Abstract
Coxsackievirus A21 (CAV21), like human rhinoviruses (HRVs), is a causative agent of the common cold. It uses the same cellular receptor, intercellular adhesion molecule 1 (ICAM-1), as does the major group of HRVs; unlike HRVs, however, it is stable at acid pH. The cryoelectron microscopy (cryoEM) image reconstruction of CAV21 is consistent with the highly homologous crystal structure of poliovirus 1; like other enteroviruses and HRVs, CAV21 has a canyon-like depression around each of the 12 fivefold vertices. A cryoEM reconstruction of CAV21 complexed with ICAM-1 shows all five domains of the extracellular component of ICAM-1. The known atomic structure of the ICAM-1 amino-terminal domains D1 and D2 has been fitted into the cryoEM density of the complex. The site of ICAM-1 binding within the canyon of CAV21 overlaps the site of receptor recognition utilized by rhinoviruses and polioviruses. Interactions within this common region may be essential for triggering viral destabilization after attachment to susceptible cells.
Enteroviruses, parechoviruses, and human rhinoviruses (HRVs) are closely related viruses belonging to the family Picornaviridae. Based on genome organization and RNA sequences, the genera Rhinovirus and Enterovirus are more closely related to each other than to the rest of the picornaviruses (15). Whereas HRVs are labile at acid pH, enteroviruses and parechoviruses are stable over a wider pH range. In general, the serotypes within each genus have greater sequence similarity to each other than they have to viruses in other genera. Coxsackieviruses, polioviruses, some echoviruses, and some other viruses are all classified as enteroviruses but are differentiated by their pathogenic properties. Historically, coxsackieviruses have been subdivided into 23 serotypes belonging to group A and 6 serotypes belonging to group B (32, 42). These groups are differentiated primarily by the type of lesions observed on infected newborn mice. More recently, coxsackieviruses have been further subdivided and assigned to distinct “clusters” within the Enterovirus genus based on evolutionary kinship (genotypes). Accordingly, coxsackie A virus serotypes 1, 11, 13, 15, 17, and 18 to 24 and poliovirus serotypes 1 to 3 have been combined into cluster C, although they cause different diseases (15, 38). All cluster C coxsackie A viruses produce common cold-like symptoms, clinically indistinguishable from those of HRV infections. Furthermore, coxsackievirus A13 (CAV13), CAV18, and CAV21 have been shown to utilize as their cellular receptor intercellular adhesion molecule 1 (ICAM-1, or CD54) (9, 43), the same cell surface molecule as that used by the major group of HRVs to gain entry into cells (13, 49). HRV infections cause the up-regulation of ICAM-1 and, hence, the recruitment of leukocytes to sites of inflammation (37). Whether a similar up-regulation occurs for CAV21 is not known at this time.
Many viruses utilize accessory cell surface molecules for cell recognition or cell entry (11). For instance, CAV21 can also bind to decay-accelerating factor (DAF, or CD55), although without causing an infection (44). The ICAM-1 and DAF binding sites appear to be nonoverlapping although spatially close to each other (45). The structures of ICAM-1 and DAF are totally different. The extracellular component of ICAM-1 consists of five tandem immunoglobulin superfamily (IgSF) domains, whereas DAF is comprised of four short consensus repeat motifs and a serine-threonine-rich region that links the protein via a glycosyl-phosphotidylinositol linker to the plasma membrane.
The CAV21 RNA genome has about 80% amino acid sequence identity to that of polioviruses, which utilize CD155 as their receptor (4, 19, 53), but only about 50% identity to that of HRVs (24). Although there is a slight preference for codon usage resembling that of HRVs (24), these sequence comparisons do not explain the receptor preference for CAV21 and related cluster C coxsackieviruses. Similarly, the minor group of HRVs has no obvious phylogenetic separation from the major group of HRVs, yet all minor-group HRVs utilize the low-density lipoprotein receptor to initiate infection (21).
The crystal structures of various HRVs and enteroviruses have been determined at nearly atomic resolution. These include HRV14 (41) and HRV16 (16, 35), both of which utilize ICAM-1 as a receptor; HRV1A (26) and HRV2 (51), which do not bind ICAM-1; coxsackievirus B3 (33), which utilizes the coxsackievirus-adenovirus receptor as well as DAF; CAV9 (20), which utilizes an integrin as a receptor (39); and all three poliovirus serotypes (22, 29, 54), which utilize the poliovirus receptor, CD155 (4, 19, 53). However, the three-dimensional atomic resolution structure of CAV21 has yet to be determined.
