Abstract
Background
Clinical studies have reported rising pre-treatment HIV drug resistance during antiretroviral treatment (ART) scale-up in Africa, but representative data are limited. We estimated population-level drug resistance trends during ART expansion in Uganda
Methods
We analyzed data from the population-based open Rakai Community Cohort Study conducted at agrarian, trading, and fishing communities in southern Uganda between 2012 and 2019. Consenting participants aged 15–49 were HIV tested and completed questionnaires. Persons living with HIV (PLHIV) provided samples for viral load quantification and virus deep-sequencing. Sequence data were used to predict resistance. Population prevalence of class-specific resistance and resistance-conferring substitutions were estimated using robust log-Poisson regression.
Findings
Data from 93,622 participant-visits, including 4,702 deep-sequencing measurements, showed that the prevalence of NNRTI resistance among pre-treatment viremic PLHIV doubled between 2012 and 2017 (PR:1.98, 95%CI:1.34–2.91), rising to 9.61% (7.27–12.7%). The overall population prevalence of pre-treatment viremic NNRTI and NRTI resistance among all participants decreased during the same period, reaching 0.25% (0.18% - 0.33%) and 0.05% (0.02% - 0.10%), respectively (p-values for trend = 0.00015, 0.002), coincident with increasing treatment coverage and viral suppression. By the final survey, population prevalence of resistance contributed by treatment-experienced PLHIV exceeded that from pre-treatment PLHIV, with NNRTI resistance at 0.54% (0.44%−0.66%) and NRTI resistance at 0.42% (0.33%−0.53%). Overall, NNRTI and NRTI resistance was predominantly attributable to rtK103N and rtM184V. While 10.52% (7.97%−13.87%) and 9.95% (6.41%−15.43%) of viremic pre-treatment and treatment-experienced PLHIV harbored the inT97A mutation, no major dolutegravir resistance mutations were observed.
Interpretation
Despite rising NNRTI resistance among pre-treatment PLHIV, overall population prevalence of pre-treatment resistance decreased due to treatment uptake. Most NNRTI and NRTI resistance is now contributed by treatment-experienced PLHIV. The high prevalence of mutations conferring resistance to components of current first-line ART regimens among PLHIV with viremia is potentially concerning.
Introduction
Antiretroviral therapy (ART) suppresses human immunodeficiency virus (HIV) replication in persons living with HIV (PLHIV),1 which slows disease progression2 and prevents viral transmission.3 With increased uptake of ART as well as other interventions such as voluntary medical male circumcision, HIV incidence has fallen by nearly 40% globally since 2010.4
Viral resistance to ART threatens the clinical and public health impact of treatment scale-up5,6. Drug resistance can be acquired when an individual infected with a susceptible virus develops resistance following treatment. This is more common when treatment adherence is intermittent,7 but can occur despite high adherence.8 Throughout sub-Saharan Africa, the epicenter of the global HIV epidemic4, the majority of patients who remain viremic despite being on treatment with first-line regimens harbor resistance to at least one component of that regimen1. Viral genomes with resistance-conferring mutations can be transmitted to HIV seronegative individuals, increasing the risk of first-line treatment failure approximately five-fold.9
Through 2018, preferred first-line HIV ART regimens relied on a combination of nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitor (NNRTI)10. During the scale-up of ART, an increase in the prevalence of NNRTI resistance among pre-treatment PLHIV has been observed globally11. A systematic meta-regression estimated a 1.3% annual increase in NNRTI resistance in eastern Africa, reaching 10% by 2016. These findings have been corroborated by recent cross-sectional studies and World Health Organization (WHO) surveys1,12,13 and prompted a shift in recommendations to dolutegravir (DTG, an integrase strand transfer inhibitor [INSTI]) given in combination with NRTIs (e.g. Tenofovir disoproxil fumarate [TDF] and Lamivudine [3TC]) for first-line ART. Further, long-lasting injectable INSTIs (e.g. cabotegravir [CAB]) in combination with an NNRTI (rilpivirine [RPV]) are currently being rolled out throughout sub-Saharan Africa14.
The vast majority of data on the prevalence of HIV ART resistance in sub-Saharan Africa is derived from the population of PLHIV who report to healthcare clinics or hospitals1,11–13. Clinic-based studies are subject to biases in the population of PLHIV who are engaged and retained in care, which is not universal15. Specifically in eastern and southern Africa, only 95% of women and 91% of men are aware of their HIV status and of those only 92% and 86% are on ART, respectively4. Further, clinic-based studies are able to estimate only the prevalence of ART resistance among PLHIV and not among the general population. The latter is the epidemiologically relevant parameter to inform the risk of exposure to HIV with ART resistance among seronegative individuals16.
General population-based studies, in which all individuals, regardless of HIV serostatus, are recruited to participate, can address these shortcomings. This design also allows for the accurate estimation of the overall prevalence of resistance and the relative contributions from different groups, such as pre-treatment and treatment-experienced PLHIV. A recent cross-sectional population-based study of drug resistance in KwaZulu-Natal, South Africa found very low (<1%) levels of resistance to INSTIs prior to DTG roll-out, but observed rtM184V (3TC resistance), rtK65R (TDF resistance), and rtK70E (TDF resistance) in 32.6%, 12.0%, and 6.2% of treatment-experienced PLHIV. Further, the rtE138A mutation, which confers resistance to RPV, was observed in in 6.5% and 7.9% of ART-experienced and -naïve PLHIV, respectively, in this setting17. Cross-sectional studies, however, are unable to assess temporal trends in the prevalence of resistance among PLHIV and the general population and do not capture the overall decrease in the prevalence of viremic HIV during ART scale-up. Conversely, longitudinal population-based cohort designs enable more precise monitoring of resistance evolution and a dynamic evaluation of the risks posed to current and future ART regimens. This study design is particularly useful in the context of rapidly changing population sizes of pre-treatment and treatment-experienced viremic PLHIV as has been observed globally in recent decades during expansion of treatment and prevention programs15.
Here, we analyzed HIV deep-sequence data collected from 3,407 PLHIV as part of a general population-based open cohort study in southern Uganda spanning a nine-year period of intense ART scale-up and declines in HIV incidence18–20. Our validated deep-sequencing protocol21 allows for the identification and quantification of drug resistance mutations present in a minority of the viral population within a given PLHIV, which can be selected for upon treatment initiation but are missed by consensus sequencing methods22–24. We estimated the prevalence of NNRTI, NRTI, and protease inhibitor (PI) resistance among the entire study population and among PLHIV as well as the temporal dynamics of viral resistance-conferring mutations among pre-treatment and treatment-experienced PLHIV.
Methods
Study design and participant selection
The Rakai Community Cohort Study (RCCS) is an open population-based census and cohort study conducted at approximately 18–24 month intervals (appendix pp 2–3) in agrarian (HIV prevalence 9–26%25), semi-urban trading (11–21%25), and Lake Victoria fishing (38–43%25) communities in southern Uganda.20 At each survey round, households in participating communities are censused and residents aged 15–49 capable of providing informed written consent (or assent if under 18) are invited to participate. Consenting participants are administered a structured questionnaire that obtains sociodemographic, behavioral, and health information, including self-reported past and current ART use. Voluntary HIV testing of participants is conducted using a rapid test algorithm26 and venous blood samples are taken for viral quantification and sequencing.
