Abstract
Previous genetic and biochemical analyses have indicated that the Epstein-Barr virus EBNA-2 amino terminus is important for primary B-lymphocyte growth transformation and may be involved in self-association. We now report that EBNA-2 has at least two domains, amino acids 1 to 60 and 96 to 210, which independently mediate homotypic association, 1 to 60 with 1 to 60 and 96 to 210 with 96 to 210. EBNA-2 self-association is likely to be critical to the ability of EBNA-2 to interact simultaneously with multiple cellular transcription factors, coactivators, and histone acetyltransferases through its RBPJκ binding and acidic activating domains.
Epstein-Barr virus (EBV) infection of B lymphocytes usually results in limited virus gene expression and virus-induced lymphocyte proliferation (for a review, see reference 20). EBV nuclear proteins EBNA-LP and EBNA-2 are the first proteins expressed (for a review, see reference 16). EBNA-LP and EBNA-2 up-regulate transcription of cell and viral genes and are required for lymphocyte immortalization. EBNA-2 stably associates with a cellular sequence-specific DNA binding protein, RBPJκ, and stimulates transcription from promoters with RBPJκ binding sites (8, 11, 30). The transcriptional effects of EBNA-2 are mediated by an acidic transactivating domain that interacts with TFIIB, TFIIH, TBP, p300, CBP, and a novel nuclear protein, p100, that is a scaffolding protein for c-myb and pim-1 (6, 18, 23–25, 27). EBNA-LP specifically coactivates through the EBNA-2 acidic domain and can coactivate a minimal promoter that has multiple upstream Gal4 DNA binding sites when a fusion of the EBNA-2 acidic domain with the Gal4 DNA binding domain is expressed in the same cell (9, 19).
Most recombinant EBV reverse genetic data are consistent with a model in which the critical EBNA-2 domains mediate association with RBPJκ at specific promoters and the recruitment of transcription factors to those promoters (Fig. 1) (3, 5, 10, 28, 29). Deletion or mutation of DNA encoding these domains results in an EBV that is unable to activate transcription or transform primary B lymphocytes into lymphoblastoid cell lines (LCLs). Deletions of most other parts of the EBNA-2 open reading frame have only small or moderate effects on EBNA-2-mediated transcriptional activation and on primary B-lymphocyte transformation. One anomalous result is that EBV recombinants with a deletion of the entire polyproline domain cannot transform primary B lymphocytes, although EBV recombinants with a deletion of the amino terminus through most of the polyproline domain or of the unique sequence carboxyl terminal to the polyproline domain have substantial transforming activity. These latter data are consistent with the possibility that the EBNA-2 amino terminus may have a critical role in transformation that is provided by the domain amino terminal to the polyproline domain, the polyproline domain, and the domain carboxyl terminal to the polyproline domain.
FIG. 1.
Two separate domains in the EBNA-2 amino terminus mediate homotypic association. (A) Schematic of previously identified EBNA-2 domains and the deletion mutations used in this study. EBNA-2 of the W91 EBV strain (5) is comprised from amino to carboxyl terminus of a 58-unique-amino-acid sequence, 37 amino acids that are mostly prolines, a 185-unique-amino-acid sequence, a 56-amino-acid sequence that mediates interactions with RBPJκ and PU.1, a 25-amino-acid RG domain, 61 unique amino acids of uncertain function, a 40-amino-acid acidic transcriptional activating domain, and a 21-amino-acid carboxyl terminus that includes a nuclear localization sequence. F194 is EBNA-2 amino acids 1 to 194, followed by an in-frame Flag epitope and a nonsense codon. F194 d59-93 is F194 with a deletion of all but two amino acids of the polyproline domain. F194 d97-122, F194 d2-95, and F194 d2-88 are F194 with deletions of amino acids 97 to 122, 2 to 95, and 2 to 88, respectively. (B and C) Experiments were conducted to investigate whether EBNA-2 associates with EBNA-2 residues 1 to 194 in B lymphoblasts. (B) BJAB cells were transfected with pSG5 (21), which expresses wild-type EBNA-2 (pSGWE2), and pSG5 that expresses F194 (pSGF194). (C) BJAB cells that had been converted to stable F194 expression cells were transfected with pSGWE2 or with pSG5 expressing the indicated EBNA-2 deletion derivatives. After 24 h, cells were lysed in nonionic detergent, the nuclear and cytoskeletal residue was removed by centrifugation, and the lysate was split for immunoprecipitation either with PE2 antibody to an epitope in the EBNA-2 acidic activating domain, followed by protein G-Sepharose (PE2ip), or with M2 monoclonal antibody-conjugated Sepharose 4B (M2ip). Immunoprecipitates were rendered soluble in SDS sample buffer, run on denaturing polyacrylamide gels, and immunoblotted with PE2 or M2 monoclonal antibody. Immunoprecipitates were visualized by secondary antibody and enhanced chemiluminescence. The estimated sizes of standard protein markers are indicated.
