Abstract
Although the immediate-early proteins of both herpes simplex virus (HSV) and cytomegalovirus (CMV) are known to modify promyelocytic leukemia (PML) (ND10) bodies in the nucleus of the host cell, it has been unclear whether lytic infection with gamma herpesviruses induces a similar effect. The PML protein is induced by interferon, involved in major histocompatibility complex class I presentation, and necessary for certain types of apoptosis. Therefore, it is likely that PML bodies function in an antiviral capacity. SUMO-1 modification of PML is known to be required for the formation of PML bodies. To examine whether Epstein-Barr virus (EBV) lytic replication interferes with PML bodies, we expressed the EBV immediate-early genes BZLF1 (Z) and BRLF1 (R) in EBV-positive cell lines and examined PML localization. Both Z and R expression resulted in PML dispersion in EBV-positive cells. Z but not R expression is sufficient to disrupt PML bodies in EBV-negative cell lines. We show that dispersion of PML bodies by Z requires a portion of the transcriptional activation domain of Z but not the DNA-binding function. As was previously reported for the HSV-1 ICP0 and CMV IE1 proteins, Z reduces the amount of SUMO-1-modified PML. We also found that Z itself is SUMO-1 modified (through amino acid 12) and that Z competes with PML for limiting amounts of SUMO-1. These results suggest that disruption of PML bodies is important for efficient lytic replication of EBV. Furthermore, Z may potentially alter the function of a variety of cellular proteins by inhibiting SUMO-1 modification.
Epstein-Barr virus (EBV) is a member of the human herpesvirus family of viruses and infects approximately 90% of the world's population. EBV is responsible for the clinical syndrome infectious mononucleosis (66) and is found in various tumors, including nasopharyngeal carcinoma and Burkitt's lymphoma (66, 88). Upon infection of the host, EBV initially replicates within epithelial cells in the oropharynx (44, 52, 66, 71) and subsequently infects B cells trafficking through the pharynx. In B cells, the virus usually converts to a latent form and persists indefinitely in the host (44, 52, 66, 71). During latency, only a small subset of EBV-encoded proteins are expressed. Occasionally, the latent virus within B cells switches back to the lytic mode of replication (66). Virus released from B cells then reinfects pharyngeal epithelial cells, resulting in the secretion of infectious virus into the saliva.
The first EBV genes expressed during the lytic form of viral replication are the immediate-early genes BZLF1 (Z) and BRLF1 (R) (44). The Z and R proteins function as transcriptional activators and induce expression of the next tier of EBV genes, the early genes (14, 15, 16, 28, 32, 33, 36, 41, 53, 64, 67, 68, 75, 78). The early genes encode the viral proteins required for EBV DNA replication (44). Viral DNA replication is followed by EBV late-gene expression and packaging of the virus (44).
Viruses often manipulate their host's cellular environment in order to create favorable conditions for viral replication and survival. Virally induced changes include modulation of host cell cycle progression, signal transduction cascades, and transcriptional functions, among others. Notably, a number of viruses, including herpes simplex virus type 1 (HSV-1) and cytomegalovirus (CMV), disperse nuclear structures known as promyelocytic leukemia (PML, also called nuclear domain 10 [ND10]) bodies. The viral proteins responsible for PML body dispersion in HSV-1 and CMV are the immediate-early proteins HSV-1 ICP0 and CMV IE1 (3, 4, 23, 40, 46, 57). Interestingly, ICP0 mutants (containing alterations within the RING finger) which cannot disperse PML bodies also fail to activate gene expression and cannot replicate efficiently, suggesting a link between PML bodies and viral replication (21).
PML-containing nuclear bodies are made up of several proteins, including CREB-binding protein (CBP), Sp100, Rb, Daxx, ISG20, and the small ubiquitin-related modifier SUMO-1 (6, 31, 48, 74, 87). However, only the PML protein is known to be absolutely essential for the formation of these bodies (86). The PML protein was initially identified in patients with acute PML who harbored a translocation resulting in a fusion protein between the retinoic acid receptor α and PML protein (18, 30, 34, 38, 58, 62). This RARα-PML fusion protein prevents PML protein from localizing in nuclear bodies (20, 45, 80), which normally appear as 10 to 30 discrete dots (depending upon cell type) within the nucleus. Treatment with retinoic acid leads to degradation of the RARα-PML fusion protein, proper PML protein localization, and cancer remission (34, 58). The wild-type PML protein is also involved in major histocompatibility complex (MHC) class I presentation (85) and is required for certain types of apoptosis (10, 24, 37, 51, 63, 77, 79, 87). In addition, PML protein expression is induced by type I and II interferons (27, 50). Therefore, it is likely that PML protein functions in an antiviral as well as antitumor capacity.
The PML protein was recently shown to be covalently modified by the SUMO-1 protein (8, 60, 72). SUMO-1 is an 11.5-kDa protein that is homologous to ubiquitin (8, 39, 49, 54, 56, 70); however, SUMO-1 does not function in the same manner as ubiquitin. SUMO-1 becomes covalently attached to a variety of proteins, including PML, RanGAP1, and IκB, and serves to localize and/or stabilize these proteins (8, 17, 55, 60, 72). The SUMO-1 modification of PML protein is required for its localization to nuclear bodies (60, 86). SUMO-1 modification of PML protein is also required for the localization of several other proteins, including Sp100, Daxx, CBP, and ISG20, into PML bodies (86).
HSV-1 and CMV were both recently shown to disrupt PML bodies by inhibiting the formation of SUMO-1-modified species of PML protein (13, 22, 59). Interestingly, the CMV immediate-early protein IE1 (as well as the IE2 protein) is itself SUMO-1 modified (35, 59). The ability of both alphaherpesviruses (HSV) and betaherpesviruses (CMV) to disperse PML bodies in the host cell suggests that this function may be universally important for efficient herpesvirus replication, including gammaherpesviruses such as EBV. However, to our knowledge the effect of lytic EBV infection on PML bodies has not been previously examined, probably due to the difficulty of inducing efficient lytic EBV infection in vitro.
In this study, we have examined the ability of lytic EBV infection to disperse PML bodies. We show that induction of the lytic form of EBV infection using either expression vectors for Z or R or an adenovirus vector that expresses Z leads to dispersion of PML bodies in EBV-positive cells. In EBV-negative cells, expression of the immediate-early protein Z alone but not R alone is sufficient to disperse PML bodies. The dispersal of PML bodies is coincident with the loss of high-molecular-weight (SUMO-1-modified) isoforms of PML bodies. In transfection assays, we show that the first 86 amino acids of Z (which encode the transcriptional activation domain as well as a replication function) are required for PML body dispersion. We have also discovered that Z itself is SUMO-1 modified at amino acid 12. Additionally, we demonstrate that Z is able to outcompete PML bodies for SUMO-1 modification and that competition for SUMO-1 modification requires the lysine at amino acid 12. The disruption of PML bodies by all classes of herpesviruses suggests that this function is important for efficient lytic replication.
MATERIALS AND METHODS
Cell lines.
