Abstract
Hypertrophic cardiomyopathy (HCM) is characterized by thickening of the left ventricular wall, diastolic dysfunction, and fibrosis, and is associated with mutations in genes encoding sarcomere proteins. While in vitro studies have used human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) to study HCM, these models have not examined the multicellular interactions involved in fibrosis. Using engineered cardiac microtissues (CMTs) composed of HCM-causing MYH7-variant hiPSC-CMs and wild-type fibroblasts, we observed cell-cell cross-talk leading to increased collagen deposition, tissue stiffening, and decreased contractility dependent on fibroblast proliferation. hiPSC-CM conditioned media and single-nucleus RNA sequencing data suggested that fibroblast proliferation is mediated by paracrine signals from MYH7-variant cardiomyocytes. Furthermore, inhibiting epidermal growth factor receptor tyrosine kinase with erlotinib hydrochloride attenuated stromal activation. Last, HCM-causing MYBPC3-variant CMTs also demonstrated increased stromal activation and reduced contractility, but with distinct characteristics. Together, these findings establish a paracrine-mediated cross-talk potentially responsible for fibrotic changes observed in HCM.
HCM hiPSC-derived cardiomyocytes activate fibroblast proliferation and alter tissue function via aberrant paracrine signaling.
INTRODUCTION
Familial hypertrophic cardiomyopathy (HCM) affects between 1 in 200 and 1 in 500 people and is one of the leading causes of sudden cardiac death in young people (1–3). Pathologic findings associated with the clinical presentation of HCM include thickening of the left ventricular wall, diastolic dysfunction, and fibrosis. The extent of fibrosis is a key determinant in patient outcome, with as much as 25 to 50% of the myocardium being replaced with fibrotic tissue remodeling in end-stage HCM (4–7). Mutations in genes encoding numerous sarcomere proteins that regulate cardiac contractility have been identified as causes of HCM. Population studies suggest that approximately 60% of patients with inherited HCM harbor at least one mutation in the gene encoding β-myosin heavy chain, MYH7, or the gene encoding cardiac myosin binding protein-C, MYBPC3 (8, 9). However, it remains unclear how the intrinsic changes in such proteins contribute to fibrotic remodeling.
Previous studies of fibrosis in HCM have shown up-regulation of inflammatory cytokines and fibrosis-related miRNAs in patient plasma, as well as elevated transforming growth factor–β (TGF-β) pathway enrichment in both human tissue and genetic mouse models of HCM (10–13). Interventional studies in mouse models indicate that TGF-β neutralizers (13), angiotensin II receptor inhibitors (12, 14, 15), a myosin adenosine triphosphatase inhibitor (16), and reactive oxygen species (ROS) scavengers (12, 17) reduce fibrosis. While these studies demonstrate the multifactorial nature of fibrosis, the mechanistic understanding of how different cell types in the myocardium coordinate and contribute to tissue remodeling remains limited.
Recent advances in human induced pluripotent stem cell (hiPSC) and CRISPR-Cas9 technologies allow for the study of genetic mutations in human in vitro models. Investigators have used these approaches to study patient-derived hiPSC-derived cardiomyocytes (hiPSC-CMs) and to genetically introduce point mutations in sarcomere genes to mimic HCM in hiPSC-CMs and have shown that these cell lines exhibit features of HCM, including cardiomyocyte hypertrophy (18–22), increased contractility (18, 23–25), dysregulation of calcium cycling (19, 20, 23, 26, 27), impaired relaxation (18, 20, 23), sarcomere disorganization (20, 23), decreased myosin in the super relaxed state (18), and altered cardiomyocyte energetics (18, 21, 26, 27). Whereas these studies have demonstrated the ability of CRISPR-Cas9–edited hiPSC-CMs to recapitulate cardiomyocyte-autonomous aspects of HCM, whether these HCM-variant cardiomyocytes can recapitulate aspects of myocardial remodeling and fibrosis observed in vivo remains unclear.
Here, we set out to examine whether an engineered three-dimensional (3D) cardiac tissue model, incorporated with stromal cells (fibroblasts) can recapitulate the pathological extracellular matrix (ECM) remodeling associated with HCM. To study the interactions between hiPSC-CMs and stromal cells within a 3D, mechanically loaded environment, we adapted a previously described cardiac microtissue (CMT) model (28). Using hiPSC-CMs harboring a common MYH7 point mutation (R403Q+/−), we show that HCM-variant CMTs exhibit increased collagen deposition and tissue stiffening, leading to decreased contractility. Single-nucleus RNA sequencing (snRNA-seq) reveals alterations in expression of paracrine factors, as well as an activation of ECM and inflammatory signaling in the stromal cell population. Using conditioned media assays, we demonstrate that paracrine factors from HCM-variant CMs activate stromal cells, that epidermal growth factor (EGF) alone is sufficient to stimulate stromal cell proliferation, and that inhibiting EGF receptor (EGFR) signaling attenuates stromal cell proliferation and ECM remodeling. We show that the stromal activation and hypocontractility phenotypes are also present in CMTs harboring an HCM-causing MYBPC3 truncation (MYBPC3t/+) and that inhibiting EGFR signaling improves contractility in these tissues. Overall, this study offers an approach to modeling and studying the tissue-level changes associated with HCM and provides insights into the critical cross-talk between cardiomyocytes and stromal cells that together induce a fibrotic phenotype in HCM.
RESULTS
MYH7-variant CMTs model fibrotic phenotype in vitro
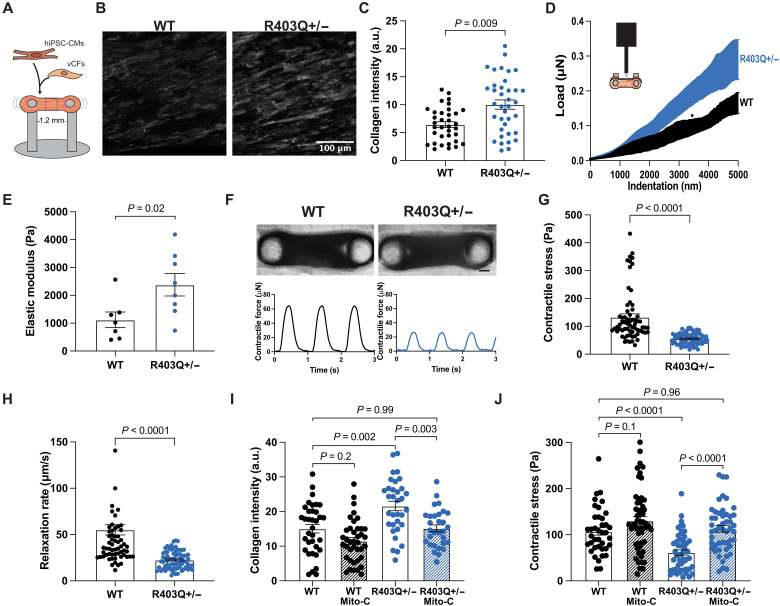
In HCM, excessive collagen deposition leads to a stiffer ECM, which can impair the function of healthy cardiomyocytes (29), but it remains unknown why increased collagen deposition occurs (4–7). Using an adapted version of previously described engineered CMTs (28) composed of hiPSC-CMs and wild-type ventricular cardiac fibroblasts (vCFs) supplemented with ascorbic acid, a cofactor for collagen synthesis (30), we aimed to examine whether the fibrotic activation seen in HCM could be recapitulated in vitro. CMTs were made with isogenic healthy wild-type (WT) hiPSC-CMs or MYH7-variant (R403Q+/−) hiPSC-CMs in matched batches for 7 days (Fig. 1A) and then were assessed for collagen deposition by immunofluorescence staining for collagen type I. R403Q+/− CMTs exhibited a 40% increase in collagen I content compared to WT CMTs (Fig. 1, B and C, and fig. S1A) with no associated increase in alpha-smooth muscle actin (fig. S2A). We validated this increased collagen content in R403Q+/− CMTs via hydroxyproline assay, showing a 75% increase in total collagen content (fig. S2B). Next, to investigate whether increased collagen deposition altered tissue stiffness, we characterized CMT stiffness through nanoindentation (Fig. 1D) and confirmed that R403Q+/− CMTs displayed a higher stiffness (2.3-fold increase in elastic modulus) than WT CMTs (Fig. 1E), as calculated from the slope of each respective force-displacement curve (Fig. 1D). This increase is comparable to that found in an animal model of HCM, in which MYH7-variant native porcine tissue exhibited a 3.4-fold increase in modulus at 9% stretch (29).
Fig. 1. MYH7-variant CMTs exhibit increased collagen content, leading to hypocontractility.
(A) Schematic of CMT made with hiPSC-CMs and vCFs in a fibrin-based gel. (B) Representative images of maximum intensity z-projections of CMTs fixed and stained for collagen type I on day 7. Scale bar, 100 μm. (C) Mean fluorescence intensity of collagen type I in CMTs: WT (n = 35), R403Q+/− (n = 37) (N = 3). (D) Schematic of nanoindentation of CMT and average load-displacement curves from nanoindentation of CMTs. (E) Quantification of the elastic modulus: WT (n = 7), R403Q+/− (n = 8) (N = 3). (F) Representative bright-field images of WT (left) and R403Q+/− (right) CMTs with representative contractile stress traces overtime. Scale bar, 150 μm. (G) Contractile stress generation in CMTs: WT (n = 65), R403Q+/− (n = 73) (N = 4). (H) Contractile relaxation rate of CMTs: WT (n = 65), R403Q+/− (n = 73) (N = 4). (I) Mean fluorescence intensity of collagen type I in CMTs made with mitomycin-C (Mito-C)–treated vCFs (n > 15) (N = 2). (J) Contractile stress of CMTs made with Mito-C–treated vCFs (n > 37) (N = 4). Individual CMTs (n) across all independent experiments (N) are shown with means ± SEM.
After characterizing compositional and mechanical changes to the ECM in HCM-variant CMTs, we investigated the functional effects of increased collagen content, as the presence of fibrosis in patients with HCM has been found to be associated with systolic dysfunction (31). We found that the stiffer R403Q+/− CMTs were less able to shorten (contract) during systole (Fig. 1, F and G, and fig. S1B), consistent with being a stiffer tissue, and showed a decrease in relaxation rate (Fig. 1H). To ensure that these differences in contractility were not biased by potential changes in CMT size, we calculated the final dimensions of WT and HCM CMTs. There were no differences in R403Q+/− CMT dimensions (width, length, and thickness) compared to WT CMTs (fig. S3, A to C). Contractile force (not normalized to CMT cross-sectional area) was also reduced in mutant CMTs (fig. S3D), indicating an overall reduction in contractile force production as well as contractile stress.
The observed hypocontractility of HCM CMTs is in contrast to the hypercontractile phenotype reported for MYH7-variant (18) hiPSC-CMs when cultured alone on 2D substrates. When cultured on 2D substrates alone as opposed to in a cocultured 3D tissue, our R403Q+/− hiPSCs-CMs exhibited a hypercontractile phenotype, as demonstrated by single-cell traction force microscopy (fig. S4). Together, these findings suggest that excessive collagen deposition impairs the contractile function of HCM-variant 3D tissue constructs even while the hiPSC-CMs within those tissues are generating higher forces.
To understand the nature of this ECM remodeling and impaired contractility in HCM tissue, we interrogated the potential contributing factors within our model. Our model was able to bring out differences in collagen content of HCM tissues while previous studies had not observed this effect. One contributing factor in our model appears to be the presence of ascorbic acid, an enzymatic cofactor important for collagen biosynthesis, that is present in normal human tissues and plasma. While R403Q+/− CMTs exhibited a hypocontractile phenotype without ascorbic acid, the presence of ascorbic acid exacerbated the effect (fig. S5). These findings are consistent with the model that collagen deposition contributes to the fibrotic response.
