Abstract
Previous studies have shown the presence of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8) DNA in endothelial cells, in keratinocytes in the basal layer of the epidermis overlying plaque-stage nodular lesions of cutaneous Kaposi's sarcoma (KS), and in the epithelial cells of eccrine glands within KS lesions. We infected primary cell cultures of human keratinocytes with KSHV/HHV8. At 6 days post infection, transcription of viral genes was detected by reverse transcriptase PCR (RT-PCR), and protein expression was documented by an immunofluorescence assay with an anti-LANA monoclonal antibody. To determine whether the viral lytic cycle was inducible by chemical treatment, KSHV/HHV8-infected keratinocytes were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA) and RT-PCR was performed to confirm the transcription of lytic genes such as open reading frame 26, (which encodes a capsid protein). Finally, to assess infectious viral production, other primary human cells (human umbilical vein endothelial cells), were infected with concentrated supernatant of KSHV-infected, TPA-induced keratinocytes and the presence of viral transcripts was confirmed by RT-PCR. The uninfected keratinocytes senesced 3 to 5 weeks after mock infection, while the KSHV/HHV8-infected keratinocytes continued to proliferate and to date are still in culture. However, 8 weeks after infection, viral genomes were no longer detectable by nested PCR. Although the previously KSHV/HHV8-infected keratinocytes still expressed epithelial markers, they acquired new characteristics such as contact inhibition loss, telomerase activity, anchorage-independent growth, and changes in cytokine production. These results show that KSHV/HHV8, like other herpesviruses, can infect and replicate in epithelial cells in vitro and suggest that in vivo these cells may play a significant role in the establishment of KSHV/HHV8 infection and viral transmission.
Kaposi's sarcoma (KS) is a multicentric vascular neoplasm involving the skin and mucosal surfaces and in aggressive cases may involve visceral organs and lymph nodes. KS lesions contain distinctive proliferating spindle cells, activated endothelial cells, fibroblasts, smooth muscle cells, and infiltrating inflammatory cells (35, 39). Four different epidemiologic forms of Kaposi's sarcoma have been described, i.e., the classic, endemic, iatrogenic, and AIDS-related forms (22, 47).
The gamma-2 herpesvirus KSHV, also known as human herpesvirus 8 (HHV8), has been detected in >95% of KS patients with all clinical forms of KS (22, 34, 47). KSHV/HHV8 DNA sequences are also present in primary effusion lymphomas (PEL) (8), a subset of B-cell lymphomas occurring more frequently in AIDS patients and in a significant percentage of patients with multicentric Castelman's disease (51). By in situ hybridization (5) and immunohistochemistry (7, 15), the results of several studies have documented the presence of KSHV/HHV8 in spindle and endothelial cells in KS lesions. Reed et al. (38) by analysis of the expression of the latently expressed v-cyclin gene, noted the presence of KSHV/HHV8 DNA in scattered keratinocytes of the epidermis overlying cutaneous lesions. Positive signals were also detected in eccrine ductular epithelial cells and in spindle and endothelial cells lining the well-formed blood vessels within and surrounding KS lesions (38). Similar results were described by Foreman et al. (20), who detected KSHV/HHV8-positive signals by in situ PCR in basal keratinocytes in the area immediately above some of the later-stage (i.e., plaque or tumor) KS lesions. Epstein-Barr virus (EBV), the human herpesvirus most closely related to KSHV/HHV8, has been detected both serologically and histologically in epithelial cells (42, 49, 56). Epithelial cells of many tissues, including salivary glands and kidneys, support the replication of cytomegalovirus, another herpesvirus that can also infect human keratinocytes (58).
In this study we investigated whether KSHV/HHV8 was capable of infecting primary cultures of human keratinocytes in vitro and analyzed the biological consequences of this infection. We demonstrate that KSHV/HHV8 can establish a productive infection in primary human keratinocytes and that viral particles purified from the supernatant of infected keratinocytes can infect other primary cells, such as human endothelial cells. Infection by KSHV/HHV8 resulted in alterations of the keratinocytes, including the development of a spindle-shaped cell morphology similar to that observed in other immortalized cell lines of epithelial origin (24), and the ability to form colonies in soft agar. We also observed an induced telomerase activity, which is one of the characteristic factors of transformed human cells (27).
While rodent cells from several normal tissues can undergo spontaneous neoplastic transformation after variable periods in culture (46), human cells are rather resistant in this respect (14). To exclude the possibility of spontaneous immortalization, the infection experiment was repeated eight times, and four out of eight KSHV-infected cultures did not senesced and to date are still proliferating. Moreover, if such cells are the outcome of spontaneous transformation, they might also be expected to arise in uninfected cultures; however, these were never observed and the uninfected cultures did not proliferate longer than 3 to 5 weeks.
MATERIALS AND METHODS
Cell cultures.
Primary normal human epidermal keratinocytes (NHEK) derived from newborn foreskin were obtained from Clonetics Corp. (San Diego, Calif.). Primary cultures of keratinocytes were established from histologically normal adult body skin. The homogeneity of the cultures was examined by immunohistochemistry staining using cytokeratin-specific antibodies for epithelial cells (CK AE1/AE3 and CAM 5.2). Four separate cultures of keratinocytes from Clonetics and four separate cultures derived from skin biopsy samples were used for the experiments.
Cells were grown in low-calcium, serum-free Keratinocyte-SFM medium (GIBCO BRL, Gaithersburg, Md.) supplemented with 100 U of penicillin per ml, 100 μg of streptomycin per ml, 2 mM glutamine, bovine pituitary extract (30 μg/ml), and recombinant epidermal growth factor (0.2 ng/ml) (GIBCO BRL). According to the manufacturer, this medium is selective for propagation of human keratinocytes.
BC-3, a KSHV/HHV8-positive, EBV-negative primary effusion lymphoma (PEL)-derived cell line (2), was cultured in RPMI 1640 supplemented with 20% fetal bovine serum. Primary human umbilical vein endothelial cells (HUVECs) were isolated from umbilical cord veins by collagenase treatment as described previously (6). Cells were grown in gelatin-coated flasks containing M199 medium (Bio-Whittaker, Walkersville, Md.) supplemented with 10 ng of vascular endothelial growth factor (VEGF) (Perprotech Inc., Rockville, Md.) per ml, 10 ng of basic fibroblast growth factor (bFGF) (R&D Systems Inc., Minneapolis, Minn.), per ml, and 10 U of heparin (Sigma, St. Louis, Mo.) per ml.
Viral infection.
