Abstract
Recently, self-replicating and self-limiting RNA vaccines (RNA replicons) have emerged as an important form of nucleic acid vaccines. Self-replicating RNA eventually causes lysis of transfected cells and does not raise the concern associated with naked DNA vaccines of integration into the host genome. This is particularly important for development of vaccines targeting proteins that are potentially oncogenic. However, the potency of RNA replicons is significantly limited by their lack of intrinsic ability to spread in vivo. The herpes simplex virus type 1 protein VP22 has demonstrated the remarkable property of intercellular transport and provides the opportunity to enhance RNA replicon vaccine potency. We therefore created a novel fusion of VP22 with a model tumor antigen, human papillomavirus type 16 E7, in a Sindbis virus RNA replicon vector. The linkage of VP22 with E7 resulted in a significant enhancement of E7-specific CD8+ T-cell activities in vaccinated mice and converted a less effective RNA replicon vaccine into one with significant potency against E7-expressing tumors. These results indicate that fusion of VP22 to an antigen gene may greatly enhance the potency of RNA replicon vaccines.
Recently, self-replicating RNA vaccines (RNA replicons) have emerged as an important strategy to enhance the potency of nucleic acid vaccines for cancer immunotherapy (for a review, see reference 19). RNA replicon vaccines may be derived from alphavirus vectors, such as Sindbis virus (13), Semliki Forest virus (3, 4), or Venezuelan equine encephalitis virus (27) vectors. These vaccines are self-replicating and self-limiting and may be administered as either RNA or DNA, which is then transcribed into RNA replicons in transfected cells or in vivo (4, 18). Self-replicating RNA eventually causes lysis of transfected cells (33). These vectors therefore do not raise the concern associated with naked DNA vaccines of integration into the host genome. This is particularly important for development of vaccines targeting proteins that are potentially oncogenic, such as the human papillomavirus (HPV) E6 and E7 proteins.
One limitation on the potency of RNA replicon vaccines is their inability to spread in vivo. We reasoned that a strategy that facilitates the spread of antigen may significantly enhance the potency of RNA replicon vaccines. One potential strategy to enhance the spread of antigen is the employment of the herpes simplex virus type 1 (HSV-1) protein VP22, which has demonstrated the remarkable property of intercellular transport and is capable of distributing protein to many surrounding cells (10). Previously, VP22 has been linked to p53 (26) or thymidine kinase (8), facilitating the spread of linked protein to surrounding cells in vitro. We therefore investigated the use of VP22 linked to a model antigen (HPV-16 E7) in the context of a Sindbis virus-derived RNA replicon vaccine and explored whether it led to enhancement of antigen-specific immune responses and antitumor effects.
HPV-16 E7 was used as a model tumor antigen for our vaccine development because HPVs, particularly HPV-16, are associated with most cervical cancers. HPV E7 is expressed in most HPV-containing cervical cancers and is important in the induction and maintenance of cellular transformation. Therefore, vaccines targeting E7 protein may provide an opportunity to prevent and treat HPV-associated cervical malignancies. Our data indicated that a Sindbis virus RNA vaccine encoding HSV-1 VP22 linked to E7 significantly increased activation of E7-specific CD8+ T cells, resulting in potent antitumor immunity against E7-expressing tumors.
MATERIALS AND METHODS
Plasmid DNA constructs and preparation.
The Sindbis virus RNA replicon vector SINrep5 has been described previously (5). SINrep5 was kindly provided by Charles M. Rice at the Washington University School of Medicine, St. Louis, Mo. For the generation of pcDNA3-VP22, VP22 was subcloned from pVP22/myc-His (Invitrogen, Carlsbad, Calif.) into the unique EcoRV and BamHI cloning sites of the pcDNA3.1(−) expression vector (Invitrogen). The generation of pcDNA3-E7 has been described previously (6). For the generation of pcDNA3-VP22/E7, VP22 was subcloned from pcDNA3-VP22 into the unique EcoRV and BamHI cloning sites of pcDNA3-E7. For the generation of SINrep5-VP22, SINrep5-E7, and SINrep5-VP22/E7, DNA fragments encoding VP22, HPV-16 E7, and chimeric VP22/E7 were isolated by digesting pcDNA3-VP22, pcDNA3-E7, and pcDNA3-VP22/E7, respectively, with XbaI and PmeI restriction enzymes. These isolated DNA fragments were further cloned into the corresponding XbaI and PmeI sites of the SINrep5 vector to generate SINrep5-VP22, SINrep5-E7, and SINrep5-VP22/E7 constructs. The accuracy of these constructs was confirmed by DNA sequencing.
In vitro RNA preparation.
The generation of RNA transcripts from SINrep5-VP22, SINrep5-E7, SINrep5-VP22/E7, and SINrep5 was performed using the protocol described by Mandl et al. (24). SpeI was used to linearize DNA templates for the synthesis of RNA replicons from SINrep5-VP22, SINrep5-E7, SINrep5-VP22/E7, and SINrep5. RNA vaccines were transcribed in vitro and capped using SP6 RNA polymerase and capping analog from the in vitro transcription kit (Life Technologies, Rockville, Md.) according to the vendor's manual. After synthesis, DNA was removed by digestion with DNase I. Synthesized RNA was then purified by precipitation. RNA concentration was determined by optical density measured at 260 nm. The integrity and quantity of RNA transcripts were further checked using denaturing gel electrophoresis (24). The purified RNA was divided into aliquots to be used for vaccination in animals and for transfection of a BHK21 cell line. The protein expression of the transcripts was characterized by transfection of the RNA into BHK21 cells using electroporation with the Cell-Porator electroporation system (Life Technologies) according to the vendor's manual and characterized using Western blot analysis.
Cell lines.
Baby hamster kidney (BHK21) cells were obtained from the American Type Culture Collection (Manassas, Va.) and grown in Glasgow minimum essential medium supplemented with 5% fetal bovine serum, 10% tryptose phosphate broth, 2 mM glutamine, and antibiotics. Cells were kept at 37°C in a humidified 5% CO2 atmosphere and were passaged every 2 days. The production and maintenance of TC-1 cells have been described previously (20).
RNA vaccination.
Six- to eight-week-old female C57BL/6 mice from the National Cancer Institute (Frederick, Md.) were purchased and kept in the oncology animal facility of the Johns Hopkins Hospital (Baltimore, Md.). All animal procedures were performed according to approved protocols and in accordance with recommendations for the proper use and care of laboratory animals. Mice were vaccinated intramuscularly with 1 μg of various SINrep5 RNA vaccines in the right hind leg.
Cytotoxic T-lymphocyte (CTL) assays.