DG75 is an EBV-negative Burkitt's lymphoma cell line, and Akata is an EBV-positive Burkitt's lymphoma cell line. B-cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. A549 is a human lung carcinoma line. HeLa is a cervical carcinoma cell line. D98/HE-R-1 is an EBV-positive epithelial cell line formed by the fusion of an HeLa subclone (D98) with the EBV-positive Burkitt's lymphoma cell line P3HR/1 (29). Epithelial cell lines were maintained in Dulbecco's modified Eagle's medium H supplemented with 10% fetal calf serum.
Akata cell induction.
Akata cells, at 2 × 106 cells/ml, were incubated in the presence or absence of 100 μg of anti-human immunoglobulin G (IgG; Sigma) per ml for 3 h.
Adenovirus construction and infection.
The Z and R cDNAs were cloned into a shuttle vector (under the control of the CMV promoter) which contains a loxP site, the left adenovirus terminal repeat, and a packaging signal. This vector was recombined (in a cell line expressing the phage P1 Cre protein) into the loxP site of an adenovirus lacking the E1 and E3 genes as well as a packaging sequence to create adenovirus-Z and adenovirus-R, as previously described (81). A control vector (adenovirus-LacZ), containing the lacZ gene, was made in the same manner.
HeLa cells were plated at a cell density of 106 cells per 100-mm plate. Cells were infected with no adenovirus (mock infection), adenovirus-LacZ, adenovirus-R, or adenovirus-Z at a multiplicity of infection of 50. The cells were harvested at 48 h postinfection.
Plasmids.
The CMV-Z expression vector contains the Z cDNA downstream of the CMV immediate-early promoter in the pHD1013 vector (64). The SV-Z expression vector contains a genomic clone downstream of the simian virus 40 (SV40) immediate-early promoter in the SVpIE vector (25). The Z311 expression vector contains a mutation in the DNA-binding domain of Z in which amino acid 185 is altered (abolishing DNA binding), downstream of the CMV immediate-early promoter, in the pHD1013 vector (28, 42). Z86–245 (previously referred to as RAZΔR) has the codons for amino acids 2 to 86 deleted (gift of Joseph Pagano) downstream of the CMV promoter (26). Z(S186A) contains a genomic clone mutated at amino acid 186 downstream of the SV40 immediate-early promoter in the SVpIE vector (25). Z25–131 has a deletion of the codons for the first 24 amino acids of Z and is downstream of the SV40 promoter (7). Z131–245 has a deletion of the codons for the first 130 amino acids of Z and is downstream of the SV40 promoter (7). ZΔ25–42 contains the Z cDNA with a deletion of the codons for amino acids 25 to 42 (gift of Alain Sergeant) downstream of the CMV immediate-early promoter in the pHD1013 vector. Zta (m12/13), referred to as Zm12/13 in this paper, contains mutations at amino acids 12 and 13 (from KF to AA) in the pcDL-SRα296 vector (gift of Diane Hayward) (69). In experiments with Zm12/13, a plasmid containing wild-type Z in the pcDL-SRα296 vector was used (69). PRTS-15 contains R downstream of the SV40 promoter in the pSG5 vector (gift of Diane Hayward). The PML protein vector contains the PML gene downstream of the CMV promoter in the CMX vector (19). The HA-SUMO-1 vector contains the hemagglutinin (HA)-tagged SUMO-1 gene downstream of the CMV promoter in the pHSS10B vector (55).
DNA purification.
Plasmid DNA was purified through Qiagen columns as described by the manufacturer (Qiagen).
DNA transfection.
DNA (20 μg) was transfected into cells by electroporation with a Zapper electroporation unit (Medical Electronics Shop, University of Wisconsin) at 1,500 V as described (76). All cells were resuspended in RPMI 1640 medium prior to electroporation.
Protein preparation.
Cells were washed twice with phosphate-buffered saline (PBS), resuspended in ELB buffer (0.25 M NaCl, 0.1% NP-40, 50 mM HEPES [pH 7], 5 mM EDTA, protease inhibitors), and freeze-thawed twice. The lysed cells were centrifuged, and the supernatant was used for immunoblot analysis.
For SUMO-1-modified proteins, the cells were washed twice with PBS and resuspended in a 1:3 mixture of buffer I (5% sodium dodecyl sulfate [SDS], 0.15 M Tris-HCl [pH 6.8], 30% glycerol) and buffer II (25 mM Tris-HCl [pH 8.3], 50 mM NaCl, 0.5% NP-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors). The cells were briefly sonicated and centrifuged, and the resulting supernatant was used for immunoblot analysis.
Immunoblot analysis.
Immunoblot analysis was performed to detect the PML, Z, and HA-SUMO-1 proteins as follows. Briefly, 10 to 100 μg of protein was loaded in each lane, and SDS-polyacrylamide gel electrophoresis was performed. The proteins were transferred onto nitrocellulose (Protran), blocked in 1× PBS–5% milk–0.1% Tween 20, and incubated in primary antibody for 1 to 2 h at room temperature or overnight at 4°C (monoclonal anti-EBV Zebra [1:100] from Argene; monoclonal anti-PML [PG-M3] [1:100] from Santa Cruz; anti-HA probe [Y-11] [1:100] from Santa Cruz). The membrane was washed in PBS–0.1% Tween 20, incubated in secondary antibody for 30 to 60 min at room temperature (goat anti-mouse Ig-horseradish peroxidase [1:10,000] conjugate from Promega; goat anti-rabbit Ig-horseradish peroxidase conjugate [1:10,000] from Promega), and washed, and the results were visualized with the ECL chemiluminescence kit (Amersham) according to the manufacturer's instructions.
Immunocytochemistry.
Cells were grown on glass coverslips, rinsed in 1× Tris-saline (TS; 10 mM Tris-HCl [pH 8], 150 mM NaCl), and fixed in 100% methanol for 10 min at −20°C. The cells were then rehydrated in cold 1× TS for 5 min, incubated in incubation mix (1× TS, 0.3% bovine serum albumin [BSA], 5% goat serum) at room temperature for 10 min, incubated in primary antibody diluted in incubation mix for 1 h at 37°C (monoclonal anti-EBV Zebra [1:50], polyclonal anti-Z M47 [1:500], from Erik Flemington; monoclonal anti-PML protein [PG-M3] [1:100], from Santa Cruz; polyclonal anti-PML protein [1:1,000], from Gary Hayward; or monoclonal anti-EBV R [1:50] from Argene). Cells were washed four times with 1× TS–0.5% BSA for 5 min each and then incubated in secondary antibody diluted in incubation mix for 45 min at 37°C (goat anti-mouse Ig-fluorescein isothiocyanate conjugate [1:100], from Sigma; goat anti-rabbit Ig-indocarbocyanine conjugate [1:100], from Jackson Labs). Cells were washed as before, mounted in Anti-fade medium (Molecular Probes), and viewed on a Zeiss Axioskop or Zeiss confocal microscope.
RESULTS
Lytic EBV replication disperses PML bodies.