Concentrated supernatant and cell-free lysates from 4 × 108 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced BC-3 cells were used as a source of KSHV/HHV8 for the infection of eight NHEK cultures: four derived from skin biopsy samples and four from Clonetics. Concentrated supernatant and cell-free lysates from KSHV-infected, TPA-induced NHEK cultures were used as a source of KSHV/HHV8 for the infection of two HUVEC cultures. Primary keratinocytes supplied by Clonetics are specified to be at passage 3. The cells were split once before the infection; therefore, all primary cell cultures (Clonetics and skin biopsy sample) we used were infected at passage 4. To release the virus in the medium, cells were treated for 48 h with 20 ng of TPA (Sigma) per ml and centrifuged and supernatant was collected. The cell pellet was lysed by three freeze-thaw cycles, centrifuged to eliminate cell debris, pooled with the supernatant, filtered through a 0.45-μm-pore-size filter, and concentrated at 4°C overnight with 7% polyethylene glycol 6000. The precipitate was collected by centrifugation at 15,000 × g for 2 h, resuspended in a small amount of phosphate-buffered saline (PBS), and purified through a 25% sucrose cushion (26,000 rpm for 3 h in a Beckman SW27 rotor). The pellet was allowed to soak overnight at 4°C in 1 ml of PBS with 0.1% bovine serum albumin, and the suspension was treated with DNase (1 U/ml; Promega) and RNase (5 μg/ml; Quiagen), layered over a 5 to 45% sucrose gradient in PBS, and centrifuged at 14,000 rpm for 1 h at 10°C in a Beckman SW41 rotor. The visible band at the center of the gradient was collected, dialyzed against PBS, and then frozen at −70°C. Since no protocol for determining the PFU of KSHV currently exists, viral DNA from purified particles was extracted with phenol-chloroform and the optical density was measured by spectrophotometry. From the optical density value, the DNA quantity was assessed in micrograms per milliliter, it was converted to moles per milliliter, and the number of molecules of viral genome was calculated from the number of moles. We estimated a yield in the range of 1.5 × 1010 to 2 × 1010 KSHV/HHV8 genome equivalents (DNA molecules) per ml of concentrated supernatant, depending on the viral preparation. Human primary keratinocytes were infected at passage 4 with 5 to 10 viral genome equivalents/cell.
Supernatant and cell pellet lysates from 106 KSHV-infected and TPA-induced NHEK cells were concentrated and purified as described above and used to infect two parallel primary HUVEC cultures.
Cells were incubated for 1 h at 37°C with purified viral particles, and then the specific medium was added: Keratinocyte-SFM in the keratinocyte cultures or M199 medium in the HUVEC cultures. After incubation for 48 h, the monolayers were washed twice to eliminate the inoculum, fresh medium was added, and the cells were maintained at 37°C. Viral infection experiments were repeated eight times.
PCR amplification.
To calculate the number of viral DNA molecules in infected keratinocytes, semiquantitative PCR was performed using the purified viral DNA from BC-3 cells as a standard. A fragment of 233 bp was amplified with KS330233 BamHI primers (10) and quantitated in serial dilutions from 100 to 10−8 amol/μl by electrophoresis on an ethidium bromide agarose gel followed by Southern blot hybridization with an internal probe. The range of DNA concentration measurable by regular PCR was from 100 to 10−5 amol/μl.
Genomic DNA was extracted from the 106 KSHV/HHV8-infected and uninfected NHEK cells by the phenol-chloroform method (45). The presence of KSHV/HHV8-specific DNA in infected NHEK cells was verified by amplifying the 233-bp fragment with KS330233 primers. BC-3 cells and uninfected keratinocytes were used as positive and negative controls, respectively. PCR products were loaded onto a 1.5% agarose gel, transferred to a nylon membrane, and hybridized with the same internal probe used for the standard DNA. Radiolabeling of the probe was performed with the RediPrime DNA-labeling system (Amersham International, Little Chalfont, England) and [32P]dCTP as specified by the manufacturer. Because the molar quantity of the standard viral DNA was known, the actual number of viral DNA molecules present in KSHV-infected keratinocytes was calculated by comparing the intensity of the band with that of the standard viral DNA. The total quantity of cellular genomic DNA extracted from 106 cells was quantitated, 100 ng was used for PCR, and this was multiplied back to obtain the total number of viral DNA molecules.
RT-PCR.
Total RNA was extracted from 106 infected and uninfected keratinocytes with Tri-reagent (Molecular Research Center, Inc., Cincinnati, Ohio) as specified by the manufacturer, and the extracted RNA was treated with RNase-free DNase (Boeringher Mannheim, Indianapolis, Ind.). RNA was reverse transcribed with the reverse transcription system (Promega, Madison, Wis.), and PCR was performed. The cellular β-actin gene was amplified from each sample as a control. The KSHV/HHV8 genes open reading frame 72 (ORF 72) (encoding v-cyclin) (18), ORF K12 (encoding kaposin), and ORF 26 (encoding the minor capsid protein) (10) were used as reverse transcriptase PCR (RT-PCR) targets. The primers used for K12 detection were 5′-GGATAGAGGCTTAACGGTGTTTGTG-3′ and (reverse) 5′-TGCAACTCGTGTCCTGAATGC-3′, with the following amplification protocol: 94°C for 3 min, then 35 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min. RT-PCR was also performed targeting the VEGF (12) and bFGF transcripts in infected and uninfected keratinocytes. The primers used to amplify bFGF were 5′-GGCCACTTCAAGGACCCCAAG-3′ and reverse 5′-TCAGCTCTTAGCAGACA-3′ with the following protocol: 94°C for 3 min, then 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min.
PCR products were loaded onto 1.5% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled internal probe.
Nested PCR.
The first PCR was performed with the following set of outer primers to amplify a fragment of 327 bp in the KSHV/HHV8 major capsid protein (MCP) gene: 5′-AGGCAACGTCAGATGTGAC-3′ and 5′-GAAATTACCCACGAGATCGC-3′. The conditions used for amplification were a 10-min denaturation at 94°C followed by 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, with a single 5-min extension at 72°C. The inner primers used for nested PCR were 5′-CATGGGAGTACATTGTCAGGACCTC-3′ and 5′-GGAATTATCTCGCAGGTTGCC-3′, and the condition for amplification were a 10-min denaturation at 94°C followed by 35 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 45 s, with a single 5-min extension at 72°C to amplify a 212-bp fragment. The enzyme used for the reaction was Taq Gold polymerase from Perkin-Elmer. Standard viral DNA was diluted 1/10 from 2 to 2 × 10−8 amol./μl.
IFA.
