Abstract
IgA vasculitis (IgAV) is a pediatric disease with skin and systemic manifestations. Here, we conducted genome, transcriptome, and proteome-wide association studies in 2,170 IgAV cases and 5,928 controls, generated IgAV-specific maps of gene expression and splicing from blood of 255 pediatric cases, and reconstructed myeloid-specific regulatory networks to define disease master regulators modulated by the newly identified disease driver genes. We observed significant association at the HLA-DRB1 (OR=1.55, P=1.1×10−25) and fine-mapped specific amino-acid risk substitutions in DRβ1. We discovered two novel non-HLA loci: FCAR (OR=1.51, P=1.0×10−20) encoding a myeloid IgA receptor FcαR, and INPP5D (OR=1.34, P=2.2×10−9) encoding a known inhibitor of FcαR signaling. The FCAR risk locus co-localized with a cis-eQTL increasing FCAR expression; the risk alleles disrupted a PRDM1 binding motif within a myeloid enhancer of FCAR. Another risk locus was associated with a higher genetically predicted levels of plasma IL6R. The IL6R risk haplotype carried a missense variant contributing to accelerated cleavage of IL6R into a soluble form. Using systems biology approaches, we prioritized IgAV master regulators co-modulated by FCAR, INPP5D and IL6R in myeloid cells. We additionally identified 21 shared loci in a cross-phenotype analysis of IgAV with IgA nephropathy, including novel loci PAID4, WLS, and ANKRD55.
Introduction
IgA vasculitis (IgAV), also known as Henoch-Schönlein purpura (HSP), is the most common form of childhood vasculitis, affecting 10–27 children per 100,000 per year1. IgAV is characterized by IgA deposition in blood vessel walls in the skin, joints, gastrointestinal tract, and the glomerular mesangium2,3. IgAV can lead to serious complications, including kidney failure4. The exact pathogenic mechanisms of IgAV remain unclear, resulting in the general lack of targeted treatments and preventive strategies5. The contribution of common genetic variants to IgAV risk has not been systematically assessed, although the HLA region has been implicated in small studies6–9.
Herein, we report the largest genetic study of IgAV involving 8,098 individuals of European and East Asian ancestries (2,170 cases and 5,928 controls). By combining genome-wide association study (GWAS), transcriptome-wide association study (TWAS), and proteome-wide association study (PWAS) approaches, our study implicated myeloid cells as central to the disease pathogenesis, and prioritized HLA-DRB1, FCAR, INPP5D, and IL6R as candidate causal genes for IgAV. To assess the transcriptional consequences of the newly identified risk alleles, we performed blood RNA sequencing of 255 pediatric cases with IgAV and built a disease context-specific atlas of expression and splicing quantitative trait loci (QTLs). Using methods proposed for cancer10,11, we reconstructed a myeloid-specific regulatory network based on blood single cell transcriptomes and identified proteins representing candidate master regulators of IgAV-related myeloid cell state whose activity is modulated by the candidate causal genes. In aggregate, our comprehensive genetic and systems biology approach defined new pathogenic mechanisms and provided strong genetic support for novel therapeutic targets in IgAV.
Results
Genome-wide Association Study (GWAS)
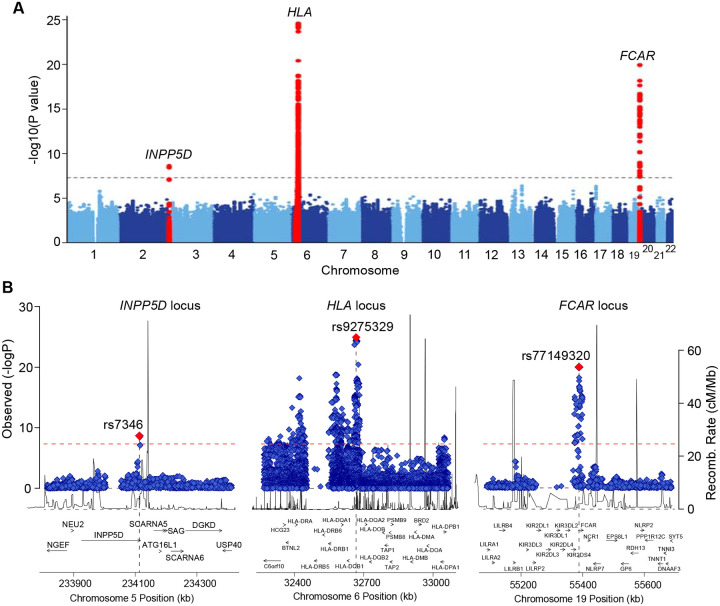
We performed a GWAS meta-analysis of three large case-control cohorts (two newly genotyped and one previously published cohort7) comprising a total of 8,098 individuals (2,170 IgAV cases and 5,928 controls). The study cohorts are summarized in Table S1 with detailed descriptions provided in the Methods. In each cohort, the controls were recruited from the same ancestral populations and geographic regions as the cases and had no history of kidney or autoimmune diseases. Of the three cohorts, two were of European ancestry (1,025 cases and 3,173 controls), and one cohort was of East-Asian ancestry (1,145 cases and 2,755 controls). All cohorts were genotyped with high-density SNP arrays, ancestrally matched using principal component-based methods12, and imputed using ancestry-specific genome sequence reference panels13 (Fig. S1A–D). In the meta-analysis, we observed negligible genomic inflation, consistent with the absence of systematic biases (λ=1.02, Fig. S1E). We detected three genome-wide significant loci (P<5×10−8, Fig. 1), including the HLA locus (OR=1.55, P=1.1×10−25) and two novel non-HLA loci, on chr.2q37.1 (INPP5D, OR=1.34, P=2.2×10−09) and chr.19q13.42 (FCAR, OR=1.51, P=1.0×10−20). Conditional analysis identified two independent signals within the HLA region, but no secondary signals at the non-HLA loci (Table S2). We also detected 14 suggestive loci based on P<1×10−5 , including the TNFSF13 and IL6R loci (Table S3).
Figure 1. GWAS meta-analysis for IgAV identified 3 genome-wide significant regions.
(A) Manhattan plot with genome-wide significant loci highlighted in red; the dotted horizontal line indicates a genome-wide significance threshold (P=5×10–8). (B) Regional plots for the INPP5D locus (left), the HLA locus (middle), and the FCAR locus (right). The x-axis shows the physical position in kilobases (kb, hg19 coordinates) and includes known gene annotations; the left y-axis presents -log10 p-values for association statistics and the right y-axis shows the recombination rates; the dotted horizontal line indicates a genome-wide significant threshold of 5×10−8.
Transcriptome-wide Association Study (TWAS) and Proteome-wide Association Study (PWAS)
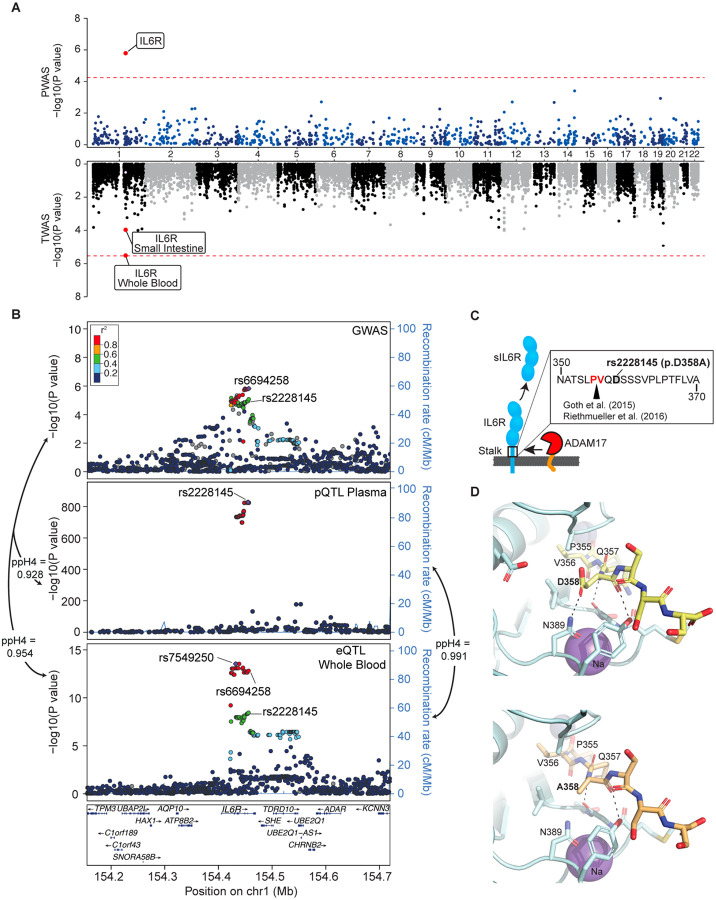
TWAS14 and PWAS15 represent powerful approaches to identify causal genes for complex traits by testing the effects of genetically determined gene or protein expression on a disease risk. Our PWAS (see Methods) identified a single proteome-wide significant signal for soluble IL6R, with higher genetically determined plasma levels associated with greater disease risk (P=1.6×10−6, Fig. 2A, Supplementary Data 1). TWAS provided no signals of transcriptome-wide significance although the IL6R signal was suggestive in blood and small intestine tissues (Supplementary Data 2).
