Abstract
To determine if mutations of an immunodominant HLA-restricted cytomegalovirus (CMV) peptide sequence occur in nature, the sequence corresponding to the HLA A∗0201-specific peptide CMVpp65495–503 was determined in 50 human CMV isolates. Rare mutations were detected; 6 of 50 were silent mutations at the amino terminus of the peptide, while 3 of 50 were mutations of the native methionine residue to isoleucine (M499I). The observed M499I mutation in three isolates decreased cytolytic targeting.
CMVpp65, a tegument protein of human cytomegalovirus (CMV), is the main viral antigen found in peripheral blood mononuclear cells after viral infection. More recent investigations have shown it to be an immunodominant target of the cellular immune system (1, 10, 12, 15). CMVpp65 is introduced into the cell during CMV infection and subsequently traffics to the nucleus, due to its nuclear localization signals (5, 13). Various resultant peptides which are presumably generated via proteasomal antigen processing serve as recognition targets in the context of HLA class I molecules, and the surface-displayed peptide complex serves as a target of cytotoxic T-lymphocyte (CTL)-mediated lysis (6).
The CMVpp65495–503 peptide NLVPMVATV is a prevalent CTL target in the context of HLA A∗0201 (2, 14, 15). It is known that most high-affinity HLA-A∗0201 binding peptides are between 9 and 11 amino acid residues long and are characterized by invariant residues (anchors) at positions 2 (leucine) and 9 (valine or leucine) (3, 7, 8). These residues contact the major histocompatibility complex (MHC) molecule in the X and F pockets of the peptide-binding groove, and their sequence integrity is mandatory for efficient MHC binding (11). Whether an HLA A∗0201-binding peptide could tolerate amino acid substitutions at positions other than anchors and still be competent for MHC binding is a concern for peptide-based vaccine development. The validity of using a peptide vaccine against a viral protein sequence relies on its sequence stability among horizontally detected human CMV isolates. Selection pressure exerted by drug selection or by immune response mechanisms has been shown to cause sequence variability and immune escape for both viral proteins and tumor antigens. Although CMVpp65 is conserved in the laboratory isolates that have been sequenced, little information is available from natural isolates. In addition, it would be of interest to determine whether, in spite of a mutation, the presentation of the peptide-MHC complex could still be recognized by a CTL clone specific to the native CMVpp65495–503 peptide.
CMV isolates (n = 50) were obtained from a bone marrow transplant (BMT) population at the City of Hope National Medical Center by using specimens that include mouthwash, blood culture, bronchoalveolar lavage (BAL) isolates, or plasma (Table 1). The CMV isolates were obtained from healthy BMT subjects at approximately day 35 post-BMT as part of preemptive therapy surveillance (17). The isolates were obtained using shell vial culture, were grown on fibroblasts (MRC-5 cells), and were then passed twice to ensure stable growth of the isolate before being frozen in liquid nitrogen prior to PCR. In addition, plasma from BMT subjects with known positivity for CMV DNA was used for sequence analysis. For all samples, the virus sequence was obtained from PCR performed as previously described (4, 16). In brief, 100 μl of cell lysate or plasma was digested with proteinase K, denatured for 10 min at 94°C, and spun briefly to remove precipitate. Then 10 μl was used in a PCR. The primers and the position on the CMVpp65 gene were the following: MP1 (1242 to 1262), 5′ CTCGTAACCACCGAGCGCAAG 3′; and AP4 (1677 to 1700), 5′ TCAACCTCGGTGCTTTTTGGGCG 3′. The amplification product was 458 bp.
TABLE 1.