The RCCS is administered by the Rakai Health Sciences Program (RHSP) and has received ethical approval from the Uganda Virus Research Institute’s Research and Ethics Committee (HS540), the Uganda National Council for Science and Technology (GC/127/08/12/137), and the Johns Hopkins School of Medicine (IRB00217467). Participants provided written informed consent at each survey round.
We used survey data from 19 RCCS surveys conducted between November 5, 1994 and November 4, 2020, and HIV viral load and sequence data from five rounds conducted between August 10, 2011 and November 4, 2020. Participants with serologically confirmed HIV infection were considered pre-treatment during a given round if they reported never having taken ARTs at that round and all prior rounds in which they participated. PLHIV were considered treatment-experienced during a given round if they reported using ART at that round or any earlier rounds. Recommended first-line ART regimens in RCCS communities are presented in appendix p 4. Herein, study rounds were referred to by the year of the median interview date (appendix p 2). Reporting of this study adheres to the STROBE guidance27.
HIV viral load quantification
HIV viral load was measured on serum/plasma samples using the Abbott real-time m2000 assay (Abbott Laboratories) at the Rakai health Sciences Program (Kalisizo, Uganda). Viral load measurements were conducted primarily among PLHIV in fishing communities in the 2012 survey round and for all PLHIV in later survey rounds. Viral loads ≥ 1000 copies/mL were considered viremic. Pre-treatment PLHIV in the 2012 round with missing viral load were imputed (appendix pp 4–5). In the rare instance where viral load measurements were missing for PLHIV in subsequent survey rounds, these observations were dropped from the analysis.
HIV deep sequencing
Full-length HIV deep sequencing was conducted through the Phylogenetics and Networks for Generalized HIV Epidemics in Africa consortium (PANGEA-HIV).28,29 As described elsewhere,19 for the 2012 and 2014 surveys sequencing on Illumina MiSeq and HiSeq platforms using an amplicon-based approach30 was attempted for participants who self-reported never having been on ART and either had a missing viral load or were known to be viremic (appendix p 21). All viremic participant-visits, regardless of treatment status, in the 2015 through 2019 survey rounds, as well as select 2012 and 2014 participant-visits, were sequenced using the veSEQ-HIV protocol, which involves oligo-nucleotide bait enrichment of HIV from pooled metagenomic libraries prepared without virus-specific PCR.31 Our high-throughput implementation of the veSEQ-HIV protocol incorporates quantitative positive controls consisting of a serial dilution HXB2 cultured virus diluted in pooled human plasma from donors testing negative for HIV, as well and negative plasma controls included with every batch of samples processed. Sequencing quality was monitored from total counts of HIV reads detected in the quantitative controls, their PCR duplication rates and median insert sizes. Where contaminations were detected either by the presence of HIV reads in the negative control, or HXB2 reads present in the samples, sequencing runs were repeated. Sequencing for the 2019 round was only conducted for samples collected through May 17, 2019 (N = 171/453, 37.7% of viremic participant visits). Consensus sequences were generated using shiver32 and subtyped by identifying the most similar reference sequence and using the Recombination Identification Program.33
Identification of drug resistance mutations
A validated bioinformatic pipeline, drmSEQ, was used to identify amino acid substitutions associated with reduced susceptibility to ART and predict individual drug and drug class susceptibilities at the University of Oxford Nuffield Department of Medicine.34 Paired-end reads were trimmed of adapters, primers, and low-quality bases with trimmomatic35 and then filtered to remove pol hypermutated sequences and non-HIV pol sequences. Duplicate reads introduced by PCR were removed using Picard MarkDuplicates36 and unique reads were locally aligned to 142 HIV subtype references using blastx.37 A manually-curated codon-restricted multiple-alignment of the references was used to lookup coordinates and mutations relative to HXB2 (GenBank: K03455.1). Only mutations supported by a minimum of 10 PCR-deduplicated reads and by ≥5% of reads spanning the corresponding site were considered.38 These thresholds are based on HIV read counts after removal of non-unique PCR duplicate reads. Importantly, the same thresholds were used in a previous validation and demonstrated comparable sensitivity to a gold standard clinical assay34. Amino acid substitutions were scored according to the Stanford University HIV Drug Resistance Database. Scores were summed to predict susceptibility to 25 HIV drugs (appendix p 13).39–41 A score ≥30 (intermediate/high-level) for a given drug was categorized as resistant. Resistance was not predicted if less than half of the relevant positions for a given drug had fewer than 10 reads. Samples in which there was insufficient sequencing coverage for one or more drug within a class were not assigned a resistance categorization for that class. Samples with resistance to at least one drug within each class were categorized as resistant to that class.
Outcome measures
The primary outcomes of this study were the prevalence of viremic PLHIV with INSTI, NNRTI, NRTI, and or PI resistance among all participants, regardless of HIV serostatus, in each survey round. We also estimated the population prevalence of NNRTI, NRTI, and PI resistance contributed by viremic pre-treatment and treatment-experienced PLHIV and the population prevalence of multi-class resistance. We further estimated the prevalence of NNRTI, NRTI, and PI resistance and individual resistance-conferring viral mutations specifically among viremic pre-treatment and treatment-experienced PLHIV in each survey round. Given the greater prevalence of viremic HIV in fishing communities, among men, and among younger age groups in the RCCS19,20,25 we evaluated the association between these variables and resistance in bivariate analyses. Stratified estimates were generated for covariates identified as significant in bivariate analyses. Given the availability of sequence data, we restricted prevalence estimates that include treatment-experienced PLHIV to the 2015 and 2017 survey rounds and use the 2017 survey as an end-point for pre-treatment PLHIV. For context, we estimated the prevalence of PLHIV, viremic PLHIV (2014 and later due to missing viral load data), viremic PLHIV pre-treatment, and viremic treatment-experienced PLHIV among participants in each round (2014 and later).
Statistical methods
Statistical analyses were conducted in R v.4.4.1.42 Prevalence was estimated using Poisson regression with a log-link and robust (sandwich) standard errors43 which were fit with general estimating equations using geepack v.1.3.11 to account for repeated measures44. Correlation structures were chosen by minimizing the Quasi Information Criterion (QIC). We used inverse probability weighting to account for missing sequence data among viremic study participants. Sampling weights were calculated based on availability of a viral load measurement (True/False), log10 copies/mL where available, community type (agrarian/fishing/trading), age category ((14,24]/(24,34]/(34,49]), and sex (M/F) stratified by survey round. Emmeans v. 1.10.4 was used to calculate prevalence within strata45. 95% confidence intervals and p-values (0.05) were calculated using the Wald method. p-values were calculated using the stats package in R. Data analysis and visualization was done using tidyverse v.2.0.0,46 ggplot2 v.3.5.1,47 cowplot v.1.1.3,48 patchwork v. 1.2.049, and ggpattern v.1.1.150. Readxl v.1.4.351 and haven v.2.5.4.952 were used to parse data files. See appendix pp 5–17 for detailed methods.