Splenocytes were harvested 2 weeks after RNA vaccination and cultured with the major histocompatibility complex (MHC) class I-restricted E7 peptide (amino acids [aa] 49 to 57) (11) in a total volume of 2 ml of RPMI 1640 supplemented with 10% (vol/vol) fetal bovine serum, 50 U of penicillin-streptomycin per ml, 2 mM l-glutamine, 1 mM sodium pyruvate, and 2 mM nonessential amino acids in a 24-well tissue culture plate for 6 days. TC-1 tumor cells were used as target cells. Splenocytes were mixed with TC-1 cells (104 per well) at various effector/target (E/T) ratios (1:1, 5:1, 15:1, and 45:1) in a final volume of 200 μl. After 5 h of incubation at 37°C, 50 μl of the cultured medium was collected for CTL assays. Cytolysis was determined by quantitative measurements of lactate dehydrogenase (LDH) using CytoTox96 nonradioactive cytotoxicity assay kits (Promega, Madison, Wis.) according to the manufacturer's protocol. The percentage of lysis was calculated from the following equation: 100 × (A − B)/(C − D), where A is the reading of experimental-effector signal value, B is the effector spontaneous background signal value, C is maximum signal value from target cells, and D is the target spontaneous background signal value.
Enzyme-linked immunosorbent assay (ELISA).
For the determination of gamma interferon (IFN-γ) in the supernatant of cultured splenocytes, splenocytes were harvested 2 weeks after vaccination and cultured with 1 μg of E7 peptide (aa 49 to 57) containing MHC class I epitope (11) per ml or 10 μg of E7 peptide (aa 30 to 67) containing MHC class II peptide (29) per ml in a total volume of 2 ml of RPMI 1640 supplemented with 10% (vol/vol) fetal bovine serum, 50 U of penicillin and streptomycin per ml, 2 mM l-glutamine, 1 mM sodium pyruvate, and 2 mM nonessential amino acids, in a 24-well tissue culture plate for 6 days. The supernatants were harvested and assayed for the presence of IFN-γ using ELISA kits (Endogen, Woburn, Mass.) according to the manufacturer's protocol.
For the detection of HPV-16 E7-specific antibodies in the sera, a direct ELISA was used as described previously (31). Briefly, a 96-microwell plate was coated with 100 μl of 0.5 μg of bacterium-derived HPV-16 E7 proteins per ml and incubated at 4°C overnight. The wells were then blocked with phosphate-buffered saline (PBS) containing 20% fetal bovine serum. Sera were prepared from mice on day 21 postimmunization, serially diluted in PBS, added to the ELISA wells, and incubated at 37°C for 2 h. After washing with PBS containing 0.05% Tween 20, the plate was incubated with a 1/2,000 dilution of a peroxidase-conjugated rabbit anti-mouse immunoglobulin G (IgG) antibody (Zymed, San Francisco, Calif.) at room temperature for 1 h. The plate was washed, developed with 1-Step Turbo TMB-ELISA (Pierce, Rockford, Ill.), and stopped with 1 M H2SO4. The ELISA plate was read with a standard ELISA reader at 450 nm.
Intracellular cytokine staining and flow cytometry analysis.
Splenocytes from vaccinated mice were collected 2 weeks after vaccination and incubated with 1 μg of E7 peptide (aa 49 to 57) per ml in a total volume of 2 ml of culture medium for 6 days. Cell surface marker staining of CD8 and intracellular cytokine staining for IFN-γ as well as FACScan analysis were performed using conditions described previously (6). Phycoerythrin-conjugated CD8 antibody, fluorescein isothiocyanate (FITC)-conjugated anti-IFN-γ antibody, and the immunoglobulin isotype control antibody (rat IgG1) were all purchased from PharMingen (San Diego, Calif.). Analysis was performed on a Becton Dickinson FACScan with CELLQuest software (Becton Dickinson Immunocytometry Systems, Mountain View, Calif.).
In vivo tumor protection experiments.
For the tumor protection experiment, mice (five per group) were immunized intramuscularly with 1 μg of SINrep5-VP22, SINrep5-E7, SINrep5-VP22/E7, or SINrep5 (no insert) RNA vaccine per mouse. Fourteen days after immunization, mice were injected intravenously with 104 TC-1 tumor cells per mouse through the tail vein. Three weeks later, mice were euthanatized. The lung weight and number of pulmonary nodules in each mouse were evaluated and counted by experimenters in a blinded fashion.
In vivo antibody depletion experiments.
The procedure for in vivo antibody depletion has been described previously (20, 32). In brief, mice (five per group) were vaccinated with 1 μg of self-replicating SINrep5-VP22/E7 RNA per mouse intramuscularly and challenged with 104 TC-1 tumor cells per mouse via tail vein injection 2 weeks after vaccination. Depletions were started 1 week prior to tumor challenge. Monoclonal antibody (MAb) GK1.5 (7) was used for CD4 depletion, MAb 2.43 (28) was used for CD8 depletion, and MAb PK136 (17) was used for NK1.1 depletion. Flow cytometry analysis revealed that >95% of the appropriate lymphocyte subset was depleted while other subsets were at normal levels. Depletion was terminated on day 21 after tumor challenge.
In vitro cell death analysis.
BHK21 cells (107) were transfected with 4 μg of SINrep5, SINrep5-E7, SINrep5-VP22, or SINrep5-VP22/E7 RNA transcripts as described above. Untreated BHK21 cells or BHK21 cells electroporated without SINrep5 RNA were used as controls. Transfected BHK21 cells were collected 24 h after transfection. The percentages of apoptotic and necrotic BHK21 cells were analyzed using annexin V apoptosis detection kits (PharMingen) according to the manufacturer's protocol, followed by flow cytometry analysis.
In vivo cell death assay.
Mice were immunized with self-replicating SIN5-VP22/E7 RNA injected intramuscularly into the right leg. Normal saline without self-replicating RNA was also injected intramuscularly into the left leg as a control. Mice were sacrificed 4 days after immunization. For the detection of apoptotic cells, a modified terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed (15). The formalin-fixed and paraffin-embedded tissue was cut into 5-μm slices. After deparaffinization, the tissue was placed into a plastic jar containing 0.1 M citrate buffer, pH 6.0, and microwave irradiation was performed for 1 min. Then, the slides were immersed in 0.1 M Tris-HCl containing 3% bovine serum albumin and 20% normal bovine serum, pH 7.5, for 30 min at room temperature. After that, the slides were rinsed twice with PBS at room temperature. The slides were then incubated with 50 μl of TUNEL reaction mixture that was provided in an in situ cell detection kit (Roche Diagnostics GmbH) for 60 min at 37°C in a humidified chamber. Each slide was rinsed three times in PBS for 5 min. Subsequently, 0.3% H2O2 in methanol was added for 10 min at room temperature in order to eliminate endogenous peroxidase. Fifty microliters of Converter-POD (Roche Diagnostics GmbH) was added and incubated for 30 min at 37°C in a humidified chamber. The slides were rinsed again three times in PBS at room temperature for 5 min, followed by addition of 50 μl of diaminobenzidine substrate solution and incubation for 3 min at room temperature. Finally, the slides were washed extensively in tap water and counterstained with Mayer's hematoxylin. Apoptotic index is used as a measure of the extent of apoptosis in the stained slides following inspection under a light microscope. Apoptotic index is defined as percentage of apoptotic cells and bodies per 100 cells (21).
