Abstract
A novel adenovirus system for analyzing the adenovirus entry pathway has been developed that contains green fluorescent protein bound to the encapsidated viral DNA (AdLite viruses). AdLite viruses enter host cells and accumulate around the nuclei and near the microtubule organizing centers (MTOC). In live cells, individual AdLite particles were observed trafficking both toward and away from the nucleus. Depolymerization of microtubules during infection prevented AdLite accumulation around the MTOC; however, it did not abolish perinuclear localization of AdLite particles. Furthermore, depolymerization of microtubules did not affect AdLite motility and did not affect gene expression from wild-type adenovirus and adenovirus-derived vectors. These data revealed that adenovirus intracellular motility and nuclear targeting can be supported by a mechanism that does not rely on the microtubule network.
To successfully infect cells, adenoviruses must reach the nucleus, where viral gene expression can begin and the genomes can replicate. Adenoviruses are nonenveloped; the viral particle consists of an icosahedral protein capsid surrounding the 36-kb linear double-stranded DNA genome and associated DNA binding proteins (reviewed in reference 40). Human adenoviruses infect predominantly epithelial cells and can trigger respiratory and gastrointestinal tract ailments of a mild course in the majority of cases (reviewed in reference 23). Adenoviruses have been extensively used, both in molecular biology and gene therapy fields, as tools to deliver a variety of molecules into cells (reviewed in reference 41). Recently, adenoviruses have received additional attention as a model system to study the mechanisms of viral entry into eukaryotic cells (15, 16, 52). Adenoviruses bind at the cell membrane and are internalized by receptor-mediated endocytosis (5, 48, 53, 54). Subsequently, adenoviruses are thought to be released from early endosomes into the cytoplasm by a process that requires an activity of the virus-encoded protease (10, 18); viral capsids are gradually dismantled by a combination of step-wise dissociation and proteolytic degradation (16). Finally, liberated viral cores are thought to enter the nucleus via nuclear pores to initiate the replicative cycle (19).
The exact mechanisms of adenovirus movement to the nucleus are not completely understood. The size of an infective particle is 80 to 90 nm in diameter, which is most likely above the limit of free diffusion in the cytoplasm (52 nm in diameter) (27, 33). Because adenoviruses traverse the distance of 5 to 50 μm between the cell membrane and the nucleus of an infected cell within 30 to 60 min following binding to the cell membrane, it has long been suspected that adenoviruses use one of the cytoskeleton-based transport systems of the host cell to travel toward the nucleus. Evidence has accumulated that microtubules are involved in this transit. Adenoviruses are found in close proximity to microtubules in the infected cells and can bind to purified microtubules in vitro (12, 28, 31, 50). However, depolymerization of microtubules in cells infected with adenoviruses does not impede their infective properties (12). Recently, using video microscopy, two groups have studied the intracellular movement of adenoviruses using adenovirus particles whose capsids have been covalently linked to a fluorochrome (25, 46). Capsid-labeled adenoviruses were observed to move along linear pathways both toward and away from the microtubule organizing centers (MTOC), where minus ends of microtubules are nucleated, and this motility was inhibited in the presence of the microtubule depolymerizing reagent nocodazole (46). In addition, the motility toward, but not away from, the MTOC was reported to be partially inhibited by the overexpression of dynamitin, a protein required for the motor function of dynein (1, 7, 46). Thus, it was proposed that adenoviruses interact with both plus end- and minus end-directed microtubule motors and that microtubule-dependent transport in association with a minus end-directed, dynein-like motor contributes to the localization of adenoviruses to the nucleus of infected cells (46).
The goal of our experiments was to examine the mechanisms of adenovirus nuclear targeting using a different experimental approach. We sought to follow the ultimate adenovirus genome deposition into the nucleus, and therefore we have developed fluorescently labeled adenovirus particles with the DNA genomes marked with green fluorescent protein (GFP)-DNA binding protein fusions (termed AdLite particles). DNA labeling by GFP-DNA binding protein fusions have been successfully used to follow, in real time, sister chromatid separation in yeast (29, 43). We have chosen to label the DNA of the core and not the capsid of the viral particles for the following two reasons. First, the capsid proteins must interact with the host machinery on the way to the nucleus, and modifications of the capsid may impair these interactions. Second, as the capsid is progressively dismantled during the entry process, following the fate of labeled capsid may not directly reflect the fate or route of the genome. Using AdLite particles, we confirm that in live cells, adenoviruses move along linear pathways both toward and away from the nucleus as well as in other, more random directions. However, we find that AdLite particle motility is also observed under conditions when microtubules are depolymerized. In addition, we demonstrate that microtubule depolymerization does not affect transgene or early gene expression from adenovirus. Our data reveal that adenoviruses can utilize microtubule-independent mechanisms to target the nuclei of the infected cells and suggest that the mechanisms of cytoplasmic transit and nuclear targeting are more diverse than previously thought.
MATERIALS AND METHODS
Cloning of AdLite genomes and AdLite virus production.
DNA fragments containing 56 and 112 tet operator (teto) tandem repeats were excised from pRS306tetO2×56 and pRS306tetO2×112 (29) with BamHI and BglII restriction enzymes, gel purified, and cloned into the BamHI and BglII sites of the transfer vector (pΔE1sp1B) (6), generating pAd/teto56/Ad and pAd/teto112/Ad. Using homologous recombination in Escherichia coli BJ5183 (8), pAd/teto56/Ad and pAd/teto112/Ad were recombined with pAIM33 containing a modified E1, E3-defective Ad5 genome bearing a lacZ cassette in the former E1 region, an insertion destroying the internal SpeI site, and SpeI sites flanking the terminal repeats, thus generating pAdteto49, pAdteto77, pAdteto112, and pAdteto119 containing 49, 77, 112, and 119 teto sites, respectively. To generate Adteto(N), plasmids pAdteto(N) were linearized with SpeI and used to transfect 293 cells, as described previously (30). Lysates from 293 cells containing replicated Adteto viruses were used to infect a 293 cell line stably expressing a TetR-GFP fusion protein (see below). Newly replicated genomes were bound by TetR-GFP to produce TetR-GFP-loaded adenovirus particles (AdLite particles), which were purified by standard cesium chloride gradients (9), with the omission of Freon extraction, which was found to diminish the fluorescent properties of the AdLite particles.
Generating 293 cell line expressing TetR-GFP.
293 cells were cotransfected with plasmids encoding a TetR-eGFP fusion protein under the control of the cytomegalovirus immediate-early promoter (details can be provided upon request) and the neomycin gene and placed under selection (300 μg of neomycin per ml). The resulting neomycin-resistant clones were trypsinized, pooled, and resuspended at 106 cells/ml in phosphate-buffered saline (PBS)–1% fetal calf serum (FCS) containing propidium iodide (Molecular Probes) at a final concentration of 1 μg/ml. Cells were analyzed immediately using a FACS Vantage SE (Becton Dickinson) equipped with an Ar-Ion laser tuned to 200 mW at 488 nm. Enhanced GFP-positive and propidium iodide-negative cells were sorted into FCS. GFP-positive cells were then collected by centrifugation, resuspended in normal medium, and replated as a pool. Cells were expanded, fluorescence-activated cell sorter (FACS) sorted for the second time, and replated at a low density to allow growth from single cells; the resulting clones were trypsinized individually and expanded. Cells from individual clones were plated on coverslips and were found to be highly fluorescent. Further culture of clones was carried out with the omission of neomycin, without loss of the fluorescence. AdLite viruses were generated either in pooled cells after the second FACS sorting or in clones stably expressing TetR-GFP; both preparation types were found to have similar fluorescent properties.
Cell culture, infections, and drug administration.
A549 and HeLa cells were grown in Dulbecco's modified Eagle's medium–10% FCS, and 293 cells were grown in minimal essential medium alpha with 10% newborn calf serum. Unless indicated otherwise, A549 or HeLa cells were plated 1 day before the experiment; prior to infection, cells were exposed to drugs or appropriate solvents in Dulbecco's modified Eagle's medium for 1 h at 37°C. For infection, cells were placed on ice, and fresh medium (± drugs) containing the virus was added for 1 to 2 h at 4°C with gentle agitation; medium containing unbound virus was removed and replaced with fresh medium (± drugs). Drugs were made as 1,000× stocks in dimethyl sulfoxide (DMSO) (nocodazole and vinblastine; Sigma) (taxol; Molecular Probes) or in 100% ethanol (colchicine; Sigma). In all experiments, control cells were treated with appropriate concentrations of DMSO or ethanol solvents.
Immunofluorescence.