Figure 2. Effects of the IL6R locus.
(A) Miami plot for PWAS and TWAS analyses. The red lines indicate the Bonferroni adjusted significance thresholds (5.6×10−5 for PWAS and 2.5×10−6 for TWAS). (B) Regional association plots for the IL6R locus. SNPs are plotted by position (hg19, 250kb window) versus −log10(P-values) from GWAS of IgAV (top), pQTL of plasma sIL6R levels (middle) and eQTL of IL6R expression in whole blood (bottom). The purple diamond highlights the most significant SNP for each association. SNPs are color-coded to reflect their LD with this SNP using LocusZoom with HapMap CEU reference. The missense variant IL6R p.Asp358Ala (rs2228145) is indicated in each plot. (C) The p.Asp358Ala substitution in relation to the cleavage site of the membrane bound IL6R. (D) Structure of the ADAM17 catalytic domain/IL-6R complex. The structure of ADAM17 (pale cyan ribbon render) was modeled with bound peptide 355-PVQDSSS-361 (yellow sticks in upper panel) or 355-PVQASSS-361 (pale orange sticks in lower panel) corresponding to a segment of the stalk region of IL-6R including the cleavage site (P355/V356). Residues of ADAM17 within 5 angstroms of D358 or A358 are shown as sticks. Polar interactions between residue 358 and ADAM17 are noted by dashed lines. The change D358A eliminates the interaction with N389 of ADAM17, potentially reducing the affinity of IL-6R for ADAM 17. Zinc and Sodium ions are shown as slate and purple spheres, respectively. Images generated with PyMOL (Molecular Graphics System, Version 2.0, Schrödinger, LCC).
HLA-DRB1 Locus
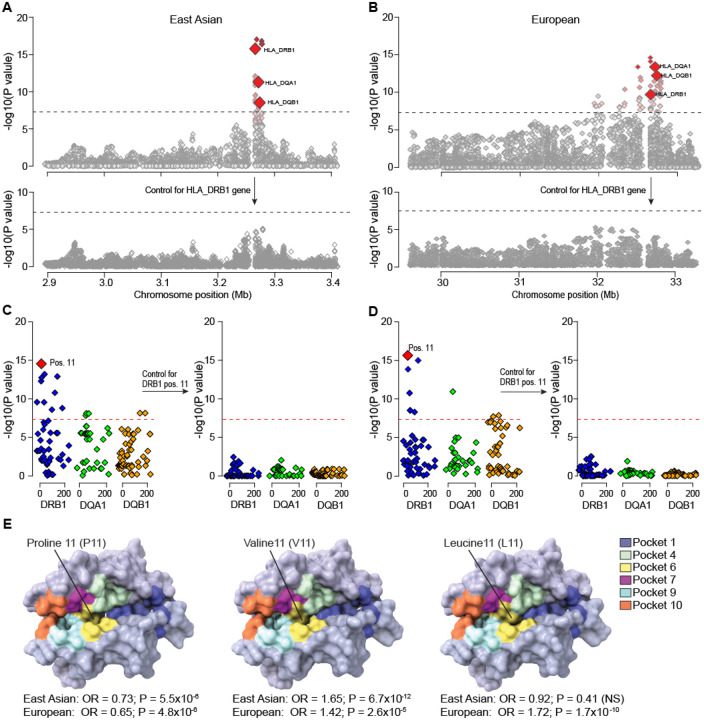
Using ancestry-specific reference panels, we imputed classical HLA alleles and individual amino-acid polymorphisms at the HLA class I and II genes (Methods). In the East Asian cohort, gene-based analysis of classical HLA alleles demonstrated the strongest association at HLA-DRB1 (P=1.5×10−16, Fig. 3A, Table S4). Conditioning the association on DRB1 classical alleles eliminated the entire HLA signal (Fig. 3A). Among the polymorphic sites in DRβ1, position 11 was most significant (P=2.9×10−15) and accounted for the entire signal in the conditional analysis (Fig. 3C and Table S5). Specifically, valine at DRβ1 position 11 conveyed the strongest risk (OR=1.65, P=3.4×10−12, Table S6). This substitution tags the HLA-DRB1*04 classical allele (also genome-wide significant, OR=1.62, P=3.9×10−10). In contrast, proline at DRβ1 position 11, tagging HLA-DRB1*1501, conveyed a protective effect (OR=0.73, P=4.5×10−6, Table S6). The same residues with concordant effects have been previously reported in the GWAS for IgA nephropathy (IgAN)16, suggesting shared HLA susceptibility between IgAV and IgAN.
Figure 3. Fine mapping of the HLA region in East Asian and European ancestry cohorts.
(A) Regional plot for the East Asian cohort with bi-allelic and multi-allelic association statistics for all imputed variants and classical HLA alleles; the strongest association signal was for HLA-DRB1 (upper panel); the results for HLA-DQA1 and HLA-DQB1 are highlighted for reference; after controlling for HLA-DRB1 classical alleles (black arrow), there was no residual association across the entire 3-Mb region (lower panel). (B) Regional plot for the European cohorts with bi-allelic and multi-allelic association statistics for all imputed variants and classical HLA alleles; after controlling for HLA-DRB1 classical alleles, there were no significant associations in the region (lower panel). The dotted horizontal line indicates the genome-wide significance threshold (α=5×10−8). (C) East Asian analysis of polymorphic amino acid positions within DRβ1 (blue), DQα1 (green) and DQβ1 (orange) using conditional-haplotype tests; the horizontal dash line indicates the genome-wide significance level. The most strongly associated polymorphic site was position 11 in DRβ1 (left panel); after controlling for this position, there was no residual association (right panel). (D) European analysis of polymorphic amino acid positions within DRβ1 (blue), DQα1 (green) and DQβ1 (orange); consistent with East Asian analysis, the DRβ1 amino acid position 11 (left panel) provided the strongest signal; no additional independent positions were found upon conditioning on this position (right panel). (E) Structure of the DRα1-DRβ1 complex. The protective variant (P11, left) and risk variants (V11, middle and L11, right) are shown in surface model viewed into the peptide binding groove. Models are colored in a background of light blue shade, with individual peptide amino acid binding pockets shaded according to the provided color key.
Consistent with the East Asian cohort analysis, HLA-DRB1 was also genome-wide significant in European cohorts (Fig. 3B, Table S7). Similarly, the analysis of polymorphic sites identified the strongest association of DRβ1 position 11, with no other independent signals after conditioning on position 11 (Fig. 3D and Table S8). We observed that leucine at position 11 was a strong risk substitution in Europeans (OR=1.72, P=1.7×10−10, Table S6). This substitution is rare in East Asian (control freq. 0.03) compared to European populations (control freq. 0.11), explaining ancestry-specificity of this signal. Consistent with the findings in East Asians, we also replicated the risk effect of valine (OR=1.42, P=2.6×10−5) and the protective effect of proline (OR=0.65, P=4.8×1−6) in Europeans.
Next, we performed detailed analysis of the potential impact of the associated amino acid risk residues on the structure and potential function of DRβ1. Notably, valine at position 11 is in tight LD with histidine at position 13 (r2=0.99) and tyrosine at position 96 (r2=0.98) forming a distinct DRβ1 risk haplotype (V11-H13-Y96). The second (European-specific) DRβ1 risk haplotype (L11-F13-E96) is formed by leucine at position 11, in LD with phenylalanine at position 13 (r2=0.99) and glutamic acid at position 96 (r2=0.99). The positions 11 and 13 map to the P6 and P4 pockets of the DRβ1 antigen-binding groove, while the position 96 is at the surface-exposed on the DR-β2 domain (Fig. S2 and S3). Comparisons of protein structures between the protective and risk haplotypes showed two features. First, the crevasse between pockets 6 and 4 with the protective haplotype (Fig. S2B left) is less accessible compared to the two risk haplotypes (Fig. S2B middle and right). Second, the protective substitution with arginine at position 13 provides a distinct electronegative character to the binding groove (Fig. S2C left). These landscape and electrostatic changes may alter which peptides are accommodated by DRβ117. Moreover, the position 96 is involved in the DRβ1 interactions with the HLA-DM complex in the thymic antigen presenting cells18, and thus substitutions at this position may additionally alter the presentation of self-antigens for clonal deletion19,20 (Fig. S3). As shown in Fig. S3B1–3, residue 96 is in the binding-interface with HLA-DM β-chain. All three residues Q96 (protective), E96 and Y96 (both risk) showed differing degrees of association with DM and the aqueous environment. As this is one of several residues that contribute to the interface with DM, future studies will be needed to define the mechanism of this association. Additionally, HLA-DO, a nonclassical MHC-II protein that appears to promote tolerance to self-antigens21,22, binds to HLA-DM through this same interface to competitively inhibit interaction with DRβ123. Thus, residues such as 96 could modulate interaction of HLA-DM with other factors involved in antigen presentation.