Role of the funding source
The funders had no role in study design, collection, analysis, and interpretation of data; and no role in the writing of the report and decision to publish.
Results
Study population
Between August 10, 2011 and November 4, 2020, a total of 43,361 people participated in the RCCS, of whom 7,923 (18.27%) were PLHIV. Of 93,622 participant-visits about a fifth were from PLHIV (table 1). Over the analysis period, the median age of study participants remained stable whereas the age of PLHIV increased slightly (appendix pp 18–19). Viral load measurements were available for 1,959/3,498 (56.00%) of PLHIV in the 2012 survey and 13,962/14,008 (99.67%) PLHIV in later survey rounds. A total of 46 participant-visits from the 2014–2019 surveys with missing viral loads were dropped from subsequent analyses. Among participant-visits from PLHIV in 2014–2019, 26.29% were contributed by viremic PLHIV and of those 79.81% were pre-treatment viremic. After imputation of missing viral loads (Methods), 56.75% of PLHIV in the 2012 survey were identified as pre-treatment viremic.
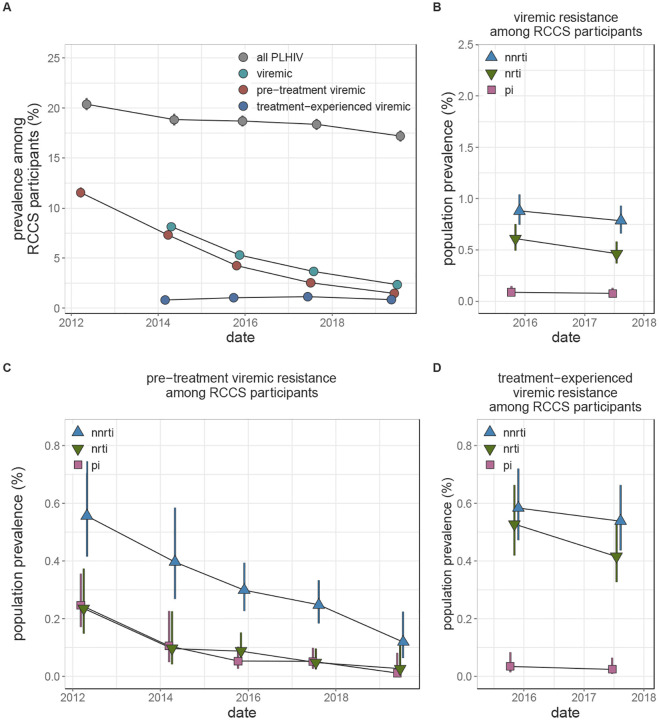
HIV seroprevalence among participants decreased from 20.38% (95% CI 19.78% - 20.99%) in the 2012 survey to 17.20% (95% CI 16.68% - 17.74%) in the 2019 survey (figure 1A, appendix p 20). Concurrent with an increase in the proportion of PLHIV reporting ever having been on treatment from 25.04% (2012) to 85.25% (2019, appendix p 21), HIV viremia among all participants decreased significantly from 8.14% (2014, 95% CI 7.75% - 8.55%) to 2.34% (2019, 95% CI 2.14% - 2.57%). These declines were driven by a nearly nine-fold (prevalence ratio (PR) 0.13, 95% CI 0.11 – 0.15) decrease in the prevalence of pre-treatment viremia among participants over the study (figure 1A, appendix p 20). The prevalence of treatment-experienced viremia remained stable at around 1% of participants.
Figure 1: Longitudinal trends in HIV seroprevalence and population prevalence of viremic HIV drug resistance among Rakai Community Cohort Study participants, 2012–2019.
(A) Estimated prevalence of all HIV, viremic HIV, viremic pre-treatment HIV, and viremic treatment-experienced HIV in each round. Due to missing viral load data, prevalence of viremic HIV and viremic treatment-experienced HIV were not estimated in the 2012 survey. For some estimates confidence bands do not extend beyond point. (B-D) Estimated population prevalence of all viremic (B), pre-treatment viremic (C), and treatment-experienced viremic (D) NNRTI, NRTI, and PI resistance among all study participants. Estimates were generated using Poisson regression with robust standard errors with survey round as a predictor variable. Generalized estimating equations with correlation structure selection by Quasi Information Criterion value (A: independent, B: independent, C: independent, D: exchangeable (NNRTI and PI), independent (NRTI)) were used to account for repeat participants across study rounds. Error bars indicate the Wald 95% confidence interval for the mean value. For clarity, points are jittered along the x-axis. PLHIV = people living with HIV. NNRTI = non-nucleoside reverse transcriptase inhibitors (blue upwards facing triangles). NRTI = nucleoside reverse transcriptase inhibitors (green downwards facing triangles). PI = integrase inhibitors (pink squares).
Identification of resistance genotypes in deep-sequence data
Deep-sequence based identification of drug resistance mutations (DRMs) was attempted on 4,525/5,724 (79.51%) of viremic participant visits (appendix p 22). Attempted genotyping did not vary by participant age, sex, or community type of residence. The veSeq-HIV sequencing protocol was used for 44.99% of all sequenced viremic participants and the vast majority (99.37%) of those in the 2015 through 2019 survey rounds (appendix p 23). Among samples from viremic participant-visits on which deep-sequence based genotyping was attempted, sufficient data were available to reliably genotype 4,072/4,525 (90.01%, appendix pp 24–25) viruses from 3,407 PLHIV for at least one drug. Sequencing success did not depend on age, community, type or sex, (p-values ≥ 0.37) but was more likely among samples with higher viral load and those sequenced with veSeq-HIV (p-values = 0.0005). Among sequenced participant-visits successfully genotyped for all INSTIs (n=2,578, appendix pp 26–32), NNRTIs (n=3,050), NRTIs (n=3,009) or PIs (n=3,520) <1%, 12.46%, 6.75%, and 1.88% had predicted resistance, respectively (appendix pp 33–34). Given the minimal INSTI resistance we did not estimate the prevalence of INSTI resistance.
Population prevalence of viremic resistance
In 2017, the population prevalence of viremic NNRTI, NRTI, and PI resistance among all participants, regardless of HIV serostatus, was 0.79% (95% CI 0.66% - 0.93%), 0.46% (95% CI 0.37% - 0.58%), and 0.08% (95% CI 0.04% - 0.13%), respectively. These levels were stable compared to 2015 (figure 1B, appendix p 35). In stratified analyses, NNRTI and NRTI resistance was more than three-times as common in fishing as compared to agrarian or trading communities and most prevalent among people aged 25–34 years old (p-values 0.0001, appendix pp 36–38).