Fibrotic remodeling is dependent on stromal cell proliferation
It has been shown that in addition to elevated collagen production, mice with the R403Q+/− mutation also have increased proliferation of nonmyocytes that contribute to fibrotic activation in HCM (13). This raises the hypothesis that increased cell proliferation of noncardiomyocytes contributes to the fibrotic phenotype observed in the HCM-variant CMTs. To test this hypothesis, we pretreated vCFs with mitomycin-C (Mito-C), a DNA cross-linker, to disrupt the cell cycle and proliferation of vCFs in CMTs. We found that inhibiting vCF proliferation in CMTs reduced collagen deposition in R403Q+/− tissues back to near-normal levels but did not have a significant effect on collagen content in WT tissues compared to tissues with untreated vCFs (Fig. 1I), suggesting that differential proliferation of stromal cells plays a substantial role in the fibrotic state of HCM-variant tissue. This effect was further demonstrated functionally, as inhibiting proliferation of vCFs in CMTs restored the contractility of HCM-variant tissues to a level comparable to that of WT tissues (Fig. 1J). Together, these data suggest that cell proliferation of vCFs in R403Q+/− CMTs contributes to increased collagen deposition and the ensuing reduced contractile output.
snRNA-seq shows altered ligand-receptor interactions in R403Q+/− CMTs
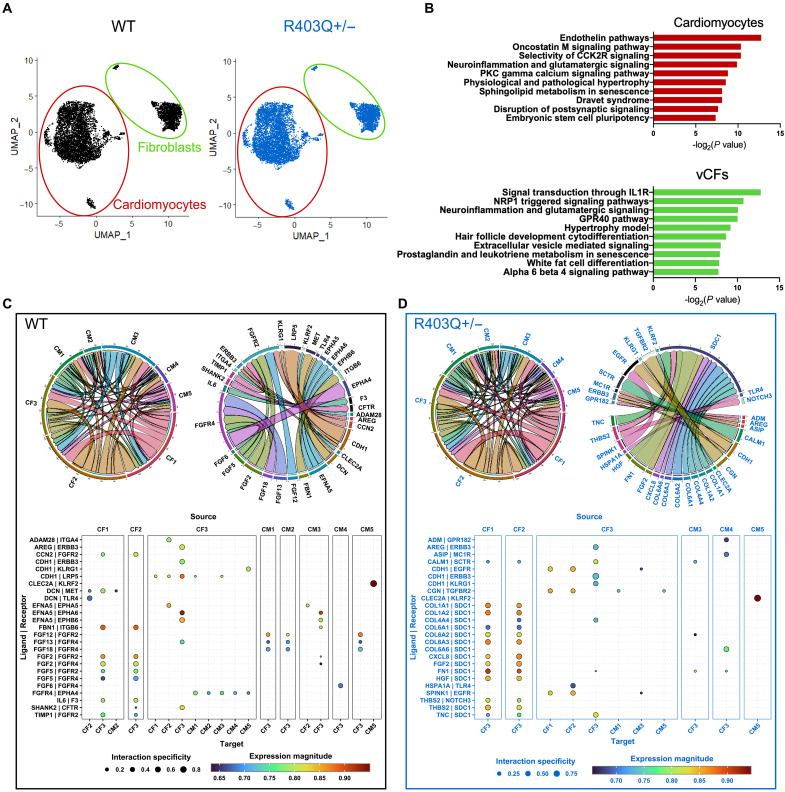
To gain a deeper understanding of the interactions between cardiomyocytes and stromal cells in tissues that lead to stromal activation, we performed snRNA-seq on hiPSC-CM and vCF-containing CMTs cultured for 7 days. Visualization of snRNA-seq data using Uniform Manifold Approximation and Projection (UMAP) confirmed the presence of cardiomyocyte and fibroblast populations in CMTs (Fig. 2A and fig. S6). Compared to WT CMTs, R403Q+/− CMTs had more nuclei identified as fibroblasts, as observed in cardiac fibroblasts cluster 1 (CF1) and cardiac fibroblasts cluster 2 (CF2), and roughly the same number of cardiomyocyte nuclei across clusters (Fig. 2A and fig. S6, B and C). Consistent with our other studies, these data suggest that fibroblasts appear to be more prevalent within R403Q+/− CMTs, while hiPSC-CMs proportion remain unchanged. Analysis also showed a transcriptional shift in both cell types (fig. S7). The top pathways associated with genes up-regulated in both cardiomyocyte and fibroblast cell populations within R403Q+/− tissues included pathways related to hypertrophy, senescence, and inflammatory cytokine signaling (Fig. 2B).
Fig. 2. snRNA-seq shows altered ligand-receptor interactions in R403Q+/− CMTs.
(A) UMAP of snRNA-seq of WT (black, left) and R403Q+/− (blue, right) tissues on day 7. (B) Top 10 up-regulated pathways in R403Q+/− compared to WT in CM population (top) and fibroblast population (bottom) using the WikiPathway database. (C) Chord plots of ligand-receptor interactions between cell population clusters (left) and specific ligand-receptors (right) for WT CMTs. Arrows indicate paracrine interactions between ligand and receptor across cell clusters. The arrow width and corresponding circumferential segment represent the number of interactions (left) and magnitude of expression for interaction (right). Expression levels of the top 25 ligand-receptor interactions from source cell cluster (large columns) to target cell (narrow columns) in WT CMTs. (D) Chord plots of ligand-receptor interactions between cell population clusters (left) and specific ligand-receptors (right) for R403Q+/− CMTs. Expression levels of the top 25 ligand-receptor interactions from source cell cluster (large columns) to target cell (narrow columns) in WT CMTs.
To better understand this signaling, we next looked at the number of ligand-receptor interactions between all cell populations in both the WT and R403Q+/− tissues. Overall, the number of ligand-receptor interactions appeared comparable between WT and R403Q+/− tissues and mainly involved fibroblasts as a highly active paracrine cell population as both source and target (Fig. 2, C and D). Comparison of the top-ranked ligand-receptor interactions indicated that paracrine communication is altered in R403Q+/− compared to WT tissues (Fig. 2, C and D). Notably, paracrine signaling to cardiac fibroblasts cluster 3 (CF3) was more active in R403Q+/− CMTs. This fibroblast population is characterized by increased expression of genes related to epithelial-mesenchymal transition, as noted by the high expression of cadherin 6 (CDH6) (fig. S5G). While much of the top 25 ligand-receptor interactions in WT CMTs involved fibroblast growth factor (FGF) family signaling, the top 25 up-regulated interactions in R403Q+/− CMTs revealed an up-regulated receptor, syndecan-1 (SDC1), a membrane protein that participates in cell proliferation, cell migration, and cell-matrix interactions. EGF family receptors, receptor tyrosine-protein kinase erbB-3 (ERBB3) and EGF receptor (EGFR), were also involved in top ligand-receptor interactions within R403Q+/− CMTs, including that between epithelial cadherin (CDH1) and EGFR. Furthermore, syndecan-1 has been shown to modulate cancer cell phenotype via EGFR signaling, suggesting that there may be some interplay between these signaling pathways (32). Overall, these data support the possibility that the ECM remodeling seen in HCM CMTs is mediated via inflammatory paracrine signaling that differentially activates the fibroblast population.
R403Q+/− cardiomyocytes drive activation of stromal cells via paracrine signaling
Stromal cells are primarily responsible for myocardial matrix remodeling and are activated by mechanical signals, such as pressure overload (33, 34), or signaling molecules, such as TGFβ1 (13, 35, 36), angiotensin-II (37, 38), and other cytokines (10, 39, 40). Given the observation that tissues containing HCM-variant cardiomyocytes stimulate proliferation-mediated stromal activation and up-regulation of inflammatory signaling, we set out to determine whether paracrine factors released by the R403Q+/− hiPSC-CMs may be responsible for this effect. We collected conditioned media from density- and time-matched 2D cultures of WT and R403Q+/− hiPSC-CMs, which was concentrated with a 3-kDa filter to isolate signaling proteins and remove metabolic waste. After resuspending the concentrated conditioned medium in low-glucose media containing 0.1% serum, serum-starved vCFs were cultured in hiPSC-CM conditioned medium for 2 days (Fig. 3A). While hiPSC-CMs alone are typically not as mature as hiPSC-CMs in coculture with stromal cells (41, 42), as in our CMTs, this method enabled the isolation of the effect of hiPCM-CM paracrine signaling on vCFs.
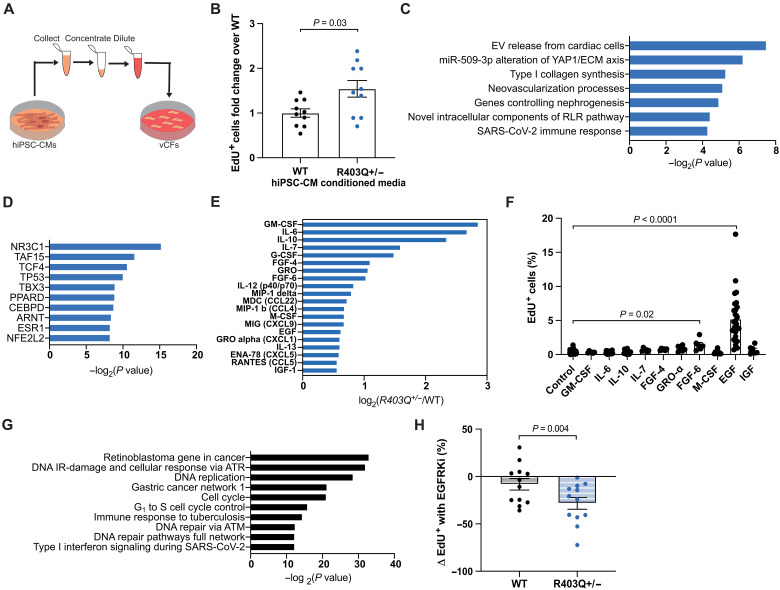
Fig. 3. MYH7-variant hiPSC-CMs drive fibrotic activation of stromal cells via paracrine signaling of EGF.
(A) Schematic of conditioned media experiments. (B) Average fold change of EdU+ vCFs after incubation with EdU from hours 20 to 24 of culture in hiPSC-CM conditioned media normalized to WT: WT (n = 10), R403Q+/− (n = 10) (N = 5). (C) Top pathways up-regulated in vCFs given R403Q+/− hiPSC-CM conditioned media compared to WT hiPSC-CM conditioned media, determined from genes (average fold change >1.15 and P value <0.01 from N = 3) input into the WikiPathway database. EV, extracellular vesicle; RLR, RIG-I-like receptor. (D) Transcription factors associated with genes up-regulated in vCFs given R403Q+/− hiPSC-CM conditioned media compared to WT hiPSC-CM conditioned media (average fold change >1.15 and P value <0.01 from N = 3) input into the ChEA 2022 database. (E) Log2 of the fold change of the top 15 cytokines differentially secreted in R403Q+/− compared to WT hiPSC-CM conditioned media. (F) Average percentage of EdU+ vCFs after incubation with EdU from hours 20 to 24 of culture in 100 ng/ml of 10 different human recombinant proteins (n > 5) (N > 2). (G) Top 10 pathways up-regulated in vCFs given rhEGF (100 ng/ml) compared to serum starvation, determined from genes (fold change >1.3 and P value <0.01) input into the WikiPathway database. (H) Change in proliferation rate of serum-starved vCFs given WT or R403Q+/− hiPSC-CM conditioned media with 1 μM erlotinib hydrochloride (EGFRKi) compared to 0.01% DMSO for 24 hours: All conditions (n = 9) (N = 3). Individual sample wells (n) across all independent experiments (N) are shown with means ± SEM unless otherwise noted.