ICAM-1 is a cell surface molecule that normally binds to leukocyte function-associated antigen 1 to provide adhesion between leukocytes and endothelial cells. Apart from being recruited by viral pathogens, such as HRVs and some coxsackieviruses, as a receptor, ICAM-1 also binds to erythrocytes that have been infected by the malarial parasite Plasmodium falciparum (3, 5, 34). The structure of the two amino-terminal domains of ICAM-1, designated D1D2, has been determined to nearly atomic resolution, in both partially unglycosylated (3) and fully glycosylated (7, 28) forms. The amino-terminal domain D1 was found to have an “intermediate” type of IgSF fold, whereas the second domain, D2, has a constant type 2 IgSF structure (8, 18, 52). Amino acid sequence analyses (48) predict that the other three domains also have IgSF-type folds.
The structures of glycosylated and unglycosylated, two- and five-domain ICAM-1, when complexed with HRV16 (28, 36) or HRV14 (28), have been determined to a resolution of about 28 Å by cryoelectron microscopy (cryoEM). By fitting the known X-ray structures of ICAM-1 and HRVs into the envelope of the cryoEM reconstructions of virus-receptor complexes, it has been possible to determine the nature of the virus-receptor interactions. Similar results were obtained for the complex formed between poliovirus and CD155 (4, 19, 53), although the structure of CD155 was based upon homology modeling. In every case, the receptor is a long, thin, flexible molecule that binds into the canyon, an ∼15-Å-deep surface depression on rhinoviruses and enteroviruses. These results are consistent with the prediction that receptor molecules would be sufficiently thin to bind to the more conserved residues in the canyon, at a site that is less accessible to neutralizing antibodies (40, 41).
We report here a cryoEM image reconstruction, at a resolution of 26 Å, of CAV21 complexed with its primary receptor, ICAM-1, in which all five domains of the receptor molecule are visible. Although domain D1 of ICAM-1 binds to a similar site in the CAV21 canyon vicinity as in HRVs and overlaps the site of binding of CD155 to poliovirus, the orientation of the receptor molecule relative to the virus is different for each type of virus. In contrast, ICAM-1 binds similarly to different HRV serotypes regardless of whether it is glycosylated or unglycosylated or is present as two- or five-domain structures.
MATERIALS AND METHODS
Sample preparation.
CAV21, strain Kuykendall, was obtained from the American Type Culture Collection, Manassas, Va. (VR-850). The virus was propagated in H1-HeLa cells two times, and CAV21-specific, infectious cDNA was prepared and sequenced (E. Rieder and E. Wimmer, unpublished data). CAV21 derived from the infectious cDNA was propagated in 3.6 × 107 HeLa cells/ml, suspended in Dulbecco minimal Eagle medium (Life Technologies) with 10% bovine serum. The cells were infected with CAV21 at a multiplicity of infection of 20. After adsorption at room temperature for 1 h under mild agitation, the cells were diluted to a concentration of 3 × 106 cells/ml with fresh medium. They were allowed to grow for 9.5 h, after which time they were harvested and lysed with 1% Nonidet P-40. They were then briefly homogenized and centrifuged at 4,000 × g to separate the components. DNase (0.5 mg/ml) was added to permit digestion of nucleic acids for 1 h, followed by the addition of 0.5 mg of trypsin per ml for 10 min to digest protein. Sucrose gradient and CsCl isopycnic ultracentrifugations were used to purify the virus. Five-domain, fully glycosylated ICAM-1 lacking the cytoplasmic and transmembrane domains was kindly provided by J. M. Greve (14). Plaque reduction assays, used to determine whether the soluble receptor blocked H1-HeLa cell infection, showed 100% inhibition at 0.46 mg of ICAM-1 per ml.
CryoEM.
A 50-μl sample of purified CAV21 (6.2 mg/ml) was incubated with or without (Table 1) 20 μl of 46.3-mg/ml five-domain ICAM-1 (an excess of about nine receptor molecules per binding site on the virus) for 35 min on ice. CryoEM specimens were prepared from 3.5-μl aliquots that were applied to perforated carbon support electron microscope grids and vitrified in liquid ethane (2). Micrographs were recorded on Kodak SO-163 film in a Philips CM200 field emission gun (FEG) microscope at a nominal magnification of 50,000 and a dose level of 18.5 e−/Å2. Micrographs were taken at different underfocus levels (Table 1) so that useful contrast transfer function corrections could be achieved uniformly for all resolutions. The micrographs were digitized on a Zeiss PHODIS microdensitometer at 7-μm intervals but, for computational reasons, the pixels were averaged to produce 14-μm pixels (2.80-Å spacing at the specimen). Particles of the virus-receptor complex were boxed from scanned micrographs with a radius of 130 pixels (364 Å).
TABLE 1.