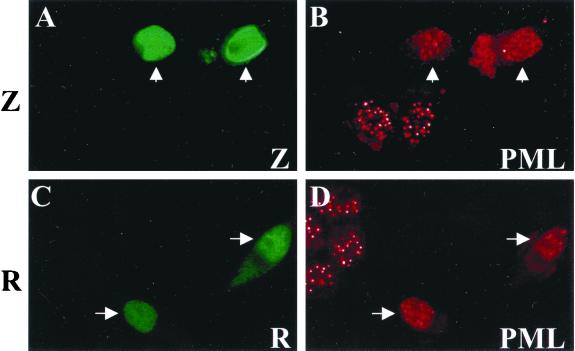
Expression of either the EBV Z or R immediate-early protein in EBV-positive cell lines is sufficient to trigger lytic EBV replication (14, 15, 43, 65, 67, 68, 75, 81, 82). To investigate whether lytic EBV replication induces disruption of PML bodies, we used expression vectors to express the Z and R proteins in the latently infected EBV-positive cell line D98/HE-R1 (Fig. 1). In cells that are positively expressing Z (Fig. 1A), there is a coincident dispersal of PML bodies (Fig. 1B). Expression of the other immediate-early protein, R, also induced relocalization of PML bodies in these EBV-positive cells (Fig. 1C and D). These experiments were repeated several times with similar results. Therefore, lytic EBV replication induces dispersal of PML bodies.
FIG. 1.
EBV lytic replication disrupts PML bodies. EBV-positive cells (D98/HE-R-1) were transfected with expression plasmids for either Z (A and B) or R (C and D). At 48 h posttransfection, cells were fixed and immunostained with either anti-Z (monoclonal) (A), anti-R (monoclonal) (C), or anti-PML protein (polyclonal) (B and D) antibodies. Arrows indicate cells that are positively expressing Z or R.
Z but not R expression disperses PML bodies.
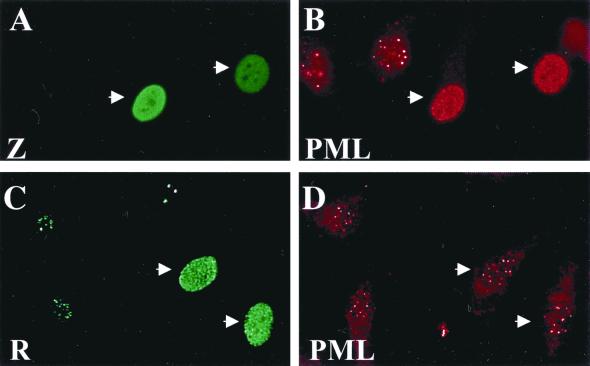
The two other herpesvirus family members known to disperse PML bodies, HSV-1 and CMV, do so via their immediate-early proteins ICP0 and IE1, respectively (3, 23, 40, 46, 57). To examine whether an EBV immediate-early protein is also responsible for PML body relocalization, we expressed either Z or R in EBV-negative cells and examined the resulting PML body distribution (Fig. 2). Expression of Z in EBV-negative HeLa cells resulted in a redistribution of PML protein from PML bodies to a diffuse microspeckled pattern (Fig. 2A and B). However, expression of R in HeLa cells did not affect the localization of PML protein (Fig. 2C and D). Additionally, expression of the EBV early protein BMRF1 had no effect on PML protein localization (data not shown). These experiments were repeated several times with results identical to those presented. Therefore, Z expression is sufficient to disrupt PML bodies.
FIG. 2.
Z but not R expression disrupts PML bodies. EBV-negative (HeLa) cells were transfected with expression plasmids for either Z (A and B) or R (C and D). At 48 h posttransfection, cells were fixed and immunostained with either anti-Z (monoclonal) (A), anti-R (monoclonal) (C), or anti-PML protein (polyclonal) (B and D) antibodies.
Transactivation domain of Z is necessary for dispersal of PML protein.
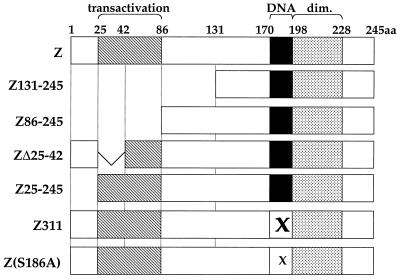
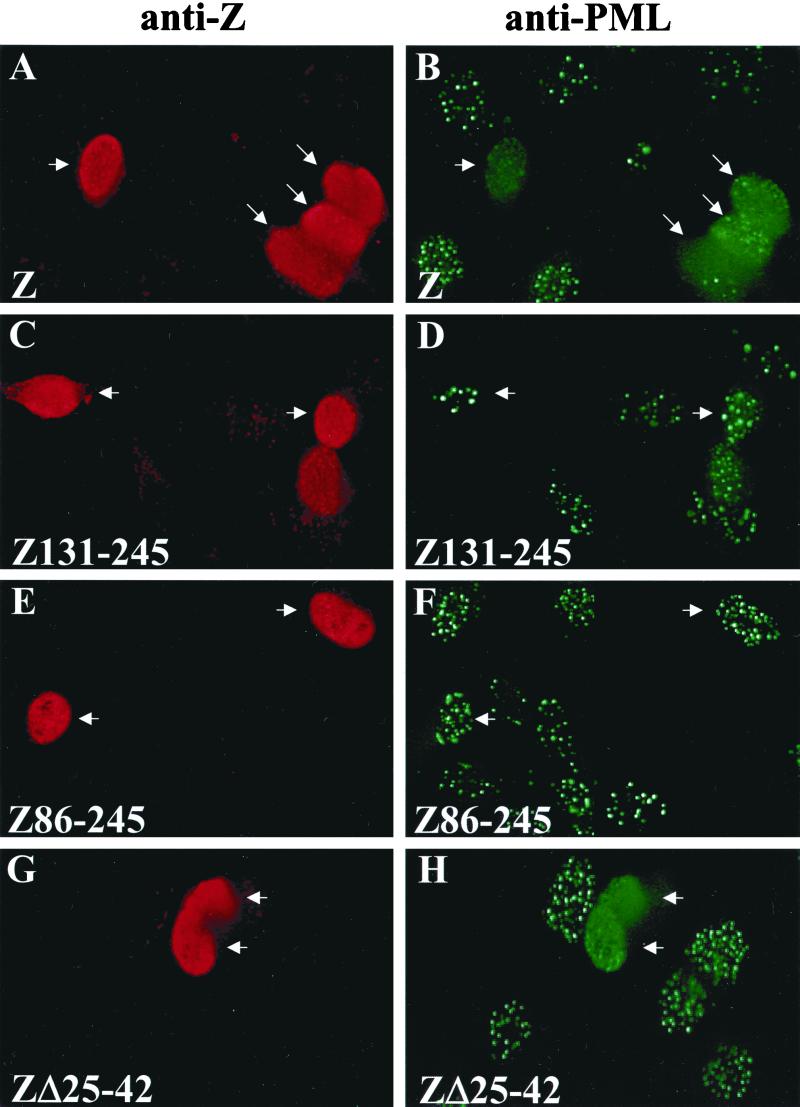
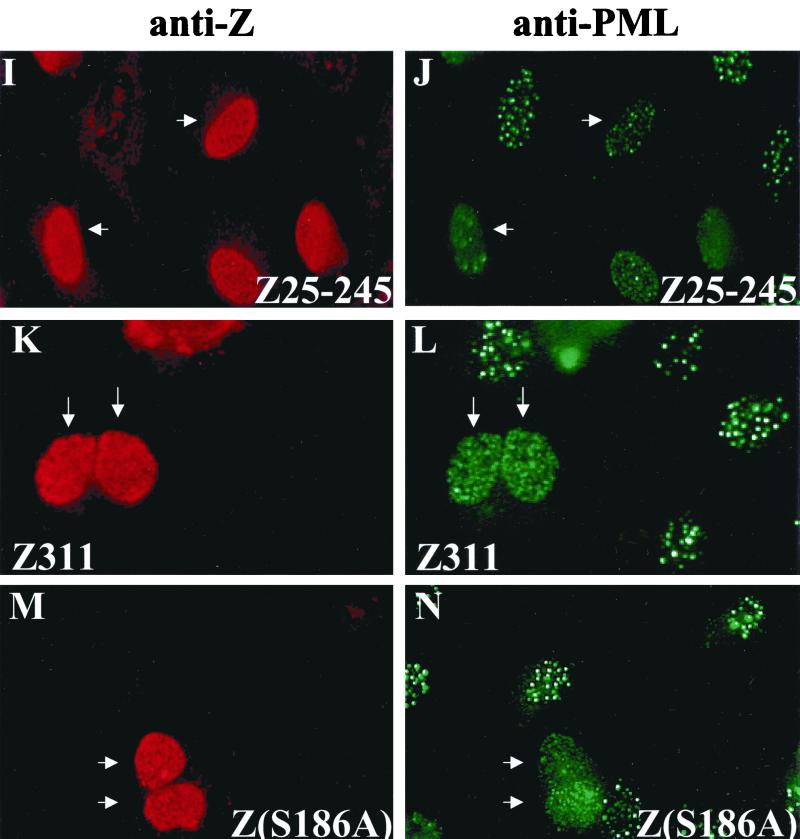
The Z protein has functional domains for transcriptional transactivation, DNA binding, and homodimerization (25, 47, 61). To determine which region(s) of the Z protein is required for PML protein dispersal, we expressed various Z mutants (Fig. 3) in EBV-negative HeLa cells and examined the resulting PML protein localization (Fig. 4). Deletion of the first half of Z (Z131–245) rendered the protein unable to disperse PML bodies (Fig. 4C and D). More specifically, amino acids 1 to 86 of the activation domain of Z are necessary for PML dispersal (Fig. 4E and F). ZΔ25–42 retained the ability to disperse PML bodies (Fig. 4G and H). Deletion of the first 24 amino acids of Z (Z25–245) partially but not completely blocked the ability of the protein to disrupt PML bodies (Fig. 4I and J). Z constructs containing mutations within the DNA-binding domain of Z, including Z311 (mutation at codon 185 which renders the protein unable to bind to DNA) and Z(S186A) (mutation at codon 186 which prevents protein from inducing lytic EBV replication) retained the ability to disrupt PML bodies (Fig. 4K, L, M, and N). Therefore, amino acids 1 to 24, which were previously shown to have a replication function distinct from the Z transcriptional function (69), and the second half of the activation domain of Z (amino acids 42 to 86) are necessary for Z to disperse PML bodies fully. However, direct DNA binding is not required.