A549 or HeLa cells were plated on glass coverslips (12 by 12 mm) at 2 × 105/well in 6-well dishes 1 day before infection. At the indicated times postinfection, cells were fixed in 3% paraformaldehyde-PBS, pH 7.0, for 15 min, rinsed three times with PBS, permeabilized with PBS–0.1% Triton X-100 (PBT) for 5 to 15 min, blocked in 5% bovine serum albumin (BSA)-PBT for 30 to 60 min, incubated with primary antibody for 15 min in 5% BSA-PBT, washed three times with PBT, incubated with secondary antibody for 15 min in 5% BSA-PBT, washed two times with PBT and two times with PBS, and mounted in 50% glycerol-PBS–10 mM Tris (pH 9.0)–4% n-propyl gallate (Sigma); all incubations were done at room temperature. The primary antibodies used were mouse anti-tubulin (clone DM1A; Sigma), mouse anti-adenovirus (Fitzgerald), and mouse anti-E1A (Calbiochem), and the secondary antibodies used were DTAF or Cy3-conjugated donkey anti-mouse (Jackson Laboratories), all at 1:100. DNA was stained with Hoechst dye (Sigma). Images were acquired with a cooled charge-coupled device (CCD) camera (Spot II; Diagnostic Instruments) mounted on an Axiovert microscope (Zeiss) and equipped with a 63×/1.4 lens; filters were from Chroma Tech. Images were processed using the Adobe Photoshop program.
DNA in situ hybridization.
The in situ hybridization protocol was adapted from the method of Greber et al. (17). A549 cells grown on glass coverslips for 1 day were infected with AdLite particles at a 104 multiplicity of infection (MOI) in particles per cell. At 1 h after infection, cells were fixed in prechilled (−20°C) methanol for 6 min at −20°C, postfixed for 30 min at room temperature in 1% paraformaldahyde, and washed three times with PBS. Coverslips were inverted (cells down) over 10 μl of the probe in hybridization buffer placed on a glass slide, sealed with rubber cement, denatured at 93°C for 3 min on a heating block, and then incubated in a humidified chamber for 2 h at 50°C. Coverslips were gently removed and washed with 50% formamide–2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) three times for 5 min each at 50°C and with 0.1× SSC three times for 5 min each at 60°C, rinsed twice with PBS at room temperature, and mounted in n-propyl gallate mounting medium (see above). Microscopy and image processing were performed as described above. The teto-specific rhodamine-labeled RNA probe was prepared as previously described (14), with the following modifications. teto-specific RNA was in vitro transcribed from pRS306tetO2×7 containing seven teto sites (350 bp), and amino-allyl UTP (Sigma) was incorporated at a 1:5 ratio to the UTP (Sigma). Purified RNA containing amino-allyl UTP was labeled with NHS-rhodamine [(5,6)-carboxytetramethylrhodamine N-hydroxysuccinimidyl ester] (Pierce). The hybridization buffer used contained 59% formamide, 7.3% dextran sulfate, 0.74× SSC, and 125 μg of tRNA per ml.
Video microscopy.
A549 cells were plated on glass coverslips (12 by 12 mm) 1 day before infection. On the day of the experiment, cells were incubated for 1 h at 37°C in the presence of nocodazole or DMSO solvent (control); these reagents were maintained in the media throughout the experiment. Cells were infected with AdLite viruses at 104 particles/cell for 1 h at 4°C, unbound virus was removed, and the cells were transferred to 37°C to initiate infection. After approximately 30 min, individual coverslips were removed, washed with PBS (± drugs), placed on a glass slide, and sealed with petrolatum to prevent evaporation. Slides were mounted on a fluorescent Axiovert microscope (Zeiss) equipped with a 63×/1.4 lens; images were acquired with a cooled CCD camera (Quantix; Photometrix) using the IPLab acquisition program (Scanalytics). Typically, 10 to 20 images were taken per series at 3- to 5-s intervals using a 1- to 1.5-s exposure time. The image series were processed using the National Institutes of Health image program to reconstitute the paths and rates of moving AdLite particles.
Western and Southern blot analysis, luciferase assay, and GFP analysis.
Western blot analysis was performed by standard methods. Equal numbers of virus particles were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and either stained with Coomassie blue to confirm equal protein loading or transferred to a nitrocellulose membrane and probed with anti-GFP antibody (1:500) (Clontech). For Southern analysis, viral DNA was isolated from AdLite particles by treatment with proteinase K (0.5 μg/ml) (Sigma) in 0.5% SDS for 30 min at 56°C, phenol-chloroform extraction, and alcohol precipitation of the DNA. DraI cuts at approximately 2.6 kb from the left end of an insertless vector genome. Thus, a DraI digestion and Southern blot with a teto probe can reveal the size of a teto insert. Viral and plasmid DNAs were digested with DraI, and the resulting fragments were resolved on a 1% agarose gel, transferred, and probed with a teto DNA probe. The teto probe was prepared by digestion of pAdteto56Ad with XhoI, releasing a 350-bp DNA fragment containing seven tandem teto repeats which was gel purified and labeled with [32P]dCTP using the Prime-It II Random Primer labeling kit (Stratagene). Luciferase assays were performed by standard methods.
RESULTS
Construction and molecular characterization of AdLite viruses.
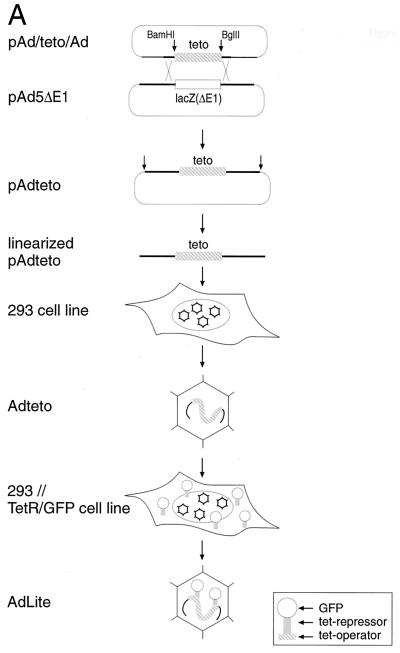
The strategy employed to generate AdLite viruses is summarized in Fig. 1A. Briefly, multimerized teto DNA sequences (56 and 112 tandem repeats of the teto) were inserted in place of the E1A gene in a transfer vector that contains stretches of adenovirus genome upstream and downstream from the teto insertion site. The resulting Ad/teto/Ad fragment was introduced into a plasmid-borne E1A-defective adenovirus type 5 genome using homologous recombination in E. coli (8) to produce various pAdteto plasmids. pAdteto plasmids were digested with SpeI, and the released viral genomes were introduced into 293 cells to generate stocks of Adteto. Aliquots of these stocks were then used to infect a 293 cell line derivative that stably expresses a TetR-GFP fusion protein (see Materials and Methods for additional details). TetR-GFP localizes to both the cytoplasm and the nucleus, despite lacking classical nuclear localization signal sequences (data not shown). In this cell line, replicated Adteto genomes were bound by TetR-GFP and packaged into viral particles which we called AdLite particles. TetR-GFP-loaded AdLite particles were purified by a standard cesium chloride gradient and further characterized.
FIG. 1.