Electron microscopy data collection and processing
| Sample | Underfocus (μm)a | No. of:
|
Correlation coefficientc | Resolution (Å)d | |
|---|---|---|---|---|---|
| Micrographs | Particlesb | ||||
| CAV21+ICAM-1 | 1.22 to 1.80 | 10 | 176 (580) | 0.294 (0.073) | 26 |
| CAV21 | 1.40 to 1.93 | 5 | 390 (1,616) | 0.473 (0.081) | 21 |
Determined from the phase-contrast transfer function of the microscope.
Number of particles included in the three-dimensional map (total number of boxed particles).
 |
Highest resolution at which the correlation between two independent three-dimensional reconstructions falls below 0.5.
A cryoEM reconstruction image of HRV16 served as an initial model for a particle orientation search using the polar-Fourier transform method (1). The final resolution for both the native and the complexed CAV21 density maps (Table 1) was determined by splitting the image data randomly into two equally sized groups and determining at what resolution the average agreement of phases was less than 45° and the correlation coefficient was less than 0.5. Although many particle images were used in the reconstructions, the final resolution extended at best to only about 21 Å. The original Fourier-Bessel reconstruction program (10), which was later modified (2), was rewritten for parallel processing on an IBM SP2 computer with 16 nodes to permit the calculations to be accomplished in a reasonable time.
Difference map calculations and model fitting.
CryoEM image reconstructions were made for native CAV21 and for CAV21 complexed with five-domain ICAM-1 (Fig. 1). The size of each map was scaled to that of a map of poliovirus 1 obtained from X-ray diffraction coordinate data (Protein Data Bank accession number 2PLV) and calculated to approximately the same resolution. The maps were scaled by comparing the densities within the protein shell (108- to 144-Å radii). A difference map was calculated by subtracting the virus map from the virus-receptor map.
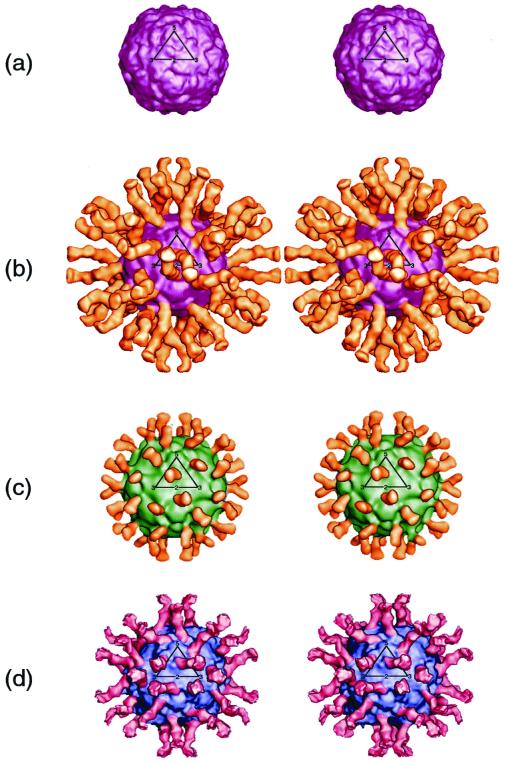
FIG. 1.
Stereoview of cryoEM reconstructions showing native CAV21 (a), complex of CAV21 with ICAM-1 domains D1 to D5 (b), complex of HRV16 with ICAM-1 domains D1 and D2 (c), and complex of poliovirus 1 (Mahoney strain) with CD155 domains D1 to D3 (d). CAV21 is purple, HRV16 is green, poliovirus 1 is blue, ICAM-1 is yellow, and CD155 is red. In panel b, five domains of ICAM-1 can be seen; in panel c, only two domains of ICAM-1 can be seen. All three domains of CD155 are observed in panel d. One icosahedral asymmetric unit is outlined in black on each of the four image reconstructions.