Several lines of evidence are consistent with the possibility that the EBNA-2 amino terminus may be involved in self-association and that self-association may be important for transcriptional activation and primary B-lymphocyte transformation. First, purified recombinant EBNA-2 is a salt-stable oligomer of about 440 kDa, and EBNA-2 from EBV-infected cells sediments in sucrose gradients at 13S and 34S, indicative of complexes that are much larger than monomeric EBNA-2 (7, 26). Second, EBNA-2 residues 122 to 344 can interact with residues 1 to 428 in yeast two-hybrid tests (26). Third, EBNA-2 interacts with several sites at all EBNA-2-responsive promoters and mediates transcriptional activation by recruiting multiple factors through its acidic transactivating domain. EBNA-2-responsive promoters have one or more RBPJκ sites, and at least one other sequence-specific DNA binding protein is required for EBNA-2 responsiveness. For example, the LMP1 promoter has two RBPJκ sites and also requires EBNA-2 interaction with Pu.1/Spi1 at a specific site, while the Cp promoter has one RBPJκ site and also requires CBF2 (13, 14, 17). Thus, there appears to be a requirement for several EBNA-2 molecules to be in close proximity to enable interactions among transcription factors that are necessary for appropriate promoter regulation. Fourth, the activity of the transcription factors, coactivators, and histone acetylases that can interact with the EBNA-2 acidic domain would be enhanced by the coordinated assembly of these factors at promoters. This may require more than a few EBNA-2 molecules.
The experiments described here focus on the potential role of the EBNA-2 amino terminus in enabling self-association. To investigate this possibility, an oligonucleotide encoding a Flag epitope followed by a nonsense codon was fused in frame to the 3′ end of the EBNA-2 amino-terminal 194 codons. The resulting gene fusion was inserted into the BamHI site in the pSG5 expression vector (21) to make pSGF194. The interaction of the Flag epitope-tagged EBNA-2 amino-terminal 194-amino-acid fusion protein (F194) with wild-type EBNA-2 was then investigated by transfection of BJAB cells with pSGF194 and with pSG5 expressing wild-type EBNA-2 (pSGWE2). After 24 h, the transfected BJAB cells were lysed in 1% NP-40 buffer (10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 150 mM NaCl, 3% glycerol, 1 mM phenylmethylsulfonyl fluoride, 5-μg/ml leupeptin, 10-μg/ml aprotinin), and half was used to immunoprecipitate F194 with M2 anti-Flag monoclonal antibody (Sigma Chemical Co.) and protein G-Sepharose (Pharmacia Corporation), while the other half was used to immunoprecipitate wild-type EBNA-2 with PE2 monoclonal antibody. PE2 monoclonal antibody recognizes an epitope in residues 424 to 464 of the EBNA-2 acidic domain (Fig. 1A and B). F194 was detected in the immunoprecipitate by blotting with M2 antibody, and the extent of wild-type EBNA-2 coprecipitation was determined by immunoblotting with PE2. Wild-type EBNA-2 abundantly coprecipitated with F194 and did not precipitate with M2 antibody from lysates of BJAB cells transfected with only pSGWE2 (Fig. 1B and data not shown). Immunoprecipitation of wild-type EBNA-2 with PE2 antibody from lysates of BJAB cells cotransfected with pSGF194 and pSGWE2 resulted in immunoprecipitation of wild-type EBNA-2 and abundant coprecipitation of F194 (Fig. 1B). As expected, F194 was not precipitated with PE2 antibody from lysates of cells that were transfected with only pSGF194 (data not shown). These data indicate that EBNA-2 residues 1 to 194 readily associate with wild-type EBNA-2.