Sequencing of the CMVpp65495–503 coding sequence in patient isolates
| Specimen ID (source) | HLA type | CMVpp65 epitope mutation |
|---|---|---|
| 405AE (plasma) | A∗0201 | None |
| 802R (plasma) | A∗0201 | None |
| 831AE (plasma) | A∗0201 | None |
| 288AD (plasma) | A∗0201 | None |
| 370AB (plasma) | A∗0201 | None |
| 643AD (plasma) | A∗0201 | None |
| 953AC (plasma) | A∗0201 | None |
| 703AB (plasma) | A∗0201 | None |
| 319AE (plasma) | A∗0201 | None |
| 188AC (plasma) | A∗0201 | None |
| 851AA (plasma) | A∗0201 | None |
| 818AC (plasma) | A∗0201 | AAC to AAT (silent) |
| 599AA (plasma) | A∗0201 | AAC to AAT (silent) |
| 552AD (plasma) | A∗0201 | AAC to AAT (silent) |
| 626U (blood culture) | A∗0201 | None |
| 481W (blood culture) | A∗0201 | None |
| 81Q (blood culture) | A∗0201 | AAC to AAT (silent) |
| 56T (mouthwash) | A∗0201 | None |
| 174R (mouthwash) | A∗0201 | AAC to AAT (silent) |
| 374AC (BAL) | A∗0201 | None |
| 847X (BAL) | A∗0201 | None |
| 924Z (BAL) | A∗0201 | None |
| 584W (BAL) | A∗0201 | None |
| 854W (BAL) | A∗0201 | None |
| 637W (BAL) | A∗0201 | None |
| 954W (BAL) | A∗0201 | None |
| 425T (BAL) | A∗0201 | None |
| 764T (BAL) | A∗0201 | None |
| 355Q (BAL) | A∗0201 | None |
| 108Q (BAL) | A∗0201 | None |
| 634S (BAL) | A∗0201 | None |
| 694V (BAL) | A∗0201 | ATG to ATA (Met to Ile) |
| 559AE (plasma) | A29 | None |
| 669AE (plasma) | A11 | None |
| 281R (blood culture) | A3 | None |
| 664R (blood culture) | A24 | None |
| 278R (blood culture) | A11 | ATG to ATA (Met to Ile) |
| 805R (blood culture) | A26 | None |
| 115V (mouthwash) | A24 | None |
| 740W (BAL) | A1 | None |
| 741W (BAL) | A31 | None |
| 853W (BAL) | A11 | None |
| 499AB (BAL) | A11 | None |
| 956R (BAL) | A11 | None |
| 517R (BAL) | A26 | None |
| 519 (BAL) | A1 | None |
| 630P (BAL) | A24 | None |
| 989P (BAL) | A24 | None |
| 875Z (BAL) | A32 | AAC to AAT (silent) |
| 317W (BAL) | A11 | ATG to ATA (Met to Ile) |
For plasma specimens, an additional nested PCR was used with the following secondary primers: RAP1 (1434 to 1445), 5′ GGATTCCGACAACGAAATCCAC 3′; and AP5 (1584 to 1605), 5′ ATACGCTTCCAATTCGGCGAA 3′. The amplification product was 171 bp.
The sequencing of CMVpp65465–503 was performed on amplification products that were gel purified using the Qiaquick gel extraction kit (Qiagen, Valencia, Calif.). The sequencing was carried out on an ABI Prism 377 Fluorescent DNA sequencer using the Big Dye Terminator Cycle Sequencing kit supplied by PE Applied Biosystems (Foster City, Calif.).
As shown in Table 1, 50 samples were sequenced through the site of the CMVpp65495–503 peptide, 32 of which were from HLA A∗0201 BMT recipients. In the CMV sequence from the HLA A∗0201-defined population, the CMVpp65495–503 peptide NLVPMVATV was silently mutated (AAC to AAT) in the amino-terminal position (P1, asparagine) in 5 of 32 specimens (16%). Only 1 of 32 mutations (3%) resulted in an amino acid change; this was at the P5 site with methionine changed to isoleucine (M499I; ATG to ATA). Of 18 CMV isolates occurring in a population with a wide range of HLA types other than HLA A∗0201, no new types of mutations other than the ones previously described for healthy CMV-seropositive subjects were detected; one was a silent mutation, while two exhibited the P5 mutation M499I. Thus, an amino acid mutation occurred in only 3 of 50 isolates and was not preferentially associated with the HLA A∗0201 allele. Finally, we observed that the sequenced region of these CMV isolates overlapped with the B∗0702-specific CTL epitope (TPRVTGGGAM) and with the B∗4402 epitope (EFFWDANDIY). No significant mutation frequency was observed, and those mutations, observed in known laboratory strains as well, were mainly silent (data not shown).