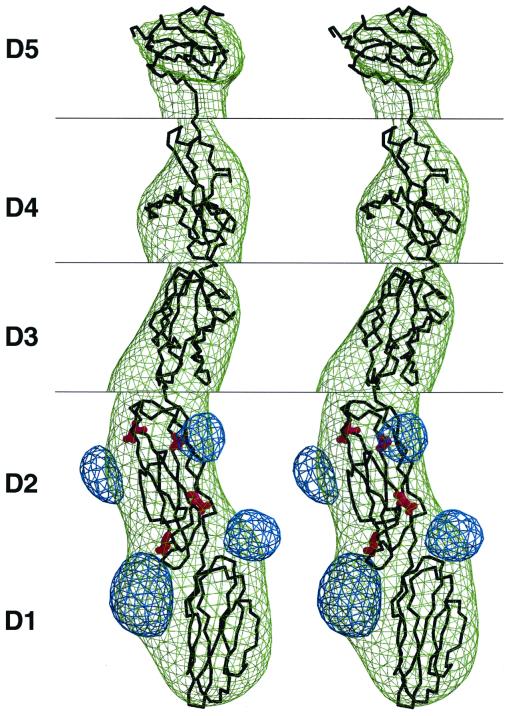
The known structure of the first two ICAM-1 domains (28) was fitted into the difference map (Fig. 2). The slight variation in the elbow angle, about 6° (28), between domains D1 and D2 for different crystallographic determinations did not significantly impact the quality of fit. This was also true for the fitting of the ICAM-1 structure into the HRV–ICAM-1 reconstruction density map (28). The fitted structure was then used to compute electron density corresponding to an unglycosylated ICAM-1 D1D2 structure. A further difference map between the cryoEM map and the calculated map identified the sites of carbohydrate attachment to the D1D2 domains of the ICAM-1 molecule. These sites acted as fiducial marks to obtain the best fit of the ICAM-1 structure to the difference map. The absence of available structures with sufficient similarity in amino acid sequence to domains D3, D4, and D5 of ICAM-1 made it difficult to find useful homologous structures to fit the remaining ICAM-1 density. The closest homologous structure (25) found for domain D3 was neural cellular adhesion molecule domain D1 (Protein Data Bank accession number 2NCM [50]). In the absence of any better homologous structures, the previously modeled D1D2 domains of CD155 (19) were used to model the fourth and fifth ICAM-1 domains.
FIG. 2.
Stereoview of difference map between CAV21–ICAM-1 and CAV21. The ICAM-1 difference density (green) has been fitted with the known structure (black) of ICAM-1 (D1D2) and homologous models of D3, D4, and D5 (see the text). Shown also is the difference density (blue) between the observed glycosylated ICAM-1 structure and the unglycosylated ICAM-1 structure. The N-linked Asn carbohydrate sites are indicated in red. The correspondence of the observed locations of the glycosylation sites and their known positions on the ICAM-1 backbone acted as a marker for accurate fitting of the ICAM-1 molecule. Because the height of the density decreases approximately in proportion to the distance of each domain from the virus center, the contour level is shown at 2.00 ς around domains D1 and D2, 1.30 ς around D3, 0.80 ς around D4, and 0.65 ς around domain D5; the ς value is the root-mean-square difference from the mean density of the map, which in this case included large areas of zero density outside the cryoEM image.
Structure of CAV21.
The atomic coordinates of poliovirus 1 were used as a basis for building a model of CAV21. Residue replacement and energy minimization were performed with the program SEGMOD (30). The footprint of the ICAM-1 molecule on the CAV21 surface was defined by the residues on the viral surface that have any atoms within 4 Å of any atom in the receptor.
RESULTS AND DISCUSSION
Structure of ICAM-1.
The 180-Å-long structure (Fig. 1 and 3) of the complete extracellular component of five-domain ICAM-1 bound to CAV21 is clearly visible in the reconstruction. However, only the first three domains were visible in an earlier reconstruction of HRV16 complexed with five-domain ICAM-1 (28). The difference may partially reflect the use of about 200 particle images in the present reconstruction, compared to only 43 in the earlier reconstruction. In addition, the use of an electron microscope fitted with an FEG provided better image conditions because the electron beam was brighter and more coherent than in conventional instruments. The importance of the data collected with the FEG was confirmed in that an earlier reconstruction of the CAV21–ICAM-1 complex, obtained with a Philips 420 electron microscope (data not shown), did not show the complete ICAM-1 molecule. Some flexibility of the articulated ICAM-1 molecule is apparent because the height of the density decreases in successive domains. If the mean height of the density in the viral capsid coat is arbitrarily set to a value of 10, the densities in domains 1, 2, 3, 4, and 5 are 10, 10, 6, 3, and 1, respectively. Although the density progressively decreases toward the C terminus of ICAM-1, it is surprising that the long ICAM-1 molecule is sufficiently rigid for all domains to remain visible even after the icosahedral averaging inherent in the reconstruction technique has been applied. It was reported (47) that ICAM-1 is kinked between domains 2 and 3, but this orientation does not appear in the present reconstruction. Kirchhausen et al. (27) suggested that ICAM-1 is slightly kinked between domains 3 and 4, an observation that is approximately consistent with the structure reported here (Fig. 1 and 3). All the various receptor molecules utilized by picornaviruses are long, thin, and articulated at hinges between domains. Although ICAM-1 appears to be more rigid than expected, its properties are consistent with the requirement that the receptor be a molecule able to flex sufficiently to recognize additional sites on the viral surface once the first receptor has been bound. The apparent preference for long, thin, hinged receptor molecules may be essential for the recruitment of successive receptors to the viral surface after the initial recognition event.
