Abstract
Steroid hormone metabolism by the gut microbiome has multiple implications for mammalian physiology, but the underlying mechanisms and broader significance of this activity remains largely unknown. Here, we isolate a novel human gut bacterium, Clostridium steroidoreducensT strain HCS.1, that reduces cortisol, progesterone, testosterone, and related steroid hormones to 3β,5β-tetrahydrosteroid products. Through transcriptomics and heterologous enzyme profiling, we identify and biochemically characterize the C. steroidoreducens OsrABC reductive steroid hormone pathway. OsrA is a 3-oxo-Δ1-steroid hormone reductase that selectively targets the Δ1-bond present in synthetic steroid hormones, including the anti-inflammatory corticosteroids prednisolone and dexamethasone. OsrB is a promiscuous 3-oxo-Δ4-steroid hormone reductase that converts steroid hormones to 5β-dihydrosteroid intermediates. OsrC is a 3-oxo-5β-steroid hormone oxidoreductase that reduces 5β-intermediates to 3β,5β-tetrahydro products. We find that osrA and osrB homologs predict steroid hormone reductase activity in diverse gut bacteria and are enriched in Crohn’s disease fecal metagenomes. These studies thus identify the basis of reductive steroid hormone metabolism in the gut and establish a link between inflammatory disease and microbial enzymes that deplete anti-inflammatory corticosteroids.
INTRODUCTION
Steroid hormones encompass a broad class of biologically active molecules that play crucial roles in diverse physiological processes. Corticosteroids, like cortisol, are involved in regulating inflammation, immune response, and metabolism,1 while sex steroids, including estrogens, androgens, and progestins, regulate reproductive functions and secondary sexual characteristics.2 Due to their wide-ranging effects, both natural and synthetic steroid hormones are commonly used in medical therapies to manage conditions such as autoimmune diseases, hormone deficiencies, and cancers.
The gut microbiome mediates diverse phenotypes through the modification of host- and diet-derived metabolites, including various steroids. Research on microbiome steroid metabolism has primarily focused on bile acids, products of which are important for multiple host phenotypes.3–5 However, while less studied, steroid hormones have also been identified as an important class of substrates in the gut. These molecules interact with gut microbes after entering the gastrointestinal tract via therapeutic oral or rectal administration or through excretion in bile.6 Bile is likely a particularly important source of intestinal steroid hormones, as 9–22% of endogenous cortisol,7,8 10–15% of testosterone,9 20–30% of corticosterone,10 and up to 30% of progesterone11 are eliminated from the body through this route.
Gut microbes generate multiple products from steroid hormones, including cortisol and corticosterone derivatives that serve as fecal biomarkers for stress in animal research studies.12,13 In addition to passing into feces, intestinal microbial steroid hormone products can reenter the bloodstream through enterohepatic circulation. In humans, this is evidenced by the rectal administration of cortisol leading to an increase in specific circulating cortisol derivatives in a microbiome-dependent manner.14,15 Other studies provide evidence that microbial products of progesterone metabolism similarly enter systemic circulation.6,16
The impact of microbial steroid hormone metabolism has been linked to several aspects of mammalian biology. Microbial inactivation of orally administered steroids, including side-chain cleavage of synthetic corticosteroids, reduces the bioavailability of these drugs.17,18 Microbial pathways that dehydroxylate corticosterone and convert steroid precursors to androgens generate metabolites that contribute to hypertension in animal models19,20 and promote castration-resistant prostate cancer,21,22 respectively. Bacterial metabolisms that alter the concentration of steroid hormones with distinct biological activities thus have diverse consequences for mammalian biology.
Previous studies have reported that some gut bacteria reduce the Δ4-bond in steroid hormones, generating 5β-steroid derivatives.23 This activity decreases the anti-inflammatory and androgenic properties of glucocorticoids and androgens, respectively, and converts progestins into a neuroactive form.24 While it stands to reason that Δ4-steroid hormone reduction may have implications for host biology, the molecular basis and the broader significance of activities in the gut microbiome remains unknown.
Here, we describe the isolation and characterization of a novel steroid hormone-metabolizing gut bacterium, Clostridium steroidoreducens HCS.1. By employing a multidisciplinary approach, integrating genomics, transcriptomics, and metabolomics, we characterize the osrABC reductive steroid hormone pathway. These findings provide new insights into the diversity of steroid hormone metabolism in the gut microbiome and its potential impact on host health.
RESULTS
Clostridium steroidoreducens is a novel steroid hormone-enriched gut bacterium
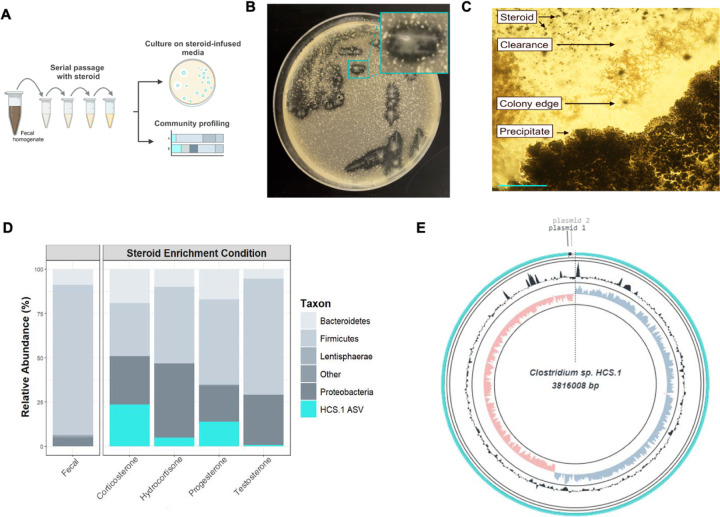
To select for members of the gut microbiome with metabolic capabilities that provide a selective advantage in the presence of steroid hormones, we passaged a human fecal sample in a nutritionally limited base medium supplemented with individual corticosteroids (cortisol or corticosterone) or sex steroids (progesterone or testosterone) (Figure 1A). We isolated strains from the final enrichment passages and cultivated them on steroid hormone-infused solid media. We identified one strain, HCS.1, that cleared insoluble cortisol or progesterone from the solid media and accumulated a white precipitate indicative of a potential reaction product at colony centers on progesterone-infused media (Figure 1B and 1C). Consistent with HCS.1 possessing a selective advantage in the presence of steroid hormones, 16S rRNA amplicon sequencing of the final enrichment passages revealed an amplicon sequence variant matching the HCS.1 16S rRNA sequence was enriched from below the limit of detection in the fecal inoculum to 0.7–23.6% of the microbial community following cortisol, corticosterone, progesterone, or testosterone enrichment (Figure 1D).
Figure 1. Clostridium steroidoreducens is a novel steroid-enriched species.
(A) Schematic overview of steroid enrichment and strain isolation experiments. (B) HCS.1 strain colonies on cortisol-infused media. (C) HCS.1 strain colonies on progesterone-infused media. Scale bar, 200 μm. (D) Microbial community profile of the final steroid hormone enrichment passage based on16S rRNA amplicon sequencing. (E) Circular representation of the HCS.1 genome.
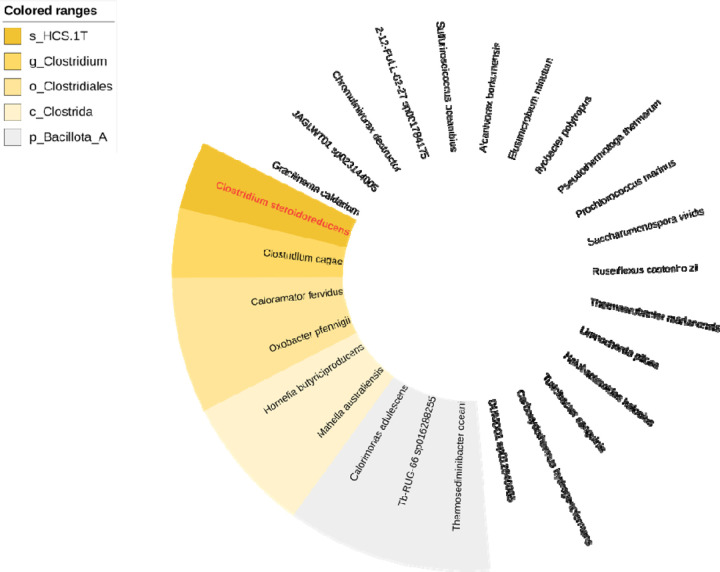
To facilitate further strain characterization, we sequenced and assembled HCS.1 DNA into 3 circularized contigs, comprising a genome and two plasmids, that contain 3,816,008 base pairs with 28.4% G + C content, 3595 protein-coding genes, and 111 RNA genes (Figure 1E). Phylogenetic analyses revealed that HCS.1 was closely related to Clostridium chrysemydis but represents a novel species of the genus Clostridium, based on an average nucleotide identity (ANI) of 92.09% to the closest reference genome and accepted taxonomic assignment criteria (Extended Data Figure 1).25 In recognition of steroid hormone reductase activities detailed below, we assigned HCS.1 the species name Clostridium steroidoreducens.
