Abstract
Mucins are a major component of the innate defense system in the airways and their biological functions are important to consider in pulmonary disease research. However, the available mucus models for basic research relevant to the lung can be difficult to acquire in sufficient quantity to conduct such studies. Here, we present a new strategy to isolate airway mucins from pig trachea at the milligram to gram scale for use in pulmonary disease research. Using this protocol, we were able to isolate mucins with minimal DNA contamination consisting of ~70% by weight protein. Compared to porcine gastric mucins extracted with the same procedure, the porcine tracheal extract possessed significantly greater O-linked glycoprotein (mucin) content. Particle tracking microrheology was used to evaluate the biophysical properties of porcine trachea mucins. We found porcine tracheal mucins formed a much tighter mesh network and possessed a significantly greater microviscosity compared to lab extracted porcine gastric mucins. In comparison to mucus harvested from human airway tissue cultures, we found porcine tracheal mucins also possessed a greater microviscosity suggesting these mucins can form into a gel-like material at physiological total solids concentrations. These studies establish an accessible means to isolate airway mucins from porcine trachea at large scale for use in pulmonary disease research.
INTRODUCTION
Mucus lines the epithelium of mammalian organs including the stomach, eyes, respiratory tract, and reproductive tract.1 The mucus gel lining these tissues serves as a protective barrier against foreign particulates as well as providing lubrication and hydration.2–4 Together, mucins secreted at epithelial surfaces function to form the mucus gel network through electrostatic interactions, entanglement, and polymerization via disulfide bonding within cysteine rich domains of the mucin monomer.2,5,6 Mucus gels produced in distinct tissues possess unique biomolecular and biophysical properties depending on their precise functions. For example in the gut, a mucus gel layer exists with a loosely cross-linked, microbe-rich mucus layer overlying a more densely crosslinked, microbe-free mucus layer to physically separate the epithelium from the microbiome and other potentially pathogenic microbes.7,8 Prior studies have shown airway mucins can self-organize into strands and sheets to support mucociliary function and airway clearance.9,10 In addition, each mucin varies in its O-linked glycosylation pattern which contain different functional groups such as terminal sulfates, sialic acid, and fucose.6,11 Prior work has established these terminal functional groups directly impact the ability of airway mucins to neutralize viral pathogens and prevent infection.12–14 Thus, the structure and function of mucins is highly tailored to the tissue from which they arise.
Historically, mucins have been purified from animal tissues, such as bovine submaxillary mucins (BSM), porcine small intestinal mucins (PSIM), and porcine gastric mucins (PGM).15 These animal derived mucins are unique in their physicochemical properties, such as containing regionally specific mucins such as MUC2 and MUC5AC in the gastrointestinal tract.7 There are two commercially available mucins BSM and PGM, which will herein be referred to as BSMC and PGMC for clarification between lab-purified mucins and commercially purified mucins. While these commercial mucins are available in bulk making them convenient for use, a previous report has shown that these mucins contain significant amounts of DNA as well as other contaminants and may be partially degraded due to their processing.16 Mucins in the airway possess a unique composition, predominantly composed of MUC5B and MUC5AC,2,5 that is distinct from other mucosal tissues. As a result, previously reported lab extracted and commercial mucins do not provide suitable mucin/mucus models for airway disease research. Airway derived mucins have been primarily sourced from human patient samples (e.g. sputum17, mucus collected from endotracheal tubes18,19) or from human airway tissue cultures grown at air-liquid interface.20–22 This presents limitations in their broader use as patient-derived airway mucus is not widely available and tissue culture derived airway mucus is not easy to produce in large quantities. Motivated by this, we report a scalable extraction protocol, adapted from previous work,23,24 to isolate mucins from the porcine trachea. This establishes an approach to isolate airway mucins in an accessible and scalable manner which could be adopted broadly by researchers with interests in mucin-driven lung disease mechanisms.
