Abstract
The initial step in pre-mRNA splicing involves formation of a spliceosome commitment complex (CC) or E-complex by factors that serve to bind and mark the exon-intron boundaries that will undergo splicing. The CC component U1 snRNP assembles at the 5′-splice site (ss), whereas SF1, U2AF2, and U2AF1 define the 3′-ss of the intron. A PRP40 protein bridges U1 snRNP with factors at the 3′-ss. To determine how defects in CC components impact cancers, we analyzed human gastrointestinal (GI) cancer patient tissue and clinical data from cBioPortal. cBioPortal datasets were analyzed for CC factor alterations and patient outcomes in GI cancers (bowel, stomach, esophagus, pancreas, and liver). In addition, co-expression datasets were used to determine the splicing targets of the CC. Our analysis found that frequency of genetic changes was low (1%-13%), but when combined with changes in expression levels, there was an overall surprisingly high incidence of CC component (>30%) alterations in GI cancers. Colon cancer patients carrying BRAF driver gene mutations had high incidences of CC alterations (19%-61%), whereas patients with APC, KRAS, or TP53 gene mutations had low (<5%) incidences of CC alterations. Most significantly, patients with mutations in CC genes exhibited a consistent trend of favorable survival rates, indicating that mutations that impair or lower CC component expression favor patient survival. Conversely, patients with high CC expression had worse survival. Pathway analysis indicates that the CC regulates specific metabolic and tumor suppressor pathways. Metabolic pathways involved in cell survival, nutrition, biosynthesis, autophagy, cellular movement (invasion), or immune surveillance pathways correlated with CC factor upregulation, whereas tumor suppressor pathways, which regulate cell proliferation and apoptosis, were inversely correlated with CC factor upregulation. This study demonstrates the versatility of in silico analysis to determine molecular function of large macromolecular complexes such as the spliceosome CC. Furthermore, our analysis indicates that therapeutic lowering of CC levels in colon cancer patients may enhance patient survival.
Keywords: Commitment complex, E-complex, bowel cancer
Introduction
Alternative splicing generates vast protein diversity from the limited number of human genes. 1 Dysregulation of alternative splicing in aging or cancer cells results in production of aberrant splice variants.2-7
The spliceosome assembles at exon-intron junctions of newly synthesized pre-mRNAs. Multiple dynamic interactions progressively form the early, intermediate, and advanced states of the spliceosome complex which sequentially carries out transesterification reactions, leading to removal of introns. During the initial stage, the exon-intron junctions are defined by the spliceosome E-complex, also known as the commitment complex (CC)8-10 (Figure 1A). snRNP U1, a multiplex of snRNA and at least 10 proteins, assembles at the 5′-splice sequence (5′-ss) and interacts with RNA Pol II. SF1 binds to the branch point sequence within the intron, near the 3′-ss, and co-operatively interacts with U2AF2.11-14 U2AF2 (U2AF65) and U2AF1 (U2AF35) assemble at the pyrimidine tract within the intron and at the 3′-end splice site sequence, respectively.13,15 Interaction of SF1 with U2AF2 stabilizes the SF1-U2AF2-U2AF1 complex. The PRP40 protein family member (PRPF40A, PRPF40B, or TCERG1) connects snRNP U1 and phosphorylated C-terminal domain of Pol II with SF1. Thus, PRP40 proteins bridge the snRNP U1 and Pol II at 5′-ss with SF1-U2AF2-U2AF1 complex at 3′-ss, allowing 5′ and 3′ sites of the introns to be brought in close proximity. U2 snRNP subsequently displaces SF1 from the transient CC, and further exchange of factors allows progression of the spliceosome complex into more mature states that culminate in excision and ligation reactions to generate mRNA.