We next investigated the parts of EBNA-2 required for association with F194 by transiently expressing wild-type EBNA-2 or EBNA-2 deletion mutants in BJAB cells that stably express F194 (Fig. 1C; the EBNA-2 deletion mutants are denoted above each lane). After 24 h, cells transfected with pSG5-based plasmids that express wild-type EBNA-2 or deletion derivatives were lysed in nonionic detergent. Half of the lysate was used to immunoprecipitate F194 with M2 antibody conjugated to Sepharose 4B (Sigma) (Fig. 1C, lower panel), while half was used to immunoprecipitate wild-type or mutant EBNA-2 with PE2 antibody and protein G-Sepharose, as a control for the level of EBNA-2 expression (Fig. 1C, upper panel). The efficiency of immunoprecipitation of wild-type or mutant EBNA-2 compared to that of the cell lysate was about 30% (data not shown). Immunoprecipitation of F194 with M2 antibody resulted in precipitation of about 40% of F194 and coprecipitation of about 30% of wild-type EBNA-2 (compare the amount of WE2 in the immunoblots shown in the upper and lower panels of Fig. 1C). This confirms that wild-type EBNA-2 efficiently associates with F194. Nearly as much EBNA-2 d361-425 and only slightly reduced amounts of EBNA-2 d59-93 coprecipitated with F194, indicating that these mutations do not affect or only modestly affect association with F194 in this assay (Fig. 1C, lower panel). In contrast, EBNA-2 d2-88, d2-95, and d97-122 were severely impaired in F194 association, and EBNA-2 d2-289 did not coprecipitate with F194 (Fig. 1C, lower panel). These data indicate that EBNA-2 amino acids 290 to 484 do not associate with EBNA-2 amino acids 1 to 194, that the polyproline domain that corresponds to amino acids 59 to 95 is not itself critical for association with amino acids 1 to 194, and that amino acids 2 to 88 and 97 to 122 are important for high-level association of wild-type EBNA-2 with EBNA-2 amino acids 1 to 194.
These results were confirmed and extended in experiments examining the ability of wild-type or deletion mutant EBNA-2 to associate with wild-type or deletion mutant F194 in BJAB cells that were transiently transfected with expression vectors for these proteins (Fig. 2; results from similar experiments not shown). Figure 2 shows the efficiency of wild-type or deletion mutant EBNA-2 immunoprecipitation with PE2 (EBNA-2 specific) antibody in lanes marked P compared to the efficiency of coprecipitation of EBNA-2 with Flag-tagged wild-type or deletion mutant F194 using M2 (Flag-specific) antibody in lanes marked M. The results strongly support the theory that EBNA-2 residues 1 to 58 and 97 to 122 are both essential parts of separate domains that mediate homotypic interaction. Deletion of either domain from F194 decreased wild-type EBNA-2 association with F194 (Fig. 2). Furthermore, deletion of amino acids 2 to 58 and part or all of the polyproline domain from F194 decreased but did not completely prevent EBNA-2 with a deletion of amino acids 2 to 58 from associating with F194 (Fig. 2). Moreover, the association of EBNA-2 with F194 that had a deletion of amino acids 2 to 58 was completely dependent on EBNA-2 not having a deletion of amino acids 97 to 122 (Fig. 2). Similarly, deletion of either domain from EBNA-2 rendered EBNA-2 less efficient in binding to wild-type F194 (Fig. 2). EBNA-2 with a deletion of either domain was dependent on the presence of the other domain in F194 for association with F194 (Fig. 2). EBNA-2 d97-122, for example, did not associate at all with F194 d2-88 or F194 d2-95. Thus, deletion of one homotypic interaction domain results in total dependence on the other domain. In contrast, deletion of most of the polyproline domain had little effect on EBNA-2 d59-93 association with F194 or F194 d59-93 (Fig. 2).
FIG. 2.