MATERIALS & METHODS
PTM & PGM extraction from porcine tissue
Porcine trachea mucins (PTM) and porcine gastric mucins (PGM) were extracted based on a previously described protocol.23 Briefly, tissue samples were dissolved in 0.1 M NaOH overnigh. The porcine small intestine was filled with NaOH whereas the porcine trachea tissue was submerged in NaOH to allow for the solubilization of the mucus layer. Mucins were then removed from solution by lowering the pH to 4.0 with 1 M HCl (gel phase), then centrifuged at 3500 rpm for 20 minutes to allow for removal of the supernatant. The resulting pellet was resuspended in deionized water and the pH was adjusted to 8.0 to allow for resolubilization of the mucins (sol phase). The pH was altered to repeat the gel-sol cycle three times for further mucin extraction. Mucins were then purified by addition of DNase I (10 U/mL) at 21 C overnight. Following DNA removal, the gel-sol cycle was repeated 4 times through pH cycling and centrifugation for further mucin purification. Supernatant was then dialyzed (100 kDa) in deionized water for 72 hours, and then frozen at −80C overnight prior to lyophilization. The lyophilized extracted mucins were solubilized at 2% or 4% (w/v) in a physiological buffer containing 154 mM NaCl, 3 mM CaCl2, and 15 mM NaH2PO4, pH 7.4 prior to usage for further biochemical and biophysical analysis.
BCi-NS1.1 cell culture & mucus collection
The immortalized BCi-NS1.1 human airway epithelial cell line was supplied by Ronald Crystal (Weill Cornell Medical College) and cultured as previously described.25,26 Briefly, BCi-NS1.1 cells were first expanded in a flask with Pneumacult-Ex Plus medium (no. 05040, StemCell Technologies) until confluent. Cells were then seeded (1 × 104 cell/cm2) on rat tail collagen type 1-coated permeable Transwell membranes (12 mm; no. 38023, StemCell Technologies) until confluent. After confluence, only basal Pneumacult-ALI medium was provided in the basolateral compartment (no. 05001, StemCell Technologies) for 4 weeks to allow for polarization to occur at the air-liquid interface (ALI). Culturing at ALI allowed for the formation of an in vivo pseudostratified mucociliary epithelium. Mucus was collected through a 30 minute PBS wash, and concentrated using 100k MWCO amicon filters, and stored at −80C until usage.
Mass DNA quantification assay
The mass DNA of extracted mucins was measured according to a previously established protocol.24 In brief, 30 μL of 20% (w/v) 3,5-diaminobenzoic acid was added to solubilized mucin samples, incubated at 60°C for 1 hour, and then the reaction stopped with addition of 1 mL of 1.76 M HCl. The reaction was stopped with the addition of 1 mL of 1.76 M HCl. The fluorescence intensity was measured at 390/530 nm (ex./emis.).
Protein quantification assay
Protein concentration was quantified using a bicinchoninic acid (BCA) assay (no. 23225, ThermoFisher) as described by the manufacturer. In brief, a BCA working reagent was prepared, of which 200 μL was added to 15 μL of each sample, and then incubated for 30 minutes at room 37°C. Absorbance values of the samples were measured at 562 nm and compared to BCA standards.
Relative O-linked glycoprotein content assay
O-linked glycoprotein (mucin) concentration was determined using a cyanoacetamide (CNA) reagent protocol as previously established.24 In brief, 200 μL of CNA was mixed with 1 mL of 0.15 M NaOH to create the CNA reagent. 60 μL of the CNA reagent were mixed with 50 μL of solubilized mucin samples (2% of 4% w/v) and incubated for 30 minutes at 100°C. The fluorescence intensity of the resulting samples was measured at 336/383 nm (ex./emis.) and compared to a serially diluted bovine submaxillary mucin (BSM; Sigma-Aldrich) standard curve.
Sialic acid concentration assay
Sialic acid concentration was measured for extracted and commercial mucins by utilizing a modified Warren method centered on the thiobarbituric acid reaction (Sigma-Aldrich, MAK314) and following the manufacturer protocol. Briefly, bound sialic acid was hydrolyzed, then oxidized to form formylpyruvic acid which formed a measurable colored solution with the addition of thiobarbituric acid. The fluorescence of the colored solution was measured at 555/585 nm (ex./emis.) and compared to the sialic acid standard curve.