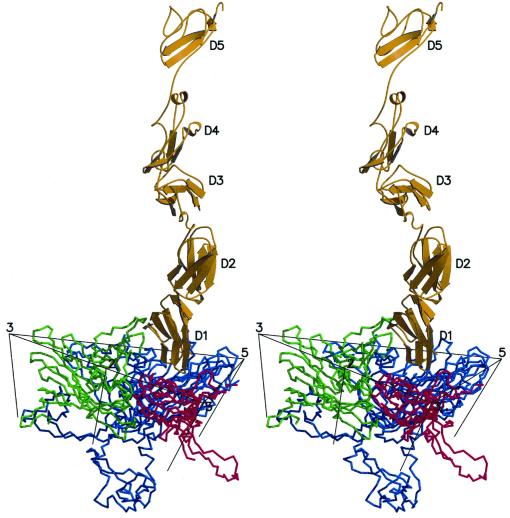
FIG. 3.
Stereoview of a ribbon diagram of one molecule of ICAM-1 (yellow) bound to one icosahedral asymmetric unit of CAV21. Ribbons of VP1, VP2, and VP3 are shown in blue, green, and red, respectively. Icosahedral symmetry axes surrounding the site of receptor attachment are also shown.
The glycosylation sites on ICAM-1 helped improve the accuracy by which the known ICAM-1 D1D2 structure could be fitted to the cryoEM density (Fig. 2). The footprint of ICAM-1 on the homology-modeled structure of CAV21 is complementary in shape and charge to the ICAM-1 binding surface (Fig. 4). There is no evidence for the formation of any hydrogen bonding interactions.
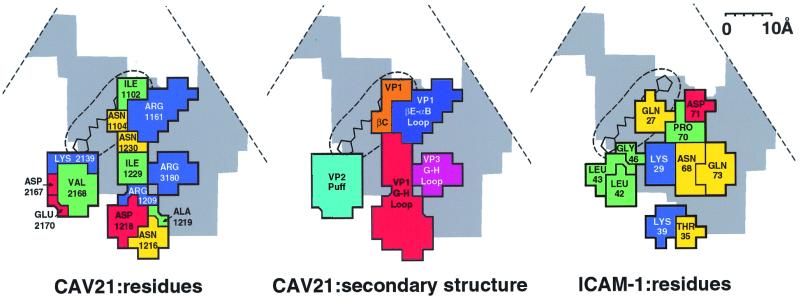
FIG. 4.
Road map of the ICAM-1 footprint on the surface of CAV21, showing the surface amino acids (left) and exposed peptide segments that are in contact with ICAM-1 (middle). The partial outline of the triangular, icosahedral asymmetric unit is indicated. The canyon is shown in gray, and the buried VP1 hydrophobic pocket is shown by the circular broken line, with a representative drug bound. Also shown are the amino acids of ICAM-1 in contact with the CAV21 surface (right). Amino acids (left and right panels) are blue (basic), red (acidic), green (hydrophobic), and yellow (polar). Virus residues (left panel) are numbered from 1001, 2001, and 3001 in VP1, VP2, and VP3, respectively. Note the charge complementarity and matching of hydrophobic regions between the CAV21 and ICAM-1 binding surfaces. In the middle panel, the peptide segments are differentiated by color.
Location of the receptor binding site.
The interactions of ICAM-1 with the surface of CAV21 span between the “north” and “south” canyon rims (Fig. 4), involving βC and the loop from βE to αB of viral protein 1 (VP1) on the north rim, the GH loops of VP1 and VP3 on the floor, and the “puff” on the south rim. The puff is a variably sized insertion in VP2 of picornaviruses (41).
The footprint of ICAM-1 on the surface of CAV21 and on HRV14 and HRV16 of the major-group rhinoviruses and the footprint of CD155 on the surface of poliovirus 1 have a common core of somewhat conserved residues (Fig. 5 and 6). However, the orientations of the receptor molecules are different for each virus-receptor complex. Whereas in CAV21 the receptor leans very slightly to the east-southeast (Fig. 1b), as viewed in the conventional orientation with the fivefold axis of the icosahedral asymmetric unit in the north, in HRVs the ICAM-1 receptor molecule leans to the southwest and approaches the icosahedral twofold axis much more closely (Fig. 1c). However, the first domain of CD155 lies almost tangential to the poliovirus surface, with the subsequent domains pointing eastward (Fig. 1d). In contrast, the orientations of ICAM-1 (glycosylated or unglycosylated, two domains or five domains) with any of the HRVs are almost exactly the same. Similarly, in three studies of CD155 with poliovirus (4, 19, 53), the orientations of the receptor were shown to be the same. This result is surprising, as the amino acid sequence differences between CAV21 and poliovirus are fewer than the differences between HRV14 and HRV16.
FIG. 5.