An immunofluorescence assay (IFA) was performed using serum from a KS patient as a source of primary antibody or a monoclonal antibody (MAb) against ORF 73 (Advance Biotechnology, Inc., Columbia, Md.) for latent antigens and a monoclonal antibody against ORF 59 (generous gift from Bala Chandran) for lytic antigens.
The cells were detached with trypsin, counted, spotted on slides (105/slide), air dried, and fixed in acetone for 10 min. The slides were blocked with 20% normal goat serum in PBS for 30 min and then incubated overnight at 4°C with the primary antibody. Fluorescein isothiocyanate-conjugated goat anti-mouse and anti-human sera were used as secondary antibodies (ICN/Cappel, Aurora, Ohio). Staining controls included omission of the primary antibody and staining of noninfected keratinocytes. Cell nuclei were counterstained with DAPI (4′,6-diamino-2-phenylindole dihydrochloride; Boehringer Mannheim).
Growth in soft agar.
Uninfected and KSHV/HHV8-infected keratinocytes were trypsinized and resuspended in medium at concentrations of 103, 104, and 105 cells/ml. A 5-ml volume of 0.6% melted agar, made up by mixing 9 parts of Keratinocyte-SFM standard medium and 1 part of 6% agar, was poured into each 60-mm plate to form a bottom layer. Then 2 ml of 0.6% melted agar-medium was added to 1 ml of the cell suspension, and 1 ml of this mixture was poured over the bottom layer to form a top layer. Each serial dilution was plated in triplicate. Cells were fed every week with 3 ml of 0.4% agar-medium and routinely observed by microscopy for colony formation.
Telomerase activity.
The enzymatic activity was measured by using a telomeric repeat amplification protocol (4) (TRAP assay kit; BD PharMingen, San Diego, Calif.).
Cytokine assay.
Cytokines were measured by enzyme-linked immunosorbent assays in a commercial laboratory (Cytokine Core Laboratory, Baltimore, Md.).
Immunohistochemistry staining.
Cells were processed by a standard method of immunohistochemistry. Briefly, the cells were fixed in phosphate-buffered formalin and embedded in paraffin. Deparaffinized sections were treated with 0.1% pepsin (Sigma) for 7 min and were reacted overnight at 4°C with the MAbs (Boeringer-Mannheim and DAKO, Carpinteria, Calif.) listed in Table 1. The sections were rinsed in PBS, and the bound antibodies were localized by the avidin-biotin-peroxidase method (Elite kit; Vector Laboratories, Burlingame, Calif.) using diaminobenzidine as the chromogen. Sections stained with normal mouse serum were used as negative controls.
TABLE 1.
Immunohistochemistry analysis of keratinocytes 3 months after infection with KSHV/HHV8
| MAb | Staining of keratinocytes |
|---|---|
| MNF 116 | + |
| AE1-AE3 | + |
| CAM 5.2 | − |
| Vimentin | + |
| S100 protein | − |
| HMB 45 | − |
| A 103 | − |
| CD31 | − |
| CD34 | − |
| Type 4 collagen | − |
| CD1 A | − |
Cell proliferation assay.
Cells were plated in a T-25 flask (5 × 105/flask), allowed to grow for 3 days, harvested with trypsin, diluted in trypan blue to assess viability, and counted in a Burker hemocytometer chamber.
RESULTS
KSHV can infect primary human keratinocytes.
To determine whether primary human keratinocytes were susceptible to KSHV/HHV8 infection, 106 cells were exposed to purified viral particles. The presence of KSHV/HHV8-specific DNA was confirmed by performing a PCR with the KS330233 primers 6 days postinfection on 100 ng of total DNA purified from infected cells (Fig. 1A). The results showed that without TPA treatment the number of viral DNA molecules/100 ng of template was 500 to 600, depending on the particular culture. The number of KSHV genomes in the cultures was small, suggesting that only about 5 to 6% of the cells in these culture were infected, assuming one copy per infected cell. This estimate was calculated as described in Materials and Methods.
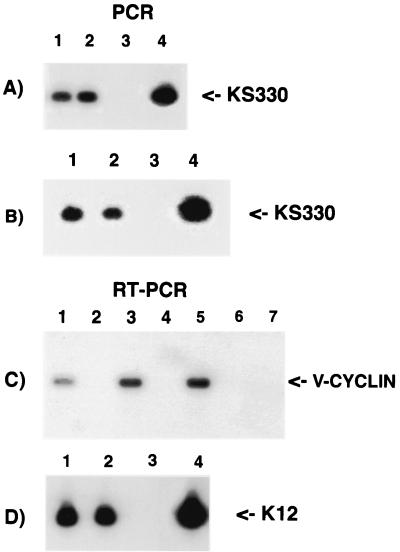
FIG. 1.
PCR and RT-PCR of 106 keratinocytes infected with KSHV/HHV8. (A) PCR with KS330 primers to test for the presence of viral DNA 6 days postinfection. Lanes: 1 and 2, two different cultures infected in parallel; 3, uninfected keratinocytes; 4, positive control (BC-3 cells). (B) PCR with KS330 primers to test the presence of KSHV/HHV8 genomes in infected keratinocytes 25 days postinfection. Lanes: 1 and 2, two different cultures infected in parallel; 3, uninfected keratinocytes; 4, BC-3 cells. (C) RT-PCR 6 days postinfection with primers specific for v-cyclin. Lanes: 1 and 3, RNA from two cell cultures infected in parallel; 2 and 4, RNA controls without reverse transcription showing no viral DNA contamination; 5, positive control (BC-3 cells); 6, uninfected keratinocytes; 7, primers with no template. (D) RT-PCR 6 days postinfection with primers specific for ORF K12. Lanes: 1 and 2, RNA from two cell cultures infected in parallel; 3, primers with no template; 4, positive control (BC-3 cells).