Disulfide bond concentration assay
For extracted mucins the disulfide bond concentration was determined using a previously described protocol.17 In brief, 8 M Guanidine hydrochloric acid was added to 50 – 70 μL of sample for a final volume of 500 μL. 10% (v/v) of 500 mM iodoacetamide was added and samples sat at room temperature for 1 hour. 10% (v/v) of 1 M DTT was added, and samples incubated at 37°C for 2 hours. Samples were then filtered and buffer exchanged in a 7 KDa MWCO Zebra desalting column with 50 mM Tris-HCl (pH 8.0). As a standard, solutions of L-cysteine with concentration ranging from 0 μm – 5000 μM were prepared. Samples were diluted 1:1 with 2 mM monobromobimane in a flat black plate 96-well plate and incubated in the dark at room temperature for 15 minutes before reading the fluorescence at 395/490 nm (ex./emis.).
Multiple particle tracking and microrheology analysis
Using a previously established protocol,17 nanoparticle (NP) probes were prepared for use in multiple particle tracking experiments by modifying 100 nm carboxylate-modified polystyrene NP (ThermoFisher) with a polyethylene glycol (PEG) coating to render these particles non-adheseive to mucus. We then constructed custom microscopy chamber consisting of a vacuum grease coated O-ring that was then filled with 20 μL of the mucin / mucus sample of interest and 1 μL of muco-inert NPs (~0.002% w/v) before being sealed with a coverslip. Slides were then incubated at room temperature for 30 minutes in the dark prior to fluorescence imaging (Zeiss Confocal LSM 800, 63x water-immersion objective) to allow for sample equilibration. NP diffusion was imaged for 10 seconds at 33.3 frames per second and then tracked and analyzed using a custom MATLAB code that calculated the mean squared displacement, , for each particle. The MSD values were used to estimate the microrheological properties of the gel through the Stokes-Einstein relation, , where is the thermal energy, is the radius, and is the complex Laplace frequency. The frequency-dependent complex modulus (G*) was calculated as, where is substituted for is the complex number, and is the frequency. The pore size was estimated from the G’ using the following expression: .
RESULTS
Biochemical properties of mucin extracted from porcine trachea
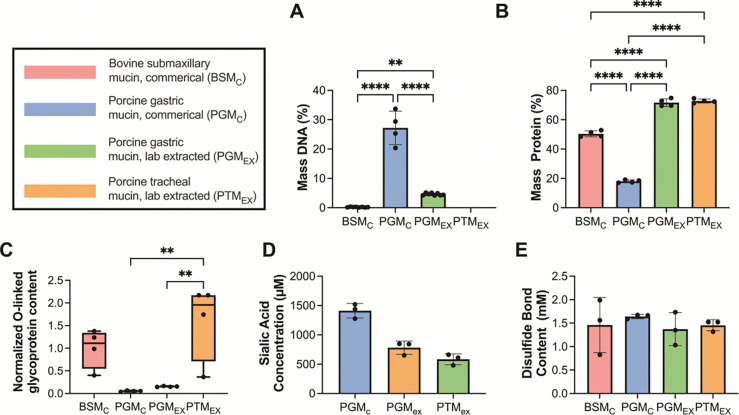
To better understand the composition of the resulting extracted mucin from porcine trachea (PTMEX), a series of biochemical assays were performed to assess DNA, total protein, O-linked glycoprotein (mucin), sialic acid, and disulfide bond content (Fig. 1). For comparison, we conducted these analyses on commercial mucins (BSMC and PGMC) as well as a lab extracted PGM harvested using the same protocol (PGMEX). The resulting measurements revealed that both extracted mucins, PTMEX and PGMEX, contained minimal DNA, comparable to BSMC, and significantly lower DNA than PGMC (Fig. 1A). Similarly, both extracted mucins contained a mass protein of ~70%, which was significantly higher than both BSMC and PGMC (Fig. 1B). Measured O-linked glycoprotein (mucin) content normalized to that measured in BSMC, PTMEX yielded a higher O-linked glycoprotein content compared to both PGMC and PGMEX (Fig. 1C). Sialic acid concentration was similar in PGMEX and PTMEX (Fig. 1D). In comparison to PGMC, the extracted mucins (PGMEX and PTMEX) contained less sialic acid on average but these differences were not significantly different. The modified Warren assay which our sialic acid protocol employs has been reported to have difficulty hydrolysis of BSMC mucins and as such, sialic acid content for BSMC was not included in our results.27 Disulfide bond content was comparable between all mucin types (Fig. 1E).
Figure 1. Biochemical characterization of mucins extracted from porcine trachea.