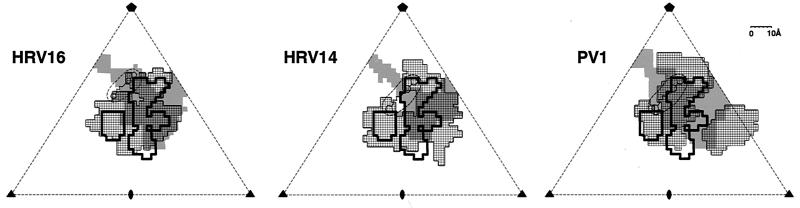
Comparison of the footprint of ICAM-1 on CAV21 (thick black outline) with the footprints of ICAM-1 on HRV16 and HRV14 and of CD155 on poliovirus 1 (PV1) (areas covered by small squares). Gray shading shows the outline of the canyon, and the circular broken line shows the site of the pocket factor underneath the canyon. Scale bar, 10 Å.
FIG. 6.
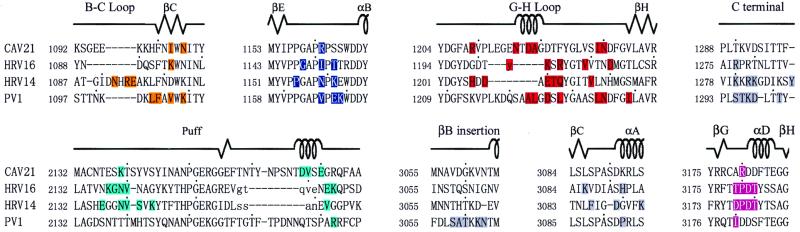
Amino acid sequence alignments for CAV21, HRV16, HRV14, and poliovirus 1 (PV1) in the regions that form the receptor-virus interactions. Amino acids that have been identified as participating in receptor interactions are colored to match the corresponding peptide segments shown in the middle panel of Fig. 4. Lowercase letters represent amino acids in the puff for which there was no alignment with the other sequences; the residues could lie anywhere in this area. Dashes represent deletions of amino acids relative to the alignment.
The utilization of similar locations around the canyon by all of the currently studied receptors bound to picornaviruses (Fig. 5) suggests that this region provides an essential function in picornavirus infections. The presence of the receptor attachment site within the canyon was predicted (41) as a position protected from host immune surveillance. This notion was questioned when it was found that the site of binding of a neutralizing antibody extended beyond the rims and into the canyon (46), thus demonstrating that the receptor and antibody sites overlapped. Nevertheless, because selected neutralizing antibody escape mutations lie outside the canyon and because residues inside the canyon are more conserved than those outside the canyon, the binding affinity of the receptor probably is slightly dominated by residues in the canyon. This preference is enhanced by avidity caused by attachment of multiple receptor molecules to attain cell entry.
Receptor binding to CAV21, HRVs, and poliovirus 1 is localized within the canyon to a site adjacent to a hydrophobic pocket within the VP1 β barrel containing an as-yet-unidentified “pocket factor” (Fig. 4 and 5) (12, 40). It has been suggested that the pocket factor stabilizes the virus during transit between hosts but is displaced by the competition of the receptor for its overlapping binding site (17, 35, 40). Thus, binding of receptor would destabilize the virion and initiate uncoating. This hypothesis is consistent with the observed degradation of HRVs in the presence of soluble ICAM-1 (23) and the possible absence of the pocket factor in the poliovirus-CD155 complex (4).
Virus stability.
The difficulty of determining incubation times and temperatures needed to obtain successful virus-receptor cryoEM images is related to the destabilizing effect of the receptor on the virus. Thus, it is significant that kinetic analyses have shown, both for HRVs (6) and for polioviruses (31), that there are two distinct modes of binding whose relative abundance varies with temperature. The binding modes observed in the cryoEM reconstructions are likely to be the most stable intermediates, although these may be different depending upon the virus-receptor complex. Kolatkar et al. (28) argue that the complexes seen for HRVs are an initial event, consistent with kinetic data (6). In view of the probable loss of the pocket factor from the poliovirus-receptor complex (4) and the more extensive contact of the receptor with the viral surface, it is possible that the poliovirus-receptor complex represents an intermediate stage of picornavirus-receptor interaction. Thus, the CAV21, HRV, and poliovirus interactions with their receptors might represent sequential steps in similar processes for each virus.
ACKNOWLEDGMENTS
We thank Sharon Wilder and Cheryl Towell for help in the preparation of the manuscript.
This work was supported by National Institutes of Health grants to M.G.R. (AI11219) and E.W. (AI32100, AI39485, and AI15122); a National Institutes of Health program project grant to M.G.R., R.J.K., T.S.B., and others (AI45976); and a National Science Foundation shared instrument grant to T.S.B., M.G.R., and others (BIR-9112921). Support was also provided by a Purdue University reinvestment grant.