INPP5D Locus
The lead SNP (rs7346) at chr.2q37.1 locus is a synonymous variant in INPP5D (encoding Inositol Polyphosphate-5-Phosphatase D, a.k.a. SHIP-1) and is in tight LD with rs9247 (r2=0.99), a missense variant p.His1157Tyr in INPP5D (Table S9). Although there is no clear deleterious prediction for this substitution by PolyPhen2, CADD and REVEL, this variant is located within a compositionally biased region of SHIP-1 that is enriched in charged residues, and the substitution of histidine (positively charged) with tyrosine (neutral) may alter its binding energy. SHIP-1 is expressed in myeloid cells, where its activation is mediated by tyrosine phosphorylation. Once phosphorylated, SHIP-1 functions as a negative regulator of FcαR signaling, inhibiting myeloid cell proliferation, chemotaxis, and degranulation24,25. Phosphorylation of SHIP-1 also occurs by simultaneous engagement of IgA and IgE Fc receptors, suppressing allergic responses26. In mice, deletion of Inpp5d leads to autoinflammatory disease with neutrophilia and gastrointestinal involvement27,28. In GWAS, this locus has previously been associated with allergy29,30, and increased neutrophil31, eosinophil32,33, and monocyte counts31. In our meta-PheWAS across major biobanks involving 786,138 individuals (Fig. S4A and Supplementary Data 3), the IgAV-risk allele at this locus was additionally associated with hypothyroidism (OR=1.05, P=2.5×10−10) and dermatitis herpetiformis (a skin rash due to IgA response against epidermal transglutaminase and involving FcαR-mediated neutrophil degranulation34, OR=1.95, P=8.7×10−7).
FCAR Locus
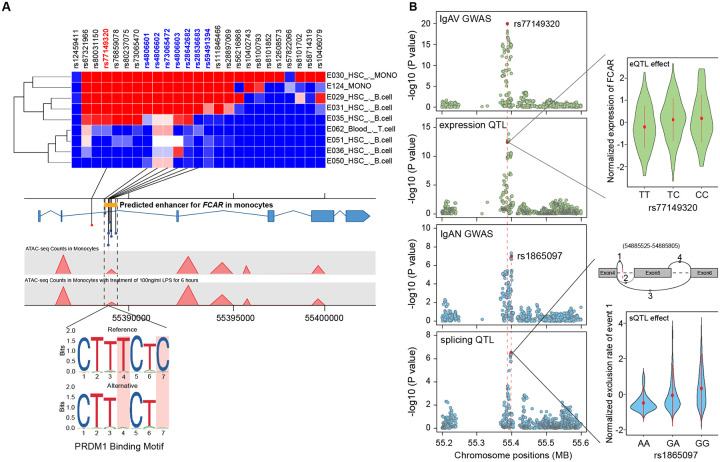
The index SNP at 19q13.42 (rs77149320) maps to the intronic region of FCAR, encoding IgA receptor FcαR (CD89), a transmembrane glycoprotein expressed specifically on myeloid cells. FcαR interacts with IgA immune complexes triggering phagocytosis, cytotoxicity, and release of inflammatory mediators. In prior GWAS, this locus has been associated with IgAN16 and inflammatory bowel disease35. In our meta-PheWAS, the most significant association of this locus was with higher risk of “nephritis and nephropathy with pathological lesion” (Fig. S4B). The index SNP is in tight LD (r2>0.8) with seven variants intersecting a putative cis-regulatory region in myeloid cells defined by ABC models36, confirmed by FUN-LDA functional scores37, and intersecting open chromatin by monocyte-specific ATAC-seq in 25 healthy donors38 (Fig. 4A, Tables S10). Two of the seven top risk alleles in LD (rs4806602-G and rs73065472-T) disrupt the predicted binding motif for PRDM139, a transcriptional repressor triggered by infections via Toll-like receptor and STAT signaling (Fig. 4A). PRDM1 activates NF-κB/TNF-R signaling by repressing NLRP240, maintains effector functions of B- and T-cells40,41, and promotes myeloid cell differentiation41. In agreement with these findings, our network-based differential activity analysis demonstrated higher regulatory activity of PRDM1 in IgAV cases versus controls (FDR=0.0037, see below).
Figure 4. Functional annotations of the FCAR locus.
(a) The heatmap of FUN-LDA functional scores for the lead SNP (rs77149320, red) and its high LD proxies (r2>0.8, only SNPs and cell types with positive scores are depicted). The 7 SNPs in LD (names in blue) intersected an ABC model-predicted intronic enhancer of FCAR. This regulatory region was further confirmed by an ATAC-seq peak in monocytes before and after LPS treatment. The prioritized SNPs were analyzed against transcription factor binding motifs predicted by JASPAR, and two risk alleles (rs4806602-G and rs73065472-T in perfect LD and two base pairs apart) were found to disrupt the PRDM1 binding motif; (b) Co-localization of the FCAR locus between GWAS for IgAV and blood eQTL for FCAR in IgAV (top two panels), and GWAS for IgAN and blood sQTL for FCAR in IgAV (bottom two panels). The eQTL effect shown in the right upper panel (x-axis: genotypes, y-axis: normalized expression of FCAR). The sQTL effect shown in the right bottom panel (x-axis: genotypes, y-axis: normalized exclusion rate of chr19:54885525–54885805 region based on hg38 annotations, the splice model depicted above).
IL6R Locus
Soluble IL6R (Interleukin 6 receptor) reached a proteome-wide significance in PWAS and was supported by a suggestive signal in GWAS. Interestingly, the top GWAS risk allele (rs6694258-A, OR=1.21, P=1.49×10−6, Table S3) was associated with higher plasma protein levels of IL6R, but lower IL6R transcript levels of in blood (Fig. 2B). The GWAS locus co-localized with both expression QTL (posterior probability 95%) and protein QTL (posterior probability 93%) supporting a shared casual haplotype (Fig. 2B and Supplementary Data 4). Four SNPs in strong LD with rs6694258 (r2≥0.8) intersected IL6R enhancers predicted by ABC model36 and confirmed by ATAC-seq in monocytes38 (Fig. S5 and Table S10). The observed protein QTL effect of this locus was particularly strong, with the top SNP (rs4129267) alone explaining 25.9% of variance in plasma IL6R levels. The second top variant (rs2228145, r2=0.998 with rs4129267), is a missense variant resulting in p.Asp358Ala substitution in the stalk region, close to the ADAM17 cleavage site of IL6R (between position 355 and 356; Fig. 2C)42, and demonstrated to increase sIL6R levels in prior studies43–45. We modeled IL6R-ADAM17 structures based on published studies46 to demonstrate that Asp358 forms a hydrogen bond with Asn389 of ADAM17, whereas p.Asp358Ala substitution in IL6R results in the loss of this interaction (Fig. 2D and Fig. S6). This may decrease affinity between IL6R and ADAM17, potentially allowing for faster release upon cleavage and higher turnover rate. Prior studies showed that a peptide harboring the Asp358Ala substitution is cleaved more rapidly by ADAM17 than the peptide with the Asp358 residue45. In our Meta-PheWAS of rs2228145, the risk allele for IgAV was additionally associated with a higher risk of asthma and urticaria, and a reduced risk of atrial fibrillation/flutter and coronary syndrome (Fig. S4C and Supplementary Data 5).
Shared and distinct loci with IgAN
To examine the genetic relationship between IgAV and IgAN, we tested genome-wide genetic correlation using results from the latest GWAS for IgAN16. We observed a significant positive genetic correlation between IgAV and IgAN (rg=0.77, P=2.8×10−12) also present after excluding the HLA region (rg=0.69, P=0.016). To better understand the shared and distinct loci between IgAN and IgAV, we performed a cross-phenotype GWAS (see Methods). We observed 21 genome-wide significant loci including 3 novel loci, near PAID4, WLS, and ANKRD55 (Fig. S7, Table S11). All three IgAV-risk loci (HLA, INPP5D, and FCAR loci) became more significant in the cross-phenotype analysis (Fig. S7), but at the FCAR locus significant heterogeneity was detected between IgAV and IgAN (I2=94.98, p<0.0001). While the INPP5D locus perfectly co-localized between the two traits (posterior probability of shared causal variant 0.99), the FCAR locus did not colocalize (posterior probability of different causal variants 0.98, Table S12), implying distinct genetic mechanisms between IgAV and IgAN at this locus.
Maps of blood expression and splicing QTLs in IgAV
To better interpret the effects of IgAV-risk alleles, we performed blood RNA-seq in a cohort of 255 IgAV cases and generated genome-wide maps of disease-context specific expression and splicing QTLs. In total, we identified 7,594 cis-eGenes (11,597 independent eQTLs) at FDR<0.05 (Supplementary Data 6 and 7). Among them, 1,345 eGenes were IgAV context-specific when compared to GTEx v7 whole blood dataset (Fig. S8). These IgAV-specific eGenes were enriched in immune-related pathways and mouse phenotypes, including TNF and interleukin signaling (Fig. S8). We also identified 1,192 sGenes (Supplementary Data 8), including 226 IgAV-specific sGenes enriched in the biological processes related to immunity, such as leukocyte and lymphocyte activation and regulation of immune system responses (Table S13). The eQTL and sQTL results were then used to investigate the consequences of IgAV-risk alleles.