CTL assay using DCs incubated with BHK21 cells transfected with various SINrep5 RNAs.
CTL assays using dendritic cells (DCs) incubated with BHK21 cells transfected with various SINrep5 RNAs were performed using a protocol similar to one described previously (1, 2) with modifications. Briefly, DCs were generated by culture of bone marrow cells in the presence of granulocyte-macrophage colony-stimulating factor as described previously (22). These DCs were characterized by flow cytometry analysis for DC markers as described previously (30). BHK21 cells (107) were transfected with 4 μg of various self-replicating SINrep5 RNA vectors via electroporation, and transfected BHK21 cells were collected 16 to 20 h after electroporation. The levels of E7 protein expression in BHK21 cells transfected with SINrep5-E7 or SINrep5-VP22/E7 RNA transcripts were determined by ELISA and standardized (data not shown). Transfected BHK21 cells (3 × 105) were then coincubated with 105 bone marrow-derived DCs at 37°C for 24 h. These prepared DCs were then used as target cells (T), while the Db-restricted E7-specific CD8+ T cells (30) served as effector cells (E). CTL assays were performed with effector cells and target cells (104 per well) mixed together at various ratios (1:1, 3:1, 9:1, and 27:1) in a final volume of 200 μl. After a 5-h incubation at 37°C, 50 μl of the cultured media was collected to assess the amount of LDH in the cultured media as described above. DCs coincubated with untransfected BHK21 cells, transfected BHK21 cells alone, untreated DCs alone, and CD8+ T cells alone were included as negative controls.
RESULTS
Generation of self-replicating RNA transcripts.
Generation of plasmid DNA constructs and subsequent preparation of self-replicating SINrep5 RNA constructs were performed as described in Materials and Methods. The SINrep5 vector contains the genes encoding Sindbis virus RNA replicase and the SP6 promoter (5). A schematic diagram of RNA transcripts derived from SINrep5, SINrep5-VP22, SINrep5-E7, and SINrep5-VP22/E7 DNA constructs using SP6 RNA polymerase is shown in Fig. 1. The level of expression of E7 in cells transfected with various E7-containing RNA constructs was characterized using Western blot analysis and ELISA. BHK21 cells transfected with self-replicating SINrep5-E7 and SINrep5-VP22/E7 expressed similar amounts of E7 protein (data not shown).
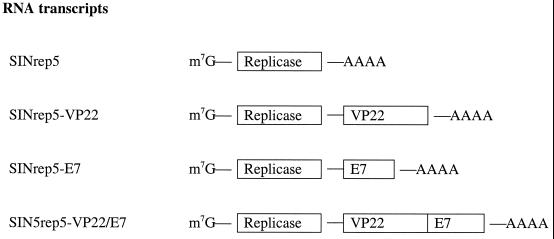
FIG. 1.
SINrep5 self-replicating RNA transcripts. The schematic diagram depicts the SINrep5 self-replicating RNA transcripts. A methylated m7G cap is located at the 5′ end of the mRNA, followed by a sequence responsible for the self-replication (replicase), the gene of interest (i.e., E7, VP22, or VP22/E7), and a polyadenylated tail (AAAA).
Vaccination with self-replicating SINrep5-VP22/E7 RNA enhances IFN-γ-secreting E7-specific CD8+ T-cell activity.
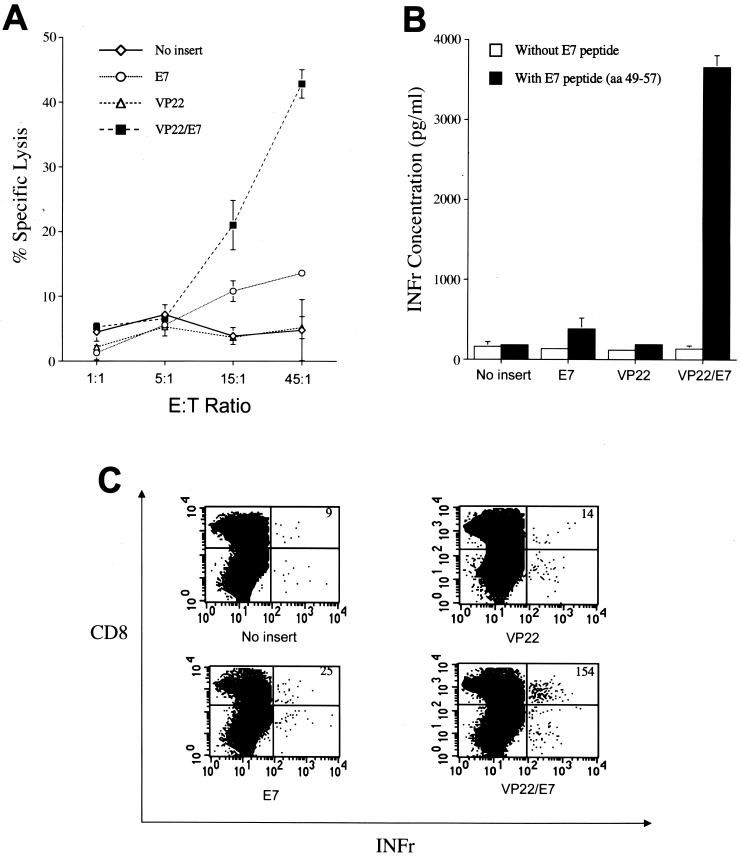
To determine the quantity of E7-specific CD8+ T-cell responses generated by the SINrep5-VP22/E7 RNA vaccine, CTL assays were used. As shown in Fig. 2A, splenocytes from mice vaccinated with various self-replicating SINrep5 RNA vaccines were cultured with the E7 peptide (aa 49 to 57) for 6 days and used as effector cells while TC-1 tumor cells served as target cells. Vaccination with SINrep5-VP22/E7 RNA generated a significantly higher percentage of specific lysis than that with the other SINrep5 RNA vaccines (P < 0.001, one-way analysis of variance [ANOVA]). The ability of SINrep5-VP22/E7 RNA to generate specific lysis was found to be approximately three times that of self-replicating SINrep5-E7 RNA ([42.8 ± 2.2]% versus [13.6 ± 0.3]%, E/T ratio of 45/1, P < 0.001).
