Abstract
In this review, we revisit the pivotal role of fibroblast growth factor receptor 3 (FGFR3) in bladder cancer (BLCA), underscoring its prevalence in both non‐muscle‐invasive and muscle‐invasive forms of the disease. FGFR3 mutations in up to half of BLCAs play a well‐established role in tumorigenesis, shaping distinct tumor initiation patterns and impacting the tumor microenvironment (TME). Emphasizing the importance of considering epithelial‐mesenchymal transition profile and TME status, we revisit their relevance in predicting responses to immune checkpoint inhibitors in FGFR3‐mutated BLCAs. This writing highlights the initially promising yet transient efficacy of the FGFR inhibitor Erdafitinib on FGFR3‐mutated BLCA, stressing the pressing need to unravel resistance mechanisms and identify co‐targets for future combinatorial studies. A thorough analysis of recent preclinical and clinical evidence reveals resistance mechanisms, including secondary mutations, epigenetic alterations in pathway effectors, phenotypic heterogeneity, and population‐specific variations within FGFR3 mutational status. Lastly, we discuss the potential of combinatorial treatments and concepts like synthetic lethality for discovering more effective targeted therapies against FGFR3‐mutated BLCA.
Keywords: Bladder Cancer, Erdafitinib, FGFR inhibition, FGFR3 mutations, Resistance to Erdafitinib, Tumor Microenvironment
Abbreviations
- AL
et al, in Latin et alias, and others
- APOBEC
Apolipoprotein B mRNA editing enzyme, catalytic polypeptide‐like
- BAIAP2L1
BAI1‐associated protein 2‐like 1
- BCG
Bacillus Calmette‐Guérin
- BLCA
Bladder cancer
- BLCAs
Bladder Cancers
- BMP
Bone Morphogenetic Protein
- CBL
Casitas B‐lineage Lymphoma
- CI
Confidence Interval
- CPIs
Checkpoint Inhibitors
- CRC
Colorectal Cancer
- ctDNA
Circulating Tumor DNA
- CTLA‐4
Cytotoxic T‐Lymphocyte Antigen 4
- Dusp‐6
Dual Specificity Phosphatase 6
- EGFR
Epidermal Growth Factor Receptor
- EMT
Epithelial‐Mesenchymal Transition
- ERBB2
Erb‐B2 Receptor Tyrosine Kinase 2
- ERBB3
Erb‐B2 Receptor Tyrosine Kinase 3
- ERK
Extracellular signal‐regulated kinase
- FDA
Food and Drug Administration
- FGF
Fibroblast Growth Factor
- FGFR
Fibroblast Growth Factor Receptor
- FGFR2
Fibroblast Growth Factor Receptor 2
- FGFR3
Fibroblast Growth Factor Receptor 3
- HIF‐1α
Hypoxia‐Inducible Factor 1‐alpha
- IGF1R
Insulin‐like Growth Factor 1 Receptor
- Ihh
Indian Hedgehog
- MAPK
Mitogen‐Activated Protein Kinase
- MEK
Mitogen‐Activated Protein Kinase Kinase
- MIBC
Muscle‐invasive bladder cancer
- MYC
Myelocytomatosis Oncogene
- NEDD4
Neural Precursor Cell Expressed Developmentally Down‐Regulated Protein 4
- NGS
Next‐Generation Sequencing
- NMBIC
Non‐Muscle‐Invasive Bladder Cancer
- NMIBC
Non‐muscle‐invasive bladder cancer
- NRG1
Neuregulin 1
- ORR
Objective Response Rate
- PD‐1
Programmed Cell Death Protein 1
- PDGFR
Platelet‐Derived Growth Factor Receptor
- PD‐L1
Programmed Death‐Ligand 1
- PDX
Patient‐Derived Xenograft
- PFS
Progression‐Free Survival
- PI3K
Phosphoinositide 3‐Kinase
- PLCγ1
Phospholipase C gamma 1
- RFS
Recurrence‐free survival
- ROS
Reactive Oxygen Species
- RT
Radiotherapy
- RTK
Receptor Tyrosine Kinase
- SEF
Similar Expression to FGF
- shRNA
Short Hairpin RNA
- SNP
Single nucleotide polymorphism
- SPRY
Sprouty
- STAT
Signal Transducer and Activator of Transcription
- T3
Thyroid Hormone
- TACC3
Transforming acidic coiled‐coil‐containing protein 3
- TGF‐β
Transforming Growth Factor Beta
- TME
Tumor Microenvironment
- UC
Urothelial carcinoma
- VEGF
Vascular Endothelial Growth Factor
- VEGFR
Vascular Endothelial Growth Factor Receptor
- VEGFR2
Vascular Endothelial Growth Factor Receptor 2
- YAP/TAZ
Yes‐associated Protein/Transcriptional coactivator with PDZ‐binding motif
1. INTRODUCTION
Bladder cancer (BLCA) accounts for the 10th most common cancer globally, with more than 550,000 newly diagnosed cases and over 220,000 annual deaths worldwide [1, 2]. Male gender increases the risk for BLCA (4:1), making it the 6th most common cancer among all cancer types worldwide [1, 2]. According to the American Cancer Society prediction, one out of every twenty‐eight men will develop BLCA during their lifetime (source: https://www.cancer.org/). Nevertheless, the biological females are diagnosed at more advanced and deadlier stages of BLCA [2].
Around 95% of BLCAs are urothelial cancer, originating from urothelial cells within one of the cellular layers of bladder epithelium [2]. Based on the tumor's local invasion, BLCA is classified into non‐muscle‐invasive disease (NMIBC) and muscle‐invasive disease (MIBC), accounting for ∼80% and ∼20% of BLCAs, respectively [1, 3]. With the emergence of knowledge about genetic alterations in BLCA, the association of these alterations with different subtypes has been unveiled [1].
The five‐year survival rate for NMIBC stands at approximately 70%‐96% [3, 4]. However, depending on the MIBC disease stage, it drops from 38% to a meager 6% in metastatic BLCA [5].
The diagnosis and management of BLCA places substantial economic pressure on healthcare systems as, particularly MIBC, is the costliest cancer type per patient, with estimated costs in the US reaching approximately $6 billion annually [6].
2. FGFR3 ALTERATIONS AND THEIR ROLE IN BLCA AND BEYOND
Fibroblast Growth Factor Receptor (FGFR) alterations, and in particular, FGFR3 mutations, are one of the most frequent alterations in BLCA, associated with ∼60% of the NMIBCs and at least 15% of the MIBCs [1, 2]. Figure 1 illustrates two BLCA categories, emphasizing their correlation with distinct stages and diverse subtypes, all within FGFR3 status.
FIGURE 1.
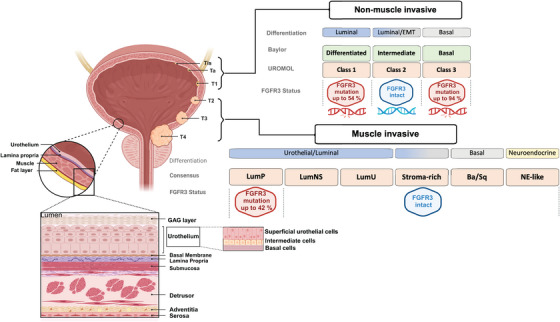
Two Categories of bladder cancer (BLCA) are attributed to Baylor, UROMOL, and Consensus subtyping systems, highlighting their correlation with different stages of local advancement and FGFR3 status. Differentiation‐based color scheme showing features associated with the respective classification. Baylor and UROMOL define NMIBC: Luminal (blue), Luminal‐to‐basal (blue‐grey), and Basal (grey). MIBC is defined by consensus class: Urothelial‐to‐Luminal (blue), Luminal‐to‐basal (blue‐grey), Basal (grey), and neuroendocrine (light yellow). FGFR3 status among subtypes is highlighted in blue (intact FGFR3) or red (mutated FGFR3), with the frequency of FGFR3 alterations in %. Abbreviations: Ba/Sq, basal/squamous; FGFR3, Fibroblast Growth Factor Receptor LumP , luminal papillary, LumNS , luminal nonspecified, LumU, luminal unstable, MIBC, muscle‐invasive bladder cancer, NE, neuroendocrine, NMIBC, non‐muscle‐invasive bladder cancer,. Figure generated in Biorender.
In 1999 Cappellen et al. [7] described recurrent activating FGFR3 in BlCA and cervical cancer. In 2006, Bernard‐Pierrot et al. [8] showed that mutated FGFR3b (S249C), the predominant isoform in epithelial cells, has oncogenic properties as demonstrated by its ability to transform NIH‐3T3 cells, and when transformed cells implanted in immunocompromised mice formed tumors. Bernard‐Pierrot et al. [8] showed that FGFR3 knockdown and FGFR chemical inhibition in BLCA MGH‐U3 cells, harboring the same mutated isoform, suppressed cell proliferation and anchorage‐independent growth. Later, Tomlinson et al. [9] reported similar findings in another BLCA cell line (97‐7) with the same mutation. Apart from mutation or overexpression, FGFR3 can also be activated in BLCA through chromosomal rearrangements that generate constitutively activated fusion genes [10]. Specifically, FGFR3‐transforming acid coiled‐coil 3 (TACC3) fusions resulting from 4p16.3 rearrangements and a t(4;7) translocation generating an FGFR3‐BAI1‐associated protein 2‐like 1 (BAIAP2L1) fusion were identified in several BLCA cell lines and tissue samples [10]. These fusion proteins have lost their Phospholipase C gamma 1 (PLCγ1) binding site. They can transform NIH‐3T3 cells. They can induce activation of the ERK pathway, but not the PLCγ1, in immortalized normal human urothelial cells, suggesting their potential as targets for FGFR‐targeted therapy in BLCA [10].
In 2014, during a Cancer Genome Atlas project study, a comprehensive genetic analysis of 131 urothelial carcinomas discovered FGFR3 alterations as one of the genetic hallmarks of urothelial bladder carcinoma [11]. More recently, FGFR3 mutations have been suggested as one of the potential therapeutic targets in advanced BLCA [12]. The consensus classification of BLCA had concluded that the high frequency of FGFR3 mutations, translocations, and FGFR3 activation signature in advanced BLCA could suggest that these tumors may be collectively responsive to FGFR‐targeted therapeutics [12]. The authors reiterated the results of studies that showed a considerable group of MIBC patients who either harbored FGFR3 mutations or were identified with high FGFR3 expression signatures could benefit from FGFR3‐targeted therapies [12]. Of note, later, the benefit of FGFR3 inhibition has been limited to patients with FGFR3 mutations and not those with solely high FGFR3 expression [13, 14].
Komura et al. integrated the three subtyping systems, Consensus [12], Baylor [15], and UROMOL [16], in BLCA categories (NMIBC vs. MIBC) in which the subtyping was not originally described [17]. The variations in subtype frequencies could be associated with either NMIBC or MIBC. For example, the class I, differentiated (vs. basal), and LumP subtypes were more prevalent in NMIBC than in MIBC [17]. However, most subtyping systems could not distinguish between NMIBC and MIBC in an absolute manner [17]. The UROMOL class 3 subtype, with the highest frequency of FGFR3 alterations, was mostly confined to NMIBC [17].
Kinase domain mutations were predominantly enriched among basal/squamous subtypes [17]. Gene Set Enrichment Analysis (GSEA) also revealed that basal/squamous and epithelial signatures characterized MIBC. Moreover, Komura et al. [17] showed that FGFR3 alterations in MIBC, in addition to luminal subtypes, can involve epithelial subtypes, including both the basal and squamous types.
Of note, the association of FGFR3 alterations with the induction of basal markers in this category warrants further investigations of whether resistance mechanisms and pathways activated by FGFR3 and, consequently, co‐targets to tackle these resistance mechanisms differ between NMIBC and MIBC.
A significant proportion, up to 50% of BLCA patients, including a subset of those with advanced disease, carry FGFR3 mutations [2]. These are primarily point mutations, followed by less frequent structural variants (∼14:1 ratio) involving fusion with other genes, such as TACC3 [3, 18]. To investigate this further, we consulted cBioPortal to query 4,732 samples from 3,993 patients across 19 non‐redundant studies (date of data retrieval: September 10th, 2023). Our analysis revealed three prominent mutational hotspots within the FGFR3 coding sequence (the reference is the FGFR3c sequence), namely S249, R248, and Y373 (Figure 2). Interestingly, these hotspots do not involve the tyrosine kinase domain but induce a conformational change that leads to constitutive protein activity and triggers its downstream signaling output [7, 19, 20]. Further, following the approach described previously [21], we manually curated the dataset, cross‐referenced patient and sample IDs, and retained only the earliest samples. Our curation corroborated that the three most frequently occurring FGFR3 mutations in BLCA were S249C (APOBEC‐type motif [22]), Y373C, and R248C, accounting for 49%, 13%, and 12% of the samples, respectively. These findings are, in principle, consistent with previous reports [23].
FIGURE 2.
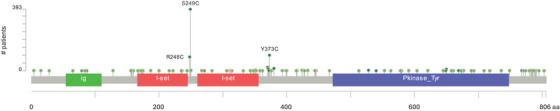
Schematic displays the FGFR3 amino acid residues subjected to mutational occurrence in BLCA. Querying 4732 BLCA samples in 19 studies. Based on the query, the figure was generated in cBioPortal and modified for clarity. Abbreviations: BLCA, Bladder Cancer; cBioPortal, cBioPortal for Cancer Genomics; FGFR3, Fibroblast Growth Factor Receptor 3.