Based on these maps, we observed strong colocalization between the FCAR locus for IgAV and FCAR cis-eQTL (posterior probability of shared causal variant of 0.96). IgAV-risk allele was associated with increased blood FCAR mRNA expression (Fig. 4B). Differential gene expression analysis confirmed higher FCAR gene expression in IgAV cases compared to controls (fold change=1.3, P=6.84×10−06). In contrast, the distinct FCAR association signal for IgAN did not co-localize with neither eQTL nor GWAS for IgAV, but it did co-localize strongly with an sQTL for FCAR (posterior probability of shared causal variant of 0.99). One of the top IgAN-risk alleles (rs1865097-A) is a deep intronic variant associated with lower exclusion rate of the chr19:54885525–54885805 region (Fig. 4B). This variant, located at the end of the alternative splicing event (chr19:54885805) functions as a cryptic splice acceptor site. The protective exclusion of chr19:54885525–54885805 region results in the transcript excluding exon 5 of FCAR encoding the transmembrane domain.
IgAV transcriptional regulators in myeloid cells
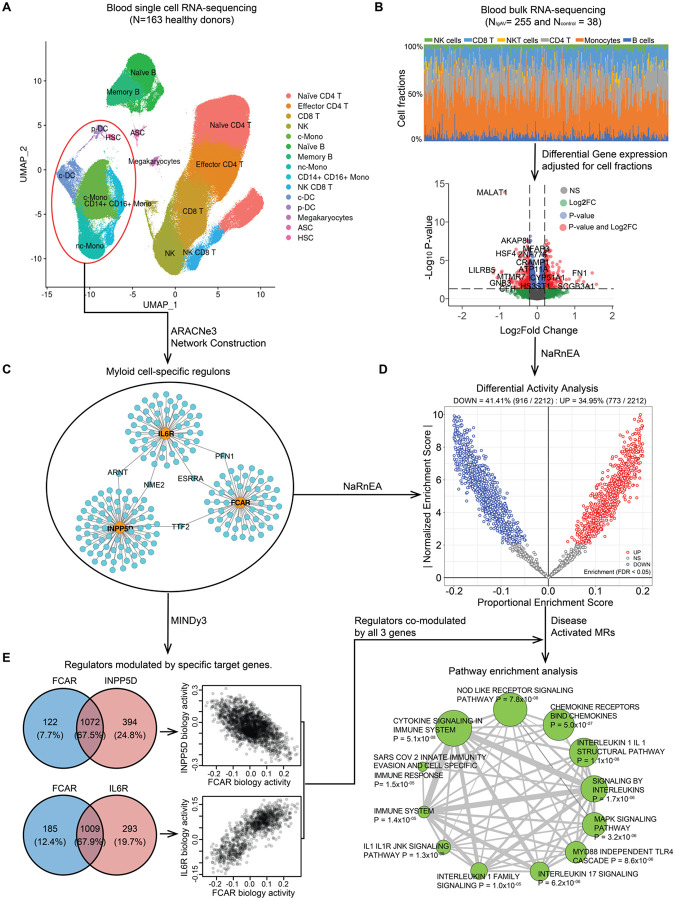
Given that all identified candidate causal genes are expressed specifically in a myeloid cell lineage, we generated a myeloid-specific transcriptome-wide regulatory gene network based on blood single-cell RNA-seq data from 163 reference individuals (Figs. 5, S9 and S10, and Methods). To overcome the sparseness of single-cell gene expression matrix, we created a pseudo-bulk gene expression profile for the myeloid cell cluster. We then used a mutual-information-based network reconstruction method, ARACNe3, to identify 812,995 significant pairwise regulatory relationships between 2,212 transcriptional regulators and their targets. Next, we generated the differential gene expression signature using the bulk RNA-seq data from our dataset of 255 IgAV patients and 38 healthy controls controlling for age, sex, genetic ancestry, and CIBERSORTx47-deconvolved cell fractions (Fig. 5B). Based on the myeloid-specific ARACNe3 regulons (Fig. 5C) and the adjusted differential gene-expression signatures, we inferred differentially activated regulators in IgAV cases compared to controls using the NaRnEA algorithm48. This analysis identified 773 regulators with increased activity and 916 with decreased activity in IgAV cases compared to controls based on FDR<0.05 (Fig. 5D, and Supplementary Data 9).
Figure 5. Master regulators of IgA vasculitis modulated by the GWAS candidate genes.
(a) the UMAP plot for the reference single cell RNA-seq data from 163 healthy individuals. The blood cells were clustered and annotated into 15 different immune cell types highlighted in different colors; (b) Deconvolution and differential gene expression analysis based on blood bulk RNA-seq data from 255 IgAV cases and 38 health controls. The upper panel shows the distribution of different cell type fractions across all 293 individuals. The volcano plot (lower panel: x-axis denotes the log2 Fold Change and the y-axis indicates the adjusted P values) with differentially expressed genes in red (Padjusted<0.05 and log2 Fold Change>0.2) after adjusting for cell type fractions; (c) The myeloid regulons reconstructed transcriptome-wide based on the myeloid lineage cells (circled in red above) using ARACNe3 software (a subset of only 3 regulons shown to illustrate this step). (d) Differential activity analysis based on the myeloid regulons and cell fraction-adjusted differential gene expression signature in IgAV cases compared to controls. The x-axis indicates the proportional enrichment score (effect size for gene set enrichment that was normalized by set’s size, parameterization, and gene expression signature). The y-axis shows the normalized score compared to the expectation under the null hypothesis. The red and blue colors indicate the regulators with higher and lower activity, respectively, in IgAV cases compared to healthy controls. (e) Regulators co-modulated by the GWAS candidate genes: 1,072 regulators co-modulated by FCAR and INPP5D with mostly opposed effects on their biological activities (upper panel) and 1,009 regulators co-modulated by FCAR and IL6R with direction consistent effects on their biological activities (lower panel). The targets of the top master regulators co-modulated by all 3 genes were enriched in multiple immune related pathways based on FDR< 0.05 (depicted as enrichment map with a node size proportional to the statistical significance of enrichment and edge thickness proportional to gene set overlaps).
The regulators activated in the disease state were significantly enriched in the TGFβ signaling pathway (Table S14). We have previously identified genetic support for the involvement of transcriptional regulators LITAF, HDAC7, RUNX2, and RUNX3 in the determination of IgA levels49. In IgAV cases, both LITAF and HDAC7 were inferred to have increased activity in myeloid cells (FDR=1.5×10−10 and 4.1×10−8, respectively), while RUNX2 and RUNX3 to have suppressed activity compared to controls (FDR=1.5×10−3 and 5.9 ×10−14, respectively). These observations are direction-consistent with the previously reported genetic effects of these regulators on IgA levels49, supporting global activation of the regulatory programs that increase IgA production in IgAV.
Master regulators modulated by IgAV candidate genes.
Next, we hypothesized that FCAR, INPP5D, and IL6R transcripts may modulate the activity of proteins representing Master Regulators (MRs) of IgAV-related myeloid cell state. To test this hypothesis, we used MINDy50, a new algorithm that identifies potential modulators of transcriptional interactions by analyzing statistical dependencies between a transcription factor and its targets conditioned on the expression level of a candidate modulator. By applying this approach to all myeloid regulators using GWAS candidate genes as potential modulators and myeloid-specific ARACNe3 regulons, we identified 1,194 regulators modulated by FCAR (Supplementary Data 10), 1,466 by INPP5D (Supplementary Data 11), and 1,302 by IL6R (Supplementary Data 12) at FWER<0.05 (Fig. 5E). A total of 1,072 regulators were co-modulated by both FCAR and INPP5D with a strong negative correlation in biological activity, indicating that the regulator activities enhanced by FCAR were generally repressed by INPP5D (Spearman’s r=−0.52, P<2.2×10−16). This phenomenon is fully consistent with the established role of INPP5D as a negative regulator of FcαR signaling. Moreover, a total of 1,009 regulators were co-modulated by FCAR and IL6R, with strong positive correlation of their biological activity (Spearman’s r=0.78, P<2.2×10−16), suggesting concordant effects on gene regulation in myeloid cells. Lastly, to identify MRs affected by our genetic risk loci, we intersected the 50 most activated and 50 most inactivated IgAV MRs with the genes co-modulated by FCAR, INPP5D, and IL6R transcripts. This nominated 28 high-priority MRs (Table S15), targets of which were significantly enriched in key immune-related pathways, including cytokine, chemokine, and Nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathways (Fig. 5E).
Discussion
By combining hypothesis-free genome, transcriptome, and proteome-wide association approaches with gene expression and splice QTL analyses and network-based analyses, our study uncovered new genetic mechanisms of IgAV. We fine-mapped HLA-DRB1, FCAR, INPP5D, and IL6R as the candidate causal genes for IgAV and characterized their effects on the disease-related regulons in myeloid cells. We also demonstrated shared genetic susceptibility between IgAV and IgAN and extensively characterized the shared and distinct susceptibility loci in combined and separate analyses of both conditions.
Our fine-mapping studies revealed that the HLA signal was largely explained by the polymorphic position 11 in DRβ1, which is in tight LD with additional substitutions at positions 13 and 96. Our haplotype-based analyses defined a protective haplotype (P11-R13-Q96) and two risk haplotypes (L11-F13-E96 and V11-H13-Y96). While positions 11 and 13 mapped to the surface of pockets 4 and 6 of peptide binding groove of DRβ1 potentially changing antigen binding affinity, position 96 may be involved in the interaction between DRβ1 and HLA-DM/DO, potentially affecting antigenic peptide loading to DRβ1. Notably, the same haplotypes have a direction-consistent effect in IgAN, suggesting that the same antigen presentation defect is shared by both conditions.