Specifically, to investigate whether vCFs have a differential proliferative response to R403Q+/− hiPSC-CM conditioned media, as we had observed in R403Q+/− CMTs, we examined the level of DNA synthesis via nuclear incorporation of the thymidine analog, 5-ethynyl-2′-deoxyuridine (EdU). vCFs exhibited a 1.6-fold higher percentage of EdU incorporation when cultured for 24 hours in R403Q+/− hiPSC-CM conditioned media as compared to WT hiPSC-CM conditioned media (Fig. 3B). These results indicate that proliferation of stromal cells is activated via paracrine factor signaling from HCM-variant hiPSC-CMs.
We next sought to investigate downstream signaling of vCFs in response to paracrine factor signaling from cardiomyocytes. RNA-seq of vCFs treated with R403Q+/− hiPSC-CM conditioned media revealed an increase in gene expression in pathways relating to the release of extracellular vesicles (EVs) from cardiomyocytes, alterations in the yes-associated protein 1 (YAP1)/ECM signaling axis, and type I collagen synthesis (N = 3) (Fig. 3C). These findings confirm that paracrine signaling from mutant cardiomyocytes drives stromal activation. In addition, transcription factors associated with genes more highly expressed in vCFs treated with R403Q+/− hiPSC-CM conditioned media include ones known to regulate cell proliferation, including nuclear receptor subfamily 3 group C member 1 (NR3C1) and T-box transcription factor (TBX3), and inflammation, including NR3C1, peroxisome proliferator activated receptor delta (PPARD), CCAAT enhancer binding protein delta (CEBPD), and nuclear factor-erythroid 2 related factor 2 (NFE2L2) (Fig. 3D). Notably, tumor protein p53 (TP53), which is highly expressed in other HCM in vitro models (22, 24), was also up-regulated in vCFs treated with the mutant cardiomyocyte conditioned media.
R403Q+/− hiPSC-CMs differentially release inflammatory cytokines
Having demonstrated that R403Q+/− hiPSC-CMs act on vCFs through paracrine-mediated mechanisms, we sought to identify the paracrine factors differentially secreted by HCM-variant hiPSC-CMs to activate these pathways. A cytokine array, performed on conditioned media from R403Q+/− versus WT hiPSC-CMs, showed increased release of granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-6 (IL-6), IL-10, FGF-4, GRO, FGF-6, M-CSF, EGF, chemokine (C-X-C motif) ligand 1 (CXCL1, also known as GROα), and insulin-like growth factor 1 (IGF-1) in conditioned medium from R403Q+/− hiPSC-CMs compared to WT (Fig. 3E). Notably, this profile includes many cytokines associated with the senescence-associated secretory phenotype (SASP) (43). SASP can be induced in response to DNA damage resulting from cell stress, such as oxidative stress, which is up-regulated in R403Q+/− hiPSC-CMs (24, 27).
After identifying the cytokines that are differentially released by R403Q+/− hiPSC-CMs, we investigated the effects of these cytokines on proliferation of vCFs. We selected 10 from the top 15 differentially released cytokines based on scientific literature, and cultured serum-starved vCFs for 24 hours in recombinant human (rh) protein (100 ng/ml) for each cytokine. Only rhFGF-6 and rhEGF significantly increased the proliferation of vCFs by a factor of 3.7-fold and 13.5-fold, respectively (Fig. 3F). To see whether this effect was still notable at a more physiologically relevant concentration, we lowered the concentration to 10 ng/ml and saw that only rhEGF increased the proliferation of vCFs by a factor of 7.9-fold (fig. S8A). In addition, when we administered rhEGF to serum-starved vCFs, gene expression pathways related to DNA replication, cell cycle progression, and inflammation were up-regulated (Fig. 3G).
Given the uniquely strong effect of rhEGF on stromal cell proliferation and its prevalence in R403Q+/− hiPSC-CM conditioned media, we examined whether inhibiting EGFR signaling in vCFs attenuates the proliferative effects in response to R403Q+/− hiPSC-CM conditioned media. We pretreated vCFs with 1 μM erlotinib hydrochloride, a US Food and Drug Administration–approved EGFR tyrosine kinase inhibitor (EGFRKi) for the treatment of non–small cell lung cancer and pancreatic cancer (44). We determined the concentration based on a dose-response curve of rhEGF-treated vCFs given different erlotinib hydrochloride concentrations (fig. S8B). Following this, vCFs were treated with conditioned media from WT and R403Q+/− hiPSC-CMs supplemented with 0.01% dimethyl sulfoxide (DMSO) or 1 μM erlotinib hydrochloride (erlotinib). Treatment with erlotinib reduced the proliferation rate of vCFs in response to R403Q+/− hiPSC-CM conditioned media by 28%, whereas it did not have a significant impact on the proliferation rate of vCFs cultured in WT hiPSC-CM conditioned media (Fig. 3H). Together, these data indicate that EGF expression is up-regulated in R403Q+/− hiPSC-CMs and that EGFR signaling is both necessary and sufficient to drive stromal proliferation.
Blocking EGFR signaling attenuates pathological changes in MYH7-variant tissues
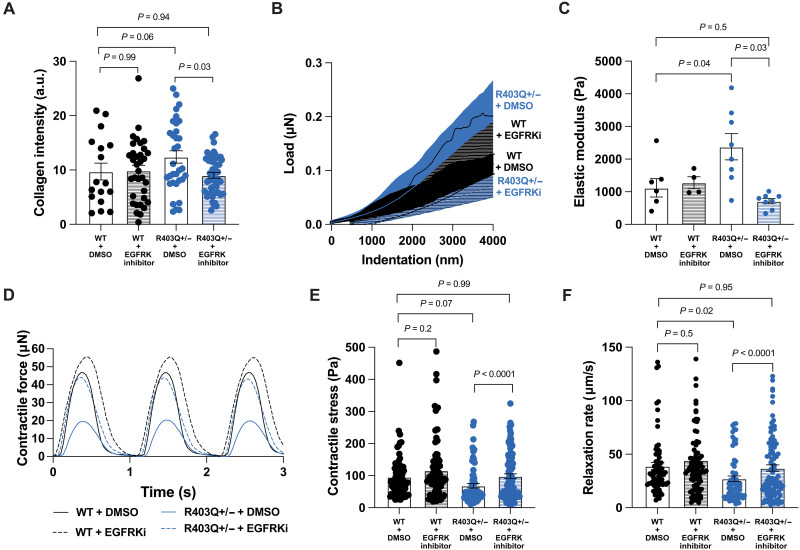
Having established that inhibiting EGFR signaling reduced the effect of R403Q+/− hiPSC-CM paracrine-mediated activation of vCFs cultured in conditioned media, and that EGFR signaling is up-regulated in R403Q+/− CMTs, we hypothesized that inhibiting EGFR signaling attenuates stromal activation in 3D tissues. To test this hypothesis, WT and R403Q+/− CMTs were treated with either 1 μM erlotinib hydrochloride or 0.01% DMSO for 7 days. Erlotinib had no effect on the collagen content in WT CMTs as compared to WT CMTs treated with DMSO. However, erlotinib reduced the collagen content of R403Q+/− CMTs by 27% (Fig. 4A). Consistent with our collagen deposition results, treatment with erlotinib significantly decreased the stiffness of R403Q+/− tissues by a factor of 3.3 (elastic moduli of 2379 ± 1141 Pa versus 714 ± 228 Pa) (Fig. 4, B and C). Together, these observations suggest that the increased collagen deposition and stiffness in R403Q+/− CMTs can be attributed to an increase in EGFR signaling. Given that treatment with erlotinib prevented the development of a fibrotic phenotype in R403Q+/− CMTs, we postulated that the contractile properties of mutant CMTs treated with erlotinib could be rescued to WT levels. R403Q+/− CMTs treated with erlotinib showed an increase in contractility to a level comparable to WT CMTs (Fig. 4, D and E). These effects were also observed in the diastolic relaxation rates (Fig. 4F). Together, these findings suggest that EGFR signaling is contributing to stromal activation in HCM-variant CMTs.
Fig. 4. Blocking EGFR signaling attenuates pathological changes in MYH7-variant tissues.
(A) Mean fluorescence intensity of collagen type I in CMTs treated with 1 μM EGFRKi or 0.01% DMSO: WT + DMSO (n = 17), WT + EGFRKi (n = 34), R403Q+/− + DMSO (n = 33), and R403Q+/− + EGFRKi (n = 45) (N = 3). (B) Average load-displacement curves from nanoindentation of CMTs. (C) Quantification of elastic modulus: WT + DMSO (n = 7), WT + EGFRKi (n = 4), R403Q+/− + DMSO (n = 8), and R403Q+/− + EGFRKi (n = 8) (N = 2). DMSO groups in (B) and (C) are presented in Fig. 1 (D and E). (D) Representative contractile stress traces overtime for WT and R403Q+/− CMTs with and without EGFRKi. (E) Contractile stress of CMTs treated with and without EGFRKi: WT + DMSO (n = 71), WT + EGFRKi (n = 90), R403Q+/− + DMSO (n = 69), and R403Q+/− + EGFRKi (n = 98) (N = 8). (F) Contractile relaxation rates of CMTs treated with and without EGFRKi: WT + DMSO (n = 71), WT + EGFRKi (n = 90), R403Q+/− + DMSO (n = 69), and R403Q+/− + EGFRKi (n = 98) (N = 8). Individual CMTs (n) across all independent experiments (N) are shown with means ± SEM.
Stromal activation and hypocontractility are preserved in MYBPC3t/+ CMTs
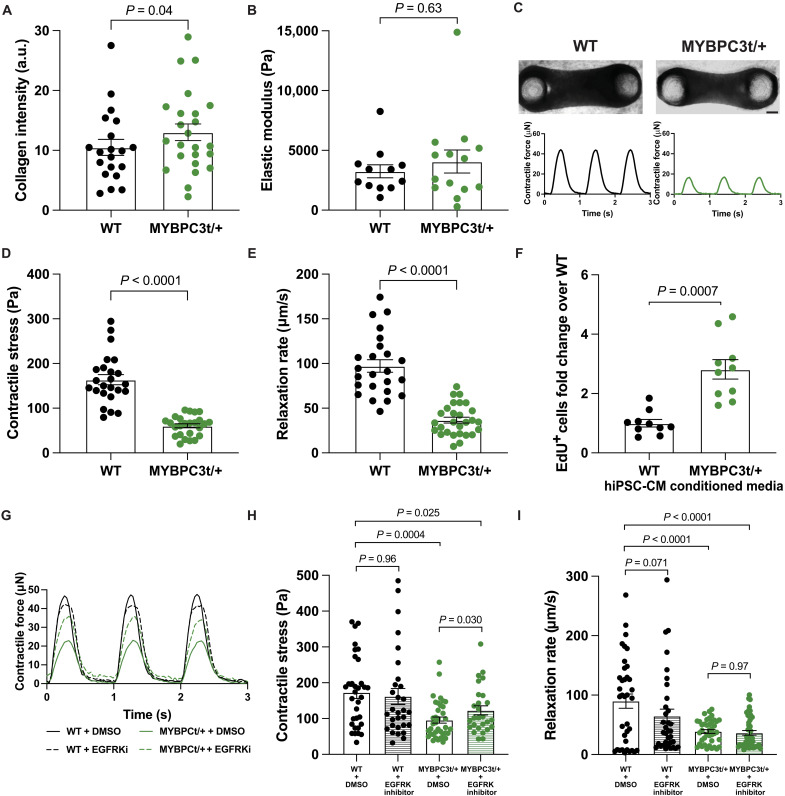
We next sought to assess whether the phenotypes observed in the MYH7-variant CMTs were present in CMTs made with hiPSC-CMs harboring another HCM-causing mutation, a MYBPC3 truncation (MYBPC3t/+). Like R403Q+/− CMTs, MYBPC3t/+ CMTs demonstrated an increase in collagen type I content quantified via immunofluorescence staining (Fig. 5A and fig. S9A). This increase in collagen content, however, was not as pronounced in the MYBPC3 variant and did not show a difference by hydroxyproline assay (fig. S10A). Consistent with these findings, an increase in CMT stiffness was not evident compared to WT CMTs (Fig. 5B). However, immunofluorescent staining for alpha-smooth muscle actin, a marker of fibroblast activation, was markedly increased in MYBPC3t/+ CMTs (fig. S10, B and C). Because the R403Q+/− and MYBPC3t/+ hiPSC-CMs used in this study have the same genetic background, we also repeated our CMT experiments on a distinct patient-derived MYBPC3-variant line, MYBPC3 T3330G+/−, to determine whether evidence of fibrotic activation was present in an unrelated HCM-associated line. Outputs were normalized to each respective WT control line to minimize noise emerging from batch-to-batch variability as observed in R403Q+/− and MYBPC3t/+ experiments (figs. S1 and S9). These patient-derived CMTs showed an increase in stiffness (fig. S10D) and alpha-smooth muscle actin intensity levels (fig. S10, E and F). Overall, while the nature of the response was slightly different, each of these three HCM lines demonstrated evidence of stromal activation.
