Abstract
Many of the events required for productive herpes simplex virus type 1 (HSV-1) infection occur within globular nuclear domains called replication compartments, whose formation appears to depend on interactions with cellular nuclear domains 10 (ND10). We have previously demonstrated that the formation of HSV-1 replication compartments involves progression through several stages, including the disruption of intact ND10 (stage I to stage II) and the formation of PML-associated prereplicative sites (stage III) and replication compartments (stage IV) (J. Burkham, D. M. Coen, and S. K. Weller, J. Virol. 72:10100–10107, 1998). In this paper, we show that some, but not all, PML isoforms are recruited to stage III foci and replication compartments. Genetic experiments showed that the recruitment of PML isoforms to stage III prereplicative sites and replication compartments requires the localization of the HSV-1 polymerase protein (UL30) to these foci but does not require polymerase catalytic activity. We also examined the stages of viral infection under conditions affecting ND10 integrity. Treatment with factors that increase the stability of ND10, arsenic trioxide and the proteasome inhibitor MG132, inhibited viral disruption of ND10, formation of replication compartments, and production of progeny virus. These results strengthen the previously described correlation between ND10 disruption and productive viral infection.
Herpes simplex virus type 1 (HSV-1) carries out gene expression, DNA replication, and DNA encapsidation in globular nuclear domains designated replication compartments (53, 55). These domains contain the essential viral DNA replication proteins (the origin-binding protein, the single-stranded-DNA-binding protein, the helicase-primase subunits, and the polymerase subunits [34, 36, 55]) and are usually visualized by antibodies either against ICP8, the single-stranded-DNA-binding protein, or UL42, the polymerase processivity subunit. The formation of replication compartments is mediated in part by interactions with nuclear structures called ND10 (nuclear domains 10), promyelocytic leukemia bodies, or PODs (17). The function of ND10 has not yet been defined for cellular or viral growth. Proteins found in ND10 have been associated with the control of cellular growth, cell cycle regulation, transcription, and apoptosis (11, 12, 24, 27, 46, 71). In the case of the herpesviruses, viral DNA is deposited at ND10 and immediate-early transcripts can be detected at sites adjacent to ND10 (42). Furthermore, replication compartments formed after transfection with the seven essential HSV-1 replication proteins localize adjacent to ND10 (36, 74).
ND10 are dynamic structures which are disrupted during mitosis and respond to environmental stimuli including interferon treatment, heat shock, treatment with heavy metals, and viral infection (44, 64, 65). The most extensively studied ND10 protein, PML, is expressed as a fusion with retinoic acid receptor α in individuals with acute promyelocytic leukemia (31, 56). In this disease, disruption of ND10 correlates with loss of growth control (24) and reformation of ND10 correlates with recovery of growth control. This may indicate that PML and ND10 play a role in the control of cell division.
During the course of HSV-1 infection, ND10 become disrupted, presumably through the action of the viral immediate-early regulatory protein ICP0 (21, 40). ICP0 alone is able to induce the disruption of ND10 (21, 41), and during infection, it appears to be required for the proteasome-dependent disappearance of high-molecular-weight forms of two ND10 proteins, PML and Sp100 (20). Some of these high-molecular-weight forms of PML and Sp100 have been shown to be covalently modified by the ubiquitin-like modifier SUMO-1 (32, 49, 62). The disruption of ND10 and the apparent degradation of modified forms of ND10 proteins may be one of several complex strategies herpesviruses have evolved to intervene in host cell regulatory processes.
In this study, we explored many aspects of the formation of replication compartments and their relationship to ND10. We have previously demonstrated that the formation of HSV-1 replication compartments involves progression through several stages, including the disruption of intact ND10 (stage I to stage II) and the formation of PML-associated prereplicative sites (stage III) and replication compartments (stage IV) (7). We and others have shown that PML is recruited to stage III (7) and stage IV replication compartments (7, 53). In cells transfected with the seven replication proteins, an ND10 protein was also observed in replication compartments (36). Since HSV-1 infection has been shown to cause the degradation of some forms of PML (20), we set out to examine the identity of the isoforms that are recruited to replication compartments during infection. We demonstrate here that only some isoforms of PML are recruited to HSV-1 replication compartments. We took a genetic approach to show that recruitment of a PML isoform(s) to stage III prereplicative sites and replication compartments requires the localization of the HSV-1 polymerase protein (UL30) at viral foci but does not require the polymerase to be catalytically active. We have also explored the effect of various environmental stimuli known to affect ND10 on the establishment of replication compartments and the production of viral progeny. Our results strengthen the established correlation between ND10 disruption and productive viral infection. Models for the relationships among PML, ND10, and viral infection are discussed.
MATERIALS AND METHODS
Cells.
African green monkey kidney fibroblasts (Vero; American Type Culture Collection), several Vero derivative cell lines, human esophageal carcinoma cells (HEp-2; American Type Culture Collection), and human osteosarcoma cells (U2OS; American Type Culture Collection) were propagated in Dulbecco's modified Eagle's (DME) medium supplemented with 10% fetal bovine serum (Atlanta Biologicals) and penicillin-streptomycin solution (Sigma) (69). G418-resistant B3 cells containing the HSV-1 UL30 gene were described previously (29). Cell lines expressing various PML isoforms from cDNA (described below) were propagated in DME medium containing 1 mg of G418 (Geneticin; GIBCO Laboratories, Grand Island, N.Y.) per ml.
Viruses.
Strain KOS was used as the wild-type HSV-1. Numerous KOS-derived mutants were used in this study. Several mutants with changes in the catalytic subunit of the polymerase were used. Viable mutants include V462A, AraAr9, 615.8, F891C, PAAr5, Y7, and YD12 (13, 14, 26, 29, 37, 57). Mutants that do not make a polymerase protein that is detectable by Western blot analysis (data not shown) include the null virus HP66 and ΔX17, ΔX14, ΔS1.1, and 7E4A (38). Mutants that make protein but are still nonviable include 6C4, E460D, G464V, and both tsC4 and tsC7 at 39.5°C (13, 26, 38).
Reagents and antibodies. (i) Antibodies recognizing PML.
PG-M3 is a monoclonal antibody that recognizes the human PML protein (25) (Santa Cruz Biotechnology, Santa Cruz, Calif.). 5E10, a monoclonal antibody that recognizes PML (65), was kindly provided by L. de Jong (E. C. Slater Instituut, University of Amsterdam, Amsterdam, The Netherlands). Rat anti-PML R-n and R-m, two polyclonal antibodies that recognize PML, were kindly provided by T. Sternsdorf and H. Will (Heinrich-Pette-Institut fur Experimentelle Virologie und Immunologie, Universität Hamburg, Hamburg, Germany).
(ii) Antibodies recognizing HSV-1 polymerase.
Polyclonal antibodies M1 and αPol recognize the catalytic subunit of the HSV-1 polymerase, UL30. M1 was prepared by K. Weisshart against a fusion protein containing a segment from the middle of HSV-1 Pol; αPol was a kind gift from D. Dorsky (University of Connecticut Health Center) (15).
(iii) Antibodies recognizing the major viral DNA-binding protein ICP8 (UL29).
39S, a monoclonal antibody that recognizes ICP8 (60), was provided by M. Zweig (National Cancer Institute), and αICP8, a rabbit polyclonal antibody that recognizes ICP8 (59), was a generous gift of W. Ruyechan (State University of New York at Buffalo).
(iv) Secondary antibodies.
Goat antibodies conjugated with either fluorescein isothiocyanate or Texas Red and directed against rabbit, mouse, or rat immunoglobin were obtained from Cappel, Organon Teknika Corporation (Durham, N.C.).
(v) Other reagents used for immunofluorescence assay (IF).
Glycerol gelatin and 1,4-diazobicyclo-[2.2.2]octane were obtained from Sigma.
Transfection of mammalian cells.
Vero cells were transfected with various plasmids using Lipofectamine-plus (Gibco BRL). Cells were plated in 60-mm-diameter tissue culture dishes at a density of 106 cells/plate approximately 24 h prior to transfection. Cells were transfected by following the manufacturer's instructions.
Isolation of cell lines stably expressing PML splice variants.
Four PML cDNA clones on a simian virus 40 promoter expression plasmid were generously provided by M. Fagioli (Perugia, Italy). Clones PML1-[3,4,5,6,7], PML1-[3,4,7], and PML3-[3,4,6,7] were previously described (26), whereas clone PML3-[3,7] was not previously reported (Fagioli et al., unpublished data). Vero cells were stably cotransfected with a plasmid bearing the G418 resistance gene and a 10-fold excess of one of the PML cDNA clones. Cells were allowed to recover for 24 h posttransfection before the addition of 1 mg of G418 per ml to the medium. Individual colonies were twice cloned by single-colony isolation and screened for PML expression by indirect IF.
Indirect IF.