The prevalence of NNRTI and NRTI resistance contributed by pre-treatment viremic PLHIV decreased 2.3-fold (PR 0.44, 95% CI 0.29 – 0.68) and 5-fold (PR 0.21, 95% CI 0.09 – 0.47) between the 2012 and 2017 surveys (p-values < 0.0001, figure 1C, appendix p 38), concurrent with the observed decline in population prevalence of pre-treatment viremia. Consequently, in the 2017 survey round, treatment-experienced viremic PLHIV contributed 68.46% (95% CI 59.49% - 75.44%) and 89.61% (79.92% - 94.62%) of all NNRTI and NRTI resistance, respectively. Specifically, the population prevalence of resistance to NNRTIs and NRTIs contributed by treatment-experienced viremic PLHIV in the 2017 survey was 0.54%, 95% CI 0.44%−0.66% and 0.42%, 0.33%−0.53% as compared to 0.25%, 0.18%−0.32% and 0.05%, 0.02%−0.1% (appendix pp 39 – 40) contributed by pre-treatment viremic PLHIV.
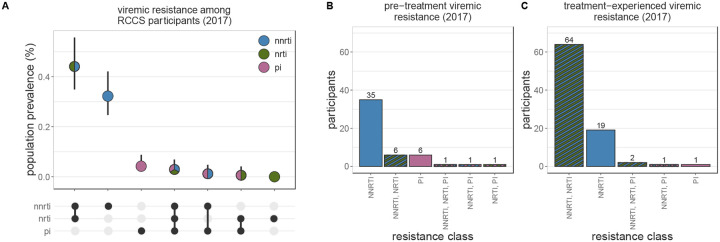
Resistance profiles varied considerably by treatment status (figure 2 and appendix p 41). Among pre-treatment viremic PLHIV with available genotype for NNRTIs, NRTI, and PIs the majority with any resistance were NNRTI mono-resistant (2017 survey n=35, 70%). In contrast, among treatment-experienced viremic PLHIV with any resistance, NNRTI/NRTI dual-class resistance was the most common profile (2017 survey n=64, 73.56%). Among all participants, the most common forms of viremic resistance were NNRTI/NRTI dual-class resistance (2017: 0.44%, 95% CI 0.35% - 0.56%) and NNRTI mono-resistance (2017: 0.32%, 95% CI 0.25% - 0.42%), consistent with a dominant contribution from treatment-experienced viremic PLHIV. Other resistance profiles were extremely rare (<0.1%).
Figure 2: Patterns of multi-class resistance in Rakai Community Cohort Study, 2017.
(A) Estimating population prevalence of NNRTI, NRTI, and PI mono-resistance and NNRTI/NRTI, NNRTI/PI, NRTI/PI, and NNRTI/NRTI/PI multi-class resistance among all RCCS study participants. Estimates were generated using Poisson regression with robust standard errors with survey round as a predictor variable. General estimating equations with the best fit correlation structure by QIC value (NNRTI, NRTI, PI mono-resistance and NNRTI/NRTI and NNRTI/PI multi-class resistance: independent, NRTI/PI and NNRTI/NRTI/PI: exchangeable) were used to account for repeated measures from the same participant Error bars indicate the Wald 95% confidence interval for the mean value. (B) Multi-class resistance profiles among 50 pre-treatment viremic 2017 participant-visits with genotype data for all NNRTIs, NRTIs, PIs, and resistance to at least one of these drug classes. (C) Multi-class resistance profiles among 87 treatment-experienced viremic 2017 participant-visits with genotype data for all NNRTIs, NRTIs, PIs, and resistance to at least one of these drug classes. NNRTI = non-nucleoside reverse transcriptase inhibitors. NRTI = nucleoside reverse transcriptase inhibitors. PI = integrase inhibitors.
Prevalence of resistance among pre-treatment viremic PLHIV
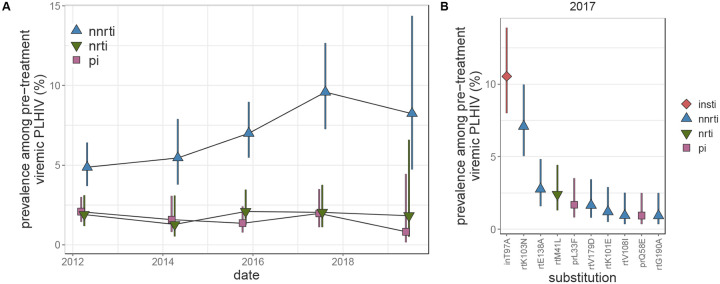
Between the 2012 and 2017, NNRTI resistance among pre-treatment viremic PLHIV increased by a factor of 1.98 (95% CI: 1.34–2.91), reaching 9.61% (95% CI: 7.27% - 12.7%) (figure 1A and appendix pp 42). This increasing trend did not vary by sex, age, type of community of residence, or sequencing approach (appendix pp 43–44).The prevalence of NRTI and PI resistance remained stable and below 2.1% over the same time period.
Among pre-treatment viremic PLHIV, the most prevalent resistance-associated mutation was inT97A (figure 3B and appendix pp 46–48), an INSTI-resistance (particularly Elvitegravir53) mutation, detected in ~10% of pre-treatment viremic participants in the 2012 through 2017 survey rounds and in 20% (95% CI 13.89% - 28.8%) in the partial 2019 survey data. The most common NNRTI-resistance mutation was rtK103N, found in 7.1% (95% CI 5.05% - 9.97%) of pre-treatment viremic PLHIV in the 2017 survey, a 4.26-fold (95% CI 2.11 – 8.59) increase compared to the 2012 survey. The next most prevalent NNRTI mutation, rtE138A, which is associated with 2.5-fold reduced susceptibility to RPV54, was present in only 2.77% (95% CI 1.59% – 4.85%) of pre-treatment viremic PLHIV and its prevalence remained stable compared to the 2012 survey (p-value = 0.49). NRTI resistance mutations were rare compared to NNRTI mutations. Genotypes associated with intermediate/high-level INSTI resistance were identified in 16 pre-treatment viremic participant-visits (<1%), the majority of which harbored inE92G (n=13), which confers resistance to Elvitegravir, a drug that is not routinely used in Uganda (appendix p 4). Mutations conferring intermediate/high-level resistance to DTG were not observed.
Figure 3: Longitudinal trends in HIV drug resistance among pre-treatment viremic Rakai Community Cohort Study participants, 2012–2019.
(A) Estimated prevalence of NNRTI, NRTI, and PI resistance among pre-treatment viremic PLHIV. For visual clarity, points are jittered along the x-axis. (B) Prevalence in the 2017 survey of the 10 most prevalent substitutions in pre-treatment viremic PLHIV sorted by prevalence. Estimates were generated using Poisson regression with robust standard errors with survey round as a predictor variable. General estimating equations with the best fit correlation structure by QIC value (NNRTI: exchangeable, NRTI: exchangeable, PI: AR1, substitutions: independent to ensure convergence) were used to account for repeated measures from the same participant Error bars indicate the Wald 95% confidence interval for the mean value within each category. PLHIV = people living with HIV. NNRTI = non-nucleoside reverse transcriptase inhibitors. NRTI = nucleoside reverse transcriptase inhibitors. PI = integrase inhibitors.