We evaluated whether natural CMV isolates that contained the native or mutated form of the CMVpp65495–503 sequence equally triggered recognition by clonal CTL. Viral isolates were propagated in MRC-5 cells, which naturally express the HLA A∗0201 allele, and were used as targets in a chromium release assay (CRA) using a CTL clone (3-3F4) derived from an HLA A∗0201 donor (2). The CMV isolates were passaged at least three times as infected cells into a fresh monolayer of fibroblast, to obtain enough target cells to do the analysis. Table 2 shows the results of a CRA at an effector/target ratio of 30, using 25 different CMV isolates, among which 6 had a base pair mutation. The percent lysis of infected cells with viral isolates containing the native CMVpp65495–503 sequence ranged from 7.40 to 25.70%. Similarly, for the isolates with a silent mutation (P1, asparagine), the range of lysis was 11.20 to 24.30%. In contrast, the group bearing the M499I mutation ranged only from 5.7 to 6.5% lysis, which was significantly lower than that found for the other two groups when analyzed with the nonparametric Mann-Whitney U test (P < 0.0015). The assays were controlled with targets including MRC-5 cells infected or not infected with the CMV Toledo strain at a multiplicity of infection of 5 (positive control), a lymphoblast line (Epstein-Barr virus-transformed HLA A∗0201 B lymphocytes) pulsed with the CMVpp65495–503 peptide (positive control) or with no peptide (negative control), and CMV Toledo-infected fibroblasts of mismatched HLA type (negative control). The results indicate that the M499I mutation does result in reduced lysis, suggesting that this mutation could affect CTL recognition of CMV strains with the altered sequence.
TABLE 2.
Percent lysis in CMV isolate-infected MRC-5 target cells
| Virus (source) or control | pp65495–503 mutation (type) | % Lysis (E/T = 30)a |
|---|---|---|
| Viruses | ||
| 584W (BAL) | None | 7.4 |
| 637W (BAL) | None | 10.7 |
| 854W (BAL) | None | 10.8 |
| 924Z (BAL) | None | 11.1 |
| 954W (BAL) | None | 12.6 |
| 499AB (BAL) | None | 13.0 |
| 805R (blood culture) | None | 13.0 |
| 355Q (BAL) | None | 16.3 |
| 853W (BAL) | None | 16.4 |
| 56T (mouthwash) | None | 18.4 |
| 374AC (BAL) | None | 18.5 |
| 481W (blood culture) | None | 19.3 |
| 740W (BAL) | None | 20.0 |
| 847X (BAL) | None | 20.7 |
| 741W (BAL) | None | 21.1 |
| 956R (BAL) | None | 22.0 |
| 281R (blood culture) | None | 23.0 |
| 115V (mouthwash) | None | 24.7 |
| 630P (BAL) | None | 25.7 |
| 81Q (blood culture) | AAC to AAT (silent) | 11.2 |
| 174R (mouthwash) | AAC to AAT (silent) | 16.9 |
| 875Z (BAL) | AAC to AAT (silent) | 24.3 |
| 317W (BAL) | ATG to ATA (Met to Ile) | 5.7 |
| 694V (BAL) | ATG to ATA (Met to Ile) | 5.8 |
| 278 (blood culture) | ATG to ATA (Met to Ile) | 6.5 |
| Controls | ||
| MRC-5 (A∗0201) cells | 2.9 | |
| MRC-5 (Toledo) cells | 57.2 | |
| LCL-A2 (A∗0201) cells | 4.3 | |
| LCL-A2 (pp65495–503) cells | 82.7 | |
| Fibroblasts (A11) | 1.0 | |
| Fibroblasts (A11 Toledo) | 3.4 |
E/T, effector/target ratio.