EBNA-2 residues 2 to 88 or 97 to 122 are required for homotypic association with F194 residues 2 to 88 or 97 to 122, respectively. BJAB cells were cotransfected with pSGWE2 or deletion mutants and with pSGF194 or deletion mutants. Cell lysates were divided for immunoprecipitation with PE2 (EBNA-2 specific) or M2 (Flag epitope F194 specific) antibody and with protein G-Sepharose. M2 immunoprecipitates (lanes M) and PE2 immunoprecipitates (lanes P) were separated in denaturing polyacrylamide gels, and wild-type EBNA-2 or its deletion derivatives were detected with horseradish peroxidase-conjugated PE2 followed by enhanced chemiluminescence.
Similar experiments investigating the extent to which wild-type or deletion mutant EBNA-2 and F194 coimmunoprecipitate from in vitro-translated mixtures of EBNA-2 and F194 yielded similar results (Fig. 3A to C). Wild-type EBNA-2, EBNA-2 d59-93, and EBNA-2 d412-483 efficiently coprecipitated with F194 using M2 antibody to precipitate F194, whereas EBNA-2 d2-95 was markedly deficient (∼15% of the wild-type result) in coprecipitation with F194 and EBNA-2 d97-122 was slightly deficient (∼60% of the wild-type result). F194 also efficiently coprecipitated with EBNA-2 or EBNA-2 d59-93 using PE2 antibody to precipitate EBNA-2, whereas F194 was deficient in coprecipitation with EBNA-2 d2-95 and slightly deficient in precipitation with EBNA-2 d97-122. The efficiency of F194 coprecipitation with EBNA-2 d412-483 could not be assessed, since the PE2 epitope is absent from this mutant as a consequence of the deletion. In vitro-translated EBNA-2 d97-122 was variable in its difference from wild-type EBNA-2 in association with F194 and does not differ in the experiment whose results are shown in Fig. 3.
FIG. 3.
Homotypic association of EBNA-2 amino acids 1 to 194. (A to C) Coimmunoprecipitation of in vitro-translated F194 and in vitro-translated wild-type EBNA-2 or deletion mutants is dependent on amino acids 2 to 95 and 97 to 122 but not on amino acids 59 to 93. Wild-type EBNA-2 or deletion mutant EBNA-2 d59-93, d2-95, d97-122, or d412-483 and F194 were synthesized in an in vitro-coupled transcription and reticulocyte lysate translation (TNT T7) system (Promega) by using 0.5 μg of the relevant pSG5 expression plasmids and [35S]methionine labeling. The products were separated on 10% polyacrylamide gels and were fluorographed (A). An equal portion was immunoprecipitated with PE2 (B) or M2 (C) antibody. The immunoprecipitates were separated on a 10% polyacrylamide gel, visualized by fluorography, and quantified using a PhosphorImager and Image Quant software (Molecular Dynamics). The efficiency of F194 immunoprecipitation was relatively constant. (D and E) 35S-labeled F194 or F194 deletion mutants were electrophoresed in native, nondenaturing (ND) (D) or denaturing polyacrylamide gels (E). The gels were subjected to fluorography.
An expectation from these experiments is that EBNA-2 residues 1 to 194 will self-associate into dimeric or higher-order structures. We therefore compared the behavior of F194 in native and denaturing polyacrylamide gels with the behavior of luciferase, a protein known to be a dimer under nondenaturing conditions. As expected, in vitro-translated luciferase had an apparent size of about 75 kDa under denaturing gel conditions and an apparent size of about 150 kDa in a nondenaturing gel (Fig. 3D and E). In parallel, F194 and F194 d59-93 had apparent sizes of about 40 and 25 kDa, respectively, in denaturing gels and of 360 to 400 and 240 kDa, respectively, in nondenaturing gels. These data indicate that F194 can self-associate and form discrete homo-oligomers comprised of six to eight monomers. Deletion of the polyproline domain in F194 d59-93 did not reduce the extent of oligomerization but did reduce the efficiency of oligomer formation, compatible with the notion that the polyproline domain is not per se required for self-association but can affect the homotypic association of adjacent domains. Consistent with their role in overall stable homotypic association of F194, specific oligomers of F194 d2-88, d2-95, and d97-122 were not well resolved in nondenaturing gels. Based on apparent migration in nondenaturing versus denaturing gel conditions, the extent of F194 homotypic association is greater than would be compatible with the tetramer-sized 13S EBNA-2 complexes from LCLs and more compatible with the size of 34S complexes from LCLs (7) or from Sf9 cells infected with a baculovirus expressing EBNA-2 (26). Intermolecular association between the amino-terminal 194 residues of EBNA-2 is likely to be an important factor in the large size of EBNA-2 complexes.