A multi‐omic analysis of 124 NMIBC and 265 MIBC patient samples and 35 adjacent normal tissue samples by Komura et al. [17] has provided insights into the differential FGFR3 mutational status among the Asian and Western populations.
In the Asian population, FGFR3 SNPs, including Q29H, G65R, L164V, T450M, and A720S, appeared enriched at the germline level, though these mutations were not enriched in BLCA patients, and no established associations with clinical outcomes among BLCA patients were identified [17].
Unlike the Western population, FGFR3 mutations within the kinase domain, specifically K650E and T757P, were more prevalent yet showed no conclusive changes in clinical outcomes [17]. Notably, these mutations were more recurrent in MIBCs [17]. Additionally, Komura et al. [17] discovered two novel FGFR3 fusions involving NSD2 and SPON2.
Regarding the frequency of FGFR3 alterations among subtypes, 54% and 94% of class I and class III UROMOL subtypes, respectively, and 42% of the MIBC LumP (consensus) subtype harbored these alterations [17].
Mutant FGFR3 is shown to have transforming capacity in a context‐dependent and variant‐specific manner [24]. As unraveled by di Martino et al. [24], variants could exhibit distinctive downstream signaling output and differential impacts on proliferation. For instance, in a human urothelial cellular model, FGFRS249C but not FGFRK652E could activate the PLCγ1 and induce cellular proliferation (see Figure 3 for a simplified illustration of FGFR3 signaling). Conversely, in a mouse embryonic fibroblast cell line, all three FGFR3 variants could activate the PLCγ1 and exhibited cell transforming capacity [24].
FIGURE 3.
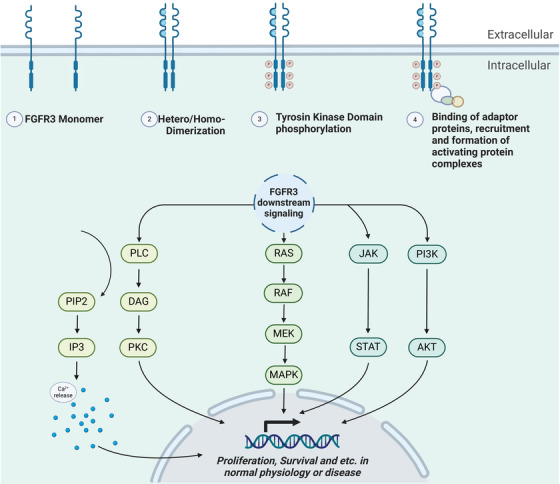
Oversimplified illustration of FGFR3 signaling. Upon Fibroblast Growth Factors (FGFs) binding to the extracellular domain of FGFRs, these receptors, such as FGFR3, go through conformational changes, which subsequently trigger their dimerization and transphosphorylation of the intracellular C‐terminal tyrosine kinase domains. Different tyrosine residues at the intracellular C‐terminal fragment of the FGFR can trigger distinctive downstream signaling pathways. When FGFRs become active, they may, via their essential adaptor, FRS2α, recruit complexes from the RAS or PI3K/AKT pathways, initiating signaling output through the ERK or AKT pathway. Active FGFRs may also play a role in activating the JAK/STAT pathway. Furthermore, FGFRs, by activating PLCγ, induce the hydrolysis conversion of phosphatidylinositol‐4,5‐biphosphate (PIP2) into inositol triphosphate (IP3), resulting in Calcium ion influx or via diacylglycerol (DAG), trigger the protein kinase C (PKC) signaling. Note that FGFR3 is also shown to be involved in nonliganded activation, which is not acknowledged in this schematic. The figure was generated in Biorender. Abbreviations: AKT, Protein Kinase B; DAG, Diacylglycerol; ERK: Extracellular Signal‐Regulated Kinase; FGFs, Fibroblast Growth Factors; FGFRs, Fibroblast Growth Factor Receptors; FGFR3, Fibroblast Growth Factor Receptor 3; FRS2α, Fibroblast Growth Factor Receptor Substrate 2α; IP3, Inositol Triphosphate; JAK/STAT, Janus Kinase/Signal Transducer and Activator of Transcription; PIP2, Phosphatidylinositol‐4,5‐bisphosphate; PI3K, Phosphoinositide 3‐ Kinase; PLCγ: Phospholipase C Gamma; PKC, Protein Kinase C; RAS, Rat Sarcoma.
Indeed, FGFR3 is frequently mutated or overexpressed in BLCA. Assessing mutation status and receptor expression levels may help define the patient population that could benefit from FGFR‐targeted therapies [25].
Of note, Shi et al. [26] have recently shown that human FGFR3S249C alone can initiate luminal‐like papillary tumors in a transgenic mouse model with higher incidence among male mice. Interestingly, the formed tumors exhibited a similar expression profile as class I and class III human BLCA [26]. Both subtypes belonged to NMIBC and are known to be enriched with FGFR3 mutations [26]. The authors reported that FGFR3S249C‐induced tumor initiation correlates with FGFR3 tissue expression levels [26]. As the authors reiterated, in previous studies, other FGFR3 variants, such as K644E, the murine equivalent of human K652E, had failed to induce tumors without other cooperating factors [26, 27].
Today, when patients relapse on immune checkpoint inhibitors, they may be eligible for FGFR3‐targeted therapy [2]. Therapeutics targeting FGFR3 encompass a range of approaches in terms of medicinal chemistry, including multi‐kinase inhibitors, selective kinase inhibitors, and monoclonal antibodies directed against FGFR3 [28, 29, 30]. The first two categories have made more significant progress in the clinical trial phases [30]. Selective FGFR inhibitors like erdafitinib and rogaratinib are advanced in clinical trials [3, 28, 29]. In particular, erdafitinib, which has shown selectivity towards FGFRs and high potency, has been granted FDA approval for metastatic urothelial carcinoma with FGFR3 and FGFR2 alterations and is currently being explored in other BLCA settings [3, 14, 28, 29, 31–36]. In one study leading to accelerated FDA approval, erdafitinib's median duration of response in patients with advanced BLCA was shown to be 5.6 months (95% CI, 4.2 to 7.2) [14].
Recently, erdafitinib has been clinically explored among different groups of FGFR mutated BLCA patients [14, 31‐35, 37]. For instance, among high‐risk NMIBC patients who harbored FGFR3 or FGFR2 genetic alterations and were refractory to BCG therapy, oral erdafitinib was shown to have more favorable outcomes in terms of recurrence‐free survival (RFS) compared to intravesical chemotherapy (median RFS: ∼16.9 months vs. ∼11.6 months) [38]. Moreover, a novel drug delivery system for site‐specific and sustained intravesical delivery of erdafitinib is also being developed [39].
Clinically available compounds targeting FGFRs are pan‐FGFR inhibitors with varying degrees of activity against each FGFR isoform. However, this broad FGFR targeting comes at the cost of adverse events. For instance, the FGFR1 activity of these compounds is suggested to be associated with hyperphosphatemia, while anti‐FGFR4 targeting is linked to diarrhea [40].
Nail damage, alopecia, dry mouth, stomatitis, blurred vision, central serous retinopathy, and hand‐foot syndrome are recurrent adverse events observed among patients receiving FGFR inhibitors [30]. While these adverse events are often below grade 3 toxicities, they may necessitate temporary dose withdrawal or dose reduction until the adverse event is resolved [14, 30‐34, 36‐39, 40–46].
Hyperphosphatemia is rather a shared adverse event caused by different FGFR inhibitors. At least 60% of patients treated with FGFR inhibitors experience some extent of hyperphosphatemia.
FGF23‐FGFR1‐klotho axis controls phosphate levels. When FGF23 binds to FGFR1, it inhibits 25‐hydroxyvitamin D 1α‐hydroxylase, leading to the breakdown of 1,25‐dihydroxyvitamin D (1,25(OH)2D), and blocks sodium‐phosphate co‐transporters in the proximal renal tubules, reducing phosphate reabsorption [30, 40, 47]. FGFR inhibitors block this pathway, preventing the breakdown of 1,25(OH)2D and the inhibition of sodium‐phosphate co‐transporters, which increases both 1,25(OH)2D levels and phosphate reabsorption in the kidneys. By disrupting the FGF23‐FGFR1‐klotho axis that typically limits phosphate reabsorption, FGFR inhibitors cause hyperphosphatemia through increased intestinal phosphate absorption due to higher 1,25(OH)2D levels and increased renal phosphate reabsorption and decreased excretion [30, 40, 47].
Maintaining a low‐phosphate diet and prescribing phosphate binders can prevent and manage hyperphosphatemia, reducing therapy discontinuation risk [30]. While interventions like phosphate binders and dietary phosphate restriction are necessary to manage the common hyperphosphatemia caused by FGFR inhibitors, these measures can paradoxically lead to hypophosphatemia in some patients. Unlike hyperphosphatemia, which typically is low‐grade, hypophosphatemia resulting from overcorrection may manifest as a higher‐grade (grade ≥3) adverse event [48].
Interestingly, serum phosphate levels have emerged as a robust pharmacodynamic marker for the FGFR inhibitor erdafitinib [14, 49]. Increases in serum phosphate levels, a consequence of FGFR inhibition by erdafitinib, were found to strongly correlate with positive clinical responses and improved outcomes in treated patients [14, 49, 50]. As such, based on pharmacodynamic and pharmacokinetic modeling, a target serum phosphate threshold of 5.5 mg/dL (1.8 mmol/L) has been defined, indicative of an effective erdafitinib dose capable of achieving the desired pharmacologic effect [14, 49, 50]. A study proposes initiating erdafitinib at a dose of 8 mg, followed by an escalation to 9 mg between Days 14 and 21. While minimizing dose‐limiting interventions, this approach maximized the proportion of patients achieving the target serum phosphate concentrations of 5.5‐7 mg/dL [50].
In theory, selective FGFR3 inhibitors should effectively inhibit FGFR3 while enhancing the therapeutic safety profile. Efforts in this regard have been made, and several FGFR3‐targeting monoclonal antibodies with enhanced selectivity for FGFR3 have been preclinically and clinically explored [51, 52, 53, 54, 55]. FGFR inhibitor toxicity is not exclusively due to their pan‐FGFR effects but can be associated with their targeting potential towards other proteins, such as VEGFR2 [56]. Efforts are being made to develop inhibitors with enhanced selectivity towards FGFR3 over VEGFR2 [56].
In the following lines, we revisit FGFR3, a less‐understood receptor tyrosine kinase, beyond its role in BLCA.
FGFR3 is one of the four conventional members of the FGFR family [57]. In 1986, Drohan's lab isolated the coding sequence of FGF1, one of the cognate ligands of FGFRs, from the human brain cDNA library [58]. Today, we acknowledge at least eighteen human FGF ligands (twenty‐two unique FGFs in rodents and humans) that bind to different FGFR isoforms, triggering diverse signal transduction processes and resulting in distinct cellular outcomes [29, 57, 59]. Four FGFR receptors are encoded from four different chromosomes, and FGFR3 is encoded from chromosome 4 [29, 60]. FGFR3 can be activated by FGF1, FGF2, FGF4, FGF5, FGF6, FGF8, FGF9, FGF16, FGF17, FGF18, FGF19, FGF20, FGF21, or FGF23 in human or mouse [3, 57]. FGFR3, like the other three members of the related Receptor Tyrosine Kinase family (RTK), has functions beyond its role in cancer, from early embryogenesis to cell homeostasis throughout mammalian cell life [57]. FGFR3 possesses three extracellular domains where ligand binding occurs, a juxta‐membranous domain and an intracellular domain where the tyrosine kinase domain is positioned [29, 57]. Upon ligand binding, FGFR3 undergoes activation and dimerization [57, 61, 62]. Notably, despite conjecture, the mechanisms and extent of FGFR3's involvement in heterodimerization events remain poorly understood. Following ligand binding and the initiation of dimerization, FGFRs may experience transphosphorylation of their tyrosine kinase domains, and subsequently, via their C‐terminal tails, they facilitate the binding and activation of adaptor proteins, setting in motion downstream signaling cascades [3, 57] (Figure 3). Notably, nonliganded FGFR3 has basal dimerization capacity that can be augmented in some mutant forms of FGFR3 [61, 62].
FGFRs, including FGFR3, can be genetically altered in solid and liquid tumors [63, 64, 65, 66, 67, 68], and therefore, endeavors have been dedicated to therapeutically targeting such alterations [69, 70, 71, 72]. Apart from BLCA, where FGFR3 mutations are prevalent, other human cancers can be identified with these mutations, albeit at a much lower frequency. These cancer types, among others, include uterine and cervical cancers [73, 74], glioblastoma [75], multiple myeloma [76], penile cancer [77], non‐melanoma skin cancer [78], melanoma [79], small bowel cancer [80] and non‐small cell lung cancer [81, 82].
FGFR3 has attracted attention not only in the context of cancer. Interestingly, activating FGFR3 mutations, particularly S249C, can be recurrently found in benign skin conditions such as seborrheic keratoses [83, 84].
FGFR3 has been extensively examined in bone repair,‐remodeling, and skeletal disorders [85, 86, 87, 88, 89, 90, 91, 92, 93]. Herein, unlike in aforementioned cancers, FGFR3's role is rather suppressive. In particular, FGFR3 can restrict the proliferation of chondrocytes, and its signaling can lie behind skeletal disorders such as achondroplasia and thanatophoric dysplasia [94]. Interestingly, activating FGFR3 germline mutations have been linked to skeletal disorders in humans and animal models [29, 87‐89, 95–97]. FGFR3 negatively influences chondrocyte balance, viability, and differentiation by sabotaging the related autophagy machinery in achondroplasia [85]. FGFR3 can suppress sheep growth plate chondrocyte proliferation by limiting telomerase activity during bone elongation in a Thyroid hormone‐dependent (T3) manner [86]. Interestingly, in addition to several FGF ligands, FGFR3 is the target of T3 [86].