C. steroidoreducens possesses broad steroid hormone reductase activity
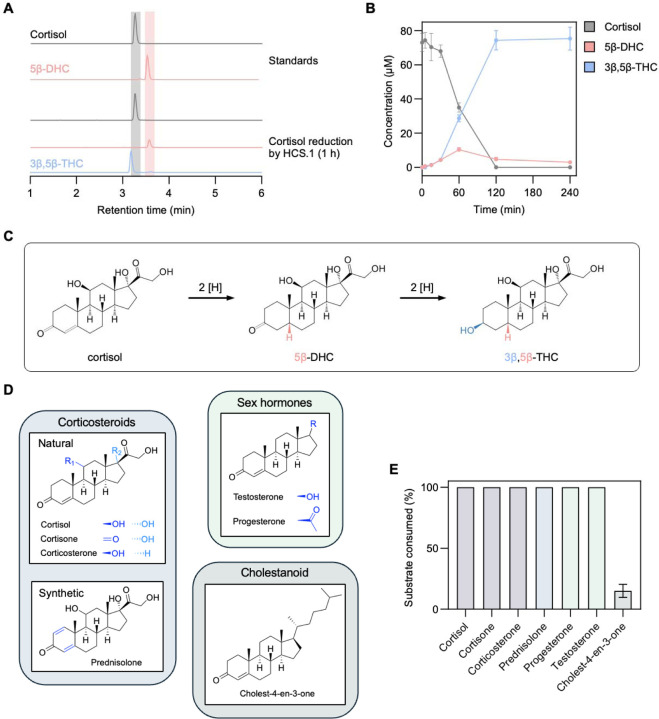
To determine whether the C. steroidoreducens HCS.1 steroid clearance phenotype was indicative of metabolic activity, we employed an LC-MS-based assay to track the fate of cortisol or progesterone in C. steroidoreducens HCS.1 culture. We observed that both steroid hormones were fully depleted from the media, coinciding with the emergence of a minor and major product. By comparing to compound reference standards, we confirmed that major and minor products corresponded to 5β-dihydro- and 3β,5β-tetrahydro-steroid derivatives, respectively (Figure 2A, Extended Data Figure 2).
Figure 2. C. steroidoreducens HCS.1 possesses promiscuous 3-oxo-Δ4-beta steroid hormone reductase activity.
(A) Products formed from C. steroidoreducens HCS.1 incubation with cortisol. (B) Time-course analysis of cortisol metabolism by C. steroidoreducens HCS.1. (C) Proposed pathway for cortisol reduction by C. steroidoreducens HCS.1. DHC and THC stand for dihydrocortisol and tetrahydrocortisol, respectively. (D) Steroid substrates tested for C. steroidoreducens HCS.1. (E) Measured C. steroidoreducens HCS.1 steroid substrate consumption.
Tracking C. steroidoreducens HCS.1 cortisol metabolism over time, we observed that 5β-dihydrocortisol transiently accumulated, peaking at 60 minutes before decreasing to less than 2% of the total corticosteroid present by 120 minutes (Figure 2B). By contrast, 3β,5β-tetrahydrocortisol steadily accumulated following the introduction of cortisol, exceeding 98% of the corticosteroid present by 120 minutes (Figure 2B). These results suggest that, in contrast to previously characterized bacterial steroid dehydroxylation26 and side chain-cleaving27 activities, C. steroidoreducens HCS.1 exclusively reduces cortisol, converting it to 3β,5β-tetrahydrocortisol via a 5β-dihydrosteroid intermediate (Figure 2C).
To address the specificity of C. steroidoreducens HCS.1 steroid utilization, we next tested the strain’s activity on a panel of steroids with variable functional groups at multiple positions on the sterol core (Figure 2D). We found that C. steroidoreducens tolerated substitutions at C1, C11, C17 positions, exhibiting activity on distinct corticosteroids (corticosterone, cortisone, prednisolone) and sex steroids (progesterone, testosterone) (Figure 2E). In contrast to these polar steroids, the hydrophobic cholesterol-derivative cholestenone was a poor substrate (Figure 2E). These results establish C. steroidoreducens as a steroid hormone-reducing gut bacterium with broad substrate specificity.
Fe-S flavoenzyme family OsrB is a 3-oxo-Δ4-steroid hormone reductase
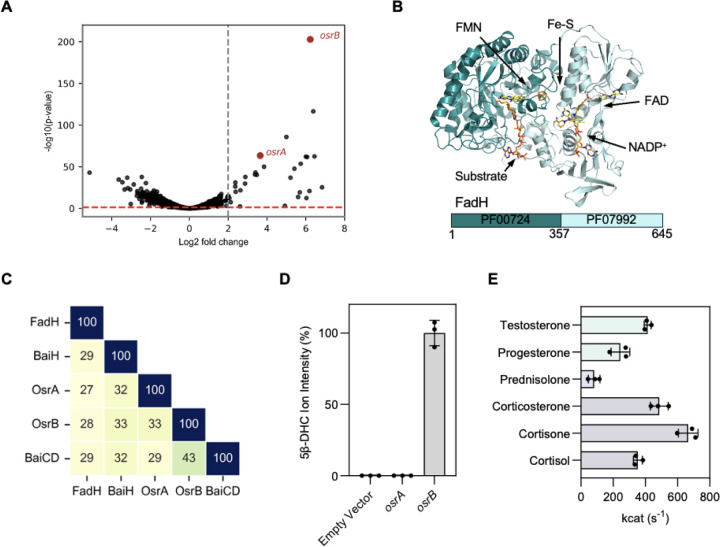
We next sought to identify the mechanism of steroid hormone reduction by C. steroidoreducens. As bacterial reductases are often induced in the presence of their substrate,26,28 we employed a transcriptomics-based approach to identify candidate steroid hormone reductases in C. steroidoreducens. We performed RNA-seq analysis on C. steroidoreducens cells cultivated in the presence or absence of cortisol and identified 30 genes that were induced >2-fold when cortisol was present (Supplementary Table 1). Two of the most highly induced genes, which we renamed osrA and osrB (oxosteroid reductase A and B), encoded proteins annotated as fadH-like 2,4-dienoyl-CoA reductases (Figure 3A, Supplementary Table 1).
Figure 3. OsrB is a 3-oxo-Δ4-steroid hormone reductase.
(A) Gene expression of C. steroidoreducens HCS.1 in the presence versus absence of cortisol. Gray and red dashed lines indicate genes with statistical significance and >2-fold induction in response to cortisol, respectively. (B) Crystal structure of Fe-S flavoenzyme 2,4-dienoyl-CoA reductase (FadH) bound to ligands (PDB code: 1PS9). (C) Percent sequence identity of OsrA and OsrB to Fe-S flavoenzymes FadH and bile acid reductases BaiH and BaiCD. (D) Conversion of cortisol to 5β-dihydrocortisol (DHC) by E. coli expressing osrA or osrB versus an empty vector control. (E) Rate of reduction of indicated steroid hormones by purified OsrB.
E. coli 2,4-dienoyl-CoA reductase is the best characterized member of the “Fe-S flavoenzyme family” of oxidoreductases that contain a conserved N-terminal substrate-binding domain (PF00724) and a C-terminal NAD(P)H cofactor-binding domain (PF07992) (Figure 3B).29 Divergent members of the Fe-S flavoenzyme family are widespread in gut bacteria and possess distinct substrate specificities for host- and diet-derived metabolites.30 Consistent with OsrA and OsrB representing novel Fe-S flavoenzyme subtypes with distinct substrates, we observed that these enzymes exhibited remote sequence homology to previously characterized Fe-S flavoenzymes, including Clostridium scindens Fe-S flavoenzymes, BaiCD and BaiH, which reduce bile acid intermediates structurally related to steroid hormones (Figure 3C).29,31
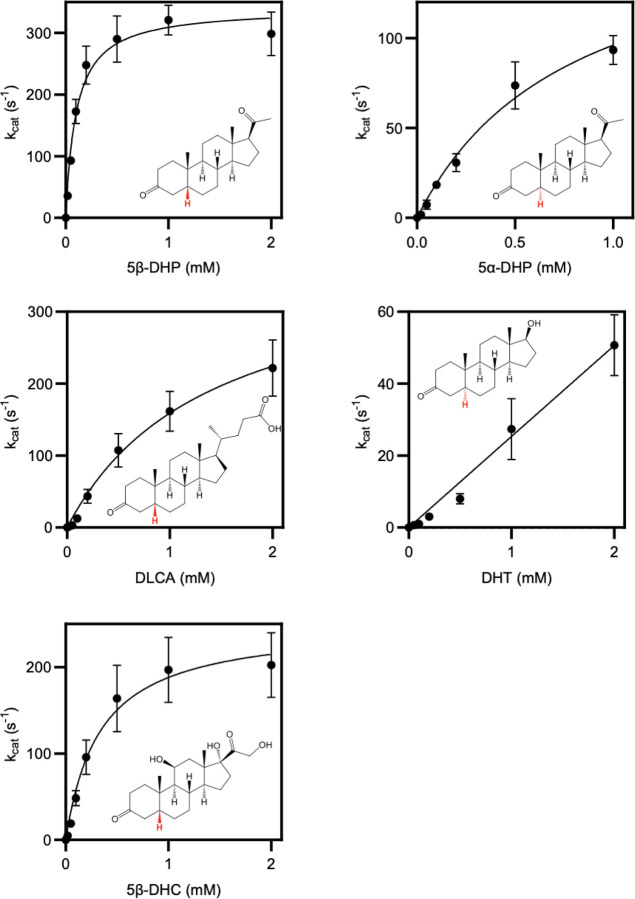
To test steroid reductase activity of OsrA and OsrB, we heterologously produced the enzymes in anaerobically cultured E. coli cells. We found cells expressing osrB, but not osrA, reduced cortisol to 5β-dihydrocortisol (Figure 3D). Studies with anaerobically purified OsrB revealed that NADH and NADPH were poor electron donors for OsrB. Using the artificial electron donor methyl viologen, we observed that OsrB similarly reduced a variety of steroid hormones substrates (Figure 3E). These results thus establish OsrB as a promiscuous 3-oxo-Δ4-steroid hormone reductase that uses a presently unidentified electron donor.