Fig. 5. Stromal activation and hypocontractility are preserved in MYBPC3t/+ CMTs.
(A) Mean fluorescence intensity of collagen type I in CMTs: WT (n = 20) and MYBPC3t+/− (n = 24) (N = 3). (B) Quantification of the elastic modulus of day 7 CMTs: WT (n = 12) and MYBPC3t/+ (n = 14) (N = 3). (C) Representative bright-field images of WT (left) and MYBPC3t/+ (right) CMTs with representative contractile stress traces over time. Scale bar, 150 μm. (D) Contractile stress generation in CMTs: WT (n = 24) and MYBPC3t/+ (n = 25) (N = 3). (E) Contractile relaxation rate of CMTs: WT (n = 24) and MYBPC3t/+ (n = 25) (N = 3). (F) Average relative percentage of EdU+ vCFs after incubation with EdU from hours 20 to 24 of culture in hiPSC-CM conditioned media normalized to WT: WT (n = 10) and MYBPC3t/+ (n = 10) (N = 2). (G) Representative contractile stress traces overtime for WT and MYBPC3t/+ CMTs. (H) Contractile stress of day 6 CMTs treated with and without EGFRKi: WT + DMSO (n = 33), WT + EGFRKi (n = 30), MYBPC3t/+ + DMSO (n = 37), and MYBPC3t/+ + EGFRKi (n = 27) (N = 4). (I) Contractile relaxation rates of CMTs treated with and without EGFRKi: WT + DMSO (n = 33), WT + EGFRKi (n = 30), MYBPC3t/+ + DMSO (n = 37), and MYBPC3t/+ + EGFRKi (n = 27) (N = 4). Individual CMTs (n) across all independent experiments (N) are shown with means ± SEM.
Similar to MYH7-variant CMTs, engineered tissues generated from MYBPC3t/+ hiPSC-CMs also exhibited the functional phenotypes of hypocontractility and reduced relaxation rate (Fig. 5, C to E, and fig. S9B). MYBPC3t/+ CMTs were more compact than WT CMTs (smaller width), while the length and thickness remained similar to WT CMTs (fig. S11, A to C). Despite this difference, contractile force (not normalized to CMT cross-sectional area) was also reduced in mutant CMTs (fig. S11D), indicating an overall reduction in contractile force production as well as contractile stress. In addition, when cultured on 2D substrates alone, MYBPC3t/+ were hypercontractile (fig. S12), similar to R403Q+/− hiPSC-CMs.
We next investigated the role of paracrine signaling from MYBPC3t/+ hiPSC-CMs in stromal activation. vCFs exhibited a 3.1-fold higher percentage of EdU incorporation when cultured for 24 hours in MYBPC3t/+ conditioned media as compared to WT hiPSC-CMs (Fig. 5F), similar to the stromal cell proliferation induced by MYH7-variant hiPSC-CMs. We then treated MYBPC3t/+ CMTs with erlotinib, an EGFRK inhibitor, to specifically interrogate the role of EGFR signaling. MYBPC3t/+ CMTs treated with erlotinib also showed improved contractility, although not quite back to levels comparable to WT CMTs (Fig. 5, G and H), but diastolic relaxation rates remained low (Fig. 5I). Nonetheless, these findings suggest that EGFR signaling contributes to stromal activation by MYBPC3-variant cardiomyocytes as well.
DISCUSSION
Despite fibrosis being a key factor driving the clinical outcome of patients with HCM, there is a paucity of in vitro models that study fibrosis in HCM and other cardiomyopathies. To address this gap, we present an in vitro 3D tissue-engineered genetic model that reflects fibrotic phenotypes of HCM. Previous studies using CMTs to study the contractility of HCM-variant hiPSC-CMs did not report the fibrotic activation because the CMTs were engineered without stromal cells or with growth-arrested stromal cells (20, 23, 24, 26). One recent study used CMTs with proliferative stromal cells and showed that contractile dysfunction in MYH7-variant tissues is independent of p53 cell death pathways but did not report on stromal interactions in this system (22). Thus, the inclusion of proliferative cardiac fibroblasts, the key effector cells of fibrotic remodeling, was a critical factor that enabled our study of stromal activation.
In addition to coculturing cardiac fibroblasts with HCM-variant hiPSC-CMs, including ascorbic acid in the culture medium, as occurs in human tissue and plasma, allowed for collagen deposition in the model. Ascorbic acid functions as an enzymatic cofactor to hydroxylate procollagen to form a stable triple helix, allowing collagen biogenesis and deposition (45). Unlike mouse cells, human cells lack gulonolactone oxidase and, therefore, cannot synthesize ascorbic acid; hence, ascorbic acid deficiency leads to scurvy and impaired wound healing (45). Thus, ascorbic acid was required to support collagen secretion by human cardiac fibroblasts and enabled a comprehensive examination of the HCM fibrotic phenotype in vitro.
While our findings highlight the importance of studying the interactions of cardiomyocytes and fibroblasts in the context of cardiomyopathies, further investigation is needed to fully understand the broad applicability of our findings. Three HCM mutant lines investigated in this study demonstrated activation of the stromal compartment leading to tissue changes modeling fibrotic remodeling, but each with different characteristics ranging from increased collagen deposition to alpha-smooth muscle actin expression. Additional studies are required to more fully characterize the activated phenotype of these fibroblasts and the similarities and differences of these phenotypes depending on HCM variant. In future studies, a more complete panel of fibrotic markers should be assessed, including periostin, vimentin, and more extensive evaluation of fibroblast and matrix changes. Whether the differences in stromal responses among the HCM mutants stem from fundamental biology arising from the mutations themselves, or other sources of variances between the hiPSC clones, remains to be determined by future work.
Our study suggests that EGFR signaling may play a central role in the development of HCM caused by the R403Q+/− mutation in MYH7. HCM-variant hiPSC-CMs activated EGF-related pathways in stromal cells, causing an increase in stromal cell proliferation, collagen deposition, tissue stiffness, and a reduction in tissue contractility in HCM-variant CMTs. When HCM-variant CMTs were treated with EGFRKi, these pathological alterations did not occur. The findings are consistent with those described in other fibrosis settings, including stromal activation via parenchyma-induced paracrine factors (46) and increased stromal cell proliferation (13), and appear to be reliant on phosphoinositide 3-kinase–Akt signaling (47). EGFR signaling has been previously implicated in stromal activation in other pathologies, such as skin (48), lung (48), renal (49), and myocardial fibrosis (50). Some studies have also shown that EGFR activation is required for TGFβ-driven stromal activation (39, 49, 51). Furthermore, antagonism of EGFR has been shown to reduce myocardial inflammation, fibrosis, apoptosis, and dysfunction in angiotensin II–induced cardiac hypertrophy (52) and diabetic cardiomyopathy (53). By antagonizing EGFR in our genetic model, we demonstrate its important role in stromal activation of HCM. EGFR has been a therapeutic target for numerous diseases involving stromal activation in other organ systems, which could be adapted to cardiomyopathies in the future. However, additional studies are warranted to more clearly confirm and identify the specific EGF ligands and receptors that contribute to the pathological phenotype in our model.
Further, the mechanisms that induce EGFR signaling in HCM have yet to be elucidated. Our study suggests that EGF production by cardiomyocytes may be part of a broader response to stress, perhaps mediated through the increased oxidative or mechanical stresses in HCM-associated mutations. Previous studies have reported that hypoxic stress causes cardiomyocytes to release heparin-binding EGF-like growth factor (HB-EGF), an EGFR ligand that is mitogenic for fibroblasts (54). In addition, EGFR inhibition decreases cardiac remodeling in a diabetic mouse model by attenuating oxidative stress (55). Thus, cardiomyocytes with HCM-causing mutations may secrete EGFR ligands differently due to their inefficient energy consumption (18, 26, 56) and increase in oxidative stress (24, 56). An alternative potential mechanism for EGF secretion could be through mechanical pathways due to the intrinsic hypercontractility of HCM hiPSC-CMs. Mechanical stretch has been found to activate EGFR phosphorylation (57) in cardiomyocytes. Likewise, mechanical stress can also mediate ROS formation by proteins such as NADPH oxidase (NOX2) through the X-ROS mechanotransduction pathway (58), which could increase EGFR ligand secretion. Furthermore, NOX2-deficient mice in an angiotensin II infusion model showed decreased interstitial myocardial fibrosis (59), indicating that NOX2 is required to activate myocardial fibrotic remodeling in certain pathologies. Understanding these stress activation mechanisms could provide crucial insight into specific therapeutic targets upstream of stromal activation and fibrosis in HCM.
In summary, we show an in vitro model for recapitulating early fibrotic changes observed in HCM. This model required a focus on interactions between mutation-carrying hiPSC-CMs and normal human cardiac fibroblasts within a 3D engineered tissue context to reveal a previously unappreciated paracrine signaling pathway. The use of more advanced 3D culture models provides an important approach in identifying root causes of complex processes such as fibrosis and underscores the need to continue to refine these technologies.