FIG. 2.
Immunologic responses generated by self-replicating RNA vaccines. (A) E7-specific T-cell cytotoxic activity. Mice were immunized with various SINrep5 self-replicating RNA vaccines via intramuscular injection. Splenocytes were collected after 14 days. Vaccination with VP22/E7 RNA generated significantly higher percentages of specific lysis than did that with the other RNA vaccines (P < 0.001, one-way ANOVA). (B) IFN-γ concentration secreted by E7-specific CD8+ T-cell precursors measured by ELISA. Mice were immunized, and their splenocytes were collected and cultured as described above. Splenocytes obtained from mice vaccinated with self-replicating VP22/E7 secreted the highest concentration of IFN- γ compared to those from mice vaccinated with the other RNA vaccines (P < 0.001, one-way ANOVA). (C) Flow cytometry analysis of IFN-γ-secreting E7-specific CD8+ cells in mice vaccinated with various recombinant RNA vaccines. Mice were immunized, and their splenocytes were collected and cultured as described above. Splenocytes cultured in vitro with E7 peptide (aa 49 to 57) for 6 days were stained for both CD8 and intracellular IFN-γ. The number of IFN-γ-secreting CD8+ T cells, indicated in the upper right-hand corner of each panel, was determined using flow cytometry. The VP22/E7-vaccinated group generated a higher number of E7-specific IFN-γ-secreting CD8+ cells than did groups vaccinated with other recombinant RNA vaccines.
We performed an indirect ELISA using supernatant from cultured splenocytes in order to determine the level of IFN-γ secreted from splenocytes pulsed with MHC class I-restricted E7 peptide. As a negative control, an ELISA was also performed without peptide. Supernatants in the culture medium were collected to detect the IFN-γ concentration. As shown in Fig. 2B, splenocytes from mice vaccinated with SINrep5-VP22/E7 RNA secreted the highest concentration of IFN-γ (3,655 ± 149 pg/ml) compared to splenocytes from mice vaccinated with other RNA vaccines (183 ± 15 pg/ml for no insert, 382 ± 30 pg/ml for E7, and 190 ± 16 pg/ml for VP22; P < 0.001, one-way ANOVA).
We performed intracellular cytokine staining with flow cytometry analysis for IFN-γ and CD8 markers to determine the subset of IFN-γ-secreting lymphocytes. Splenocytes from naive or vaccinated groups of mice were incubated with the MHC class I (H-2 Db)-restricted E7 peptide (aa 49 to 57) (29) for detecting E7-specific CD8+ T cells. As shown in Fig. 2C, vaccination with SINrep5-VP22/E7 RNA generated a higher number of IFN-γ-secreting E7-specific CD8 T cells than those for the other vaccination groups (P < 0.01, one-way ANOVA). These results indicated that fusion of VP22 to E7 significantly enhances IFN-γ-secreting E7-specific CD8+ T-cell activity. The physical linkage of VP22 with E7 was important for the enhancement of E7-specific CD8+ T-cell-mediated immunity because vaccination of mice with VP22 RNA mixed with E7 RNA did not generate a significant increase of E7-specific CD8+ T-cell activity over that with vaccination with E7 RNA alone (data not shown).
We also performed an indirect ELISA for IFN-γ secretion generated by splenocytes pulsed with MHC class II-restricted E7 peptide and intracellular cytokine staining with flow cytometry analysis for IFN-γ and CD4 marker. We found no significant differences in IFN-γ-secreting CD4+ T-cell activities among mice vaccinated with various SINrep5 RNA vaccines (data not shown). In addition, we performed an ELISA for anti-E7 antibodies using sera from vaccinated mice. We did not observe any significant differences in anti-E7 antibody titers among mice vaccinated with various SINrep5 RNA constructs (data not shown).
Vaccination with self-replicating SINrep5-VP22/E7 RNA protects mice against the growth of E7-expressing tumors.
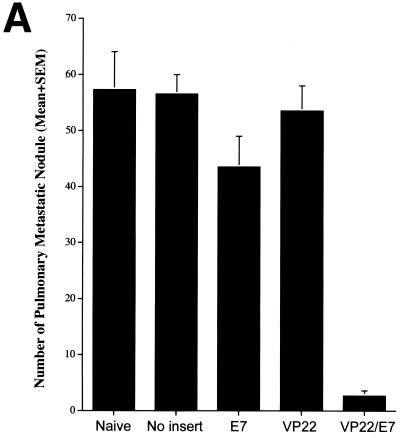
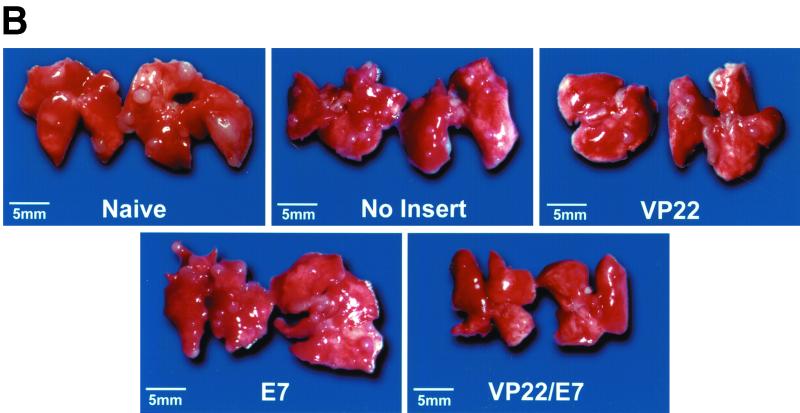
To determine whether vaccination with the self-replicating SINrep5-VP22/E7 RNA protected mice against E7-expressing tumors, we performed an in vivo tumor protection experiment. Mice received vaccination and tumor challenge as described in Materials and Methods. The number of pulmonary nodules and mean lung weight were assessed 21 days after tumor challenge. As shown in Fig. 3A, there were fewer pulmonary nodules in mice vaccinated with the self-replicating VP22/E7 RNA vaccine than in mice vaccinated with other RNA vaccines (P < 0.001, one-way ANOVA). Representative gross photographs of the lung tumors are shown in Fig. 3B. Our results demonstrated that the self-replicating SINrep5-VP22/E7 RNA vaccine protected mice from intravenous tumor challenge even at the low dosage of 1 μg/mouse. The physical linkage of VP22 with E7 was essential for the antitumor effect because vaccination of mice with 1 μg of VP22 RNA per mouse mixed with E7 RNA did not generate significant tumor protection compared to vaccination with E7 RNA alone (data not shown).