Short chain dehydrogenase OsrC is a 3-oxo-5β-steroid hormone oxidoreductase
We next sought to identify the C. steroidoreducens enzyme responsible for reduction of the 3-oxo group on the 5β-dihydrosteroid intermediate generated by OsrB. Microbial bile acid oxidoreductases with specificity for 3α-, 3β-, 7α-, 7β-, 12α- and 12β-hydroxyl groups have been previously identified.32–34 As these characterized steroid oxidoreductases are members of the short chain dehydrogenase (SDR) enzyme superfamily, we reasoned the C. steroidoreducens enzyme was likely related to this family. An analysis of the C. steroidoreducens genome identified 12 genes with SDR domains. However, none were induced by cortisol or exhibited high sequence similarity to previously characterized bile acid oxidoreductases.
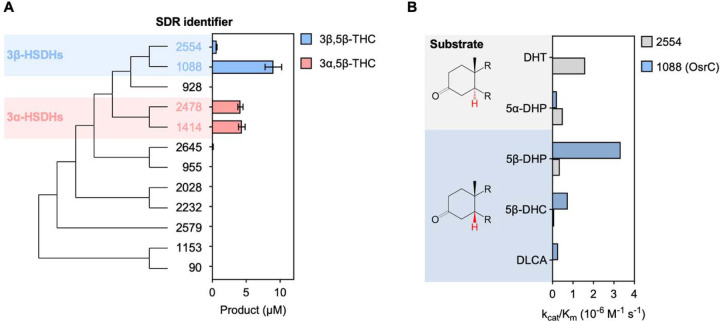
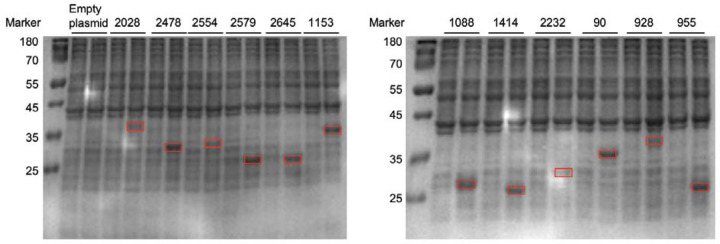
As these analyses failed to identify obvious candidates, we next performed an unbiased screen of SDR-containing C. steroidoreducens proteins for 3-oxo-5β-steroid hormone reductase activity. We confirmed soluble expression of all 12 SDRs in E. coli and tested the activity of overexpressing E. coli strains on 5β-dihydrocortisol (Figure 4A, Extended Data Figure 3). We identified two SDRs (BLEONJ_2554 and BLEONJ_1088) that produced 3β,5β-tetrahydrocortisol and two others (BLEONJ_2478 and BLEONJ_1414) that yielded 3α,5β-tetrahydrocortisol (Figure 4A).
Figure 4. OsrC is a 3-oxo-5β-steroid hormone oxidoreductase.
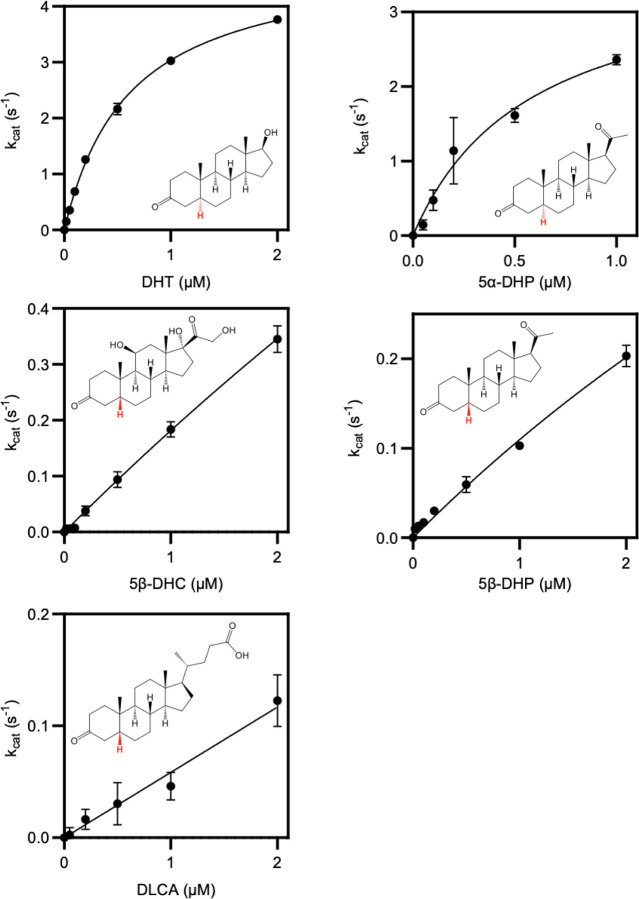
(A) Phylogenetic analysis of C. steroidoreducens HCS.1 SDR domain-containing protein sequences. Product formed from 5β-dihydrocortisol by E. coli strains overexpressing SDR domain-containing proteins are shown with their respective gene identifiers. THC stand for tetrahydrocortisol. (B) Kinetic parameters of reduction of indicated steroid hormones by purified SDR domain-containing proteins. Abbreviations stand for dihydrotestosterone (DHT), dihydroprogesterone (DHP), dihydrocortisol (DHC), and dehydrolithocholic acid (DLCA).
Studies with anaerobically purified BLEONJ_2554 and BLEONJ _1088 revealed divergent substrate specificities. BLEONJ_2554 showed a pronounced preference for 5α-steroids and exhibited weak activity that did not follow classical Michaelis-Menten kinetics with 5β-steroid substrates (Figure 4B, Extended Data Figure 4). Conversely, gene BLEONJ _1088 displayed a preference for 5β-steroid hormones and accommodated multiple functional groups at the C9 or C17 positions (Figure 4B, Extended Data Figure 5). We further found that BLEONJ_1088 exhibited a preference for steroid hormones relative to the comparable bile acid derivative lithocholic acid. We thus conclude that BLEONJ_1088 is a 3-oxo-Δ4-steroid hormone reductase and, on this basis, renamed it OsrC.
Fe-S flavoenzyme family OsrA is a 3-oxo-Δ1-steroid hormone reductase active on synthetic corticosteroids
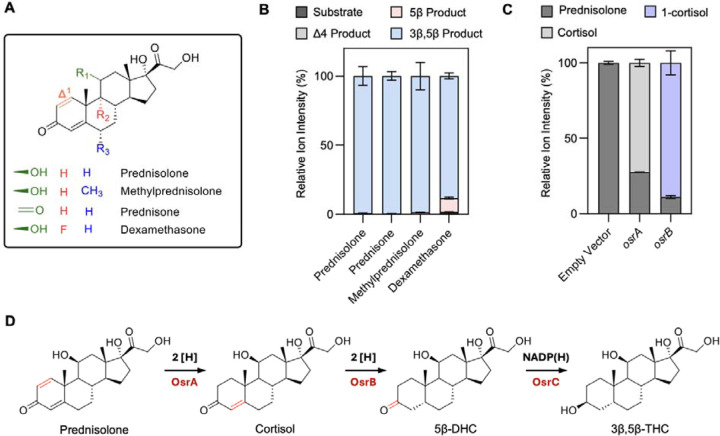
Synthetic corticosteroids possess potent anti-inflammatory properties and are used to treat a range of pathologies, including inflammatory bowel disease.35 The synthetic corticosteroids drugs dexamethasone, prednisone, prednisolone, and methylprednisolone contain a Δ1-bond that is absent in natural corticosteroids and which significantly extends their half-life (Figure 5A).36 As our initial screen of steroids identified prednisolone as a substrate for C. steroidoreducens (Figure 1), we sought to address the molecular basis of synthetic corticosteroid metabolism. We first tested C. steroidoreducens activity on additional synthetic corticosteroids dexamethasone, prednisone, and methylprednisolone and found that all were reduced to 3β,5β-tetrahydrocortisol derivatives, indicating that the bacterium possesses both Δ1- and Δ4-steroid hormone reductase activities (Figure 5B).
Figure 5. OsrA is a 3-oxo-Δ1-reductase essential for complete reduction of synthetic corticosteroids.
(A) Structure of synthetic corticosteroids used in assays. (B) Products formed from synthetic corticosteroids following incubation with C. steroidoreducens HCS.1 cells. (C) Percent prednisolone conversion to cortisol following incubation of E. coli cells with osrA- and osrB-expressing plasmids or an empty vector control. (D) C. steroidoreducens HCS.1 steroid reduction pathway identified in this study.