FIG. 3.
Structures of wild-type and mutant Z proteins. The transactivation (hatched), DNA-binding (DNA; black), and dimerization (dim.; stippled) domains are indicated. The amino acid (aa) numbers are noted at the top. Χ, mutated amino acid.
FIG. 4.
Mapping the region of Z required for PML body dispersion. HeLa cells were transfected with 20 μg of DNA. At 48 h posttransfection, cells were fixed and immunostained with anti-Z (polyclonal) antibody (A, C, E, G, I, K, and M) and anti-PML protein (monoclonal) antibody (B, D, F, H, J, L, and N). (A and B) wild-type Z. (C and D) Z131–245. (E and F) Z86–245. (G and H) ZΔ25–42 (I and J) Z25–245. (K and L) Z311. (M and N) Z(S186A). Arrows point to cells that are positively expressing Z.
Z induces loss of high-molecular-weight isoforms of PML protein.
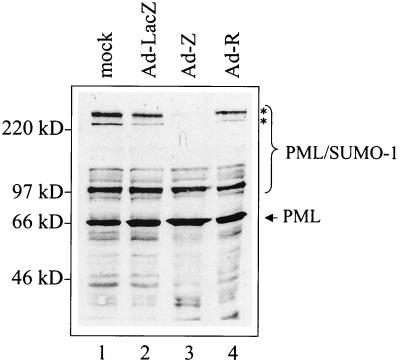
The PML protein has a molecular mass of approximately 70 kDa; however, several higher-molecular-weight isoforms of PML protein exist. These higher-molecular-weight isoforms are modified by SUMO-1, a small ubiquitin-like protein that is covalently linked to PML protein (8, 66, 72). To investigate whether Z disperses PML protein by inducing a loss of SUMO-1-modified PML protein isoforms, we expressed Z in EBV-negative A549 cells via an adenovirus construct and performed an immunoblot with an anti-PML antibody (Fig. 5). Cells that expressed Z but not the control protein LacZ lost the high-molecular-weight forms of PML protein (Fig. 5). R expression did not affect PML protein levels or SUMO-1 modification. This Z-induced loss of SUMO-1-modified PML protein isoforms was also detected in other cell types, including HeLa and D98/HE-R1 (data not shown).
FIG. 5.
Z expression induces a loss of PML protein isoforms. EBV-negative A549 cells were mock infected (lane 1) or infected with an adenovirus expressing either LacZ (lane 2), Z (lane 3), or R (lane 4). Cells were harvested 48 h postinfection, and Western blot analysis was performed with anti-PML protein monoclonal antibody. PML isoforms lost after Z expression are indicated by asterisks.
Z is modified by SUMO-1.
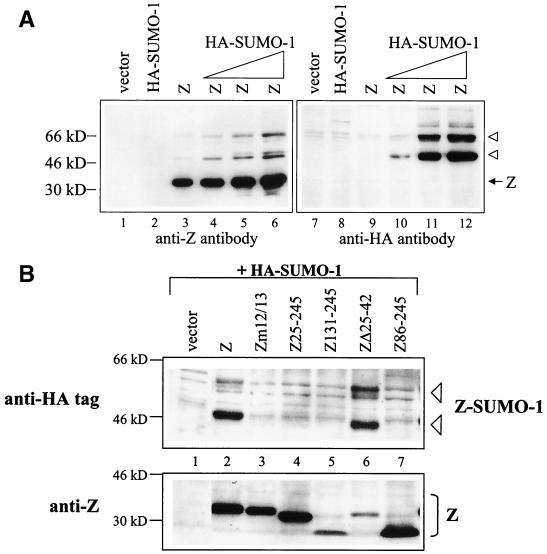
The SUMO-1 modification of PML protein targets PML protein to nuclear bodies (60, 86). To examine whether Z is modified by SUMO-1, we expressed Z and HA-tagged SUMO-1 proteins in DG75 cells, harvested these cells in a rapidly denaturing buffer which has been shown previously to prevent loss of SUMO-1 (17), and performed immunoblot analysis with anti-Z and anti-HA antibodies (Fig. 6A). Using these harvesting conditions, the anti-Z antibody detected at least two larger bands (at approximately 50 and 66 kDa), and possibly a third larger band, in addition to the normal Z band (35 kDa) (Fig. 6A, left panel, lane 3). Increasing amounts of transfected HA-SUMO-1 increased the intensity of these larger bands (Fig. 6A, left panel, lanes 4 to 6), indicating that increased levels of SUMO-1 lead to increased levels of Z/SUMO-1. To ensure that the 50 and 66-kDa Z bands were indeed modified by the HA-SUMO-1 protein, we probed an identical blot with anti-HA antibody (Fig. 6A, right panel). Lanes 10 to 12 clearly show that the 50- and 66-kDa bands are HA-SUMO-1-modified Z. These results indicate that Z is SUMO-1 modified and that there are at least two forms of SUMO-1-modified Z.