Cells were grown on glass coverslips prior to infection. Cells on coverslips were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min, washed in PBS, and permeabilized in 1.0% Triton X-100 in PBS for 10 min. The coverslips were again washed in PBS and pretreated with 3% normal goat serum in PBS for several minutes. The antibodies PG-M3, αICP8, and αPol and the secondary antibodies were used at a dilution of 1:200 in 3% normal goat serum in PBS. The rat anti-PML antibodies and the M1 antibody were used at a dilution of 1:100; the 5E10 antibody, a hybridoma supernatant, was used undiluted. Cells were stained with the primary antibodies for 30 min. Coverslips were washed six times with PBS between primary and secondary antibody treatments. Cells were then stained with secondary antibodies for 30 min. Coverslips were then washed extensively in PBS and mounted in glycerol gelatin containing 2.5% 1,4-diazabicyclo-[2.2.2]octane to retard bleaching.
Imaging.
Imaging was performed on a Zeiss Axiovert 135 laser scanning confocal microscope equipped with an argon-krypton laser. Texas Red was excited at 568 nm; fluorescein isothiocyanate was excited at 488 nm. Emissions were collected separately, and the channels were overlaid by computer for the dual images. Images were collected with either a 63× Neofluar lens or a 100× Zeiss apochromat lens and arranged and labeled using Adobe Photoshop 5.0.
Western blotting.
Protein expression was examined by Western blot analysis. Detection of Pol protein was carried out by infecting HEp-2 cells with various viruses bearing UL30 mutations at a multiplicity of infection (MOI) of 10 and collecting infected cells at 8 h postinfection. Cells were lysed in sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) loading buffer, sonicated, boiled for 5 min, and loaded onto an SDS–8% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane, and Pol was detected as described below. For detection of PML, cells were grown on 60-mm tissue culture plates, transiently transfected with one of the PML cDNA plasmids 18 h prior to collection, and infected with 100 PFU of KOS per cell 6 h prior to collection. Cells were washed once with Tris-buffered saline and scraped into 1 ml of TBS supplemented with leupeptin, pepstatin, and EDTA. Cells were then pelleted and resuspended in 50 to 100 μl of 5× SDS-PAGE loading buffer supplemented with protease inhibitors. PML samples to be examined with the PG-M3 antibody were resuspended in buffer lacking β-mercaptoethanol (BME), as its presence interferes with the antibody's ability to recognize the protein (data not shown). Each sample was sonicated to decrease the viscosity of the solution. Samples were then boiled for 5 min and promptly loaded onto SDS–10% polyacrylamide gels. When the dye front had run off the gel, the proteins were transferred to a nitrocellulose membrane at 350 mA for 2 h. The membrane was blocked with 5% nonfat dry milk in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 80) for 2 h and incubated in the primary antibodies overnight. The αPol antibody was used at 1:10,000, 5E10 was used at 1:500, and PG-M3 was used at 1:5,000. Membranes were then incubated in secondary antibodies at a 1:10,000 dilution for 1 to 2 h, washed, and developed with alkaline phosphatase color detection (Promega).
RESULTS
Only some isoforms of PML are recruited to replication compartments.
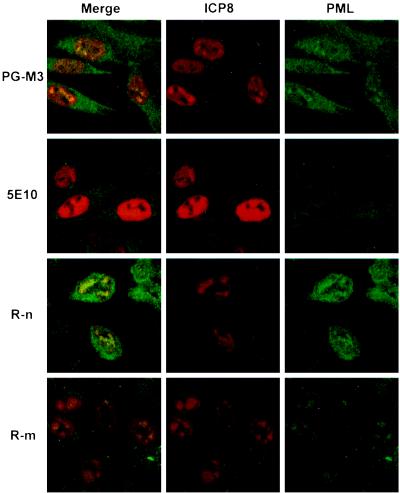
We and others have previously observed that PML and the ND10 antigen recognized by Mab138 can be recruited to stage III prereplicative sites (7) and into replication compartments (7, 35, 53). Since several forms of PML have been shown to be degraded after HSV-1 infection (20), we set out to determine which isoforms of PML are recruited to replication compartments by examining HSV-1-infected cells with several different PML antisera. The PML antisera used include the PG-M3 monoclonal antibody (25), monoclonal antibody 5E10 (65), and two polyclonal antibodies of rat origin, R-m and R-n. Each antibody was tested by indirect IF for the ability to stain ND10 in uninfected HEp-2 cells and for the ability to stain replication compartments in infected cells. We found that all of the antibodies stained uninfected HEp-2 cells in a pattern identical to that previously described for ND10 (data not shown). In cells infected with HSV-1, the polyclonal rat antibody R-n and the monoclonal antibody PG-M3 showed PML staining in replication compartments; the polyclonal rat antibody R-m also showed faint PML staining in replication compartments; the monoclonal antibody 5E10, however, did not show PML staining in replication compartments (Fig. 1). This result suggests that only some isoforms of PML are recruited to replication compartments. Boulware and Weber (6a) recently published a report suggesting that PG-M3 cross-reacts with a viral protein if used undiluted. To control for possible cross-reaction with viral proteins found in replication compartments, HSV-1-infected Vero cells were stained with PG-M3, which does not recognize monkey PML. In infected Vero cells, PML staining was not seen in replication compartments using the PG-M3 antibody at the dilution used in this study (reference 6a and data presented below). Thus, we concluded that the PG-M3 staining observed in replication compartments in HEp-2 cells reflects the presence of PML at these sites. The observation that 5E10 staining was not seen in replication compartments whereas PG-M3 and rat polyclonal antibody staining was observed supports our proposal that only some isoforms of PML are recruited to replication compartments (7).
FIG. 1.
Indirect IF examination of HSV-1-infected cells using four anti-PML antibodies. At 6 h postinfection with KOS, HEp-2 cells were stained with a polyclonal antibody against the major HSV-1 DNA-binding protein ICP8 (shown in red) and one of the following antibodies against PML: monoclonal antibody PG-M3 or 5E10 or rat polyclonal antibody R-n or R-m (shown in green).
PG-M3 and 5E10 react differently with PML encoded by four cDNA clones.
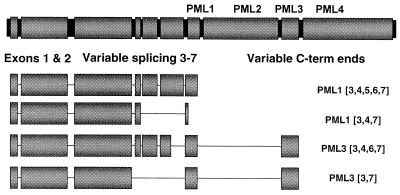
Although PML is encoded by a single-copy gene in human cells, multiple forms of PML are observed due to splice site variation and posttranslational modification (23, 47, 49, 62). The PML gene consists of nine exons in three domains: (i) a relatively invariant N-terminal portion consisting of exons 1 and 2, (ii) a variably spliced middle region consisting of exons 3 through 7, and (iii) four distinct C-terminal exons named PML1, PML2, PML3, and PML4 (23) (shown in Fig. 2). Thus, PML can exist in 16 different forms for each of the four C termini (23). Furthermore, the protein is known to be phosphorylated and covalently modified by the small, ubiquitin-like protein SUMO-1 (49, 62). The SUMO modification can occur at three sites in the protein: two of the sites are found in the invariant N terminus, and the third site is found in the variably spliced region (and therefore is not present in all clones) (16, 32). We set out to examine whether the heterogeneity of PML isoforms is responsible for the differential staining of replication compartments by the antibodies PG-M3 and 5E10.
FIG. 2.
Genomic PML and the PML splice variants used in this study. PML is diagrammed showing three major domains: the invariant N-terminal portion consisting of exons 1 and 2, the variably spliced middle region consisting of exons 3 through 7, and four distinct C termini called PML1 (continuous with exon 7), PML2, PML3, and PML4. The four representative cDNA clones used in this study are shown: PML1-[3,4,5,6,7], which contains the PML1 C terminus and all of variably spliced exons 3 through 7 (predicted to encode a protein of approximately 67 kDa); PML1-[3,4,7], which contains the PML1 C terminus and exons 3, 4, and 7 (predicted to encode a protein of approximately 48 kDa; this isoform contains a premature stop codon which generates a smaller C-terminal (C-term) end than the PML1-[3,4,5,6,7] isoform described above); PML3-[3,4,6,7], which contains the PML3 C terminus and exons 3, 4, 6, and 7 (predicted to encode a protein of approximately 65 kDa); and PML3-[3,7], which contains the PML3 C terminus and exons 3 and 7 (predicted to encode a protein of approximately 54 kDa.