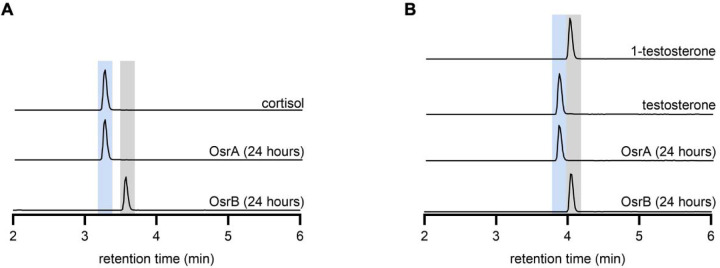
Considering the similarity of the Δ1-reduction to the OsrB catalyzed Δ4-reduction, we next tested the activity of OsrA and OsrB and found that the two enzymes generated distinct cortisol and testosterone isomers from prednisolone and the synthetic androgen boldenone, respectively (Figure 5C, Extended Data Figure 6). Based on comparison to reference standards, we establish that OsrA and OsrB products reflected Δ1- and Δ4-steroid hormone reductase activities, respectively (Extended Data Figure 6). These results demonstrate that OsrA functions as a Δ1-steroid hormone reductase that acts in conjunction with OsrB and OsrC to reduce synthetic steroid hormones to 3β,5β-reduced products (Figure 5E).
Steroid hormone reductase activities are common in gut bacteria and correlate with the distribution of osrA and osrB homologs
We next sought to address the breadth of steroid hormone reductase activity in the gut microbiome. We performed BLASTp searches of OsrA, OsrB, and OsrC in the Unified Human Gastrointestinal Genome catalog of representative genomes and metagenome-assembled genomes, which includes 4,644 prokaryotic species that colonize the human gastrointestinal tract.37 These searches identified homologs with high sequence homology to OsrA, OsrB, and OsrC in 2, 59, and 90 genomes, respectively (Supplementary Table 2). Genomes encoding osrABC homologs included gram-positive bacterial species from multiple taxa, primarily from the Erysipelotrichaceae and Lachnospiraceae families.
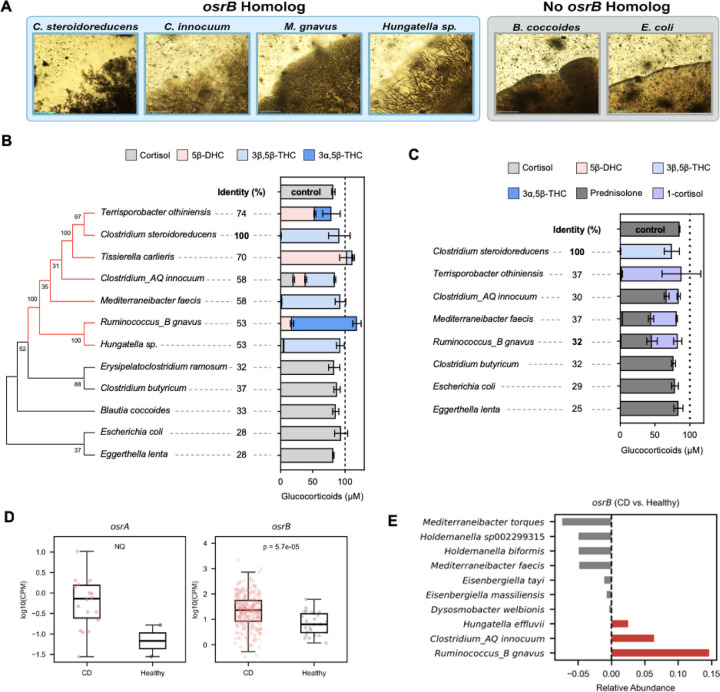
To determine the association of osrABC homologs with observed C. steroidoreducens phenotypes, we selected 117 gut bacteria strains for experimental characterization. We tested these strains on solid media on steroid clearance/precipitate accumulation and assayed a select subset for cortisol and prednisolone activity. From these studies we identified 29 strains from 14 species with a steroid clearance/precipitate accumulation phenotype and 6 species isolates with steroid hormone reductase activity (Figure 6A–6C and Supplementary Table 3).
Figure 6. Steroid reductase activities are widespread in gut bacteria and elevated in active Crohn’s disease.
(A) Representative images of gut bacteria grown on progesterone-infused media, showing steroid clearance/precipitate accumulation-positive (blue background) and -negative (gray background) colonies. Scale bar, 200 μm. (B) Corticosteroids produced by gut bacterial isolates after incubation with cortisol. The protein with the highest sequence identity to OsrB encoded by each genome was used to generate the tree. (C) Corticosteroids produced by gut bacterial isolates after incubation with prednisolone. Identity refers to the sequence identity of the protein with the highest sequence identity to OsrA encoded by each genome. (D) reads mapping to osrA and osrB homologs in metagenomes from healthy and Crohn’s disease (CD) patients. Only metagenomes with at least one read mapping to a gene are included in the analysis. NQ refers to the not quantified statistical difference, due to the low number of healthy metagenomes with reads mapping to osrA. CPM refers to copies per million. (E) Difference in osrB homolog abundance for taxa showing the greatest changes in relative abundance between healthy and CD metagenomes.
Comparing strain genotypes to observed phenotypes revealed several patterns. First, presence of an osrB homolog in a genome strongly predicted steroid clearance/precipitate accumulation and steroid hormone Δ4-reductase activity (Figure 6B and Supplementary Table 3). Second, the absence of osrA homologs tracked with a consistent lack of steroid hormone Δ1-reductase activity (Figure 6C and Supplementary Table 3). Third, while the presence of an osrC homolog tracked with production of 3β,5β-tetrahydrocortisol, absence of an osrC homolog was not predictive of fate of the C3 functional group. Indeed, osrC-negative strains varied in their major cortisol product, generating either 5β-dihydrocortisol, 3α,5β-tetrahydrocortisol, or 3β,5β-tetrahydrocortisol (Figure 6B and Supplementary Table 3). These results provide evidence that osrA and osrB specifically confer Δ1- and Δ4 -steroid hormone reductase activities, respectively, while osrC likely represents one of multiple evolutionarily distinct 3-oxo-5β-steroid hormone oxidoreductases.
osrB is prevalent in human fecal metagenomes and associated with active Crohn’s disease
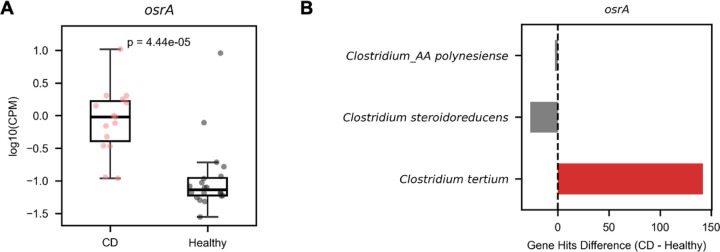
Having established the relevance of osrABC homologs for steroid reductase activity in gut bacteria, we next sought to determine the prevalence of the pathway in the human gut. We focused our analysis on osrA and osrB, since these homologs reliably predicted steroid hormone reductase activities of assayed strains. We recruited reads to osrA and osrB homologs in a collection of 1,491 previously published healthy human fecal metagenomes. We detected at least one read mapping to osrA and osrB homologs in 2.2% and >98.9% of samples, respectively (Supplementary Table 4). Within most metagenomes multiple osrB homologs recruited many reads. By contrast, the majority osrA reads recruited to Clostridium tertium osrA homologs, often with only one or two reads per metagenome (Supplementary Table 4). These analyses demonstrate that osrB homologs are common in the gut but that osrA homologs are confined to bacteria that colonize the gut at a low relative abundance.
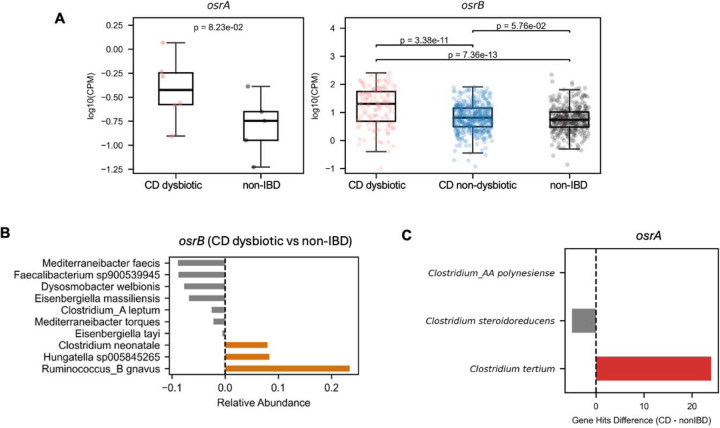
Considering that glucocorticoids possess potent anti-inflammatory activities and natural and synthetic variants, including cortisol and prednisolone, are rectally and orally administered for the treatment of inflammatory bowel disease, we reasoned that OsrA and OsrB activity could be clinically relevant in this patient population. We analyzed 314 metagenomes from the Lewis et al.38 study of active Crohn’s disease patients, including a subset treated with corticosteroids (Supplementary Table 5). We observed osrB homologs were elevated in Crohn’s disease patient relative to a healthy control population (Figure 6D). Further scrutiny revealed that the increased abundance osrB homologs from Ruminococcus_B gnavus and Clostridium_AQ innocuum, two taxa previously associated with Crohn’s disease inflammation,39,40 was the primary driver of this association (Figure 6E).
osrA homologs similarly exhibited elevated abundance in Crohn’s disease metagenomes, but their low prevalence coupled with the relatively small sample size of this dataset complicated statistical analysis of the significance of this relationship (Figure 6D). To address this issue, we expanded our dataset to include 1537 metagenomes from multiple separate studies that included Crohn’s disease and control populations. Analysis of this larger dataset confirmed that osrA homologs were significantly elevated in Crohn’s disease patient metagenomes and revealed that the osrA from Clostridium tertium was the primary driver of this association (Extended Data Figure 7A and 7B, Supplementary Table 5).
