Abstract
Increasing evidence suggests that the generation of cytotoxic T-lymphocyte (CTL) responses specific for a diversity of viral epitopes will be needed for an effective human immunodeficiency virus type 1 (HIV-1) vaccine. Here, we determine the frequencies of CTL responses specific for the simian immunodeficiency virus Gag p11C and HIV-1 Env p41A epitopes in simian-human immunodeficiency virus (SHIV)-infected and vaccinated rhesus monkeys. The p11C-specific CTL response was high frequency and dominant and the p41A-specific CTL response was low frequency and subdominant in both SHIV-infected monkeys and in monkeys vaccinated with recombinant modified vaccinia virus Ankara vectors expressing these viral antigens. Interestingly, we found that plasmid DNA vaccination led to high-frequency CTL responses specific for both of these epitopes. These data demonstrate that plasmid DNA may be useful in eliciting a broad CTL response against multiple epitopes.
Of the many potential peptide epitopes generated by a virus replicating in a host cell, only a very limited number induce high-frequency immunodominant effector CD8+ cytotoxic T-lymphocyte (CTL) responses (32). A high-affinity interaction between the peptide and major histocompatability complex (MHC) class I molecule is a necessary but not sufficient condition for eliciting an immunodominant CTL response. In human immunodeficiency virus type 1 (HIV-1) infection of humans and simian immunodeficiency virus (SIV) infection of rhesus monkeys, the CTL responses are usually highly focused on a small number of viral peptide epitopes, as demonstrated by the oligoclonal Vβ repertoires of CD8+ T lymphocytes during primary infection (7, 15, 22).
Recent studies have demonstrated that virus-specific CD8+ CTLs are crucial for containing HIV-1 replication in humans and SIV replication in rhesus monkeys (5, 13, 17, 20, 21, 26). Strategies that can elicit HIV-1-specific CTL responses are therefore receiving considerable attention in the effort to develop an AIDS vaccine. However, recent vaccination studies of nonhuman primates suggest that single viral epitope-specific CTL responses may not be sufficient to block infection with pathogenic SIV (11, 31). These observations suggest that the generation of broad CTL responses specific for multiple viral epitopes may be critical for the development of an effective AIDS vaccine. It has been assumed, however, that achieving this goal might be difficult because of the natural bias in antiviral immune responses toward focusing CTL responses on a limited number of immunodominant epitopes (32). In this study, we demonstrate that vaccination with plasmid DNA, but not vaccination with a live recombinant vector or infection with simian-human immunodeficiency virus (SHIV), elicits potent CTL responses against both dominant and subdominant epitopes in rhesus monkeys. These findings suggest a potential advantage of DNA vaccination over other vaccination modalities in generating CTL responses against multiple epitopes.
SIV infection of rhesus monkeys expressing the MHC class I allele Mamu-A∗01 elicits a CTL response specific for the immunodominant SIV Gag p11C epitope (CTPYDINQM) (1, 19). It has also previously been shown that infection of Mamu-A∗01+ rhesus monkeys with SHIV-89.6 and SHIV-HXBc2 elicits an immunodominant CTL response specific for the p11C epitope as well as a weak CTL response specific for the subdominant HIV-1 Env p41A epitope (YAPPISGQI) (9). The use of fluorochrome-labeled tetrameric MHC class I-peptide complexes has allowed the quantitative analysis of these epitope-specific CD8+ CTL populations by flow cytometry (2, 16).