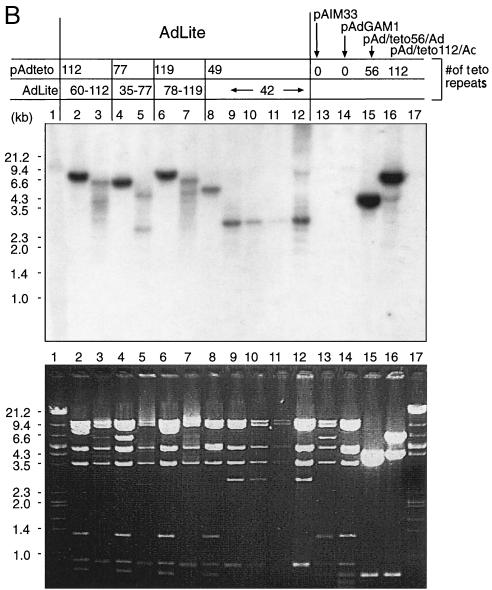
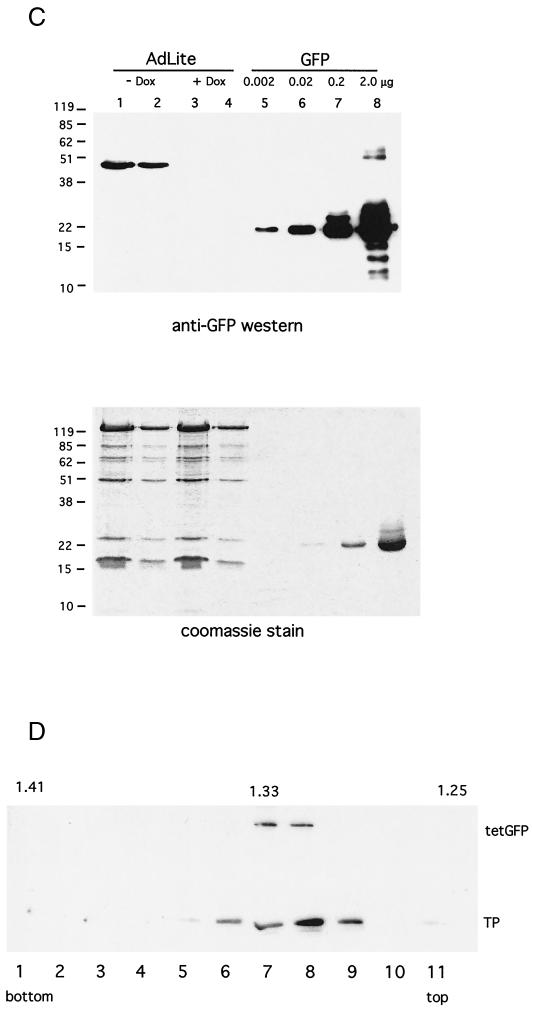
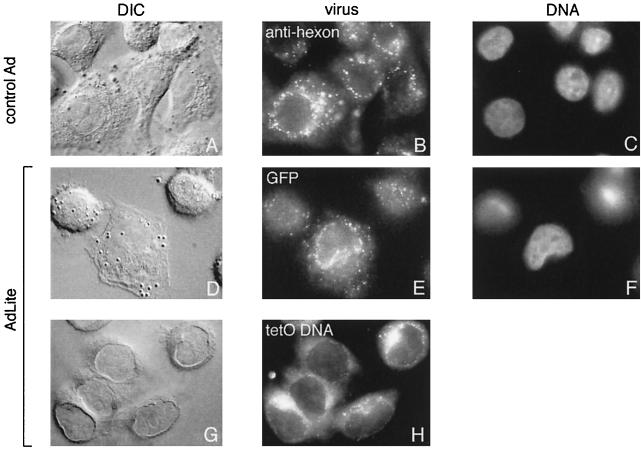
Construction and molecular characterization of AdLite viruses. (A) Strategy applied to generate AdLite viruses. A total of 56 or 112 teto sites (gray box) were cloned into the BamHI and BglII sites in the polylinker of the transfer vector pΔE1sp1B (see Materials and Methods), which contains stretches of adenovirus genome (thick black lines) upstream and downstream from the deleted E1A region, generating pAd/teto/Ad. pAd/teto/Ad was homologously recombined in E. coli BJ5183 with pAdDE1, which contains the full adenovirus genome (thick black lines) and either a lacZ or a Gam1 coding sequence (white box; see panel B) in place of the E1A region, generating pAdteto carrying the teto fragment in place of the E1A region. Linearized pAdteto was used to transfect 293 cells, and lysates from 293 cells containing replicated Adteto viruses (hexagons) were used to infect a 293 cell line stably expressing the TetR-GFP fusion protein (see inset). Newly replicated genomes were bound by TetR-GFP to produce TetR-GFP-loaded adenovirus particles called AdLite viruses. (B) Southern analysis of DraI-digested plasmids used to generate AdLite and/or Dra-I digested viral DNA obtained from AdLite particles, probed with teto probe (top panel). Lower panel, Ethidium bromide-stained gel. Plasmids and AdLite viruses are indicated (lanes 2 to 16). Lanes 1 and 17, lambda DNA marker. pAd/teto56/Ad and pAd/teto112/Ad (lanes 15 and 16) contain 56 and 112 teto sites, respectively, and are donors of teto sites in homologous recombination with two acceptor plasmids containing E1- and E3-defective adenovirus genomes and either the lacZ gene (pAIM33; lane 13) or the Gam1 gene (pAdGam1; lane 14). Products of homologous recombination between pAd/teto112/Ad and pAdGAM1 are designated pAdteto112, pAdteto77, and pAdteto119 (lanes 2, 4, and 6) and contain 112, 77, and 119 teto sites, respectively. When these plasmids were used to produce viruses, the resulting AdLite particles contained mixed populations of genomes with 60 to 112, 35 to 77, and 78 to 119 teto sites, respectively (lanes 3, 5, and 7). pAdteto49 (lane 8) is a product of homologous recombination between pAIM33 and pAd/teto56/Ad and contains 49 teto sites; resulting Adlite genomes contain 42 teto sites (shown are four different viral preparations of AdLite42; lanes 9 to 12). (C) Western analysis of GFP content of AdLite particles. AdLite was grown in 293 clone 9 cells in either the absence or presence of DOX. Virus was purified by a CsCl gradient (with or without DOX). Equal numbers of adenovirus particles were loaded per lane, blotted, and probed with anti-GFP antibody (top) or stained with Coomassie blue (bottom). Lane 1, AdLite, 8 × 1010 particles; lane 2, AdLite, 2.7 × 1010 particles; lane 3, AdLite, 8 × 1010 particles grown and purified in the presence of DOX; lane 4, AdLite, 2.7 × 1010 particles grown and purified in the presence of DOX; lanes 5 to 8, pure GFP standard. The TetR-GFP fusion protein is approximately 50 kDa. The estimated number of TetR-GFP molecules per AdLite particle is 20 (see text). (D) TetR-GFP remains associated with viral DNA during early stages of infection. A549 cells infected for 30 min with AdLite at 104 particles per cell were lysed by three freeze-thaw cycles, and the lysate was mixed with 1.33 g of CsCl per cm3 in 10 mM HEPES and centrifuged in a vTi65 rotor at 60,000 rpm for 20 h. The gradient was fractionated, resolved by SDS-polyacrylamide gel electrophoresis, and immunoblotted for either TetR-GFP (top panel) or TP (bottom panel). Densities of specific fractions (in g/cm3) are listed at the top of the figure, and fraction numbers are listed at the bottom, with fraction number 1 being the bottom of the gradient.
Our initial expectation was that incorporation of a higher number of teto sequences into the adenovirus genome would result in a higher number of TetR-GFP molecules bound per genome and thus in a brighter fluorescence of AdLite particles. Southern analysis of the DNA genomes obtained from several batches of AdLite viruses revealed the presence of heterogeneity among the viral genomes: the number of teto repeats ranged from 35 to 119 (Fig. 1B, lanes 3, 5, 7, and 9 to 12), which differs from the original number of 56 and 112 teto sites in the inserts that were used for the homologous recombination and the original Adteto plasmids (Fig. 1B, lanes 15 and 16). This indicates that a partial excision-recombination of the teto insert occurred during the production of AdLite particles. Surprisingly, when immunoblotting was used to examine the amount of encapsidated TetR-GFP, very similar quantities of TetR-GFP were detected in all AdLite virus preparations independently of the number of teto repeats present in their genomes (results not shown and see below).
We next evaluated the fluorescent properties of AdLite viruses. Drops of AdLite preparations were placed on glass slides and examined using fluorescence microscopy. Images of different virus preparations were acquired with a cooled CCD camera using conditions that would capture the whole dynamic range of fluorescence. We found that the fluorescence of individual particles was comparable in each batch of AdLite and among the different batches (data not shown), consistent with our estimation that they contained comparable quantities of TetR-GFP.
To evaluate the specificity of TetR-GFP binding to teto sequences in AdLite particles during propagation in 293 cells expressing the TetR-GFP fusion, we propagated control viruses lacking the teto sequences in these cells. The purified viruses were not fluorescent (results not shown), indicating that the loading of fluorescent molecules of AdLite was dependent on binding between teto sequences within their genomes and TetR-GFP fusion proteins and not due to nonspecific entrapment of TetR-GFP molecules during virus growth. As a further demonstration of the specificity of the TetR-GFP loading, we grew and purified AdLite viruses in the presence of the tetracycline analogue doxycycline (DOX). When complexed with the antibiotic, the tet repressor affinity for operator sequences is abolished (20, 21). Thus, if TetR-GFP loading in AdLite viruses is due to specific teto-TetR interactions, then AdLite viruses grown in the presence of DOX should not incorporate TetR-GFP. We found this to be so, with no detectable TetR-GFP incorporated into AdLite viruses when the viruses were grown and purified in the presence of DOX (compare Fig. 1C, lanes 1 and 2 [no DOX] to lanes 3 and 4 [with DOX]). The quantity of GFP encapsidated in AdLite could be calculated by comparison with defined quantities of purified GFP loaded on the same blot and revealed that approximately 20 TetR-GFP molecules were bound per AdLite particle. Analysis of the TetR-GFP content of a number of preparations with teto repeats ranging from 35 to 119 showed similar levels of TetR-GFP content independently of the number of teto repeats present in the virus genome (results not shown). This suggests that TetR-GFP binding is not limited by teto repeat number but by some other factor, such as the space within the capsid available for additional protein. We concluded that the fluorescence of AdLite particles depends on a specific interaction between TetR-GFP and teto sequences within the viral genomes and a maximum of 20 TetR-GFP molecules can be encapsidated per virion. We have chosen Adteto42 to characterize further; in these and following experiments we refer to Adteto42 as AdLite.
Characterization of infective properties of AdLite particles.
One justification for pursuing the AdLite system is that it allows visualization of the infecting viral DNA rather than a labeled capsid protein. An additional set of control experiments was performed to determine if the TetR-GFP label remains associated with the infecting viral DNA. To do this, we infected cells with AdLite. Cells were exposed to AdLite particles at 10,000 particles/cell at 4°C, the unbound virus was removed by extensive washing, and infection was allowed to proceed at 37°C for 30 min. The cells were then harvested and lysed to release the viral material that had been internalized, and the material was fractionated by density on a CsCl step gradient. Under the conditions used, free TetR-GFP protein released from the infecting virion would band at the density of free protein (<1.3 g/cm3) (35), while virion- or subvirion-associated material would band at densities substantially greater than this.