Prevalence of resistance among treatment-experienced viremic PLHIV
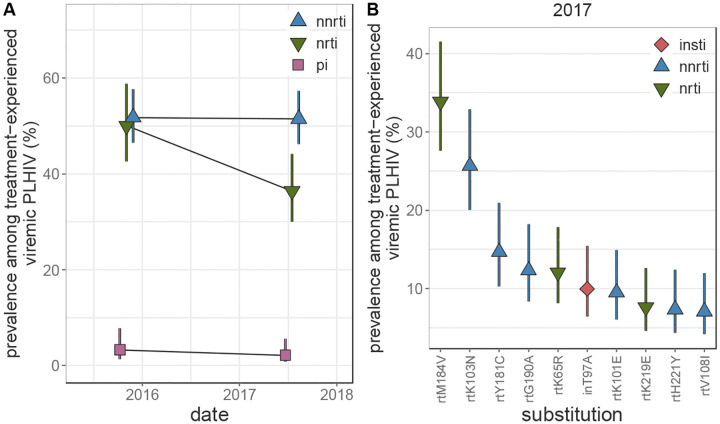
Prevalence of NNRTI and NRTI resistance was substantially higher among treatment-experienced participants as compared to pre-treatment PLHIV. In 2017, 51.49% (95 CI 46.24%−57.34%) and 36.46% (95% CI 30.06%−44.22%) of treatment-experienced participants with viremia harbored NNRTI and NRTI resistant viruses, respectively (figure 4A and appendix p 49). NRTI resistance was 1.62 (95% CI 1.03 – 2.56, appendix pp 50–51) times more common among participants aged 25–34 years compared to 15–24 year-olds (p-value = 0.037). While NNRTI resistance remained stable between the 2015 and 2017 survey rounds, NRTI resistance decreased by more than quarter (prevalence ratio 0.73%, 95% CI 0.58 – 0.92, p-value = 0.0084). Only 2.13% (95% CI 0.81%−5.63%) of treatment-experienced viremic participants in the 2017 survey round had viruses with PI resistance.
Figure 4: Longitudinal trends in HIV drug resistance among treatment-experienced viremic Rakai Community Cohort Study participants, 2015–2017.
(A) Estimated prevalence of NNRTI, NRTI, and PI resistance among treatment-experienced viremic PLHIV. For visual clarity, points are jittered along the x-axis. (C) Prevalence of the 10 most prevalent drug resistance mutations in treatment-experienced viremic PLHIV in the 2017 survey round, sorted by prevalence. Estimates were generated using Poisson regression with robust standard errors with survey round as a predictor variable. General estimating equations with the best fit correlation structure by QIC value (NNRTI and PI: exchangeable, NRTI: independent, mutations: independent to ensure convergence) were used to account for repeated measures from the same participant Error bars indicate the Wald 95% confidence interval for the mean value within each category. PLHIV = people living with HIV. NNRTI = non-nucleoside reverse transcriptase inhibitors. NRTI = nucleoside reverse transcriptase inhibitors. PI = integrase inhibitors.
The resistance-associated mutations observed among treatment-experienced viremic participants differed considerably compared to pre-treatment viremic participants (appendix pp 46, 52). NRTI resistance among treatment-experienced viremic participants in the 2017 survey was most frequently due to the rtM184V (33.89%, 95% CI 27.61% - 41.59%), rtK65R (12.07% 95% CI 8.14% - 17.9%), and rtK219E (7.64%, 95% CI 4.6% −12.67%) substitutions, which were rarely observed among pre-treatment PLHIV. On the contrary, the most prevalent NNRTI-associated substitution among treatment-experienced viremic participants was rtK103N (25.68%, 95% CI 20.03 – 32.92%), however rtY181C (14.67%, 95% CI 10.27% - 20.95%) and rtG190A (12.34%, 95% CI 8.33% - 18.27%) were also frequently observed. inT97A was observed at a similar prevalence as among pre-treatment viremic participants (9.96%, 95% CI 6.41 −15.48%). Only four participant-visits contributed by viremic treatment-experienced PLHIV (<1%) harbored INSTI resistance mutations, which were each observed only once (inG163K, inG163R, inR263K, inS147G) and not associated with DTG resistance.
Discussion
In this study, we report on trends in HIV drug resistance from a longitudinal, population-based cohort in southern Uganda between 2012 and 2019, a period marked by the substantial expansion of ART programs. Despite a doubling in the prevalence of NNRTI resistance among pre-treatment PLHIV, we observed an overall decline in the population prevalence of pre-treatment HIV drug-resistant viremia, alongside increasing ART uptake and viral suppression among PLHIV. By the end of the analysis period, the population prevalence of NNRTI resistance was 0.78% and NRTI resistance was 0.46%, with most resistance stemming from dual-class NNRTI/NRTI resistance in treatment-experienced viremic individuals. Notably, dual-class resistance remained relatively uncommon among pre-treatment viremic PLHIV, and resistance trends for NRTIs and PIs in this group remained stable throughout the ART scale-up, despite a substantial burden of NRTI resistance in treatment-experienced individuals. We also observed a relatively high background prevalence of the INSTI-resistance mutation inT97A. Overall, these findings provide important insights into the evolving dynamics of HIV drug resistance during ART scale-up in a high-burden East African population and may help guide future surveillance and HIV epidemic control efforts in the region.
Consistent with previous studies, we observed an increase in the prevalence of NNRTI resistance among viremic pre-treatment individuals living with HIV, supporting the recent shift to DTG-based regimens1,11,12. However, a key finding of this population-based analysis is that, concurrent with this rise in NNRTI resistance, we observed a substantial decline in the overall population prevalence of pre-treatment HIV, likely driven by both increased treatment initiation and declining HIV incidence18–20. Importantly, this decrease in pre-treatment HIV has outpaced the rise in NNRTI resistance, resulting in a more than 50% reduction in the population prevalence of pre-treatment HIV with NNRTI resistance over the study period. By the end of the survey period, most viremic individuals with NNRTI-resistant HIV were those with prior treatment experience.
We also find a lower burden of NNRTI and NRTI resistance among viremic treatment-experienced PLHIV in this study as compared to clinic-based studies1,12. This is likely because our population-based study design includes PLHIV who remain viremic because they are not actively engaged in care, despite past treatment exposure. In comparison, clinic-based studies may disproportionately enroll people who remain viremic due to sub-optimal adherence or are in more advanced stages of disease, and thus more likely to have drug resistance, whereas population-based sampling includes people lost to clinic-based care and no longer using treatment altogether.
Both NNRTI and NRTI therapies remain important components of the current and future ART landscape. Current first-line DTG-regimens incorporate two NRTIs (e.g. TDF and 3TC) and DTG treatment failure is more likely among those with NRTI resistance55,56. We here observe rtM184V (>1000-fold reduced susceptibility to 3TC57) and rtK65R (five-fold reduced susceptibility to TDF58) in 34% and 12% of viremic treatment-experienced PLHIV. In contrast, these mutations were observed only rarely among pre-treatment PLHIV. Further, we identify a number of mutations associated with reduced susceptibility to the RPV (e.g. rtK101E, rtE138A, rtY181C, and rtG190A), an NNRTI given in combination with CAB as part of long-lasting injectable therapies, in 10–12% of viremic treatment-experienced PLHIV.