To determine whether the viral DNA was also transcribed, RT-PCR assays were performed. Reactions using primers specific for v-cyclin and K12 showed positive results in all infected cultures, documenting the presence of viral gene transcripts (Fig. 1C and D show two parallel infected cultures). The infected cells were maintained in culture and passaged every 3 to 5 days depending on the particular culture. After 4 weeks, the infected cell cultures were tested again by PCR with several primer sets scattered along the viral genome. Figure 1B shows PCR using KS330233 primers for amplification of viral DNA from two cultures infected in parallel. These analyses showed that the number of viral DNA molecules/100 ng of template was about 250, suggesting that at this time postinfection only about 2 to 3% of the cells contained detectable viral DNA, assuming one copy per infected cell. Comparison between the number of viral DNA in the cultures 6 days and 4 weeks postinfection indicated that the rate of decline was about 50 to 60% during this period. These experiments were repeated six additional times with similar results. After 8 weeks in culture, we could no longer detect any KSHV/HHV8 DNA by regular PCR. To ascertain whether the viral genome was not detectable because direct PCR was not sensitive enough, we performed nested PCR with outer and inner MCP primer sets. These primer sets amplify a region in the major capsid protein gene. Figure 2A shows nested PCR-Southern blot analyses of serial dilutions from 2 × 10−3 to 2 × 10−8 amol of viral DNA per μl extracted from purified KSHV/HHV8 particles. This analysis was used as a standard to quantitate the viral DNA concentration and to measure the sensitivity of the method. The lower detection limit was 2 × 10−6 amol/μl or approximately 1.2 KSHV equivalent molecules. Figure 2B shows the results of a nested PCR-Southern blot analysis of DNA extracted from three separate cultures of keratinocytes, 8 weeks postinfection. All nested PCRs that we performed with the keratinocyte cultures 8 weeks postinfection were negative. This means that the copy number of viral DNA relative to total cellular genome was smaller then the detection limit. Nested PCR is a very sensitive assay, and by this method we have been able to detect 1.2 KSHV DNA molecules in our standard. Therefore, it is unlikely that there are any viral genomes in the proliferating keratinocytes 8 weeks postinfection.
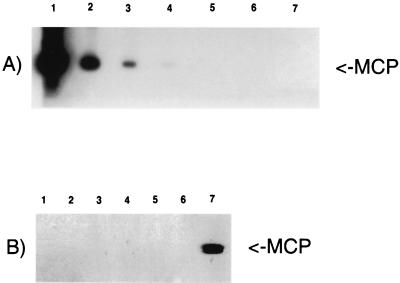
FIG. 2.
Nested PCRs with MCP outer and inner sets of primers. (A) Quantification by nested PCR of viral DNA extracted from purified particles. Lanes: 1 to 6, 10-fold serial dilutions from 2 × 10−3 to 2 × 10−8 amol/μl; 7, negative control (primers with no template). (B) Nested PCR of 106 keratinocytes 8 weeks after infection with KSHV. Lanes: 1 to 3, three keratinocytes cultures independently infected; 4 and 5, uninfected keratinocytes; 6, negative control (primers with no template); 7, positive control (BC-3 cells).
Expression of viral latent nuclear antigen.
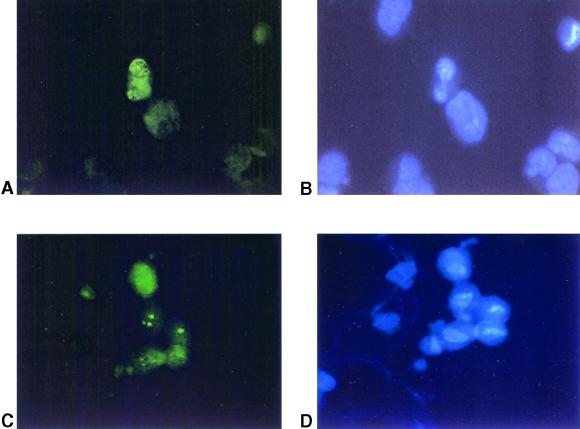
To further document the in vitro infection of primary keratinocytes with KSHV, the expression of latent viral antigens was examined by immunofluorescence. Serum from a KS patient (data not shown) or a MAb against ORF 73 was used as the primary antibody, and in both cases a punctate nuclear pattern was detected in 2 to 5% of cells in the infected culture (Fig. 3A and B). This result demonstrated the expression of viral proteins in KSHV/HHV8-infected keratinocytes. The IFA has been performed in all infected cultures 6 days and 4 weeks postinfection, and similar results have been obtained.
FIG. 3.
IFA of KSHV/HHV8-infected keratinocytes. (A and B) Detection of latent viral antigen. (A) Cells stained with anti-LANA MAb. Positive cells show speckled nuclear pattern. (B) Cells counterstained with DAPI to localize the nuclei. (C and D) Detection of lytic viral antigen after TPA induction. (C) Cells stained with MAb 11D1, raised against the ORF 59 product, DNA replication protein. Positive cells show characteristic nuclear staining. (D) Cells counterstained with DAPI.
Induction of KSHV lytic replication and viral serial transmission.
To determine whether KSHV/HHV8 could establish a productive infection in keratinocytes, infected cells were treated with TPA to induce lytic replication of KSHV/HHV8 and release viral particles into the medium (41). After 48 h of TPA treatment, induction of lytic gene transcription was detected by RT-PCR analysis of ORF 26 (encoding capsid protein) (Fig. 4A). Lytic gene expression was also confirmed by immunostaining of TPA-induced cells with a MAb against the ORF 59 product, a DNA-replication protein. The percentage of induced cells that expressed viral lytic antigens never exceeded 40% of the positive infected cells (Fig. 3C and D).
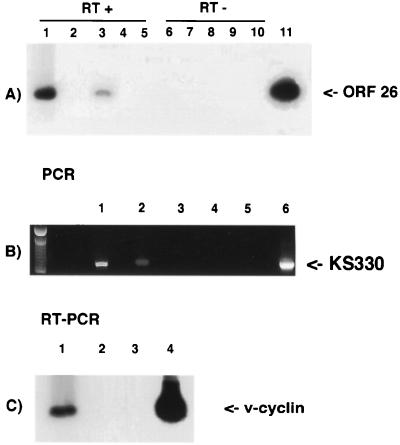
FIG. 4.
(A) RT-PCR analysis of 106 KSHV/HHV8-infected keratinocytes after treatment with TPA for 48 h. Primers are specific for ORF 26, a late lytic gene encoding a capsid protein. Lanes: 1 and 3, two different cultures infected in parallel and TPA induced; 2 and 4, the same cultures without TPA induction; 5, uninfected keratinocytes; 6 to 10, the same samples without reverse transcription; 11, positive control (BC-3 cells). (B) PCR analysis to confirm the release of viral progeny in the supernatants of TPA-induced keratinocytes. Viral DNA was amplified with KS330 primers. Lanes: 1 and 2, supernatants from two parallel cultures infected and TPA induced; 3 and 4, supernatants from uninfected keratinocytes; 5, primers with no template; 6, positive control (BC-3 cells). (C) Serial transmission. RT-PCR analysis of HUVECs infected with KSHV/HHV8 particles obtained from TPA-stimulated keratinocytes. DNA was amplified with primers specific for v-cyclin. Lanes: 1, KSHV/HHV8-infected HUVECs; 2, control with no RT; 3, primers with no template; 4, positive control (BC-3 cells).