FIG. 3.
Tumor protection mediated by various SINrep5 self-replicating RNA vaccines. (A) Mice (five per group) were immunized intramuscularly with 1 μg of SINrep5-VP22, SINrep5-E7, SINrep5-VP22/E7, and SINrep5 (no insert) RNA per mouse. Two weeks after vaccination, mice were challenged with TC-1 tumor cells via intravenous tail vein injection at a dose of 104 cells/mouse. Mice were monitored twice a week and sacrificed at day 21 after tumor challenge. Lungs were dissected from mice 35 days after vaccination with the various RNA vaccines. The mean number of lung tumor nodules was used as a measurement of the effectiveness of the various self-replicating RNA vaccines at controlling HPV-16 E7-expressing tumor growth. There were fewer mean pulmonary nodules in mice vaccinated with the self-replicating VP22/E7 RNA vaccine than in mice vaccinated with the other RNA vaccines (P < 0.001, one-way ANOVA). SEM, standard error of the mean. (B) Representative gross pictures of the lung tumors in each vaccinated group. There are multiple grossly visible lung tumors in unvaccinated control mice and mice vaccinated with SINrep5, SINrep5-E7, or SINrep5-VP22 RNA. The lung tumors in the SINrep5-VP22/E7 RNA-vaccinated group cannot be seen at the magnification provided in this figure.
CD8+ T cells and NK cells play an important role in the antitumor effect generated by vaccination with SINrep5-VP22/E7 RNA.
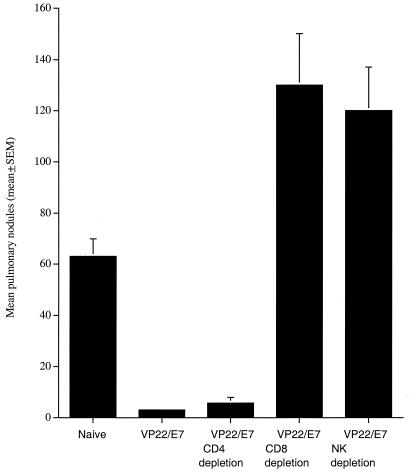
To determine the types of lymphocytes that are important for protection against E7-expressing tumor cells, we performed in vivo antibody depletion experiments. As shown in Fig. 4, the mean number of pulmonary tumor nodules from mice depleted of CD8+ T cells (130 ± 20) and NK1.1 cells (120 ± 17) was significantly higher than those of nondepleted (2.0 ± 0.6) and CD4+ T-cell-depleted (5.7 ± 2.2) groups (P < 0.001, one-way ANOVA). In comparison, the mean number of pulmonary tumor nodules from mice depleted of CD4+ T cells resembled that obtained from nondepleted mice. These antibody depletion experiments suggested that CD8+ T and NK cells are both important for the antitumor immunity generated by the SINrep5-VP22/E7 RNA vaccine.
FIG. 4.
Effect of lymphocyte subset depletions on the potency of self-replicating SINrep5-VP22/E7 RNA vaccine. Mice were immunized with 1 μg of self-replicating SINrep5-VP22/E7 RNA per mouse via intramuscular injection. Two weeks after vaccination, mice were challenged with 104 TC-1 cells/mouse intravenously via tail vein. Depletions were initiated 1 week prior to tumor challenge and terminated 21 days after tumor challenge. Three weeks after tumor challenge, mice were sacrificed. Depletion of CD8+ T cells and NK1.1 cells resulted in similar mean numbers of pulmonary nodules, significantly greater than those generated in the nondepleted group. The mean number of lung nodules from mice depleted of CD4+ T cells resembled results obtained from nondepleted mice. SEM, standard error of the mean.
Self-replicating RNA vaccines induce apoptosis in transfected cells and in vaccinated mice.
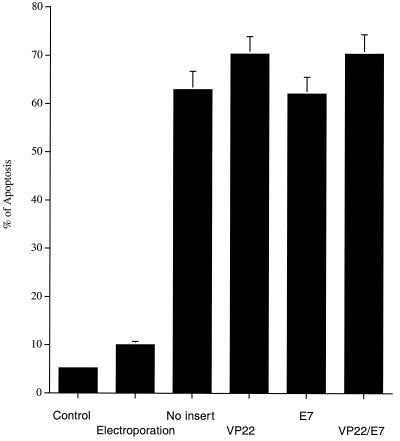
RNAs transcribed in vitro from various plasmid SINrep5 RNA vaccines were transfected into BHK21 cells via electroporation. Electroporated BHK21 cells without RNA and untreated BHK21 cells were used as controls. The percentage of apoptosis among the transfected BHK21 cells was highest 24 h after transfection (data not shown). Twenty-four hours after transfection, all of the BHK21 cells transfected with any of the SINrep5 RNA constructs generated similar levels of apoptosis (up to 70%), significantly higher than that generated by the two control groups (Fig. 5).
FIG. 5.
Flow cytometry analysis to demonstrate apoptotic changes in BHK21 cells transfected with various SINrep5 RNA vaccines. The percentage of apoptosis in the transfected BHK21 cells was determined 24 h after transfection. Note that BHK21 cells transfected with SINrep5 RNA vaccines generated a higher percentage of apoptosis than did cells for the other two control groups. No statistical differences could be found in the apoptotic percentages generated by the various SINrep5 RNA vaccines.