To determine whether identified associations extended an independent dataset, we evaluated 569 Crohn’s disease patient metagenomes collected as part of Integrative Human Microbiome Project (Supplementary Table 6). We observed a similar association between elevated abundance of osrA and osrB homologs and microbiome dysbiosis scores used as a proxy for active Crohn’s disease in this study (Extended Data Figure 8A–8C, Supplementary Table 6).41 Underscoring the relevance of these observations to active Crohn’s disease, metagenomes from this study with microbiome dysbiosis score consistent with inactive Crohn’s disease exhibited intermediate osrB homolog levels between dysbiotic Crohn’s disease and control non-IBD populations (Extended Data Figure 8A, Supplementary Table 6).
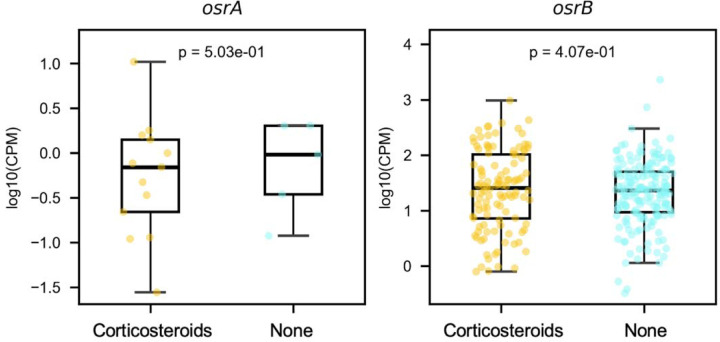
We further investigated the relationship between microbial steroid reductases and corticosteroid treatment in Crohn’s disease patients, as reported in the Lewis et al.38 study. Our analysis revealed that corticosteroid therapy was associated with an increase in osrA homolog prevalence, as these homologs were detected in 9.6% of corticosteroid-treated patients compared to only 2.4% in untreated patients. This suggests a potential selective pressure favoring bacteria that metabolize synthetic corticosteroids in patients receiving these therapies. Interestingly, within the metagenomes where osrA homologs were present, their abundance did not significantly differ between treated and untreated patients, suggesting that the effect of corticosteroid treatment may relate to increased colonization of bacteria with osrA homologs (Extended Data Figure 9).
DISCUSSION
In this study, we characterize Clostridium steroidoreducens HCS.1, a previously uncharacterized gut bacterium that encodes a novel reductive pathway, OsrABC, for the metabolism of steroid hormones. Our work expands on previous studies of microbial steroid metabolism by demonstrating the widespread prevalence and activity of OsrB and OsrC in the gut microbiota, which catalyze the reduction of natural steroid hormones into their 3β5β-tetrahydro derivatives. Notably, the OsrB homologs are prevalent in Crohn’s disease-associated bacterial communities, implicating these enzymes in both health and disease contexts.
One of the most compelling aspects of this work is the link between microbial steroid metabolism and chronic inflammatory conditions, particularly Crohn’s disease. Our data indicate that osrB homologs are enriched in pro-inflammatory taxa such as Clostridim_AQ innocuum and Ruminococcus_B gnavus, both of which have previously been associated with Crohn’s disease.39,40 Pro-inflammatory gut microbes often exhibit a competitive advantage in inflammatory conditions and induce inflammation to generate conditions favorable for their growth.41 This suggests that these microbes may leverage steroid hormone metabolism to gain a competitive advantage in the inflamed gut environment. Specifically, OsrB-mediated depletion of endogenous anti-inflammatory glucocorticoids may represent an adaptive strategy employed by Clostridim_AQ innocuum and Ruminococcus_B gnavus to perpetuate inflammation and support their growth.
In a clinical context, the OsrABC reductase pathway may also have significant implications for glucocorticoid therapies, which are commonly administered to manage inflammatory bowel disease. Our findings suggest that the OsrABC reductase pathway could modulate the effective dose of administered glucocorticoids by degrading these anti-inflammatory compounds. This underscores the importance of further investigations into the microbial impact on drug bioavailability in relation to both the efficacy and dosing of steroid therapies in patients.
Beyond the clinical considerations, our study highlights the broader significance of gut microbial steroid metabolism in human health. Notably, a concurrently published manuscript independently identifies the 3-oxo-Δ4-steroid hormone reductase activity of OsrB homologs, along with the characterization of additional novel gut bacterial enzymes that metabolize progestins.42 Together, these findings represent a crucial step forward in delineating the broader landscape of microbial steroid hormone metabolism and its potential clinical implications.
MATERIALS AND METHODS
Steroid hormone enrichment culture
For each enrichment sample, 15 mM steroid hormone (cortisol, corticosterone, progesterone, or testosterone) suspensions were prepared in 1 mL basal growth medium (Difco M9 minimal salts, 20 mM acetate, 20 mM formate, tryptone 0.01% w/v, Bacto yeast extract 0.01% w/v, trace vitamins and minerals, MgSO4 4.09% w/v; pH 6.5). Homogenized fecal samples were pelleted and washed 3x in phosphate buffer saline (PBS), then resuspended in 1 mL saline. 20 μL cell suspension was added to each enrichment culture condition and incubated for 72 hours. After 72 hours, 20 μL of each culture was used to inoculate fresh media supplemented with its respective compound. Cultures were passaged a total of 4 times. After the final passage, a portion of each condition was preserved in 20% glycerol and frozen at −80°C. The remaining culture was pelleted and processed for 16S rRNA sequencing.
Isolation of HCS.1
Preserved stocks of enrichment culture samples were plated onto fresh brain heart infusion (BHI) agar and incubated for 4 days at 37 °C under anaerobic conditions (5% H2, 10% CO2, 85% N2). Distinct colonies were passaged to confirm purity and identified by 16S rRNA V4-V5 variable region sequencing. Purified isolates of HCS.1 were stored at −80 °C in a 20% glycerol suspension. Frozen glycerol stocks were deposited in the DFI Symbiotic Bacterial Strain Bank Repository (https://dfi.cri.uchicago.edu/biobank/).
Steroid clearance assay
To prepare steroid clearance assays, progesterone and cortisol amounts for a final concentration of 12 mM or 16 mM, respectively, were sterilized by suspension in 70% v/v ethanol, followed evaporation at room temperature for 2 hours. Dried steroid powders were sifted into autoclaved BHI agar and stirred rapidly, shortly before pouring into plates. Solid plates were stored under anaerobic conditions at 25 °C for at least 24 hours prior to use.
To test HCS.1 steroid clearance, solid BHI plates were incubated at 37 °C for 2 days. After 2 days, single colonies were picked and suspended in 200 μL PBS. Aliquots of the cell suspension were spread onto solid steroid plates and incubated anaerobically for 5 days at 37 °C. To test steroid clearance of other bacterial strains, solid BHI plates were incubated at 37 °C for 4 days. After 4 days, single colonies from each isolate were picked and suspended in 200 μL PBS. 2 μL aliquots were spotted onto solid progesterone plates, dried, and incubated anaerobically for 3 days at 37 °C.
16S rRNA sequencing and analysis
Cells from final steroid enrichment passages were collected by centrifugation and their genomic DNA extracted using the QIAamp PowerFecal Pro DNA kit (Qiagen). Briefly, samples were suspended in a bead tube (Qiagen) along with lysis buffer and loaded on a bead mill homogenizer (Fisherbrand). Samples were then centrifuged, and the supernatant was resuspended in a reagent that effectively removed inhibitors. DNA was then purified routinely using a spin column filter membrane and quantified using Qubit. The 16S rRNA variable V4-V5 region was amplified using universal bacterial primers, 564F and 926R. Amplicons were purified using magnetic beads, then quantified and pooled at equimolar concentrations. The Qiagen QIAseq one-step amplicon library kit was used to ligate Illumina sequencing-compatible adaptors onto amplicons. Reads were sequenced on an Illumina MiSeq platform to generate 2 × 250 base pair reads, with 5,000–10,000 reads per sample. Amplified 16S rRNA amplicons were processed through the dada1 pipeline in R. Forward reads were trimmed at 210 bp and reverse reads were trimmed at 150 bp, to remove low-quality nucleotides. Chimeras were detected and removed using default parameters. Amplicon sequence variants between 300 and 360 bp in length were taxonomically assigned to the genus level using the RDP Classifier (v2.13) with a minimum bootstrap confidence score of 80.