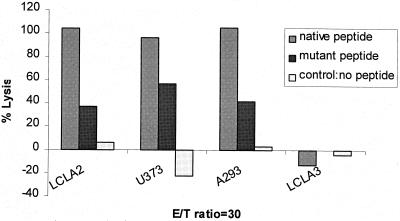
To test whether the difference among isolates for target specificity was determined by peptide sequence versus other factors such as slower growth of the viral isolate in culture, we compared the recognition of a synthetic native peptide (NLVPMVATV) to the M499I peptide (NLVPIVATV) using a CMVpp65-specific CRA. The synthetic peptides were used at 100 μM to load the HLA A∗0201-presenting cell lines LCL-A2, U373MG (glioblastoma-derived cell line), and A293 (human kidney cell line) as described previously (2). LCL-A3 cells were used as an HLA-mismatched control (a gift from S. Chatterjee and J. Sun). As shown in Fig. 1, the native peptide was always superior to the mutant peptide for sensitizing target cells for lysis. This difference in lysis was seen in all three cell lines used as target cells.
FIG. 1.
The CRA result, using an effector/target ratio of 30, is shown with various target cells presenting the HLA A∗0201 allele, U373, A293, LCL-A2, and a control cell (LCL-A3). The target cells were loaded with 100 μM synthetic native peptide (NLVPMVATV) or synthetic mutant peptide (NLVPIVATV). The CTL clone is specific for CMVpp65495–503 (3-3F4) and was incubated for 4 h with peptide-sensitized cells as previously described (2).
In summary, a mutation at amino acid 499 (M→I) of CMVpp65, even though it may reduce the ability of the viral isolate to be recognized, is not found preferentially in HLA A∗0201-bearing patients. Interestingly, the P5 methionine has been shown to be critical for T-cell recognition using this particular clone (9). P5 is intolerant to most substitutions, and based on these data (9), it is unlikely that an epitope containing the M499I mutant could induce CTL function. However, we cannot rule out the possibility that the CTLs generated by mutant isolates were not more efficient at targeting other peptides of the virus. The results of this study imply either that the specific CTL response to this virus is incapable of inducing a strong selective pressure for a single mutation or that the other allele-specific CTL responses to CMVpp65 or to other proteins make such selection unlikely. It is possible that a peptide-based vaccine strategy for CMV is not likely to be subverted by such induced mutations. Only testing of such a vaccine will determine whether vaccine-induced CTL function could result in selective pressure for induction of virus mutants.
Acknowledgments
This work was supported by U.S. Public Health Service grants PO1-CA30206, 1RO1-CA77544, 1RO1-AI43267, R21-AI44313, and 1PO1-CA30206 from the National Institutes of Health and grant 6616-98 from the Leukemia Society of America (D.J.D.); partial support was from NIH grants P30-CA33572 and NSF-BIR9602945 for the City of Hope DNA sequencing shared resource laboratory.