As this study pre-dates the scale-up of DTG, we do not observe major DTG resistance-conferring mutations. Approximately 10% of viremic participants harbored inT97A, which is a polymorphic mutation most common in subtype A and in isolation confers two-fold resistance to EVG but not to other INSTIs59. The observed prevalence of inT97A in this study is an order of magnitude higher than in a population-based cohort in South Africa17 and about twice as prevalence as globally-sampled INSTI-naïve PLHIV59. Further, we observe a significant increase in the prevalence of inT97A among pre-treatment viremic PLHIV in the 2019 survey round. As inT97A is repeatedly selected for in subjects failing DTG therapy60 and in combination with other mutations (e.g. inG140S and inQ148H) can significantly increase DTG resistance61–63, we recommend continued monitoring.
There are important limitations of this work. Due to unknown HIV serostatus among non-participating residents of RCCS communities, we did not generalize our results beyond study participants. Younger individuals, men, and residents of trading communities are less likely to participate in RCCS surveys.20 Further, only self-reported treatment status was available, which may have led to the misclassification of some participants. Prior work in this cohort demonstrated that 11% of self-reported ART-naïve participants had antivirals in their blood.64 Given the significant differences observed in the mutational profiles of pre-treatment as compared to treatment-experienced PLHIV and the consistency of these results with estimates of the fitness impact of mutations in the absence of treatment,65 we expect minimal misclassification bias. While we lack data on individual-level ART regimens, first-line therapy in this setting is highly consistent across individuals. As this work is based on sequencing of viral RNA, we could only identify resistance among viremic PLHIV. Consequently, our population prevalence estimates are an underestimate as some PLHIV with resistance may be suppressed through second-line therapy or were transiently suppressed following treatment initiation. The latter may be more pronounced in recent survey rounds as treatment scale-up has increased the proportion of recent treatment initiators.
Despite the population-based study design, viral load data and sequence data was available for only a subset of participants due to budgetary and logistical constraints. We consequently restrict analyses to survey rounds where sufficient data is available to generate reliable inferences and use imputation to account for missing viral load data. Despite this missingness, deep-sequence data was available for 4,072 participant-visits, which is considerably more than a recent population-based study in South Africa (n=1,097),17 clinic-based studies in sub-Saharan Africa (n=972),12 and WHO surveys in Uganda (n=372)1. Further, we utilized detailed demographic data on survey participants to account for the role of potential biases in sequence data availability. However, we cannot rule out potential residual biases in our estimates.
In summary, this study adds critical context to our understanding of the HIV epidemic in southern Uganda and to the impact of treatment expansion on the population burden of HIV resistance. We show that following ART scale-up, most resistance is contributed by treatment-experienced PLHIV, which may inform interventions aimed at reducing transmitted HIV resistance. The relatively high prevalence of NRTI and NRTI-resistance among treatment-experienced PLHIV and of inT97A among all viremic PLHIV is concerning in light of the roll-out of DTG+ TDF+3TC and CAB+RPV regimens in sub-Saharan Africa. Overall, these findings stress the importance of continued viral sequence-based monitoring of resistance mutations among PLHIV, particularly those with previous treatment exposure, during the roll-out of novel HIV ART regimens.
Supplementary Material
Research in context.
Evidence before the study
We searched PubMed for studies matching the keywords “hiv” “resistance” “longitudinal” “cohort” “population” published since 2004 (the beginning of antiretroviral therapy (ART) availability in sub-Saharan Africa) and identified 50 studies. We excluded 34 studies not based in sub-Saharan Africa, five studies primarily concerned with infection with other pathogens (e.g. HBV, M. tuberculosis), two studies concerned with insulin resistance, one sequencing-methods paper, and one paper concerned with host susceptibility to HIV infection. The remaining seven studies were not population-based meaning that the study population was not all persons but e.g. people living with HIV enrolled in care at a given clinic. Population-based cohort are essential for monitoring HIV drug resistance in both treated and untreated individuals, including those people who may go undetected in clinical settings, capturing evolutionary dynamics of resistance in real-world conditions.
Added value of this study
We estimated the prevalence of drug resistance over five consecutive survey rounds of a population-based open-cohort study in southern Uganda between 2012 and 2019 during a period of intense treatment scale-up. We show that among the entire population regardless of HIV status, 0.8% and 0.5% of individuals harbor viremic resistance to non-nucleoside reverse transcriptase inhibitors (NNRTIs) and nucleoside-reverse transcriptase inhibitors (NRTIs), respectively, of which the majority is dual-class NNRTI/NRTI resistance. Despite a two-fold increase in the prevalence of NNRTI resistance among pre-treatment viremic PLHIV, the overall prevalence of pre-treatment viremic resistance in the entire population decreased by more than 50% due to increased treatment initiation and population viral load suppression. The majority of resistance in recent survey rounds was contributed by treatment-experienced PLHIV. Among treatment-experienced viremic PLHIV, we observe a substantial burden of mutations that confer resistance to the NNRTI and NRTI components of dolutegravir and cabotegravir based regimens e.g. rtM184V (34%) rtY181C (15%), rtG190A (12%), rtK65R (12%), and rtK101E (9.5%). The integrase strand transfer inhibitor (INSTI) resistance mutation inT97A was observed in about a tenth of viremic PLHIV.
These results provide the first longitudinal population-based estimates of temporal trends in the prevalence of drug resistance during ART program expansion in a high-burden setting. Further, they provide critical insight into the landscape of prevalent drug resistance substitutions circulating in this population.
Implications of all the available evidence
Scale-up of HIV treatment has increased the prevalence of drug resistance mutations among viremic people living with HIV in sub-Saharan Africa. The relatively high prevalence of NNRTI resistance has prompted a recent shift to first-line regimens including dolutegravir (an INSTI) in combination with NRTIs. The high prevalence of mutations conferring resistance to components of current first-line regimens in our population warrants continued monitoring of treatment failures and the prevalence of drug resistance in high burden settings.
Acknowledgements
We thank the Rakai Health Sciences Program and the participants of the Rakai Community Cohort Study for making this research possible. We thank members of the PANGEA-HIV consortium, the Johns Hopkins University Infectious Disease Dynamics group, and the NIAID Laboratory of Immunoregulation International HIV/STD Section at the Johns Hopkins University School of Medicine for helpful feedback on this work. This work was supported by the National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) (U01AI075115, R01AI087409, U01AI100031, R01AI110324, R01AI114438, K25AI114461, R01AI123002, K01AI125086, R01AI128779, R01AI143333, R21AI145682, R01AI155080, ZIAAI001040), NIH National Institute of Child Health and Development (R01HD050180, R01HD070769, R01HD091003), NIH National Heart, Lung, and Blood Institute (R01HL152813), the Fogarty International Center (D43TW009578, D43TW010557), the Johns Hopkins University Center for AIDS Research (P30AI094189), the Bill & Melinda Gates Foundation (OPP1084362, INV-007573, INV-035619, INV-060259, INV-075093), the U.S. President’s Emergency Plan for AIDS Relief through the Centers for Disease Control and Prevention (NU2GGH000817), and in part by the Division of Intramural Research, NIAID, NIH.