(A-E) Commercially available BSM (BSMC), commercially available PGM type III (PGMC), laboratory extracted PGM (PGMEX), and laboratory extracted PGM (PGMEX) were characterized to determine (A) % mass DNA content, (B) % mass protein, (C) O-linked glycoprotein content normalized to BSMC, (D) sialic acid content, and (E) disulfide bond content. Bars indicate mean with error bars for standard deviations and individual measurements are shown as data points. Data set analyzes for statistical significance with Kruski-Wallis statistical test: *p<0.05, **p<0.01, and ****p<0.0001. Comparisons are not significant (p > 0.05) unless noted otherwise.
Microrheological properties of mucin extracted from porcine trachea
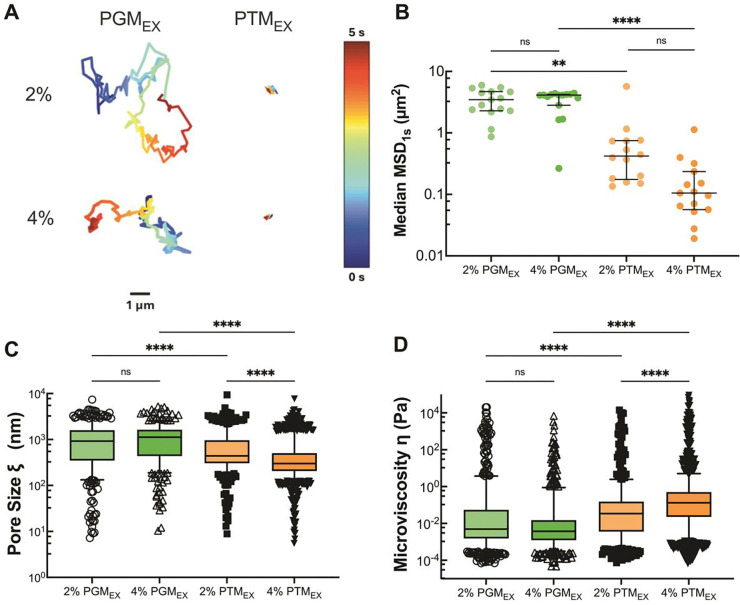
We next used particle tracking microrheology to evaluate the viscoelastic properties of PGMEX and PTMEX prepared at physiological solids concentration of 2% and 4% w/v. Based on representative trajectories and measured mean squared displacement at a time scale of 1 s (MSD1s), NP were highly mobile within PGMEX whereas NP diffusion appeared significantly constrained within PTMEX (Fig. 2A,B). The mean pore sizes for PGMEX were ~1.5–2.5 fold larger than PTMEX where average pore sizes were ~1 μm in PGMEX and ~500–750 nm in PTMEX (Fig. 2C). The mean microviscosity for PGMEX was similar at both 2% and 4% w/v concentration. In comparison to PGMEX, PTMEX possessed a 1.5 greater microviscosity at 2% w/v and a ~13-fold greater microviscosity at 4% w/v PTMEX (Fig. 2D). Additional studies were conducted to compare the microrheological properties of PGMC and PGMEX where we found PGMC formed a tighter network overall presumably due to the high DNA content in this mucin source (Fig. S1). Similar studies were conducted comparing PTMEX and BSMC which showed PTMEX possessed smaller pore sizes and greater microviscosity (Fig. S2).
Figure 2. Microrheology of porcine gastric and tracheal mucins.
(A) Representative trajectories of NP diffusion of 100 nm NP in 2% and 4% w/v PTMEX and PGMEX. Trajectory colors change as a function of time with 0 s indicated by dark blue and 5 s indicated by dark red. Scale bar = 1 μm. (B) Calculated median MSD at a time scale of 1 second (MSD1s) for solubilized mucins. Each data point represents the median calculated MSD1s in each video with at least 5 videos from 3 technical replicates. Black lines indicate interquartile range. (C) Estimated pore size (ξ) from NP diffusion. (D) Estimated microviscosity (ƞ) from NP diffusion. Datasets in (B,C,D) analyzed with Kruski-Wallis test with Dunn’s test for multiple comparison: ns = not significant, **** p < 0.0001, ** p < 0.01.