FIG. 6.
Z is modified by SUMO-1. (A) EBV-negative DG75 cells were transfected with vector, HA-SUMO-1 (10 μg), Z (10 μg), or Z (10 μg) plus HA-SUMO-1 (1, 5, or 10 μg) expression plasmids. Cells were harvested 48 h posttransfection using conditions which preserve SUMO-1 modification, and Western blot analysis was performed with anti-Z (monoclonal) (left panel) or anti-HA (right panel) antibodies. Arrowheads indicate HA-SUMO-1-modified Z proteins. (B) EBV-negative DG75 cells were transfected with 5 μg of HA-SUMO-1 plus 10 μg of either vector, Z, Zm12/13, Z25–245, Z131–245, ZΔ25–42, or Z86–245 expression plasmids. Zm12/13 contains mutations that alter amino acids 12 and 13 (69). Cells were harvested 48 h posttransfection using conditions which preserve SUMO-1 modification, and Western blot analysis was performed with anti-HA (monoclonal) (top panel) or anti-Z (bottom panel) antibodies.
We subsequently used the same technique to map the domain(s) of Z that is required for SUMO-1 modification. As shown in Fig. 6B, cotransfection of the HA-SUMO-1 plasmid with a series of wild-type and mutant Z plasmids induced SUMO-1 modification of all constructs containing the first 24 amino acids of Z but did not modify any mutant in which these residues were deleted. Since there is only one lysine in the first 24 amino acids of Z (residue 12), we examined a Z construct that contained mutations at amino acids 12 and 13 and found that this mutant did not become SUMO-1 modified. Therefore, Z is SUMO-1 modified on the lysine residing at amino acid position 12.
Z and PML protein compete for limiting levels of SUMO-1.
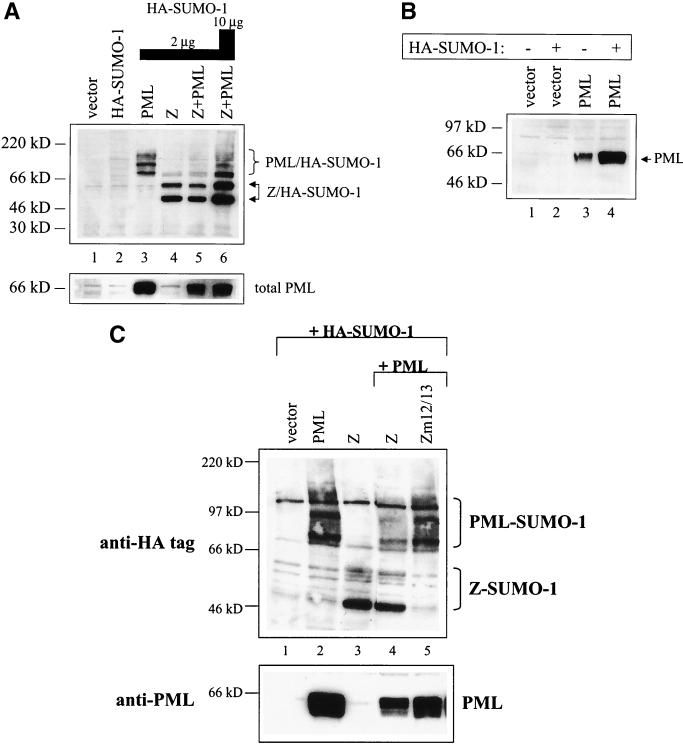
As evident from Fig. 6, Z is very efficiently modified by SUMO-1. If SUMO-1 is limiting in cells, then a protein that is strongly modified by SUMO-1 could potentially disperse PML protein by competing with PML protein for limiting levels of SUMO-1 protein. To examine whether Z is able to compete with PML protein for SUMO-1 modification, we expressed PML protein and HA-SUMO-1, with or without Z, in DG75 cells. The cells were harvested so as to preserve SUMO-1 modification, and we performed immunoblot analysis with an anti-HA antibody (Fig. 7A). The SUMO-1 modification of PML protein was decreased in the presence of Z (compare lanes 3 and 5), while the SUMO-1 modification of Z remained unchanged in the presence of PML protein (compare lanes 4 and 5). When the level of cotransfected HA-SUMO-1 plasmid was increased to 10 μg, SUMO-1 modification of PML protein in the presence of Z was partially rescued. Additionally, the total level of PML protein was slightly decreased by Z. A decrease in total PML protein levels in the presence of Z is not unexpected, however, since Fig. 7B shows that SUMO-1 greatly stabilizes PML protein. As shown in Fig. 7C, the Z SUMO-1 modification site, lysine 12, is required for inhibition of PML protein SUMO-1 modification. These results suggest that the level of SUMO-1 is limiting in cells and that Z outcompetes PML protein for SUMO-1 modification.
FIG. 7.
Z competes with PML protein for SUMO-1. (A) EBV-negative DG75 cells were transfected with vector, HA-SUMO-1 (10 μg), PML protein (10 μg) plus HA-SUMO-1 (2 μg), Z (10 μg) plus HA-SUMO-1 (2 μg), or Z (10 μg) plus PML protein (10 μg) plus HA-SUMO-1 (2 or 10 μg) expression plasmids. Cells were harvested 48 h posttransfection, and Western blot analysis was performed with anti-HA antibody (upper panel). The blot was reprobed with anti-PML protein monoclonal antibody to check for total PML protein levels (lower panel). (B) EBV-negative DG75 cells were transfected with vector, HA-SUMO-1 (10 μg), PML protein (5 μg), or PML protein (5 μg) plus HA-SUMO-1 (2 μg). Cells were harvested 48 h posttransfection, and Western blot analysis was performed with an anti-PML protein monoclonal antibody. (C) EBV-negative DG75 cells were transfected with HA-SUMO-1 along with either vector, PML, Z, Z plus PML protein, or Zm12/13 plus PML protein expression plasmids. Ten micrograms of each plasmid was used. Cells were harvested 48 h posttransfection, and Western blot analysis was performed with anti-HA antibody (upper panel). The blot was reprobed with anti-PML protein monoclonal antibody to check for total PML protein levels (lower panel).