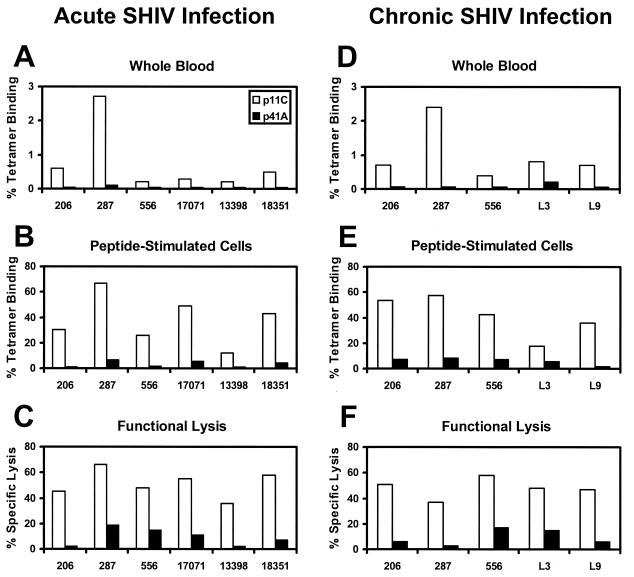
We first evaluated the relative frequencies of p11C- and p41A-specific CTLs during infection of eight Mamu-A∗01+ rhesus monkeys with SHIV-89.6, SHIV-89.6P, or SHIV-HXBc2 (23, 24). As shown in Fig. 1A to C, all SHIV-infected monkeys developed high-frequency CTL responses specific for the p11C epitope, as measured by previously described tetramer staining (16) and chromium release functional lysis (9) assays. One month after infection, tetramer staining of freshly obtained whole-blood specimens revealed that 0.2 to 2.7% of CD3+ CD8+ peripheral blood lymphocytes (PBL) specifically bound the Mamu-A∗01/p11C tetramer (Fig. 1A). In vitro stimulation of PBL with p11C expanded the tetramer-positive cell population to 12.1 to 66.7% of CD8+ T lymphocytes (Fig. 1B), and these cells exhibited potent functional p11C-specific CTL activity (Fig. 1C). In contrast, these same monkeys developed uniformly weak CTL responses specific for the p41A epitope. Staining with the Mamu-A∗01/p41A tetramer was barely detectable in both freshly obtained whole blood (0.0 to 0.1%; Fig. 1A) and p41A-stimulated PBL (0.7 to 6.6%; Fig. 1B). Specific functional lysis of p41A-pulsed target cells was also weak (Fig. 1C). Analysis of PBL from SHIV-infected monkeys 9 months into chronic infection, shown in Fig. 1D to F, demonstrated that the relative CTL epitope dominance of p11C and subdominance of p41A persisted over time. Furthermore, analysis of PBL from a subset of these SHIV-infected animals 2 years into chronic infection exhibited a similar epitope dominance hierarchy (data not shown).
FIG. 1.
Immunodominance of the Gag p11C CTL epitope and subdominance of the Env p41A CTL epitope in acute and chronic SHIV infection of rhesus monkeys. Monkeys were screened and selected for the presence of the Mamu-A∗01 MHC class I allele (1, 19). Animals were infected intravenously with SHIV-89.6 (Mm 206, 287, 556, and 17071), SHIV-89.6P (Mm 13398 and 18351), or SHIV-HXBc2 (Mm L3 and L9). CTL responses specific for the Mamu-A∗01-restricted SIV Gag p11C (CTPYDINQM) and HIV-1 Env p41A (YAPPISGQI) epitopes were measured (A to C) 1 month and (D to F) 9 months after infection. Epitope-specific CTL responses were quantitated by three independent methods. (A and D) Tetramer staining of freshly isolated PBL. PBL in whole blood were stained directly with fluorochrome-labeled Mamu-A∗01/p11C or Mamu-A∗01/p41A tetramers as previously described (16). The percent CD3+ CD8+ T cells that stain positively with each tetramer are shown. (B and E) Tetramer staining of PBL stimulated with peptide for 12 days. (C and F) Functional specific lysis. The cytotoxicity of peptide-stimulated PBL was measured using standard 51Cr-release assays at effector: target ratios of 5:1 as previously described (9). Results shown here are representative of assays performed at least three times.