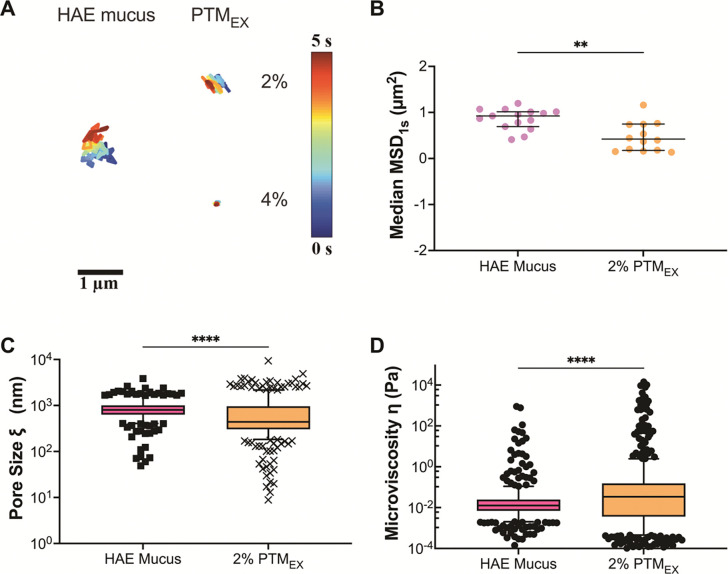
To determine how the microrheological properties of PTMEX compared to a traditionally used airway mucus model, we isolated airway mucus from differentiated human airway epithelial tissue cultures. Prior studies by our group have shown mucus collected from HAE cultures possess a total solids concentration of 2–4% w/v.26 Considering this, the microrheological properties of 2% and 4% w/v PTMEX were compared to HAE mucus. NP trajectories in HAE mucus were observed to be comparable to 2% PTMEX whereas diffusion was much more restricted in 4% PTMEX (Fig. 3A). Given the qualitative similarities, we quantitatively compared measured MSD1s, pore size, and microviscosity for 2% PTMEX and HAE mucus. While similar in magnitude, we found a significant decrease in MSD1s and pore size, indicative of tighter mesh network in 2% PTMEX (Fig. 3B,C). PTMEX also possessed a greater microviscosity compared to HAE mucus (Fig. 3D).
Figure 3. Microrheology of porcine tracheal mucins and mucus collected from human airway tissue cultures.
measurements resulting from 100 nm nanoparticles in BSMC, and PTMEX. A: Representative trajectories for diffusion of 100 nm PS-NP in 2% solubilized mucins. Trajectory colors change as a function of time with 0 s indicated by dark blue and 5 s indicated by dark red. Scale bar = 1 μm. B: Calculated median MSD at a time scale of 1 second (MSD1s) for solubilized mucins. Each data point represents the median calculated MSD1s in each video with at least 5 videos from 3 technical replicates. Black lines indicate interquartile range. C: Estimated pore size (ξ) from NP diffusion. D: Estimated microviscosity (ƞ) from NP diffusion. Datasets in (B,C,D) analyzed with Mann-Whitney test: **** p < 0.0001, ** p < 0.01.
DISCUSSION
We have developed a method for the extraction of porcine trachea mucins based on a previously established protocol by Sharma et al. Using a pH cycling method, airway mucins can be extracted at large scale (up to g) without the need for harsh chemical or enzymatic treatment. Similar to commercially available BSMC, the extracted mucins, PTMEX and PGMEX, contained minimal DNA contaminants. PTMEX contained the highest degree of extracted O-linked glycoproteins (mucin) compared to BSMC, PGMC, and PGMEX. We also quantified the content of terminal sialic acid glycans as these are relevant to many homeostatic and disease-associated processes in the airway. PTMEX and PGMEX both possessed lower sialic acid content compared to PGMC, although this is not necessarily indicative of issues with mucin glycoprotein integrity. Given PGMEX possessed less sialic acid compared to its commercial counterpart, it is possible that our extraction method reduces free and/or mucin-associated sialic acid content.