Sample preparation for whole genome sequencing
To prepare HCS.1 for whole genome sequencing, 10 mL BHI broth was inoculated with cells from a single bacterial colony and incubated anaerobically at 37 °C for 48 hours. The culture was centrifuged at 4000 x g for 10 minutes. The resulting pellet was resuspended, washed in phosphate buffer saline (PBS), and re-centrifuged.
Whole genome sequencing library preparation: Illumina short reads
Samples for Illumina short sequencing were extracted using the QIAamp PowerFecal Pro DNA kit (Qiagen), as described in the preceding subsection. Libraries were prepared using 200 ng of genomic DNA using the QIAseq FX DNA library kit (Qiagen). Briefly, DNA was fragmented enzymatically into shorter fragments and desired insert size was achieved by adjusting fragmentation conditions. Fragmented DNA was end repaired and ‘A’s’ were added to the 3’ends to stage inserts for ligation. During ligation step, Illumina compatible Unique Dual Index (UDI) adapters were added to the inserts and prepared library was PCR amplified. Amplified libraries were cleaned up, and QC was performed using Tapestation 4200 (Agilent Technologies). Libraries were sequenced on an Illumina NextSeq 1000/2000 to generate 2×150bp reads.
Whole genome sequencing library preparation: Oxford Nanopore long reads
Samples for Nanopore and Illumina hybrid assemblies were extracted using the high molecular weight NEB Monarch Genomic DNA Purification Kit. DNA was QC’ed using genomic Tapestation 4200. Nanopore libraries were prepared using the Rapid Sequencing Kit (SQK-RAD114) and sequenced on MinION R10.4.1 flow cells. Nanopore reads were base-called using ONT Guppy basecalling software version 6.5.7+ca6d6af, minimap2 version 2.24-r1122, and was demultiplexed using ONT Guppy barcoding software version 6.5.7+ca6d6af using local HPC GPU. N50 of the nanopore long read is 7077 base pairs, the average read length is 4529.4 base pairs, while the average read quality is 15.6, which is typical of Nanopore reads. Hybrid assembly was performed with both nanopore and Illumina short reads using Unicylcer v0.5.0.43,44
Taxonomic classification of Clostridium steroidoreducens sp. nov. Strain HCS.1^T^
The classification of strain HCS.1 as a novel species, Clostridium steroidoreducens sp. nov. was performed using GTDB-Tk (version 2.3.2)45 on the KBase platform. Genome quality assessment, phylogenetic placement, and taxonomic classification were performed according to GTDB guidelines. The HCS.1 genome was uploaded to the KBase website and analyzed using the GTDB-Tk classify workflow, which assigns genomes to the closest known taxa based on conserved marker genes. Strain HCS.1 was classified within the genus Clostridium, but did not match any known species in the Genome Taxonomy Database (GTDB, version r207). Phylogenetic placement within the GTDB bacterial tree confirmed that the strain represented a distinct lineage, supporting its designation as a new species.
Transcriptomic analysis of HCS.1
To prepare HCS.1 samples for transcriptomic analysis, six 50 mL Lysogeny broth cultures were inoculated with bacteria cells andshaken anaerobically at 37 °C for 48 hours. After 48 hours, cortisol powder was added to 3 cultures, to a final concentration of 8 mM. All 6 cultures were incubated for an additional 4 hours, then pelleted at 4000 x g for 10 minutes. The resulting pellets were flash-frozen in a dry ice/ethanol bath and stored at −80 °C until ready for subsequent processing. Cell pellets were thawed and total RNA from biological replicates extracted using the Maxwell RSC instrument (Promega). Extracted RNA was quantified using Qubit, and integrity was measured using TapeStation (Agilent Technologies). Libraries from ribo-depleted samples were constructed using the NEB’s Ultra Directional RNA library prep kit for Illumina. First, up to 500 ng total RNA was subjected to ribosomal RNA depletion (for bacteria) using NEBNext rRNA depletion kit. Ribosomal -RNA depleted samples were fragmented based on RNA integrity number (RIN). Post cDNA synthesis, Illumina compatible adapters were ligated onto the inserts and final libraries were QC’ed using TapeStation (Agilent technologies). Libraries were normalized using library size and final library concentration (as determined by Qubit). Library concentration (ng/ul) was converted to nM to calculate dsDNA library concentration. Equimolar libraries were then pooled together at identical volumes to ensure even read distribution across all samples. Normalized libraries were then sequenced on Illumina’s NextSeq 1000/2000 at 2×100bp read length.
High-quality reads were mapped to the circularized hybrid assembled genome of HCS.1 (NCBI: CP170704), using Bowtie2 (v.2.4.5), and sorted with Samtools (v1.6). Read counts were generated using featureCounts (v2.0.1) with Bakta annotations.46 Gene expression was quantified as the total number of reads uniquely aligning to the reference genome, binned by annotated gene coordinates. Differential gene expression and quality control analyses were performed using DESeq2 in R with Benjamini–Hochberg false discovery rate adjustment applied for multiple testing corrections.47
Bacterial culture steroid reductase assay
A complete list of strains used in this study is provided in Supplementary Table 2. Strains were incubated under anaerobic conditions (85% N2, 10% CO2, 5% H2) at 37 °C in an anaerobic chamber (Coy Laboratory). Liquid brain-heart infusion (BHI) broth supplemented with 100 μM steroids from 10 mM stocks in methanol was used for growth. Cultures were grown anaerobically with a 1% (v/v) inoculum from a pre-culture and supplemented with steroids after 4 hours of incubation during the exponential growth phase. Bacterial cultures were extracted by the addition of 9 volumes of methanol supplemented with 0.5 μM methylprednisolone as an internal standard for LC-MS analysis.
LC-MS-Q-TOF analysis of steroids
Extracted samples were vortexed and centrifuged twice at 21,000 × g for 15 minutes, with the supernatant transferred to new tubes after each centrifugation step. The methanol fraction was filtered through 0.2 μm nylon membrane filters prior to LC-MS analysis. Samples were analyzed using an Agilent 6540 UHD Q-TOF mass spectrometer coupled to an Agilent 1200 Infinity LC system. Separation was performed on a XBridge C18 column (2.1×100mm, 3.5 μm particle size) using 0.1% aqueous formic acid and acetonitrile with 0.1% formic acid as mobile phases. The separation gradient ranged from 20% to 100% acetonitrile over 4 minutes at 50 °C with a flow rate of 0.5 mL/min. Mass spectra were acquired in negative ion mode for glucocorticoids ([M+FA-H]−) or positive ion mode for all other steroids ([M+H]⁺), with an ion spray voltage of 3500 V and a nozzle voltage of 2000 V. The source temperature was set to 300 °C, and the gas flow rate was 8 L/min. Data were processed and visualized using MassHunter software version 10.
Molecular biology
Gene transformations were performed by Gibson assembly using 2x NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, NEB, E2621X). Primers were designed using SnapGene (see Supplementary Table 7), incorporating 20 bp flanking regions complementary to a linearized expression vector (pMCSG53) and the gene of interest from the HCS.1 genome. PCR, cloning, and transformation were performed according to the protocols provided on the NEB website. The Gibson assembly reaction was incubated at 50 °C for 1 hour and then transformed into E. coli XL1-Blue competent cells according to the manufacturer’s protocol. Transformed cells were plated on Luria-Bertani (LB) agar plates containing 100 μg/mL carbenicillin, and successful transformations were confirmed using sequencing primers specific for the backbone vector (see Supplementary Table 7). Positive colonies were validated and sequenced by the University of Chicago Genomics Facility. The final constructs were then transformed into chemically competent E. coli Rosetta™ (DE3) competent cells (Novagen) according to NEB protocols. Transformed cells were plated on LB agar plates supplemented with 100 μg/mL carbenicillin.
Protein production in E. coli
Protein production in E. coli Rosetta cells was performed under aerobic conditions for all short-chain dehydrogenases (SDRs) and under anaerobic conditions for OsrA and OsrB. Cultures were grown in either 2x YT medium (20 g/L tryptone, 10 g/L yeast extract, and 5 g/L NaCl) or TB medium (12 g/L tryptone, 24 g/L yeast extract, 4 mL/L glycerol, 9. 4 g/L K2HPO4, and 2.2 g/L KH2PO4) supplemented with 0.5% (w/v) glucose and 1 mM ferric ammonium citrate, respectively.
Induction of protein expression was initiated during the exponential phase when optical densities (OD600) reached 0.4–0.6 by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cultures were incubated for 3–5 hours at 37 °C with shaking at 200 rpm. Cells were harvested by centrifugation at 4500 × g for 20 minutes. Cell pellets were then frozen at −80 °C for storage prior to subsequent experimental use. Protein production was confirmed by SDS-PAGE analysis.
Purification of heterologous produced proteins
Cell Lysis and Protein Purification.
Frozen cell pellets were supplemented with 0.1 mg/mL DNase and lysed either aerobically (for SDRs) or anaerobically (for OsrA and OsrB) using a Thermo Spectronic French pressure cell at 1,100 PSI. The crude cell extract was centrifuged at 75,600 x g for 30 minutes, followed by filtration through a 0.2 μm nylon membrane (Fisher Scientific) before being applied to a purification system.
Aerobic purification of SDRs.