Funding
National Institutes of Health and the Gates Foundation
Funding Statement
National Institutes of Health and the Gates Foundation
Footnotes
Declaration of interests
We declare no competing interests.
Data sharing
Code to reproduce all analyses and visualizations as well as de-identified resistance and limited patient metadata are available at https://github.com/m-a-martin/rccs_hiv_resistance_r15_r19. Due to privacy concerns we are unable to share individual-level data on community of residence.
References
- 1.HIV Drug Resistance Report 2021. Geneva: World Health Organization, 2021. [Google Scholar]
- 2.Trickey A, Sabin CA, Burkholder G, et al. Life expectancy after 2015 of adults with HIV on long-term antiretroviral therapy in Europe and North America: a collaborative analysis of cohort studies. Lancet HIV 2023; 10: e295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen MS, Chen YQ, McCauley M, et al. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N Engl J Med 2016; 375: 830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UNAIDS. AIDSinfo. https://aidsinfo.unaids.org. [Google Scholar]
- 5.Carr A, Mackie NE, Paredes R, Ruxrungtham K. HIV drug resistance in the era of contemporary antiretroviral therapy: A clinical perspective. Antivir Ther 2023; 28: 13596535231201162. [DOI] [PubMed] [Google Scholar]
- 6.Phillips AN, Stover J, Cambiano V, et al. Impact of HIV Drug Resistance on HIV/AIDS-Associated Mortality, New Infections, and Antiretroviral Therapy Program Costs in Sub–Saharan Africa. J Infect Dis 2017; 215: 1362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethi AK, Celentano DD, Gange SJ, Moore RD, Gallant JE. Association between Adherence to Antiretroviral Therapy and Human Immunodeficiency Virus Drug Resistance. Clin Infect Dis 2003; 37: 1112–8. [DOI] [PubMed] [Google Scholar]
- 8.Bangsberg DR, Charlebois ED, Grant RM, et al. High levels of adherence do not prevent accumulation of HIV drug resistance mutations: AIDS 2003; 17: 1925–32. [DOI] [PubMed] [Google Scholar]
- 9.Beck IA, Levine M, McGrath CJ, et al. Pre-treatment HIV-drug resistance associated with virologic outcome of first-line NNRTI-antiretroviral therapy: A cohort study in Kenya. eClinicalMedicine 2020; 18: 100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Updated recommendations on first-line and second-line antiretroviral regimens and postexposure prophylaxis and recommendations on early infant diagnosis of HIV: interim guidelines. Supplement to the 2016 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Geneva: World Health Organization, 2018. [Google Scholar]
- 11.Gupta RK, Gregson J, Parkin N, et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 2018; 18: 346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowell TA, Danboise B, Parikh A, et al. Pretreatment and Acquired Antiretroviral Drug Resistance Among Persons Living With HIV in Four African Countries. Clin Infect Dis 2021; 73: e2311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ntamatungiro AJ, Kagura J, Weisser M, Francis JM. Pre-treatment HIV-1 drug resistance in antiretroviral therapy-naive adults in Eastern Africa: a systematic review and meta-analysis. J Antimicrob Chemother 2022; 77: 3231–41. [DOI] [PubMed] [Google Scholar]
- 14.Margolis DA, Gonzalez-Garcia J, Stellbrink H-J, et al. Long-acting intramuscular cabotegravir and rilpivirine in adults with HIV-1 infection (LATTE-2): 96-week results of a randomised, open-label, phase 2b, non-inferiority trial. The Lancet 2017; 390: 1499–510. [DOI] [PubMed] [Google Scholar]
- 15.The path that ends AIDS: UNAIDS Global AIDS Update 2023. Geneva: Joint United Nations Programme on HIV/AIDS, 2023. [Google Scholar]
- 16.Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford; New York: Oxford University Press, 1991. [Google Scholar]
- 17.Kemp SA, Kamelian K, Cuadros DF, et al. HIV transmission dynamics and population-wide drug resistance in rural South Africa. Nat Commun 2024; 15: 3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagaayi J, Chang LW, Ssempijja V, et al. Impact of combination HIV interventions on HIV incidence in hyperendemic fishing communities in Uganda: a prospective cohort study. Lancet HIV 2019; 6: e680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monod M, Brizzi A, Ssekubugu R, et al. Growing gender disparity in HIV infection in Africa: sources and policy implications. .
- 20.Grabowski MK, Serwadda DM, Gray RH, et al. HIV Prevention Efforts and Incidence of HIV in Uganda. N Engl J Med 2017; 377: 2154–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogel JM, Wilson EA, Piwowar Manning E, et al. HIV drug resistance in a community randomized trial of universal testing and treatment: HPTN 071 (PopART). J Int AIDS Soc 2022; 25. DOI: 10.1002/jia2.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chabria SB, Gupta S, Kozal MJ. Deep Sequencing of HIV: Clinical and Research Applications. Annu. Rev. Genomics Hum. Genet. 2014; 15: 295–325. [DOI] [PubMed] [Google Scholar]
- 23.Li JZ, Paredes R, Ribaudo HJ, et al. Low-Frequency HIV-1 Drug Resistance Mutations and Risk of NNRTI-Based Antiretroviral Treatment Failure: A Systematic Review and Pooled Analysis. JAMA 2011; 305: 1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simen BB, Simons JF, Hullsiek KH, et al. Low-Abundance Drug-Resistant Viral Variants in Chronically HIV-Infected, Antiretroviral Treatment–Naive Patients Significantly Impact Treatment Outcomes. J Infect Dis 2009; 199: 693–701. [DOI] [PubMed] [Google Scholar]
- 25.Chang LW, Grabowski MK, Ssekubugu R, et al. Heterogeneity of the HIV epidemic in agrarian, trading, and fishing communities in Rakai, Uganda: an observational epidemiological study. Lancet HIV 2016; 3: e388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagulire SC, Opendi P, Stamper PD, et al. Field evaluation of five rapid diagnostic tests for screening of HIV-1 infections in rural Rakai, Uganda. Int J STD AIDS 2011; 22: 308–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elm EV, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. Strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ 2007; 335: 806–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pillay D, Herbeck J, Cohen MS, et al. PANGEA-HIV: phylogenetics for generalised epidemics in Africa. Lancet Infect Dis 2015; 15: 259–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abeler-Dörner L, Grabowski MK, Rambaut A, Pillay D, Fraser C. PANGEA-HIV 2: Phylogenetics And Networks for Generalised Epidemics in Africa. Curr Opin HIV AIDS 2019; 14: 173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gall A, Ferns B, Morris C, et al. Universal Amplification, Next-Generation Sequencing, and Assembly of HIV-1 Genomes. J Clin Microbiol 2012; 50: 3838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonsall D, Golubchik T, de Cesare M, et al. A Comprehensive Genomics Solution for HIV Surveillance and Clinical Monitoring in Low-Income Settings. J Clin Microbiol 2020; 58: e00382–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wymant C, Blanquart F, Golubchik T, et al. Easy and accurate reconstruction of whole HIV genomes from short-read sequence data with shiver. Virus Evol 2018; 4. DOI: 10.1093/ve/vey007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siepel AC, Halpern AL, Macken C, Korber BTM. A Computer Program Designed to Screen Rapidly for HIV Type 1 Intersubtype Recombinant Sequences. AIDS Res Hum Retroviruses 1995; 11: 1413–6. [DOI] [PubMed] [Google Scholar]
- 34.Fogel JM, Bonsall D, Cummings V, et al. Performance of a high-throughput next-generation sequencing method for analysis of HIV drug resistance and viral load. J Antimicrob Chemother 2020; 75: 3510–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broad Institute Picard tools. Https://Broadinstitute.Github.Io/Picard/. 2016. https://broadinstitute.github.io/picard/%5Cnhttp://broadinstitute.github.io/picard/.
