 1 μg of vIRF-2/GST protein completely inhibited both PKR autophosphorylation and substrate phosphorylation (lanes 5 to 6).
FIG. 6.
vIRF-2 inhibits PKR autophosphorylation and phosphorylation of histone 2A (H2A) and eIF-2α in vitro. (A) Protein kinase assay of PKR (p68) immunoprecipitated by rabbit polyclonal antibody from control HeLa cells in the absence of H2A substrate (lane 1) and in the presence of H2A and ATF2/GST (2 μg) or vIRF-2/GST (400 ng). (B) Phosphorylation of H2A by PKR immunoprecipitated from mock-infected (−) or VSV-infected (+) cells. Increased amounts of purified vIRF-2/GST protein were added to each reaction as indicated. (C) Phosphorylation of eIF-2α by PKR immunoprecipitated by monoclonal antibody from control and poly(I-C) (pIC)-stimulated HeLa cells. Reactions were performed at the absence or presence of 500 ng of of each recombinant protein indicated at the top. The levels of phosphorylated PKR and eIF-2α were quantitated and are plotted below in the corresponding panel. Representative samples of at least three independent experiments are shown.
We also examined the effect of vIRF-2 on the ability of PKR to phosphorylate a PKR-specific substrate, eIF-2α. HeLa cells or dsRNA-transfected (for 2 h) HeLa cells were lysed in high-salt buffer, and PKR was immunoprecipitated with a monoclonal antibody. Phosphorylation of eIF-2α substrate was analyzed in the presence and absence of vIRF-2 (Fig. 6C). Phosphorylation of eIF-2α by PKR in unstimulated cells was significantly enhanced in cells stimulated with dsRNA (Fig. 6, lanes 1 and 2), while autophosphorylation of PKR was only marginally increased after poly (I-C) stimulation (lanes 1 and 2). When phosphorylation was done in the presence of GST/vIRF-2 protein, both PKR autophosphorylation and eIF-2α phosphorylation were greatly inhibited in poly(I-C)-treated and untreated cells (lane 4). In contrast, addition of the same amounts of IRF-3/GST or ATF2/GST had no effect (lanes 5 to 8). The same degree of inhibition by vIRF-2/GST was observed with H2A as a substrate (data not shown).
It was shown that PKR has to homodimerize in order to be autophosphorylated and catalytically active (24). Whether vIRF-2 prevents the homodimerization and consequently the autophosphorylation of PKR, or whether it binds to the catalytic domain and functions as pseudosubstrate, remains to be determined.
DISCUSSION
We have shown in this study that vIRF-2 is a latency-associated nuclear antigen of about 20 kDa which can be detected in all HHV-8-positive B-cell lymphoma lines. In contrast, only low levels of vIRF-1 expression can be detected in unstimulated BCBL-1 cells, and its expression can be further stimulated by by TPA treatment (29, 31). Since vIRF-2 expression was observed in all pleural effusion lymphomas tested, the potential use of vIRF-2 as a marker of HHV-8 infection is worth consideration. Two other HHV-8-encoded genes that are constitutively expressed in BCBL-1 cells are those encoding v-cyclin and LANA (11, 20, 28, 34). These genes were identified as class I latent genes that may contribute to HHV-8 induced malignancy (38). Both of the encoded proteins were shown to inactivate tumor suppressor proteins. LANA was shown recently to bind p53, repress its transcription activity, and protect cells against cell death (12), and v-cyclin was found to bind retinoblastoma protein (7). We have observed in this study that vIRF-2 interacts with PKR and inhibits its antiviral activity. Since PKR plays a critical role in the IFN-mediated antiviral and anticellular responses, these data indicate that by encoding vIRF-2, HHV-8 creates a mechanism by which it can directly target the key effector of the antiviral response PKR.
We have shown previously that vIRF-1, vIRF-2, and vIRF-3 can inhibit virus-mediated activation of IFN-α and IFN-β promoters by inhibiting the transactivating activities of cellular IRFs, namely, IRF-1, IRF-3, and IRF-7 (3, 29; unpublished results), thus functioning as dominant negative mutants of the cellular IRFs. All of these vIRFs also inhibited the IFN-mediated stimulation of promoters of IFN-induced genes (ISG). This inhibition occurs presumably by interference with the assembly or function of the transcription complex ISGF3 that contains IRF-9 plus activated STAT-1 and STAT-2 and binds to the IFN-sensitive response elements present in the promoters of ISGs (3, 4, 16, 47). The ability to bind PKR and modulate its activity may be a unique property of vIRF-2, since no interaction between vIRF-1 and PKR was detected and it is not known whether vIRF-3 can bind PKR. Several other DNA viruses have developed distinct mechanisms to circumvent PKR action (reviewed in reference 19). Adenovirus and Epstein-Barr virus encode small RNAs that bind to and inhibit PKR function; reovirus and vaccinia virus encode proteins that sequester dsRNA. Herpes simplex virus type 1, vaccinia virus, and hepatitis C virus (HCV) also encode proteins that bind to PKR and prevent its activation. It was shown that HCV-encoded nonstructural protein NS5A binds directly to the catalytic domain of PKR and that cells overexpressing NS5A are resistant to apoptosis. It was suggested that the NS5A-mediated disruption of apoptosis might confer the oncogenic potential to HCV (14). Therefore, a disruption of PKR activity may inhibit not only its antiviral effect but also its physiological functions and contribute to virus-induced pathogenicity. Thus, the effect of vIRF-2 on PKR in combination with the effects of LANA and v-cyclin may play a role in the maintenance of malignancy during persistent viral infection.