MATERIALS AND METHODS
hiPSC culture
hiPSCs used in this study were from the PGP1 parent line available through the Coriell Institute (GM23338) from which a subclone was generated with a green fluorescent protein (GFP) tagged to endogenous titin (60). Of note, the subclone used in this study was later found to harbor a frameshift between GFP and titin in one allele that did not affect titin expression. A separate HCM-variant line, established in Toepfer et al. (18), was created by introducing a heterozygous point mutation into MYH7 of the PGP1 parent line using CRISPR-Cas9 genome editing to change the arginine to a glutamine on amino acid 403 (R403Q+/−). A second HCM-variant line, established in Toepfer et al. (61), was created by introducing a truncating mutation to MYBPC3 (MYBPC3t/+), in which two independent lines yielded the same phenotype. All clones were characterized by sequencing within 200 base pairs of the edit sequence. In addition, an HCM patient–derived hiPSC line, SCVIi001-A, was generously provided by Greenstone Biosciences (60). This line is a noncoding variant, c.3330 + 2 T > G (MYBPC3 T3330G+/−), created from blood mononuclear cells isolated from a 60-year-old male patient with HCM.
hiPSC-CM differentiation and purification
hiPSCs were maintained in mTeSR1 (StemCell) on Matrigel (Thermo Fisher Scientific) diluted 1:80 in Dulbecco’s modified Eagles’ medium (DMEM)/F-12 (Thermo Fisher Scientific) and split using Accutase (Sigma-Aldrich) at 70 to 90% confluence. hiPSCs were differentiated into hiPSC-CMs by small-molecule, monolayer-based manipulation of the Wnt-signaling pathway as previously described (28, 62) at 80 to 95% confluence, and cells were cultured in RPMI (Thermo Fisher Scientific) with 1:50 B-27 without insulin (Thermo Fisher Scientific) and with 1× GlutaMAX (Thermo Fisher Scientific) until the end of differentiation. On day 0, cells were cultured in 12 μM (6 μM for SCVIi001-A) CHIR99021 (Tocris) for 24 hours (48 hours for SCVIi001-A), activating Wnt signaling. On day 3, cells were cultured with 5 μM IWP4 (Tocris), inhibiting Wnt signaling, for 48 hours. After 9 days, cells were cultured in RPMI supplemented with 1:50 B-27 with insulin (Thermo Fisher Scientific) and 1× GlutaMAX. After 11 to 13 days of differentiation initiation, hiPSC-CMs were purified using RPMI, no glucose media (Thermo Fisher Scientific) with 4 mM Sodium DL Lactate solution (Sigma-Aldrich) for 4 days, with media changed every other day. Following selection, cells were replated and maintained in RPMI with 1:50 B-27 supplement with insulin and 1× GlutaMAX on fibronectin (10 μg/ml; Thermo Fisher Scientific)–coated plates until day 30+. hiPSC-CMs were frozen in 50% RPMI with 1:50 B-27 with insulin, 40% fetal bovine serum (FBS) (Sigma-Aldrich, cat. F0926 lot: 17H392), and 10% DMSO (Thermo Fisher Scientific) on days 20 to 35 after a 1-hour incubation in 5 μM Y-27632 and stored in liquid nitrogen until further use. If cryo-preserved cells were used, they were thawed and cultured as a monolayer for 3 to 5 days before use, with media changes every other day to remove hiPSC-CMs that did not survive cryopreservation.
Flow cytometry assessment of cryopreserved hiPSC-CMs was performed to determine postthaw viability and purity. Three distinct batches of cryopreserved hiPSC-CMs were thawed onto Matrigel-coated tissue culture dishes in RPMI containing B-27 with insulin, as described above. Two to seven days after thawing, hiPSC-CMs were stained with Sytox Red (Thermo Fisher Scientific) for 30 min on ice for viability measurements. For purity measurements, hiPSC-CMs were fixed in suspension with 4% paraformaldehyde (PFA) for 15 min on ice, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 15 min on ice, washed with phosphate-buffered saline (PBS), and then stained for cardiac troponin T (Clone 13-11, Thermo Fisher Scientific) followed by goat anti-mouse Alexa Fluor 647 secondary (Thermo Fisher Scientific). Measurements were collected on a BD FACSMelody (BD Biosciences) and analyzed with the free online platform Floreada.io. Cryopreserved cells had an 81.1 ± 1.9% viability and 94.6 ± 1.4% purity, determined from flow cytometry (fig. S13).
Stromal cell culture
Normal human vCFs were purchased from Lonza (Cat. CC-2904). vCFs were thawed, cultured in Cardiac Fibroblast Growth Media (FGM)–3 (Lonza, CC-4526), and passaged using 0.25% Trypsin-ethylenediaminetetraacetic acid (EDTA). Cells were thawed and passaged using 0.25% Trypsin-EDTA (Thermo Fisher Scientific) and cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin (P/S). vCFs were used for assays three to seven passages after thawing.
Cardiac microtissues
Polydimethylsiloxane (PDMS) microtissue devices with tissue wells each containing two cylindrical micro-pillars with a spring constant of 2.68 μN/μm and spherical caps 1.2 mm apart were casted from a 3D printed mold (Protolabs). The surface of devices was treated with slight modifications to a previously reported protocol (28). One to three days before tissue seeding, devices were plasma treated for 60 s, treated with 0.01% poly-l-lysine (ScienCell) for 1.5 hours followed by 0.1% glutaraldehyde (EMS) for 15 min, washed three times with deionized (DI) water, and soaked in DI water at 4°C until use. Immediately before seeding, devices were soaked in 70% ethanol for 15 min, dried, and sterilized under ultraviolet (UV) light for 15 min. After sterilization, 2% Pluronic F-127 (Sigma-Aldrich) was added to each well below the pillar caps and incubated for 30 min at room temperature to prevent CMTs from attaching to the bottom surface of the PDMS devices.
A total of 60,000 cells per tissue, consisting of 90% hiPSC-CMs and 10% vCFs, were mixed in an ECM solution consisting of human fibrinogen (4 mg/ml; Sigma-Aldrich), 10% Matrigel (Corning), 0.4 units of thrombin (Sigma-Aldrich) per milligram of fibrinogen, 5 μM Y-27632 (Tocris), and aprotinin (0.033 mg/ml; Sigma-Aldrich). In Mito-C experiments, vCFs were pretreated with Mito-C (10 μg/ml; EMD Millipore 475820) for 2 hours before tissue seeding. The cell-ECM mixture was pipetted into each tissue well, and after gel polymerization for 10 min, tissue maintenance growth media containing high-glucose DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Sigma-Aldrich), 1% P/S (Thermo Fisher Scientific), 1% Nonessential Amino Acids (Thermo Fisher Scientific), 1% Glutamax (Thermo Fisher Scientific), 5 μM Y-27632, aprotinin (0.033 mg/ml), and L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (150 μg/ml) was added unless otherwise noted. Y-27632 was removed 2 days following seeding, and the growth media were replaced every other day. CMT culture media were supplemented with 1 μM erlotinib hydrochloride (Sigma-Aldrich) or 0.01% DMSO in EGFRKi experiments.
Immunofluorescence and confocal imaging
Day 7 CMTs were fixed with 4% PFA (Thermo Fisher Scientific) in 1× PBS (Thermo Fisher Scientific) at room temperature for 25 min. CMTs were rinsed three times with PBS and blocked with 2% bovine serum albumin (BSA) (Sigma-Aldrich) in PBS at room temperature for 1 hour. Primary antibody, anti-collagen I (Abcam, ab138492, RRID: AB_2861258, 1:400), or anti–alpha-smooth muscle actin (Abcam, ab7817, RRID: AB_262054, 1:1000), was applied in 2% BSA in PBS at 4°C overnight. CMTs were then incubated in secondary antibody Goat anti-Rb immunoglobulin G (IgG) (H + L) highly cross-adsorbed secondary antibody directly conjugated to Alexa Fluor 568 (Thermo Fisher Scientific 11036, RRID: AB_10563566, 1:500) and 4′,6-diamidino-2-phenylindole (DAPI, 1:1000) (Invitrogen) in 2% BSA at room temperature for 1 hour. Primaries and secondaries were washed in PBS three times over 30 min.
Immunofluorescence images of CMTs were acquired with a Leica HC FLUOTAR L 25×/0.95 W immersion objective on an upright Leica TCS SP8 multiphoton microscope equipped with a Spectra-Physics Insight DS+ laser tuned to 950 nm excitation and nondescanned emission detection at 680 nm. Z-stacks of ~150 to 300 μm thicknesses were acquired at a z-step size of 1.5 μm.
Image analysis
Maximum intensity z-projections were taken of the z-stacks. The background noise of maximum intensity projections was subtracted in ImageJ using a 50-pixel rolling ball radius. Average intensities of collagen in a 328 × 328 μm center region of the tissue were calculated.
Hydroxyproline assay for total collagen content
CMTs were fixed in 4% PFA in PBS at room temperature for 25 min. Collagen content was measured using the Total Collagen Assay Kit (Abcam, Cat. ab222942). Briefly, three fixed tissues were placed in 10 μl of DI water per tissue and homogenized with an ultrasonic probe homogenizer for 30 s. Equal parts 10 N NaOH was added to the sample homogenate and the mixture was placed at 99°C for 1 hour. The sample was placed on ice for 5 min, and the remaining NaOH was neutralized with 10 N HCl in a 1:1 ratio. The sample was centrifuged at 10,000g for 5 min, and the supernatant was removed and placed in a new tube. The supernatant, along with the provided control of type I collagen from rat tail tendon, was processed following the manufacturer’s protocol (Abcam, Cat. ab222942). Hydroxyproline content was measured based on the absorbance at an optical density of 560 nm on a microplate reader, and final values were calculated based on a standard curve created using the known 0 to 10 μg per well samples.
Nanoindentation
CMTs treated with either 0.01% DMSO or 1 μM erlotinib hydrochloride (Sigma-Aldrich) every other day were measured on days 7 to 9. To prevent contraction during measurements, tissues were incubated in a 70 mM potassium chloride relaxation buffer for 30 min before nanoindentation. Nanoindentation characterization was performed using a Piuma Nanoindenter system (Optics 11, Westwood, MA) with a spherical indentation probe with a diameter of approximately 100 μm and a cantilever spring constant of k ~ 0.5 N/m. Extra PDMS surrounding cardiac tissues was removed manually using a razor blade for the indentation probe to gain better access to the microtissues before measurements. Tissue samples were immersed in media, and the tests were performed at an indentation depth of 10 μm and displacement speed of 5 μm/s at room temperature without fixation. The elastic moduli of these tissues were calculated using the built-in Piuma software by fitting force-indentation curves to the established Hertzian contact mechanics model, assuming a Poisson’s ratio of 0.5 for incompressible materials. Measurements were taken in at least four locations per tissue and averaged.
Contraction measurements
On day 7 (day 6 for MYBPC3t/+ EGFRKi experiments), time-lapse videos of the tissue contraction were acquired at 30 frames per second using a 4× objective on a Nikon Eclipse Ti (Nikon Instruments Inc.) with an Evolve EMCCD Camera (Photometrics), equipped with a temperature and CO2 equilibrated environmental chamber. CMTs were electrically stimulated at 1 Hz using a C-Pace EP stimulator (IonOptix). A custom MATLAB script, available in a previous publication (60), was used to measure the deflection of the pillars through edge tracking in two stages; first with pixel-scale cross-correlation, and then refined at a sub-pixel resolution. Maximum contractile force and stress were then calculated based on the tracked deflection of the pillars from the resting position and the measured pillar spring constant of 2.68 μN/μm, as described previously (28).
Polyacrylamide gel and traction force microscopy
Polyacrylamide gels of 7.9 kPa stiffness were made by adjusting the concentrations of acrylamide and bisacrylamide stock solution (Bio-Rad Laboratories, Hercules, CA), as previous described (61). A 0.2-μm fluorescent beads solution (1:1000, Bangs Laboratories Inc.) was added to the gels, and they were coated with acrylic N-hydroxysuccinimide ester (acrylic-NHS; 4 mg/ml) and photoinitiator [2 hydroxy-4′-2(hydroxyethyl)-2 methylpropane; 0.5 mg/ml] and cured for 2 min at 30 W using an OmniCure S Series Lamp Spot UV (Excelitas Technologies). Gels were washed three times in 1× PBS and coated in fibronectin (0.1 mg/ml; Corning) overnight. Gels were then washed and UV sterilized for 15 min before seeding 60k hiPSC-CMs per gel. Media were changed every other day, and gels were imaged on day 3. Videos of bead motion near the substrate surface, distributed in and around the contact region of a single cell, were acquired at a frame rate of 30 frames per second at 30× on a Nikon Eclipse Ti (Nikon Instruments Inc.) equipped with a temperature and CO2 equilibrated environmental chamber and an Evolve EMCCD Camera (Photometrics) by using a 560-nm laser illumination wavelength. hiPSC-CMs were electrically stimulated at 1 Hz using a C-Pace EP stimulator (IonOptix).