- 37.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 2004; 32: W20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji H, Enns E, Brumme CJ, et al. Bioinformatic data processing pipelines in support of nextgeneration sequencing-based HIV drug resistance testing: the Winnipeg Consensus. J Int AIDS Soc 2018; 21: e25193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee S-Y. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 2003; 31: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shafer RW. Rationale and Uses of a Public HIV Drug Resistance Database. J Infect Dis 2006; 194: S51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu TF, Shafer RW. Web Resources for HIV Type 1 Genotypic-Resistance Test Interpretation. Clin Infect Dis 2006; 42: 1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Australia, 2020. https://www.R-project.org/ (accessed Dec 11, 2020). [Google Scholar]
- 43.Zou G. A Modified Poisson Regression Approach to Prospective Studies with Binary Data. Am J Epidemiol 2004; 159: 702–6. [DOI] [PubMed] [Google Scholar]
- 44.Halekoh U, Højsgaard S, Yan J. The R Package geepack for Generalized Estimating Equations. J Stat Softw 2006; 15. DOI: 10.18637/jss.v015.i02. [DOI] [Google Scholar]
- 45.Lenth RV. emmeans: Estimated Marginal Means, aka Least-Squares Means. 2024. https://CRAN.R-project.org/package=emmeans.
- 46.Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. J Open Source Softw 2019; 4: 1686. [Google Scholar]
- 47.Wickham H, Chang W, Henry L, et al. ggplot2: Elegant Graphics for Data Analysis. SpringerVerlag; New York, 2021. http://ggplot2.org (accessed Feb 8, 2022). [Google Scholar]
- 48.Wilke CO. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. 2020. https://CRAN.R-project.org/package=cowplot.
- 49.Lin Pedersen T. patchwork: The Composer of Plots. 2023. https://cran.r-project.org/web/packages/patchwork/index.html.
- 50.FC M, Davis TL, ggplot2 authors. ggpattern: ‘ggplot2’ Pattern Geoms. 2022. https://github.com/coolbutuseless/ggpattern. [Google Scholar]
- 51.Wickham H, Bryan J. readxl: Read Excel Files. 2023. https://readxl.tidyverse.org.
- 52.Wickham H, Miller E, Smith D. haven: Import and Export ‘SPSS’, ‘Stata’ and ‘SAS’ Files. 2023. https://github.com/tidyverse/haven.
- 53.Margot NA, Ram RR, White KL, Abram ME, Callebaut C. Antiviral activity of HIV 1 integrase strand transfer inhibitors against mutants with integrase resistance associated mutations and their frequency in treatment naïve individuals. J Med Virol 2019; 91: 2188–94. [DOI] [PubMed] [Google Scholar]
- 54.Azijn H, Tirry I, Vingerhoets J, et al. TMC278, a Next-Generation Nonnucleoside Reverse Transcriptase Inhibitor (NNRTI), Active against Wild-Type and NNRTI-Resistant HIV-1. Antimicrob Agents Chemother 2010; 54: 718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loosli T, Hossmann S, Ingle SM, et al. HIV-1 drug resistance in people on dolutegravir-based antiretroviral therapy: a collaborative cohort analysis. Lancet HIV 2023; 10: e733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schramm B, Temfack E, Descamps D, et al. Viral suppression and HIV-1 drug resistance 1 year after pragmatic transitioning to dolutegravir first-line therapy in Malawi: a prospective cohort study. Lancet HIV 2022; 9: e544–53. [DOI] [PubMed] [Google Scholar]
- 57.Tisdale M, Kemp SD, Parry NR, Larder BA. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3’-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci 1993; 90: 5653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wainberg MA, Miller MD, Quan Y, et al. In vitro Selection and Characterization of HIV-1 with Reduced Susceptibility to PMPA. Antivir Ther 1999; 4: 87–94. [DOI] [PubMed] [Google Scholar]
- 59.Tzou PL, Rhee S-Y, Descamps D, et al. Integrase strand transfer inhibitor (INSTI)-resistance mutations for the surveillance of transmitted HIV-1 drug resistance. J Antimicrob Chemother 2020; 75: 170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao K, Rhee S-Y, Chu C, et al. Treatment Emergent Dolutegravir Resistance Mutations in Individuals Naïve to HIV-1 Integrase Inhibitors: A Rapid Scoping Review. Viruses 2023; 15: 1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huik K, Hill S, George J, et al. High-level dolutegravir resistance can emerge rapidly from few variants and spread by recombination: implications for integrase strand transfer inhibitor salvage therapy. AIDS 2022; 36: 1835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang WW, Cheung PK, Oliveira N, Robbins MA, Harrigan PR, Shahid A. Accumulation of Multiple Mutations In Vivo Confers Cross-Resistance to New and Existing Integrase Inhibitors. J Infect Dis 2018; 218: 1773–6. [DOI] [PubMed] [Google Scholar]
- 63.George JM, Kuriakose SS, Dee N, et al. Rapid Development of High-Level Resistance to Dolutegravir With Emergence of T97A Mutation in 2 Treatment-Experienced Individuals With Baseline Partial Sensitivity to Dolutegravir. Open Forum Infect Dis 2018; 5: ofy221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grabowski MK, Reynolds SJ, Kagaayi J, et al. The validity of self-reported antiretroviral use in persons living with HIV: a population-based study. AIDS 2018; 32: 363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kühnert D, Kouyos R, Shirreff G, et al. Quantifying the fitness cost of HIV-1 drug resistance mutations through phylodynamics. PLOS Pathog 2018; 14: e1006895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code to reproduce all analyses and visualizations as well as de-identified resistance and limited patient metadata are available at https://github.com/m-a-martin/rccs_hiv_resistance_r15_r19. Due to privacy concerns we are unable to share individual-level data on community of residence.