In mice where chondrocyte FGFR3 was knocked out, there was a notable increase in osteoblast count and bone formation [98]. This effect was further elucidated through chondrocyte‐osteoblast co‐culture experiments, which demonstrated that the absence of FGFR3 in chondrocytes stimulated the differentiation and mineralization of osteoblasts, accompanied by the up‐regulation of genes including Ihh, Bmp2, Bmp4, Bmp7, Wnt4, and Tgf‐β1, along with a down‐regulation of Nog expression [98]. Conversely, FGF18, a high‐affinity ligand for FGFR3, has shown protective effects in osteoarthritis [93, 99]. Of note, a variant of FGFR3 has been reported in Camptodactyly, Tall Stature, and Hearing Loss (CATSHL) Syndrome, which counteract the wild‐type FGFR3 function in a dominant‐negative manner [100, 101, 102].
Due to the complexity of FGFs and FGFRs signaling, the full picture of FGFR3's role in bone and cartilage pathogenesis remains to be elucidated.
This duality of the FGFR3 role has also been reported in cancer. For instance, in colorectal cancer (CRC), the FGFR3 role seems Janus‐faced [103]. Due to aberrant alternative splicing and activation of cryptic splice sequences, FGFR3 was reported to be frequently inactivated in CRC [103]. Conversely, FGFR3 overexpression in CRC has been associated with a more invasive phenotype [104]. A recent study on Hepatocellular carcinoma (HCC) deciphers truncated FGFR3's role in cancer, or at least in HCC [105]. FGFR3∆7‐9, lacking the IgG III domain involved in FGF binding, exhibits increased affinity for FGF compared to wild‐type FGFR3 [105]. FGFR3∆7‐9 promotes HCC cell proliferation, migration, and metastasis both in vitro and in vivo [105]. Interestingly, FGFR3∆7‐9 directly interacts with TET2, a tumor suppressor, phosphorylating it at Y1902, thereby promoting TET2 ubiquitination and degradation and unleashing the AKT pathway and its oncogenic subsequences, which is no longer suppressed by the TET2‐PTEN axis [105].
In one study in pancreatic cancer, FGFR3 has been shown to have a tumor‐suppressing role in cells with epithelial features while exhibiting oncogenic properties in cells with mesenchymal characteristics [106].
FGFRs present a high degree of homology to other known RTKs like vascular endothelial growth factor receptor (VEGFR) and platelet‐derived growth factor receptor (PDGFR) [107]. In terms of signaling output, a great extent of similarities can be observed with other RTKs, such as the renowned Epidermal Growth Factor Receptor (EGFR). Those common features are not limited to the mechanism of action, but these receptors also share downstream effectors and, as such, shared cellular events [3, 57, 108, 109, 110]. FGFR3 activation yields at least four distinct pathway activities, including the RAS‐RAF‐MEK‐MAPK, PI3K/AKT, Calcium signaling, and Signal Transducer and Activator of Transcription (STAT) pathways (Figure 3) [24, 29, 92, 106, 111–113]. These pathways are known to be significantly influenced by receptor tyrosine kinases (RTKs) like EGFR as well [92, 113–117]. As such, it comes as no surprise that regulators of FGFR3 signaling are also shared between FGFR3 and EGFR. Indeed, CBL, SPRY, Dusp‐6, and SEF are among the known regulators of FGFR3 signaling that have been previously studied in the context of EGFR signal transduction and regulation [3, 118–122].
In BLCA with aberrant FGFR3 activation due to translocation and point mutation, the role of the FGFR3/MYC feedback loop has been unveiled [123]. FGFR3 upregulates MYC expression by increasing MYC mRNA levels via p38α MAPK activation. Additionally, FGFR3 stabilizes the MYC protein by AKT‐mediated phosphorylation of GSK3β, preventing MYC degradation by the proteasome. As such, a positive feedback loop was discovered where activated FGFR3 upregulates MYC levels, which in turn upregulates FGFR3 expression, and disrupting this FGFR3/MYC loop decreased bladder cancer cell viability in vitro and tumor growth in vivo [123].
Still, we do not fully grasp the extent of FGFR3 signaling and its unique impact. As FGFR3 shares similarities with other receptors like EGFR, these commonalities might help us uncover more about FGFR3's function and vulnerabilities.
3. MUTATED FGFR3: SENSITIVITY AND RESISTANCE TO TARGETED THERAPIES IN BLCA
Unsurprisingly, the resistance mechanism to FGFR3 inhibition in BLCA resembles those observed in EGFR‐targeted therapeutics. Indeed, just as resistance to some EGFR inhibitors can arise due to the emergence of resistant clones with secondary mutations at the gatekeeper T790 site of EGFR, secondary mutations in gatekeeper FGFR3 residue have also been reported in response to FGFR inhibition.
Resistant mechanisms are not limited to on‐target secondary mutations but can also involve bypassing mechanisms [124, 125, 126]. In the case of FGFR3‐mutated BLCA under FGFR inhibition therapy, cells may find ways to bypass signaling through EGFR in an AKT‐dependent manner [127].
Moreover, FGFR3 has a pivotal role in shaping the TME landscape and, as such, response to therapies that impact TME.
In the following paragraphs, we delve into some recent preclinical and clinical highlights of discoveries of mechanisms that contribute to mutated FGFR3 BLCA resistance to approved targeted therapies.
3.1. Mutated FGFR3 and TME
Before introducing new cancer immunotherapy approaches, treatment options in BLCA were mainly limited to platinum‐based chemotherapeutics [2, 128]. One such approach involves immune checkpoint inhibitors, which block PD‐1 on T‐cells or the associated PD‐L1 on tumor cells. This enables the human immune system to restore its capacity to recognize and further target cancer cells. Additionally, Bacillus Calmette‐Guérin (BCG) therapy is another form of classical immunotherapy that is a standard of care for non‐muscle‐invasive bladder cancer [6]. Immunotherapies offer encouraging outcomes in a group of BLCA responder patients [2]. However, these therapies are also doomed to a rise of resistance, and predicting the responders’ subpopulations is still a challenge [36, 129].
Overall, immune checkpoint inhibition has emerged as a promising avenue for BLCA patients in addition to classical chemotherapy. However, conflicting insights exist regarding the relevance of Immune checkpoint inhibitors (CPIs) in FGFR3‐mutated BLCAs, and such a dilemma has inspired further studies [17, 130, 131].
Drawing inspiration from clinical findings that suggest an association between FGFR3 mutations and the immune desert microenvironment, preclinical studies have been undertaken to uncover related mechanisms [132]. BLCA mouse models with mutated FGFR3 have corroborated the association between mutated FGFR3 and a cold TME [26, 132, 133].
Recently, Ouyang et al. [132], through single‐cell RNA sequencing of FGFR3‐mutated BLCA in mice, have discovered that corresponding macrophages exhibit immune‐inert phenotypes. The observed phenotype was associated with serine synthesis in FGFR3‐mutated BLCA cells and was PI3K/AKT pathway‐dependent in macrophages [132]. The authors showed that PI3K inhibition reverses the immune‐inert phenotype and further synergizes with Erdafitinib in exerting an anti‐BLCA tumor effect in mice [132]. An intriguing question regarding these findings is how much of the observed effect could be independent of cold TME reversal and correspond directly to the effect of PI3K inhibition on BLCA tumor cells.
Along with preclinical and clinical studies focusing on unveiling the impact of FGFR3 on TME, a significant study by Jing et al. [134] uncovered a critical aspect of FGFR3 and, consequently, its inhibition on TME. The authors discovered in a bladder cancer mouse model that erdafitinib and infigratinib, despite effectively slowing down tumor growth, concurrently mediate the upregulation of PD‐L1 on tumor cells [134]. The authors found that active FGFR3 in BLCA is associated with the downregulation of PD‐L1 [134]. Notably, in BLCAs, upon FGFR3 inhibition, the upregulation of PD‐L1 hampers the antitumor activity of CD8+ T‐cells [134]. Mechanistically, FGFR3 plays a crucial role in regulating PD‐L1 ubiquitination through its binding to and phosphorylating the ubiquitin E3 ligase NEDD4, thereby enhancing its E3 ligase activity [134]. NEDD4, in turn, targets PD‐L1 and catalyzes its K48‐linked polyubiquitination [134]. Combining FGFR3 and PD‐1 inhibition synergistically improved the overall antitumor effect [134].
Caution is advised when directly extrapolating these preclinical findings to clinical scenarios.
Okato et al. [133] from the Kim lab have recently illuminated intriguing aspects of mutated FGFR3 and its influence on urothelial carcinoma TME. Developing an innovative transgenic murine model, they conditionally induced urothelial carcinoma through the concurrent expression of the human equivalent of T5P3R273H and FGFR3S249C. The authors found that tumors in this model exhibit high‐grade NMIBC features and, in terms of transcriptome signature, could show similarities to UROMOL class I and even LumP, mirroring their human counterparts [133]. Okato et al. [133] discovered that these urothelial carcinomas exhibited an intermediate T‐cell inflamed phenotype [135]. Upon anti‐PD1 treatment, tumors demonstrated hyperprogression, possibly attributable to the abundance of T‐regulatory cells (Tregs) within the TME [136].
Interestingly, the authors demonstrated that Erdafitinib treatment could suppress the proliferation of corresponding Treg cells in vitro and even better in vivo by targeting FGFR1 on the surface of these cells [133]. Ultimately, combining Erdafitinib with anti‐PD1 showed greater therapeutic efficacy than erdafitinib monotherapy alone [133]. Consequently, the study by Okato et al. [133] presents the non‐selectivity of erdafitinib in targeting FGFR receptors as a blessing in disguise that may extend erdafitinib's application as a Treg‐suppressor agent beyond FGFR3‐mutant BLCAs. In line with these findings, the study by Shi et al. [26] is noteworthy as it also found that FGFR3‐mutated BLCA in their mouse model exhibited a “cold” TME characterized by reduced CD8+ T cell infiltration compared to carcinogen‐induced tumors.
A reduced T‐cell infiltration and an immunologically inert microenvironment may be associated with FGFR3 alterations in BLCA [12, 137]. As such, the potential resistance of FGFR3‐mutated BLCAs to CPIs is a dilemma.
In a recent clinical trial study [138], the authors delineated MIBC subtypes by integrating the tumor's gene expression and genetic profiles, TME, and the response to CPIs. The authors discovered that genetic alterations in FGFR3 were notably enriched within a non‐responder subtype [138]. Subsequent preclinical benchwork revealed that the histone demethylase KDM5B is accountable for creating a cold TME, and inhibiting both KDM5B and mutant FGFR3 may potentially reinstate a hot immune phenotype [138].
In search for comprehensive prognosis‐related immune profiling of BLCAs, Xu et al. [139] assessed immune components within tumors in TCGA while acknowledging FGFR3 status in their analysis. The authors categorized two risk groups, low and high, based on immune profiles, with the high‐risk group characterized by elevated levels of neutrophils, macrophages, follicular helper cells, CD4 and CD8 T‐cells, as well as increased expression of immune checkpoint markers such as PD‐L1, PD‐1, CTLA‐4, LAG‐3, and TIM‐3. Notably, the high‐risk tumors were associated with a reduced frequency of FGFR3 mutations [139].
Exploring whether the abundance of infiltrated T‐cells alone can be a reliable prognostic and predictive marker extends beyond FGFR3 mutations and BLCA.
A significant study by Wang et al. [131] sheds light on this matter in BLCA, providing evidence that the interplay between T‐cell infiltration and stroma‐associated epithelial‐mesenchymal transition (EMT) markers in the TME collectively establishes predictive value for the response to CPIs. The same group further delves into unraveling the relationship between the response to CPIs and the FGFR3 status based on two preceding clinical cohorts [140]. They found no significant difference between FGFR3‐mutated and wild‐type patients regarding the response to immunotherapy [140]. Despite observing a decrease in the T‐cell component, they note that tumors with FGFR3 alterations exhibit lower expression of TGF‐B in stromal components [140]. According to their previous findings in BLCA and other reports on different cancer types, the low TGF‐B expression in a tumor counteracts the cold TME, which is associated with low T‐cell infiltration within the tumor [140]. Therefore, low TGF‐B expression counteracts the lack of response to CPIs, thereby rendering these tumors with supposedly cold TME responsive to CPIs [140]. Unlike T‐cell status, lower TGF‐B expression is a positive predictive marker for CPIs [131]. As such, Wang et al. [140] hypothesize that the cumulative impact of FGFR3 mutations on T‐cell abundance and the presence of stroma‐related EMT lies behind consistent response among two distinctive FGFR3 statuses. Of note, independent clinical studies have corroborated Wang's hypothesis. For instance, in a real‐world comparison of response to CPIs among FGFR3‐altered (n = 17) vs. wild‐type (n = 86) metastatic BLCAs, Rose et al. [130] found no significant difference in clinical outcomes. Moreover, they concluded that the sum of T cell gene expression status, immunosuppressive stroma, and related markers might be determinant of response to CPIs.