To determine if productive viral replication had occurred, the supernatant from parallel cultures of KSHV/HHV8-infected, TPA-induced keratinocytes was collected and concentrated as described in Materials and Methods. Virus was purified on a sucrose gradient, and the presence of KSHV/HHV8 genome was confirmed by PCR analysis using primers KS330 as shown in Fig. 4B. To demonstrate that the isolated viral particles were infectious, new primary cultures of HUVECs were infected with the virus purified from the infected keratinocytes. At 6 days after infection, we performed an RT-PCR amplification using primers within the v-cyclin gene of KSHV, and positive RT-PCR demonstrated that viral transmission in the HUVEC culture was achieved (Fig. 4C). At 2 weeks postinfection, the HUVECs acquired the typical spindle shape observed in KSHV/HHV8-infected endothelial cells (reference 19 and data not shown).
Effect of KSHV infection on the survival and growth properties of keratinocytes.
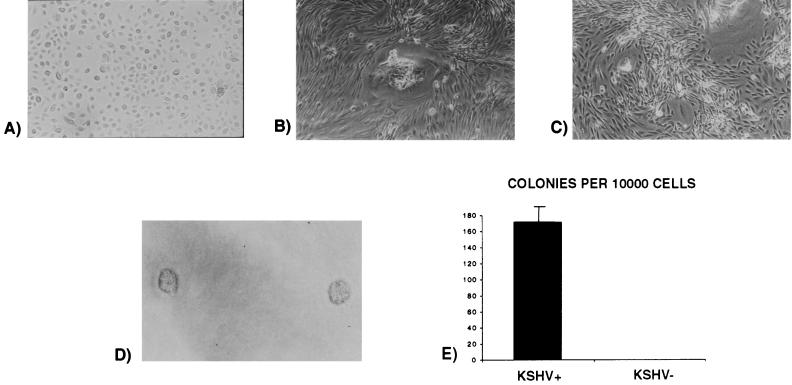
Infected and uninfected cells were passaged at the same rate. After seven passages (3 to 5 weeks, depending on the particular culture) the uninfected cell cultures (Fig. 5A) stopped proliferating and trypsinized cells did not adhere to fibronectin-coated flasks as well as the majority of the cells in the infected cultures. However, a new population of cells in the same infected flask began to proliferate after developing adherent foci (Fig. 5B). In these new cultures derived from adherent foci, KSHV/HHV8 was still detectable by PCR 4 weeks postinfection, and the cells showed a spindle-shaped morphology, loss of contact inhibition (Fig. 5C), and independence of growth factors specific for keratinocytes (epidermal growth factor and bovine pituitary extract). These cells were also capable of forming colonies in soft agar (Fig. 5D). Of 104 cells plated, 168 colonies (range, 105 to 256) were obtained; thus, only∼2% of the cells plated resulted in colony formation (Fig. 5E).
FIG. 5.
Spindle shape acquisition of KSHV/HHV8 infected-keratinocytes as shown by phase contrast microscopy. (A) Primary human keratinocytes. (B) Infected keratinocytes after 4 weeks in culture (focus formation). (C) Infected keratinocytes after 4 weeks in culture (loss of contact inhibition). (D) Infected keratinocyte-forming colonies in soft agar. (E) Number of colonies grown in agar after plating 104 cells per plate (experiments were done in triplicate).
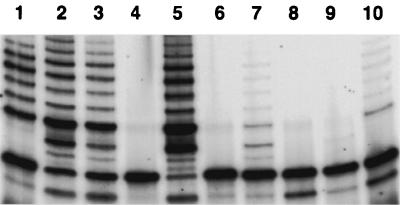
Several studies have demonstrated that telomerase is essential to maintain telomere length and the ability of cells to proliferate indefinitely (27). Although the viral genome was no longer detectable by nested PCR after 8 weeks in culture, the previously KSHV/HHV8-infected keratinocytes continued to proliferate and to date are still in culture. Therefore, we measured the telomerase activity of all cultures previously infected with KSHV/HHV8 and maintained in culture for 8 weeks and one culture of mock-infected keratinocytes, using a TRAP. We could not test any mock-infected cells after 8 weeks in culture because, as we stated above, uninfected cells did not survive longer then 3 to 5 weeks, depending on the particular culture. Only the cell cultures previously positive for KSHV/HHV8 exhibited telomerase activity, which was not detectable in mock-infected keratinocytes (Fig. 6).
FIG. 6.
Telomerase activity in previously KSHV/HHV8-infected keratinocytes after 8 weeks in culture. Lanes: 1 to 3, telomerase-positive cell lines (positive controls); 4, positive control after heat treatment; 5, telomeric DNA; 6, normal keratinocytes (negative control); 7 and 10, two parallel cultures of previously infected keratinocytes showing the presence of telomerase activity; 8 and 9, same cultures after heat treatment.
Immunohistochemistry.
Since the long-term-proliferating keratinocytes established from cultures previously infected by KSHV/HHV8 displayed changes in their morphology, it was important to exclude the possibility that these cells represented contamination with other cell types in the original cultures. By immunohistochemical staining we investigated whether the long-term-proliferating keratinocytes established from cultures previously infected by KSHV/HHV8 still retained the markers specific for keratinocytes or expressed other cellular surface molecules. Panels of antibodies specific for keratinocytes (AE1-AE3 mixture, CAM 5.2, MNF116 [37]), endothelial cells (CD31, CD34), dendritic cells (CD1A), melanocytes (S100-protein, HMB 45), and fibroblasts (type 4 collagen) were tested. Immunohistochemical detection showed positive staining for the AE1-AE3 mixture and for cytokeratin 5, 6, 8, 17, and 19 (MNF116 antibody), while molecular markers for other cell types were negative, except for vimentin, which is expressed by most spindle cells (23) (Table 1).
Human cytokine production.
Recently it has been demonstrated that inflammatory cytokines can modulate KSHV/HHV8 replication (9). Since there were differences in the proliferative capacity of uninfected keratinocytes and previously KSHV/HHV8-infected long-term-proliferating keratinocytes, possibly due to higher production or responsiveness to endogenous cytokines, we analyzed the cytokine expression pattern of both uninfected and previously KSHV/HHV8-infected keratinocytes 3 months postinfection. The latter cells expressed higher levels of IL-6 and lower levels of IL-8 than did the uninfected controls (Table 2). The levels of other cytokines (macrophage inflammatory protein 1α [MIP-1α], MIP-1β, gamma interferon (IFN-γ), tumor necrosis factor alpha [TNF-α], soluble TNF-R1, soluble TNF-R2, and RANTES) were not significantly different in uninfected and previously KSHV/HHV8-infected cells.
TABLE 2.