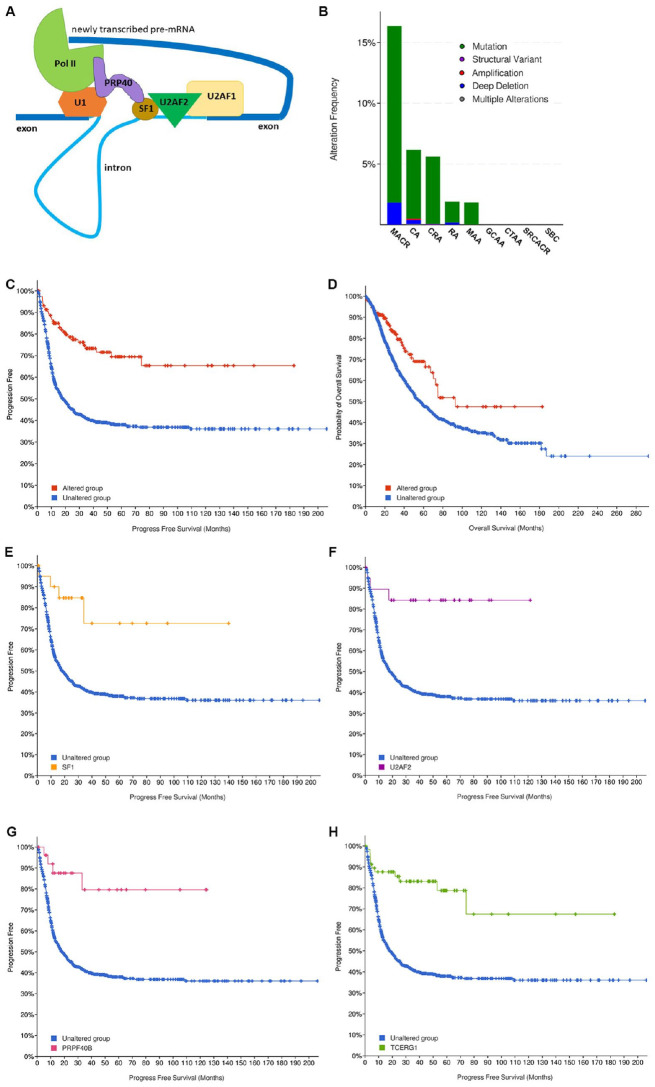
Figure 1.
(A) Components of the CC/E-complex. Thick and thin blue lines represent exons and introns, respectively. snRNP U1 (U1) binds to the 5′-splice site and also interacts with RNA Polymerase II (Pol II). SF1 binds to the branch point sequence within the intron and co-operatively interacts with U2AF2. U2AF2 and U2AF1 assemble at the pyrimidine tract within the intron and at the 3′-end splice site sequence, respectively. PRP40-family protein (PRPF40A, PRPF40B, or TCERG1) serves as a bridge between U1, Pol II, and SF1. (B) Incidence of genetic alterations in CC factors (U2AF1, U2AF2, SF1, PRPF40A, PRPF40B, and TCERG1) in different types of bowel cancers. All non-redundant studies were included (6523 patients; 17 studies). Key: green represents mutations, purple represents structural variant, red represents amplification, dark blue represents deep deletion, and gray represents multiple alterations. MACR: mucinous adenocarcinoma of the colon and rectum; CA: colon adenocarcinoma; CRA: colorectal adenocarcinoma; RA: rectal adenocarcinoma; MAA: mucinous adenocarcinoma of the appendix; GCAA: goblet cell adenocarcinoma of the appendix; CTAA: colonic type adenoma of the appendix; SRCACR: signet ring cell adenocarcinoma of the colon and rectum; SBC: small bowel cancer. (C) Progression-free survival (q = 3.05e-8) and overall survival (D) (q = 3.474e-3) of patients with genetic alterations in CC genes. Data obtained from 1342 and 2782 patients, respectively. Blue line represents unaltered group (patients without alterations in any CC factor); red line represents patients with at least 1 alteration in the 6 queried CC genes. (E) Progression-free survival of patients with genetic alterations in individual components of the CC: SF1; (F) U2AF2; (G) PRPF40B, and (H) TCERG1. Blue lines represent progression-free survival curves of patients without alterations in any of the 6 CC factors. Other colored line represents survival of patients with genetic alterations in SF1 (orange) (q = 0.016); U2AF2 (purple) (q = 5.369e-3); PRPF40B (red) (q = 2.886e-3); and TCERG1 (green) (q = 5.973e-6).
Previous studies on SF1 function used a mouse strain deficient for SF1 expression 16 and demonstrated that congenital reduction of SF1 decreased development of testicular tumors in Ter mice or intestinal polyp development in ApcMin/+ mice.16,17 Thus, lower SF1 levels in mouse tissues impeded tumor development. SF1 is known to bind to pre-mRNA during the initial splicing events. To identify the pre-mRNA targets of SF1, 2 studies on SF1 experimentally isolated its mRNA targets from HeLa cells2,18 and Caenorhabditis elegans. 2 In these studies, ectopic expression of SF1 allowed for the direct “pull-down” of associated mRNA and allowed for subsequent RNA-Sequencing and analysis of pathways regulated by SF1. These studies did not however consider the interactions of SF1 with other CC components as part of a transient macromolecular spliceosome complex. These studies spurred our examination of how changes in SF1 and associated CC partners impact human cancers. Therefore, our objective was to examine first, the profile of changes in CC factor expression in human cancers, focusing on cancers of the gastrointestinal (GI) tract and especially bowel cancer. Our second objective was to evaluate how changes in CC factor expression in bowel cancers correlate with clinical features, notably patient survival. Third, sequential driver gene mutations in APC, KRAS, TP53, and BRAF are known to drive progression of bowel cancer from the early adenoma stage to late adenocarcinoma stage.19-21 Thus, we sought to examine how CC factor expression correlated with each of the driver gene mutations and consequent patient outcomes. Finally, our objective was to determine the mRNA targets of the macromolecular CC so as to determine the major pathways that are regulated by the CC in GI cancers.