This analysis revealed that at 30 min after the initiation of viral entry, a typical time point in our analysis of AdLite by microscopy, all detectable TetR-GFP is associated with a peak of material centering around 1.31 to 1.33 g/cm3 (Fig. 1D). The same fractions were analyzed for viral terminal protein (TP) which, because it is covalently linked to the viral DNA termini, is an indicator of the position of the viral DNA (36, 37, 39). We found that all of the TetR-GFP signal is present in fractions containing TP (Fig. 1D). We concluded that at 30 min postinfection, TetR-GFP was still present in a complex that also contained the covalently bound TP, and by inference, the viral DNA. This provides firm evidence that the GFP signal monitored by microscopy is indeed from GFP associated with viral DNA.
We next compared the kinetics of infection of AdLite and control adenoviruses at the cellular level. A549 cells were infected with AdLite or control E1A-deficient adenoviruses and fixed at defined time points following infection. The distribution of AdLite viruses was visualized by virtue of GFP fluorescence, and the distribution of the control viruses was visualized by immunofluorescence using antibodies against the capsid protein hexon. We found that the distribution of both GFP signal and hexon was comparable at all time points examined. At 0 min after infection, both signals were found exclusively at the cell periphery; at 15 to 30 min after infection, both signals were in the cytoplasm (data not shown). Finally, at 30 to 60 min after infection, both signals had begun to accumulate around the nuclear membrane (Fig. 2A to F), suggesting that both control and AdLite viruses had reached the nuclear membrane. Next, using fluorescent in situ hybridization with a teto DNA-specific probe, we analyzed the distribution of AdLite viral genomes and compared that to the distribution of TetR-GFP. We found that the distribution of AdLite genomes was similar to the distribution of GFP signal at all time points examined (Fig. 2G and H and data not shown), demonstrating that the GFP signal indeed reflects the location of AdLite viral DNA. Taken together, these data show that AdLite viruses accumulate around the nucleus with kinetics similar to those of control adenoviruses and suggest that both AdLite and control viruses utilize similar entry pathways to target the nuclei. Surprisingly, at later time points following infection, we were unable to detect the GFP signal within the nucleus (data not shown), suggesting that TetR-GFP is removed from the viral DNA at the nuclear membrane. Therefore, using AdLite, we have concentrated on studying the mechanisms of adenoviruses targeting the nuclear membrane; the mechanisms of nuclear entry are addressed later using expression studies (see below).
FIG. 2.
Comparison of the distribution of AdLite and control adenoviruses in A549 cells 1 h after infection. A549 cells were infected with AdLite (D to H) or control (A to C) adenovirus at 104 virus particles/cell and fixed for 1 h after infection. The distribution of control adenoviruses was detected with anti-hexon antibodies (B). The distribution of AdLite was detected by fluorescence of TetR-GFP (E) and by DNA hybridization in situ with fluorescently labeled teto DNA-specific probe (H). Both control and AdLite viruses accumulate around the nuclei of the infected cells. Panels A, D, and G, differential interference contrast microscopy (DIC) images of infected cells (note that the morphology of cells in panel G is affected by the in situ hybridization procedure); panels C and F, Hoechst dye counterstaining of the nuclei.
Codistribution of AdLite viruses and microtubules.
Previous publications have suggested that microtubules are involved in intracellular trafficking of adenoviruses because adenovirus particles have been observed in close proximity to the microtubules in the infected cells (12, 31). Therefore, we first examined the codistribution of AdLite viruses and microtubules in infected A549 and HeLa cells. We found that AdLite particles accumulated in a striking pattern around the spindle poles in mitotic cells (Fig. 3A to C) and at the MTOC near the nucleus in nonmitotic cells (Fig. 3D to F and J to L). Similar accumulation around MTOC and spindle poles was observed with control E1A-deficient adenoviruses immunodetected with anti-hexon antibodies (data not shown). These data confirm the previous findings that adenoviruses can be found in close proximity to the microtubules in vivo.
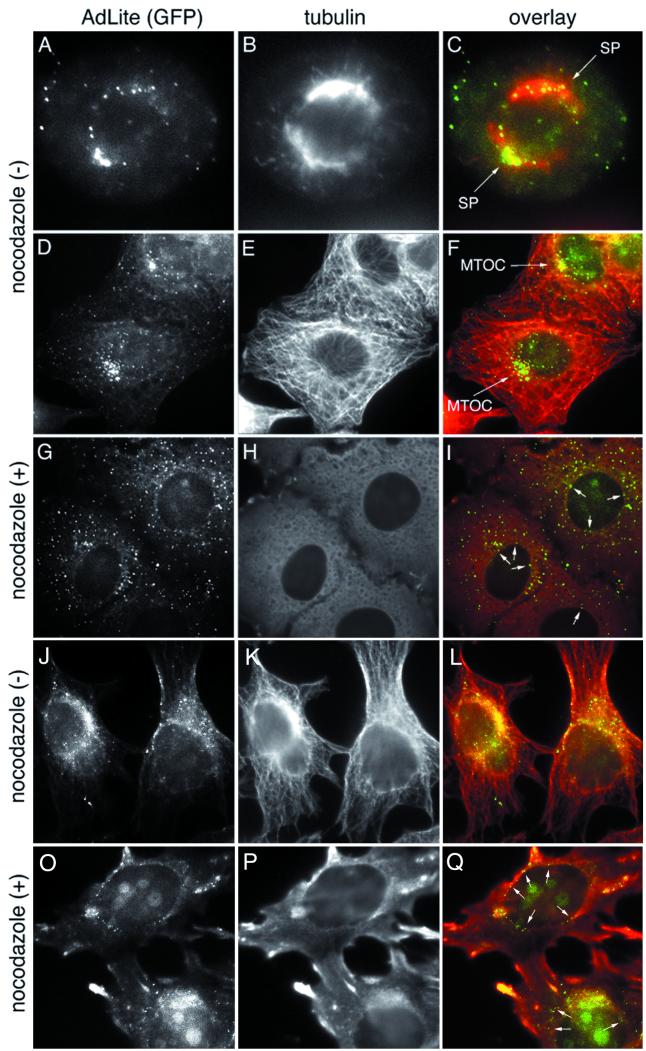
FIG. 3.
Effect of nocodazole on the distribution of AdLite viruses in infected A549 and HeLa cells. A549 cells were infected with AdLite at 104 virus particles/cell in the absence (A to F) or presence (G to I) of 2 μM nocodazole and fixed at 20 min (A to C) or 30 min (D to I) after infection. HeLa cells were infected with AdLite in the absence (J to L) or presence (O to Q) of 20 μM nocodazole and fixed at 60 min after infection. AdLite viruses were detected by fluorescence of TetR-GFP (A, D, G, J, and O) and microtubules were detected by immunolabeling (B, E, H, K, and P); in the overlay, AdLite viruses are shown in green and yellow and microtubules are shown in red (C, F, I, L, and Q). In A549 cells, in the absence of nocodazole, AdLite accumulates around the spindle poles (SP) in mitotic cells (A to C) or around the nuclei and MTOC in nonmitotic cells (D to F). In nocodazole-treated A549 cells (G to I), microtubules are completely depolymerized (H) and AdLite viruses remain dispersed in the cytoplasm, although some AdLite particles accumulate around the nucleus (I; arrows); similar AdLite distribution was observed in A549 cells treated with higher nocodazole concentrations (2 to 20 μM). In HeLa cells, in the absence of nocodazole, AdLite accumulates around the nuclei and the MTOC (J to L). In nocodazole-treated HeLa cells (O to Q), microtubules are grossly depolymerized (P); some AdLite particles were observed to colocalize with these partially depolymerized masses of tubulin. However, like in A549 cells, some AdLite particles also accumulate around the nucleus (Q; arrows).
The observed accumulation of AdLite at the MTOC and spindle poles, where minus ends of microtubules are nucleated, could suggest that the viruses are transported to this location in association with the microtubule minus end-directed molecular motors or that they have an affinity for the minus ends of the microtubules. To test whether microtubules function in the transport of AdLite particles, we performed the infection in the presence of nocodazole to depolymerize microtubules. We tested the effect of several concentrations of nocodazole that either cause partial (0.08 μM) or complete (2 to 20 μM) depolymerization of microtubules (also see Fig. 5D). The cells were fixed at defined time points after infection, and the codistribution of AdLite and microtubules was evaluated by immunofluorescence. We found that in A549 cells, AdLite viruses did not accumulate at specific sites in cells in which the microtubules have been completely depolymerized with nocodazole (Fig. 3G and H), suggesting that intact microtubules are required for AdLite accumulation at MTOC. In HeLa cells, which appear more resistant to the nocodazole treatment, some AdLite was found to colocalize with partially depolymerized masses of tubulin (Fig. 3M to O). However, in both A549 and HeLa cells, AdLite viruses still reached the nuclear membrane in the presence of nocodazole (Fig. 3I and O, arrows). We quantitated the number of AdLite particles around the nucleus of infected A549 and HeLa cells at 1 h after infection in the absence or presence of nocodazole and found that similar numbers of AdLite viruses accumulated around the nucleus in control cells and in cells in which microtubules have been depolymerized (Table 1). These data suggest that intact microtubules are required for accumulation of adenoviruses at MTOC but that these cytoskeletal elements are not obligatory for the targeting of AdLite viruses to the nuclear membrane.