Production of human cytokines by keratinocytes 3 months after infection with KSHV/HHV8
| Cytokine | Cytokine concn (pg/ml) produced by:
|
|
|---|---|---|
| Uninfected keratinocytes | Infected keratinocytes | |
| MIP-1α | <1 | 28.93 |
| MIP-1β | 4.83 | 0.18 |
| IL-6 | 172.07 | 910.94 |
| IL-8 | 504.86 | 20.85 |
| IFN-γ | 4.80 | <1 |
| TNF-α | 3.7 | 3.2 |
| sTNF-R1 | 572.84 | 434.64 |
| sTNF-R2 | <1 | 49.2 |
| RANTES | 168.33 | 74.79 |
VEGF and bFGF gene transcription.
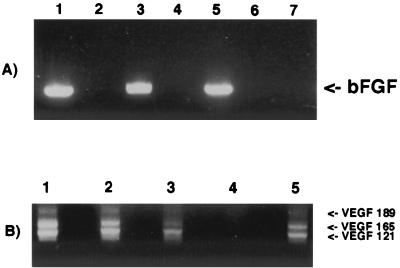
VEGF and bFGF play a key role in the growth of KS lesions (13, 32, 44), and normal human keratinocytes are a prominent source of VEGF (55). To evaluate the levels of VEGF and bFGF transcripts in the previously KSHV/HHV8-infected long-term-proliferating keratinocytes, we performed RT-PCR assays after 3 months of infection. Figures 7A (bFGF) and B (VEGF) show no significant differences in the transcription levels of the previously KSHV/HHV8-infected, long-term-proliferating keratinocytes and of normal primary keratinocytes.
FIG. 7.
Transcription levels of bFGF and VEGF genes in 106 infected and uninfected keratinocytes. (A) bFGF. Lanes: 1 and 3, two parallel cultures of previously KSHV/HHV8-infected keratinocytes, 3 months after the infection; 5, normal primary keratinocytes; 2, 4, and 6, controls with no RT; 7, primers with no template. (B) VEGF. Lanes: 1 and 2, previously KSHV/HHV8-infected keratinocytes in culture for 3 months; 3, normal primary keratinocytes; 4, primers with no template; 5, positive control (BC-3 cells). No differences in transcription were seen.
DISCUSSION
This report describes the in vitro infection of primary human keratinocytes with KSHV/HHV8. Other authors have shown, by either in situ hybridization or in situ PCR, the presence of KSHV/HHV8 DNA and RNA in scattered keratinocytes in the epidermis overlying KS lesions (20, 38). These observations suggest that different human cells, other then endothelial cells and lymphocytes, are permissive for KSHV/HHV8 infection and replication. Moreover, detection of infectious viral particles in the saliva of KSHV/HHV8-infected patients suggests that the epithelial cells in the oral mucosa or in salivary glands are a source of the virus (36, 54). Other studies have demonstrated that the human embryonal-kidney epithelial cell line 293 and the owl monkey kidney OMK 637 cell line (21, 40) can support KSHV/HHV8 infection in vitro. Since these cell lines are of epithelial origin but are immortalized, we decided to assay primary human epithelial cells, in particular keratinocytes, for their susceptibility to viral infection and to study the biological consequence of the infection. In the work presented here we demonstrate that KSHV/HHV8 can indeed infect and replicate in primary human keratinocytes. On treatment with TPA, PEL-derived cell lines and in vitro-infected endothelial cells undergo the complete program of KSHV/HHV8 gene expression, resulting in viral replication and release of mature virions (19, 29, 35, 50). In primary keratinocytes, KSHV/HHV8 conforms to this pattern as well. We show that in latently KSHV/HHV8-infected keratinocytes, TPA treatment can induce the viral lytic cycle and lead to the production of mature virions that can be serially transmitted to other primary human cells such as HUVECs. As previously reported for endothelial cells (19), the cells surviving the initial viral infection continued to proliferate. We have continued to propagate these cells, and they are still in culture, although the rate of proliferation 9 months postinfection is lower then it was 3 months postinfection. On the other hand, uninfected controls senesced after seven passages in culture (3 to 5 weeks, depending on the particular culture). The long-term-proliferating keratinocytes displayed several changes: modification of cell morphology with acquisition of a spindle shape, described also in several cell lines of epithelial origin (24), loss of contact inhibition, and acquisition of anchorage-independent growth. The presence of telomerase activity in keratinocytes 8 weeks after infection with KSHV/HHV8 provides additional evidence of a role for this virus in the long-term survival of keratinocytes in culture (27). These results are consistent with those obtained with KSHV/HHV8-infected primary endothelial cells (19). Similar observations were made for KSHV-infected dermal microvascular endothelial cells (35), although these cells were already immortalized by infection with a recombinant retrovirus and telomerase activity could not be tested. Examination of the phenotype of KSHV/HHV8-infected cells by immunohistochemical analyses excluded infection and expansion of other contaminating cell types, since all long-term-proliferating keratinocytes were cytokeratin positive, especially with MAb MNF116, which stains mainly basal keratinocytes (37). All other cellular markers were negative except vimentin. Phenotypic changes, such as vimentin expression, resemble an epithelial-to-mesenchymal transition that can occur when epithelial cells undergo malignant transformation such as in human squamous carcinoma (1, 23, 31, 53).
Cytokines produced by epidermal keratinocytes may play a significant role as regulators in inflammation and host immune response. Long-term-proliferating keratinocytes exhibited a different pattern of human cytokine production from normal primary human keratinocytes. Interestingly, expression of human interleukin-6 IL-6, a multifunctional cytokine that plays a role in neovascularization and wound healing and has also been implicated in the pathogenesis of KS (30, 33), was highly upregulated. By contrast, expression of IL-8, a proinflammatory cytokine that plays a role in the host defense mechanism through its effects on neutrophil activation (3), was downregulated. These results suggest that viral factors may play a primary role in modification of the biologic behavior of different cell types. Since bFGF and VEGF are potent angiogenic growth factors that are highly expressed in KS spindle cells and in primary KS lesions (16, 17, 43, 57), we investigated the transcript levels of bFGF and VEGF genes. No significant differences were found between infected and uninfected cells, suggesting that neither bFGF nor VEGF is particularly involved at this stage in the proliferating mechanisms of the previously KSHV/HHV8-infected keratinocytes. We hypothesize that the ability of KSHV/HHV8 to replicate in primary keratinocytes, in particular in mucosal sites, is probably one of the first steps in clinical infection. From there, the virus may reach the endothelial cells locally or via circulating hematopoietic cells and contribute to the formation of the vascular lesions typical of KS. This hypothesis is in line with the infection modalities of the closest related known human herpesvirus, EBV, which is associated with lymphoid and epithelial lesions (25, 26, 28). EBV infects epithelial cells in vivo (49, 52, 56), transforms epithelial cells, and inhibits cell differentiation (48). Several studies have also demonstrated that nasopharyngeal carcinoma cell lines and keratinocyte cell lines can be infected with EBV by cell-to-cell contact (11). Recently it has been shown that mucosal shedding of KSHV/HHV8 occurred in men who were infected with this virus but did not have KS. This suggests that oral-oral contact might play a significant role in transmission (36).