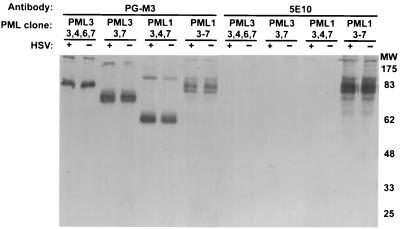
We obtained four human cDNA clones of PML from Marta Fagioli. Each clone expresses a different version of the variably spliced domain (exons 3 through 7) (shown in Fig. 2). Two express the C-terminal exon PML1, and two express the C-terminal exon PML3. To test whether the 5E10 and PG-M3 PML antibodies react specifically with these PML clones, Vero cells were transiently transfected with the cDNA plasmids and assayed for PML by Western blotting as described in Materials and Methods. The PML endogenous to Vero cells is not detected by either antiserum and therefore does not interfere with this assay. Omission of BME from the loading buffer is necessary for the PG-M3 antibody to react with PML (data not shown): presumably, disulfide bonds within the protein are required for antibody recognition. Replica Western blots of lysates of cells transfected with the four PML cDNA clones performed with PG-M3 and 5E10 are shown in Fig. 3. The PG-M3 antibody reacts with cells transfected with each of the four PML cDNA clones (Fig. 3). Each of the PML splice variants migrated slightly slower on SDS-PAGE than expected for the predicted size, which may be due to differences in migration conditions with the omission of BME. Each lane in Fig. 3 contains high-molecular-weight proteins that likely represent multimers of PML. The PG-M3 antibody was raised against exons 1 and 2 (25) and presumably recognizes this portion of the protein in each clone. In contrast, the 5E10 monoclonal antibody reacts only with the PML1-[3-7] clone (Fig. 3). PML1-[3-7] contains all of the variably spliced exons, 3, 4, 5, 6, and 7, and is the only clone that contains exon 5 (Fig. 2 and 3). Thus, it seems that the presence of exon 5 allows the protein to be recognized by 5E10. In summary, the Western blots in Fig. 3 demonstrate that two PML antibodies, PG-M3 and 5E10, react differently against the four isoforms of PML. This difference may be due to the recognition of a particular exon or to a conformational difference in the protein due to the presence of that exon.
FIG. 3.
Western blots of Vero cells transfected with four PML cDNA clones and reacted with antibodies PG-M3 or 5E10. Vero cells were transfected with PML cDNA clones PML3-[3,4,6,7], PML3-[3,7], PML1-[3,4,7], and PML1-[3–7] (referred to as PML1-[3,4,5,6,7] in the legend to Fig. 2) 18 h prior to collection. Cells (+) were infected with 100 PFU of KOS virus per cell 6 h prior to collection. At collection, cells were subjected to SDS-PAGE in the absence of BME and analyzed by Western blotting as described in Materials and Methods. MW, molecular weight (103).
Since Everett et al. reported that HSV-1 infection causes the proteasome-dependent loss of higher-molecular-weight forms of PML (20), we examined the isoforms encoded by the PML clones in cells infected with 100 PFU of wild-type HSV-1 per cell 24 h after transient transfection with the PML expression vectors. We did not observe a reliable difference between the levels of PML in infected and uninfected cells by either antibody (Fig. 3). Although these results appear to contradict those obtained by Everett et al. (20), it is possible that the overexpression of PML in this experiment hampered either demodification of PML or viral infection in general.
Only one of the four tested PML cDNA clones localizes to replication compartments.
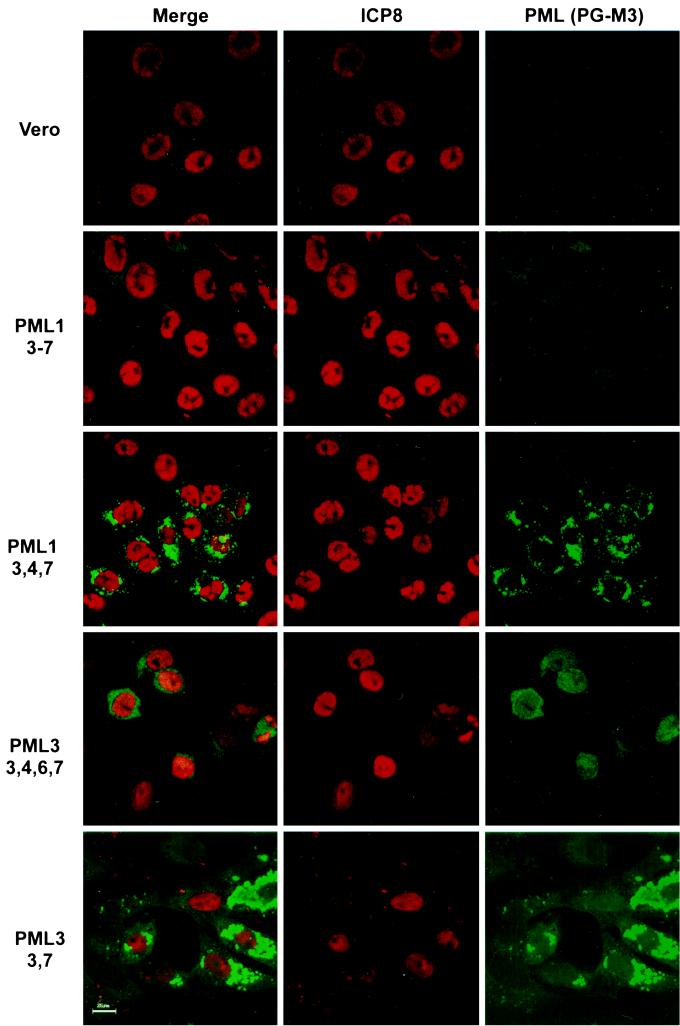
We concluded from the results presented in Fig. 1 and 3 that only some isoforms of PML are recruited to replication compartments. We set out to identify the isoform(s) of PML recruited to viral replication compartments by testing the four representative PML cDNA clones for recruitment to viral replication compartments. We constructed cell lines by stably transfecting Vero cells with each of the PML clones. Stable cell lines would express the PML isoforms in nearly 100% of the cells. Furthermore, the use of stable cell lines would circumvent possible problems or difficulties caused by transient transfections. Cells transfected with the PML3-[3,7] clone stopped expressing PML early during selection. Failure to establish a stably expressing line may indicate that overexpression of this isoform of PML is toxic to cells. Indeed, overexpression of at least one isoform of PML is known to retard cell growth and even cause apoptosis (46, 54). To establish the behavior of this isoform during HSV infection, cells were transiently transfected and then superinfected at 24 h to examine recruitment of PML to replication compartments (see below).
Cell lines containing PML1-[3-7], PML1-[3,4,7], and PML3-[3,4,6,7] and cells transiently transfected with PML3-[3,7] were infected with KOS and examined for PML recruitment to replication compartments. Only cells expressing PML3-[3,4,6,7] showed PML staining in replication compartments (Fig. 4). As can be seen in Fig. 4, the cell lines containing PML1-[3-7] and PML1-[3,4,7] did not exhibit PML staining in replication compartments although they did show PML staining in ND10 prior to infection (data not shown). PML3-[3,7] protein also failed to be recruited to replication compartments (Fig. 4); this PML isoform was primarily cytoplasmic prior to infection (data not shown). These experiments indicate that the PML3-[3,4,6,7] splice variant of PML can be recruited to replication compartments but PML1-[3-7], PML1-[3,4,7], and PML3-[3,7] cannot. This experiment demonstrates that only one of the differentially spliced isoforms of PML tested can be recruited efficiently into replication compartments in the PML-expressing cell lines. However, at this time we cannot conclude which endogenously expressed PML isoforms are actually recruited into replication compartments during infection. It is possible that in our experiment, overexpression of PML altered the modification state which, in turn, resulted in degradation or recruitment. Further experiments with more sensitive reagents are required to determine which endogenous PML isoforms are recruited to replication compartments during infection; however, we have clearly demonstrated that various PML isoforms behave differently from one another during infection.
FIG. 4.
Localization of various isoforms of PML following infection. Vero cells; Vero cells stably transfected with PML cDNA clones PML1-[3–7], PML1-[3,4,7], and PML3-[3,4,6,7]; and Vero cells transiently transfected with PML3-[3,7] were infected with KOS at an MOI of 10 PFU/cell for 6 h. All infected cells were prepared for indirect IF with a polyclonal antibody against the HSV-1 major DNA-binding protein ICP8 and monoclonal antibody PG-M3 against the cellular ND10 protein PML. Bar, 25 μm.
HSV-1 Pol requirement for PML recruitment to replication compartments.
PML is recruited to viral replication compartments and also to a subset of prereplicative sites that we have named PML-associated prereplicative sites (35). PML-associated prereplicative sites, also called stage III foci, do not colocalize with sites of cellular DNA synthesis as do other prereplicative sites (35, 68). We showed, however, that in cells infected with a Pol (UL30) null virus, sites similar to stage III foci containing ICP8 form but they do not recruit PML (7). Thus, we concluded that the recruitment of PML to PML-associated prereplicative sites requires the presence of the HSV-1 DNA polymerase (7). PML could be recruited to stage III foci in the presence of polymerase inhibitors, indicating that polymerase activity per se is not required for PML recruitment (35). In this study, we took a genetic approach to this problem in order to determine whether the recruitment of PML to replication compartments depends on a particular protein domain on the Pol protein, on a biochemical activity of Pol not affected by pharmacological polymerase inhibitors, or merely on the presence of the intact Pol protein. In order to test which feature of the polymerase protein is required for PML recruitment, we examined several HSV-1 Pol mutants which we have divided into three classes.