Using particle tracking microrheology, we found PTMEX forms a tighter network structure than PGMEX with decreased diffusivity, reduced pore size, and increased microviscosity. This is most likely due to the larger fraction of O-linked glycoproteins (mucins) in PTMEX which are the primary structural units of mucus gels. It should be noted mucus gels composed of PGMEX are typically prepared at acidic pH.28 A comparison of pH-dependent behavior of PTMEX and PGMex gel formation would be interesting to consider in future work. We also found PTMEX and HAE culture derived mucus possessed similar network structure with a decreased pore size and increased overall microviscosity observed for PTMEX. These studies suggest that PTMEX can form into a gel with similar physical structure to tissue culture-derived airway mucus and may be suitable as a model to study airway mucin function. Overall, this study establishes a new method for extraction of airway mucins with physiologically relevant biochemical and biophysical properties. This extraction method fills an important gap in the pulmonary research field to provide a more broadly accessible mucin source that is representative of airway mucus.
LIMITATIONS OF THIS STUDY
We did not perform bulk rheological analyses to confirm formation of a gel using PTMEX and we plan to perform these measurements in future work. It should be noted we did not measure the total solids content or other biochemical properties of the HAE mucus used in this study. A study comparing PTMEX and HAE mucus with matched total solids concentration would be helpful in comparing these mucus sources in future work.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by the NIH (EB030834, HL160540, and HL160540-S1 awarded to S.Y.) and NSF GRFP (GRFP DGE1840340 awarded to S.Y.).
BIBLIOGRAPHY
- 1.Bansil R., and Turner B.S. (2006). Mucin structure, aggregation, physiological functions and biomedical applications. Current Opinion in Colloid & Interface Science 11, 164–170. 10.1016/j.cocis.2005.11.001. [DOI] [Google Scholar]
- 2.Song D., Cahn D., and Duncan G.A. (2020). Mucin Biopolymers and Their Barrier Function at Airway Surfaces. Langmuir. 10.1021/acs.langmuir.0c02410. [DOI] [PubMed] [Google Scholar]
- 3.Cone R.A. (2009). Barrier properties of mucus. Advanced Drug Delivery Reviews 61, 75–85. 10.1016/j.addr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Lai S.K., Wang Y.-Y., Wirtz D., and Hanes J. (2009). Micro- and macrorheology of mucus. Advanced Drug Delivery Reviews 61, 86–100. 10.1016/j.addr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornton D.J., Rousseau K., and McGuckin M.A. (2008). Structure and Function of the Polymeric Mucins in Airways Mucus. Annual Review of Physiology 70, 459–486. 10.1146/annurev.physiol.70.113006.100702. [DOI] [PubMed] [Google Scholar]
- 6.Wagner C.E., Wheeler K.M., and Ribbeck K. (2018). Mucins and Their Role in Shaping the Functions of Mucus Barriers. Annu Rev Cell Dev Biol 34, 189–215. 10.1146/annurev-cellbio-100617-062818. [DOI] [PubMed] [Google Scholar]
- 7.Johansson M.E., Sjövall H., and Hansson G.C. (2013). The gastrointestinal mucus system in health and disease. Nature reviews Gastroenterology & hepatology 10, 352. 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson M.E.V., Larsson J.M.H., and Hansson G.C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proceedings of the National Academy of Sciences 108, 4659–4665. 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pino-Argumedo M.I., Fischer A.J., Hilkin B.M., Gansemer N.D., Allen P.D., Hoffman E.A., Stoltz D.A., Welsh M.J., and Abou Alaiwa M.H. (2022). Elastic mucus strands impair mucociliary clearance in cystic fibrosis pigs. Proc Natl Acad Sci U S A 119, e2121731119. 10.1073/pnas.2121731119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostedgaard L.S., Moninger T.O., McMenimen J.D., Sawin N.M., Parker C.P., Thornell I.M., Powers L.S., Gansemer N.D., Bouzek D.C., Cook D.P., et al. (2017). Gel-forming mucins form distinct morphologic structures in airways. Proc. Natl. Acad. Sci. U.S.A. 114, 6842–6847. 10.1073/pnas.1703228114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symmes B.A., Stefanski A.L., Magin C.M., and Evans C.M. (2018). Role of mucins in lung homeostasis: regulated expression and biosynthesis in health and disease. Biochem Soc Trans 46, 707–719. 