The filtered extract was applied to an ÄKTA pure system (Cytiva) using a 1 mL Strep-Tactin®XT 4Flow® column (Iba Lifesciences). The column was equilibrated with 10 volumes of equilibration buffer (100 mM Tris/HCl, pH 8.0, 150 mM NaCl) at 1 mL/min and 4°C. Proteins were loaded via a 5 mL loop, followed by washing of non-specifically bound proteins. Elution was performed with 5 mL elution buffer (100 mM Tris/HCl, pH 8.0, 150 mM NaCl, and 50 mM biotin). Eluted proteins were collected in 1 mL fractions and were concentrated using Pierce™ Protein concentrators (10 kDa), desalted, and either used directly or transferred to storage buffer (50 mM Tris/HCl, pH 7.5, 10% (w/v) glycerol, and 50 mM NaCl) using PD-10 desalting columns (Cytiva) before storage at −80 °C.
Anaerobic purification of OsrB.
Anaerobic purification was performed in an anaerobic chamber. A Strep-Tactin®XT 4Flow® gravity column (Iba Lifesciences) was used with a WET FRED system (Iba Lifesciences) to maintain a constant flow rate of ~1 mL/min, adjusted using a lab jack stand (LABALPHA). The column was equilibrated with anaerobic equilibration buffer (100 mM Tris/HCl, pH 8.0, 150 mM NaCl) and elution was performed with anaerobic elution buffer (100 mM Tris/HCl, pH 8.0, 150 mM NaCl, and 50 mM biotin). Eluted proteins were collected in 1 mL tubes, concentrated and desalted using Pierce™ Protein Concentrators PES, 10K MWCO, 0.5 mL, at 10,000 × g in a microcentrifuge. Proteins were either used directly for enzymatic assays or transferred to anaerobic storage buffer (50 mM Tris/HCl, pH 7.5, 10% (w/v) glycerol, and 50 mM NaCl) and frozen at −80 °C.
Whole-cell assays of heterologous enzymes
The activity of heterologously expressed proteins was assessed under either aerobic conditions (for SDRs) or anaerobic conditions (for OsrA and OsrB). E. coli Rosetta cells were grown in media as described above, supplemented with 100 μM steroids (prepared from 10 mM stock solutions in methanol) using a 1% (v/v) inoculum. Protein production was induced with 1 mM IPTG at an OD of 0.4–0.7 and cultures were incubated overnight at 37 °C without shaking. Reactions were quenched by the addition of 9 volumes of methanol containing 0.5 μM methylprednisolone as internal standard (IS). LC-MS samples were prepared as described previously.
Enzymatic assays with purified SDRs
The kinetic properties of purified SDRs were determined using reaction mixtures in 96-well plates with a total volume of 100 μL. The reaction mixture contained 25 mM Tris/HCl (pH 7.0), 1 mM NADPH, 10 μM to 1 mM steroids (diluted in 20 mM hydroxypropyl-β-cyclodextrin), and 0.01 to 0.5 mg/mL protein, depending on the enzyme activity. Enzyme activity was monitored by measuring the reduction of NADPH at 340 nm using a plate reader (BioTek, Cytation 5) at 37°C. A NADPH standard curve was analyzed under identical conditions with an extinction coefficient of 1398 M−1 for quantitation.
Enzymatic assays with purified OsrA and OsrB
The substrate preferences of OsrA and OsrB were analyzed under anaerobic conditions using a 100 μL reaction mixture containing 50 mM Tris/HCl (pH 7.0), 200 μM methyl viologen, mM steroids (from 10x stock solutions in methanol), and 50 μg/mL protein. Enzyme activity was monitored by measuring electron donor reduction at 605 nm using a plate reader (BioTek, Epoch 2) at 37°C. Quantification was performed using an electron donor standard curve generated under the same conditions with an extinction coefficient of 1689 M−1 for quantitation.
Phylogenetic tree construction
Genome metadata were retrieved from a local UHGG database and used to map genome IDs to species names. A comparative analysis was performed using BLASTp (version 2.15.0+) to search for homologs of the target sequence against the UHGP-100 database, limiting results to the top 20000 hits. The BLASTp output was processed to map genome IDs to species names and format the sequences in FASTA format, removing duplicates to ensure data quality. Additional sequences were appended to the data set as needed. Sequence alignment was performed using Clustal Omega (version 1.2.2)48 with output formatted as FASTA. Header sanitization was performed to remove special characters, and duplicate sequences were filtered out using custom Python scripts to maintain alignment integrity. Phylogenetic analysis was performed using IQ-TREE (version 2.3.6)49 with automatic model selection to determine the best-fitting substitution model based on the data. The reliability of the phylogenetic trees was assessed using 1,000 ultrafast bootstrap replicates to assess branch support. The final phylogenetic trees were visualized and interpreted using the Interactive Tree of Life (iTOL)50 to explore the evolutionary relationships among the identified protein sequences.
Metagenomics
To identify relevant sequences of OsrA or OsrB, a BLAST search was first conducted against the UHGP-100 database, applying a 49% similarity threshold based on experimental evidence indicating that this threshold effectively identifies relevant homologs while minimizing false positives. For OsrB, the identified protein sequences were used to construct a phylogenetic tree using Clustal Omega for alignment and IQ-TREE with the “mtest” model selection and 1,000 bootstrap replicates as described above. Based on the initial analysis, 25 sequences that were not phylogenetically related to OsrB were manually excluded. This curation step ensured that only sequences relevant to the target enzymes were retained for downstream analysis.
After phylogenetic filtering, genome information was traced back using the UHGP protein IDs. The corresponding genomes were downloaded from the Unified Human Gastrointestinal Genome (UHGG) database using FTP links provided in the metadata file. The genomes were then used for further analysis, where each protein sequence was screened against the respective genome using tblastn with a stringent e-value threshold of 1e−200 to ensure high specificity. The best nucleotide sequence was selected for each protein based on coverage and bit score and subsequently compiled into a combined FASTA file. Metagenomic samples were downloaded from the Sequence Read Archive (SRA). Reads were quality trimmed to remove adapter sequences using TrimGalore with default settings,51 and potential human contamination was removed by mapping the reads to the human reference genome (T2T-CHM13v2.0) using Bowtie2 (version 2.5.3) and removing the mapped reads with Samtools (version 1.61.1).52,53 Samples were then mapped to the gene reference datasets for osrA and osrB using Bowtie2 (version 2.5.3), and copies per million (CPM values) were calculated for each gene in each sample. Samples with total read counts below 1,000,000 were excluded.
Metagenomic data were filtered to ensure quality for statistical analyses. Zero values were replaced with 1e−6 for statistical assessment. A 99th percentile filter was applied to CPM values for each gene (osrA and osrB) to remove extreme outliers. Normality was assessed using the Shapiro-Wilk test; if both groups were normal (p > 0.05), a two-sided Welch’s t-test was used to determine if there was any significant difference between the groups, regardless of direction. If normality was not met, a two-sided Mann-Whitney U test was applied. This conservative approach ensured that differences were detected without assuming the direction of the effect, providing flexibility in hypothesis testing. Analyses were performed using Python with Scipy, Pandas, and Seaborn libraries.
Extended Data
Extended Data Figure 1. Phylogenetic analysis supporting assignment of HCS.1 within the Clostridium genus.
Tree generated from alignment of conserved marker genes.
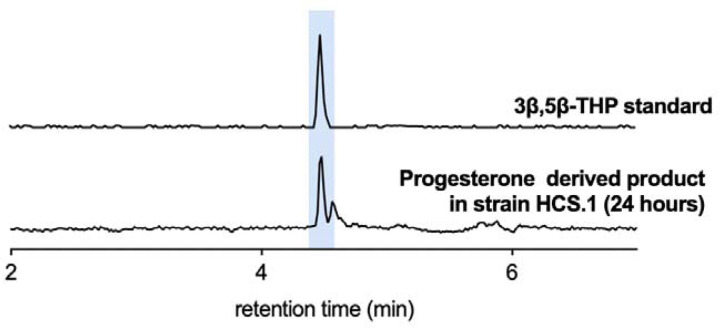
Extended Data Figure 2. Product of C. steroidoreducens HCS.1 incubation with progesterone.
Major progesterone product formed by C. steroidoreducens HCS.1. Comparison with reference standards confirms 3β-,5β-tetrahydroprogesterone (THP) production.
Extended Data Figure 3. 12% SDS-PAGE of E. coli lysates expressing SDR domain-containing proteins.
SDS-PAGE analysis showing the heterologous production of C. steroidoreducens SDR domain containing proteins in E. coli, pre- and post-isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. Red boxes highlight expressed protein.
Extended Data Figure 4. Reaction rates of enriched BLEONJ_2554 on indicated substrates.
Reaction rates for the 3’-reduction of 5-reduced steroids by anaerobically purified short-chain dehydrogenase BLEONJ_2554.
Extended Data Figure 5. Reaction rates of enriched OsrC on indicated substrates.
Reaction rates for the 3’-reduction of 5-reduced steroids by anaerobically purified short-chain dehydrogenase OsrC.
Extended Data Figure 6. OsrA and OsrB products from Δ1- and Δ4-steroid hormone substrates.
(A) Products following prednisolone incubation with purified OsrA or OsrB. Comparison to a cortisol reference standard confirm that OsrA generates the Δ1-reduced product and show that OsrB produces a distinct cortisol isomer. (B) Products following boldenone incubation with purified OsrA or OsrB. Comparison to reference standards confirm that OsrA and OsrB generate Δ1- and Δ4-reduced products, respectively.