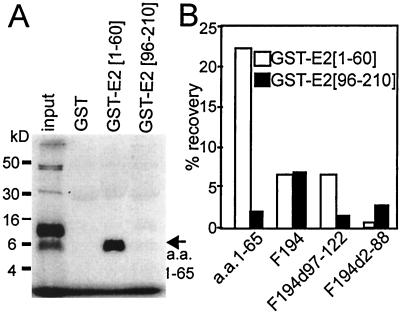
To further delineate the role of residues amino (1 to 58) or carboxyl (96 to 194) terminal to the polyproline domain in the homotypic association for EBNA-2 residues 1 to 194, the ability of in vitro-translated polypeptides consisting of residues 1 to 194, 1 to 65, 89 to 194, 96 to 194, or 1 to 194 with a deletion of residues 97 to 122 to bind to glutathione transferase (GST)–EBNA-2 1-60 or GST–EBNA-2 96-210 fusion proteins was compared. A surprising initial observation was that the polypeptide for the in vitro-translated residues 1 to 65 required boiling in sodium dodecyl sulfate (SDS) sample buffer to behave like a monomer in standard denaturing gels. For the input lane shown in Fig. 4A, in vitro translation product and added SDS sample buffer were not boiled and most of the polypeptide runs as a dimer with a small amount of larger products. In vitro-translated polypeptide for residues 1 to 65 bound specifically and at a high level to GST–EBNA-2 1-60 (Fig. 4A and B). The homotypic association of sequences amino terminal to the polyproline domain was also observed in yeast two-hybrid experiments. Fusions of EBNA-2 1-58 to the Gal4 DNA binding domain or to the Gal4 acidic domain did not activate Gal4-dependent β-galactosidase expression in Y 190 (2), whereas introduction of both plasmids resulted in uniform, high-level β-galactosidase expression in all doubly transformed colonies (data not shown). The binding of the polypeptide for residues 1 to 65 to GST–EBNA-2 1-60 was much greater than the binding of the polypeptide for residues 1 to 194 to GST–EBNA-2 1-60, compatible with the possibility that there may be some intramolecular interactions in the polypeptide for residues 1 to 194 that inhibit the interaction of its domain for residues 1 to 65 with GST–EBNA-2 1-60 (Fig. 4B). As expected, the polypeptide for residues 1 to 194 bound to GST–EBNA-2 1-60 and to GST–EBNA-2 96-210. Deletion of residues 2 to 88 from the polypeptide for residues 1 to 194 abrogated binding to GST–EBNA-2 1-60 but only reduced binding to GST–EBNA-2 96-210. The polypeptide for residues 89 to 194 (F194 d2-88) consistently bound to GST–EBNA-2 96-210 and failed to significantly bind to GST–EBNA-2 1-60, confirming the presence of a self-associating domain on the carboxyl-terminal side of the polyproline domain. Moreover, as expected from previous in vivo association data, deletion of residues 97 to 122 from the polypeptide for residues 1 to 194 markedly decreased binding to GST–EBNA-2 96-210. Thus, the polypeptide for residues 1 to 65, which is amino terminal to the polyproline domain, homotypically binds to the bacterially expressed polypeptide for residues 1 to 60, and the polypeptide for residues 89 to 194, which is carboxyl terminal to the polyproline domain, homotypically binds to the bacterially expressed polypeptide for residues 96 to 210, further confirming the existence of independent self-associating domains on either side of the EBNA-2 polyproline domain.
FIG. 4.