Consistent with Wang et al. findings [140], the EMT signatures were significantly down‐regulated among FGFR3‐altered samples in Komura et al. study [17]. This study analyzed samples from 72 patients included in CPI therapy, and distinctive immune microenvironments between FGFR3 mutant vs. wild‐type tumors were discovered [17]. In line with previous clinical and preclinical reports, Komura et al. [17] also found different markers of activated TME being associated with FGFR3 intact tumors as opposed to FGFR3 altered patients. Moreover, they uncovered significant heterogeneity among FGFR3‐altered patients and corresponding subtypes concerning immune cell components and response to CPIs. Importantly, in line with Guercio et al.’s report [23] and Wang et al.’s hypothesis [140], Komura et al.’s study [17] does not find an inverse relationship between FGFR3 mutations and response to CPIs.
Indeed, while Komura et al. [17] findings of lower T‐cell infiltration among FGFR3‐mutated BLCAs could suggest a lower response to CPIs, overall, no significant difference was observed between the two FGFR3 statuses. Moreover, when responses to CPI were stratified into subtypes, unlike presumptions, the LumP subtype with FGFR3 mutations showed ∼50% ORR vs. ∼5% among the same subtype with wild‐type FGFR3 [17].
Further substantial evidence corroborating the effectiveness of CPIs in FGFR3‐mutated BLCA is derived from cohort 2 of the phase III THOR trial [37]. Although in this trial the pembrolizumab arm (n = 176) did not demonstrate a more favorable outcome with statistical significance than the erdafitinib arm (n = 175) [37].
3.2. Resistance mechanisms to mutated FGFR3 inhibition
Due to evidence on FGFR3‐mutated BLCA addiction to this oncogene, interest had risen towards FGFR3 inhibition as a therapeutic approach and further exploration of related resistance mechanisms. In 2013, Herrera‐Abreu et al. [126] conducted a parallel RNA interference kinome/phosphatome screen in five cell lines carrying FGFR3 fusion or point mutations to identify potential co‐targets alongside FGFR3 inhibition. The cell line panel under examination comprised both cell types demonstrating partial sensitivity to FGFR3 inhibition and those intrinsically resistant to FGFR3 inhibition but reliant on EGFR [126]. Intriguingly, EGFR emerged as a co‐target in both cell types. Even in cell lines inherently resistant to FGFR3, a notable sensitivity to combined EGFR/FGFR3 targeting was observed [126]. Later, a kinome‐wide shRNA screen reported both EGFR and PI3K as co‐targets with fusion FGFR3 in RT112 cells [127].
Another recent study [141] has revealed a resistance mechanism to erdafitinib in BLCA cells harboring the fusion FGFR3‐TACC3. The authors discovered that Erdafitinib exerts cytotoxic effects on these cells by inducing incomplete autophagy and elevating reactive oxygen species (ROS) levels [141]. However, resistance could develop due to the activity of P4HA2, which stabilizes its positive transcriptional regulator, HIF‐1α, whose signaling output, in turn, yields reduced lethal ROS levels and, consequently, counteracts the cytotoxic effects of erdafitinib [141].
Recently, Pettitt et al. [142] have published their findings on the attempt to unravel resistance mechanisms in two BLCA cellular models with native fusion FGFR3‐TACC3 mutation. They generated several resistant subclones from parental cells primarily sensitive to FGFR3 inhibition (PD173074) [142]. They addressed resistance mechanisms by whole exome sequencing and bulk‐RNA sequencing. Their findings imply the emergence of very heterogeneous phenotypic switches [142]. Moreover, even if a homogenous upregulation of a gene was identified, it did not sensitize all sub‐clones to targeting that gene [142]. They identified an irreversible resistance mechanism caused by HRASG12S in only one subclone. Still, 10% of cells remained HRAS wild type in that subclone [142]. Of note, unlike the preclinical and clinical reports on the rise of secondary mutations within the FGFR3 gatekeeper or molecular break residue in BLCA cells with FGFR3 point mutations, no such secondary mutations were identified in the Pettitt et al. study [142]. All resistant derivatives showed upregulated expression of YAP/TAZ targets [142]. However, upregulation was associated with sensitivity to the YAP inhibitor CA3 in only one cell line [142]. Resistance could also be accompanied by upregulation of EGFR, ERBB2, ERBB3, and MET [142]. However, targeting these proteins did not restore sensitivity to FGFR3 inhibition [142]. These results contradict Weickhardt et al.’s findings [124], where co‐targeting ERBB3 potentiated the effect of FGFR inhibition in BLCA cells with acquired resistance to FGFR inhibition. Of note, two cellular models in Weickhardt et al. also harbored FGFR3 fusion. However, a different FGFR inhibitor (BGJ398: Infigratinib) was tested in the Weickhardt et al. study [124].
In two subclones of the same parental cell line, upregulated expression of IGF1R was associated with response to sunitinib [142]. Other signature modulations in resistant subclones included cell motility and adhesion, IFNγ response, and EMT signature [142]. One needs to consider that clinical reports about resistance mechanisms to FGFR inhibition among BLCA patients come from cohorts enriched with point mutations rather than FGFR3 fusions (reviewed below). Therefore, mutation‐specific resistance may lie behind the discrepancy between Pettitt et al. [142] and recent clinical findings [23, 143].
A recent study by Hosni et al. [125] shows that neuregulin 1 (NRG1) secreted from a subset of stromal cells, the undifferentiated adipocyte precursors, mediates resistance to erdafitinib in FGFR3‐mutated BLCA mouse xenografts in a HER3‐dependent manner [125]. As opposed to Pettitt et al. study [142], HER3 (ERBB3) targeting by Pertuzumab could restore sensitivity to FGFR3 inhibition (Erdafitinib [125] vs. PD173074 [142]), and combined erdafitinib and pertuzumab treatment led to increased survival in mice with FGFR3‐mutated BLCA xenografts [125].
In a hypothesis‐driven approach, Wang et al. [144] have recently demonstrated that quisinostat, an experimental histone deacetylase inhibitor, synergizes with Erdafitinib through suppressing FGFR3 protein translation and heparin‐binding growth factor, a BLCA‐relevant mitogen.
Often, BLCA patients of both categories who relapse on other therapeutics would be eligible candidates for treatment with Erdafitinib [2, 34‐38, 128]. Despite the initially encouraging outcomes, up to 45.6% ORR [143], and a median PFS of up to 5.6 months upon erdafitinib treatment [34, 143], justify a clear need to enhance the effectiveness of these treatments.
A comparison between erdafitinib and pembrolizumab in patients with advanced or metastatic urothelial carcinoma carrying FGFR3 mutations has been conducted [37]. Although the study revealed no statistically significant difference in overall survival between erdafitinib and pembrolizumab arms, the OS in the immunotherapy arm was slightly higher (median OS of 10.9 and 11.1 months, respectively, hazard ratio 1.18; 95% confidence interval 0.92‐1.51; P = 0.18) [37]. The median PFS for erdafitinib was 4.4 months, compared to 2.7 months for pembrolizumab (hazard ratio 0.88; 95% CI 0.70‐1.10) [37]. The objective response rate (ORR) was 40% for erdafitinib and 21.6% for pembrolizumab, with a relative risk of 1.85 (95% CI 1.32‐2.59) [37]. The median duration of response was 4.3 months for erdafitinib and 14.4 months for pembrolizumab [37].
In a recent study conducted by Facchinetti et al. [143], researchers examined the tissue and circulating tumor DNA (ctDNA) of twenty‐one BLCA patients with FGFR mutations who had progressed on different FGFR inhibitors (19 out of 21 had FGFR3 mutations) [143]. The study revealed that secondary FGFR3 mutations within its kinase domain were present in 37% of patients who developed resistance to various FGFR inhibitors [143]. Two critical resistance spots, among others, were identified: the gatekeeper residue FGFR3 V555 and the molecular brake N540, which play a crucial role in drug binding to the FGFR3 molecule [143]. These secondary mutations reduce the targeted FGFR3 affinity for the corresponding compounds [143]. Furthermore, alongside secondary FGFR3 mutations, the study identified genetic alterations among the PI3K–mTOR pathway effectors [143]. The EGFR hyperphosphorylation could also be associated with resistance to FGFR inhibition in a patient [143]. Co‐targeting PI3K (by pictilisib) and EGFR (with gefitinib) in the presence of corresponding alterations was shown to lead to enhanced inhibitory effects in FGFR3‐mutated patient‐derived xenograft (PDX) model [143]. These results are in line with those of Lang et al. [145] who report that treatment of FGFR3‐mutant PDX models of MIBC and upper urinary tract urothelial carcinoma with combined FGFR/EGFR inhibitors was more efficient than anti‐FGFR3 treatment alone.
Facchinetti et al. [143] evaluated various selective FGFR3 inhibitors in a Ba/F3 cellular model, revealing that erdafitinib exhibited a better potency against cells expressing FGFR3:TACC3 with both wild‐type or mutant FGFR3 kinase domain.
An essential point that the study of Facchinetti et al. [143] prompts our attention to is the fact that several patients developed resistance to FGFR3 inhibition without a clear molecular explanation.
Guercio et al. [23] prospectively analyzed collected patient data co‐mapped with respected NGS data in one of the largest FGFR2/FGFR3‐altered BLCA cohorts, comprising 414 annotated samples. While male patients exhibited a higher frequency in both FGFR3 wild‐type and mutant BLCA cases, female patients demonstrated a higher frequency in the mutant group than in the wild‐type group [23]. It deserves reiteration that in line with Wang et al.’s hypothesis [140] and contrary to previous assumptions regarding FGFR3‐mutated tumors’ lack of responsiveness to CPIs, Guercio et al. did not observe such an inverse relationship [23].
The FGFR3 status was dynamic according to the disease progression [23]. The FGFR3 alterations were more frequent among NMIBC and upper tract lesions than MIBC or metastatic samples [23]. Of note, among 27 patients with existing matched metastatic tumor samples, seven patients were identified with the same FGFR3 alteration, which was limited to either the primary or the metastatic tumor and not both [23]. An important conclusion that can be drawn from this finding is that considering the primary tumor's FGFR3 status for therapeutic decisions can be misleading. Guided by early NGS results, the clinician might treat a non‐existing altered FGFR3 entity in a metastatic patient [23].
Recurrent mutations in cell cycle genes CDKN2A and CDKN1A were observed alongside FGFR3 mutations [23]. Genetic alterations in ERBB2, TP53, and RB1 were found to show a mutually exclusive trend with FGFR3 mutations, whereas CDKN2A, CDKN2B, and KDM6A displayed cooccurring tendencies [23]. In line with some other cancer types and therapeutic contexts [146], TP53 mutations at the baseline were associated with poorer clinical outcomes in FGFR3‐altered patients [23]. Notably, baseline alterations in PIK3CA, TSC1, and ERBB2 were not associated with further resistance to erdafitinib [23].
Interestingly, Pettitt et al. [142] also discovered that within subclones with acquired resistance to erdafitinib, ERBB2 overexpression did not sensitize to ERBB2 targeting.
Thirty‐two metastatic BLCA patients (FGFR3/2 altered) treated with erdafitinib were examined in the respective cohort [23]. Among them, before treatment, 87% harbored a point mutation, predominantly the FGFRS249C variant (59%), 13% had FGFR3‐TACC3 fusions, and patients with Y373C and R248C variants each exhibited 9% frequency [23]. Interestingly, all three patients with FGFR3Y373C showed objective responses [23]. FGFR3‐fusion patients showed poorer response to erdafitinib; three of four showed disease progression as the best response [23]. The response rate to erdafitinib was 40%, the overall survival was six months, and the median progression‐free survival was 2.8 months [23].
Like the renowned mechanism of acquired resistance to anti‐EGFR therapies, namely secondary T790M at the EGFR gatekeeper residue [147], Guercio et al. [23] found secondary FGFR3 mutations potentially involving similar FGFR3 residues and negatively affecting the drug binding in response to erdafitinib treatment. These secondary mutations (along with other mutations) found in two patients were N540S or V553M [23].
Interestingly, the study [23] reports acquired TP53 mutations among five patients in response to erdafitinib. Considering the association of TP53 mutations at baseline with a poorer response to erdafitinib, one might question the inclusion of patients with concurrent FGFR3 and TP53 mutations in FGFR3‐targeted therapies. However, perhaps we need to learn more about the role of TP53 mutations when they cooccur with FGFR3 mutations. The TP53 mutations have shown a trend of mutual exclusivity with FGFR3 mutations [3, 23], while they can still cooccur. Concurrent TP53 and FGFR3 mutations do not necessarily render BLCA cells tolerant to FGFR3 inhibition [148]. An example of this notion is the high sensitivity of UMUC14 cells to FGFR3 inhibition [148], which concomitantly harbors FGFR3S249C and T5P3R2807, a variant linked to cooperation with the AKT pathway [149]. The TP53 mutations are distinctive regarding loss of function, dominant negative effect, and even gain of function phenotypes [150, 151, 152, 153, 154]. Moreover, at least one preclinical study has shown that combined PD‐1 and FGFR inhibition may lead to enhanced therapeutic effects in transgenic mice with concomitant FGFR3 and TP53 mutations in their BLCA tumors [133]. As such, more detailed studies are needed to unveil the differential impact of distinctive TP53 variants on FGFR3 signaling and vulnerability to FGFR3 targeted therapies.
In one patient, an AKT1 mutation was found after Erdafitinib therapy. Overall, secondary mutations were observed in six patients. An intriguing question remains concerning other patients who also develop resistance, with no genetic findings [23].