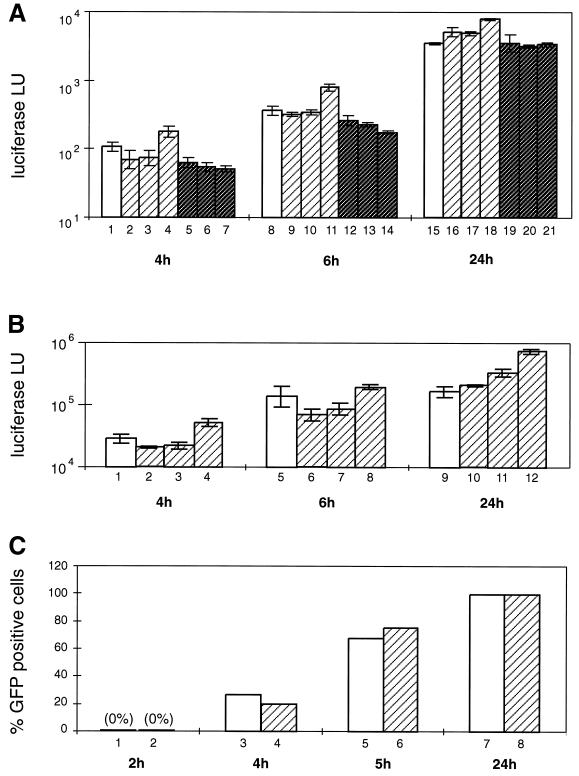
FIG. 5.
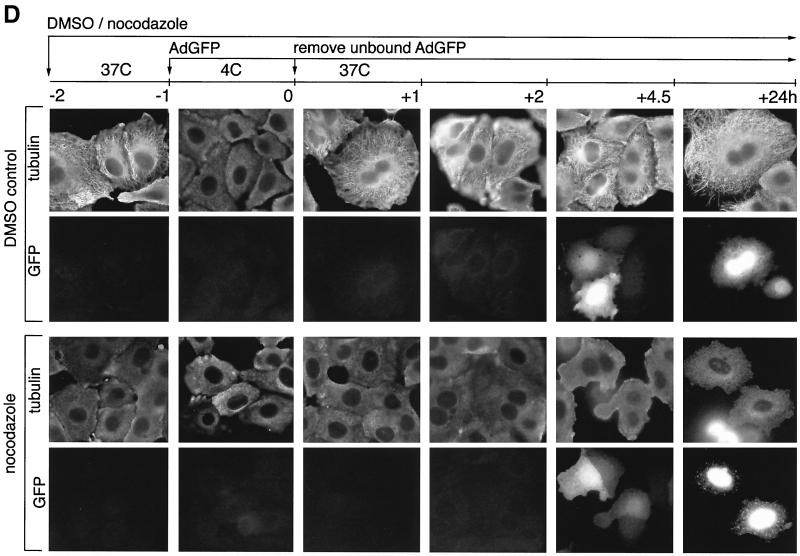
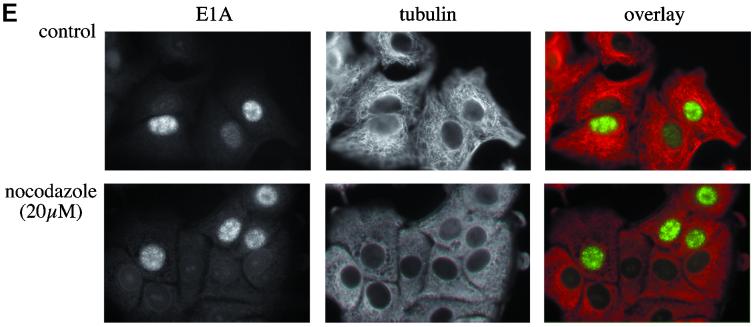
Effect of nocodazole on gene expression from adenovirus-derived vectors. A549 (A and C to E) or HeLa (B) cells were treated for 1 h at 37°C with DMSO solvent (control), with nocodazole to depolymerize microtubules, or with taxol to stabilize microtubules. Subsequently, cells were placed on ice and incubated at 4°C for 1 to 2 h with recombinant adenoviruses carrying luciferase (AdLuc) or GFP (AdGFP) genes (A to D) or with the wild-type adenovirus (E) in the continuous presence of the drugs. The unbound viruses were removed, and fresh medium containing DMSO, nocodazole, or taxol was added to the cells, which were then shifted to 37°C to initiate the infection. (A) Time course of luciferase expression from AdLuc (103 MOI) in A549 cells in the absence (white bars) or presence of various concentrations of nocodazole (0.2, 2, and 20 μM) (striped bars) or taxol (0.1, 1, and 10 μM) (gray striped bars). Cells were lysed at 1-h intervals, and luciferase activity (LU) was measured in triplicate, with the background subtracted, and averaged (standard deviations are indicated). Luciferase activity was first detectable at 4 h after infection and increased similarly over time both in the absence and presence of nocodazole or taxol. Similar results were obtained for different MOI (101 to 104 particles/cell) in three different experiments. (B) Time course of luciferase expression from AdLuc (104 MOI) in HeLa cells in the absence (white bars) or presence (striped bars) of nocodazole (0.2, 2, and 20 μM). Luciferase activity was measured as described for panel A and increased similarly over time both in the absence and presence of nocodazole. (C) Time course of GFP expression from AdGFP. Cells were infected with AdGFP at 104 MOI in the absence (white bars) or presence (striped bars) of nocodazole (20 μM). Cells were trypsinized at 1-h intervals, and the percentage of GFP-positive cells was estimated by FACS analysis. GFP expression is first detectable at 4 h after infection and is not significantly changed in the presence of nocodazole. (D) Evaluation of microtubule integrity in cells infected with AdGFP in the absence (top two image rows) or continuous presence (bottom two image rows) of nocodazole (20 μM). Cells were infected with AdGFP at 103 particles/cell; the time schedule of the drug and virus administration is indicated at the top. Infected cells were fixed at 1-h intervals and immunolabeled with anti-tubulin antibodies. Double images (microtubule staining and GFP) representative of each time point are shown. In control cells, microtubules are intact, except after the cold treatment. In nocodazole-treated cells, microtubules are depolymerized at all time points. In both control and nocodazole-treated cells, GFP expression begins approximately 4.5 h after infection, independent of the state of microtubule integrity (see 4.5-h time point). (E) Expression of E1A from the wild-type adenovirus in the presence or absence of nocodazole (virus and nocodazole administration were as described for panel D). Cells were fixed and double labeled with anti-E1A (green) and anti-tubulin (red) antibodies. Double images are shown from the time point at 5.5 h after infection. E1A is expressed both in control and nocodazole-treated cells, where microtubules are depolymerized.
TABLE 1.
Effect of nocodazole on perinuclear accumulation of AdLite particles in A549 and HeLa cells 1 h after infection
| Cell type | Presence of nocodazole | No. of AdLite particles (± SD) per nucleus (total particles/no. of cells)a |
|---|---|---|
| A549 | − | 17.4 ± 6.6 (347/20) |
| + | 16.2 ± 8.1 (324/20) | |
| HeLa | − | 20.2 ± 5.5 (404/20) |
| + | 19.5 ± 4.6 (410/21) |
Cells were photographed in the optical plane that preserved the contour of the nucleus the best. AdLite particles were counted within approximately 0.3 to 0.5 μm of the nuclear periphery in 20 to 21 cells per experimental condition, in six random optical fields per experiment (two independent experiments).
Motility of AdLite viruses in live cells.
To further examine whether microtubules participate in adenovirus intracellular motility we used video microscopy to follow the movement of AdLite viruses in live, infected cells under conditions in which microtubules were either intact or depolymerized with nocodazole. We chose to monitor the movement of AdLite viruses in infected A549 cells and not in HeLa cells, because A549 cells have a larger cytoplasmic area and therefore better optical properties than HeLa cells; furthermore, A549 cells respond more reliably to complete depolymerization of the microtubules with nocodazole (Fig. 3). AdLite intracellular movement was monitored beginning at 30 min after initiation of the infection, which corresponds to the time when viruses are thought to escape from the endosomes and traverse the cytoplasm toward the nucleus (19). To avoid phototoxicity, AdLite-infected cells were typically filmed for a maximum of 20 to 30 s, which permitted following the AdLite distribution during 10 to 15 consecutive frames. We found that in control cells, AdLite viruses moved both toward and away from the nucleus as well as in other directions (Fig. 4A and B). The rates of AdLite particle movement averaged 18.90 ± 6.67 μm/min (Table 2), with elementary movements observed up to 60.00 μm/min at room temperature (data not shown). Similar velocities were observed independently of the direction of migrating AdLite particles (Table 2). When cells were incubated in the presence of 20 μM nocodazole, AdLite particle movement occurred along similar paths and directions as in control cells (Fig. 4C). In the presence of 20 μM nocodazole, the rates of the AdLite particle movement averaged 15.82 ± 5.82 μm/min (Table 2), with elementary movements observed up to 60.32 μm/min (data not shown). These data demonstrate that nocodazole-sensitive microtubules are not required for AdLite motility.