REFERENCES
- 1.Boppana S B, Britt W J. Recognition of human cytomegalovirus gene products by HCMV-specific cytotoxic T cells. Virology. 1996;222:293–296. doi: 10.1006/viro.1996.0424. [DOI] [PubMed] [Google Scholar]
- 2.Diamond D J, York J, Sun J, Wright C L, Forman S J. Development of a candidate HLA A∗0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood. 1997;90:1751–1767. [PubMed] [Google Scholar]
- 3.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee H G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 4.Gallez-Hawkins G M, Tegtmeier B R, ter Veer A, Niland J C, Forman S J, Zaia J A. Evaluation of a quantitative plasma PCR plate assay for detecting cytomegalovirus infection in marrow transplant recipients. J Clin Microbiol. 1997;35:788–790. doi: 10.1128/jcm.35.3.788-790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallina A E, Percivalle E, Simoncini L, Revello M G, Gerna G, Milanesi G. Human cytomegalovirus pp65 lower matrix phosphoprotein harbours two transplantable nuclear localization signals. J Gen Virol. 1996;77:1151–1157. doi: 10.1099/0022-1317-77-6-1151. [DOI] [PubMed] [Google Scholar]
- 6.Gyulai Z, Endresz V, Burian K, Pincus S, Toldy J, Cox W I, Meri C, Plotkin S, Berencsi K. Cytotoxic T lymphocyte (CTL) responses to human cytomegalovirus pp65, IE1-Exon4, gB, pp150, and pp28 in healthy individuals: reevaluation of prevalence of IE1-specific CTLs. J Infect Dis. 2000;181:1537–1546. doi: 10.1086/315445. [DOI] [PubMed] [Google Scholar]
- 7.Hunt D F, Henderson R A, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox A L, Appella E, Engelhard V H. Characterization of peptides bound to the class I MHC molecule HLA- A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 8.Jardetzky T S, Lane S W, Robinson R A, Madden D R, Wiley D C. Identification of self peptides bound to purified HLA-B27. Nature. 1991;353:326–329. doi: 10.1038/353326a0. [DOI] [PubMed] [Google Scholar]
- 9.La Rosa, C., R. Krishnan, S. Markel, J. P. Schneck, R. Houghten, C. Pinilla, and D. J. Diamond. Enhanced immune activity of CTL epitope analogues derived from positional scanning synthetic combinatorial libraries. Blood, in press. [DOI] [PubMed]
- 10.McLaughlin-Taylor E, Pande H, Forman S J, Tanamachi B, Li C R, Zaia J A, Greenberg P D, Riddell S R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 11.Parker K C, Bednarek M A, Hull L K, Utz U, Cunningham B, Zweerink H J, Biddison W E, Coligan J E. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 12.Riddell S R, Rabin M, Geballe A P, Britt W J, Greenberg P D. Class I MHC-restricted cytotoxic T-lymphocyte recognition of cells infected with human cytomegalovirus does not require endogenous viral gene expression. J Immunol. 1991;146:2795–2804. [PubMed] [Google Scholar]
- 13.Schmolke S, Drescher P, Jahn G, Plachter B. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J Virol. 1995;69:1071–1078. doi: 10.1128/jvi.69.2.1071-1078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solache A, Morgan C L, Dodi A J, Morte C, Scott I, Baboonian C, Zal B, Goldman J, Grundy J E, Madrigal J A. Identification of three HLA-A∗0201-restricted cytotoxic T cell epitopes in the cytomegalovirus protein pp65 that are conserved between eight strains of the virus. J Immunol. 1999;163:5512–5518. [PubMed] [Google Scholar]
- 15.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Plachter B, Sissons J G. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaia J A, Gallez-Hawkins G, Churchill M A, Morton-Blackshere A, Pande H, Adler S P, Schmidt G M, Forman S J. Comparative analysis of human cytomegalovirus a-sequence in multiple clinical isolates using polymerase chain reaction and restriction fragment length polymorphism assays. J Clin Microbiol. 1990;28:2602–2607. doi: 10.1128/jcm.28.12.2602-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaia J A, Schmidt G M, Chao N J, Rizk N W, Nademanee A P, Niland J C, Horak D A, Lee J, Gallez-Hawkins G, Kusnierz-Glaz C R, et al. Preemptive ganciclovir based solely on asymptomatic pulmonary cytomegalovirus infection in allogeneic bone marrow transplant recipients. Biol Blood Marrow Transplant. 1995;1:88–93. [PubMed] [Google Scholar]