Enfortumab vedotin (EV) is an antibody‐drug conjugate (ADC) targeting Nectin‐4, a cell adhesion molecule expressed in BLCA cells [155, 156]. Its mechanism involves the monoclonal antibody binding to Nectin‐4 on cancer cells, leading to internalization and release of the cytotoxic monomethyl auristatin E, which disrupts the microtubule network, inducing cell cycle arrest and apoptosis [155, 156].
Erdafitinib and EV have distinct mechanisms of action and distinct toxicity profiles, providing a rationale for evaluating their combination in metastatic urothelial carcinoma patients with FGFR2/3 genetic alterations. An ongoing phase I dose‐escalation and expansion study is assessing the safety, pharmacokinetics, and antitumor activity of combining erdafitinib (8 mg/day) with EV (1 or 1.25 mg/kg) in patients progressing after platinum‐based chemotherapy and/or PD‐1/L1 inhibitors [157]. Preliminary data from the first two dose levels indicate the combination is feasible, with common toxicities such as hyperphosphatemia. All evaluable patients achieved a partial response. Dose escalation continues to identify the maximum tolerated dose for the expansion phase [157].
Table 1 summarizes the key preclinical and clinical associations with FGFR3 mutations and targeting.
TABLE 1.
Overview of recent key preclinical and clinical findings on FGFR3 mutations in BLCA.
| Recent findings on FGFR3 mutations in BLCA | Preclinical | Clinical |
|---|---|---|
| Related to TME and CPIs | ||
| Immune‐inert phenotype in FGFR3 mutated tumors in mice | ✓ | |
| Downregulation of PD‐L1 in FGFR3 active status | ✓ | |
| Impact on anti‐tumor activity of CD8+ T‐cells | ✓ | |
| Improved antitumor effect upon combining FGFR3 and PD‐1 targeting | ✓ | |
| Reduced T‐cell infiltration and inert microenvironment associated with FGFR3 mutations in BLCA patients | ✓ | |
| FGFR3 mutation impacts response to CPIs by both T‐cell infiltration and stroma‐associated EMT markers | ✓ | |
| Cold tumors not always in inverse relationship with responsiveness to CPIs in FGFR3‐altered BLCA patients | ✓ | |
| Related to FGFR3 inhibition and resistance | ||
| EGFR and PI3K identified as co‐targets | ✓ | |
| Sensitivity in FGFR3 resistant cell lines when co‐targeting EGFR | ✓ | |
| bypassing mechanisms through effectors of AKT pathway | ✓ | |
| Heterogenous phenotypic switches in FGFR3‐resistant subclones derived from the same cell | ✓ | |
| Upregulation of various genes including IGF1R, EGFR, ERBB2, ERBB3, and MET | ✓ | |
| Quisinostat synergizes with Erdafitinib by suppressing FGFR3 protein translation | ✓ | |
| Resistance to Erdafitinib in FGFR3 mutant cells induced by P4HA2 and HIF‐1α | ✓ | |
| NRG1‐HER3 axis mediates resistance to Erdafitinib and sensitizes to HER3‐targeting in FGFR3 mut BLCA mouse xenografts | ✓ | |
| Secondary mutations at gatekeeper residue sites | ✓ | |
| HRAS mutation was found in some FGFR3‐resistant sub‐clones | ✓ | |
| Resistance hotspot mutations on residues V555 and N540 | ✓ | |
| Genetic alterations within AKT pathway effectors | ✓ | |
| Poorer outcome upon baseline co‐occurrence of FGFR3 and TP53 mutations | ✓ |
Different phenomena discovered preclinically or clinically concerning the impact of FGFR3 alterations on TME, response to CPIs, and response and resistance to FGFR inhibition are summarized.
Abbreviations: BLCA, Bladder Cancer; CPIs, Checkpoint Inhibitors; FGFR3, Fibroblast Growth Factor Receptor 3; FGFR, Fibroblast Growth Factor Receptor; TME, Tumor Microenvironment.
4. DISCUSSION AND CONCLUSIONS
As the revisit of recent findings in this writing suggests, FGFR3 mutations in BLCA may not serve as conclusive predictive markers for lack of response to CPIs. Considering the EMT status of the TME and the tumor subtype, more sophisticated algorithms are needed to establish effective consensus for FGFR3‐mutated BLCA response to CPIs.
Moreover, a combination of FGFR1/3 inhibition and CPIs holds promise for further clinical exploration in BLCA.
The state of the art regarding the predictive and prognostic value of mutational profile of FGFR3 altered tumors is still in its infancy. An important lesson from the study by Guercio et al. [23] is that an updated NGS is essential before starting the FGFR3 inhibition therapy.
Whether innate or acquired, resistance to FGFR3 inhibitors in BLCA is not an isolated phenomenon limited to FGFR3 inhibitors targeting BLCA or even cancer therapy. We have drawn valuable lessons from the HIV field, where combined treatments, referred to as “drug cocktails,” have led to more favorable and durable clinical outcomes [158]. In the realm of cancer targeting, mainly since the New Era of Personalized Medicine in 1999 [159], the focus has primarily been on exploring monotherapies, with a recurrent pursuit of more effective combined treatments. One prominent example of such an approach is the combined inhibition of RAF and MEK in targeting BRAFV600E mutant melanoma [160]. It is worth noting that upfront combinatorial treatments, if tolerable, not only may enhance and deepen the initial response but also have the potential to significantly delay the emergence of acquired resistance over the long term. For instance, the combination of EGFR and VEGF targeting demonstrated a delay in developing secondary T790M EGFR mutations and enhanced the response in EGFR L858R/T790M‐positive cancer cells [161]. Since then, more evidence has supported the argument of upfront combinatorial treatments in EGFR mutant cancer [162] and beyond [163]. Piro Lito introduced the fitness threshold model, which suggests that cancer cells exposed to combinatorial treatments with a higher threshold, as opposed to monotherapy, may be at a greater risk of experiencing unfavorable outcomes [164].
In 2009, Nobel Prize laureate William G. Kaelin Jr. adopted Theodosius Dobzhansky's concept of synthetic lethality as a conceptual framework for cancer target discovery [165]. This approach has effectively widened targeting options in all areas of cancer. As such, exploring synthetically lethal (co)targets with FGFR3 holds promise in BLCA. As the revisit of precedent studies into unraveling different resistance mechanisms in FGFR3‐mutated BLCA treated with different FGFR inhibitors revealed, targeting the differentially expressed genes has not often proven effective in producing a meaningful response. It is worth noting that genetic screens, such as CRISPR screens, can be highly beneficial in tackling the significant heterogeneity observed in resistance mechanisms against FGFR inhibition by directly discovering valid co‐targets.
An intriguing aspect of both preclinical and clinical studies aiming to unravel the resistance mechanism to FGFR3 inhibition is the absence of findings at the genotype or transcriptome level in a subset of samples. Hence, the question could be investigated by delving into less‐explored facets of FGFR3's direct or indirect influence, such as its interactions with non‐coding RNAs.
In summary, mutant FGFR3 has demonstrated its significance as an oncogenic element and a promising target in BLCA. On the other hand, short‐lasting clinical benefits and the rise of resistance in all patients upon FGFR3 inhibition monotherapy underscore the need for the discovery of FGFR3 co‐targets and the uncovering of its unexplored functions and vulnerabilities in BLCA.
AUTHOR CONTRIBUTIONS
MN and MP conceived and conceptualized the concept of this writing and authored the manuscript with all other co‐authors’ scientific and authoring contributions. KK generated most of the figures with contributions and supervision from MN and MP. All authors critically revised the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING
Not applicable.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
ACKNOWLEDGMENTS
Not applicable.
Noeraparast M, Krajina K, Pichler R, Niedersüß‐Beke D, Shariat SF, Grünwald V, et al. FGFR3 alterations in bladder cancer: Sensitivity and resistance to targeted therapies. Cancer Commun. 2024;44:1189–1208. 10.1002/cac2.12602
Contributor Information
Maxim Noeraparast, Email: maxim.noeparast@gmail.com.
Martin Pichler, Email: Martin.Pichler@uk-augsburg.de.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Tran L, Xiao J‐F, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104‐21. 10.1038/s41568-020-00313-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dyrskjøt L, Hansel DE, Efstathiou JA, Knowles MA, Galsky MD, Teoh J, et al. Bladder cancer. Nat Rev Dis Prim. 2023;9:58. 10.1038/s41572-023-00468-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ascione CM, Napolitano F, Esposito D, Servetto A, Belli S, Santaniello A, et al. Role of FGFR3 in bladder cancer: Treatment landscape and future challenges. Cancer Treat Rev. 2023;115:102530. 10.1016/j.ctrv.2023.102530 [DOI] [PubMed] [Google Scholar]
- 4. The American Cancer Society medical and editorial content team . Can bladder cancer be found early? Am Cancer Soc. 2016:1‐25. [Google Scholar]
- 5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7‐34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 6. Williams SB, Howard LE, Foster ML, Klaassen Z, Sieluk J, De Hoedt AM, et al. Estimated Costs and Long‐term Outcomes of Patients With High‐Risk Non‐Muscle‐Invasive Bladder Cancer Treated With Bacillus Calmette‐Guérin in the Veterans Affairs Health System. JAMA Netw Open. 2021;4:e213800. 10.1001/jamanetworkopen.2021.3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre‐Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18‐20. 10.1038/12615 [DOI] [PubMed] [Google Scholar]
- 8. Bernard‐Pierrot I, Brams A, Dunois‐Lardé C, Caillault A, Diez de Medina SG, Cappellen D, et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006;27:740‐7. 10.1093/carcin/bgi290 [DOI] [PubMed] [Google Scholar]
- 9. Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007;26:5889‐99. 10.1038/sj.onc.1210399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams S V, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795‐803. 10.1093/hmg/dds486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315‐22. 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, et al. A Consensus Molecular Classification of Muscle‐invasive Bladder Cancer. Eur Urol. 2020;77:420‐33. 10.1016/j.eururo.2019.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pal SK, Rosenberg JE, Hoffman‐Censits JH, Berger R, Quinn DI, Galsky MD, et al. Efficacy of BGJ398, a Fibroblast Growth Factor Receptor 1‐3 Inhibitor, in Patients with Previously Treated Advanced Urothelial Carcinoma with FGFR3 Alterations. Cancer Discov. 2018;8:812‐21. 10.1158/2159-8290.CD-18-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loriot Y, Necchi A, Park SH, Garcia‐Donas J, Huddart R, Burgess E, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med. 2019;381:338‐48. 10.1056/NEJMoa1817323 [DOI] [PubMed] [Google Scholar]
- 15. Volkmer J‐P, Sahoo D, Chin RK, Ho PL, Tang C, Kurtova A V, et al. Three differentiation states risk‐stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci U S A. 2012;109:2078‐83. 10.1073/pnas.1120605109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hedegaard J, Lamy P, Nordentoft I, Algaba F, Høyer S, Ulhøi BP, et al. Comprehensive Transcriptional Analysis of Early‐Stage Urothelial Carcinoma. Cancer Cell. 2016;30:27‐42. 10.1016/j.ccell.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 17. Komura K, Hirosuna K, Tokushige S, Tsujino T, Nishimura K, Ishida M, et al. The Impact of FGFR3 Alterations on the Tumor Microenvironment and the Efficacy of Immune Checkpoint Inhibitors in Bladder Cancer. Mol Cancer. 2023;22:185. 10.1186/s12943-023-01897-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole‐genome and whole‐exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459‐63. 10.1038/ng.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Webster MK, Donoghue DJ. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 1997;13:178‐82. 10.1016/s0168-9525(97)01131-1 [DOI] [PubMed] [Google Scholar]
- 20. Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233‐7. 10.1038/ng0696-233 [DOI] [PubMed] [Google Scholar]
- 21. Vaeyens F, Hetzel J‐P, Mernberger M, Eggermont C, Olsen C, Maes K, et al. Variant‐Specific Landscape of Mutual Exclusivity Among BRAF, EGFR, and KRAS Oncogenes in Human Cancer. MedRxiv. 2023:23297089. 10.1101/2023.10.21.23297089 [DOI] [Google Scholar]
- 22. Shi M‐J, Meng X‐Y, Lamy P, Banday AR, Yang J, Moreno‐Vega A, et al. APOBEC‐mediated Mutagenesis as a Likely Cause of FGFR3 S249C Mutation Over‐representation in Bladder Cancer. Eur Urol. 2019;76:9‐13. 10.1016/j.eururo.2019.03.032 [DOI] [PubMed] [Google Scholar]