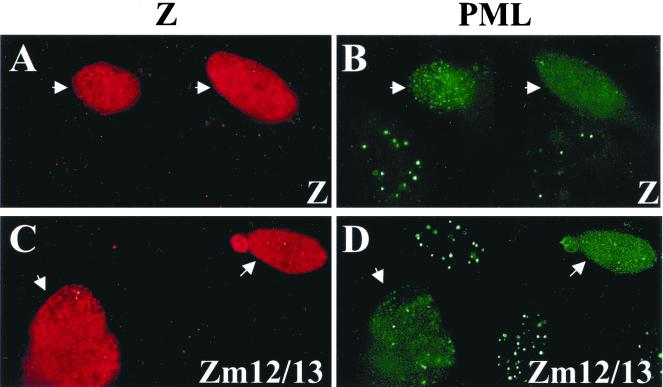
Loss of the SUMO-1 modification site of Z does not prevent PML body disruption.
To determine if SUMO-1 modification of Z is required for the disruption of PML bodies, we expressed the Z mutant lacking the SUMO-1 site, Zm12/13, in HeLa cells and performed immunocytochemistry to determine the localization of PML protein in these transfected cells (Fig. 8). Surprisingly, the Zm12/13 mutant retained the ability to disperse PML bodies (Fig. 8B and C). These data suggest that while competition for SUMO-1 may play a role in Z-induced PML body dispersion, Z must also use another mechanism to disperse PML bodies.
FIG. 8.
Z mutant that cannot be SUMO modified disperses PML bodies. HeLa cells were transfected with 20 μg of DNA. At 48 h posttransfection, cells were fixed and immunostained with anti-Z polyclonal antibody (A and C) and anti-PML protein monoclonal antibody (B and D). (A and B) Wild-type Z. (C and D) Zm12/13.
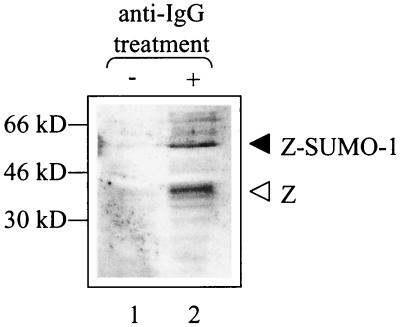
Induction of lytic replication in Akata cells leads to rapid SUMO-1 modification of Z.
To examine whether lytic replication would yield levels of SUMO-1-modified Z comparable to the levels obtained with transfected plasmids, we induced EBV-positive Akata cells to undergo lytic replication via cross-linking of the surface IgG by anti-IgG. Three hours after induction, the cells were harvested so as to preserve SUMO-1 modification, and immunoblot analysis was performed with an anti-Z antibody (Fig. 9). While control cells (no anti-IgG) did not express Z, the anti-IgG-treated cells expressed Z as well as higher-molecular-weight forms of Z corresponding to SUMO-1-modified species. Therefore, physiological levels of Z and SUMO-1 are sufficient to produce high levels of SUMO-1-modified Z.
FIG. 9.
Z is SUMO-1 modified in Akata cells after induction with anti-IgG cross-linking. Akata cells were induced to undergo lytic replication via anti-IgG cross-linking for 3 h. Cells were harvested using conditions which preserve SUMO-1 modification, and Western blot analysis was performed with anti-Z polyclonal antibody. The open arrowhead indicates unmodified Z; the solid arrowhead indicates SUMO-1-modified Z.
DISCUSSION
The EBV immediate-early protein Z is required for expression of the EBV early genes and plays an essential role in replication mediated through the lytic origin of replication (oriLyt). In addition to these direct roles in lytic viral replication, increasing evidence suggests that Z manipulates the host cell environment through a variety of different mechanisms, presumably to provide favorable conditions for viral replication. For example, Z inhibits cell cycle progression (12), activates the p38 kinase and c-Jun N-terminal kinase signaling cascades (1), and alters p53 (84) and CBP function (2). Here we show that the Z protein also induces dispersion of nuclear PML bodies, an ability it shares with at least two other immediate-early proteins in the herpesvirus family.
PML body dispersion by Z requires amino acids 1 through 86, containing the Z transactivation domain and an essential replication function, and is associated with loss of PML protein SUMO-1 modification. We also found that Z is SUMO-1 modified over residue 12, suggesting that SUMO-1 modification may play a role in targeting Z to specific domains within the nucleus or moderating its stability or protein-protein interactions. SUMO-1 modification of Z may also provide a mechanism for PML body dispersion, since Z competes with PML protein for SUMO-1. However, unmodified Z retains the ability to disperse PML bodies, indicating that another mechanism must also exist for PML body dispersion. Given that deletion of amino acids 1 to 86 totally prevents dispersal of PML bodies by Z while deletion of amino acids 1 to 25 only partially prevents dispersion, it appears that the transcriptional activator region of Z encodes a function (as yet undefined) required for full PML body dispersion.
PML bodies have become an increasingly intense area of study because, although their specific function remains uncertain, they clearly play an important role in tumor suppression. PML bodies are made up of several proteins, some of which have known functions (CBP and Rb), and some of which have unknown functions (Sp100 and PML protein). Since some of the components are transcriptional regulators and PML protein itself is structurally similar to a transcription factor, it has been suggested that PML bodies play a role in regulating cellular transcription. Although PML bodies themselves are devoid of any nucleic acid, nucleic acids can be found along their periphery (9).
SUMO-1 is another component of PML bodies. SUMO-1 is found covalently attached to at least two PML body proteins, PML and Sp100. Although structurally similar to ubiquitin, SUMO-1 does not target proteins for degradation. Instead, SUMO-1 stabilizes proteins and can act to guide them to their appropriate locations within the nucleus. The SUMO-1 modification of PML protein appears to be of utmost importance—PML protein must be SUMO-1 modified in order to reside within PML bodies, and if PML protein is not present in these bodies, other PML body members do not localize properly (86). Therefore, PML protein seems to be the main organizer of the PML body.
We have demonstrated in this study that EBV lytic replication disperses PML bodies, transforming nuclear dots into tiny microspeckles. When either immediate-early protein, Z or R, is expressed in cells latently infected with EBV, PML protein becomes dispersed, presumably because expression of either protein activates the entire EBV lytic cascade. In EBV-positive cells, the redistributed protein appears to form larger aggregates than in EBV-negative cells, potentially due to the formation of EBV replication centers containing PML protein (11). However, in EBV-negative cells, only Z is capable of dispersing PML bodies. Therefore, R likely indirectly disperses PML bodies in EBV-positive cells by activating the expression of Z.
Our data show that Z is SUMO-1 modified and that at least two different SUMO-1-modified isoforms exist. Since the SUMO-1 modification of PML protein is responsible for its localization into discrete nuclear bodies, SUMO-1 modification may likewise target Z to PML bodies. We have been unable to show that Z colocalizes with PML bodies (data not shown). However, it is possible that transient localization of Z into PML bodies does occur prior to the disruption of PML bodies.