Two novel non-HLA risk loci, FCAR and INPP5D, converge on the same pathway in myeloid cells, highlighting the central role of the FcαR (CD89) signaling in the pathogenesis of IgAV. In prior studies, elevated serum levels of IgA1-CD89 complexes have been reported in children with IgAV51 and IgAN52. However, our results demonstrate distinct genetic mechanisms underlying genetic associations of the FCAR locus in IgAV versus IgAN. In IgAV, the risk alleles disrupt a binding motif of PRDM1 in a monocyte-specific FCAR enhancer and are associated with increased FCAR gene expression. In IgAN, the risk alleles are associated with a lower rate of a splicing event that excludes a transmembrane domain of FcαR, leading to increased expression of the full-length isoform.
In contrast, the risk variant in INPP5D encoding SHIP-1, a known negative regulator of FCAR signaling25,26, had the same effect in IgAV and IgAN. The inhibition of SHIP-1 is known to generate molecular profiles consistent with activation of inflammasome53 and increased phagocytosis54,55. We hypothesize that p.His1169Tyr substitution may disrupt the function of SHIP-1, but further studies are needed to understand the precise effect of this variant.
Our study demonstrated that the PWAS approach could effectively implicate a new causal gene when GWAS by itself is underpowered to establish a significant association. The PWAS signal for soluble IL6R in IgAV was supported by a colocalization between GWAS and protein QTL. About 85% of circulating sIL6R is generated by proteolysis56, and one of the top protein QTL variants was p.Asp358Ala, a substitution mapping to the proteolytic cleavage site in the stalk region of IL6R57, and previously associated with elevated serum levels of sIL6R58 and reduced membrane-bound IL6R59,60. Once cleaved, sIL6R can interact with circulating IL6 and activate gp130, a signal transducer expressed on multiple cell types, resulting in systemic pro-inflammatory effects58–60. In aggregate, these results support a pathogenic role of sIL6R and suggest an opportunity to repurpose Tocilizumab, a monoclonal antibody blocking IL6R, as a potential new treatment for IgAV.
We additionally characterized the pattern of global transcriptional dysregulation in circulating immune cells of children with IgAV. Our methods for reverse engineering of myeloid-specific regulatory networks using single-cell transcriptomics allowed us to define the key transcriptional regulators in IgAV. To further differentiate regulatory disease drivers from reactive changes, we prioritized differentially active regulators that were also co-modulated by the transcripts supported by genetic evidence. Among the 50 most differentially activated and 50 most inactivated regulators, 28 were significantly co-modulated by FCAR, INPP5D, and IL6R; moreover, their targets were significantly enriched in key immune-related pathways. These regulators appear to drive the disease process, and thus may represent attractive therapeutic targets.
Lastly, our cross-phenotype GWAS highlights shared genetic susceptibility between IgAV and IgAN, including strong genome-wide genetic correlation and shared HLA-DRB1 risk and protective haplotypes. Most of the known IgAN-risk loci were associated with higher risk of IgAV, including the TNFSF13 locus encoding APRIL. Phase III clinical trials of APRIL inhibition are underway for IgAN, and our results provide the first genetic support for APRIL’s involvement also in IgAV. Despite these commonalities, clear differences in the disease associations were detected at the FCAR and IL6R loci, and these differences could potentially contribute to the unique systemic manifestations of IgAV.
METHODS
We performed a GWAS meta-analysis of three large case-control cohorts comprising a total of 8,098 individuals (2,170 IgAV cases and 5,928 controls). The study cohorts are summarized in Table S1 with detailed descriptions as follows:
Beijing Cohort:
The cases and controls were recruited from the clinics of Peking University First Hospital, Beijing, China. We used standard diagnostic criteria for IgAV that included a palpable purpura in association with at least one of the following: diffuse abdominal pain, arthritis or arthralgia, renal involvement (hematuria and/or proteinuria), and/or IgA deposition in a biopsy specimen (skin, intestinal tract, or kidney). The controls had no history of purpura or kidney disease. All participants provided informed consent to participate in genetic studies. Genomic DNA was extracted from whole blood samples using standard procedures and genotyping was performed with Illumina Omni 2.5, Omni 2.5 Exome, and Omni ZhongHua chips. The analysis of intensity clusters and genotype calls were performed in Illumina Genome Studio software 2.0. All SNPs were called on forward DNA strand and the overlapping SNPs across the three platforms were extracted for further analysis. The standard quality control (QC) included per-SNP genotyping rate >95%, per-individual genotyping rate >90%, MAF>0.01, and HWE test P>1×10−5 in controls. We excluded duplicates and cryptically related individuals based on the estimated pairwise kinship coefficients >0.05 using KING software61. Gender of each sample was imputed based on the analysis of sex chromosome markers using PLINK v1.962 and individuals with gender mismatch against study records were excluded. The ancestry was evaluated using principal component analyses (PCA)12 using 46,494 high-quality (genotype rate > 99%), common (MAF>1%), and independent (pairwise r2<0.05) markers. The regions of high LD were excluded from this analysis (see below). Ancestry outliers were excluded based on visual inspection of PCA plots. The final PCA produced only two significant PCs by Tracy-Widom test and well-matched distribution of cases and controls (Supplementary Figure 1a). After QC, the final dataset comprised of 3,900 individuals (1,145 cases and 2,755 controls) genotyped with 722,465 markers with an overall genotyping rate 0.99. We carried out imputation analysis with Minimac363 and pre-phasing with Eagle v2.4.164 using East Asian reference populations from 1000 Genomes Project (Phase 3). A total of 6,210,008 high quality common markers (imputation R2>0.8 and MAF>0.01) were imputed and used in downstream analyses.
GIGA-kids (Genomics of IgA-related disorders in kids) Cohort:
This cohort included a multicenter GIGA-kids Study, as well as participants recruited by the Columbia University CKD Biobank and additional cases and controls referred through the GIGA-Europe network including collaborating sites in Italy, Poland, and France. The IgAV diagnostic criteria were the same as for the Beijing cohort and included a palpable purpura in association with at least one of the following: diffuse abdominal pain, arthritis or arthralgia, renal involvement (hematuria and/or proteinuria), and IgA deposition in a biopsy specimen (skin, intestinal tract, or kidney mesangium). The controls had no history of purpura or kidney disease. All participants provided informed consent to participate in genetic studies. DNA of whole blood samples was extracted using standard procedures and genotyped using Illumina MEGA chips. The analysis of intensity clusters and genotype calls were performed using Illumina Genome Studio 2.0; all SNPs were called on forward DNA strand. Standard QC filters included per-SNP genotyping rate >95%, per-individual genotyping rate >90%, MAF >0.01, and HWE test P>1×10−5 in controls as implemented in PLINK v1.962. We excluded duplicates and cryptically related individuals with pairwise kinship coefficients >0.0561. The final post-QC dataset consisted of 2,908 individuals genotyped for 515,946 markers with an average genotyping rate of 99.9%. This cohort was further divided into two non-overlapping sub-cohorts: IgAV without nephritis sub-cohort of 316 cases and 924 controls and IgAV with nephritis (IgAVN) sub-cohort of 425 cases and 1,243 controls. For ancestry principal component analysis (PCA)12, we used 153,078 high-quality independent (pairwise r2<0.05) SNPs. The PCA produced 7 significant PCs in each case-control sub-cohort, and the visual inspection of PC plots confirmed comparable genetic ancestry between cases and controls (Supplementary Figure 1b and 1c). The imputation analysis was carried out with Minimac363 after pre-phasing in Eagle V2.3464, and using 1000 Genomes Europeans (Phase 3) population reference. A total of 7,192,202 and 7,200,332 common high-quality markers (imputation R2>0.8 and MAF>0.01) were used in downstream analyses of IgAV and IgAVN sub-cohorts, respectively.
Spanish IgAV cohort:
This cohort has been described in detail before7. Briefly, the cohort comprised of 288 cases and 1,596 controls. The participants were recruited in Spain and were of European ancestry. All participants provided informed consent to participate in genetic studies. Genomic DNA was extracted from whole blood using standard methods and genotyped using Infinium HumanCore BeadChip (Illumina, Inc). The standard QC were performed as described before, followed by imputation using the 1000 Genomes Europeans (Phase 3) population reference. The final dataset included 5,662,805 high-quality markers (imputation R2>0.8 and MAF>0.01). The PCA produced 6 significant PCs and visual inspection of PC plots confirmed comparable ancestry between cases and controls (Supplementary Figure 1d).