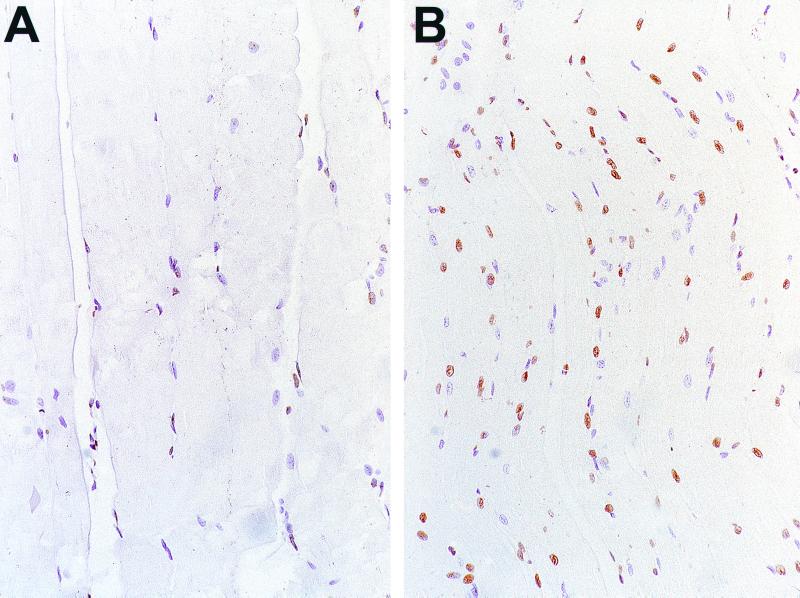
After demonstrating the extent of apoptosis in vitro, we characterized the extent of apoptosis induced by SINrep5 RNA in vivo by staining sections of muscular tissue at the injected sites using the TUNEL method. As shown in Fig. 6B, the SINrep5 RNA-injected group exhibited a higher number of apoptotic cells in the muscular tissues than did the normal saline-injected group (Fig. 6A) 4 days after immunization.
FIG. 6.
TUNEL assay to determine apoptotic cells in the skeletal muscle of vaccinated mice. (A) Mice were immunized with normal saline intramuscularly (magnification, ×160). (B) Mice were immunized with self-replicating SINrep5 RNA intramuscularly (magnification, ×100) and sacrificed 4 days after immunization. Note that injection with SINrep5 RNA induced a higher number of apoptotic cells in the muscle tissues than did injection with normal saline.
Enhanced presentation of E7 through the MHC class I pathway in DCs pulsed with SINrep5-VP22/E7 RNA-transfected cells.
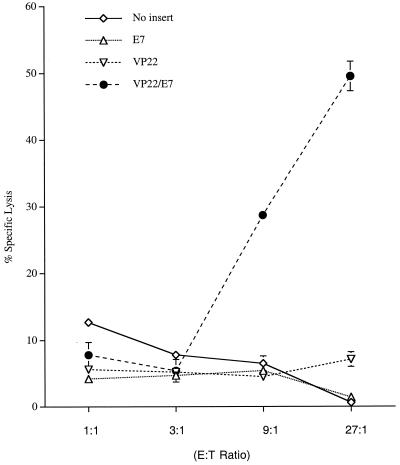
A potential mechanism for the enhanced E7-specific CD8+ T-cell immune responses in vivo is the presentation of E7 through the MHC class I pathway by uptake of apoptotic cells expressing various E7 constructs, a process known as “cross-priming.” A cross-priming experiment was performed to characterize the MHC class I presentation of E7 in DCs pulsed with apoptotic BHK21 cells transfected with various self-replicating RNAs. As mentioned above, BHK21 cells have been shown to have stable transfection efficiency and similar E7 expression levels among cells transfected with different E7-containing self-replicating RNAs. Transfected BHK21 cells were coincubated with bone marrow-derived DCs, which served as target cells. E7-specific CD8+ T cells served as effector cells. CTL assays with various E/T ratios (1:1, 3:1, 9:1, and 27:1) were performed. As shown in Fig. 7, DCs coincubated with BHK21 cells transfected with SINrep5-VP22/E7 RNA generated significantly higher percentages of specific lysis than did DCs coincubated with BHK21 cells transfected with SINrep5-E7 RNA (P < 0.001). These results suggested that DCs incubated with BHK21 cells transfected with SINrep5-VP22/E7 RNA presented E7 antigen through the MHC class I pathway more efficiently than did DCs incubated with BHK21 cells transfected with wild-type E7 RNA. Thus, the fusion of VP22 to E7 is likely able to induce enhancement of E7-specific CD8+ T-cell immune responses via a cross-priming mechanism.
FIG. 7.
CTL assays to demonstrate enhanced MHC class I presentation of E7 by bone marrow-derived DCs pulsed with apoptotic cells transfected with SINrep5-VP22/E7. BHK21 cells were electroporated with various self-replicating RNAs. The transfected BHK21 cells were cocultured with bone marrow-derived DCs. The DCs were used as target cells, and the E7-specific CD8+ T cells served as effector cells. Cytolysis was determined by quantitative measurements of LDH as described in Materials and Methods. Note that the self-replicating VP22/E7 RNA vaccines generated a significantly higher percentage of specific lysis (9:1 and 27:1 E/T ratios) than did the other RNA vaccines (P < 0.001). CTL assays shown here are from one experiment representative of two performed.
DISCUSSION
In this study, we demonstrated that linkage of VP22 with E7 can significantly enhance the potency of HPV-16 E7-expressing self-replicating RNA vaccines. RNA vaccines incorporating VP22 fused to HPV-16 E7 generated significant E7-specific CD8+ T-cell-mediated immune responses. Furthermore, the chimeric SINrep5-VP22/E7 RNA vaccine was capable of reducing the number of lethal pulmonary E7-expressing tumors.
RNA vaccines possess several advantages over existing viral vector systems designed for in vivo gene delivery, even though such viral vectors are efficient in delivering genes of interest to targeted cells. For example, immune recognition following adenovirus vector or vaccinia virus delivery inhibits repeat vaccination with the same delivery system, while retrovirus vectors display low in vitro infectivity and have potential virus-associated complications, including helper virus replication and insertional mutagenesis. In contrast, naked RNA vaccines are relatively safe and can be repeatedly administered. Furthermore, multiple naked RNA vaccines can be administered together.
The successful application of VP22 linked to a model antigen in the context of RNA vaccines suggests that proteins with similar trafficking properties may also enhance vaccine potency. For example, proteins such as bovine herpesvirus 1 VP22 homolog (14) and Marek's disease virus VP22 homolog (9) have been shown elsewhere to enhance the spread of linked protein intercellularly. Employment of such proteins linked to antigen in a manner similar to that of VP22/E7 in this study may be able to enhance the potency of self-replicating RNA vaccines.
The linkage of VP22 to E7 in the context of self-replicating RNA vaccines led to significant enhancement in E7-specific CD8+ T-cell responses compared to wild-type E7 RNA in vivo. One potential mechanism for the enhancement of E7-specific CD8+ T-cell responses in vivo is through MHC class I presentation of E7 to CTLs by cells expressing VP22/E7, a process known as “direct priming.” However, this probably is not a major contributing factor for the observed enhancement in E7-specific CD8+ T-cell responses. It has been shown elsewhere that intramuscular injection of DNA vaccines leads to uptake of DNA by muscle cells at the injected sites (25). In this study, we performed intramuscular injection of our RNA vaccines. The intramuscular injection of RNA likely also leads to uptake of RNA by muscle cells. Our data support such a conclusion (Fig. 6). However, we could not rule out the possibility that cells other than muscle cells were also transduced by the injected RNA vaccines. Muscle cells are not ideal professional antigen-presenting cells because they do not express costimulatory molecules that are important for efficient activation of T cells. Even if the various SINrep5 constructs are delivered to nonmuscle cells, self-replicating RNAs eventually result in the apoptosis of transfected cells (12) and are thus unlikely to directly present antigen in an efficient manner.