- 23. Guercio BJ, Sarfaty M, Teo MY, Ratna N, Duzgol C, Funt SA, et al. Clinical and Genomic Landscape of FGFR3‐Altered Urothelial Carcinoma and Treatment Outcomes with Erdafitinib: A Real‐World Experience. Clin Cancer Res an Off J Am Assoc Cancer Res. 2023;29:4586‐95. 10.1158/1078-0432.CCR-23-1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. di Martino E, L'Hôte CG, Kennedy W, Tomlinson DC, Knowles MA. Mutant fibroblast growth factor receptor 3 induces intracellular signaling and cellular transformation in a cell type‐ and mutation‐specific manner. Oncogene. 2009;28:4306‐16. 10.1038/onc.2009.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tomlinson DC, Baldo O, Harnden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007;213:91‐8. 10.1002/path.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi M‐J, Fontugne J, Moreno‐Vega A, Meng X‐Y, Groeneveld C, Dufour F, et al. FGFR3 Mutational Activation Can Induce Luminal‐like Papillary Bladder Tumor Formation and Favors a Male Sex Bias. Eur Urol. 2023;83:70‐81. 10.1016/j.eururo.2022.09.030 [DOI] [PubMed] [Google Scholar]
- 27. Ahmad I, Singh LB, Foth M, Morris C‐A, Taketo MM, Wu X‐R, et al. K‐Ras and β‐catenin mutations cooperate with Fgfr3 mutations in mice to promote tumorigenesis in the skin and lung, but not in the bladder. Dis Model Mech. 2011;4:548‐55. 10.1242/dmm.006874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krook MA, Reeser JW, Ernst G, Barker H, Wilberding M, Li G, et al. Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br J Cancer. 2021;124:880‐92. 10.1038/s41416-020-01157-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie Y, Su N, Yang J, Tan Q, Huang S, Jin M, et al. FGF/FGFR signaling in health and disease. Signal Transduct Target Ther. 2020;5:181. 10.1038/s41392-020-00222-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subbiah V, Verstovsek S. Clinical development and management of adverse events associated with FGFR inhibitors. Cell Reports Med. 2023;4:101204. 10.1016/j.xcrm.2023.101204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siefker‐Radtke AO, Necchi A, Park SH, García‐Donas J, Huddart RA, Burgess EF, et al. Efficacy and safety of erdafitinib in patients with locally advanced or metastatic urothelial carcinoma: long‐term follow‐up of a phase 2 study. Lancet Oncol. 2022;23:248‐58. 10.1016/S1470-2045(21)00660-4 [DOI] [PubMed] [Google Scholar]
- 32. Hanna K. Erdafitinib's Road to Approval and Use in Urothelial Carcinoma. Oncology (Williston Park). 2023;37:260‐1. 10.46883/2023.25920999 [DOI] [PubMed] [Google Scholar]
- 33. Pant S, Schuler M, Iyer G, Witt O, Doi T, Qin S, et al. Erdafitinib in patients with advanced solid tumours with FGFR alterations (RAGNAR): an international, single‐arm, phase 2 study. Lancet Oncol. 2023;24:925‐35. 10.1016/S1470-2045(23)00275-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loriot Y, Matsubara N, Park SH, Huddart RA, Burgess EF, Houede N, et al. Erdafitinib or Chemotherapy in Advanced or Metastatic Urothelial Carcinoma. N Engl J Med. 2023;389:1961‐71. 10.1056/NEJMoa2308849 [DOI] [PubMed] [Google Scholar]
- 35. Catto JWF, Tran B, Rouprêt M, Gschwend JE, Loriot Y, Nishiyama H, et al. Erdafitinib in BCG‐treated high‐risk non‐muscle invasive bladder cancer. Ann Oncol Off J Eur Soc Med Oncol. 2023. 10.1016/j.annonc.2023.09.3116 [DOI] [PubMed] [Google Scholar]
- 36. Siefker‐Radtke AO, Matsubara N, Park SH, Huddart RA, Burgess EF, Özgüroğlu M, et al. Erdafitinib versus pembrolizumab in pretreated patients with advanced or metastatic urothelial cancer with select FGFR alterations: cohort 2 of the randomized phase III THOR trial. Ann Oncol Off J Eur Soc Med Oncol. 2024;35:107‐17. 10.1016/j.annonc.2023.10.003 [DOI] [PubMed] [Google Scholar]
- 37. Siefker‐Radtke AO, Matsubara N, Park SH, Huddart RA, Burgess EF, Özgüroğlu M, et al. Erdafitinib versus pembrolizumab in pretreated patients with advanced or metastatic urothelial cancer with select FGFR alterations: cohort 2 of the randomized phase III THOR trial. Ann Oncol Off J Eur Soc Med Oncol. 2024;35:107‐17. 10.1016/j.annonc.2023.10.003 [DOI] [PubMed] [Google Scholar]
- 38. Catto JWF, Tran B, Rouprêt M, Gschwend JE, Loriot Y, Nishiyama H, et al. Erdafitinib in BCG‐treated high‐risk non‐muscle‐invasive bladder cancer. Ann Oncol Off J Eur Soc Med Oncol. 2024;35:98‐106. 10.1016/j.annonc.2023.09.3116 [DOI] [PubMed] [Google Scholar]
- 39. Vilaseca A, Jayram G, Raventos C, Shore ND, Zainfeld D, Kang TW, et al. LBA104 First safety and efficacy results of the TAR‐210 erdafitinib (erda) intravesical delivery system in patients (pts) with non–muscle‐invasive bladder cancer (NMIBC) with select FGFR alterations (alt). Ann Oncol. 2023;34:S1343. 10.1016/j.annonc.2023.10.110 [DOI] [Google Scholar]
- 40. Kommalapati A, Tella SH, Borad M, Javle M, Mahipal A. FGFR Inhibitors in Oncology: Insight on the Management of Toxicities in Clinical Practice. Cancers (Basel). 2021;13. 10.3390/cancers13122968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Necchi A, Pouessel D, Leibowitz R, Gupta S, Fléchon A, García‐Donas J, et al. Pemigatinib for metastatic or surgically unresectable urothelial carcinoma with FGF/FGFR genomic alterations: final results from FIGHT‐201. Ann Oncol Off J Eur Soc Med Oncol. 2024;35:200‐10. 10.1016/j.annonc.2023.10.794 [DOI] [PubMed] [Google Scholar]
- 42. Rodón J, Damian S, Furqan M, García‐Donas J, Imai H, Italiano A, et al. Pemigatinib in previously treated solid tumors with activating FGFR1‐FGFR3 alterations: phase 2 FIGHT‐207 basket trial. Nat Med. 2024. 10.1038/s41591-024-02934-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao D, Long X, Zhou J, Wang J. Pharmacovigilance Study of Infigratinib: A Safety Analysis of the FDA Adverse Event Reporting System. Drugs R D. 2023;23:403‐9. 10.1007/s40268-023-00439-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meric‐Bernstam F, Hollebecque A, Furuse J, Oh D‐Y, Bridgewater JA, Shimura M, et al. Safety Profile and Adverse Event Management for Futibatinib, An Irreversible FGFR1‐4 Inhibitor: Pooled Safety Analysis of 469 Patients. Clin Cancer Res an Off J Am Assoc Cancer Res. 2024;30:1466‐77. 10.1158/1078-0432.CCR-23-2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Javle M, King G, Spencer K, Borad MJ. Futibatinib, an Irreversible FGFR1‐4 Inhibitor for the Treatment of FGFR‐Aberrant Tumors. Oncologist. 2023;28:928‐43. 10.1093/oncolo/oyad149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vilaseca A, Jayram G, Raventos C, Shore ND, Zainfeld D, Kang TW, et al. LBA104 First safety and efficacy results of the TAR‐210 erdafitinib (erda) intravesical delivery system in patients (pts) with non–muscle‐invasive bladder cancer (NMIBC) with select FGFR alterations (alt). Ann Oncol. 2023;34:S1343. 10.1016/j.annonc.2023.10.110 [DOI] [Google Scholar]
- 47. Goyal R, Jialal I. Hyperphosphatemia. Treasure Island (FL): 2024. [Google Scholar]
- 48. Abou‐Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al‐Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open‐label, phase 2 study. Lancet Oncol. 2020;21:671‐84. 10.1016/S1470-2045(20)30109-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bahleda R, Italiano A, Hierro C, Mita A, Cervantes A, Chan N, et al. Multicenter Phase I Study of Erdafitinib (JNJ‐42756493), Oral Pan‐Fibroblast Growth Factor Receptor Inhibitor, in Patients with Advanced or Refractory Solid Tumors. Clin Cancer Res an Off J Am Assoc Cancer Res. 2019;25:4888‐97. 10.1158/1078-0432.CCR-18-3334 [DOI] [PubMed] [Google Scholar]
- 50. Dosne A‐G, Valade E, Stuyckens K, De Porre P, Avadhani A, O'Hagan A, et al. Erdafitinib's effect on serum phosphate justifies its pharmacodynamically guided dosing in patients with cancer. CPT Pharmacometrics Syst Pharmacol. 2022;11:569‐80. 10.1002/psp4.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martínez‐Torrecuadrada J, Cifuentes G, López‐Serra P, Saenz P, Martínez A, Casal JI. Targeting the Extracellular Domain of Fibroblast Growth Factor Receptor 3 with Human Single‐Chain Fv Antibodies Inhibits Bladder Carcinoma Cell Line Proliferation. Clin Cancer Res. 2005;11:6280‐90. 10.1158/1078-0432.CCR-05-0282 [DOI] [PubMed] [Google Scholar]
- 52. Siefker‐Radtke AO, Lugowska I, Tupikowski K, Andric ZG, Rezazadeh Kalebasty A, Curigliano G, et al. 917P ‐ Clinical activity of vofatamab (V), an FGFR3 selective antibody in combination with pembrolizumab (P) in metastatic urothelial carcinoma (mUC), updated interim analysis of FIERCE‐22. Ann Oncol. 2019;30:v365. 10.1093/annonc/mdz249.016 [DOI] [Google Scholar]
- 53. Mellado B, Castellano DE, Pang S, Urun Y, Park SH, Vaishampayan UN, et al. Interim analysis of the fierce‐21 phase 2 (P2) study of vofatamab (B‐701), a selective inhibitor of FGFR3, as salvage therapy in metastatic urothelial carcinoma (mUC). J Clin Oncol. 2019;37:4547. 10.1200/JCO.2019.37.15_suppl.4547 [DOI] [Google Scholar]
- 54. Bellmunt J, Picus J, Kohli M, Arriaga YE, Milowsky MI, Currie G, et al. FIERCE‐21: Phase 1b/2 study of docetaxel + b‐701, a selective inhibitor of FGFR3, in relapsed or refractory (R/R) metastatic urothelial carcinoma (mUCC). J Clin Oncol. 2018;36:4534. 10.1200/JCO.2018.36.15_suppl.4534 [DOI] [Google Scholar]
- 55. Qing J, Du X, Chen Y, Chan P, Li H, Wu P, et al. Antibody‐based targeting of FGFR3 in bladder carcinoma and t(4;14)‐positive multiple myeloma in mice. J Clin Invest. 2009;119:1216‐29. 10.1172/JCI38017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X, Ye C‐H, Li E‐M, Xu L‐Y, Lin W‐Q, Chen G‐H. Discovery of octahydropyrrolo [3,2‐b] pyridin derivative as a highly selective Type I inhibitor of FGFR3 over VEGFR2 by high‐throughput virtual screening. J Cell Biochem. 2023;124:221‐38. 10.1002/jcb.30357 [DOI] [PubMed] [Google Scholar]
- 57. Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215‐66. 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jaye M, Howk R, Burgess W, Ricca GA, Chiu IM, Ravera MW, et al. Human endothelial cell growth factor: cloning, nucleotide sequence, and chromosome localization. Science. 1986;233:541‐5. 10.1126/science.3523756 [DOI] [PubMed] [Google Scholar]
- 59. Karl K, Del Piccolo N, Light T, Roy T, Dudeja P, Ursachi V‐C, et al. Ligand bias underlies differential signaling of multiple FGFs via FGFR1 2023. 10.7554/elife.88144.2 [DOI] [PMC free article] [PubMed]
- 60. Keegan K, Rooke L, Hayman M, Spurr NK. The fibroblast growth factor receptor 3 gene (FGFR3) is assigned to human chromosome 4. Cytogenet Cell Genet. 1993;62:172‐5. 10.1159/000133465 [DOI] [PubMed] [Google Scholar]
- 61. Sarabipour S, Hristova K. Mechanism of FGF receptor dimerization and activation. Nat Commun. 2016;7:10262. 10.1038/ncomms10262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hartl I, Brumovska V, Striedner Y, Yasari A, Schütz GJ, Sevcsik E, et al. Measurement of FGFR3 signaling at the cell membrane via total internal reflection fluorescence microscopy to compare the activation of FGFR3 mutants. J Biol Chem. 2023;299:102832. 10.1016/j.jbc.2022.102832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Helsten T, Schwaederle M, Kurzrock R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: biologic and clinical implications. Cancer Metastasis Rev. 2015;34:479‐96. 10.1007/s10555-015-9579-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17:318‐32. 10.1038/nrc.2017.8 [DOI] [PubMed] [Google Scholar]
- 65. Dienstmann R, Rodon J, Prat A, Perez‐Garcia J, Adamo B, Felip E, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol Off J Eur Soc Med Oncol. 2014;25:552‐63. 10.1093/annonc/mdt419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010;8:1439‐52. 10.1158/1541-7786.MCR-10-0168 [DOI] [PubMed] [Google Scholar]
- 67. Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199‐213. 10.1042/BJ20101603 [DOI] [PubMed] [Google Scholar]
- 68. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next‐Generation Sequencing. Clin Cancer Res an Off J Am Assoc Cancer Res. 2016;22:259‐67. 10.1158/1078-0432.CCR-14-3212 [DOI] [PubMed] [Google Scholar]
- 69. Touat M, Ileana E, Postel‐Vinay S, André F, Soria J‐C. Targeting FGFR Signaling in Cancer. Clin Cancer Res an Off J Am Assoc Cancer Res. 2015;21:2684‐94. 10.1158/1078-0432.CCR-14-2329 [DOI] [PubMed] [Google Scholar]
- 70. Katoh M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat Rev Clin Oncol. 2019;16:105‐22. 10.1038/s41571-018-0115-y [DOI] [PubMed] [Google Scholar]
- 71. Chae YK, Ranganath K, Hammerman PS, Vaklavas C, Mohindra N, Kalyan A, et al. Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget. 2017;8:16052‐74. 10.18632/oncotarget.14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Porta R, Borea R, Coelho A, Khan S, Araújo A, Reclusa P, et al. FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit Rev Oncol Hematol. 2017;113:256‐67. 10.1016/j.critrevonc.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 73. Rosty C, Aubriot M‐H, Cappellen D, Bourdin J, Cartier I, Thiery JP, et al. Clinical and biological characteristics of cervical neoplasias with FGFR3 mutation. Mol Cancer. 2005;4:15. 10.1186/1476-4598-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yoshimoto Y, Sasaki Y, Murata K, Noda S‐E, Miyasaka Y, Hamamoto J, et al. Mutation profiling of uterine cervical cancer patients treated with definitive radiotherapy. Gynecol Oncol. 2020;159:546‐53. 10.1016/j.ygyno.2020.08.020 [DOI] [PubMed] [Google Scholar]