Viable Pol mutants.
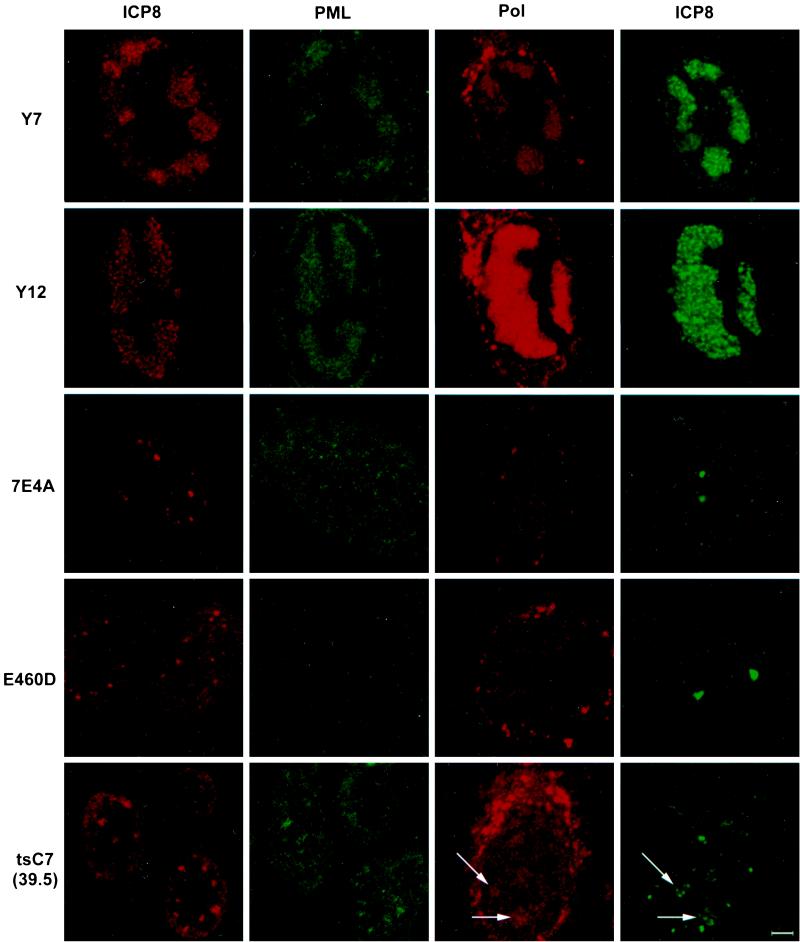
The first class of polymerase mutants included the viable Pol protein mutants that exhibited altered substrate specificity (AraAr9, 615.8, F891C, and PAAr5), severely impaired exonuclease activity (Y7 and YD12), or no apparent phenotype (V426A) (13, 14, 26, 29, 37, 57). All of the viable Pol mutants replicate on nonpermissive cells, such as Vero cells. Each of these mutants forms replication compartments on Vero cells (Table 1). In each case, the mutant Pol protein localized to replication compartments in infected cells (Table 1). Furthermore, we found that in HEp-2 cells infected with each of these mutants, PML was recruited to the replication compartments (Table 1). Figure 5 shows the staining pattern of Y7, which is typical for each of these mutants: HEp-2 cells infected with Y7 were stained for either PML or Pol and simultaneously stained for ICP8 to indicate the location of the replication compartments in the cell. In this case, both PML and Pol localized to replication compartments. These data suggest that PML recruitment is not sensitive to mutations in the active site of Pol which affect drug triphosphate binding or incorporation (AraAr9, 615.8, F891C, and PAAr5). These data also show that exonuclease activity can be severely impaired without affecting PML recruitment to replication compartments (Y7 and YD12). In both cases, PML and Pol localize to replication compartments. Interestingly, the mutant YD12 showed increased Pol protein staining in the replication compartments but did not show increased PML recruitment (Fig. 5). The increased amount of the Pol protein may be due to a high particle-to-PFU ratio for YD12 since the exonuclease defect leads to increased mutation rates within the viral stocks that could result in particles that can produce protein but are defective for plaque formation.
TABLE 1.
Summary of behavior of polymerase mutant viruses
| Virus | Growtha | Pol proteinb | Pol at focic | PML at focid |
|---|---|---|---|---|
| KOS | +++ | +++ | +++ | +++ |
| V462A | +++ | +++ | +++ | +++ |
| Arar9 | +++ | +++ | +++ | +++ |
| 615.8 | +++ | +++ | +++ | +++ |
| F891C | +++ | +++ | +++ | +++ |
| PAAr5 | +++ | +++ | +++ | +++ |
| Y7 | +++ | +++ | +++ | +++ |
| YD12 | +++ | +++ | +++++ | +++ |
| HP66 | — | — | — | — |
| ΔX17 | — | — | — | — |
| ΔX14 | — | — | — | — |
| ΔS1.1 | — | — | — | — |
| 7E4A | — | — | — | — |
| 6C4 | — | ++ | — | — |
| E460D | — | +++ | — | — |
| G464V | — | +++ | — | — |
| tsC4 | — | +++ | — | — |
| tsC7 | — | +++ | + | ++ |
Indicates ability of mutant virus to form plaques on noncomplementing cell lines. +++++, higher than wild-type levels; +++, wild-type levels; ++, slightly lower than wild-type levels; +, significantly lower than wild-type levels; —, undetectable.
Indicates ability of mutant virus to synthesize Pol protein detectable by Western blot analysis.
Indicates whether Pol protein is detectable at viral prereplicative sites.
Indicates whether PML protein is detectable at viral prereplicative sites.
FIG. 5.
Localization of ICP8, PML, and Pol in cells infected with various Pol mutant viruses. Cells were infected with wild-type or mutant HSV-1 for 6 h and double stained with antibodies against the major viral DNA-binding protein ICP8 and either the cellular ND10 protein PML (PG-M3) or the viral polymerase protein (M1). The first two columns depict the same cell labeled with antibodies recognizing ICP8 (red) or PML (green); the second two columns depict a different cell colabeled with antibodies to ICP8 (green) and Pol (red). Y7 and YD12 represent the mutants that are viable on Vero cells. 7E4A represents the mutants that made no Pol that was detectable by Western blotting. E406D represents the mutants that made Pol that was detectable by Western blotting but did not show Pol localization to prereplicative sites. At the nonpermissive temperature of 39.5°C, tsC7 made Pol that was detectable by Western blotting and showed weak localization of Pol to accumulations of ICP8 (arrows); this phenotype is seen in PAA-treated KOS-infected cells (not shown). The cytoplasmic staining observed in these experiments may be due to secondary antibody binding of the viral Fc receptors in the endoplasmic reticulum and Golgi apparatus of infected cells. Bar, 10 μm.
Nonviable Pol mutants that did not make Pol as determined by Western blot analysis.
The second class of mutants we examined consisted of nonviable Pol mutants which did not make Pol protein that was detectable by the αPol (15) antibody by Western blot analysis (data not shown): HP66, ΔX17, ΔX14, ΔS1.1, and 7E4A (38). Although ICP8-containing foci could be observed in HEp-2 cells infected with each of these null mutants, these cells did not show Pol staining in viral foci by indirect IF or exhibit PML recruitment to viral foci (Table 1). One example of this class, HEp-2 cells infected with 7E4A, is shown in Fig. 5. In this case, foci of ICP8 staining were observed; however, neither Pol nor PML was present at these foci.
Nonviable Pol mutants that make Pol protein that is detectable by Western blot assay.