A likely alternative explanation for the enhancement of E7-specific CD8+ T-cell responses by chimeric SINrep5-VP22/E7 in vivo is the so-called cross-priming effect (16), whereby exogenous proteins are presented through the MHC class I presentation pathway. We found that DCs pulsed with BHK21 cells transfected with SINrep5-VP22/E7 RNA were able to present E7 antigen through the MHC class I pathway more efficiently than were DCs pulsed with BHK21 cells transfected with SINrep5-E7 RNA in vitro (Fig. 7). Although our study suggests that transfection of cells by the SINrep5-VP22/E7 vector led to apoptosis of cells, it is not clear whether DCs take up apoptotic cells containing VP22/E7 protein or whether instead there is uptake of VP22/E7 protein when it is released after apoptosis or secreted from transduced cells. Since various SINrep5 constructs can induce cell apoptosis at similar levels (Fig. 5), the distinct enhancement in E7-specific CD8+ T-cell activity was most likely due to the linkage of E7 with VP22. Thus, our results suggested that the linkage of VP22 to E7 was capable of enhancing MHC class I presentation of the linked E7 antigen through a cross-priming effect.
In our study, we demonstrated that depletion of NK cells led to a loss in the antitumor effect generated by the VP22/E7 RNA replicon-based vaccine (Fig. 4), indicating that NK cells are necessary for the antitumor effects generated by the vaccine. However, NK cells alone are not sufficient to account for the antitumor effect generated by the VP22/E7 RNA vaccine since the various RNA replicon-based vaccines produced similar numbers of NK cells (data not shown). Our in vivo antibody depletion data suggested that CD8+ CTLs were also important for the antitumor effects generated by the SINrep5-VP22/E7 RNA vaccine (Fig. 4). Thus, our data indicate that both NK cells and E7-specific CD8+ T cells were important for the antitumor effects generated by the VP22/E7 RNA vaccine. It would be interesting to investigate if CD8+ T cells interact with NK cells in vivo to generate an antitumor effect.
While NK cells appeared to play an essential role in the antitumor effect generated by the VP22/E7 RNA replicon-based vaccine, they were not as important for the antitumor effect generated by naked VP22/E7 DNA vaccine administered intradermally through a gene gun (C.-F. Hung, personal communication). Depletion of NK1.1+ cells in mice vaccinated with naked VP22/E7 DNA only slightly decreased the antitumor effects generated by the VP22/E7 DNA vaccine. Thus, different forms or different routes of administration of nucleic acid vaccines may activate different subsets of effector cells in the vaccinated host and have different mechanisms for the antitumor effects.
Several safety issues need to be addressed before the VP22/E7 RNA vaccine can be considered for human immunizations. The first issue is related to the usage of the RNA replicon. We observed increased apoptotic changes and inflammatory responses localized at the injection sites of RNA replicon-based vaccines in mice, which eventually subsided (Fig. 6B). Furthermore, we performed pathological examination of the vital organs in the VP22/E7 RNA-vaccinated mice and did not observe any significant pathological changes. These observations alleviate the concern of potential tissue damage after administration of RNA replicon vaccines. Another issue concerns potential risks associated with the presence of HPV-16 E7 protein in host cells. E7 is a viral oncoprotein that disrupts cell cycle regulation by binding to tumor suppressor pRB protein in nuclei (23). Thus, the presence of E7 in host cells may lead to accumulation of genetic aberrations and eventual malignant transformation in the host cells. The usage of the RNA replicon vector may reduce concerns about the oncogenicity of E7 protein since cells transfected with RNA replicon vaccines will eventually undergo apoptotic changes.
In summary, our results revealed that fusion of the gene encoding HSV-1 VP22 to the HPV-16 E7 gene in an RNA replicon can generate significant E7-specific CD8+ T-cell-mediated immune responses and antitumor effects against HPV-16 E7-expressing murine tumors. Since a majority of cervical cancers express HPV E7, our vaccine can potentially be used for the control of HPV-associated tumors. The strategy of linking VP22 to an antigen gene can potentially be applied to other cancer systems and infectious diseases. The strategy may also be applied in other nonreplicating vectors to improve the potency of vaccines that are otherwise limited by the inability to spread in vivo.
ACKNOWLEDGMENTS
W.-F. Cheng and C.-F. Hung contributed equally to this paper.
We thank Drew M. Pardoll, Robert J. Kurman, Chang-Yao Hsieh, and Keertie Shah for helpful discussions. We thank Richard Roden for critical review of the manuscript.
This work was supported by grants U19 CA72108–02 and RO1 CA72631–01, the Cancer Research Institutes, and the Alexander and Margaret Stewart Trust.