REFERENCES
- 1.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 2.Baker T S, Olson N H, Fuller S D. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryoelectron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bella J, Kolatkar P R, Marlor C W, Greve J M, Rossmann M G. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belnap D M, McDermott B M, Jr, Filman D J, Cheng N, Trus B L, Zuccola H J, Racaniello V R, Hogle J M, Steven A C. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci USA. 2000;97:73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berendt A R, McDowall A, Craig A G, Bates P A, Sternberg M J E, Marsh K, Newbold C I, Hogg N. The binding site on ICAM-1 for Plasmodium falciparum-infected erythrocytes overlaps, but is distinct from, the LFA-1 binding site. Cell. 1992;68:71–81. doi: 10.1016/0092-8674(92)90207-s. [DOI] [PubMed] [Google Scholar]
- 6.Casasnovas J M, Springer T A. Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- 7.Casasnovas J M, Stehle T, Liu J, Wang J, Springer T A. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chothia C, Jones E Y. The molecular structure of cell adhesion molecules. Annu Rev Biochem. 1997;66:823–862. doi: 10.1146/annurev.biochem.66.1.823. [DOI] [PubMed] [Google Scholar]
- 9.Colonno R J. Cell surface receptors for picornaviruses. Bioessays. 1986;5:270–274. doi: 10.1002/bies.950050609. [DOI] [PubMed] [Google Scholar]
- 10.Crowther R A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc London Ser B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 11.Evans D J, Almond J W. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 1998;6:198–202. doi: 10.1016/s0966-842x(98)01263-3. [DOI] [PubMed] [Google Scholar]
- 12.Filman D J, Syed R, Chow M, Macadam A J, Minor P D, Hogle J M. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 14.Greve J M, Forte C P, Marlor C W, Meyer A M, Hoover-Litty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromeier M, Wimmer E, Gorbalenya A E. Genetics, pathogenesis, and evolution of picornaviruses. In: Domingo E, Webster R G, Holland J J, editors. Origin and evolution of viruses. San Diego, Calif: Academic Press, Inc.; 1999. pp. 287–343. [Google Scholar]
- 16.Hadfield A T, Lee W, Zhao R, Oliveira M A, Minor I, Rueckert R R, Rossmann M G. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure. 1997;5:427–441. doi: 10.1016/s0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 17.Hadfield A T, Oliveira M A, Kim K H, Minor I, Kremer M J, Heinz B A, Shepard D, Pevear D C, Rueckert R R, Rossmann M G. Structural studies on human rhinovirus 14 drug-resistant compensation mutants. J Mol Biol. 1995;253:61–73. doi: 10.1006/jmbi.1995.0536. [DOI] [PubMed] [Google Scholar]
- 18.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 19.He Y, Bowman V D, Mueller S, Bator C M, Bella J, Peng X, Baker T S, Wimmer E, Kuhn R J, Rossmann M G. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci USA. 2000;97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendry E, Hatanaka H, Fry E, Smyth M, Tate J, Stanway G, Santti J, Maaronen M, Hyypiä T, Stuart D. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure. 1999;7:1527–1538. doi: 10.1016/s0969-2126(00)88343-4. [DOI] [PubMed] [Google Scholar]
- 21.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 23.Hoover-Litty H, Greve J M. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes P J, North C, Minor P D, Stanway G. The complete nucleotide sequence of coxsackievirus A21. J Gen Virol. 1989;70:2943–2952. doi: 10.1099/0022-1317-70-11-2943. [DOI] [PubMed] [Google Scholar]
- 25.Jones D T, Taylor W R, Thornton J M. A new approach to protein fold recognition. Nature (London) 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Smith T J, Chapman M S, Rossmann M G, Pevear D C, Dutko F J, Felock P J, Diana G D, McKinlay M A. Crystal structure of human rhinovirus serotype 1A (HRV1A) J Mol Biol. 1989;210:91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- 27.Kirchhausen T, Staunton D E, Springer T A. Location of the domains of ICAM-1 by immunolabeling and single-molecule electron microscopy. J Leukoc Biol. 1993;53:342–346. doi: 10.1002/jlb.53.3.342. [DOI] [PubMed] [Google Scholar]
- 28.Kolatkar P R, Bella J, Olson N H, Bator C M, Baker T S, Rossmann M G. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lentz K N, Smith A D, Geisler S C, Cox S, Buontempo P, Skelton A, DeMartino J, Rozhon E, Schwartz J, Girijavallabhan V, O'Connell J, Arnold E. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of the three poliovirus serotypes. Structure. 1997;5:961–978. doi: 10.1016/s0969-2126(97)00249-9. [DOI] [PubMed] [Google Scholar]