FIG. 4.
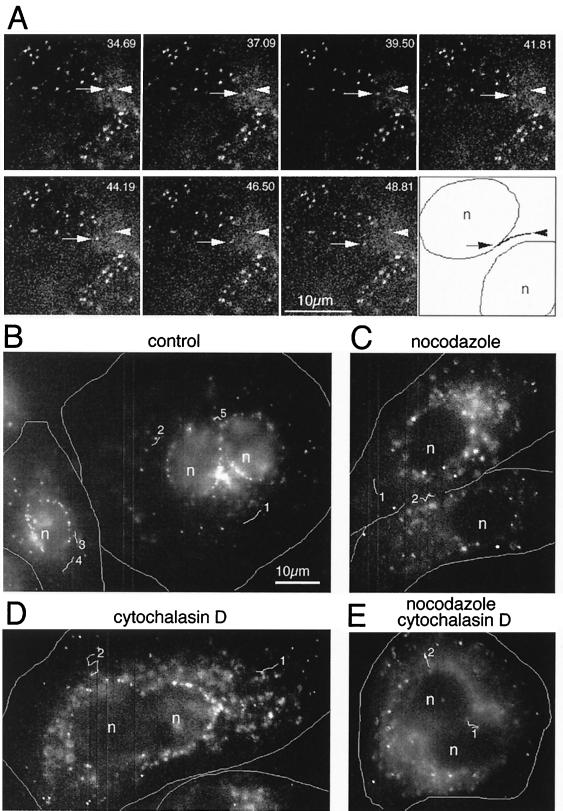
Real-time visualization of AdLite motility in live A549 cells in the absence or presence of nocodazole. A549 cells grown on coverslips were infected with AdLite viruses in the presence of DMSO solvent (control) or cytoskeleton-disrupting reagents. Coverslips were transferred onto glass slides and mounted on a fluorescence microscope, and the distribution of AdLite-associated fluorescence was filmed with a cooled CCD camera. (A) Images of two adjacent control cells infected with AdLite viruses showing the distribution of AdLite particles at approximately 2-s intervals (the exact acquisition times are shown in seconds and milliseconds). In the last panel, the two nuclei (n) are outlined and the trajectory of the particle moving toward the “upper” nucleus is reconstructed. The position of one AdLite particle moving toward the nucleus is indicated by an arrow; its initial position is indicated by an arrowhead. (B to E) Reconstitution of trajectories of AdLite particle movement in A549 cells treated with DMSO (control) (B), 20 μM nocodazole (C), 0.5 μM cytochalasin D (D), or both nocodazole and cytochalasin D (E); DMSO and nocodazole were administered before and during infection, and cytochalasin D was added after virus internalization for 15 to 30 min at 37°C. Images shown are projections of four to eight consecutive images acquired at approximately 2- to 4-s intervals. The reconstituted trajectories of moving AdLite particles are shown as white lines, with a number located at the beginning of each path. Images were acquired at approximately 30 to 90 min after infection. In both series, some AdLite particles have already reached the nuclei (n); other particles are observed to move toward the nucleus (particle 3 in B, particles 1 and 2 in D, and particles 1 and 2 in E), away from the nucleus (particle 4 in B), or in other directions.
TABLE 2.
Characteristics of AdLite motility in A549 cellsa
| Relative direction of AdLite movement | Type of average velocity measurement | Average velocity (μm/min) ± SD of cells treated withb:
|
|||
|---|---|---|---|---|---|
| Control | Nocodazole | Cytochalasin D | Nocodazole and Cytochalasin D | ||
| Toward the nucleus | Per particle | 16.9 ± 6.7 (N = 7) | 14.4 ± 5.6 (N = 13) | 25.5 ± 5.8 (N = 4) | 20.3 ± 0.6 (N = 2) |
| Per step | 18.0 ± 10.6 (n = 40) | 16.2 ± 9.0 (n = 78) | 26.3 ± 14.9 (n = 20) | 20.3 ± 11.8 (n = 18) | |
| Away from the nucleus | Per particle | 19.3 ± 7.6 (N = 9) | 15.2 ± 5.3 (N = 9) | 28.3 ± 5.1 (N = 2) | 10.6 (N = 1) |
| Per step | 17.9 ± 10.5 (n = 51) | 19.8 ± 11.0 (n = 44) | 28.3 ± 11.6 (n = 10) | 10.6 ± 5.8 (n = 9) | |
| Other directions | Per particle | 20.7 ± 12.6 (N = 6) | 17.7 ± 6.3 (N = 13) | 0 | 14.4 ± 3.8 (N = 7) |
| Per step | 18.9 ± 15.7 (n = 40) | 19.0 ± 8.7 (n = 96) | 0 | 14.1 ± 7.2 (n = 50) | |
| All directions | Per particle | 18.9 ± 8.7 (N = 22) | 15.8 ± 5.8 (N = 35) | 26.4 ± 5.2 (N = 6) | 15.1 ± 4.3 (N = 10) |
| Per step | 18.2 ± 12.3 (n = 131) | 18.2 ± 9.4 (n = 218) | 27.0 ± 13.7 (n = 30) | 15.1 ± 8.8 (n = 77) | |
Images of moving AdLite particles were acquired at 30 to 80 min after infection. For each particle, between 3 and 20 consecutive, individual steps were analyzed, and elementary velocities per step were calculated (see Materials and Methods).
N, number of particles; n, number of steps of all particles analyzed per given direction of the movement.
To evaluate whether actin microfilaments play a role in AdLite motility, cells were infected with AdLite for 15 to 30 min at 37°C to allow virus adsorption and endocytosis, a process that requires intact actin filaments (26). Subsequently, to depolymerize actin filaments, cells were exposed to cytochalasin D (0.5 μM) for 15 to 30 min at 37°C, and then the distribution of internalized AdLite particles was monitored by video microscopy. This concentration of cytochalasin caused actin to depolymerize only partially; higher concentrations of cytochalasin could not be used, as these severely affect cellular architecture and viability (data not shown). We found that in the presence of cytochalasin, AdLite motility was similar to that in control cells (Fig. 4D), with an observed velocity of 26.44 ± 5.22 μm/min (Table 2). These data demonstrate that cytochalasin D-sensitive actin filaments are not required for AdLite motility. When cells were infected with AdLite in the constant presence of nocodazole, followed by the administration of cytochalasin D at 15 to 30 min after virus internalization, AdLite particles still remained motile and moved in similar directions as control cells at 15.16 ± 4.3 μm/min (Fig. 4E; Table 2). Thus, infecting adenovirus particles remain motile even under conditions when both microtubule and actin networks are disrupted.
We also analyzed the vectorial distances that AdLite particles had traveled in the infected cells by calculating the net distance between the start and the end points during each sequence. We found that during the typical 20 to 30 s of observation, AdLite viruses migrated 2 to 6 μm both in control cells and in cells treated with nocodazole and/or cytochalasin D (data not shown). This distance corresponds to approximately 5 to 100% of the distance between the cell membrane and the nuclear membrane of all analyzed cells. We concluded that the observed motility could suffice to accomplish the perinuclear localization of adenovirus.
Effect of depolymerization of the microtubules on gene expression from the adenovirus-derived vectors.
To further test whether the microtubules participate in nuclear targeting of adenoviruses, we monitored adenovirus-dependent gene expression in the absence or presence of drugs that interfere with microtubule function. We assumed that only successful nuclear entry of these adenoviruses would result in gene expression; cytoplasmic transcription from the adenovirus genome would be unprecedented (34). Briefly, A549 or HeLa cells were treated for 1 h at 37°C with solvent (control) or with nocodazole, colchicine, or vinblastine to depolymerize microtubules or with taxol to stabilize microtubules. Subsequently, cells were placed on ice and incubated with selected adenoviruses in the absence or presence of drugs for 1 to 2 h at 4°C. The unbound viruses were then removed, and fresh medium containing the drugs was added to the cells, which were then shifted to 37°C to initiate the infection.