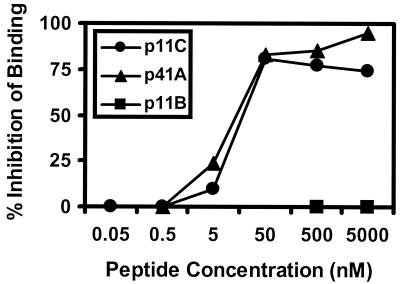
We next assessed the binding affinity of the p11C and p41A peptides for Mamu-A∗01, utilizing a quantitative in vitro binding assay (8). This assay utilized 2 × 106 C1R-Mamu-A∗01 transfectants to measure the inhibition of binding of 105 cpm of iodinated index p11C analog peptide (ATPYDINQM) by unlabeled test peptides for 4 h at 20°C in the presence of human β2m. As shown in Fig. 2, both p11C and p41A had equal, high binding affinities for Mamu-A∗01 (50% inhibitory concentration, 10 nM). Thus, the dominance of the p11C epitope and the subdominance of the p41A epitope did not simply reflect different MHC class I binding affinities. We therefore investigated the abilities of recombinant modified vaccinia virus Ankara (MVA) and plasmid DNA vaccination to elicit p11C- and p41A-specific CTL responses.
FIG. 2.
Binding affinities of p11C and p41A for Mamu-A∗01 are equivalent. Live-cell peptide binding assays were performed by measuring the inhibition of binding of the iodinated index p11C analog peptide (ATPYDINQM) (8). Varying concentrations of unlabeled p11C (●), p41A (▴), or control p11B (■) test peptides were utilized in this assay. The 50% inhibitory concentrations for both p11C and p41A were determined to be 10 nM in this assay. Results shown here are representative of assays performed three times.
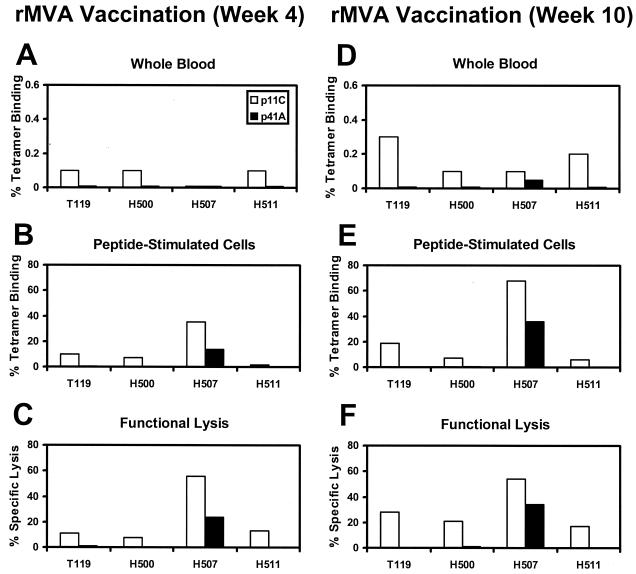
Recombinant MVA vectors expressing either SIV Gag-Pol or HIV-1 89.6 Env under control of the same early/late vaccinia virus promoter were constructed. Open reading frames of SIVmac239 Gag-Pol and HIV-1 89.6 Env were inserted adjacent to the modified H5 promoter in the previously described plasmid transfer vectors pLW-9 and pLW-17 (29, 30), and recombinant MVA vectors were produced by homologous recombination, identification by live immunostaining of infected cell foci, and clonal isolation. Efficient expression of both Gag-Pol and Env in cultured monkey cells was determined by radioimmunoprecipitation, and the production of Gag particles and surface expression and fusion competence of Env proteins were demonstrated (data not shown). Figure 3 shows the relative frequencies of p11C- and p41A-specific CTLs in four Mamu-A∗01+ rhesus monkeys vaccinated by separate injections with these vectors. The monkeys were immunized with 108 PFU of MVA-gag pol plus 108 PFU of MVA-env 89.6 by separate intramuscular (i.m.) injections. Four weeks after this immunization, p11C-specific CTLs were detected in peptide-stimulated PBL of all the monkeys by both tetramer staining (Fig. 3B) and peptide-specific functional lysis (Fig. 3C). In contrast, these same monkeys developed significantly weaker CTL responses specific for the p41A epitope, although one animal (H507) out of the four did generate a readily detectable p41A-specific response. The monkeys were boosted at week 4 with both recombinant MVA vectors, and their PBL were assessed again for CTLs at week 10. As shown in Fig. 3D to F, the relative epitope dominance persisted after the boosting immunization. Overall, these monkeys vaccinated with recombinant MVA vectors showed the same p11C epitope dominance and p41A epitope subdominance observed in SHIV infection.