In conclusion, we have shown that KSHV/HHV8, like other herpesviruses, can productively infect epithelial cells. These results indicate that keratinocytes can support KSHV/HHV8 infection and replication and are consistent with the notion that epithelial cells, particularly those in mucosal sites, are likely to be a primary site of infection from which the virus may reach the endothelial cells or can be amplified in the locally infiltrating B lymphocytes and then spread to other tissues.
ACKNOWLEDGMENTS
We thank Claudio Basilico and Jan T. Vilcek for helpful discussions, Michael Bouchard for critical reading of the manuscript, and Amy Chadburn and Elizabeth M. Hyjek for assistance with the immunohistochemistry. We also thank Iraida Sharra-Pagano and Ron Liebman for providing keratinocytes and endothelial cells, respectively, and Lily Ying for technical help.
Francesca Curreli was supported by a fellowship from FIRC (Federazione Italiana per la Ricerca sul Cancro). This work was supported by the Howard Gilman Foundation and by the Center for AIDS Research (CFAR).
REFERENCES
- 1.Andreoli J M, Trevor K T. Structural and biological consequences of increased vimentin expression in simple epithelial cell types. Cell Motil Cytoskeleton. 1995;32:10–25. doi: 10.1002/cm.970320103. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Atta-ur R, Harvey K, Siddiqui R A. Interleukin-8: An autocrine inflammatory mediator. Curr Pharm Des. 1999;5:24–53. [PubMed] [Google Scholar]
- 4.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 6.Bussolino F, Arese M, Montrucchio G, Barra L, Primo L, Benelli R, Sanavio F, Aglietta M, Ghigo D, Rola-Pleszczynski M R, et al. Platelet activating factor produced in vitro by Kaposi's sarcoma cells induces and sustains in vivo angiogenesis. J Clin Investig. 1995;96:940–52. doi: 10.1172/JCI118142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannon J S, Nicholas J, Orenstein J M, Mann R B, Murray P G, Browning P J, DiGiuseppe J A, Cesarman E, Hayward G S, Ambinder R F. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J Infect Dis. 1999;180:824–828. doi: 10.1086/314956. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 9.Chang J, Renne R, Dittmer D, Ganem D. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology. 2000;266:17–25. doi: 10.1006/viro.1999.0077. [DOI] [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Chang Y, Tung C, Huang Y, Lu J, Chen J, Tsai C. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J Virol. 1999;73:8857–8866. doi: 10.1128/jvi.73.10.8857-8866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung N, Wong M P, Yuen S T, Leung S Y, Chung L P. Tissue-specific expression pattern of vascular endothelial growth factor isoforms in the malignant transformation of lung and colon. Hum Pathol. 1998;29:910–914. doi: 10.1016/s0046-8177(98)90195-2. [DOI] [PubMed] [Google Scholar]
- 13.Cornali E, Zietz C, Benelli R, Weninger W, Masiello L, Breier G, Tschachler E, Albini A, Sturzl M. Vascular endothelial growth factor regulates angiogenesis and vascular permeability in Kaposi's sarcoma. Am J Pathol. 1996;149:1851–1869. [PMC free article] [PubMed] [Google Scholar]
- 14.DiPaolo J A. Relative difficulties in transforming human and animal cells in vitro. JNCI. 1983;70:3–8. [PubMed] [Google Scholar]
- 15.Dupin N, Grandaman M, Calvez V, Gorin i, Aubin T, Havard S, Lamy F, Leibowitch M, Hauraux J, Escande J, Augut H. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi's sarcoma. Lancet. 1995;345:761–762. doi: 10.1016/s0140-6736(95)90642-8. [DOI] [PubMed] [Google Scholar]
- 16.Ensoli B, Markham P, Kao V, Barillari G, Fiorelli V, Gendelman R, Raffeld M, Zon G, Gallo R C. Block of AIDS-Kaposi's sarcoma (KS) cell growth, angiogenesis, and lesion formation in nude mice by antisense oligonucleotide targeting basic fibroblast growth factor. A novel strategy for the therapy of KS. J Clin Investig. 1994;94:1736–1746. doi: 10.1172/JCI117521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ensoli B, Nakamura S, Salahuddin S Z, Biberfeld P, Larsson L, Beaver B, Wong-Staal F, Gallo R C. AIDS-Kaposi's sarcoma-derived cells express cytokines with autocrine and paracrine growth effects. Science. 1989;243:223–226. doi: 10.1126/science.2643161. [DOI] [PubMed] [Google Scholar]
- 18.Flore O, Gao S J. Effect of DNA synthesis inhibitors on Kaposi's sarcoma-associated herpesvirus cyclin and major capsid protein gene expression. AIDS Res Hum Retroviruses. 1997;13:1229–1233. doi: 10.1089/aid.1997.13.1229. [DOI] [PubMed] [Google Scholar]
- 19.Flore O, Rafii S, Ely S, O'Leary J J, Hyjek E M, Cesarman E. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 20.Foreman K E, Bacon P E, Hsi E D, Nickoloff B J. In situ polymerase chain reaction-based localization studies support role of human herpesvirus-8 as the cause of two AIDS-related neoplasms: Kaposi's sarcoma and body cavity lymphoma. J Clin Investig. 1997;99:2971–2978. doi: 10.1172/JCI119492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 23.Islam S, Kim J B, Trendel J, Wheelock M J, Johnson K R. Vimentin expression in human squamous carcinoma cells: relationship with phenotypic changes and cadherin-based cell adhesion. J Cell Biochem. 2000;78:141–150. doi: 10.1002/(sici)1097-4644(20000701)78:1<141::aid-jcb13>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 24.Jones J C. Epithelia: advances in cell physiology and cell culture. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1990. pp. 342–345. [Google Scholar]
- 25.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 26.Kim K I, Kim Y S, Kim H K, Chae Y S, Yoem B W, Kim I. The detection of Epstein-Barr virus in the lesions of salivary glands. Pathol Res Pract. 1999;195:407–412. doi: 10.1016/S0344-0338(99)80014-4. [DOI] [PubMed] [Google Scholar]
- 27.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi R, Takeuchi H, Sasaki M, Hasegawa M, Hirai K. Detection of Epstein-Barr virus infection in the epithelial cells and lymphocytes of non-neoplastic tonsils by in situ hybridization and in situ PCR. Arch Virol. 1998;143:803–813. doi: 10.1007/s007050050332. [DOI] [PubMed] [Google Scholar]