The cBioPortal for Cancer Genomics (cBioPortal.org) database is an open access, web-based platform that contains curated genomics data of cancer tissues of major cancer types and corresponding patient outcomes.22-24 cBioPortal stores de-identified clinical data, such as sex, age, tumor type, tumor grade, and overall and disease-free survival data, when available. We therefore examined cBioPortal for CC factor changes and patient survival outcomes especially focused on bowel and other GI tract cancers of the stomach, esophagus, pancreas, liver, and biliary tract. We report here how genetic and expression level changes of CC genes correlate with patient disease prognosis. We also report on CC alterations in bowel cancer patient cohorts with driver gene, APC, KRAS, TP53, or BRAF, mutations. Furthermore, we report on the use of cBioPortal gene co-expression datasets to define the genes and molecular pathways targeted by CC in GI cancers.
Our analysis and results highlight the use of the cBioPortal cancer database to determine, in silico, the pathways and targets of large macromolecular complexes like the CC.
Methods
CBioPortal data analysis
Queried dataset for bowel cancers simultaneously for 6 CC factors: SF1, U2AF1, U2AF2, PRPF40A, PRPF40B, and TCERG1. The 6 CC components were chosen because (a) they closely contact with SF1 and (b) together with SF1 are needed for stability of the CC macromolecule during the initial splicing step11-15 (Figure 1A). To reduce complexity in our analysis, we excluded the 5′-splice site binding complex, U1 snRNP, because mammalian U1 snRNP is a complex of U1 snRNA, 7 Sm proteins (SmB/SmB′, SmD1, SmD2, SmD3, SmE, SmF, and SmG) and 3 U1-specific proteins (U1-70K, U1-A, and U1-C).25,26
Included in our analysis were 3 different mammalian PRP40-like proteins, PRPF40A,27,28 PRPF40B, 29 and TCERG1. 30 These 3 factors participate independently in the CC and directly contact SF1.
To access genomic data: selected for bowel (cancers) and non-redundant studies (selected for 17 studies, which excluded 2 redundant studies, the TCGA, Firehose Legacy and TCGA, Nature 2012; this selected 6523 patients/6745 samples). The selected studies were simultaneously queried for 6 CC factors: SF1, U2AF1, U2AF2, PRPF40A, PRPF40B, and TCERG1. Data were obtained from headings entitled OncoPrint, Cancer Types Summary (Cancer Type Detailed), Mutual Exclusivity, Mutations, Comparison/Survival, and Clinical. Similar queries were performed for other GI cancers and other cancer types. Note that querying individually for any CC factor, for example SF1, will yield different results. That is because a simultaneous query for all 6 components of the CC compares alterations versus no alterations in any of the other 6 CC factors.
Driver gene cohorts
To establish driver gene cohorts, a free account was created with cBioPortal for Cancer Genomics at www.cbioportal.org/.
Bowel cancer non-redundant studies (17 studies) were chosen and queried for driver gene, for example, APC. Combined Study was selected, and under Mutated Genes, selected for APC. Saved selected cohort as a new study. Similar cohorts with other driver genes were created: APC cohort (4393 patients); KRAS cohort (2737 patients); TP53 cohort (4275 patients); BRAF cohort (661 patients); and BRAF cohort from TCGA PanCancer Atlas study (62 patients). Each driver gene cohort was simultaneously queried for changes in the 6 CC factors. Patients with driver gene mutations (eg, APC mutations) are designated Unaltered Group and patients with driver gene + CC factor changes (eg, APC + CC factors) are designated Altered Group. Data were obtained from sections entitled Cancer Types Summary (Cancer Type Detailed), Mutual Exclusivity, Mutations, Comparison/Survival, and Clinical. Additional details on queries and analysis pertaining to specific Results sections is included in Supplementary Methods.
Alternatively, bowel cancer non-redundant studies were chosen and queried for each driver gene. Selected the altered samples (with alterations in the queried driver gene) by clicking the “Query” next to the “Altered Samples” underneath the “Download” tab. The selected “altered samples” were simultaneously queried for the 6 CC factors. Both query protocols yielded the same results.