FIG. 3.
Immunodominance of the Gag p11C CTL epitope and subdominance of the Env p41A CTL epitope elicited by recombinant MVA vaccination of rhesus monkeys. Four Mamu-A∗01+ rhesus monkeys were immunized by separate injections with MVA vectors expressing SIV Gag-Pol or HIV-1 89.6 Env. A total of 108 PFU of each vector was inoculated i.m. at separate sites, and the animals were boosted at 4 weeks. CTL responses to the SIV Gag p11C and HIV-1 Env p41A epitopes were measured 4 weeks (A to C) and 10 weeks (D to F) after primary immunization. Epitope-specific CTLs were quantitated as described in the legend to Fig. 1 by tetramer staining of PBL in freshly isolated whole blood (A and D), tetramer staining of peptide-stimulated PBL (B and E), and functional specific lysis by peptide-stimulated PBL (C and F). Results shown here are representative of assays performed at least three times.
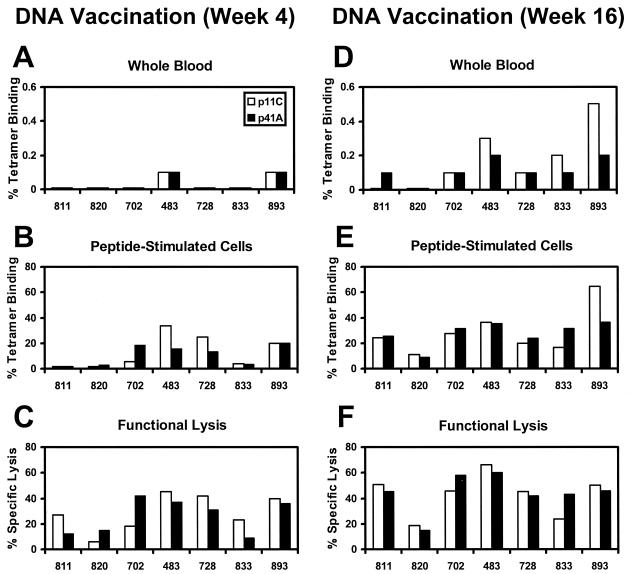
Plasmid DNA vaccines expressing either SIV Gag or HIV-1 89.6P Env in the pV1R backbone under control of the same cytomegalovirus promoter were then constructed (27). Equivalent expression of the Gag and Env proteins from these plasmids was observed in transiently transfected COS cells (data not shown). Seven Mamu-A∗01+ rhesus monkeys were immunized with 5 mg of gag DNA and 5 mg of env DNA by separate injections, and four of these monkeys also received 5 mg of an interleukin-2-immunoglobulin plasmid as an adjuvant (4). Four weeks after this immunization, comparable levels of p11C- and p41A-specific CTLs were detected in peptide-stimulated PBL of all the monkeys by both tetramer staining (Fig. 4B) and peptide-specific functional lysis (Fig. 4C). The monkeys were boosted at weeks 4 and 8 with both plasmid DNA vaccines, and their PBL were assessed again for CTLs at week 16. Following these boosting immunizations, codominance of both epitopes persisted in all of the monkeys. High frequencies of both p11C- and p41A-specific CTLs were observed by tetramer staining of freshly obtained PBL (Fig. 4D), tetramer staining of peptide-stimulated PBL (Fig. 4E), and peptide-specific functional lysis (Fig. 4F). These p41A-specific CTLs were able to lyse both peptide-pulsed target cells and virally infected target cells (data not shown). Two-tailed Mann-Whitney tests show that the DNA vaccine-elicited p41A-specific CTL responses, but not p11C-specific responses, were of significantly higher frequency than those elicited by SHIV infection (P = 0.0012 for tetramer staining; P = 0.0023 for functional lysis). Staining of these peptide-stimulated PBL using a control Mamu-A∗01/p68A tetramer consistently demonstrated a background level of staining of <0.1% of that of CD8+ T lymphocytes (data not shown).