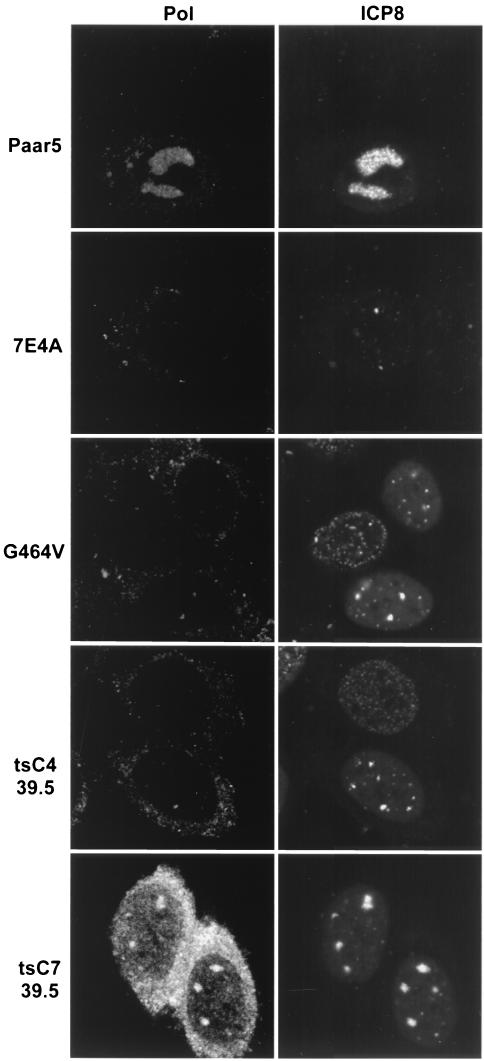
The third class of polymerase mutants examined made full-length protein that was detectable by Western blot analysis (data not shown) but were nonviable on nonpermissive HEp-2 or Vero cells (6C4, E460D, and G464V) or on HEp-2 or Vero cells at the nonpermissive temperature (tsC4, and tsC7) (13, 26, 38). At 34°C, the temperature-sensitive (ts) mutants behaved in all respects like wild-type HSV-1 (KOS) (data not shown). Under nonpermissive conditions, cells infected with each of these mutants exhibited ICP8-containing foci. HEp-2 cells infected with E460D at 37°C showed no Pol or PML recruitment into prereplicative sites (Fig. 5); thus, these mutants make Pol but it does not localize to viral foci. Cells infected with tsC7 at 39.5°C showed both Pol and PML recruitment to prereplicative sites (Fig. 5). We believe that the weak recruitment of Pol to replication compartments seen in Fig. 5 is real because of results obtained with another polyclonal antibody, αPol (shown in Fig. 6). When cells infected with tsC7 at 39.5°C and with the viable mutant PAAr5 were stained with the αPol antibody, Pol recruitment to areas of ICP8 staining was clearly observed; however, in cells infected with 7E4A or G464V at 37°C or tsC4 at 39.5°C, no Pol recruitment could be detected (Fig. 6). Thus, none of the nonviable mutants except tsC7 were able to recruit Pol or PML to stage III foci or replication compartments (Table 1).
FIG. 6.
Localization of ICP8 and Pol in cells infected with various Pol mutant viruses. Cells were infected as described in the legend to Fig. 5 but stained with the Pol antibody αPol. Paar5 represents the viable mutants that showed Pol in replication compartments. 7E4A represents the Pol-null viruses with no Pol staining of ICP8 foci. G464V and tsC4 at 39.5°C represent nonviable mutants that did not show Pol at foci of ICP8. The ts mutant tsC7 is the only nonviable mutant that showed Pol at foci of ICP8. All of these microphotographs were taken on the same day at the same brightness and contrast.
At 39.5°C, tsC7-infected cells recruit PML to stage III foci despite the fact that the polymerase protein is not catalytically active. This resembles the phenotype of cells infected with wild-type viruses and treated with phosphonoacetic acid (PAA) (7). In both situations, the Pol protein is inactive but properly localized to viral foci, and in both situations, the foci recruit PML. To rule out the possibility that tsC7 recruits Pol and PML to stage III foci at 39.5°C only because of a leaky ts phenotype, we examined tsC7 infections at 39.5°C for bromodeoxyuridine incorporation and for production of infectious virus. No DNA replication was detectable by bromodeoxyuridine staining at 6 or 24 h postinfection (data not shown). Furthermore, fewer than 50 PFU/ml were produced at 39.5°C 24 h postinfection, compared to 4 × 107 PFU/ml at 34°C, indicating that this mutant polymerase is operationally inactive at the nonpermissive temperature. In summary, both Pol and PML are located in prereplicative sites in tsC7-infected cells; PAA-treated, KOS-infected cells; and all viable-mutant-infected cells. Yet neither is present in prereplicative sites formed in cells infected with other nonviable Pol mutants. Thus, under conditions in which Pol fails to localize to viral prereplicative sites, PML also fails to be recruited.
Do factors that affect the localization of PML or the structure of ND10 affect the replication of the virus?
If ND10 and/or ND10 proteins like PML play a significant role in the life cycle of the virus, then one might expect factors affecting the structure of ND10 or the modification of ND10 proteins would also affect viral replication. We addressed this hypothesis by examining the stages of infection under two conditions known to modulate ND10: cells infected in the presence of arsenic trioxide and cells infected in the presence of the proteasome inhibitor MG132.
Arsenic trioxide.
Reports from China describing arsenic trioxide as the active ingredient in an ancient treatment for acute promyelocytic leukemia (11, 12) led to the discovery that As2O3 acts on ND10 and ND10 proteins (49, 63). Arsenic trioxide causes increased SUMO-1 modification and ND10 partitioning of PML and Sp100 (43, 49, 50, 54, 63, 75). Arsenic trioxide was subsequently shown to increase ND10 size in all cells, not only those containing the PML-retinoic acid receptor α fusion protein (2, 49, 50, 63). Following the increase in PML partitioning to ND10, some isoforms of PML are degraded (75). Because of these effects on ND10 and ND10 proteins, we decided to examine the effect of arsenic on HSV-1 infection. We hypothesized that increased ND10 protein partitioning might make ND10 more resistant to viral disruption and may therefore affect the progression of the virus from stage I to stage II of viral infection.
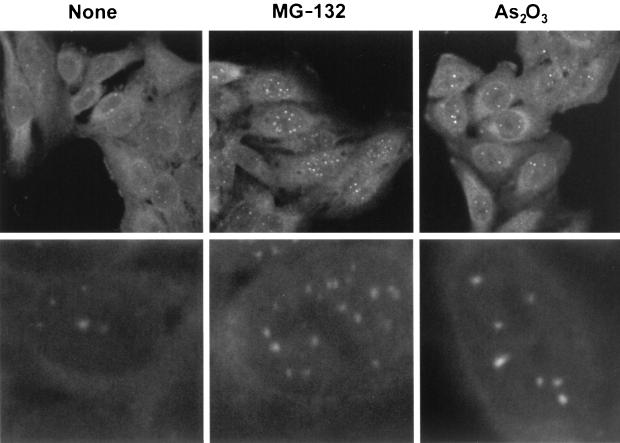
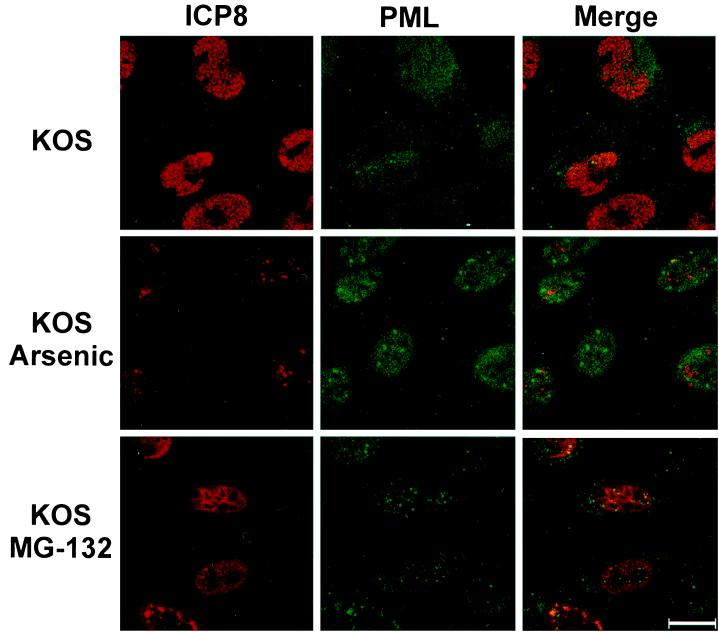
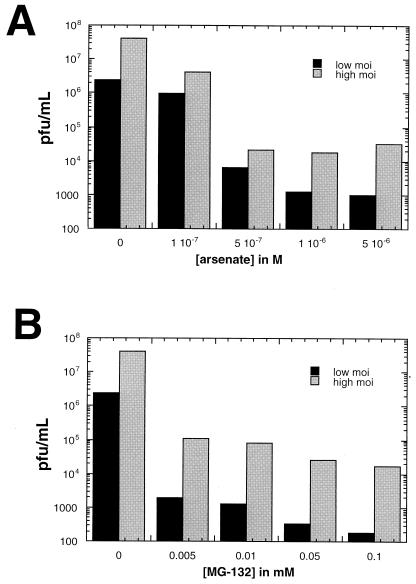
When HEp-2 cells were treated with As2O3 at a concentration of 10−6 M, ND10 increased in size but not in number (Fig. 7). A single-cell IF was performed at 6 h postinfection on cells treated with the same concentration (10−6 M) of As2O3. Figure 8 shows that cells pretreated with As2O3 were less likely to contain replication compartments and more likely to show intact ND10. In these cells, ICP8 often formed aggregates that did not colocalize with PML and did not resemble the well-organized, speckled appearance of HSV-1 replication compartments (Fig. 8). In contrast, untreated cells exhibited replication compartments in nearly every cell. At the concentration used in this assay (10−6 M), cells were still dividing at 24 h albeit with slightly longer doubling times. Virus production was measured in cells pretreated with As2O3 concentrations ranging from 1 × 10−7 to 5 × 10−6 M (Fig. 9). Cells were treated for 30 min prior to infection with HSV-1 strain KOS at an MOI of either 0.1 PFU/cell (low MOI) or 10 PFU/cell (high MOI). Virus was harvested at 24 h postinfection. Figure 9 shows that at concentrations of 5 × 10−7 and higher, virus production was decreased significantly, with the most severe decreases seen in the low-MOI infections. In these and other experiments (data not shown), we have observed that ND10 stabilization correlates with decreased viral yields. Although we cannot rule out some toxic effects of As2O3 treatment, it appears to inhibit the assembly of stage III foci and prevent the disruption of ND10. These data indicate that arsenic treatment can prevent the earliest stages of HSV-1 infection.