Genome-wide association studies (GWAS):
Within each cohort, primary association scans were performed using logistic regression under additive coding of dosage genotypes, and with adjustment for cohort-specific significant principal components of ancestry using PLINK v1.962. No substantial genomic inflation was observed in any individual cohort (lambda <1.05 consistently for each individual cohort). Subsequently, a fixed effects meta-analysis was performed to combine the results across all cohorts using METAL65. Genome-wide distribution of P-values was examined visually using quantile-quantile plots for the combined analysis and the genomic inflation factor was estimated at 1.02 for the overall meta-analysis (Supplementary Figure 2). To declare genome-wide significance, we used the generally accepted P-value threshold of 5.0×10−8, while signals with P<1.0×10−6 were considered as suggestive. For sensitivity analysis we also performed a trans-ethnic meta-analysis under a random effects model using TransMeta software66, but this analysis did not change our results or the number of significant loci. To detect independent signals at genome-wide significant loci, we also performed stepwise conditional analyses of each locus in each cohort using GCTA67, and the conditioned summary statistics were combined across cohorts using fixed effects meta-analysis65.
Transcriptome-wide association studies (TWAS):
TWAS was performed following the FUSION workflow68 based on weights developed from the European ancestry subset of the GTEx v8 tissues that were considered relevant to IgAV (kidney cortex, liver, spleen, whole blood, small intestine-terminal ileum, and EBV-transformed lymphocyte cells). Prediction models were based on elastic net modelling68. Multiple testing was accounted for by a Bonferroni adjustment for 17,126 unique number of protein-coding transcripts modelled across tissues (P < 0.05/17,126 = 2.9 × 10−6).
Proteome-wide association study (PWAS):
For PWAS, we adapted the FUSION workflow68 using plasma protein weights from the European ancestry participants of the Atherosclerosis Risk in Communities Study (ARIC) study69. These weights were based on plasma proteomic quantifications using Somascan V.4.1. Prediction models were based on elastic net modelling68. Proteome-wide significance level was defined using a Bonferroni correction for the total number of circulating proteins tested (P < 0.05/884 = 5.7×10−5).
Functional annotations of significant loci:
We first defined each GWAS locus by +/− 400kb of the genome-wide significant index SNP. We annotated all transcripts within these intervals using hg19 to create sets of positional candidate genes for each locus. Using ANNOVAR software70, we annotated all variants within the region that were in LD (r2>0.5) with the top SNP, including all known coding, splice, and 3’UTR and 5’UTR variants. We then performed FUN-LDA analysis to estimate the posterior probability of these variants being functional across various cell types37. Lastly, the variants were intersected with the predicted enhancers and mapped to regulated genes using the Activity-by-Contact (ABC) model36. The myeloid-specific regulatory regions were confirmed by the analysis of monocyte ATAC-seq peaks before and after LPS treatment38. The top prioritized variants were screened against transcription factor binding motifs using JASPAR39. To additionally prioritize target genes, we performed locus co-localization analyses with whole blood gene expression and splicing QTLs quantified from bulk RNA-seq of 255 IgAV cases. After harmonization of effect alleles, we identified all co-localized QTLs mapping to the region of the index SNP ±400 kb using Coloc package in R71. For the IL6R locus supported by PWAS, we additionally performed co-localization with pQTL summary statistics from participants of the ARIC study69. Because the pQTL signal exhibited seven conditionally independent signals (defined by the COJO-Slct algorithm67), we computed conditional pQTL summary statistics by conditioning on all other independently significant SNPs using COJO-Cond67. Subsequently, colocalization analyses were conducted for all pairwise combinations of the GWAS and conditionally independent pQTL summary statistics using an adapted version of the Giambartolomei colocalization method as implemented in the ‘coloc.fast’ function with default parameters71. Any co-localization with PP4>0.8 was considered as strong evidence in support of a shared causal variant. Only one of the seven conditionally independent sIL6R cis-pQTL signals co-localized with the IL6R GWAS and eQTL loci.
HLA imputation and testing:
We performed imputation of classical HLA alleles at two- and four-digit resolution, as well as individual amino acid polymorphisms at class I (HLA-A, -B, and -C) and class II (HLA-DQB1, -DQA1 and -DRB1) loci using SNP2HLA software72. The European and East Asian cohorts were imputed separately, using ethnicity-specific reference panels. For European reference, we used the pre-phased HLA reference dataset generated by the Type 1 Diabetes Genetics Consortium (T1DGC, 5,225 individuals)72. For our East Asian cohorts, we used the Pan-Asian HLA Reference Panel (268 individuals)73. In the association analyses, we included only common HLA alleles (MAF>0.01) that were imputed with high certainty (R2>0.8). Because frequencies of HLA alleles are known to vary by ancestry, the association testing was performed separately in European and East Asian cohorts. We used logistic regression models to test the additive effects of HLA allele dosages with adjustment for significant PCs of ancestry. For multi-allelic loci, we used the following logistic regression model:
| (1) |
where m denotes a total number of alleles at a specific multi-allelic locus, j indicates a specific allele being tested, and x, is the imputed dosage for allele j for individual i; β represents the intercept and β represents the additive effect of an allele j; P, denotes the value for k PC of individual i, n is the total number of significant PCs in the dataset, β is the effect size of principal component k. We compared log-likelihoods of the two tested models: the full model containing the test locus and relevant covariates with the reduced model without the test locus, but with the same set of covariates. The deviance was defined as −2 × log likelihood ratio, which follows a X2-distribution with m − 1 degrees of freedom, from which we calculated P-values. In addition to multi-allelic tests, we also performed bi-allelic tests of association for all imputed SNPs, classical HLA alleles, and individual amino acid residues in HLA molecules. All analyses were performed using dosage method under additive coding. Stepwise conditioning analyses across the HLA region were performed using both multi-allelic and bi-allelic coding of HLA variants. In each round of stepwise conditioning, we first included the most significant variant as the covariate in the logistic regression model. If additional independently associated markers were detected, they were included as covariates in subsequent models. We repeated these analyses until no residual associations across the entire locus were observed.
Analysis of polymorphic amino-acid sites in HLA genes:
To test the effects of individual amino acid substitution sites within the three significant HLA genes (HLA-DRB1, HLA-DQA1 and HLA-DQB1), we applied a conditional haplotype analysis using fully phased haplotypes. We tested each single amino acid position by first identifying the m possible amino-acid residues occurring at that position and then using m − 1 degrees of freedom test to derive P-values, with a single amino acid residue arbitrarily selected as a reference. For conditioning on individual amino-acid sites, we used the following procedure: by adding a new amino acid position to the model, a total of k additional unique haplotypes were generated and tested over the null model using the likelihood ratio test with k degrees of freedom. If the new position was independently significant, we further updated the null model to include all unique haplotypes created by all amino-acid residues at both positons to identify another independent position. The procedure was repeated until no statistically significant positions were observed.
Protein structure modeling:
Models of DRα1 and DRβ1 were generated based on PDB ID: 5V4M74, which harbored protective residues P11 and R13. Risk variants L11/F13 and V11/H13 were generated with COOT75. After removing the peptide, all three models were subjected to molecular dynamics minimization protocols with YASARA76. Images of DRα1 and DRβ1structures were generated with ChimeraX77. Modeling of the IL6R-ADAM17 peptide structures followed previously published methods46. Briefly, ADAM17 and N-terminal residues (CTCSPSH) of the bound TIMP3 were extracted from PDB ID: 3CKI78. Peptide residues were mutated to 355-PVQDSSS-361 or 355-PVQASSS-361 of the IL6R stalk region, and complexes were minimized in YASARA76. Models of HLA-DM-HLA-DR1 were generated with PDB ID: 4GBX79. The parent coordinates have glutamic acid at residue 96. Variants Q96 and Y96 were generated in COOT75 and minimized in YASARA76.
Meta-phenome-wide association study (Meta-PheWAS):
The meta-PheWAS for individual variants was performed in 786,138 participants across three biobanks: the UK Biobank (UKBB, N=460,363), Electronic Medical Records and Genomics-III (eMERGE-III, N=102,138) and All-of-Us (AoU, N=223,637). For meta-analysis, we harmonized coded diagnoses across biobanks by converting all available ICD-10 codes to the ICD-9-CM system. After conversions, we identified 1,816 unique phecodes. These phecodes were then tested using logistic regression adjusted for age, sex, study site, imputation batch, and 3 PCs of ancestry using the PheWAS R package80. Meta-PheWAS was performed using metagen software under fixed effects. For phenome-wide significance, we used the Bonferroni-corrected P < 2.75 × 10–5 to account for 1,816 independent phecodes tested.
Blood RNA sequencing:
Blood samples were collected using PAXGene RNA tubes from the participants of GIGA-kids study. To ensure complete lysis of blood cells and stabilization of RNA, the collected blood samples were incubated at room temperature for a minimum of 2 hours. The stabilized blood samples were further centrifuged to pellet cellular debris. Supernatant of the samples was lysed, digested, and homogenized using QuickGene RNA blood cell kit S (Catalog #: RB-S). Automated RNA extraction of lysate was done using QuickGene 810 Nucleic Acid Isolation System. RNA samples were purified with Zymo Research RNA Clean & Concentrator-5 kit (Catalog #: R1014). We use poly-A pull-down to enrich mRNAs from extracted total RNA samples with RIN>8 (200ng-1ug per sample) and performed library preparation using Illumina TruSeq RNA prep kit. Libraries were then sequenced using Illumina HiSeq2500/HiSeq4000 at Columbia University Genome Center. Sequencing generated 100bp single/pair-end reads, with a median achieved total reads per sample at 23.5M. Raw sequencing data were processed using RTA (Illumina) for base calling and bcl2fastq2 (version 2.17) for converting BCL to fastq format, coupled with adaptor trimming. RNA-seq data were aligned to the human reference genome hg38 with STAR software81. Following GTEx pipelines82, we identified and excluded low-quality samples with <10 million mapped reads, read mapping rate <0.2, intergenic mapping rate >0.3, base mismatch rate >0.01, and rRNA mapping rate >0.3. The outlier samples were further identified based on expression profile using correlation-based statistics and sex incompatibility checks following published methods83. After individual sample QC, we quantified the gene expression across samples using RNA-SeQC84 as read counts and transcripts per million (TPM). Only transcripts with TPM >0.1 and 6 read counts in at least 20% of samples were used for downstream analyses. The final post-QC dataset included 294 individuals (255 IgAV cases and 38 healthy controls) and 17,790 transcripts.