In many respects, our findings here are reminiscent of previous reports regarding the effects of HSV-1 and CMV on PML bodies. In the case of HSV-1, the immediate-early protein ICP0, like Z, disperses PML bodies. In cells infected with intact HSV, new nuclear foci appear after PML bodies are disrupted, and these new foci function as HSV-1 replication compartments that contain HSV-1 replication proteins as well as PML protein (11). This suggests a role for PML protein in viral replication. CMV also disrupts PML bodies shortly after infection. In the case of CMV, the IE1 protein is targeted to and disperses PML bodies, possibly through a mechanism involving direct protein-protein interactions between IE1 and PML protein (3). In contrast, the CMV IE2 protein is also targeted to but does not disperse PML bodies. Similar to HSV-1, CMV replication compartments (which contain IE2 as well as several other essential viral replication proteins) subsequently form adjacent to the former PML bodies (5).
Another striking similarity is the ability of all three viruses HSV-1, CMV, and EBV to decrease the relative amount of the SUMO-1-modified forms of PML protein. Although the exact mechanisms by which herpesvirus immediate-early proteins reduce the abundance of SUMO-1-modified PML protein remains unknown, in the case of HSV-1, at least, this process can be reversed by proteosome inhibitors (13, 22), suggesting that HSV-1 preferentially activates degradation of SUMO-modified PML protein. In contrast, we did not find that proteosome inhibitors reverse the effect of Z on PML protein SUMO-1 modification (unpublished data).
It is intriguing that both the Z and CMV IE1 proteins are SUMO-1 modified. Thus, if any component of the SUMO-1 modification machinery is limiting in cells, one potential mechanism by which Z (and CMV IE1) could inhibit SUMO-1 modification of PML protein would be through direct competition for a limiting factor, similar to the ability of the adenovirus E1A protein to prevent histone acetylation of cellular genes by direct competition for CBP/p300. Against this hypothesis is the finding that the CMV IE1 protein (which, in contrast to Z, interacts directly with the PML protein) inhibits SUMO modification of only a limited number of known SUMO-1 substrates (13, 22, 59). However, Z appears to be a highly efficient substrate for SUMO-1 modification, even though the Z SUMO-1 modification site (DVKFT) does not conform to the previously reported consensus sequence (I/L)KxE (73). Indeed, in cells cotransfected with Z and SUMO-1 expression vectors, Z appears to be the major SUMO-1-modified protein in the host cell (Figs. 6 and 7), and Z competed with PML protein for cotransfected SUMO-1. Alternatively, since phosphorylation of the PML protein has been shown to inhibit SUMO-1 modification (60), Z could potentially prevent PML protein SUMO-1 modification by increasing the level of PML protein phosphorylation.
To our knowledge, Z (in addition to CMV IE1 and IE2) is only the third viral protein shown to be SUMO-1 modified. As discussed above, SUMO-1 modification of Z, like PML protein, could potentially be important for its localization to PML bodies. Alternatively, SUMO-1 modification could affect the stability of Z or its interactions with other proteins. Mutation of the two SUMO-1 modification sites in the CMV IE2 protein was recently shown to inhibit its transcriptional function without affecting protein stability or nuclear localization (35). Given the increasing evidence that PML bodies regulate transcription in the cell, it is interesting that the domain of Z required for full PML body dispersion (amino acids 1 through 86) is also known to be required for Z transcriptional function (25). An increasing number of important cellular transcriptional proteins, including CBP, p53, Ap-1, Rb, Daxx, and Sp1, have been shown to be regulated by their interactions with PML bodies. Similarly, in the HSV-1 ICP0 protein, small mutations in the RING finger domain which disrupt the ability of ICP0 to disperse PML bodies considerably decrease the ability of ICP0 to transcriptionally activate early HSV-1 promoters and decrease viral replication by 10- to 100-fold (21). The ability of Z to disperse PML bodies may likewise be important for its transcriptional function and/or its well-described ability to modulate the activity of cellular transcription factors such as CBP and p53 (2, 83, 84).
At this point it remains unknown whether lytic EBV infection, like HSV-1 and CMV infection, also incorporates PML protein into replication foci. The Hayward group has shown that a specific mutation affecting only residues 12 and 13 in Z impairs Z replication function but not transactivator function (69). As shown here, this mutation also prevents SUMO-1 modification of Z. Thus, SUMO-1 modification of Z may well play a role in lytic EBV replication.
Increasing evidence suggests that a variety of PML protein-dependent functions in the cell would be inhibitory to viral replication in the host. Most importantly, PML protein is required for apoptosis induced by several different types of stimuli, including interferon (27, 50), and for inducing MHC class I presentation (85). The fact that so many different viruses encode mechanisms to disrupt PML body function suggests that this protein may have a profoundly important role in vivo for controlling viral infection.
ACKNOWLEDGMENTS
This work was supported by grants 2-R01-CA58853 and P01-CA19014 from the National Institutes of Health.
We thank Gary Hayward for the polyclonal PML antibody, Erik Flemington for polyclonal Z antibody and various Z expression plasmids, Ron Evans for the CMX-PML plasmid, Frauke Melchior for the HA-tagged SUMO-1 plasmid, Diane Hayward for the R expression and Zm12/13 plasmids, Amy Mauser for the Z adenovirus vector, Jennifer Swenson for the R adenovirus vector, the UNC Gene Therapy Center for construction of the adenovirus vectors, and the UNC Microscopy Services Laboratory for assistance with confocal microscopy.