Extended Data Figure 7. osrA levels in expanded dataset of healthy and Crohn’s disease metagenomes.
(A) Reads mapping to osrA homologs in expanded dataset of Crohn’s disease (CD) patient metagenomes relative to healthy controls. (B) Difference in osrA homolog levels in CD relative to healthy metagenomes.
Extended Data Figure 8. Association between osrA and osrB and Crohn’s disease in Integrative Human Microbiome Project metagenomes.
(A) Reads mapping to osrA and osrB homologs in Crohn’s disease (CD) metagenomes relative to non-IBD controls. CPM stands for copies per million. (B) Difference in osrB homolog levels from taxa with the most significant changes in relative abundance between CD dysbiotic and non-IBD metagenomes. (C) Difference in osrA homolog levels in CD dysbiotic relative to non-IBD metagenomes.
Extended Data Figure 9. Association between osrA and osrB and steroid usage in the Lewis et al study.
Reads mapping to osrA and osrB homologs in Crohn’s disease (CD) patient metagenomes grouped based on patient corticosteroid treatment.
Supplementary Material
ACKNOWLEDGEMENTS
Research reported in this publication was supported by funding from the National Institutes of Health (NIGMS R35GM146969 and NIDDK P30DK042086, via the University of Chicago Center for Interdisciplinary Study of Inflammatory Intestinal Disorders) and the Searle Scholars Program (to S.H.L), as well as the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer 542537779 (to C.J.).
REFERENCES
- 1.Nussey S. & Whitehead S. The adrenal gland. in Endocrinology: An Integrated Approach (BIOS Scientific Publishers, 2001). [PubMed] [Google Scholar]
- 2.Nussey S. & Whitehead S. The gonad. in Endocrinology: An Integrated Approach (BIOS Scientific Publishers, 2001). [PubMed] [Google Scholar]
- 3.Buffie C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paik D. et al. Human gut bacteria produce ΤΗ17-modulating bile acid metabolites. Nature 603, 907–912 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adlercreutz H., Martin F., Järvenpää P. & Fotsis T. Steroid absorption and enterohepatic recycling. Contraception 20, 201–223 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Decker H. A. et al. Metabolism of 4-C 14-cortisol in man: body distribution and rates of conjugation. J. Clin. Endocrinol. Metab. 16, 1137–1150 (1956). [DOI] [PubMed] [Google Scholar]
- 8.Peterson R. E., Wyngaarden J. B., Guerra S. L., Brodie B. B. & Bunim J. J. THE PHYSIOLOGICAL DISPOSITION AND METABOLIC FATE OF HYDROCORTISONE IN MAN. J. Clin. Invest. 34, 1779–1794 (1955). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandberg A. A. & Slaunwhite W. R. Metabolism of 4-C14-testosterone in human subjects. I. Distribution in bile, blood, feces and urine. J. Clin. Invest. 35, 1331–1339 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adlercreutz H. & Martin F. Biliary excretion and intestinal metabolism of progesterone and estrogens in man. J. Steroid Biochem. 13, 231–244 (1980). [DOI] [PubMed] [Google Scholar]
- 11.Migeon C. J., Paul A. C., Samuels L. T. & Sandberg A. A. Metabolism of 4-C14-corticosterone in man. J. Clin. Endocrinol. Metab. 16, 1291–1298 (1956). [DOI] [PubMed] [Google Scholar]
- 12.Möstl E. & Palme R. Hormones as indicators of stress. Domest. Anim. Endocrinol. 23, 67–74 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Palme R., Rettenbacher S., Touma C., El-Bahr S. M. & Möstl E. Stress hormones in mammals and birds: comparative aspects regarding metabolism, excretion, and noninvasive measurement in fecal samples. Ann. N. Y. Acad. Sci. 1040, 162–171 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Nabarro J. D., Moxham A., Walker G. & Slater J. D. Rectal hydrocortisone. Br. Med. J. 2, 272–274 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wade A. P., Slater J. D., Kellie A. E. & Holliday M. E. Urinary excretion of 17-ketosteroids following rectal infusion of cortisol. J. Clin. Endocrinol. Metab. 19, 444–453 (1959). [DOI] [PubMed] [Google Scholar]
- 16.Martin F., Peltonen J., Laatikainen T., Pulkkinen M. & Adlercreutz H. Excretion of progesterone metabolites and estriol in faeces from pregnant women during ampicillin administration. J. Steroid Biochem. 6, 1339–1346 (1975). [DOI] [PubMed] [Google Scholar]
- 17.Ly L. K. et al. Bacterial steroid-17,20-desmolase is a taxonomically rare enzymatic pathway that converts prednisone to 1,4-androstanediene-3,11,17-trione, a metabolite that causes proliferation of prostate cancer cells. J. Steroid. Biochem. Mol. Biol. 199, 105567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R. & Goodman A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latif S. A., Sheff M. F., Ribeiro C. E. & Morris D. J. Selective inhibition of sheep kidney 11β-hydroxysteroid dehydrogenase isoform 2 activity by 5α-reduced (but not 5β) derivatives of adrenocorticosteroids. Steroids 62, 230–237 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Honour J. W., Borriello S. P., Ganten U. & Honour P. Antibiotics attenuate experimental hypertension in rats. J. Endocrinol. 105, 347–350 (1985). [DOI] [PubMed] [Google Scholar]
- 21.N P. et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 374, (2021). [DOI] [PubMed] [Google Scholar]
- 22.Terrisse S., Zitvogel L. & Kroemer G. Effects of the intestinal microbiota on prostate cancer treatment by androgen deprivation therapy. Microb. Cell Graz Austria 9, 202–206 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokes N. A. & Hylemon P. B. Characterization of delta 4–3-ketosteroid-5 beta-reductase and 3 beta-hydroxysteroid dehydrogenase in cell extracts of Clostridium innocuum. Biochim. Biophys. Acta 836, 255–261 (1985). [DOI] [PubMed] [Google Scholar]
- 24.Penning T. M. & Covey D. F. 5β-Dihydrosteroids: Formation and Properties. Int. J. Mol. Sci. 25, 8857 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 26.McCurry M. D. et al. Gut bacteria convert glucocorticoids into progestins in the presence of hydrogen gas. Cell 187, 2952–2968.e13 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devendran S., Mythen S. M. & Ridlon J. M. The desA and desB genes from Clostridium scindens ATCC 35704 encode steroid-17,20-desmolase. J. Lipid Res. 59, 1005–1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maini Rekdal V. et al. A widely distributed metalloenzyme class enables gut microbial metabolism of host- and diet-derived catechols. eLife 9, e50845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard P. A., Liang X., Schulz H. & Kim J.-J. P. The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase. J. Biol. Chem. 278, 37553–37560 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Little A. S. et al. Dietary- and host-derived metabolites are used by diverse gut bacteria for anaerobic respiration. Nat. Microbiol. 9, 55–69 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funabashi M. et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang D.-J., Ridlon J. M., Moore D. R., Barnes S. & Hylemon P. B. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta 1781, 16–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edenharder R. & Schneider J. 12 beta-dehydrogenation of bile acids by Clostridium paraputrificum, C. tertium, and C. difficile and epimerization at carbon-12 of deoxycholic acid by cocultivation with 12 alpha-dehydrogenating Eubacterium lentum. Appl. Environ. Microbiol. 49, 964–968 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doden H. L. & Ridlon J. M. Microbial Hydroxysteroid Dehydrogenases: From Alpha to Omega. Microorganisms 9, 469 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doden H. et al. Metabolism of Oxo-Bile Acids and Characterization of Recombinant 12α-Hydroxysteroid Dehydrogenases from Bile Acid 7α-Dehydroxylating Human Gut Bacteria. Appl. Environ. Microbiol. 84, e00235–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ardizzone S. & Bianchi Porro G. Comparative tolerability of therapies for ulcerative colitis. Drug Saf. 25, 561–582 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Yang R. & Yu Y. Glucocorticoids are double-edged sword in the treatment of COVID-19 and cancers. Int. J. Biol. Sci. 17, 1530–1537 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almeida A. et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 39, 105–114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis J. D. et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 18, 489–500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ha C. W. Y. et al. Translocation of Viable Gut Microbiota to Mesenteric Adipose Drives Formation of Creeping Fat in Humans. Cell 183, 666–683.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall A. B. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lloyd-Price J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arp G. et al. Gut Bacteria Encode Reductases that Biotransform Steroid Hormones. bioRxiv 2024.10.04.616736 (2024) doi: 10.1101/2024.10.04.616736. [DOI] [Google Scholar]
- 43.Wick R. R., Judd L. M., Gorrie C. L. & Holt K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wick R. R., Judd L. M., Gorrie C. L. & Holt K. E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genomics 3, e000132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaumeil P.-A., Mussig A. J., Hugenholtz P. & Parks D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O S. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genomics 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mi L., W H. & S A. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen L.-T., Schmidt H. A., von Haeseler A. & Minh B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I. & Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Krueger F. et al. FelixKrueger/TrimGalore: v0.6.10. Zenodo doi: 10.5281/zenodo.7598955 (2023). [DOI] [Google Scholar]
- 52.Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.H L. et al. The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 25, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.