FIG. 4.
Codominance of the Gag p11C and Env p41A CTL epitopes elicited by DNA vaccination of rhesus monkeys. Seven Mamu-A∗01+ rhesus monkeys were immunized by separate injections with two purified pV1R plasmids expressing either SIVmac239 Gag or HIV-1 89.6P Env. A total of 5 mg of each plasmid was inoculated i.m. at separate sites, and the animals were boosted at 4 and 8 weeks. Four monkeys also received a plasmid expressing an interleukin-2-immunoglobulin fusion protein (4). CTL responses to the SIV Gag p11C and HIV-1 Env p41A epitopes were measured 4 weeks (A to C) and 16 weeks (D to F) after primary immunization. Epitope-specific CTLs were quantitated as described in the legend to Fig. 1 by tetramer staining of PBL in freshly isolated whole blood (A and D), tetramer staining of peptide-stimulated PBL (B and E), and functional specific lysis by peptide-stimulated PBL (C and F). Tetramer staining of freshly isolated PBL and peptide-stimulated PBL utilizing a control Mamu-A∗01/Pol p68A (STPPLVRLV) tetramer was consistently <0.1% (data not shown). Results shown here are representative of assays performed at least three times.
These data demonstrate that the p41A-specific CTL responses that are of low frequency in the setting of SHIV infection and recombinant MVA vaccination are of high frequency when elicited by plasmid DNA vaccination. However, the exact mechanism underlying these differences remains unclear. The immunodominance of the p11C epitope does not simply reflect inadequate Env protein expression in SHIV-infected cells, since CTLs capable of lysing vaccinia-env-infected target cells are detectable in PBL of SHIV-infected rhesus monkeys (28). In addition, efficient expression of both Gag and Env was observed for both the MVA and DNA constructs. The dominance of the p11C epitope and the subdominance of the p41A epitope are also not explained by differences in MHC-peptide affinity. Moreover, the epitope hierarchy is not a viral isolate-specific phenomenon, since a similar epitope dominance pattern was observed in SHIV-IIIB-, SHIV-89.6-, and SHIV-89.6P-infected animals. It is possible that CTL epitope immunodominance may, in part, reflect the intracellular processing or presentation of these peptides. SHIV- and MVA-infected cells produce a number of viral proteins that might compete or interfere with the proteolysis, transport, or MHC class I binding of a particular epitope (3, 32). Virally infected cells may also have nonspecific effects on the generation of CTL epitopes. In contrast, plasmid DNA-transfected antigen-presenting cells express only the antigenic protein encoded by the vaccine, providing a much simpler system for the processing and presentation of particular peptide epitopes.
It has previously been shown that CTL responses against multiple epitopes can be elicited by a multiepitope DNA vaccine containing nine dominant epitopes (12). Other reports have shown that DNA vaccines can elicit CTL responses specific for dominant and subdominant epitopes in a pattern similar to that observed in viral infection (6, 10, 18). Our results extend these observations by demonstrating that vaccination with plasmid DNA can alter the natural epitope dominance pattern and can elicit high-frequency CTL responses to an epitope that is weak in the context of infection or vaccination with a live recombinant vector.
The generalizability of this study may be limited by the fact that it examines only two epitopes and compares inherently different expression systems and immunization methods. However, the results suggest that DNA vaccines may offer a potential advantage over recombinant live vector vaccines in their ability to elicit high-frequency CTL responses to certain epitopes that are subdominant in the context of infection. DNA vaccines may therefore have an important benefit in diversifying and broadening the CTL response. We have recently described the efficacy of these DNA vaccine-elicited multiepitope CTL responses in controlling viremia and preventing disease progression after a highly pathogenic viral challenge (5). In the development of candidate HIV-1 vaccines, there is growing interest in strategies that utilize DNA to prime CTL responses (11, 14, 25). The present study provides a rationale for the use of plasmid DNA as a method of priming a broad CTL response against multiple viral epitopes.