In vitro-translated EBNA-2 polypeptide for residues 1 to 65 binds to bacterially expressed GST–EBNA-2 1-60, and in vitro-translated EBNA-2 polypeptide for residues 89 to 194 binds to bacterially expressed GST–EBNA-2 96-210. (A) Results of a typical experiment in which EBNA-2 polypeptide for residues 1 to 65 was in vitro translated in the presence of [35S]methionine and was then incubated with bacterially expressed GST GST–EBNA-2 1-60 or GST–EBNA-2 96-210 that had been adsorbed onto glutathione-conjugated Sepharose beads. Bound proteins were identified by electrophoresis in a 15- to 18% polyacrylamide gel and fluorography. (B) Summary of several similar experiments evaluating the binding of in vitro-translated [35S]methionine-labeled EBNA-2 polypeptide for residues 1 to 65, F194, F89-194 (F194 d2-88), or F194 d97-122 to GST–EBNA-2 1-60 or GST–EBNA-2 96-210. [35S]methionine binding was calculated using a PhosphorImager and Image Quant software. The percent binding shown is an average of two independent experiments with complete data sets. Binding to GST in those experiments was negligible.
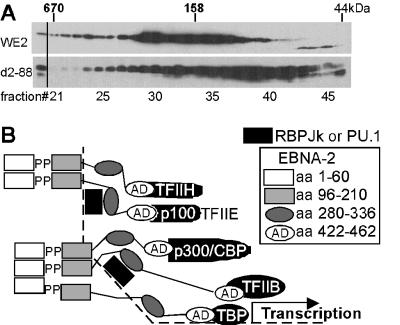
The importance of the EBNA-2 2-88 domain for in vivo complex formation by EBNA-2 was investigated by comparing the sizes of the complexes formed by EBNA-2 d2-88 with the sizes of those formed by wild-type EBNA-2 in BJAB cells (7). As is evident in Fig. 5, the sedimentation properties of the smaller wild-type EBNA-2 complexes are those expected for EBNA-2 dimers or tetramers (6). EBNA-2 d2-88 was more heterogenous in sedimentation, with most of the protein having the size expected for monomers or dimers. While not excluding the possibility that these differences in sedimentation could be due to differences in association with other proteins, no associated proteins have been identified for this domain of EBNA-2 and the simplest interpretation is that residues 2 to 88 are important for self-association in vivo.
FIG. 5.
EBNA-2 complexes. (A) The sucrose gradient sedimentations of wild-type EBNA-2 and EBNA-2 with a deletion of amino acids 2 to 88 are compared. Nonionic detergent (1% NP-40) isotonic salt extracts from BJAB cells expressing EBNA-2 wild type or d2-88 were applied to a 10 to 30% sucrose gradient in 50 mM sodium phosphate buffer (pH7.4) 0.1% NP-40, and 1 mM dithiothreitol. The gradient was run for 16 h at 35,000 rpm and 4°C in an SW41 rotor (Beckman Corp). The gradient were collected from the bottom as 52 0.23-ml fractions. Fractions 21 to 46, which contained the peaks of 670- and 44-kDa markers, respectively, were precipitated with ethanol, and the proteins were run in 10% SDS-polyacrylamide gels. Wild-type EBNA-2 and EBNA-2 d2-88 were identified by immunoblotting using horseradish peroxidase-conjugated PE2 antibody. The positions of bovine thyroglobulin (670), bovine gamma globulin (158), and chicken ovalbumin (44) are indicated along the top. (B) Model for the role of the EBNA-2 homotypic association domains in mediating cooperative transcriptional interactions at promoters that have nearby binding sites for cellular sequence-specific DNA binding proteins. EBNA-2 oligomers are postulated to enable the EBNA-2 acidic domain to simultaneously recruit basal and activated transcription factors, p100, and p300 (also called CBP) and PCAF to promoters to achieve transcriptional up-regulation.
The data presented here indicate that EBNA-2 has at least two independent domains that mediate homotypic association. One domain is within the amino-terminal 58 amino acids and associates with itself in yeast, indicative of a direct association. This domain may also be able to form higher-order structures, as was observed with partial denaturation of the in vitro translation product. The second domain is within residues 89 to 210 and is dependent on residues 97 to 122. Interestingly, deletion of the polyproline domain between amino acids 59 and 95 and the consequent approximation of the two homotypic association domains did not affect self-association, in vitro or in vivo, but did inhibit the ability to maintain higher-order structures in nondenaturing gels.