The stress vector fields were generated by using an open-source package of Fiji plugins (63) and were interpolated with a custom Matlab script (61). Root mean squares of the magnitudes of single-cell stress vectors were computed using the formula: Root mean squares of the magnitudes of single-cell stress vectors = √1(|S1|2 + |S2|2 + … + |Sn|2), in which Sn is a single-stress vector. Root mean squares of the magnitudes of single-cell stress vectors were plotted over time and smoothed using a custom Matlab script. Contractile stress was calculated as the difference between local maximum and minimum root mean squares of the magnitudes of single-cell stress vectors and was averaged over three to four contraction periods for each cell.
Bulk RNA-seq
vCFs were trypsinized, centrifuged, and flash frozen after 2 days in conditioned media (N = 3), serum starvation, or rhEGF (100 ng/ml). Cells were homogenized in TRIzol Reagent (Life Technologies Inc., Grand Island, NY) with TissueLyzer II (QIAGEN Inc., Valencia, CA) and RNA was isolated by conventional methods. RNA went through two rounds of mRNA purification (polyA-selection) using Dynabeads mRNA DIRECT Kit (Invitrogen, Carlsbad, CA). Double-stranded cDNA was generated using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). The cDNA products were used to construct libraries with the Nextera XT DNA Sample Preparation Kit (Illumina Inc., San Diego, CA). Libraries were paired-end sequenced with a read length of 75 base pairs (75PE) on the Illumina NextSeq 500. Reads were aligned to the hg38 Human Genome using Spliced Transcripts Alignment to a Reference (64). Data were analyzed as previously described (65) and normalized to the total number of reads per kilobase of exon per million (RPKM).
Genes that were up-regulated in rhEGF-treated vCFs compared to serum-starved vCFs with a P value <0.01 and fold change greater than 1.3 were input into Enrichr (66, 67). For conditioned media experiments, gene expression values were averaged over three experimental repeats. Genes that had an average fold-change expression greater than 1.15 with a P value <0.01 for all three experiments were input into Enrichr. The top up-regulated pathways and transcription factors associated with the top up-regulated genes were obtained using WikiPathways (68) and ChEA (69) databases, respectively.
Single-nucleus RNA sequencing
CMTs were flash frozen in liquid nitrogen on day 7 and stored at −80°C. Individual nuclei were isolated from frozen tissue samples. Briefly, nuclei were isolated (70) and RNA was reverse transcribed and converted into cDNA libraries using a 10x Chromium Controller and Chromium Single Cell 3′ v3.1 reagent kit (10x Genomics). Bar-coded libraries were pooled and sequenced (Illumina NovaSeq 6000). snRNA-seq alignment and gene counts were performed using Cell Ranger 1.2 (10x Genomics), Seurat (71), and R 4.1.0 and managed via RStudio. There were 15,129 total sequenced nuclei.
For Fig. 2 and figs. S6 and S7, Seurat (version 4.0.6) and R (version 4.1.2) were used for analysis. Principal components analysis (PCA) was performed to determine the dimensionality of this dataset within Seurat and the number of principal components (PCs) was selected using both permutation-based and heuristic methods. Cluster assignment was performed in an unsupervised manner using a shared nearest-neighbor approach within Seurat. To validate the ideal number of PCs and clustering resolution in this dataset, repeat analysis was performed while varying both the dimensionality and resolution. Using the Clustree R package (72), the hierarchical evolution of cluster identity was determined. This analysis allows the user to investigate how clustering resolution affects cell identity, in an effort to assign robust classification to cell clusters which remain consistent across resolution values. For each clustering resolution, marker genes for each cluster, the gene encoding titin (TTN) for cardiomyocytes (fig. S14A), and the gene encoding fibronectin (FN1) (fig. S14B) for fibroblasts were identified to assign relevant biological function to various subclusters. Dimensionality and resolution were adjusted until each identified cluster contained a nonzero amount of expressed marker genes that were able to assign functional gene ontological terms, in this case, at cluster resolution 0.4, which was used for clustering analysis (fig. S14C). At greater resolution values, cluster subsets returned either too few genes to assign functional classifications or noncoding genes of unknown function. Dotplot projections of single nuclei were generated with UMAP coordinates, using the number of dimensions determined from PCA as described above. For the purposes of this analysis, 30 dimensions were considered for both the initial PCA, and subsequent UMAP visualization. To assign cell clusters’ specific identities, canonical marker gene expression was analyzed to assign broad cell classes. To assign cell subclusters, such as cardiomyocyte clusters 1 to 6 (CM1 to CM6), unbiased genetic markers for each population were calculated using the Wilcoxon Rank Sum test within Seurat, at which point at most the top 100 genes were used as a Gene Ontology query to assign cellular functions to each cell subcluster.
To analyze paracrine signaling between cells, we explored ligand and corresponding receptor expression across all sampled cardiac cell types. Initially, confounding clusters such as cardiomyocyte clusters that expressed high levels of mitochondrial DNA or high levels of cell cycle markers, doublets [identified using the Solo package (73)], and a small subpopulation of stromal cells with low-level TTN expression, were each excluded. LIANA package (version 0.1.7), which combines multiple ligand-receptor databases, was used to score paracrine interactions within a given dataset based on multiple analysis methods. The results were then used to generate an aggregate rank to build a consensus of abundant paracrine interactions (74). The package SCpubr (version 1.0.0.9) was then used to generate chord and paracrine interaction plots; in this context, interaction specificity is used to show enriched interactions within the LIANA consensus and expression magnitude indicates how strong a particular interaction is based on ligand and receptor gene expression levels (75).
Conditioned media experiments
hiPSC-CMs after glucose selection were trypsinized with 0.25% trypsin-EDTA and DNase (10 μg/ml; StemCell catalog no. 07469) at 37°C for 5 min; then, the cells were replated onto fibronectin (10 μg/ml)–coated (Corning) plates at a density of ~1 million cells per well in a 12-well plate in 1 ml of RPMI per well with 1:50 B-27 supplement with insulin and 1× GlutaMAX. Media were collected on ice and replenished every other day from days 30 to 40. Media were centrifuged at 2000 rpm at 4°C for 4 min to remove cell debris, the supernatant was collected on ice, and the media were frozen at −80°C. Final hiPSC-CM density was calculated by incubating cells in NucBlue (Thermo Fisher Scientific R37606) for 30 min, imaging the nuclear stain at 4×, and averaging the nuclei content across five images.
vCFs were plated at a density of 14,000 cells per well in a 24-well plate in respective media for cell culture for 2 days. The stromal cells were washed and serum starved in low-glucose DMEM with 0.1% FBS and 1% P/S for 2 days. Conditioned media from hiPSC-CMs were thawed on ice at 4°C and concentrated with a 3-kDa exclusion filter (Sigma-Aldrich) by centrifuging at 14,000g for 10 min at 4°C. Conditioned media were diluted to original volumes with low-glucose DMEM with 0.01% FBS and 1% P/S, normalized to have equal hiPSC-CM per volume of collected media, and given to serum-starved stromal cells.
EdU-click-iT cell proliferation assays
Serum-starved vCFs were supplemented with a 5×, 50 μM EdU solution (Thermo Fisher Scientific C10337) in low-glucose DMEM with 0.1% FBS (EdU working concentration of 10 μM) for 4 hours, starting 20 hours after the addition of conditioned media. Stromal cells were fixed in 4% PFA in PBS for 15 min and washed three times with PBS. Samples were washed twice with 3% BSA in PBS and incubated in a reaction cocktail using Click-iT EdU imaging kit (Thermo Fisher Scientific C10337) or 5× tris-buffered saline (Thermo Fisher Scientific), 4 mM CuSO4, 3.5 μM azide (AFDye 647 or 568, Fluoprobes), and 100 mM sodium ascorbate at room temperature for 45 min. Samples were washed once with 3% BSA in PBS, incubated in Hoechst 33342 (1:1000) (Thermo Fisher Scientific) at room temperature for 30 min, and washed three times in PBS. Images of 2D stromal cells were acquired on a Nikon Eclipse Ti (Nikon Instruments Inc.) with an Evolve EMCCD Camera (Photometrics) or on a Zeiss Axiovert 200M inverted spinning disk microscope with an ORCA-100 Camera (Hamamatsu) at 4×. Images were adjusted for brightness and contrast using ImageJ. The percentage of EdU-positive cells across 30 images per well was calculated for the total cell count and normalized to the percentage of EdU-positive cells in response to WT conditioned media to minimize the effects of batch-to-batch variability seen in other assays (figs. S1 and S9).
Cytokine array of conditioned media
Conditioned media collected from WT and R403Q+/− hiPSC-CMs were thawed on ice. Media were added to RayBio C5 Human Cytokine Antibody Array (RayBiotech), and the manufacturer’s protocol was followed. Briefly, membranes were blocked at room temperature for 30 min, followed by sample incubation at 4°C overnight. Membranes were washed with provided buffers, incubated in the provided biotinylated antibody cocktail at room temperature for 2 hours, washed with provided buffers for a second time, incubated in 1× HRP-streptavidin at room temperatures for 2 hours, and washed with provided buffers for a third time. Excess buffer was blotted, and membranes were incubated in provided Detection Buffers for 2 min. Images of membranes were acquired with an iBrightFL1000 (Invitrogen). The background signal was subtracted from the average intensity of each spot in the array measured in ImageJ. The average intensity of each spot was normalized to the positive controls, and the relative intensities of R403Q+/− conditioned media to WT conditioned media were calculated.
Recombinant protein experiments
vCFs were plated at 14k cells per well of a 24-well plate. After 2 days, vCFs were serum starved in 0.1% FBS for 2 days. Either 10 ng/ml or 100 ng/ml of recombinant human protein was added to stromal cells for EdU assays. Recombinant human proteins included EGF (PeproTech), FGF4 (PeproTech), FGF6 (PeproTech), GM-CSF (PeproTech), Gro-α (PeproTech), IGF (PeproTech), IL-6 (R&D Systems), IL-7 (PeproTech), IL-10 (PeproTech), and M-CSF (R&D Systems).
Inhibitor concentration optimization
vCFs were plated at 14k cells per well of a 24 well-plate. After 2 days, vCFs were serum starved in 0.1% FBS for 2 days. vCFs were then pretreated with varying concentrations of erlotinib hydrochloride (EGFRKi) (Sigma-Aldrich), including 0 nM, 10 nM, 100 nM, 500 nM, 1 μM, and 10 μM in the same loading concentration of DMSO for 45 min. rh-EGF (100 ng/ml; PeproTech) was then added to the cells, along with the same concentration of EGFRKi as the pretreatment. An EdU assay was performed as described above.
Statistical analysis
Samples sizes and P values are reported in each figure legend and statistical analyses were performed using GraphPad Prism 9.0. Graphs show mean ± SEM unless otherwise noted. Means are reported as mean ± SD in text. Data were assessed with a two-way analysis of variance (ANOVA) with a mixed-effect model that was corrected for multiple comparisons using a Tukey test with a 95% confidence level and adjusted for lack of sphericity with a Greenhouse-Geisser correction unless otherwise noted.
Acknowledgments
We acknowledge S. Sundaram for the development of MATLAB code to analyze pillar displacement during cardiac microtissue contraction.
Funding: This work was supported by National Science Foundation Engineering Research Center on Cellular Metamaterials grant EEC-1647837 (C.E.S., J.G.S., and C.S.C.), National Science Foundation Science and Technology Center for Engineering Mechanobiology grant CMMI-1548571 (C.S.C.), National Institute of Health 5R01HL084553 (J.G.S. and C.E.S.), National Science Foundation Graduate Research Fellowship (J.K.E., M.C.W., and X.G.), National Institute of Health National Heart Lung and Blood grant F31 HL158195-03 (J.K.E.), Sir Henry Dale Wellcome Fellowship 222567/Z/21/Z, BHF Centre of Research Excellence Intermediate Transition Fellowship RE/18/3/34214, Oxford BHF Centre of Research Excellence RE/13/1/30181 (C.N.T.), the Howard Hughes Medical Institute (C.E.S.), and the Leducq Foundation 16CVD03 (J.G.S. and C.E.S.).