Acknowledgments
We acknowledge support from grants NO1-AI-65301 (M.G.L.), AI-85343 (N.L.L.), and CA-50139 (N.L.L.).
We are grateful to Nancy Miller, Carroll Crabbs, Tavis Steenbeke, Suzanne Robinson, Meryl Forman, and Frederick Vogel for generous advice, assistance, and reagents.
REFERENCES
- 1.Allen T M, Sidney J, del Guercio M-F, Glickman R L, Lensmeyer G L, Wiebe D A, DeMars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A∗01) that binds an immunodominant CTL epitope from SIV. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 2.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 3.Anton L C, Yewdell J W, Bennink J R. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J Immunol. 1997;158:2535–2542. [PubMed] [Google Scholar]
- 4.Barouch D H, Craiu A, Kuroda M J, Schmitz J E, Zheng X X, Santra S, Frost J D, Krivulka G R, Lifton M A, Crabbs C L, Heidecker G, Perry H C, Davies M-E, Xie H, Nickerson C E, Steenbeke T D, Lord C I, Montefiori D C, Strom T B, Shiver J W, Lewis M G, Letvin N L. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc Natl Acad Sci USA. 2000;97:4192–4197. doi: 10.1073/pnas.050417697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch D H, Santra S, Schmitz J E, Kuroda M J, Fu T-M, Wagner W, Bilska M, Craiu A, Zheng X X, Krivulka G R, Beaudry K, Lifton M A, Nickerson C E, Trigona W L, Punt K, Freed D C, Guan L, Dubey S, Casimiro D, Simon A, Davies M-E, Chastain M, Strom T B, Gelman R S, Montefiori D C, Lewis M G, Emini E A, Shiver J W, Letvin N L. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Webster R B, Woodland D L. Induction of CD8+ T cell responses to dominant and subdominant epitopes and protective immunity to Sendai virus infection by DNA vaccination. J Immunol. 1998;160:2425–2432. [PubMed] [Google Scholar]
- 7.Chen Z W, Kou Z C, Lekutis C, Shen L, Zhou D, Halloran M, Li J, Sodroski J, Lee-Parritz D, Letvin N L. T cell receptor Vβ repertoire in an acute infection of rhesus monkeys with simian immunodeficiency viruses and a chimeric simian-human immunodeficiency virus. J Exp Med. 1995;182:21–31. doi: 10.1084/jem.182.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Guercio M F, Sidney J, Hermanson G, Perez C, Grey H M, Kubo R T, Sette A. Binding of a peptide antigen to multiple HLA alleles allows definition of an A2-like supertype. J Immunol. 1995;154:685–693. [PubMed] [Google Scholar]
- 9.Egan M A, Kuroda M J, Voss G, Schmitz J E, Charini W A, Lord C I, Forman M A, Letvin N L. Use of major histocompatibility complex class I/peptide/β2M tetramers to quantitate CD8+ cytotoxic T lympocytes specific for dominant and nondominant viral epitopes in simian-human immunodeficiency virus-infected rhesus monkeys. J Virol. 1999;73:5466–5472. doi: 10.1128/jvi.73.7.5466-5472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu T-M, Friedman A, Ulmer J B, Liu M A, Donnelly J J. Protective cellular immunity: cytotoxic T-lymphocyte responses against dominant and recessive epitopes of influenza virus nucleoprotein induced by DNA immunization. J Virol. 1997;71:2715–2721. doi: 10.1128/jvi.71.4.2715-2721.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanke T, Samuel R V, Blanchard T J, Neumann V C, Allen T M, Boyson J E, Sharpe S A, Cook N, Smith G L, Watkins D I, Cranage M P, McMichael A J. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J Virol. 1999;73:7524–7532. doi: 10.1128/jvi.73.9.7524-7532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishioka G Y, Fikes J, Hermanson G, Livingston B, Crimi C, Qin M, del Guercio M F, Oseroff C, Dahlberg C, Alexander J, Chesnut R W, Sette A. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J Immunol. 1999;162:3915–3925. [PubMed] [Google Scholar]