REFERENCES
- 1.Albert L M, Pearce S F, Francisco L M, Sauter B, Roy P, Silverstein R L, Bhardwaj N. Immature dendritic cells phagocytose apoptotic cells via alphavbeta5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J Exp Med. 1998;188:1359–1368. doi: 10.1084/jem.188.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert L M, Sauter B, Bhardwaj N. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature. 1998;392:86–89. doi: 10.1038/32183. [DOI] [PubMed] [Google Scholar]
- 3.Berglund P, Quesada-Rolander M, Putkonen P, Biberfeld G, Thorstensson R, Liljestrom P. Outcome of immunization of cynomolgus monkeys with recombinant Semliki Forest virus encoding human immunodeficiency virus type 1 envelope protein and challenge with a high dose of SHIV-4 virus. AIDS Res Hum Retrovir. 1997;13:1487–1495. doi: 10.1089/aid.1997.13.1487. [DOI] [PubMed] [Google Scholar]
- 4.Berglund P, Smerdou C, Fleeton M N, Tubulekas I, Liljestrom P. Enhancing immune responses using suicidal DNA vaccines. Nat Biotechnol. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- 5.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C-H, Wang T-L, Hung C-F, Yang Y, Young R A, Pardoll D M, Wu T-C. Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Res. 2000;60:1035–1042. [PubMed] [Google Scholar]
- 7.Dialynas D P, Quan Z S, Wall K A, Pierres A, Quintans J, Loken M R, Pierres M, Fitch F W. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5: similarity of L3T4 to the human Leu-3/T4 molecule. J Immunol. 1983;131:2445–2451. [PubMed] [Google Scholar]
- 8.Dilber S M, Phelan A, Aints A, Mohamed A J, Elliott G, Smith C I, O'Hare P. Intercellular delivery of thymidine kinase prodrug activating enzyme by the herpes simplex virus protein, VP22. Gene Ther. 1999;6:12–21. doi: 10.1038/sj.gt.3300838. [DOI] [PubMed] [Google Scholar]
- 9.Dorange F, El Mehdaoui S, Pichon C, Coursaget P, Vautherot J F. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J Gen Virol. 2000;81(Part 9):2219–2230. doi: 10.1099/0022-1317-81-9-2219. [DOI] [PubMed] [Google Scholar]
- 10.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 11.Feltkamp C M, Smits H L, Vierboom M P, Minnaar R P, de Jongh B M, Drijfhout J W, ter Schegget J, Melief C J, Kast W M. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur J Immunol. 1993;23:2242–2249. doi: 10.1002/eji.1830230929. [DOI] [PubMed] [Google Scholar]
- 12.Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol. 1996;70:1182–1190. doi: 10.1128/jvi.70.2.1182-1190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hariharan J M, Driver D A, Townsend K, Brumm D, Polo J M, Belli B A, Catton D J, Hsu D, Mittelstaedt D, McCormack J E, Karavodin L, Dubensky T W, Jr, Chang S M, Banks T A. DNA immunization against herpes simplex virus: enhanced efficacy using a Sindbis virus-based vector. J Virol. 1998;72:950–958. doi: 10.1128/jvi.72.2.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms J S, Ren X, Oliveira S C, Splitter G A. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J Virol. 2000;74:3301–3312. doi: 10.1128/jvi.74.7.3301-3312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haussler O, Epstein J I, Amin M B, Heitz P U, Hailemariam S. Cell proliferation, apoptosis, oncogene, and tumor suppressor gene status in adenosis with comparison to benign prostatic hyperplasia, prostatic intraepithelial neoplasia, and cancer. Hum Pathol. 1999;30:1077–1086. doi: 10.1016/s0046-8177(99)90226-5. [DOI] [PubMed] [Google Scholar]
- 16.Huang A Y, Golumbek P, Ahmadzadeh M, Jaffee E, Pardoll D, Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–965. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 17.Koo G C, Dumont F J, Tutt M, Hackett J, Jr, Kumar V. The NK-1.1(−) mouse: a model to study differentiation of murine NK cells. J Immunol. 1986;137:3742–3747. [PubMed] [Google Scholar]
- 18.Leitner W W, Ying H, Driver D A, Dubensky T W, Restifo N P. Enhancement of tumor-specific immune response with plasmid DNA replicon vectors. Cancer Res. 2000;60:51–55. [PMC free article] [PubMed] [Google Scholar]
- 19.Leitner W W, Ying H, Restifo N P. DNA and RNA-based vaccines: principles, progress and prospects. Vaccine. 1999;18:765–777. doi: 10.1016/s0264-410x(99)00271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin K-Y, Guarnieri F G, Staveley-O'Carroll K F, Levitsky H I, August T, Pardoll D M, Wu T-C. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 21.Lipponen P K, Aaltomaa S. Apoptosis in bladder cancer as related to standard prognostic factors and prognosis. J Pathol. 1994;173:333–339. doi: 10.1002/path.1711730408. [DOI] [PubMed] [Google Scholar]
- 22.Lu Z, Yuan L, Zhou X, Sotomayor E, Levitsky H I, Pardoll D M. CD40-independent pathways of T cell help for priming of CD8(+) cytotoxic T lymphocytes. J Exp Med. 2000;191:541–550. doi: 10.1084/jem.191.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukas J, Muller H, Bartkova J, Spitkovsky D, Kjerulff A A, Jansen D P, Strauss M, Bartek J. DNA tumor virus oncoproteins and retinoblastoma gene mutations share the ability to relieve the cell's requirement for cyclin D1 function in G1. J Cell Biol. 1994;125:625–638. doi: 10.1083/jcb.125.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandl C W, Aberle J H, Aberle S W, Holzmann H, Allison S L, Heinz F X. In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model. Nat Med. 1998;4:1438–1440. doi: 10.1038/4031. [DOI] [PubMed] [Google Scholar]
- 25.Pardoll D M, Beckerleg A M. Exposing the immunology of naked DNA vaccines. Immunity. 1995;3:165–169. doi: 10.1016/1074-7613(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 26.Phelan A, Elliott G, O'Hare P. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 27.Pushko P, Parker M, Ludwig G V, Davis N L, Johnston R E, Smith J F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 28.Sarmiento M, Glasebrook A L, Fitch F W. IgG or IgM monoclonal antibodies reactive with different determinants on the molecular complex bearing Lyt 2 antigen block T cell-mediated cytolysis in the absence of complement. J Immunol. 1980;125:2665–2672. [PubMed] [Google Scholar]
- 29.Tindle R W, Fernando G J, Sterling J C, Frazer I H. A “public” T-helper epitope of the E7 transforming protein of human papillomavirus 16 provides cognate help for several E7 B-cell epitopes from cervical cancer-associated human papillomavirus genotypes. Proc Natl Acad Sci USA. 1991;88:5887–5891. doi: 10.1073/pnas.88.13.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T-L, Ling L, Shih I-M, Pham T, Pai S I, Lu L, Kurman R J, Pardoll D M, Wu T-C. Intramuscular administration of E7-transfected dendritic cells generates the most potent E7-specific anti-tumor immunity. Gene Ther. 2000;7:726–733. doi: 10.1038/sj.gt.3301160. [DOI] [PubMed] [Google Scholar]
- 31.Wu T-C, Guarnieri F G, Staveley-O'Carroll K F, Viscidi R P, Levitsky H I, Hedrick L, Cho K R, August T, Pardoll D M. Engineering an intracellular pathway for MHC class II presentation of HPV-16 E7. Proc Natl Acad Sci USA. 1995;92:11671–11675. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu T C, Huang A Y, Jaffee E M, Levitsky H I, Pardoll D M. A reassessment of the role of B7–1 expression in tumor rejection. J Exp Med. 1995;182:1415–1421. doi: 10.1084/jem.182.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying H, Zaks T Z, Wang R F, Irvine K R, Kammula U S, Marincola F M, Leitner W W, Restifo N P. Cancer therapy using a self-replicating RNA vaccine. Nat Med. 1999;5:823–827. doi: 10.1038/10548. [DOI] [PMC free article] [PubMed] [Google Scholar]