Cell type deconvolution and differential gene expression analyses:
We used CIBERSOFTx47 to deconvolve immune cell type abundance from bulk blood RNA-seq of 255 IgAV cases and 38 controls. We optimized this pipeline for 6 different major immune cell types, including CD4 and CD8 T cells, NK cells, monocytes, B cells and dendritic cells, using published reference panels generated by single-cell RNA-sequencing of 25,000 blood cells from 45 healthy donors85. The deconvolution was conducted based on the default parameters. For differential gene expression, we used DEseq2 R package86. This analysis was adjusted for age, sex, ancestry PCs, and deconvoluted cell-type fractions.
Expression QTL (eQTL) mapping:
For eQTL mapping, gene counts were normalized across samples using Trimmed Mean of M-values (TMM) method87 across 255 IgAV cases, and the expression values for each gene were inverse-normal transformed. The cis-eQTLs were identified using linear regression as implemented in fastQTL88, adjusting for age, sex, top 5 ancestry PCs and 45 PEER factors89. A false discovery rate (FDR) threshold ≤ 0.05 was applied to identify genes with at least one significant cis-eQTL (eGenes). To identify the list of all significant variant-gene pairs associated with cis-eGenes, a genome-wide empirical P-value threshold was defined as the empirical P-value of the gene closest to the 0.05 FDR threshold as descripted previously90. For each gene, variants with a nominal P-value below the gene-level threshold were considered significant and included in the final list of variant-gene pairs.
Splicing QTL (sQTL) mapping:
Splicing quantification was performed based on the intron excision levels across samples computed by LeafCutter91. Introns with few counts or with insufficient variability across samples were removed as described previously82. The filtered counts were normalized using the LeafCutter91 and the resulting splicing quantification files were used for sQTL mapping. Mapping of cis-sQTLs was performed in 255 IgAV cases using FastQTL88 controlling for age, sex, top 5 ancestry PCs, and 15 PEER factors92. To identify cis-sGenes, FDR was calculated in the same manner as for cis-eQTL, and an sGene-level nominal p-value threshold was determined for all significant variant-intron excision pairs.
Single-cell sequencing and data processing:
The reference blood single cell RNA-seq dataset was constructed using samples of volunteers recruited by the Reference Ability Neural Network (RANN) study though random mailing in Northern Manhattan93. Single-cell RNA-seq libraries of peripheral blood mononuclear cells (PBMC) were constructed using the 10× Genomics Chromium Next GEM Single Cell 3’ Reagent Kits as previously described94. An estimated 20,000 cells pooled from 3–4 donors were loaded on the 10× Genomics Chromium instrument. A total of 41 sequencing libraries (163 donors) were pooled and sequenced on the Illumina NovaSeq 6000 with S4 flow cell (Illumina, San Diego, CA) using paired-end, dual-index sequencing with 28 cycles for read 1, 10 cycles for the i7 index, 10 cycles for the i5 index, and 90 cycles for read 2. Gene expression profiles were generated for a total of 984,441 cells from 163 donors. FASTQ files were preprocessed using the count command of the CellRanger software (v6.0.0, 10× Genomics) with a human reference transcriptome GRCh38–2020-A. To de-multiplex cells from pooled samples, we took advantage of SNPs in the RNA-seq reads. Cells in the filtered Unique Molecular Identifier (UMI) count matrix were grouped into four clusters (each representing one participant) based on RNA genotypes using the freemuxlet software (downloaded on May 5th, 2021 from https://github.com/statgen/popscle). Cells were excluded if freemuxlet called them as doublets (Ncells=133,117). We further excluded cells with <200 or >4500 transcripts (Ncells=1,479), percentage mitochondrial transcripts >10% (Ncells=44,737), and the percentage of ribosomal RNA >5% (Ncells=12,706). We additionally removed hemoglobin genes (Ngenes=12) and genes expressed in <= 3 cells (Ngenes=681). After QC, we included 710,654 cells and 26,970 unique transcripts from 163 donors in downstream analyses. Using the Seurat R package, we performed normalization of gene expression across all 710,654 cells using LogNormalize method, followed by identification of variable transcripts, data scaling, and principal component analysis. We performed cell type clustering analysis based on the top 15 principal components. We then annotated 16 different cell types based on known blood cell markers85, including HSC (hematopoietic stem cells), ASC (antibody-secreting cells), Megakaryocytes, p-DC (plasmacytoid dendritic cell), c-DC (conventional dendritic cells), NK (natural killer) cells, NK CD8 T-cells, transitional or precursor cells, CD14+ CD16+ (classical) monocytes, non-classical monocytes, memory B cells, naïve B cells, effector CD4 T-cells as well as Naïve CD4 T-cells (Figure 5A).
Myeloid-specific regulatory network construction:
For reverse engineering of regulatory networks in myeloid cells, we used ARACNe3 (Algorithm for the Reconstruction of Accurate Cellular Networks version 3). This is an algorithm based on mutual information (MI) that identifies direct regulatory interactions mediated by transcription factors48,95. Using ARACNe3, we reconstructed a transcriptome-wide regulatory network based on myeloid cluster transcriptome generated using our reference blood single-cell RNA-seq data from 163 donors. Briefly, after QC filtering of poor-quality cells, we used 143,192 myeloid cells with UMI/cell >=3,000 across all 163 samples. We then excluded any donors with less than 500 high-quality myeloid cells, resulting in a total of 111 donors for network analysis. To address the sparsity of single cell expression matrix, we created a pseudo-bulk expression matrix by aggregating UMI counts across all myeloid cells (Figure S11). The ARACNe3 network was inferred using this pseudo-bulk matrix based on FDR<0.05. A total of 7 iterations were performed by random selecting 63.2% of all samples to achieve >50 targets per regulator. The final myeloid regulatory network involved 26,196 genes and 2,212 regulators based on the Gene Oncology (GO) annotations95.
Master Regulator (MR) analysis:
We used NaRnEA (Nonparametric analytical Rank-based Enrichment Analysis)48 to define differential activity of regulators based on the ARACNe-3 inferred myeloid regulons and differential gene expression signatures. NaRnEA is a gene set enrichment method which leverages a null model derived under the principle of maximum entropy. For each transcription factor, NaRnEA tests for significant enrichment of its ARACNe3-infered regulons in differential gene expression signature compared to randomly selected non-overlapping gene sets. This is important, as master regulators may be differentially active but not necessarily differentially expressed. The direction of effect was derived for each regulator (activated or suppressed in IgAV cases compared to controls), and the statistical significance was defined based on FDR<0.05. The top 50 most significant activated regulators in cases were defined as “IgAV Master Regulators”.
Modulation of Master Regulators by candidate genes
To test how our GWAS candidate genes (INPP5D, FCAR, and IL6R) co-modulate IgAV Master Regulators, we performed MINDy analysis based on the myeloid regulons. MINDy is an algorithm designed to uncover modulator genes influencing transcriptional interactions. This is accomplished by the analysis of ΔMI (the difference in mutual information between a TF and its target genes) after conditioning on the highest and lowest expression of any candidate modulator. The statistical significance of a particular ΔMI for a given TF-target gene pair and a given modulator was determined based on FDR < 0.05 as descripted previously96.
Supplementary Material
Acknowledgements:
We are grateful to all study participants for their participation and contributions to this study. The study was supported by the NIH grants 1K01DK137031 (LL), 2R01DK105124 (KK, JN), 1RC2DK116690 (KK). JN and TJG are supported in part by NIH grants AI149431, DK078244, and DK082753. ATG was supported by the Ruth L. Kirschstein National Research Service Awards 5T32GM007367-43 and F30CA257765. The work of AK was supported by German Research Foundation (DFG) Project ID 431984000 (SFB 1453) and Project ID 390939984 (EXC-2189, CIBSS). The work of SMM was supported by the EQUIP Program for Medical Scientists, Faculty of Medicine, University of Freiburg. AC was supported by the NCI Outstanding Investigator award R35CA197745. The GIGA-kids Study was additionally supported by the Pediatric Nephrology Research Consortium (PNRC) and a gift from the IgA Nephropathy Foundation.
Footnotes
Disclosures:
JN is a co-founder and co-owner of and consultant for Reliant Glycosciences, LLC. BAJ and DVR are co-founders and co-owners of Reliant Glycosciences, LLC. JN and BAJ are a co-inventor on US patent application 14/318,082 (assigned to the UAB Research Foundation [UABRF] and licensed by UABRF to Reliant Glycosciences, LLC). Dr. Califano is founder, equity holder, and consultant of DarwinHealth Inc., a company that has licensed some of the algorithms used in this manuscript from Columbia University. Columbia University is also an equity holder in DarwinHealth Inc.