- 30.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J Mol Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 31.McDermott B M J, Rux A H, Eisenberg R J, Cohen G H, Racaniello V R. Two distinct binding affinities of poliovirus for its cellular receptor. J Biol Chem. 2000;275:23089–23096. doi: 10.1074/jbc.M002146200. [DOI] [PubMed] [Google Scholar]
- 32.Minor P D, Brown F, Domingo E, Hoey E, King A, Knowles N, Lemon S, Palmenberg A, Rueckert R R, Stanway G, Wimmer E, Yin-Murphy M. Picornaviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 329–336. [Google Scholar]
- 33.Muckelbauer J K, Kremer M, Minor I, Tong L, Zlotnick A, Johnson J E, Rossmann M G. Structure determination of coxsackievirus B3 to 3.5 Å resolution. Acta Crystallogr Sect D. 1995;51:871–887. doi: 10.1107/S0907444995002253. [DOI] [PubMed] [Google Scholar]
- 34.Ockenhouse C F, Betageri R, Springer T A, Staunton D E. Plasmodium falciparum-infected erythrocytes bind ICAM-1 at a site distinct from LFA-1, Mac-1, and human rhinovirus. Cell. 1992;68:63–69. doi: 10.1016/0092-8674(92)90206-r. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira M A, Zhao R, Lee W, Kremer M J, Minor I, Rueckert R R, Diana G D, Pevear D C, Dutko F J, McKinlay M A, Rossmann M G. The structure of human rhinovirus 16. Structure. 1993;1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 36.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papi A, Johnston S L. Rhinovirus infection induces expression of its own receptor intercellular adhesion molecule 1 (ICAM-1) via increased NF-κB-mediated transcription. J Biol Chem. 1999;274:9707–9720. doi: 10.1074/jbc.274.14.9707. [DOI] [PubMed] [Google Scholar]
- 38.Pulli T, Koskimies P, Hyypiä T. Molecular comparison of coxsackie A virus serotypes. Virology. 1995;212:30–38. doi: 10.1006/viro.1995.1450. [DOI] [PubMed] [Google Scholar]
- 39.Roivainen M, Hyypiä T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VPI protein by intestinal proteases. J Virol. 1991;65:4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossmann M G. Viral cell recognition and entry. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature (London) 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 42.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 43.Shafren D R, Dorahy D J, Greive S J, Burns G F, Barry R D. Mouse cells expressing human intercellular adhesion molecule-1 are susceptible to infection by coxsackievirus A21. J Virol. 1997;71:785–789. doi: 10.1128/jvi.71.1.785-789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shafren D R, Dorahy D J, Ingham R A, Burns G F, Barry R D. Coxsackievirus A21 binds to decay-accelerating factor but requires intercellular adhesion molecule 1 for cell entry. J Virol. 1997;71:4736–4743. doi: 10.1128/jvi.71.6.4736-4743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shafren D R, Dorahy D J, Thorne R F, Barry R D. Cytoplasmic interactions between decay-accelerating factor and intercellular adhesion molecule-1 are not required for coxsackievirus A21 cell infection. J Gen Virol. 2000;81:889–894. doi: 10.1099/0022-1317-81-4-889. [DOI] [PubMed] [Google Scholar]
- 46.Smith T J, Chase E S, Schmidt T J, Olson N H, Baker T S. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature (London) 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staunton D E, Dustin M L, Erickson H P, Springer T A. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 48.Staunton D E, Marlin S D, Stratowa C, Dustin M L, Springer T A. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families. Cell. 1988;52:925–933. doi: 10.1016/0092-8674(88)90434-5. [DOI] [PubMed] [Google Scholar]
- 49.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 50.Thomsen N K, Soroka V, Jensen P H, Berezin V, Kiselyov V V, Bock E, Poulsen F M. The three-dimensional structure of the first domain of neural cell adhesion molecule. Nat Struct Biol. 1996;3:581–585. doi: 10.1038/nsb0796-581. [DOI] [PubMed] [Google Scholar]
- 51.Verdaguer N, Blaas D, Fita I. Structure of human rhinovirus serotype 2 (HRV2) J Mol Biol. 2000;300:1181–1196. doi: 10.1006/jmbi.2000.3943. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Springer T A. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol Rev. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 53.Xing L, Tjarnlund K, Lindqvist B, Kaplan G G, Feigelstock D, Cheng R H, Casasnovas J M. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 2000;19:1207–1216. doi: 10.1093/emboj/19.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeates T O, Jacobson D H, Martin A, Wychowski C, Girard M, Filman D J, Hogle J M. Three-dimensional structure of a mouse-adapted type 2/type 1 poliovirus chimera. EMBO J. 1991;10:2331–2341. doi: 10.1002/j.1460-2075.1991.tb07772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]