However, vIRF-2 does not seem to share the pro-oncogenic potential of vIRF-1 (18, 27). Overexpression of vIRF-1 in NIH 3T3 cells conferred growth in soft agar, and these cells when transplanted formed tumors in nude mice. Tumor formation was associated both with vIRF-1-mediated enhancement of c-myc expression and inhibition of apoptosis (3, 18). In contrast, NIH 3T3 cells expressing vIRF-2 formed neither colonies in soft agar nor tumors in immunocompromised mice (L. Burýšek and P. M. Pitha, unpublished results). These results indicate that vIRF-2 alone is not able to induce the transformed phenotype.
The inhibition or delay of apoptosis of host cells may be an important requirement for persistent HHV-8 infection, since the virus employs several alternative mechanisms to prevent death of infected cells. It encodes a cellular homologue of Bcl-2 (8), an inhibitor of apoptosis, and employs vIRF-1 to inhibit IRF-1-mediated apoptosis (4, 47). Both of these genes are expressed during lytic infection, and their expression may allow the virus to complete a full replication cycle and delay host cell death. In addition, two latently expressed genes, encoding LANA and vIRF-2, target functions of proapoptotic proteins p53 and PKR, respectively, which may be important for maintenance of a chronic infection. It is therefore likely that the inhibition of apoptosis in combination with the effects of other HHV-8-encoded oncogenes is also important for the oncogenicity of Kaposi's sarcoma.
ACKNOWLEDGMENTS
We are grateful to J. Jefferson and S. R. Kimball (Pennsylvania State University College of Medicine, Hershey) for purified eIF-2α and to A. Hovanessian (Institute Pasteur, Paris, France) for the monoclonal antibody against PKR. We thank G. N. Barber for the rabbit polyclonal antibody against PKR, M. G. Katze for the PKR expression plasmid, J. Nicholas for the λ phage library, HBL-6 cells, and BCBL-1 cells, and J. Cannon for JSC-1 cells. We also thank M. Kellum for preparation of VSV.
This study was supported by grants CA76946 (NCI) and AI19737 (NIAID) from the National Institutes of Health to P.M.P.
REFERENCES
- 1.Alexander L, Denekamp L, Knapp A, Auerbach M R, Damania B, Desrosiers R C. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Immunol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 3.Burýšek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burýšek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden K, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 8.Cheng E H-Y, Nicholas J, Bellows D, Hayward G S, Guo H-G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong K L, Feng L, Schappert K, Meurs E, Donahue T F, Friesen J D, Hovanessian A G, Williams B R. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 11.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 12.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 13.Galabru J, Hovanessian A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 14.Gale M, Jr, Kwieciszewski B, Dossett M, Nakao H, Katze M G. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 16.Gao S J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;85:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 17.Hovanessian A G. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;9:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 18.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Kalvakolanu D V. Virus interception of cytokine-regulated pathways. Trends Microbiol. 1999;7:166–171. doi: 10.1016/s0966-842x(99)01476-6. [DOI] [PubMed] [Google Scholar]
- 20.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 22.Kostura M, Mathews M B. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langland J O, Jacobs B L. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr = 66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 25.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S B, Bablanian R, Esteban M. Regulated expression of the interferon-induced protein kinase p68 (PKR) by vaccinia virus recombinants inhibits the replication of vesicular stomatitis virus but not that of poliovirus. J Interferon Cytokine Res. 1996;16:1073–1078. doi: 10.1089/jir.1996.16.1073. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubyova B, Pitha P M. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol. 2000;74:8194–8201. doi: 10.1128/jvi.74.17.8194-8201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 32.Patel R C, Stanton P, McMillan N M, Williams B R, Sen G C. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitha P M, Burysek L, Schafer S. Critical role of Kaposi sarcoma virus (HHV8)-encoded IRFs in virus replication and pathogenicity. J Interferon Cytokine Res. 1997;17(Suppl. 2):S37. [Google Scholar]
- 34.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice A P, Kostura M, Mathews M B. Identification of a 90-kDa polypeptide which associates with adenovirus VA RNAI and is phosphorylated by the double-stranded RNA-dependent protein kinase. J Biol Chem. 1989;264:20632–20637. [PubMed] [Google Scholar]
- 36.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samuel C E. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 38.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 40.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 41.Shors S T, Beattie E, Paoletti E, Tartaglia J, Jacobs B L. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J Interferon Cytokine Res. 1998;18:721–729. doi: 10.1089/jir.1998.18.721. [DOI] [PubMed] [Google Scholar]
- 42.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 43.Tan S L, Katze M G. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J Interferon Cytokine Res. 1999;19:543–554. doi: 10.1089/107999099313677. [DOI] [PubMed] [Google Scholar]
- 44.Thomis D C, Samuel C E. Mechanism of interferon action: autoregulation of RNA-dependent P1/eIF-2 alpha protein kinase (PKR) expression in transfected mammalian cells. Proc Natl Acad Sci USA. 1992;89:10837–10841. doi: 10.1073/pnas.89.22.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams B R. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 46.Williams B R. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 47.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]