FIG. 7.
Changes in ND10 morphology following treatment with arsenic or a proteasome inhibitor. Two magnifications of HEp-2 cells stained for the cellular ND10 protein PML are shown, either with no treatment or after treatment with 0.05 mM MG-132 or 10−6 M As2O3.
FIG. 8.
Localization of ICP8 and PML in cells treated with arsenic or MG-132. HEp-2 cells were infected with KOS at an MOI of 10 for 6 h in the presence or absence of 10−6 M As2O3 or 0.05 mM MG-132. Cells were pretreated with drug for 30 min prior to infection. After fixation, cells were stained for ICP8 (shown in red) and PML (shown in green). Bar, 25 μm.
FIG. 9.
Viral yields after infection in the presence or absence of As2O3 or MG-132. (A) HEp-2 cells were pretreated for 30 min with normal medium (0) or concentrations of As2O3 ranging from 10−7 to 5 × 10−6 M. After drug treatment, the cells were infected with KOS at an MOI of 0.1 or 10 PFU/cell. Viral stocks were collected at 24 h postinfection, and titers were determined on Vero cells. This experiment was repeated several times, and the data from one representative experiment are shown. (B) HEp-2 cells were pretreated for 30 min with normal medium (0) or concentrations of MG-132 ranging from 0.005 to 0.1 mM and infected as described for panel A. Viral stocks were collected, and titers were determined as described above. This experiment was repeated several times, and the data from one representative experiment are shown.
MG-132.
High-molecular-weight forms of PML, modified by SUMO, have been reported to disappear soon after infection (9, 20, 48). Studies utilizing the proteasome inhibitor MG-132 at a concentration of 0.05 mM show that treatment with MG-132 prevents the loss of higher-molecular-weight forms of PML and inhibits viral infection at an immediate-early stage (20). Furthermore, it appears that the inhibition of viral growth correlated with the retention of ND10 in infected cells (9). In uninfected cells, MG-132 causes an increase in the size and number of ND10, so we decided to examine the effects of this proteasome inhibitor on both the structure of ND10 and the recruitment of PML to viral replication compartments.
HEp-2 cells were treated with MG-132 at a concentration of 0.05 mM, and ND10 increased in both number and size (Fig. 7). When MG-132 (0.05 mM)-treated cells were examined by IF the inhibitory effect of the proteasome inhibitor on viral infection was striking. As reported previously (9), these ND10 were resistant to disruption by HSV-1 infection. Compared to the robust replication compartments seen in untreated cells, MG-132-treated cells showed either disorganized ICP8 staining (>90%) or staining in a pattern that resembled stage III foci (<10%) (Fig. 8) but no cells contained replication compartments. Cells infected at an MOI of 0.1 exhibit intact ND10 and little ICP8 staining (data not shown). Thus, at a low MOI, MG-132 inhibits progression to stage II; at a high MOI, MG-132 inhibits disruption of ND10 in many cells and abolishes the formation of replication compartments. As shown in Fig. 9, cells were pretreated prior to infection with concentrations of MG-132 ranging from 0.005 to 0.1 mM and assayed for virus production as described above. Infected cells showed a significant decrease in virus production at all of the drug concentrations tested. Again, the most severe decreases were observed in cells infected at a low MOI (Fig. 9). At the drug concentration used for the single-cell assay (0.05 mM), cell toxicity was not evident at 6 h postinfection but was clearly evident by 24 h. Toxicity was minimal at the lowest concentration used in the virus production assay (0.005 mM) and was quite evident at the highest concentration (0.1 mM) (data not shown). Thus, although MG-132 displays toxicity at 0.05 M, the concentration used here and by others (9, 19), our experiments confirm previous reports that stabilization of ND10 correlates with inhibition of progression of viral infection (9, 19).
DISCUSSION
In this study, the complex interactions between viral and host proteins were monitored using a single-cell assay and our results suggest that the complex interactions that occur between HSV and ND10 are important for the ability of the virus to establish a productive infection. In these studies, we have investigated the interaction of HSV-1 with ND10 and ND10 proteins. The following observations have been made. (i) Some, but not all, isoforms of PML are recruited to viral replication compartments. (ii) PML is recruited to viral foci only when the catalytic subunit of the HSV-1 polymerase protein is located in viral foci. (iii) Treatment of infected cells with compounds that increase the stability of ND10 inhibits the formation of replication compartments and viral replication.
ND10 disruption is important for the progression of viral infection.
The single-cell assay and time course experiments result presented in this paper support the notion that ND10 disruption is critical for productive infection: during the first 6 h of infection, replication compartments did not readily form in the presence of ND10-stabilizing arsenic or MG-132 (9, 20). Others have found that interferon treatment, which is also known to increase the size and numbers of ND10, has a dramatic effect on the ability of the virus to disrupt ND10 and establish a productive infection (67). In addition, overexpression of the ND10 protein PML also appears to inhibit HSV-1 infection (10; H. Yamada and S. K. Weller, unpublished results). These results are all consistent with the notion that disruption of ND10 is required for the formation of stage III foci and replication compartments.
We have previously reported, however, that in cells transfected with the seven essential HSV replication genes, replication compartments form adjacent to presumably intact ND10 (36). Furthermore, Maul et al. reported (42) that in cells infected with an HSV-1 mutant lacking ICP0, viral replication compartments formed at sites adjacent to ND10. These results conflict with the observation that during infection, ND10 disruption is required for replication compartment formation. We propose three scenarios to explain these apparent discrepancies. (i) ND10 disruption is not required for replication compartment formation. (ii) Replication compartment formation during transfection or during infection with an ICP0 mutant may be fundamentally different from replication compartment formation during wild-type infection. In the transfection experiments, the HSV replication genes were expressed from the major immediate-early human cytomegalovirus promoter (36). It is possible that demodification of ND10 proteins and disruption of ND10 are required for optimal gene expression during wild-type infection but would not be required for expression from the cytomegalovirus promoter. (iii) Whether or not ND10 are actually disrupted may depend on the antisera used to detect them. We have shown, for instance, that various PML isoforms behave differently during infection. It is possible that some ND10 proteins are dispersed under certain conditions while others remain associated in ND10. Modification states of ND10 proteins may also play an important role. It will be important in the future to use several different reagents to monitor the status of ND10s during infection and under other experimental conditions, since various isoforms may undergo different fates, including degradation, disruption from ND10, or recruitment to ND10. The observations made in this study suggest that at least some isoforms of PML are dispersed from ND10 prior to formation of stage III foci and replication compartments in infected cells.
PML is recruited to viral foci only when the catalytic subunit of the HSV-1 polymerase protein is located in viral foci.
In this paper, we also show that PML is not recruited to viral foci when the catalytic subunit of the HSV-1 polymerase protein (UL30) is not located in viral foci. Seventeen HSV-1 polymerase mutants were tested for the ability to recruit PML to replication compartments. All of the mutants that showed localization of the catalytic subunit of polymerase to viral foci showed recruitment of PML to viral foci, whereas mutants whose polymerase protein failed to localize to these sites also failed to recruit PML to viral foci. This may indicate that Pol interacts directly with PML to promote recruitment, but the IF staining patterns of the two proteins are located in slightly different patterns of speckles within replication compartments. The PML microspeckles do not appear to colocalize perfectly with the ICP8 microspeckles described by Liptak et al. (34). These data suggest that if an interaction between Pol and PML occurs, it may be indirect. Another possible explanation for these results is that PML may be recruited only to a fully formed viral DNA replication complex like a replisome, a small factory of replication proteins (4, 74). The assembly of such a complex would require the viral DNA polymerase but not necessarily polymerase activity. This model is based on the observation that both tsC7-infected cells and PAA-treated, KOS-infected cells show PML recruitment to prereplicative sites but do not exhibit polymerase activity (data not shown). Thus, we hypothesize that when Pol binds to the viral replication complex at the onset of DNA replication, it causes a conformational change at the replication fork that then allows recruitment of PML to the viral foci. Taken together, these results indicate that the HSV-1 polymerase, a heterodimer of UL30 and UL42, plays two roles in the development of HSV-1 replication compartments. The first is its ability to organize viral foci, which allows the recruitment of PML, and the second is its catalytic activity, which results in the replication of viral DNA. The assembly of an active HSV-1 replisome, which would contain the essential viral DNA replication proteins, may also be part of the early-late shift in viral gene expression that occurs at the onset of DNA replication, since viral DNA replication has been implicated in the early-late shift. When recruited to viral foci, PML may have a role in the transcriptional program of the virus, since it is known to affect cellular transcription patterns (27) and cell growth (46). We conclude from these studies that the recruitment of PML depends on the presence of the HSV-1 polymerase protein in replication complexes or replisomes that form within prereplicative sites.