Genomic plus expression data
Under Bowel, TCGA PanCancer Atlas dataset (594 samples) was selected for study. Under Select Genomic Profile, Mutations, Structural Variant, and Putative copy-number alterations from GISTIC were selected. Specifically selected for mRNA Expression (mRNA expression z-scores relative to diploid samples; RNA Seq V2 RSEM) and protein/phosphoprotein level (protein level z-scores; mass spectrometry by CPTAC). The dataset was simultaneously queried for the 6 CC factors. Data were obtained from sections entitled Cancer Types Summary (Cancer Type Detailed), Mutual Exclusivity, Mutations, Comparison/Survival, Clinical, and Co-expression.
Clinical attributes and demographic data
Demographic data corresponding to the survival curves and other Clinical Attributes can be accessed by selecting Comparison/Survival tab followed by Clinical tab. Further deselection of altered group tab and selection for SF1 (or any other factor) gives the demographic data for that particular gene alteration. For example, the demographic data for patients with alteration of SF1 was performed as follows: query first for alteration in 6 CC factors (SF1, U2AF1, U2AF2, PRPF40A, PRPF40B, and TCERG1). Select for Comparison/Survival. Select for Clinical. Deselect Altered Group tab and select SF1 tab. Under Clinical Attribute select for Race Category. Altered group in bar graph represents race profile of patients with alterations in SF1, whereas Unaltered group represents race profile of patients without alterations in any of the 6 CC factors.
Co-expression data and IPA
Using TCGA, PanCancer Atlas (Colorectal Adenocarcinoma) dataset, the co-expression data (RNA, protein, or both when available) of correlated genes was accessed for each CC factor: SF1, U2AF1, U2AF2, PRPF40A, PRPF40B, and TCERG1. For example, for mRNA expression changes that correlated with SF1 mRNA, gene lists were created by selecting for: Find genes in mRNA Expression, RSEM (Batch normalized from Illumina HiSeq_RNASeq V2; 592 samples) that are correlated with SF1 mRNA Expression, RSEM (Batch normalized from Illumina HiSeq_RNASeq V2; 592 samples). A second gene list for proteins that correlated with SF1 protein expression was created by selecting: Find genes in Protein levels (mass spectrometry by CPTAC; 84 samples) that are correlated with SF1 in Protein levels (mass spectrometry by CPTAC; 84 samples). For each factor, for example, SF1, mRNA, and protein gene lists of all the correlated genes were downloaded. Only significantly co-expressed genes were retained for analysis (q-value < 0.05). For the Colorectal Adenocarcinoma (TCGA, PanCancer Atlas) study, correlated mRNA expression was obtained for each of the queried factors, whereas correlated protein data was available for all except PRPF40B.
Co-expression mRNA data for each of the 6 CC factors was also obtained from esophageal adenocarcinoma (TCGA, PanCancer Atlas), stomach adenocarcinoma (TCGA, PanCancer Atlas), pancreatic adenocarcinoma (TCGA, PanCancer Atlas), and liver hepatocellular carcinoma (TCGA, PanCancer Atlas). There was insufficient co-expression data for biliary tract (cholangiocarcinoma, TCGA, PanCancer Atlas) and was thus not used for IPA. Protein co-expression data was unavailable from these studies.
QIAGEN Ingenuity Pathway Analysis (QIAGEN IPA) (https://www.qiagen.com/us) was performed simultaneously using all the mRNA and protein co-expression gene lists (q-value < 0.5) of each CC factor obtained from the colorectal adenocarcinoma (TCGA, PanCancer Atlas) dataset.
As protein co-expression data was unavailable for the other GI cancers, IPA was performed simultaneously on mRNA co-expressed with each of the 6 CC factors from esophageal adenocarcinoma (TCGA, PanCancer Atlas), stomach adenocarcinoma (TCGA, PanCancer Atlas), and colorectal adenocarcinoma (TCGA, PanCancer Atlas). A separate IPA analysis was performed on the co-expressed gene lists from pancreatic adenocarcinoma (TCGA, PanCancer Atlas) and liver hepatocellular carcinoma (TCGA, PanCancer Atlas).