REFERENCES
- 1.Adamson A L, Darr D, Holley-Guthrie E, Johnson R A, Mauser A, Swenson J, Kenney S. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J Virol. 2000;74:1224–1233. doi: 10.1128/jvi.74.3.1224-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson A L, Kenney S. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J Virol. 1999;73:6551–6558. doi: 10.1128/jvi.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn J H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn J H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112–113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci P G. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol Cell Biol. 1998;18:1084–1093. doi: 10.1128/mcb.18.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Askovic S, Baumann R. Activation domain requirements for disruption of Epstein-Barr virus latency by ZEBRA. J Virol. 1997;71:6547–6554. doi: 10.1128/jvi.71.9.6547-6554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 9.Boisvert F-M, Hendzel M J, Bazett-Jones D P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borden K L, Campbell Dwyer E J, Salvato M S. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 1997;418:30–34. doi: 10.1016/s0014-5793(97)01344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 13.Chelbi-Alix M K, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 14.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors EB1 and EB2 are required to activate transcription from an early EBV promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Countryman J, Miller G. Activation of expression of latent Epstein-Barr virus after gene transfer with a small cloned fragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox M, Leahy J, Hardwick J M. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivors. J Virol. 1990;64:313–321. doi: 10.1128/jvi.64.1.313-321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desterro J M P, Rodriguez M S, Hay R T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 18.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML/RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 19.Doucas V, Tini M, Egan D A, Evans R M. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc Natl Acad Sci USA. 1999;96:2627–2632. doi: 10.1073/pnas.96.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyck J, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 21.Everett R, O'Hare P, O'Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagioli M, Alcalay M, Tomassoni L, Ferrucci P F, Mencarelli A, Riganelli D, Grignani F, Pozzan T, Nicoletti I, Grignani F, Pelicci P G. Cooperation between the RING + B1–B2 and coiled-coil domains of PML is necessary for its effects on cell survival. Oncogene. 1998;16:2905–2913. doi: 10.1038/sj.onc.1201811. [DOI] [PubMed] [Google Scholar]
- 25.Flemington E K, Borras A M, Lytle J P, Speck S H. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J Virol. 1992;66:922–929. doi: 10.1128/jvi.66.2.922-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furnari F B, Zacny V, Quinlivin E B, Kenney S, Pagano J S. RAZ, an Epstein-Barr virus transdominant repressor that modulates the viral reactivation mechanism. J Virol. 1994;68:1827–1836. doi: 10.1128/jvi.68.3.1827-1836.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaboli M, Gandini D, Delva L, Wang Z G, Pandolfi P P. Acute promyelocytic leukemia as a model for cross-talk between interferon and retinoic acid pathways: from molecular biology to clinical applications. Leuk Lymphoma. 1998;30:11–22. doi: 10.3109/10428199809050925. [DOI] [PubMed] [Google Scholar]
- 28.Giot J-F, Mikaelian I, Buisson M, Manet E, Joab I, Nicolas J-C, Sergeant A. Transcriptional synergy and interference between the EBV transcription factors EB1 and R require both the basic region and the activation domains of EB1. Nucleic Acids Res. 1991;19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaser R, O'Neill F J. Hybridization of Burkitt lymphoblastoid cells. Science. 1972;176:1245–1247. doi: 10.1126/science.176.4040.1245. [DOI] [PubMed] [Google Scholar]
- 30.Goddard A D, Borrow P S, Freemont P S, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 31.Gongora C, David G, Pintard L, et al. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J Biol Chem. 1997;272:19457–19463. doi: 10.1074/jbc.272.31.19457. [DOI] [PubMed] [Google Scholar]
- 32.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He L Z, Merghoub T, Pandolfi P P. In vivo analysis of the molecular pathogenesis of acute promyelocytic leukemia in the mouse and its therapeutic implications. Oncogene. 1999;18:5278–5292. doi: 10.1038/sj.onc.1203088. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann H, Floss S, Stamminger T. Covalent modification of the transactivator protein IE2–p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol. 2000;74:2510–2524. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holley-Guthrie E, Quinlivin E, Mar E, Kenney S. The Epstein-Barr virus (EBV) promoter for early antigen (EA-D) is regulated by the EBV transactivators BRLF1 and BZLF1 in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T, Strauss III J F, Maul G G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakizuka A, Miller W H, Umesono K, Warrell R P, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 39.Kamitani T, Nguyen H P, Yeh E T H. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 40.Kelly C, van Driel R, Wilkinson G W. Disruption of PML-associated bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 41.Kenney S, Holley-Guthrie E, Mar E-C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenney S, Holley-Guthrie E, Quinlivan E, Gutsch D, Zhang Q, Bender T, Giot J, Sergeant A. The cellular oncogene c-myb can interact synergistically with the Epstein-Barr virus BZLF1 transactivator in lymphoid cells. Mol Cell Biol. 1992;12:136–146. doi: 10.1128/mcb.12.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kenney S, Kamine J, Holley-Guthrie E, Lin J-C, Mar E-C, Pagano J. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J Virol. 1989;63:1729–1736. doi: 10.1128/jvi.63.4.1729-1736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 45.Koken M H M. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 47.Kouzarides T, Packham G, Cook A, Farrell P. The BZLF1 protein of EBV has a coiled-coil dimerization domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene. 1991;6:195–204. [PubMed] [Google Scholar]
- 48.LaMorte V J, Dyck J A, Ochs R L, Evans R M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lapenta V, Chiurazzi P, van der Spek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- 50.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. The acute promyelocytic leukemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 51.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E J, Chen J D. Sequestration and inhibition of daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q X, Young L S, Niedobitek G, Dawson C W, Birkenbach M, Wang F, Rickinson A B. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature. 1992;356:347–350. doi: 10.1038/356347a0. [DOI] [PubMed] [Google Scholar]
- 53.Lieberman P, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 55.Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc binding domain protein family, is rearranged during herpes virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 58.Melnick A, Licht J D. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 59.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Packham G, Economou A, Rooney C M, Rowe D T, Farrell P J. Structure and function of the Epstein-Barr virus BZLF1 protein. J Virol. 1990;64:2110–2116. doi: 10.1128/jvi.64.5.2110-2116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandolfi P P, Grignani F, Alcalay M, Mencarelli A, Biondi A, Lo Coco G, Grignani F, Pelicci P G. Structure and origin of the acute promyelocytic leukemia myl/RARα cDNA and characterization of its retinoid-binding ability and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 63.Quignon F, De Bels F, Koken M, Feunteun J, Ameisin J C, de The H. PML induces a novel caspase-independent death process. Nature Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 64.Quinlivan E, Holley-Guthrie E, Norris M, Gutsch D, Bachenheimer S, Kenney S. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 67.Rooney C, Rowe D, Ragot T, Farrell P. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J Virol. 1989;63:3109–3116. doi: 10.1128/jvi.63.7.3109-3116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rooney C, Taylor N, Countryman J, Jenson H, Kolman J, Miller G. Genome rearrangements activate the Epstein-Barr virus gene whose product disrupts latency. Proc Natl Acad Sci USA. 1988;85:9801–9805. doi: 10.1073/pnas.85.24.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 71.Sixby J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 72.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sternsdorf T, Jensen K, Will H. The nuclear dot protein sp100: characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifers. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 74.Szostecki C H, Guldner H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 75.Takada K, Shimizu N, Sakuma S, Ono Y. Transactivation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tonneguzzo F, Hayday A C, Keating A. Electric field-mediated DNA transfer: transient and stable gene expression in human and mouse lymphoid cells. Mol Cell Biol. 1986;6:703–706. doi: 10.1128/mcb.6.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torii S, Egan D A, Evans R A, Reed J C. Human Daxx regulates Fas-induced apoptosis from nuclear PNL oncogenic domains (PODs) EMBO J. 1999;18:6037–6049. doi: 10.1093/emboj/18.21.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Z-G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–271. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 80.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemic cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 81.Westphal E-M, Mauser A, Swenson J, Davis M G, Talarico C L, Kenney S C. Induction of lytic Epstein-Barr virus (EBV) infection in EBV-associated malignancies using adenovirus vectors in vitro and in vivo. Cancer Res. 1999;59:1485–1491. [PubMed] [Google Scholar]
- 82.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zerby D, Chen C-J, Poon E, Lee D, Shiehattar R, Lieberman P M. The amino-terminal C/H1 domain of CREB-binding protein mediates Zta transcriptional activation of latent Epstein-Barr virus. Mol Cell Biol. 1999;19:1617–1626. doi: 10.1128/mcb.19.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng P, Guo Y, Niu Q, Levy D E, Dyck J A, Lu S, Sheiman L A, Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]
- 86.Zhong S, Muller S, Ronchetti S, Freemont P S, Dejean A, Pandolfi P P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2753. [PubMed] [Google Scholar]
- 87.Zhong S, Salomoni P, Ronchetti S, Guo A, Ruggero D, Pandolfi P P. PML and Daxx participate in a novel nuclear pathway for apoptosis. J Exp Med. 2000;191:631–640. doi: 10.1084/jem.191.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.zur Hausen H, Schulte-Holthauzen H, Klein G, Henle G, Henle W, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1956–1958. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]