Some, but not all, isoforms of PML are recruited to viral replication compartments.
In this study, we confirmed and extended our initial observation that at least one isoform of PML is recruited to stage III foci and replication compartments in infected cells (7). Antibodies expected to recognize most isoforms of PML show PML recruitment to replication compartments (PG-M3 and two polyclonal antibodies), whereas a monoclonal antibody (5E10) that recognizes only a subset of isoforms does not show recruitment. We have shown that different splice variants of PML are recruited differently to viral replication compartments. It will be of considerable interest to pursue the relationship between the recruitment and the posttranslational modification state of PML vis-à-vis phosphorylation and modification by SUMO-1. SUMO-1 conjugation and deconjugation have been implicated in the regulation of a number of processes in yeast and higher eukaryotes, including cytokinesis and chromosome segregation (18, 66). Several recent observations indicate that HSV-1 proteins, specifically ICP0, interact with host cell proteins involved in the normal progression of the cell cycle and mitosis. For instance, it appears that ND10 proteins can be detected both at ND10 and at centromeres, suggesting a dynamic association between these two nuclear substructures (18, 19). ICP0 can localize to centromeres, cause the degradation of the centromeric protein CENP-C, and induce a specific G2/M block in the cell cycle (19). These observations provide one example of how HSV-1 has evolved complex strategies of intervention in host cell processes.
Events during early HSV-1 infection.
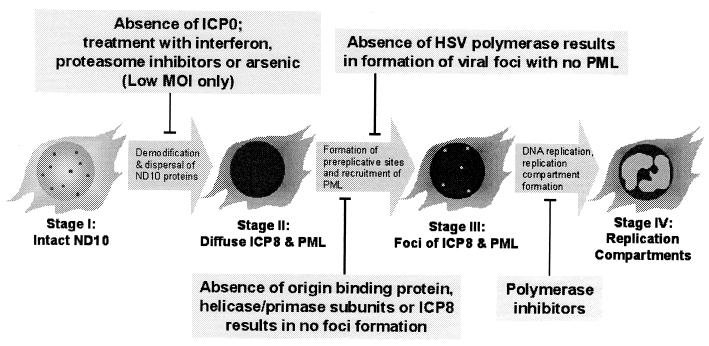
A diagram indicating the sequence of events during early HSV-1 infection is depicted in Fig. 10. Disruption of ND10 (progression from stage I to stage II) is sensitive to the MOI. At a low MOI, which likely represents in vivo infection conditions more closely than high-MOI infections, the progression from stage I to stage II is inhibited by treatment with arsenic, a proteasome inhibitor, or interferon (67). At higher MOIs, the infection can progress from stage I to stage II even in the presence of drugs. Thus, during natural infections, ND10 disruption may be required for the progression of HSV-1 infection. It remains a formal possibility, however, that ND10 disruption is a consequence of the progression of lytic HSV-1 infection rather than its catalyst.
FIG. 10.
Progression of HSV-1 infection occurs in four distinct stages. A sequence of events during the formation of replication compartments is shown along with the conditions which block progression of infection. See Discussion for a description of the diagram.
Progression from stage II to stage III involves the formation of PML-associated prereplicative sites containing ICP8 and at least one isoform of PML. We and others have previously shown that formation of foci of viral DNA replication proteins requires the presence of ICP8, UL9, and the three subunits of the helicase-primase (UL5, UL8, and UL52) during infection (7, 68). In this study, we have confirmed and extended our previous observation that HSV DNA polymerase (UL30) is required for the recruitment of PML into stage III foci. It is well established that the presence of polymerase inhibitors inhibits the formation of replication compartments (progression from stage III to stage IV) (55).
This work confirms that the correlation between ND10 disruption and productive infection is strong. ND10 stabilization may thus represent a new approach for inhibiting the viral life cycle. Several nonexclusive models can be considered for the role of ND10 in viral infection. (i) ND10 may represent nuclear defense centers that evolved to protect cells from viral infection. The observation that interferon, which protects the cell against viral infection, can increase the size and number of ND10 and can result in inhibition of viral infection supports this model (5, 28, 44, 45, 51, 67). This theory is also supported by the observation that ICP0-null mutants, which are defective for disruption of ND10 at nonpermissive MOIs, are exquisitely sensitive to interferon treatment in all cells except naturally permissive U2OS cells (45). The observation that many ND10 proteins function as transcriptional repressors (61) may also support this model, as it may be necessary to disperse transcriptional repressors in order to establish the lytic transcription program. (ii) ND10 may be sites of sequestered transcription factors and growth regulatory proteins that must be released to generate an environment conducive to efficient viral gene expression. (iii) ND10 may be centers from which cellular protein modification cascades originate. The ubiquitin-like peptide SUMO-1 that affects the localization of ND10 proteins may be such a signal. The infection-induced global loss of modified proteins like PML, Sp100, CENP-C, and DNA-PK (9, 18, 20, 22, 48, 52) and the reorganization of factors like p53 and pRb near ND10 (1, 8, 70) are consistent with a model in which HSV-1 can initiate a cellular signaling pathway that affects the growth and transcription status of the cell. The disruption of ND10 and changes in ND10 protein modification may thus be a novel pathway by which HSV-1 affects the cell cycle, cell division, and apoptosis. If the modification status of ND10 proteins signals a cascade of events leading to changes in the cell cycle status of the cell (18, 19, 58, 66), this signal may be usurped by the virus to create an environment conducive to productive infection. (iv) ND10 may be sites of deposition of nuclear proteins (39). This model is supported by the observation that many nuclear proteins localize to ND10 when overexpressed. (v) ND10 may mark sites in the cell, such as a nuclear matrix attachment site, necessary for the establishment of a productive infection (39). Mammalian DNA replication occurs in small, factory-like enzymatic centers called replisomes on the nuclear matrix (reviewed in reference 3); it is possible that HSV-1 also requires such an attachment for its DNA replication and that attachment occurs at or near the nuclear matrix-bound ND10. ND10 may need to be disrupted to allow efficient access to this site. (vi) ND10 may be the sites at which HSV-1 DNA circularization occurs. Under some circumstances, ND10 may facilitate recombination of telomeric DNA via the alternative lengthening of telomere pathway (73). A nuclear domain able to facilitate recombination may promote HSV DNA replication, since it has been proposed that only those genomes that circularize via homologous recombination, and not end joining, are capable of productive replication (72). Further experiments are necessary to distinguish between aspects of these models.
ACKNOWLEDGMENTS
We thank all members of the Weller laboratory for helpful discussions of the manuscript. We also thank Robin Pietropaolo for advice and Rik Martinez for performing the experiment described in Fig. 9. We gratefully acknowledge D. Dorsky, W. Ruyechan, M. Fagioli, L. De Jong, N. Deluca, T. Sternsdorf, and H. Will for providing reagents used in this study.
This investigation was supported by Public Health Service grants A121747 to S.K.W., AI19838 to D.M.C., and RO1DE10051 to C.B.C.H.