References:
- 1.Hetland L.E., Susrud K.S., Lindahl K.H. & Bygum A. Henoch-Schonlein Purpura: A Literature Review. Acta Derm Venereol 97, 1160–1166 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Jennette J.C. et al. 2012 revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum 65, 1–11 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Sunderkotter C.H. et al. Nomenclature of Cutaneous Vasculitis: Dermatologic Addendum to the 2012 Revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheumatol 70, 171–184 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Magistroni R., D’Agati V.D., Appel G.B. & Kiryluk K. New developments in the genetics, pathogenesis, and therapy of IgA nephropathy. Kidney Int 88, 974–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watts R.A. & Scott D.G. Epidemiology of the vasculitides. Semin Respir Crit Care Med 25, 455–64 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Mejias R. et al. Association of HLA-B*41:02 with Henoch-Schonlein Purpura (IgA Vasculitis) in Spanish individuals irrespective of the HLA-DRB1 status. Arthritis Res Ther 17, 102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Mejias R. et al. A Genome-Wide Association Study Suggests the HLA Class II Region As the Major Susceptibility Locus for IgA Vasculitis. Arthritis & Rheumatology 69(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ares O.M. et al. HLA-B and HLA-DRB1 associations with henoch-schonlein purpura. Immunology 137, 475–475 (2012). [Google Scholar]
- 9.Amoli M.M. et al. HLA-DRB1*01 association with Henoch-Schonlein purpura in patients from northwest Spain. Journal of Rheumatology 28, 1266–1270 (2001). [PubMed] [Google Scholar]
- 10.Chen J.C. et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell 159, 402–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paull E.O. et al. A modular master regulator landscape controls cancer transcriptional identity. Cell 184, 334–351 e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galinsky K.J. et al. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am J Hum Genet 98, 456–472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genomes Project C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamazon E.R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47, 1091–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandes N., Linial N. & Linial M. PWAS: proteome-wide association study-linking genes and phenotypes by functional variation in proteins. Genome Biol 21, 173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiryluk K. et al. Genome-wide association analyses define pathogenic signaling pathways and prioritize drug targets for IgA nephropathy. Nat Genet 55, 1091–1105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche P.A. & Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature 345, 615–8 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Ishina I.A. et al. MHC Class II Presentation in Autoimmunity. Cells 12(2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloan V.S. et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature 375, 802–6 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Kropshofer H. et al. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J 15, 6144–54 (1996). [PMC free article] [PubMed] [Google Scholar]
- 21.Denzin L.K., Fallas J.L., Prendes M. & Yi W. Right place, right time, right peptide: DO keeps DM focused. Immunol Rev 207, 279–92 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Jensen P.E. Antigen processing: HLA-DO--a hitchhiking inhibitor of HLA-DM. Curr Biol 8, R128–31 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Guce A.I. et al. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol 20, 90–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q. et al. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev 13, 786–91 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park R.K., Izadi K.D., Deo Y.M. & Durden D.L. Role of Src in the modulation of multiple adaptor proteins in FcalphaRI oxidant signaling. Blood 94, 2112–20 (1999). [PubMed] [Google Scholar]
- 26.Matsui T., Nunomura S., Shimokawa T., Yoshimaru T. & Ra C. Functionality of the IgA Fc receptor (FcalphaR, CD89) is down-regulated by extensive engagement of FcepsilonRI. Clin Immunol 129, 155–62 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Kerr W.G., Park M.Y., Maubert M. & Engelman R.W. SHIP deficiency causes Crohn’s disease-like ileitis. Gut 60, 177–88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park M.Y. et al. Impaired T-cell survival promotes mucosal inflammatory disease in SHIP1-deficient mice. Mucosal Immunol 7, 1429–1439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira M.A. et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet 49, 1752–1757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson A., Rask-Andersen M., Karlsson T. & Ek W.E. Genome-wide association analysis of 350 000 Caucasians from the UK Biobank identifies novel loci for asthma, hay fever and eczema. Hum Mol Genet 28, 4022–4041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M.H. et al. Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 182, 1198–1213 e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Astle W.J. et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429 e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vuckovic D. et al. The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell 182, 1214–1231 e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antiga E. et al. Dermatitis Herpetiformis: Novel Perspectives. Front Immunol 10, 1290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jostins L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fulco C.P. et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backenroth D. et al. FUN-LDA: A Latent Dirichlet Allocation Model for Predicting Tissue-Specific Functional Effects of Noncoding Variation: Methods and Applications. Am J Hum Genet 102, 920–942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calderon D. et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat Genet 51, 1494–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauluseviciute I. et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res 52, D174–D182 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulmert I., Henriques-Oliveira L., Pereira C.F. & Lahl K. Mononuclear phagocyte regulation by the transcription factor Blimp-1 in health and disease. Immunology 161, 303–313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S.J. Immunological function of Blimp-1 in dendritic cells and relevance to autoimmune diseases. Immunol Res 63, 113–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullberg J. et al. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol 152, 4958–68 (1994). [PubMed] [Google Scholar]
- 43.Rafiq S. et al. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun 8, 552–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galicia J.C. et al. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun 5, 513–6 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Flynn C.M. et al. Cathepsin S provokes interleukin-6 (IL-6) trans-signaling through cleavage of the IL-6 receptor in vitro. Sci Rep 10, 21612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riethmueller S. et al. Cleavage Site Localization Differentially Controls Interleukin-6 Receptor Proteolysis by ADAM10 and ADAM17. Sci Rep 6, 25550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newman A.M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37, 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffin A.T., Vlahos L.J., Chiuzan C. & Califano A. NaRnEA: An Information Theoretic Framework for Gene Set Analysis. Entropy (Basel) 25(2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L. et al. Genetic regulation of serum IgA levels and susceptibility to common immune, infectious, kidney, and cardio-metabolic traits. Nat Commun 13, 6859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giorgi F.M. et al. Correction: Inferring Protein Modulation from Gene Expression Data Using Conditional Mutual Information. PLoS One 11, e0163402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pillebout E. et al. Biomarkers of IgA vasculitis nephritis in children. PLoS One 12, e0188718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cambier A. et al. Soluble CD89 is a critical factor for mesangial proliferation in childhood IgA nephropathy. Kidney Int 101, 274–287 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Chou V. et al. INPP5D regulates inflammasome activation in human microglia. Nat Commun 14, 7552 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedicone C. et al. Pan-SHIP1/2 inhibitors promote microglia effector functions essential for CNS homeostasis. J Cell Sci 133(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamen L.A., Levinsohn J. & Swanson J.A. Differential association of phosphatidylinositol 3-kinase, SHIP-1, and PTEN with forming phagosomes. Mol Biol Cell 18, 2463–72 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riethmueller S. et al. Proteolytic Origin of the Soluble Human IL-6R In Vivo and a Decisive Role of N-Glycosylation. PLoS Biol 15, e2000080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goth C.K. et al. A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc Natl Acad Sci U S A 112, 14623–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schett G. Physiological effects of modulating the interleukin-6 axis. Rheumatology (Oxford) 57, ii43–ii50 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Garbers C. et al. The interleukin-6 receptor Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim Biophys Acta 1842, 1485–94 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Ferreira R.C. et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet 9, e1003444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manichaikul A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das S. et al. Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loh P.R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willer C.J., Li Y. & Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi J. & Lee S. A novel random effect model for GWAS meta-analysis and its application to trans-ethnic meta-analysis. Biometrics 72, 945–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J., Lee S.H., Goddard M.E. & Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gusev A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J. et al. Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat Genet 54, 593–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giambartolomei C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jia X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pillai N.E. et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum Mol Genet 23, 4443–51 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Ooi J.D. et al. Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature 545, 243–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emsley P., Lohkamp B., Scott W.G. & Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krieger E. et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77 Suppl 9, 114–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meng E.C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci 32, e4792 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wisniewska M. et al. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex. J Mol Biol 381, 1307–19 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Pos W. et al. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell 151, 1557–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denny J.C. et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aguet F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wright F.A. et al. Heritability and genomics of gene expression in peripheral blood. Nature Genetics 46, 430–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeLuca D.S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van der Wijst M.G.P. et al. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet 50, 493–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love M.I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson M.D., McCarthy D.J. & Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ongen H., Buil A., Brown A.A., Dermitzakis E.T. & Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stegle O., Parts L., Durbin R. & Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol 6, e1000770 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Consortium G.T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y.I. et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet 50, 151–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pillebout E. IgA Vasculitis and IgA Nephropathy: Same Disease? J Clin Med 10(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stern Y. et al. The Reference Ability Neural Network Study: motivation, design, and initial feasibility analyses. Neuroimage 103, 139–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Touil H. et al. Human central nervous system astrocytes support survival and activation of B cells: implications for MS pathogenesis. J Neuroinflammation 15, 114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Margolin A.A. et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 Suppl 1, S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giorgi F.M. et al. Inferring protein modulation from gene expression data using conditional mutual information. PLoS One 9, e109569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.