10.1042/BST20170455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iverson E., Kaler L., Agostino E.L., Song D., Duncan G.A., and Scull M.A. (2020). Leveraging 3D Model Systems to Understand Viral Interactions with the Respiratory Mucosa. Viruses 12, 1425. 10.3390/v12121425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuve S., Wang H., Jacobs J.D., Yumul R.C., Smith D.F., and Lieber A. (2008). Role of Cellular Heparan Sulfate Proteoglycans in Infection of Human Adenovirus Serotype 3 and 35. PLoS Pathog 4, e1000189. 10.1371/journal.ppat.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen M., Zhang X.-Q., Senaati H.P., Chen H.-W., Varki N.M., Schooley R.T., and Gagneux P. (2013). Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virology Journal 10, 321. 10.1186/1743-422x-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrou G., and Crouzier T. (2018). Mucins as multifunctional building blocks of biomaterials. Biomater. Sci. 6, 2282–2297. 10.1039/C8BM00471D. [DOI] [PubMed] [Google Scholar]
- 16.Marczynski M., Jiang K., Blakeley M., Srivastava V., Vilaplana F., Crouzier T., and Lieleg O. (2021). Structural Alterations of Mucins Are Associated with Losses in Functionality. Biomacromolecules 22, 1600–1613. 10.1021/acs.biomac.1c00073. [DOI] [PubMed] [Google Scholar]
- 17.Duncan G.A., Jung J., Joseph A., Thaxton A.L., West N.E., Boyle M.P., Hanes J., and Suk J.S. (2016). Microstructural alterations of sputum in cystic fibrosis lung disease. JCI Insight 1. 10.1172/jci.insight.88198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaler L., Joyner K., and Duncan G.A. (2022). Machine learning-informed predictions of nanoparticle mobility and fate in the mucus barrier. APL Bioengineering 6, 026103. 10.1063/5.0091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markovetz M.R., Subramani D.B., Kissner W.J., Morrison C.B., Garbarine I.C., Ghio A., Ramsey K.A., Arora H., Kumar P., Nix D.B., et al. (2019). Endotracheal tube mucus as a source of airway mucus for rheological study. American Journal of Physiology-Lung Cellular and Molecular Physiology 317, L498–L509. 10.1152/ajplung.00238.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonser L.R., Zlock L., Finkbeiner W., and Erle D.J. (2016). Epithelial tethering of MUC5AC-rich mucus impairs mucociliary transport in asthma. Journal of Clinical Investigation 126, 2367–2371. 10.1172/JCI84910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornton D.J., Gray T., Nettesheim P., Howard M., Koo J.S., and Sheehan J.K. (2000). Characterization of mucins from cultured normal human tracheobronchial epithelial cells. American Journal of Physiology-Lung Cellular and Molecular Physiology 278, L1118–L1128. 10.1152/ajplung.2000.278.6.L1118. [DOI] [PubMed] [Google Scholar]
- 22.Hill D.B., Vasquez P.A., Mellnik J., McKinley S.A., Vose A., Mu F., Henderson A.G., Donaldson S.H., Alexis N.E., Boucher R.C., et al. (2014). A Biophysical Basis for Mucus Solids Concentration as a Candidate Biomarker for Airways Disease. PLoS One 9, e87681. 10.1371/journal.pone.0087681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A., and Kwak J.-G. In Vitro Reconstitution of an Intestinal Mucus Layer Shows That Cations and pH Control the Pore Structure That Regulates Its Permeability and Barrier Function. [DOI] [PMC free article] [PubMed]
- 24.Yang S., and Duncan G.A. (2023). Synthetic mucus biomaterials for antimicrobial peptide delivery. Journal of Biomedical Materials Research Part A 111, 1616–1626. 10.1002/jbm.a.37559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters M.S., Gomi K., Ashbridge B., Moore M.A.S., Arbelaez V., Heldrich J., Ding B.-S., Rafii S., Staudt M.R., and Crystal R.G. (2013). Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res 14, 135. 10.1186/1465-9921-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song D., Iverson E., Kaler L., Boboltz A., Scull M.A., and Duncan G.A. (2022). MUC5B mobilizes and MUC5AC spatially aligns mucociliary transport on human airway epithelium. Sci. Adv. 8, eabq5049. 10.1126/sciadv.abq5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Colorimetric Analysis of Sialic Acid in Human Saliva and Bovine Salivary Mucin 10.1177/00220345780570111701. [DOI] [PubMed]
- 28.Wagner C.E., Turner B.S., Rubinstein M., McKinley G.H., and Ribbeck K. (2017). A rheological study of the association and dynamics of MUC5AC gels. Biomacromolecules 18, 3654–3664. 10.1021/acs.biomac.7b00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.