REFERENCES
- 1.Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci P G. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol Cell Biol. 1998;18:1084–1093. doi: 10.1128/mcb.18.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre C, Guillemin M C, Zhu J, Koken M H, Quignon F, Herve L, Chelbi-Alix M K, Dhumeaux D, Wang Z Y, Degos L, Chen Z, de Thé H. The PML and PML/RARalpha domains: from autoimmunity to molecular oncology and from retinoic acid to arsenic. Exp Cell Res. 1996;229:253–260. doi: 10.1006/excr.1996.0368. [DOI] [PubMed] [Google Scholar]
- 3.Baker T A, Bell S P. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 4.Berezney R. Visualizing DNA replication sites in the cell nucleus. Semin Cell Biol. 1991;2:103–115. [PubMed] [Google Scholar]
- 5.Birch C J, Tyssen D P, Tachedjian G, Doherty R, Hayes K, Mijch A, Lucas C R. Clinical effects and in vitro studies of trifluorothymidine combined with interferon-alpha for treatment of drug-resistant and -sensitive herpes simplex virus infections. J Infect Dis. 1992;166:108–112. doi: 10.1093/infdis/166.1.108. [DOI] [PubMed] [Google Scholar]
- 6.Boisvert F M, Hendzel M J, Bazett-Jones D P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Boulware D W, Weber P C. Cross-reactivity of the anti-PML antibody PG-M3 with the herpes simplex virus type 1 immediate early protein ICP4. J Gen Virol. 2000;81:1773–1777. doi: 10.1099/0022-1317-81-7-1773. [DOI] [PubMed] [Google Scholar]
- 7.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J Y, Li L, Fan Y H, Mu Z M, Zhang W W, Chang K S. Cell-cycle regulation of DNA damage-induced expression of the suppressor gene PML. Biochem Biophys Res Commun. 1997;240:640–646. doi: 10.1006/bbrc.1997.7692. [DOI] [PubMed] [Google Scholar]
- 9.Chelbi-Alix M K, de Thé H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 10.Chelbi-Alix M K, Quignon F, Pelicano L, Koken M H M, de Thé H. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol. 1998;72:1043–1051. doi: 10.1128/jvi.72.2.1043-1051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G Q, Shi X G, Tang W, Xiong S M, Zhu J, Cai X, Han Z G, Ni J H, Shi G Y, Jia P M, Liu M M, He K L, Niu C, Ma J, Zhang P, Zhang T D, Paul P, Naoe T, Kitamura K, Miller W, Waxman S, Wang Z Y, de Thé H, Chen S J, Chen Z. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL). I. As2O3 exerts dose-dependent dual effects on APL cells. Blood. 1997;89:3345–3353. [PubMed] [Google Scholar]
- 12.Chen G Q, Zhu J, Shi X G, Ni J H, Zhong H J, Si G Y, Jin X L, Tang W, Li X S, Xong S M, Shen Z X, Sun G L, Ma J, Zhang P, Zhang T D, Gazin C, Naoe T, Chen S J, Wang Z Y, Chen Z. In vitro studies on cellular and molecular mechanisms of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia: As2O3 induces NB4 cell apoptosis with downregulation of Bcl-2 expression and modulation of PML-RAR alpha/PML proteins. Blood. 1996;88:1052–1061. [PubMed] [Google Scholar]
- 13.Coen D M, Aschman D P, Gelep P T, Retondo M J, Weller S K, Schaffer P A. Fine mapping and molecular cloning of mutations in the herpes simplex virus DNA polymerase locus. J Virol. 1984;49:236–247. doi: 10.1128/jvi.49.1.236-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coen D M, Fleming H E, Jr, Leslie L K, Retondo M J. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J Virol. 1985;53:477–488. doi: 10.1128/jvi.53.2.477-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorsky D I, Crumpacker C S. Expression of herpes simplex virus type 1 DNA polymerase gene by in vitro translation and effects of gene deletions on activity. J Virol. 1988;62:3224–3232. doi: 10.1128/jvi.62.9.3224-3232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duprez E, Saurin A J, Desterro J M, Lallemand-Breitenbach V, Howe K, Boddy M N, Solomon E, de Thé H, Hay R T, Freemont P S. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 17.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 18.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D, Earnshaw W C, Pluta A F, Sternsdorf T, Ainsztein A M, Carmena M, Ruchaud S, Hsu W, Orr A. A dynamic connection between centromeres and ND10 proteins. J Cell Sci. 1999;112:3443–3454. doi: 10.1242/jcs.112.20.3443. [DOI] [PubMed] [Google Scholar]
- 20.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fagioli M, Alcalay M, Pandolfi P P, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci P G. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene. 1992;7:1083–1091. [PubMed] [Google Scholar]
- 24.Fagioli M, Grignani F, Ferrucci P F, Alcalay M, Mencarelli A, Nicoletti I, Pelicci P G. Effect of the acute promyelocytic leukemia PML/RAR alpha protein on differentiation and survival of myeloid precursors. Leukemia. 1994;8(Suppl. I):S7–S11. [PubMed] [Google Scholar]
- 25.Flenghi L, Fagioli M, Tomassoni L, Pileri S, Gambacorta M, Pacini R, Grignani F, Casini T, Ferrucci P F, Martelli M F, et al. Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells, and epithelia. Blood. 1995;85:1871–1880. [PubMed] [Google Scholar]
- 26.Gibbs J S, Weisshart K, Digard P, deBruynKops A, Knipe D M, Coen D M. Polymerization activity of an α-like DNA polymerase requires a conserved 3′-5′ exonuclease active site. Mol Cell Biol. 1991;11:4786–4795. doi: 10.1128/mcb.11.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guiochon-Mantel A, Savouret J F, Quignon F, Delabre K, Milgrom E, De Thé H. Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol Endocrinol. 1995;9:1791–1803. doi: 10.1210/mend.9.12.8614415. [DOI] [PubMed] [Google Scholar]
- 28.Guldner H H, Szostecki C, Grotzinger T, Will H. IFN enhances expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- 29.Hwang Y T, Liu B-Y, Coen D M, Hwang C B C. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J Virol. 1997;71:7791–7798. doi: 10.1128/jvi.71.10.7791-7798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15; 17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 32.Kamitani T, Kito K, Nguyen H P, Wada H, Fukuda-Kamitani T, Yeh E T. Identification of three major sentrinization sites in PML. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 33.LaMorte V J, Dyck J A, Ochs R L, Evans R M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcy A I, Hwang C B C, Ruffner K L, Coen D M. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among α-like DNA polymerases is involved in substrate recognition. J Virol. 1990;64:5883–5890. doi: 10.1128/jvi.64.12.5883-5890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcy A I, Yager D R, Coen D M. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J Virol. 1990;64:2208–2216. doi: 10.1128/jvi.64.5.2208-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 40.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 41.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 42.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 43.Maul G G, Negorev D, Bell P, Ishov A M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 44.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 45.Mossman K L, Saffran H A, Smiley J R. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J Virol. 2000;74:2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mu Z-M, Chin K-V, Liu J-H, Lozano G, Chang K-S. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mu Z-M, Le X F, Vallian S, Glassman A B, Chang K S. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- 48.Müller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller S, Miller W H, Jr, Dejean A. Trivalent antimonials induce degradation of the PML-RAR oncoprotein and reorganization of the promyelocytic leukemia nuclear bodies in acute promyelocytic leukemia NB4 cells. Blood. 1998;92:4308–4316. [PubMed] [Google Scholar]
- 51.Nason-Burchenal K, Gandini D, Botto M, Allopenna J, Seale J R, Cross N C, Goldman J M, Dmitrovsky E, Pandolfi P P. Interferon augments PML and PML/RAR alpha expression in normal myeloid and acute promyelocytic cells and cooperates with all-trans retinoic acid to induce maturation of a retinoid-resistant promyelocytic cell line. Blood. 1996;88:3926–3936. [PubMed] [Google Scholar]
- 52.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puvion-Dutilleul F, Venturini L, Guillemin M C, de Thé H, Puvion E. Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp Cell Res. 1995;221:448–461. doi: 10.1006/excr.1995.1396. [DOI] [PubMed] [Google Scholar]
- 54.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de Thé H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 55.Quinlan M P, Chen L B, Knipe D M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 56.Rowley J D, Golomb H M, Vardiman J, Fukuhara S, Dougherty C, Potter D. Further evidence for a non-random chromosomal abnormality in acute promyelocytic leukemia. Int J Cancer. 1977;20:869–872. doi: 10.1002/ijc.2910200608. [DOI] [PubMed] [Google Scholar]
- 57.Sacks S L, Wanklin R J, Reece D E, Hicks K A, Tyler K L, Coen D M. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann Intern Med. 1989;111:893–899. doi: 10.7326/0003-4819-111-11-893. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shelton L S G, Albright A G, Ruyechan W T, Jenkins F J. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J Virol. 1994;68:521–525. doi: 10.1128/jvi.68.1.521-525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 62.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PICI/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sternsdorf T, Puccetti E, Jensen K, Hoelzer D, Will H, Ottmann O G, Ruthardt M. PIC-1/SUMO-1-modified PML-retinoic acid receptor α mediates arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Mol Cell Biol. 1999;19:5170–5178. doi: 10.1128/mcb.19.7.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuurman N, Floore A, Middelkoop E, van Driel R, de Jong L. PML shuttles between nuclear bodies and the cytoplasm. Cell Mol Biol Lett. 1997;2:137–150. [Google Scholar]
- 65.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi Y, Iwase M, Konishi M, Tanaka M, Toh-e A, Kikuchi Y. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem Biophys Res Commun. 1999;259:582–587. doi: 10.1006/bbrc.1999.0821. [DOI] [PubMed] [Google Scholar]
- 67.Taylor J L, Unverrich D, O'Brien W J, Wilcox K W. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J Interferon Cytokine Res. 2000;20:805–815. doi: 10.1089/10799900050151076. [DOI] [PubMed] [Google Scholar]
- 68.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 69.Weller S K, Lee K J, Sabourin D J, Schaffer P A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilcock D, Lane D P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 71.Xie K, Lambie E J, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao X-D, Matecic M, Elias P. Direct repeats of the herpes simplex virus a sequence promote nonconservative homologous recombination that is not dependent on XPF/ERCC4. J Virol. 1997;71:6842–6849. doi: 10.1128/jvi.71.9.6842-6849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeager T R, Neumann A A, Englezou A, Huschtscha L I, Noble J R, Reddel R R. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 74.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu J, Koken M H M, Quignon F, Chelbi-Alix M K, Degos L, Wang Z Y, Chen Z, de Thé H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]