Author contributions: Conceptualization: J.K.E., A.C., C.N.T., J.E., C.E.S., J.G.S., and C.S.C. Methodology and investigation: J.K.E., M.C.W., M.A.M., P.E.C., A.C., L.L., X.G., J.H.L., D.M.D., J.G., J.A.L.W., O.L., and R.J.L. Supervision: J.E., C.E.S., J.G.S., and C.S.C. Writing—original draft: J.K.E., J.E., and C.S.C. Writing—review and editing: All authors.
Competing interests: C.S.C. is a founder and own shares of Satellite Biosciences, a company that is developing cell-based therapies. C.N.T. has consulted for Bristol Myers Squibb and Myokardia not related to this work. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Bulk and snRNA-seq datasets are available on Dryad Repositories (https://datadryad.org/stash/share/Ha244QOSW4fQVxzUSrhmGTGLH441H5raqvOube0P3Bk).
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
REFERENCES AND NOTES
- 1.Semsarian C., Ingles J., Maron M. S., Maron B. J., New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 65, 1249–1254 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Maron B. J., Gardin J. M., Flack J. M., Gidding S. S., Kurosaki T. T., Bild D. E., Prevalence of hypertrophic cardiomyopathy in a general population of young adults: Echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation 92, 785–789 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Maron B. J., Olivotto I., Spirito P., Casey S. A., Bellone P., Gohman T. E., Graham K. J., Burton D. A., Cecchi F., Epidemiology of hypertrophic cardiomyopathy-related death revisited in a large non-referral-based patient population. Circulation 102, 858–864 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Galati G., Leone O., Pasquale F., Olivotto I., Biagini E., Grigioni F., Pilato E., Lorenzini M., Corti B., Foà A., Agostini V., Cecchi F., Rapezzi C., Histological and histometric characterization of myocardial fibrosis in end-stage hypertrophic cardiomyopathy: A clinical-pathological study of 30 explanted hearts. Circ. Heart Fail. 9, e003090 (2016). [DOI] [PubMed] [Google Scholar]
- 5.O’Hanlon R., Grasso A., Roughton M., Moon J. C., Clark S., Wage R., Webb J., Kulkarni M., Dawson D., Sulaibeekh L., Chandrasekaran B., Bucciarelli-Ducci C., Pasquale F., Cowie M. R., McKenna W. J., Sheppard M. N., Elliott P. M., Pennell D. J., Prasad S. K., Prognostic significance of myocardial fibrosis in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 56, 867–874 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Bruder O., Wagner A., Jensen C. J., Schneider S., Ong P., Kispert E. M., Nassenstein K., Schlosser T., Sabin G. V., Sechtem U., Mahrholdt H., Myocardial scar visualized by cardiovascular magnetic resonance imaging predicts major adverse events in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 56, 875–887 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Ellims A. H., Iles L. M., Ling L. H., Hare J. L., Kaye D. M., Taylor A. J., Diffuse myocardial fibrosis in hypertrophic cardiomyopathy can be identified by cardiovascular magnetic resonance, and is associated with left ventricular diastolic dysfunction. J. Cardiovasc. Magn. Reson. 14, 76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thierfelder L., Watkins H., Macrae C., Lamas R., Yckenna W., Vosberg H.-P., Feldman J. G., Seidmant C. E., Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: A disease of the sarcomere. Cell 77, 701–712 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Morita H., Rehm H. L., Menesses A., McDonough B., Roberts A. E., Kucherlapati R., Towbin J. A., Seidman J. G., Seidman C. E., Shared genetic causes of cardiac hypertrophy in children and adults. N. Engl. J. Med. 358, 1899–1908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang L., Ellims A. H., Beale A. L., Taylor A. J., Murphy A., Dart A. M., Systemic inflammation is associated with myocardial fibrosis, diastolic dysfunction, and cardiac hypertrophy in patients with hypertrophic cardiomyopathy. Am. J. Transl. Res. 9, 5063–5073 (2017). [PMC free article] [PubMed] [Google Scholar]
- 11.Roncarati R., Viviani Anselmi C., Losi M. A., Papa L., Cavarretta E., da Costa Martins P., Contaldi C., Saccani Jotti G., Franzone A., Galastri L., Latronico M. V. G., Imbriaco M., Esposito G., de Windt L., Betocchi S., Condorelli G., Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 63, 920–927 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Afzal J., Vakrou S., Greenland G. V., Talbot C. C., Hebl V. B., Guan Y., Karmali R., Tardiff J. C., Leinwand L. A., Olgin J. E., Das S., Fukunaga R., Abraham M. R., Differences in microRNA-29 and pro-fibrotic gene expression in mouse and human hypertrophic cardiomyopathy. Front. Cardiovasc. Med. 6, 170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teekakirikul P., Eminaga S., Toka O., Alcalai R., Wang L., Wakimoto H., Nayor M., Konno T., Gorham J. M., Wolf C. M., Kim J. B., Schmitt J. P., Molkentin J. D., Norris R. A., Tager A. M., Hoffman S. R., Markwald R. R., Seidman C. E., Seidman J. G., Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-β. J. Clin. Invest. 120, 3520–3529 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim D.-S., Lutucuta S., Bachireddy P., Youker K., Evans A., Entman M., Roberts R., Marian A. J., Angiotensin II blockade reverses myocardial fibrosis in a transgenic mouse model of human hypertrophic cardiomyopathy. Circulation 103, 789–791 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada Y. J., Passeri J. J., Baggish A. L., O’Callaghan C., Lowry P. A., Yannekis G., Abbara S., Ghoshhajra B. B., Rothman R. D., Ho C. Y., Januzzi J. L., Seidman C. E., Fifer M. A., Effects of losartan on left ventricular hypertrophy and fibrosis in patients with nonobstructive hypertrophic cardiomyopathy. JACC Heart Fail. 1, 480–487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green E. M., Wakimoto H., Anderson R. L., Evanchik M. J., Gorham J. M., Harrison B. C., Henze M., Kawas R., Oslob J. D., Rodriguez H. M., Song Y., Wan W., Leinwand L. A., Spudich J. A., McDowell R. S., Seidman J. G., Seidman C. E., Heart disease: A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science 351, 617–621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marian A. J., Senthil V., Chen S. N., Lombardi R., Antifibrotic effects of antioxidant N-acetylcysteine in a mouse model of human hypertrophic cardiomyopathy mutation. J. Am. Coll. Cardiol. 47, 827–834 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toepfer C. N., Garfinkel A. C., Venturini G., Wakimoto H., Repetti G., Alamo L., Sharma A., Agarwal R., Ewoldt J. F., Cloonan P., Letendre J., Lun M., Olivotto I., Colan S., Ashley E., Jacoby D., Michels M., Redwood C. S., Watkins H. C., Day S. M., Staples J. F., Padrón R., Chopra A., Ho C. Y., Chen C. S., Pereira A. C., Seidman J. G., Seidman C. E., Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation 141, 828–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan F., Lee A. S., Liang P., Sanchez-Freire V., Nguyen P. K., Wang L., Han L., Yen M., Wang Y., Sun N., Abilez O. J., Hu S., Ebert A. D., Navarrete E. G., Simmons C. S., Wheeler M., Pruitt B., Lewis R., Yamaguchi Y., Ashley E. A., Bers D. M., Robbins R. C., Longaker M. T., Wu J. C., Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12, 101–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosqueira D., Mannhardt I., Bhagwan J. R., Lis-Slimak K., Katili P., Scott E., Hassan M., Prondzynski M., Harmer S. C., Tinker A., Smith J. G. W., Carrier L., Williams P. M., Gaffney D., Eschenhagen T., Hansen A., Denning C., CRISPR/Cas9 editing in human pluripotent stemcell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur. Heart J. 39, 3879–3892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S., Pan H., Tan C., Sun Y., Song Y., Zhang X., Yang W., Wang X., Li D., Dai Y., Ma Q., Xu C., Zhu X., Kang L., Fu Y., Xu X., Shu J., Zhou N., Han F., Qin D., Huang W., Liu Z., Yan Q., Mitochondrial dysfunctions contribute to hypertrophic cardiomyopathy in patient iPSC-derived cardiomyocytes with MT-RNR2 mutation. Stem Cell Rep. 10, 808–821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loiben A. M., Chien W. M., Friedman C. E., Chao L. S. L., Weber G., Goldstein A., Sniadecki N. J., Murry C. E., Yang K. C., Cardiomyocyte apoptosis is associated with contractile dysfunction in stem cell model of MYH7 E848G hypertrophic cardiomyopathy. Int. J. Mol. Sci. 24, 4909 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prondzynski M., Lemoine M. D., Zech A. T., Horváth A., Di Mauro V., Koivumäki J. T., Kresin N., Busch J., Krause T., Krämer E., Schlossarek S., Spohn M., Friedrich F. W., Münch J., Laufer S. D., Redwood C., Volk A. E., Hansen A., Mearini G., Catalucci D., Meyer C., Christ T., Patten M., Eschenhagen T., Carrier L., Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 11, e11115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohn R., Thakar K., Lowe A., Ladha F. A., Pettinato A. M., Romano R., Meredith E., Chen Y. S., Atamanuk K., Huey B. D., Hinson J. T., A contraction stress model of hypertrophic cardiomyopathy due to sarcomere mutations. Stem Cell Rep. 12, 71–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dainis A., Zaleta-Rivera K., Ribeiro A., Chang A. C. H., Shang C., Lan F., Burridge P. W., Liu W. R., Wu J. C., Chang A. C. Y., Pruitt B. L., Wheeler X. M., Ashley E., Silencing of MYH7 ameliorates disease phenotypes in human iPSC-cardiomyocytes. Physiol. Genomics 52, 293–303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhagwan J. R., Mosqueira D., Chairez-Cantu K., Mannhardt I., Bodbin S. E., Bakar M., Smith J. G. W., Denning C., Isogenic models of hypertrophic cardiomyopathy unveil differential phenotypes and mechanism-driven therapeutics. J. Mol. Cell. Cardiol. 145, 43–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore J., Ewoldt J., Venturini G., Pereira A. C., Padilha K., Lawton M., Lin W., Goel R., Luptak I., Perissi V., Seidman C. E., Seidman J., Chin M. T., Chen C., Emili A., Multi-omics profiling of hypertrophic cardiomyopathy reveals altered mechanisms in mitochondrial dynamics and excitation–contraction coupling. Int. J. Mol. Sci. 24, 4724 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K., Cloonan P. E., Sundaram S., Liu F., Das S. L., Ewoldt J. K., Bays J. L., Tomp S., Toepfer C. N., Marsiglia J. D. C., Gorham J., Reichart D., Eyckmans J., Seidman J. G., Seidman C. E., Chen C. S., Plakophilin-2 truncating variants impair cardiac contractility by disrupting sarcomere stability and organization. Sci. Adv. 7, 3995–4010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sewanan L. R., Schwan J., Kluger J., Park J., Jacoby D. L., Qyang Y., Campbell S. G., Extracellular matrix from hypertrophic myocardium provokes impaired twitch dynamics in healthy cardiomyocytes. JACC Basic Transl. Sci. 4, 495–505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murad S., Grove D., Lindberg K. A., Reynolds G., Sivarajah A., Pinnellt S. R., Regulation of collagen synthesis by ascorbic acid. Proc. Natl. Acad. Sci. U.S.A. 78, 2879–2882 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olivotto I., Maron B. J., Appelbaum E., Harrigan C. J., Salton C., Gibson C. M., Udelson J. E., O’Donnell C., Lesser J. R., Manning W. J., Maron M. S., Spectrum and clinical significance of systolic function and myocardial fibrosis assessed by cardiovascular magnetic resonance in hypertrophic cardiomyopathy. Am. J. Card. 106, 261–267 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Ibrahim S. A., Gadalla R., El-Ghonaimy E. A., Samir O., Mohamed H. T., Hassan H., Greve B., El-Shinawi M., Mohamed M. M., Götte M., Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol. Cancer 16, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang F. L., Fang M., Yutzey K. E., Loss of β-catenin in resident cardiac fibroblasts attenuates fibrosis induced by pressure overload in mice. Nat. Commun. 