The two EBNA-2 domains for homotypic association described here, their independent role in enabling self-association, and their joint role in enabling EBNA-2 to form high-order oligomers probably account for the stringent and imprecise requirement for the amino-terminal half of EBNA-2 for transient transactivation of responsive promoters and for primary B-lymphocyte growth transformation. Deletion of codons 19 to 33, 59 to 93, 19 to 110, 2 to 95, or 112 to 141 quite significantly affected the transformation efficiency of the respective EBV recombinants, while deletion of codons 2 to 88, 97 to 122, 143 to 230, or 231 to 280 resulted in EBV recombinants whose transforming efficiency was close to that of the wild type (5, 10, 28). The accumulated data are most consistent with a model that these residues do not mediate critical or unique interactions with cellular proteins but instead are part of two independent homotypic association domains, either of which can be sufficient for activity in transient reporter activation or primary B-lymphocyte growth transformation assays. Amino acids 1 to 60 appear to be particularly able to homotypically associate so that the effects of mutations in the other domain might be expected to be more subtle. Moreover, EBNA-2 mutations in either of the two homotypic association domains that are compatible with near wild-type transforming activity in clonal transformation assays (5, 10, 28) are marked by abnormally high-level EBNA-2 expression. Higher-level EBNA-2 expression may partially compensate for the effects of the deletion on homotypic association and lower transcriptional activation.
Association among EBNA-2 molecules is likely to be critical for assembly of the multiple transcription factors, coactivators, and histone acetylases that are required for efficient transcription (Fig. 5B). The associated EBNA-2 molecules would be able to simultaneously interact with RBPJκ, PU.1, and other cellular sequence-specific DNA binding proteins through amino acids 290 to 360 and with the p100 coactivator, TFIIB, TAF40, TFIIH, p300, and CBP through amino acids 420 to 464. EBNA-2 may also interact with other factors bound near the promoter, as has been described for the LMP1 promoter (22). By bridging among multiple factors, EBNA-2 coordinately up-regulates transcription at highly specific sites, including the c-myc promoter (1, 12, 15).
Acknowledgments
This research was supported by grant number CA47006 from the National Cancer Institute of the U.S. Public Health Service and by a grant-in-aid from the Ministry of Education, Science, and Culture of Japan.
We thank Frederick Wang for PE2 conjugated to HRP and Hiroshi Sato for his support of S.H.
REFERENCES
- 1.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. . (Erratum, 185:946.) [DOI] [PubMed] [Google Scholar]
- 2.Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J I. A region of herpes simplex virus VP16 can substitute for a transforming domain of Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1992;89:8030–8034. doi: 10.1073/pnas.89.17.8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J I, Kieff E. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J Virol. 1991;65:5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J I, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dash A B, Orrico F C, Ness S A. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 7.Grasser F A, Haiss P, Gottel S, Mueller-Lantzsch N. Biochemical characterization of Epstein-Barr virus nuclear antigen 2A. J Virol. 1991;65:3779–3788. doi: 10.1128/jvi.65.7.3779-3788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada S, Yalamanchili R, Kieff E. Residues 231 to 280 of the Epstein-Barr virus nuclear protein 2 are not essential for primary B-lymphocyte growth transformation. J Virol. 1998;72:9948–9954. doi: 10.1128/jvi.72.12.9948-9954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 12.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Jin X W, Speck S H. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm G W, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 17.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leverson J D, Koskinen P J, Orrico F C, Rainio E M, Jalkanen K J, Dash A B, Eisenman R N, Ness S A. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 19.Nitsche F, Bell A, Rickinson A. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickinson A B, Kieff E. Epstein-Barr virus. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 21.Seed B. Developments in expression cloning. Curr Opin Biotechnol. 1995;6:567–573. doi: 10.1016/0958-1669(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 22.Sjoblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsui S, Schubach W H. Epstein-Barr virus nuclear protein 2A forms oligomers in vitro and in vivo through a region required for B-cell transformation. J Virol. 1994;68:4287–4294. doi: 10.1128/jvi.68.7.4287-4294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yalamanchili R, Harada S, Kieff E. The N-terminal half of EBNA2, except for seven prolines, is not essential for primary B-lymphocyte growth transformation. J Virol. 1996;70:2468–2473. doi: 10.1128/jvi.70.4.2468-2473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 30.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]