- 75. Gött H, Uhl E. FGFR3‐TACCs3 Fusions and Their Clinical Relevance in Human Glioblastoma. Int J Mol Sci. 2022;23. 10.3390/ijms23158675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Benard B, Christofferson A, Legendre C, Aldrich J, Nasser S, Yesil J, et al. FGFR3 Mutations Are an Adverse Prognostic Factor in Patients with t(4;14)(p16;q32) Multiple Myeloma: An Mmrf Commpass Analysis. Blood. 2017;130:3027. 10.1182/blood.V130.Suppl_1.3027.3027 [DOI] [Google Scholar]
- 77. Ermakov MS, Kashofer K, Regauer S. Different Mutational Landscapes in Human Papillomavirus‐Induced and Human Papillomavirus‐Independent Invasive Penile Squamous Cell Cancers. Mod Pathol an Off J United States Can Acad Pathol Inc. 2023;36:100250. 10.1016/j.modpat.2023.100250 [DOI] [PubMed] [Google Scholar]
- 78. Czyz M. Fibroblast Growth Factor Receptor Signaling in Skin Cancers. Cells. 2019;8. 10.3390/cells8060540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Castillo P, Marginet M, Jares P, García M, Gonzalvo E, Arance A, et al. Implementation of an NGS panel for clinical practice in paraffin‐embedded tissue samples from locally advanced and metastatic melanoma patients. Explor Target Anti‐Tumor Ther. 2020;1:101‐8. 10.37349/etat.2020.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Aparicio T, Svrcek M, Henriques J, Afchain P, Lièvre A, Tougeron D, et al. Panel gene profiling of small bowel adenocarcinoma: Results from the NADEGE prospective cohort. Int J Cancer. 2021;148:1731‐42. 10.1002/ijc.33392 [DOI] [PubMed] [Google Scholar]
- 81. Suster D, Mackinnon AC, Ronen N, Mejbel HA, Harada S, Suster S. Non‐Small Cell Lung Carcinoma With Clear Cell Features and FGFR3::TACC3 Gene Rearrangement : Clinicopathologic and Next Generation Sequencing Study of 7 Cases. Am J Surg Pathol. 2024;48:284‐91. 10.1097/PAS.0000000000002167 [DOI] [PubMed] [Google Scholar]
- 82. Li J, Hu K, Huang J, Zhou L, Yan Y, Xu Z. Insights of fibroblast growth factor receptor 3 aberrations in pan‐cancer and their roles in potential clinical treatment. Aging (Albany NY). 2021;13:16541‐66. 10.18632/aging.203175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hafner C, Hartmann A, van Oers JMM, Stoehr R, Zwarthoff EC, Hofstaedter F, et al. FGFR3 mutations in seborrheic keratoses are already present in flat lesions and associated with age and localization. Mod Pathol an Off J United States Can Acad Pathol Inc. 2007;20:895‐903. 10.1038/modpathol.3800837 [DOI] [PubMed] [Google Scholar]
- 84. Hafner C, van Oers JMM, Hartmann A, Landthaler M, Stoehr R, Blaszyk H, et al. High frequency of FGFR3 mutations in adenoid seborrheic keratoses. J Invest Dermatol. 2006;126:2404‐7. 10.1038/sj.jid.5700422 [DOI] [PubMed] [Google Scholar]
- 85. Wang X, Qi H, Wang Q, Zhu Y, Wang X, Jin M, et al. FGFR3/fibroblast growth factor receptor 3 inhibits autophagy through decreasing the ATG12‐ATG5 conjugate, leading to the delay of cartilage development in achondroplasia. Autophagy. 2015;11:1998‐2013. 10.1080/15548627.2015.1091551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Smith LB, Belanger JM, Oberbauer AM. Fibroblast growth factor receptor 3 effects on proliferation and telomerase activity in sheep growth plate chondrocytes. J Anim Sci Biotechnol. 2012;3:39. 10.1186/2049-1891-3-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Brodie SG, Kitoh H, Lachman RS, Nolasco LM, Mekikian PB, Wilcox WR. Platyspondylic lethal skeletal dysplasia, San Diego type, is caused by FGFR3 mutations. Am J Med Genet. 1999;84:476‐80. [PubMed] [Google Scholar]
- 88. Bellus GA, Spector EB, Speiser PW, Weaver CA, Garber AT, Bryke CR, et al. Distinct missense mutations of the FGFR3 lys650 codon modulate receptor kinase activation and the severity of the skeletal dysplasia phenotype. Am J Hum Genet. 2000;67:1411‐21. 10.1086/316892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bellus GA, Hefferon TW, Ortiz de Luna RI, Hecht JT, Horton WA, Machado M, et al. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56:368‐73. [PMC free article] [PubMed] [Google Scholar]
- 90. Aviezer D, Golembo M, Yayon A. Fibroblast growth factor receptor‐3 as a therapeutic target for Achondroplasia–genetic short limbed dwarfism. Curr Drug Targets. 2003;4:353‐65. 10.2174/1389450033490993 [DOI] [PubMed] [Google Scholar]
- 91. Aikawa T, Segre G V, Lee K. Fibroblast growth factor inhibits chondrocytic growth through induction of p21 and subsequent inactivation of cyclin E‐Cdk2. J Biol Chem. 2001;276:29347‐52. 10.1074/jbc.M101859200 [DOI] [PubMed] [Google Scholar]
- 92. Foldynova‐Trantirkova S, Wilcox WR, Krejci P. Sixteen years and counting: the current understanding of fibroblast growth factor receptor 3 (FGFR3) signaling in skeletal dysplasias. Hum Mutat. 2012;33:29‐41. 10.1002/humu.21636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xie Y, Zinkle A, Chen L, Mohammadi M. Fibroblast growth factor signalling in osteoarthritis and cartilage repair. Nat Rev Rheumatol. 2020;16:547‐64. 10.1038/s41584-020-0469-2 [DOI] [PubMed] [Google Scholar]
- 94. Kunova Bosakova M, Varecha M, Hampl M, Duran I, Nita A, Buchtova M, et al. Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3‐related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Hum Mol Genet. 2018;27:1093‐105. 10.1093/hmg/ddy031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Martin L, Kaci N, Estibals V, Goudin N, Garfa‐Traore M, Benoist‐Lasselin C, et al. Constitutively‐active FGFR3 disrupts primary cilium length and IFT20 trafficking in various chondrocyte models of achondroplasia. Hum Mol Genet. 2018;27:1‐13. 10.1093/hmg/ddx374 [DOI] [PubMed] [Google Scholar]
- 96. Iwata T, Li CL, Deng CX, Francomano CA. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Hum Mol Genet. 2001;10:1255‐64. 10.1093/hmg/10.12.1255 [DOI] [PubMed] [Google Scholar]
- 97. Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka P V, et al. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517‐25. 10.1172/JCI6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wen X, Li X, Tang Y, Tang J, Zhou S, Xie Y, et al. Chondrocyte FGFR3 Regulates Bone Mass by Inhibiting Osteogenesis. J Biol Chem. 2016;291:24912‐21. 10.1074/jbc.M116.730093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yao X, Zhang J, Jing X, Ye Y, Guo J, Sun K, et al. Fibroblast growth factor 18 exerts anti‐osteoarthritic effects through PI3K‐AKT signaling and mitochondrial fusion and fission. Pharmacol Res. 2019;139:314‐24. 10.1016/j.phrs.2018.09.026 [DOI] [PubMed] [Google Scholar]
- 100. Ota S, Tonou‐Fujimori N, Yamasu K. The roles of the FGF signal in zebrafish embryos analyzed using constitutive activation and dominant‐negative suppression of different FGF receptors. Mech Dev. 2009;126:1‐17. 10.1016/j.mod.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 101. Sun X, Zhang R, Chen H, Du X, Chen S, Huang J, et al. Fgfr3 mutation disrupts chondrogenesis and bone ossification in zebrafish model mimicking CATSHL syndrome partially via enhanced Wnt/β‐catenin signaling. Theranostics. 2020;10:7111‐30. 10.7150/thno.45286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Toydemir RM, Brassington AE, Bayrak‐Toydemir P, Krakowiak PA, Jorde LB, Whitby FG, et al. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am J Hum Genet. 2006;79:935‐41. 10.1086/508433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jang J‐H, Shin K‐H, Park Y‐J, Lee RJ, McKeehan WL, Park J‐G. Novel Transcripts of Fibroblast Growth Factor Receptor 3 Reveal Aberrant Splicing and Activation of Cryptic Splice Sequences in Colorectal Cancer1. Cancer Res. 2000;60:4049‐52. [PubMed] [Google Scholar]
- 104. Fromme JE, Schmitz K, Wachter A, Grzelinski M, Zielinski D, Koppel C, et al. FGFR3 mRNA overexpression defines a subset of oligometastatic colorectal cancers with worse prognosis. Oncotarget. 2018;9:32204‐18. 10.18632/oncotarget.25941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jin Z, Feng H, Liang J, Jing X, Zhao Q, Zhan L, et al. FGFR3(△7‐9) promotes tumor progression via the phosphorylation and destabilization of ten‐eleven translocation‐2 in human hepatocellular carcinoma. Cell Death Dis. 2020;11:903. 10.1038/s41419-020-03089-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lafitte M, Moranvillier I, Garcia S, Peuchant E, Iovanna J, Rousseau B, et al. FGFR3 has tumor suppressor properties in cells with epithelial phenotype. Mol Cancer. 2013;12:83. 10.1186/1476-4598-12-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zheng J, Zhang W, Li L, He Y, Wei Y, Dang Y, et al. Signaling Pathway and Small‐Molecule Drug Discovery of FGFR: A Comprehensive Review. Front Chem. 2022;10:860985. 10.3389/fchem.2022.860985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel). 2017;9. 10.3390/cancers9050052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L, et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers (Basel). 2019;11. 10.3390/cancers11101618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ullah R, Yin Q, Snell AH, Wan L. RAF‐MEK‐ERK pathway in cancer evolution and treatment. Semin Cancer Biol. 2022;85:123‐54. 10.1016/j.semcancer.2021.05.010 [DOI] [PubMed] [Google Scholar]
- 111. Krejci P, Salazar L, Goodridge HS, Kashiwada TA, Schibler MJ, Jelinkova P, et al. STAT1 and STAT3 do not participate in FGF‐mediated growth arrest in chondrocytes. J Cell Sci. 2008;121:272‐81. 10.1242/jcs.017160 [DOI] [PubMed] [Google Scholar]
- 112. Nguyen HB, Estacion M, Gargus JJ. Mutations causing achondroplasia and thanatophoric dysplasia alter bFGF‐induced calcium signals in human diploid fibroblasts. Hum Mol Genet. 1997;6:681‐8. 10.1093/hmg/6.5.681 [DOI] [PubMed] [Google Scholar]
- 113. Koike M, Yamanaka Y, Inoue M, Tanaka H, Nishimura R, Seino Y. Insulin‐like growth factor‐1 rescues the mutated FGF receptor 3 (G380R) expressing ATDC5 cells from apoptosis through phosphatidylinositol 3‐kinase and MAPK. J Bone Miner Res Off J Am Soc Bone Miner Res. 2003;18:2043‐51. 10.1359/jbmr.2003.18.11.2043 [DOI] [PubMed] [Google Scholar]
- 114. de Miranda MC, Rodrigues MA, de Angelis Campos AC, Faria JAQA, Kunrath‐Lima M, Mignery GA, et al. Epidermal growth factor (EGF) triggers nuclear calcium signaling through the intranuclear phospholipase Cδ‐4 (PLCδ4). J Biol Chem. 2019;294:16650‐62. 10.1074/jbc.RA118.006961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Eggermont C, Giron P, Noeparast M, Vandenplas H, Aza‐Blanc P, Gutierrez GJ, et al. The EGFR‐STYK1‐FGF1 axis sustains functional drug tolerance to EGFR inhibitors in EGFR‐mutant non‐small cell lung cancer. Cell Death Dis. 2022;13:611. 10.1038/s41419-022-04994-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Giron P, Eggermont C, Noeparast A, Vandenplas H, Teugels E, Forsyth R, et al. Targeting USP13‐mediated drug tolerance increases the efficacy of EGFR inhibition of mutant EGFR in non‐small cell lung cancer. Int J Cancer. 2020;148:2579‐93. 10.1002/ijc.33404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Umelo I, Noeparast A, Chen G, Renard M, Geers C, Vansteenkiste J, et al. Identification of a novel HER3 activating mutation homologous to EGFR‐L858R in lung cancer. Oncotarget. 2016;7:3068‐83. 10.18632/oncotarget.6585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ren Y, Cheng L, Rong Z, Li Z, Li Y, Zhang X, et al. hSef potentiates EGF‐mediated MAPK signaling through affecting EGFR trafficking and degradation. Cell Signal. 2008;20:518‐33. 10.1016/j.cellsig.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Pinilla‐Macua I, Sorkin A. Cbl and Cbl‐b independently regulate EGFR through distinct receptor interaction modes. Mol Biol Cell. 2023;34:ar134. 10.1091/mbc.E23-02-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Reich A, Sapir A, Shilo B. Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development. 1999;126:4139‐47. 10.1242/dev.126.18.4139 [DOI] [PubMed] [Google Scholar]
- 121. Unni AM, Harbourne B, Oh MH, Wild S, Ferrarone JR, Lockwood WW, et al. Hyperactivation of ERK by multiple mechanisms is toxic to RTK‐RAS mutation‐driven lung adenocarcinoma cells. Elife. 2018;7:1‐24. 10.7554/eLife.33718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Giron P, Eggermont C, Noeparast A, Teugels E, Gutierrez G, De Grève J. The deubiquitinase USP13 as a novel therapeutic co‐target in EGFR mutant non‐small cell lung cancer, 2018.