We first evaluated the gene expression from recombinant adenoviruses that carry either luciferase (AdLuc) or eGFP (AdGFP) genes in place of the E1A region. Cells were harvested at 1-h intervals and either lysed to measure luciferase activity (Fig. 5A and B) or subjected to FACS analysis to measure the percentage of GFP-positive cells (Fig. 5C). We found that the expression of both luciferase and GFP began approximately 4 h after infection and increased over time, with similar kinetics in all control and nocodazole- or taxol-treated A549 and HeLa cells (Fig. 5A and B, respectively). Only at the earliest time point was a slight inhibition of luciferase and GFP expression observed; however, these differences disappeared over time. Similar results were obtained when A549 cells were treated with other microtubule-depolymerizing reagents, such as colchicine or vinblastine (data not shown).
To confirm that the nocodazole treatment caused a complete depolymerization of the microtubules throughout the duration of the experiment, the cells were fixed at defined time points before and after infection with AdGFP and immunolabeled with tubulin-specific antibodies (see the legend to Fig. 5D for detailed protocol). As expected, in control cells microtubules were well preserved at all time points except following the cold treatment, which causes a transient microtubule depolymerization (32) (Fig. 5D, first row of panels). In the nocodazole-treated cells, microtubules were depolymerized at all time points (Fig. 5D, third row of panels). When the tubulin-labeled cells were simultaneously evaluated for GFP expression, we found that both in control cells in which microtubules were intact and in nocodazole-treated cells in which microtubules were depolymerized, GFP expression could be observed at 4.5 h after infection (Fig. 5D, second and fourth rows of panels, respectively), consistent with the results of FACS analysis (see above).
We also evaluated the effect of microtubule depolymerization on the expression of an adenovirus early gene, E1A. A549 cells were infected with wild-type adenovirus in the presence or absence of nocodazole, fixed at defined time points, and coimmunolabeled with E1A and tubulin-specific antibodies. We found that E1A was expressed at approximately 5.5 h after infection both in cells with intact microtubules and in cells in which microtubules have been disrupted by nocodazole (Fig. 5E). To quantitate these results we estimated the percentage of E1A-positive cells infected with the wild-type adenovirus in the presence or absence of nocodazole (Table 3). We found that 35.0% of the cells expressed E1A when microtubules were depolymerized; this percentage was comparable with 33.3% of E1A-positive cells in which microtubules were intact. Taken together, these data indicate that adenovirus-directed gene expression can occur in the absence of microtubules with kinetics similar to those observed in cells in which microtubules are intact.
TABLE 3.
Effect of nocodazole on E1A expressiona
| Expt no. | % E1A-positive nuclei (no. positive/no. of cells in field)
|
|
|---|---|---|
| Control | Nocodazole | |
| 1 | 30.7 (54/176) | 34.9 (67/192) |
| 2 | 36.1 (62/172) | 35.0 (69/197) |
| Total | 33.3 (116/348) | 35.0 (136/389) |
Virus and drug administration and stainings were performed as described for Fig. 5E. Cells were immunolabeled with anti-E1A and anti-tubulin antibodies and were counterstained with a DNA dye to visualize nuclei. Fifteen random fields were photographed per slide (two independent experiments) to visualize both E1A-specific signals and the nuclei.
DISCUSSION
Following internalization by endocytosis, adenoviruses must somehow traverse the cytoplasm to reach the vicinity of the nuclear envelope. In this report we have investigated the mechanisms of nuclear targeting by adenoviruses using AdLite viruses that can be directly observed by video microscopy during infection in live cells. We found that AdLite particles infect these cells, and upon internalization, move toward and away from the nucleus at velocities averaging approximately 19 μm/min at room temperature. Motility with similar directionality toward and away from the nucleus and comparable velocities have been previously described in studies with fluorochrome-labeled adenovirus capsids (46). The motility of capsid-labeled adenoviruses was reported to be inhibited by 20 μM nocodazole, although not by lower doses of this microtubule-depolymerizing reagent. In addition, the motility toward, but not away from, the presumptive MTOC was partially inhibited by overexpression of dynamitin, a protein required for dynein function. Based on these observations, it was proposed that adenoviruses reach the nucleus by a microtubule-dependent active transport in association with a dynein-like motor (46).
Our observations of directional AdLite movements support the notion that the viruses utilize some kind of a cytoskeleton-dependent motility system. However, we do not observe any significant inhibition of AdLite motility in A549 cells in the presence of a wide range of nocodazole concentrations which cause either partial depolymerization of the microtubules (concentrations below 2 μM) or complete depolymerization (2 to 20 μM). It is formally possible that we have failed to detect microtubule-dependent motility using AdLite due to differences in image acquisition conditions required to follow AdLite particles in vivo compared with those used to follow distribution of capsid-labeled adenoviruses (46). However, it is not clear whether the capsid-labeled studies followed the adenovirus genome (and not merely the capsid) fate in the cell. Therefore, we favor the hypothesis that our live observations of AdLite particles reveal that adenovirus particles can move in infected cells in the absence of microtubules. Our observations are consistent with results in previous reports which revealed the existence of a microtubule-independent pathway during adenovirus infection by finding that treatment of cells with colchicine or vinblastine, two microtubule-severing reagents, did not significantly affect the infective properties of adenoviruses nor did they affect the accumulation of adenovirus DNA in the nuclei of the infected cells (12, 47). Using a complementary approach, we have further shown that depolymerization of microtubules with a variety of microtubule-severing reagents, including nocodazole, colchicine, and vinblastine, does not prevent transgene or early expression from adenovirus, demonstrating that successful nuclear deposition of the adenovirus genomes had occurred in the absence of intact microtubules. Thus, our live observations of AdLite particle movements in the presence of nocodazole support the possibility that a microtubule-independent motility system(s) can contribute to the nuclear targeting by the adenovirus.
The observed AdLite movements in the presence of nocodazole suggest that cytoskeletal elements distinct from microtubules participate in this motility. Microtubule-independent motility has been described in the case of vaccinia viruses, which move in infected cells using continuous polymerization of actin monomers behind the viral particles (11). In the case of adenoviruses, the actin-based network is required for the endocytosis of receptor-bound virions (26, 49). Actin reorganization has been observed following adenovirus attachment (3), but nothing is known about the interaction between free adenovirus particles released from the endosomes into the cytoplasm and actin filaments. We did not observe any significant perturbations of AdLite motility in the presence of cytochalasin D, suggesting that cytochalasin-sensitive filaments do not play a role in adenovirus vectorial transport. However, under these conditions, actin depolymerization is not complete, and it cannot be ruled out that this cytoskeletal system participates in adenovirus intracellular motility. Apart from actin, interfilaments have been indicated in adenovirus-host cell interactions. In cross-linking experiments it has been shown that adenoviruses interact with vimentin (2, 4). In addition, the vimentin-based cytoskeleton becomes rapidly reorganized and collapses around the nucleus within the first 15 to 45 min following adenovirus infection (13). It is conceivable that due to the collapse of the vimentin network, changes in the solubility of the cytoplasm occur, facilitating vectorial motility of the viral particles.
If microtubules are dispensable for successful infection by adenoviruses, then what is the significance of the apparent interaction between adenoviruses and microtubules? Adenoviruses bind to microtubules both in vivo and in vitro (12, 28, 31, 50); our observations that AdLite viruses accumulate around MTOC and spindle poles further substantiate these findings. The observed accumulation of adenoviruses around MTOC and spindle poles, where the minus ends of microtubules are nucleated, may reflect an affinity of adenoviruses for the microtubule minus ends. Indeed, in electron microscopy studies, many adenovirus particles are found at the microtubule ends; however, the polarity of these microtubules was not assessed (12). The distribution of adenoviruses around MTOC and spindle poles strikingly resembles that of the Golgi apparatus in nonmitotic and mitotic cells, respectively (38, 51). It is possible that some virus particles become sequestered within this compartment by the cell, perhaps as a cellular antiviral response rather than a viral infectious process. Thus, it remains to be determined whether adenovirus accumulation around MTOC is of functional importance to the infection process per se.
In conclusion, based on our demonstration of microtubule-independent motility of AdLite viruses in the infected cells, complemented by our findings that nuclear targeting of wild-type and recombinant adenoviruses can occur in the absence of microtubules, we propose that adenoviruses can employ microtubule-independent mechanisms to successfully infect cells. The mechanisms of cytoplasmic trafficking may therefore include both microtubule-dependent and -independent pathways, as has been implicated in the case of herpes simplex virus (42). Adenoviruses may be capable of interactions with different cytoskeletal systems of the host cell, analogous to certain types of vesicles which have been observed to travel along both microtubules and actin filaments (24). Interestingly, fluorescently labeled adenovirus mutant ts1 particles, which are endocytosed like wild-type viruses but are not released into the cytoplasm and remain trapped in the endosomes, also move along linear pathways toward and away from the nucleus (46). This observation could imply that endosomes serve as vehicles for adenovirus intracellular trafficking; however, whether any cytoskeletal elements participate in this process remains to be determined. In addition, adenoviruses may be nonspecifically swept along within intracellular channels that serve to transport macromolecules and vesicles antero- and retrogradely in the cell (27). Such a mechanism, relying on a nonselective “cytoplasmic flow,” has been previously implicated in the localization of P granules, large (5 μm in diameter) RNA-protein complexes, to the posterior pole of a Caenorhabditis elegans embryo or specific RNA molecules during oogenesis in Drosophila melanogaster (14, 22, 44, 45). Currently, no virus- or cytoskeleton-specific interacting molecules have been identified, and it thus remains an open question whether adenoviruses utilize one specific mechanism or multiple mechanisms of cytoplasmic transit.