8, 712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore-Morris T., Guimarães-Camboa N., Banerjee I., Zambon A. C., Kisseleva T., Velayoudon A., Stallcup W. B., Gu Y., Dalton N. D., Cedenilla M., Gomez-Amaro R., Zhou B., Brenner D. A., Peterson K. L., Chen J., Evans S. M., Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J. Clin. Invest. 124, 2921–2934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maione A. S., Stadiotti I., Pilato C. A., Perrucci G. L., Saverio V., Catto V., Vettor G., Casella M., Guarino A., Polvani G., Pompilio G., Sommariva E., Excess tgf-β1 drives cardiac mesenchymal stromal cells to a pro-fibrotic commitment in arrhythmogenic cardiomyopathy. Int. J. Mol. Sci. 22, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayça B., Sahin I., Kucuk S. H., Akin F., Kafadar D., Avşar M., Avci I. I., Gungor B. D. L. D. A. L., Okuyan E., Dinckal M. H., Increased transforming growth factor-β levels associated with cardiac adverse events in hypertrophic cardiomyopathy. Clin. Cardiol. 38, 371–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsutsumi Y., Matsubara H., Ohkubo N., Mori Y., Nozawa Y., Murasawa S., Kijima K., Maruyama K., Masaki H., Moriguchi Y., Shibasaki Y., Kamihata H., Inada M., Iwasaka T., Angiotensin II type 2 receptor is upregulated in human heart with interstitial fibrosis, and cardiac fibroblasts are the major cell type for its expression. Circ. Res. 83, 1035–1046 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Li L., Fan D., Wang C., Wang J. Y., Cui X. B., Wu D., Zhou Y., Wu L. L., Angiotensin II increases periostin expression via Ras/p38 MAPK/CREB and ERK1/2/TGF-β1 pathways in cardiac fibroblasts. Cardiovasc. Res. 91, 80–89 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Liu Y., He K., Hu Y., Guo X., Wang D., Shi W., Li J., Song J., YAP modulates TGF-β1-induced simultaneous apoptosis and EMT through upregulation of the EGF receptor. Sci. Rep. 7, 45523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svystonyuk D. A., Ngu J. M. C., Mewhort H. E. M., Lipon B. D., Teng G., Guzzardi D. G., Malik G., Belke D. D., Fedak P. W. M., Fibroblast growth factor-2 regulates human cardiac myofibroblast-mediated extracellular matrix remodeling. J. Transl. Med. 13, 147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beauchamp P., Jackson C. B., Ozhathil L. C., Agarkova I., Galindo C. L., Sawyer D. B., Suter T. M., Zuppinger C., 3D Co-culture of hiPSC-derived cardiomyocytes with cardiac fibroblasts improves tissue-like features of cardiac spheroids. Front. Mol. Biosci. 7, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giacomelli E., Bellin M., Sala L., van Meer B. J., Tertoolen L. G. J., Orlova V. V., Mummery C. L., Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coppé J. P., Desprez P. Y., Krtolica A., Campisi J., The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson J. R., Cohen M., Sridhara R., Chen Y. F., Williams G. M., Duan J., Gobburu J., Booth B., Benson K., Leighton J., Hsieh L. S., Chidambaram N., Zimmerman P., Pazdur R., Approval summary for erlotinib for treatment of patients with locally advanced or metastatic non-small cell lung cancer after failure of at least one prior chemotherapy regimen. Clin. Cancer Res. 11, 6414–6421 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Padayatty S. J., Levine M., Vitamin C: The known and the unknown and Goldilocks. Oral Dis. 22, 463–493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H., Tian L., Shen M., Tu C., Wu H., Gu M., Paik D. T., Wu J. C., Generation of quiescent cardiac fibroblasts from human induced pluripotent stem cells for in vitro modeling of cardiac fibrosis. Circ. Res. 125, 552–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuzmanov U., Wang E. Y., Vanderlaan R., Kim D. H., Lee S. H., Hadipour-Lakmehsari S., Guo H., Zhao Y., McFadden M., Sharma P., Billia F., Radisic M., Gramolini A., Emili A., Mapping signalling perturbations in myocardial fibrosis via the integrative phosphoproteomic profiling of tissue from diverse sources. Nat. Biomed. Eng. 4, 889–900 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Odell I. D., Steach H., Gauld S. B., Reinke-Breen L., Karman J., Carr T. L., Wetter J. B., Phillips L., Hinchcliff M., Flavell R. A., Epiregulin is a dendritic cell-derived EGFR ligand that maintains skin and lung fibrosis. Sci. Immunol. 16, eabq6691 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J., Chen J. K., Nagai K., Plieth D., Tan M., Lee T. C., Threadgill D. W., Neilson E. G., Harris R. C., EGFR signaling promotes TGFβ-dependent renal fibrosis. J. Am. Soc. Nephrol. 23, 215–224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou Y., Pan L., Shen Y., Wang X., Huang C., Wang H., Jin X., Yin C., Wang Y., Jia J., Qian J., Zou Y., Gong H., Ge J., Cardiac Wnt5a and Wnt11 promote fibrosis by the cross-talk of FZD5 and EGFR signaling under pressure overload. Cell Death Dis. 12, 877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shu D. Y., Hutcheon A. E. K., Zieske J. D., Guo X., Epidermal growth factor stimulates transforming growth factor-beta receptor type II expression in corneal epithelial cells. Sci. Rep. 9, 8079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zha Y., Li Y., Ge Z., Wang J., Jiao Y., Zhang J., Zhang S., ADAMTS8 promotes cardiac fibrosis partly through activating EGFR dependent pathway. Front. Cardiovasc. Med. 9, 797137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W., Fang Q., Zhong P., Chen L., Wang L., Zhang Y., Wang J., Li X., Wang Y., Wang J., Liang G., EGFR inhibition blocks palmitic acid-induced inflammation in cardiomyocytes and prevents hyperlipidemia-induced cardiac injury in mice. Sci. Rep. 6, 24580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munk M., Memon A. A., Goetze J. P., Nielsen L. B., Nexo E., Sorensen B. S., Hypoxia changes the expression of the epidermal growth factor (EGF) system in human hearts and cultured cardiomyocytes. PLOS ONE 7, e40243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang D., Zhong P., Hu J., Lin F., Qian Y., Xu Z., Wang J., Zeng C., Li X., Liang G., EGFR inhibition protects cardiac damage and remodeling through attenuating oxidative stress in STZ-induced diabetic mouse model. J. Mol. Cell Cardiol. 82, 63–74 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Nollet E. E., Duursma I., Rozenbaum A., Eggelbusch M., Wüst R. C. I., Schoonvelde S. A. C., Michels M., Jansen M., van der Wel N. N., Bedi K. C., Margulies K. B., Nirschl J., Kuster D. W. D., van der Velden J., Mitochondrial dysfunction in human hypertrophic cardiomyopathy is linked to cardiomyocyte architecture disruption and corrected by improving NADH-driven mitochondrial respiration. Eur. Heart J. 44, 1170–1185 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kippenberger S., Loitsch S., Guschel M., Müller J., Knies Y., Kaufmann R., Bernd A., Mechanical stretch stimulates protein kinase B/Akt phosphorylation in epidermal cells via angiotensin II type 1 receptor and epidermal growth factor receptor. J. Biol. Chem. 280, 3060–3067 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Pratt S. J. P., Lee R. M., Chang K. T., Hernández-Ochoa E. O., Annis D. A., Ory E. C., Thompson K. N., Bailey P. C., Mathias T. J., Ju J. A., Vitolo M. I., Schneider M. F., Stains J. P., Ward C. W., Martin S. S., Mechanoactivation of NOX2-generated ROS elicits persistent TRPM8 Ca2+ signals that are inhibited by oncogenic KRas. Proc. Natl. Acad. Sci. U.S.A. 117, 26008–26019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johar S., Cave A. C., Narayanapanicker A., Grieve D. J., Shah A. M., Johar S., Cave A. C., Narayanapanicker A., Grieve D. J., Shah A. M., Aldosterone mediates angiotensin II-induced interstitial cardiac fibrosis via a Nox2-containing NADPH oxidase. FASEB J. 20, 1546–1548 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Karakan M. Ç., Ewoldt J. K., Segarra A. J., Sundaram S., Wang M. C., White A. E., Chen C. S., Ekinci K. L., Geometry and length control of 3D engineered heart tissues using direct laser writing. Lab. Chip. 24, 1685–1701 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toepfer C. N., Sharma A., Cicconet M., Garfinkel A. C., Mücke M., Neyazi M., Willcox J. A. L., Agarwal R., Schmid M., Rao J., Ewoldt J., Pourquié O., Chopra A., Chen C. S., Seidman J. G., Seidman C. E., SarcTrack. Circ. Res. 124, 1172–1183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lian X., Zhang J., Azarin S. M., Zhu K., Hazeltine L. B., Bao X., Hsiao C., Kamp T. J., Palecek S. P., Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tseng Q., Duchemin-Pelletier E., Deshiere A., Balland M., Guilloud H., Filhol O., Theŕy M., Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl. Acad. Sci. U.S.A. 109, 1506–1511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Christodoulou D. C., Wakimoto H., Onoue K., Eminaga S., Gorham J. M., DePalma S. R., Herman D. S., Teekakirikul P., Conner D. A., McKean D. M., Domenighetti A. A., Aboukhalil A., Chang S., Srivastava G., McDonough B., de Jager P. L., Chen J., Bulyk M. L., Muehlschlegel J. D., Seidman C. E., Seidman J. G., 5’RNA-Seq identifies Fhl1 as a genetic modifier in cardiomyopathy. J. Clin. Invest. 124, 1364–1370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma’ayan A., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., Lachmann A., Wojciechowicz M. L., Kropiwnicki E., Jagodnik K. M., Jeon M., Ma’ayan A., Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pico A. R., Kelder T., Van Iersel M. P., Hanspers K., Conklin B. R., Evelo C., WikiPathways: Pathway editing for the people. PLOS Biol 6, 1403–1407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lachmann A., Xu H., Krishnan J., Berger S. I., Mazloom A. R., Ma’ayan A., ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nadelmann E. R., Gorham J. M., Reichart D., Delaughter D. M., Wakimoto H., Lindberg E. L., Litviňukova M., Maatz H., Curran J. J., Ischiu Gutierrez D., Hübner N., Seidman C. E., Seidman J. G., Isolation of nuclei from mammalian cells and tissues for single-nucleus molecular profiling. Curr. Protoc. 1, e132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zappia L., Oshlack A., Clustering trees: A visualization for evaluating clusterings at multiple resolutions. Gigascience 7, giy083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bernstein N. J., Fong N. L., Lam I., Roy M. A., Hendrickson D. G., Kelley D. R., Solo: Doublet identification in single-cell RNA-seq via semi-supervised deep learning. Cell Syst. 11, 95–101.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Dimitrov D., Türei D., Garrido-Rodriguez M., Burmedi P. L., Nagai J. S., Boys C., Ramirez Flores R. O., Kim H., Szalai B., Costa I. G., Valdeolivas A., Dugourd A., Saez-Rodriguez J., Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 13, 3224 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.E. Blanco-Carmona, Generating publication ready visualizations for single cell transcriptomics using SCpubr. bioRxiv 482303 [Preprint] (2022). 10.1101/2022.02.28.482303. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14