- 123. Mahe M, Dufour F, Neyret‐Kahn H, Moreno‐Vega A, Beraud C, Shi M, et al. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol Med. 2018;10:e8163. 10.15252/emmm.201708163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Weickhardt AJ, Lau DK, Hodgson‐Garms M, Lavis A, Jenkins LJ, Vukelic N, et al. Dual targeting of FGFR3 and ERBB3 enhances the efficacy of FGFR inhibitors in FGFR3 fusion‐driven bladder cancer. BMC Cancer. 2022;22:478. 10.1186/s12885-022-09478-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hosni S, Kilian V, Klümper N, Gabbia D, Sieckmann K, Corvino D, et al. Adipocyte precursor‐derived NRG1 promotes resistance to FGFR inhibition in urothelial carcinoma. Cancer Res. 2024. 10.1158/0008-5472.CAN-23-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Herrera‐Abreu MT, Pearson A, Campbell J, Shnyder SD, Knowles MA, Ashworth A, et al. Parallel RNA interference screens identify EGFR activation as an escape mechanism in FGFR3‐mutant cancer. Cancer Discov. 2013;3:1058‐71. 10.1158/2159-8290.CD-12-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang L, Šuštić T, Leite de Oliveira R, Lieftink C, Halonen P, van de Ven M, et al. A Functional Genetic Screen Identifies the Phosphoinositide 3‐kinase Pathway as a Determinant of Resistance to Fibroblast Growth Factor Receptor Inhibitors in FGFR Mutant Urothelial Cell Carcinoma. Eur Urol. 2017;71:858‐62. 10.1016/j.eururo.2017.01.021 [DOI] [PubMed] [Google Scholar]
- 128. Nössing C, Herek P, Shariat SF, Berger W, Englinger B. Advances in preclinical assessment of therapeutic targets for bladder cancer precision medicine. Curr Opin Urol. 2024;34:251‐7. 10.1097/MOU.0000000000001177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Szklener K, Chmiel P, Michalski A, Mańdziuk S. New Directions and Challenges in Targeted Therapies of Advanced Bladder Cancer: The Role of FGFR Inhibitors. Cancers (Basel). 2022;14. 10.3390/cancers14061416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rose TL, Weir WH, Mayhew GM, Shibata Y, Eulitt P, Uronis JM, et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br J Cancer. 2021;125:1251‐60. 10.1038/s41416-021-01488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wang L, Saci A, Szabo PM, Chasalow SD, Castillo‐Martin M, Domingo‐Domenech J, et al. EMT‐ and stroma‐related gene expression and resistance to PD‐1 blockade in urothelial cancer. Nat Commun. 2018;9:3503. 10.1038/s41467-018-05992-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ouyang Y, Ou Z, Zhong W, Yang J, Fu S, Ouyang N, et al. FGFR3 alterations in bladder cancer stimulate serine synthesis to induce immune‐inert macrophages that suppress T‐cell recruitment and activation. Cancer Res. 2023. 10.1158/0008-5472.CAN-23-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Okato A, Utsumi T, Ranieri M, Zheng X, Zhou M, Pereira LD, et al. FGFR inhibition augments anti‐PD‐1 efficacy in murine FGFR3‐mutant bladder cancer by abrogating immunosuppression. J Clin Invest. 2024;134. 10.1172/JCI169241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jing W, Wang G, Cui Z, Xiong G, Jiang X, Li Y, et al. FGFR3 Destabilizes PD‐L1 via NEDD4 to Control T‐cell‐Mediated Bladder Cancer Immune Surveillance. Cancer Res. 2022;82:114‐29. 10.1158/0008-5472.CAN-21-2362 [DOI] [PubMed] [Google Scholar]
- 135. Trujillo JA, Sweis RF, Bao R, Luke JJ. T Cell‐Inflamed versus Non‐T Cell‐Inflamed Tumors: A Conceptual Framework for Cancer Immunotherapy Drug Development and Combination Therapy Selection. Cancer Immunol Res. 2018;6:990‐1000. 10.1158/2326-6066.CIR-18-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Aksoylar H‐I, Boussiotis VA. PD‐1+ Treg cells: a foe in cancer immunotherapy? Nat Immunol. 2020;21:1311‐2. 10.1038/s41590-020-0801-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Song Y, Peng Y, Qin C, Wang Y, Yang W, Du Y, et al. Fibroblast growth factor receptor 3 mutation attenuates response to immune checkpoint blockade in metastatic urothelial carcinoma by driving immunosuppressive microenvironment. J Immunother Cancer. 2023;11. 10.1136/jitc-2022-006643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Robertson AG, Meghani K, Cooley LF, McLaughlin KA, Fall LA, Yu Y, et al. Expression‐based subtypes define pathologic response to neoadjuvant immune‐checkpoint inhibitors in muscle‐invasive bladder cancer. Nat Commun. 2023;14:2126. 10.1038/s41467-023-37568-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Xu P‐H, Chen S, Wang Y, Jin S, Wang J, Ye D, et al. FGFR3 mutation characterization identifies prognostic and immune‐related gene signatures in bladder cancer. Comput Biol Med. 2023;162:106976. 10.1016/j.compbiomed.2023.106976 [DOI] [PubMed] [Google Scholar]
- 140. Wang L, Gong Y, Saci A, Szabo PM, Martini A, Necchi A, et al. Fibroblast Growth Factor Receptor 3 Alterations and Response to PD‐1/PD‐L1 Blockade in Patients with Metastatic Urothelial Cancer. Eur Urol. 2019;76:599‐603. 10.1016/j.eururo.2019.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Li X, Li Y, Liu B, Chen L, Lyu F, Zhang P, et al. P4HA2‐mediated HIF‐1α stabilization promotes erdafitinib‐resistance in FGFR3‐alteration bladder cancer. FASEB J Off Publ Fed Am Soc Exp Biol. 2023;37:e22840. 10.1096/fj.202201247R [DOI] [PubMed] [Google Scholar]
- 142. Pettitt GA, Hurst CD, Khan Z, McPherson HR, Dunning MC, Alder O, et al. Development of resistance to FGFR inhibition in urothelial carcinoma via multiple pathways in vitro. J Pathol. 2023;259:220‐32. 10.1002/path.6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Facchinetti F, Hollebecque A, Braye F, Vasseur D, Pradat Y, Bahleda R, et al. Resistance to Selective FGFR Inhibitors in FGFR‐Driven Urothelial Cancer. Cancer Discov. 2023;13:1998‐2011. 10.1158/2159-8290.CD-22-1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang Z, Muthusamy V, Petrylak DP, Anderson KS. Tackling FGFR3‐driven bladder cancer with a promising synergistic FGFR/HDAC targeted therapy. Npj Precis Oncol. 2023;7:70. 10.1038/s41698-023-00417-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lang H, Béraud C, Cabel L, Fontugne J, Lassalle M, Krucker C, et al. Integrated molecular and pharmacological characterization of patient‐derived xenografts from bladder and ureteral cancers identifies new potential therapies. Front Oncol. 2022;12:930731. 10.3389/fonc.2022.930731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Stiewe T, Haran TE. How mutations shape p53 interactions with the genome to promote tumorigenesis and drug resistance. Drug Resist Updat Rev Comment Antimicrob Anticancer Chemother. 2018;38:27‐43. 10.1016/j.drup.2018.05.001 [DOI] [PubMed] [Google Scholar]
- 147. Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346‐56. 10.1038/onc.2009.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kikuchi A, Suzuki T, Nakazawa T, Iizuka M, Nakayama A, Ozawa T, et al. ASP5878, a selective FGFR inhibitor, to treat FGFR3‐dependent urothelial cancer with or without chemoresistance. Cancer Sci. 2017;108:236‐42. 10.1111/cas.13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Qin Z‐Q, Li Q‐G, Yi H, Lu S‐S, Huang W, Rong Z‐X, et al. Heterozygous p53‐R280T Mutation Enhances the Oncogenicity of NPC Cells Through Activating PI3K‐Akt Signaling Pathway. Front Oncol. 2020;10:104. 10.3389/fonc.2020.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471‐80. 10.1038/s41568-020-0262-1 [DOI] [PubMed] [Google Scholar]
- 151. Bargonetti J, Prives C. Gain‐of‐function mutant p53: history and speculation. J Mol Cell Biol. 2019;11:605‐9. 10.1093/jmcb/mjz067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov A V, et al. A dominant‐negative effect drives selection of <em>TP53</em> missense mutations in myeloid malignancies. Science (80‐). 2019;365:599 LP – 604. 10.1126/science.aax3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kotler E, Shani O, Goldfeld G, Lotan‐Pompan M, Tarcic O, Gershoni A, et al. A Systematic p53 Mutation Library Links Differential Functional Impact to Cancer Mutation Pattern and Evolutionary Conservation. Mol Cell. 2018;71:178‐190.e8. 10.1016/j.molcel.2018.06.012 [DOI] [PubMed] [Google Scholar]
- 154. Chen X, Zhang T, Su W, Dou Z, Zhao D, Jin X, et al. Mutant p53 in cancer: from molecular mechanism to therapeutic modulation. Cell Death Dis. 2022;13:1‐14. 10.1038/s41419-022-05408-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Maiorano BA, Catalano M, Maiello E, Roviello G. Enfortumab vedotin in metastatic urothelial carcinoma: the solution EVentually? Front Oncol. 2023;13:1254906. 10.3389/fonc.2023.1254906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Santini D, Banna GL, Buti S, Isella L, Stellato M, Roberto M, et al. Navigating the Rapidly Evolving Advanced Urothelial Carcinoma Treatment Landscape: Insights from Italian Experts. Curr Oncol Rep. 2023;25:1345‐62. 10.1007/s11912-023-01461-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Jain RK, Heiligh J, Kim Y, Piekarz R, Pelosof LC, Yang Y (Aaron), et al. Phase Ib trial of erdafitinib (E) combined with enfortumab vedotin (EV) following platinum and PD‐1/L1 inhibitors for metastatic urothelial carcinoma (mUC) with FGFR2/3 genetic alterations (GAs). J Clin Oncol. 2024;42:625. 10.1200/JCO.2024.42.4_suppl.625 [DOI] [Google Scholar]
- 158. Bock C, Lengauer T. Managing drug resistance in cancer: lessons from HIV therapy. Nat Rev Cancer. 2012;12:494‐501. 10.1038/nrc3297 [DOI] [PubMed] [Google Scholar]
- 159. Langreth R, Waldholz M. New era of personalized medicine: targeting drugs for each unique genetic profile. Oncologist. 1999;4:426‐7. [PubMed] [Google Scholar]
- 160. Dankner M, Rose AANN, Rajkumar S, Siegel PM, Watson IR. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene. 2018;37:3183‐99. 10.1038/s41388-018-0171-x [DOI] [PubMed] [Google Scholar]
- 161. Naumov GN, Nilsson MB, Cascone T, Briggs A, Straume O, Akslen LA, et al. Combined vascular endothelial growth factor receptor and epidermal growth factor receptor (EGFR) blockade inhibits tumor growth in xenograft models of EGFR inhibitor resistance. Clin Cancer Res an Off J Am Assoc Cancer Res. 2009;15:3484‐94. 10.1158/1078-0432.CCR-08-2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Moore S, Wheatley‐Price P. EGFR Combination Therapy Should Become the New Standard First‐Line Treatment in Advanced EGFR‐Mutant NSCLC. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2021;16:1788‐92. 10.1016/j.jtho.2021.06.004 [DOI] [PubMed] [Google Scholar]
- 163. Settleman J, Neto JMF, Bernards R. Thinking Differently about Cancer Treatment Regimens. Cancer Discov. 2021;11:1016‐23. 10.1158/2159-8290.CD-20-1187 [DOI] [PubMed] [Google Scholar]
- 164. Xue Y, Martelotto L, Baslan T, Vides A, Solomon M, Mai TT, et al. An approach to suppress the evolution of resistance in BRAF(V600E)‐mutant cancer. Nat Med. 2017;23:929‐37. 10.1038/nm.4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kaelin WG. Synthetic lethality: a framework for the development of wiser cancer therapeutics. Genome Med. 2009;1:99. 10.1186/gm99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