ACKNOWLEDGMENTS
We thank Rafal Ciosk for providing teto plasmids pRS306tetO2×7, pRS306tetO2×56, and pRS306tetO2×112 and for helpful discussions about the teto–TetR-GFP system; Ron Hay for terminal protein antiserum; Michael Glotzer for the name “AdLite” and helpful discussions; Lukas Huber for comments on the manuscript; and Karin Paiha and Peter Steinlein for help with video and fluorescence microscopy and for performing FACS analysis.
REFERENCES
- 1.Ahmad F J, Echeverri C J, Vallee R B, Baas P W. Cytoplasmic dynein and dynactin are required for the transport of microtubules into the axon. J Cell Biol. 1998;140:391–401. doi: 10.1083/jcb.140.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belin M T, Boulanger P. Cytoskeletal proteins associated with intracytoplasmic human adenovirus at an early stage of infection. Exp Cell Res. 1985;160:356–370. doi: 10.1016/0014-4827(85)90182-x. [DOI] [PubMed] [Google Scholar]
- 3.Belin M T, Boulanger P. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1993;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 4.Belin M T, Boulanger P. Processing of vimentin occurs during the early stages of adenovirus infection. J Virol. 1987;61:2559–2566. doi: 10.1128/jvi.61.8.2559-2566.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 6.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkhardt J K, Echeverri C J, Nilsson T, Vallee R B. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chartier C, Degryse E, Gantzer M, Dieterie A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotten M, Baker A, Birnstiel M L, Zatloukal K, Wagner E. Adenovirus polylysine DNA conjugates. In: Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. Current protocols in human genetics. New York, N.Y: John Wiley and Sons, Inc.; 1996. pp. 12.3.1–12.3.33. [Google Scholar]
- 10.Cotten M, Weber J M. The adenovirus protease is required for virus entry into host cells. Virology. 1995;213:494–502. doi: 10.1006/viro.1995.0022. [DOI] [PubMed] [Google Scholar]
- 11.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 12.Dales S, Chardonnet Y. Early events in the interaction of adenoviruses with HeLa cells. IV. Association with microtubules and the nuclear pore complex during vectorial movement of the inoculum. Virology. 1973;56:465–483. doi: 10.1016/0042-6822(73)90050-0. [DOI] [PubMed] [Google Scholar]
- 13.Defer C, Belin M T, Caillet-Boudin M L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glotzer J B, Saffrich R, Glotzer M, Ephrussi A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr Biol. 1997;7:326–337. doi: 10.1016/s0960-9822(06)00156-4. [DOI] [PubMed] [Google Scholar]
- 15.Greber U F, Kasamatsu H. Nuclear targeting of SV40 and adenovirus. Trends Cell Biol. 1996;6:189–195. doi: 10.1016/0962-8924(96)10016-7. [DOI] [PubMed] [Google Scholar]
- 16.Greber U F, Singh I, Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 1994;2:52–56. doi: 10.1016/0966-842x(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 17.Greber U F, Suomalainen M, Stidwill R P, Boucke K, Ebersold M W, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greber U F, Webster P, Weber J, Helenius A. The role of the adenovirus protease in virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 19.Greber U F, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 20.Hillen W, Gatz C, Altschmied L, Schollmeier K, Meier I. Control of expression of the Tn10-encoded tetracycline resistance genes. Equilibrium and kinetic investigation of the regulatory reactions. J Mol Biol. 1983;169:707–721. doi: 10.1016/s0022-2836(83)80166-1. [DOI] [PubMed] [Google Scholar]
- 21.Hinrichs W, Kisker C, Duvel M, Muller A, Tovar K, Hillen W, Saenger W. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- 22.Hird S N, Paulsen J E, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz S B. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology, third ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2149–2171. [Google Scholar]
- 24.Kuznetsov S A, Langford G M, Weiss D G. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- 25.Leopold P L, Ferris B, Grinberg I, Worgall S, Hackett N R, Crystal R G. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther. 1998;9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
- 26.Li E, Stupack D, Bokoch G M, Nemerow G R. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J Virol. 1998;72:8806–8812. doi: 10.1128/jvi.72.11.8806-8812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luby-Phelps K. Effect of cytoarchitecture on the transport and localization of protein synthetic machinery. J Cell Biochem. 1993;52:140–147. doi: 10.1002/jcb.240520205. [DOI] [PubMed] [Google Scholar]
- 28.Luftig R B, Weihing R R. Adenovirus binds to rat brain microtubules in vitro. J Virol. 1975;16:696–706. doi: 10.1128/jvi.16.3.696-706.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 30.Michou A I, Lehrmann H, Saltik M, Cotten M. Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J Virol. 1999;73:1399–1410. doi: 10.1128/jvi.73.2.1399-1410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miles B D, Luftig R B, Weatherbee J A, Weihing R R, Weber J. Quantitation of the interaction between adenovirus types 2 and 5 and microtubules inside infected cells. Virology. 1980;105:265–269. doi: 10.1016/0042-6822(80)90177-4. [DOI] [PubMed] [Google Scholar]
- 32.Moskalewski S, Thyberg J, Friberg U. Cold and metabolic inhibitor effects on cytoplasmic microtubules and the Golgi complex in cultured rat epiphyseal chondrocytes. Cell Tissue Res. 1980;210:403–415. doi: 10.1007/BF00220198. [DOI] [PubMed] [Google Scholar]
- 33.Provance D W, Jr, McDowall A, Marko M, Luby-Phelps K. Cytoarchitecture of size-excluding compartments in living cells. J Cell Sci. 1993;106:565–577. doi: 10.1242/jcs.106.2.565. [DOI] [PubMed] [Google Scholar]
- 34.Puvion-Dutilleul F, Puvion E. Sites of transcription of adenovirus type 5 genomes in relation to early viral DNA replication in infected HeLa cells. A high resolution in situ hybridization and autoradiographical study. Biol Cell. 1991;71:135–147. doi: 10.1016/0248-4900(91)90060-z. [DOI] [PubMed] [Google Scholar]
- 35.Rickwood R. Centrifugal methods for characterising macromolecules and their interactions. In: Rickwood D, editor. Preparative centrifugation, a practical approach. Oxford, United Kingdom: Oxford University Press; 1992. pp. 143–184. [Google Scholar]
- 36.Robinson A J, Younghusband H B, Bellett A J. A circular DNA-protein complex from adenoviruses. Virology. 1973;56:54–69. doi: 10.1016/0042-6822(73)90287-0. [DOI] [PubMed] [Google Scholar]
- 37.Roninson I, Padmanabhan R. Studies on the nature of the linkage between the terminal protein and the adenovirus DNA. Biochem Biophys Res Commun. 1980;94:398–405. doi: 10.1016/s0006-291x(80)80234-8. [DOI] [PubMed] [Google Scholar]
- 38.Sandoval I V, Bonifacino J S, Klausner R D, Henkart M, Wehland J. Role of microtubules in the organization and localization of the Golgi apparatus. J Cell Biol. 1984;99:S113–S118. doi: 10.1083/jcb.99.1.113s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaack J, Ho W Y, Freimuth P, Shenk T. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 1990;4:1197–1208. doi: 10.1101/gad.4.7.1197. [DOI] [PubMed] [Google Scholar]
- 40.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology, third ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2111–2148. [Google Scholar]
- 41.Shenk T. Group C adenovirus as vectors for gene therapy. In: Kaplitt M G, Loewy A D, editors. Viral vectors. San Diego, Calif: Academic Press; 1995. pp. 43–54. [Google Scholar]
- 42.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Straight A F, Belmont A S, Robinett C C, Murray A W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 44.Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strome S, Wood W B. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 46.Suomalainen M, Nakano M, Keller S, Boucke K, Stidwill R, Greber U. Microtubule-dependent plus and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svensson U, Persson R. Entry of adenovirus 2 into HeLa cells. J Virol. 1984;51:687–694. doi: 10.1128/jvi.51.3.687-694.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Huang S, Kapoor-Munshi A, Nemerow G. Adenovirus internalization and infection require dynamin. J Virol. 1998;72:3455–3458. doi: 10.1128/jvi.72.4.3455-3458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weatherbee J A, Luftig R B, Weihing R R. Binding of adenovirus to microtubules. II. Depletion of high-molecular-weight microtubule-associated protein content reduces specificity of in vitro binding. J Virol. 1977;21:732–742. doi: 10.1128/jvi.21.2.732-742.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wehland J, Henkart M, Klausner R, Sandoval I V. Role of microtubules in the distribution of the Golgi apparatus: effect of taxol and microinjected anti-alpha-tubulin antibodies. Proc Natl Acad Sci USA. 1983;80:4286–4290. doi: 10.1073/pnas.80.14.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 53.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]