- 29.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z Y, Ganju R K, Wang G F, Ona M A, Hatch W C, Zheng T, Avraham S, Gill P, Groopman J E. Cytokine signaling through the novel tyrosine kinase RAFTK in Kaposi's sarcoma cells. J Clin Investig. 1997;99:1798–1804. doi: 10.1172/JCI119344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell M J. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masood R, Cai J, Zheng T, Smith D L, Naidu Y, Gill P S. Vascular endothelial growth factor/vascular permeability factor is an autocrine growth factor for AIDS-Kaposi sarcoma Proc. Natl Acad Sci USA. 1997;94:979–984. doi: 10.1073/pnas.94.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles S A, Rezai A R, Salazar-Gonzalez J F, Vander Meyden M, Stevens R H, Logan D M, Mitsuyasu R T, Taga T, Hirano T, Kishimoto T, et al. AIDS Kaposi sarcoma-derived cells produce and respond to interleukin 6. Proc Natl Acad Sci USA. 1990;87:4068–4072. doi: 10.1073/pnas.87.11.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore P S, Chang Y. Detection of Herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and those without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 35.Moses A V, Fish K N, Ruhl R, Smith P P, Strussenberg J G, Zhu L, Chandran B, Nelson J A. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–6902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauk J, Huang M L, Brodie S J, Wald A, Koelle D M, Schacker T, Celum C, Selke S, Corey L. Mucosal shedding of human herpesvirus 8 in men. N Engl J Med. 2000;343:1369–1377. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- 37.Prieto V G, Lugo J, McNutt N S. Intermediate- and low-molecular-weight keratin detection with the monoclonal antibody MNF116. An immunohistochemical study on 232 paraffin-embedded cutaneous lesions. J Cutaneous Pathol. 1996;23:234–241. doi: 10.1111/j.1600-0560.1996.tb01472.x. [DOI] [PubMed] [Google Scholar]
- 38.Reed J A, Nador R G, Spaulding D, Tani Y, Cesarman E, Knowles D M. Demonstration of Kaposi's sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposi's sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood. 1998;91:3825–3832. [PubMed] [Google Scholar]
- 39.Regezi J A, MacPhail L A, Daniels T E, Greenspan J S, Greenspan D, Dodd C L, Lozada-Nur F, Heinic G S, Chinn H, Silverman S, Jr, et al. Oral Kaposi's sarcoma: a 10-year retrospective histopathologic study. J Oral Pathol Med. 1993;22:292–297. doi: 10.1111/j.1600-0714.1993.tb01075.x. [DOI] [PubMed] [Google Scholar]
- 40.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 42.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 43.Samaniego F, Markham P D, Gallo R C, Ensoli B. Inflammatory cytokines induce AIDS-Kaposi's sarcoma-derived spindle cells to produce and release basic fibroblast growth factor and enhance Kaposi's sarcoma-like lesion formation in nude mice. J Immunol. 1995;154:3582–3592. [PubMed] [Google Scholar]
- 44.Samaniego F, Markham P D, Gendelman R, Watanabe Y, Kao V, Kowalski K, Sonnabend J A, Pintus A, Gallo R C, Ensoli B. Vascular endothelial growth factor and basic fibroblast growth factor present in Kaposi's sarcoma (KS) are induced by inflammatory cytokines and synergize to promote vascular permeability and KS lesion development. Am J Pathol. 1998;152:1433–1443. [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sanford K K, Evans V J. A quest for the mechanism of “spontaneous” malignant transformation in culture with associated advances in culture technology. JNCI. 1982;68:895–913. [PubMed] [Google Scholar]
- 47.Schalling M, Ekman M, Kaaya E E, Linde A, Biberfeld P. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat Med. 1995;1:707–708. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- 48.Scholle F, Bendt K M, Raab-Traub N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J Virol. 2000;74:10681–10689. doi: 10.1128/jvi.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sixbey J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 50.Smith M S, Bloomer C, Horvat R, Goldstein E, Casparian J M, Chandran B. Detection of human herpesvirus 8 DNA in Kaposi's sarcoma lesions and peripheral blood of human immunodeficiency virus-positive patients and correlation with serologic measurements. J Infect Dis. 1997;176:84–93. doi: 10.1086/514043. [DOI] [PubMed] [Google Scholar]
- 51.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, D'Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 52.Takasaka N, Tajima M, Okinaga K, Satoh Y, Hoshikawa Y, Katsumoto T, Kurata T, Sairenji T. Productive infection of Epstein-Barr virus (EBV) in EBV-genome-positive epithelial cell lines (GT38 and GT39) derived from gastric tissues. Virology. 1998;247:152–159. doi: 10.1006/viro.1998.9231. [DOI] [PubMed] [Google Scholar]
- 53.van der Velden L A, Schaafsma H E, Manni J J, Ruiter D J, Ramaekers C S, Kuijpers W. Cytokeratin and vimentin expression in normal epithelium and squamous cell carcinomas of the larynx. Eur Arch Otorhinolaryngol. 1997;254:376–383. doi: 10.1007/BF01642554. [DOI] [PubMed] [Google Scholar]
- 54.Vieira J, Huang M L, Koelle D M, Corey L. Transmissible Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with history of Kaposi's sarcoma. J Virol. 1997;71:7083–7087. doi: 10.1128/jvi.71.9.7083-7087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weninger W, Rendl M, Mildner M, Tschachler E. Retinoids downregulate vascular endothelial growth factor/vascular permeability factor production by normal human keratinocytes. J Investig Dermatol. 1998;111:907–911. doi: 10.1046/j.1523-1747.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- 56.Wolf H, Haus M, Wilmes E. Persistence of Epstein-Barr virus in the parotid gland. J Virol. 1984;51:795–798. doi: 10.1128/jvi.51.3.795-798.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xerri L, Hassoun J, Planche J, Guigou V, Grob J J, Parc P, Birnbaum D, deLapeyriere O. Fibroblast growth factor gene expression in AIDS-Kaposi's sarcoma detected by in situ hybridization. Am J Pathol. 1991;138:9–15. [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu J. Cytomegalovirus infection induces expression of 60 KD/Ro antigen on human keratinocytes. Lupus. 1995;4:396–406. doi: 10.1177/096120339500400511. [DOI] [PubMed] [Google Scholar]