- 13.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent S J, Zhao A, Best S J, Chandler J D, Boyle D B, Ramshaw I A. Enhanced T-cell immunogenicity and protective efficacy of a human immunodeficiency virus type 1 vaccine regimen consisting of consecutive priming with DNA and boosting with recombinant fowlpox virus. J Virol. 1998;72:10180–10188. doi: 10.1128/jvi.72.12.10180-10188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda M J, Schmitz J E, Barouch D H, Criau A, Allen T M, Sette A, Watkins D I, Forman M A, Letvin N L. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMichael A. T cell responses and viral escape. Cell. 1998;93:673–676. doi: 10.1016/s0092-8674(00)81428-2. [DOI] [PubMed] [Google Scholar]
- 18.Milan G, Zambon A, Cavinato M, Zanovello P, Rosato A, Collavo D. Dissecting the immune response to Moloney murine sarcoma/leukemia virus-induced tumors by means of a DNA vaccination approach. J Virol. 1999;73:2280–2287. doi: 10.1128/jvi.73.3.2280-2287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller M D, Yamamoto H, Hughes A H, Watkins D I, Letvin N L. Definition of an epitope and MHC class I molecule recognized by Gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol. 1991;147:320–329. [PubMed] [Google Scholar]
- 20.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath M J. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. New Engl J Med. 1997;337:1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 21.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 22.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P, Fauci A S. Major expansion of CD8+ T cells with a predominant Vβ usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 23.Reimann K A, Li J T, Veazey R, Halloran M, Park I-W, Karlsson G B, Sodroski J, Letvin N L. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reimann K A, Li J T, Voss G, Lekutis C, Tenner-Racz K, Racz P, Lin W, Montefiori D C, Lee-Parritz D E, Lu Y, Collman R G, Sodroski J, Letvin N L. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson H L, Montefiori D C, Johnson R P, Manson K H, Kalish M L, Lifson J D, Rizvi T A, Lu S, Hu S-L, Mazzara G P, Panicali D L, Herndon J G, Glickman R, Candido M A, Lydy S L, Wyand M S, McClure H M. Neutralizing antibody-independent containment of immunodeficiency virus challenges by DNA priming and recombinant pox virus booster immunizations. Nat Med. 1999;5:526–534. doi: 10.1038/8406. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 27.Shiver J W, Davies M-E, Perry H C, Freed D C, Liu M A. Humoral and cellular immunities elicited by HIV-1 DNA vaccination. J Pharm Sci. 1996;85:1317–1323. doi: 10.1021/js9600991. [DOI] [PubMed] [Google Scholar]
- 28.Voss G, Li J, Manson K, Wyand M, Sodroski J, Letvin N L. Human immunodeficiency virus type 1 envelope glycoprotein-specific cytotoxic T lymphocytes in simian-human immunodeficiency virus-infected rhesus monkeys. Virology. 1995;208:770–775. doi: 10.1006/viro.1995.1209. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt L S, Shors S T, Murphy B R, Moss B. Development of a replication-deficient recombinant vaccinia virus vaccine effective against parainfluenza virus 3 infection in an animal model. Vaccine. 1996;14:1451–1458. doi: 10.1016/s0264-410x(96)00072-2. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt L S, Whitehead S S, Venanzi K A, Murphy B R, Moss B. Priming and boosting immunity to respiratory syncytial virus by recombinant replication-defective vaccinia virus MVA. Vaccine. 1999;18:392–397. doi: 10.1016/s0264-410x(99)00257-1. [DOI] [PubMed] [Google Scholar]
- 31.Yasutomi Y, Koenig S, Woods R M, Madsen J, Wassef N M, Alving C R, Klein H J, Nolan T E, Boots L J, Kessler J A, Emini E A, Conley A J, Letvin N L. A vaccine-elicited, single viral epitope-specific cytotoxic T lymphocyte response does not protect against intravenous, cell-free simian immunodeficiency virus challenge. J Virol. 1995;69:2279–2284. doi: 10.1128/jvi.69.4.2279-2284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yewdell J W, Bennick J R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]