Statistical analysis
The versatility of cBioPortal is that curated data can be easily accessed and groups can be compared (Group Comparison) using a suite of analysis features which allows users to compare clinical or genomic features of user-defined groups of samples with corresponding statistical analysis. The log-rank test is used to compute significance of survival curves as indicated by p < 0.05 or more importantly, q of <0.05.22-24
Often the survival curves in cBioPortal compare unequal number of patients. To demonstrate that the q-values of the survival curves are indeed significant in spite of unequal patient numbers in altered and unaltered group, we performed further analysis. An Excel-based program randomly selected and compared survival of the altered group with equal number of patients from the unaltered group and computed the q-value. This is described in Supplemental Methods and Supplemental Figures.
Result
Selection of CC factors for cBioPortal query
First, we searched for changes in the components of the CC in human GI cancers. Gastrointestinal tract cancer (bowel, stomach, esophagus, pancreas, liver) datasets from cBioPortal were simultaneously queried for alterations in 6 CC components: SF1, U2AF1, U2AF2, PRPF40A, PRPF40B, and TCERG1. The 6 CC components were chosen for our analysis because they closely contact with SF1 and each other and together with SF1 are needed for stability of the CC macromolecule during the initial splicing step (Figure 1A). To reduce complexity in our analysis, we excluded the 5′-splice site binding complex, U1 snRNP, in our queries because mammalian U1 snRNP is a complex of U1 snRNA, 7 Sm proteins (SmB/SmB′, SmD1, SmD2, SmD3, SmE, SmF, and SmG), and 3 U1-specific proteins (U1-70K, U1-A, and U1-C).25,26
Three different mammalian PRP40-like proteins, PRPF40A,27,28 PRPF40B, 29 and TCERG1, 30 characterized as having protein-protein interaction WW and FF domains, 31 have been identified in the CC and all have been reported to directly contact SF1 and were included in our analysis. The 3 PRP40 proteins were experimentally isolated from different sources and may function in a mutually exclusive manner. 32
Alteration frequency of CC factors in GI cancers
All non-redundant studies in cBioPortal for each type of GI cancer were selected (eg, under bowel cancers, our query selected 17 non-redundant studies). The first query was for genetic alterations in the 6 CC factors in each type of GI cancer. Results showed that CC factor genetic alteration rates ranged from less than 1% to 13% in cancer patients of the bowel, stomach, esophagus, pancreas, liver, and biliary tract (Table 1). Thus, there is an overall low rate of DNA alterations (mutations, deletions, gene amplifications) in the 6 CC components, in human cancers of the bowel, stomach, esophagus, pancreas, liver, and biliary tract.
Table 1.
Incidence of CC factor alterations in GI cancers from cBioPortal.
| GI cancer studies | All non-redundant cancer studies | TCGA, PanCancer Atlas | |||
|---|---|---|---|---|---|
| No. of patients | No. of studies | Genetic alteration frequency of CC | No. of patients | Genetic + expression alteration frequency of CC | |
| Bowel | 6523 | 17 | 5% | 594 | 39% a |
| Stomach | 739 | 5 | 13% | 440 | 37% |
| Esophagus | 3346 | 13 | 5% | 182 | 32% |
| Pancreas | 1233 | 12 | 5% | 184 | 33% |
| Liver | 1333 | 10 | 3% | 372 | 30% |
| Biliary tract | 1913 | 14 | <1% | 36 | 36% |
Genetic alteration rates are derived from all non-redundant studies on each type of GI cancer. Genetic plus expression level alteration frequency is derived from TCGA, PanCancer Atlas study for each type of GI cancer.
Indicates data includes mRNA and protein expression (mass spectrometry data) changes.
CC factor changes in bowel cancer
Next, we examined bowel cancer data in greater detail so as to compare the results with previous experimental studies in mice on the role of SF1 on intestinal polyp development. 16 Of 6523 patients in 17 non-redundant bowel cancer studies, 5% (or 310 patients) had genetic changes in the 6 CC factors (Table 1). Mutations were the predominant type of genetic alteration (Figure 1B, green bars) and very low frequencies of gene amplification (red bars) or deletions (blue bars) are observed. Intriguingly, multiple CC factor mutations such as in SF1 and U2AF2, co-occur in patients in a statistically significant manner (Supplementary Table 1). For example, SF1 mutations co-occur with mutations in TCERG1, PRPF40B, PRPF40A, or U2AF2 (q < 0.05). It is curious why mutations in multiple CC factors are retained in patients, considering that alteration in any one factor could be sufficient to destabilize the CC. One reason could be that the PRP40-like proteins, PRP40A, PRPF40B, or TCERG1, are not present in the same CC, and thus mutations in each are selectively retained in cancer cells. Another possibility is that genetic alteration of any single CC component does not sufficiently destabilize or adversely affect CC function, thus favoring selection of mutations in multiple components.
To summarize, analysis of 17 different bowel cancer studies revealed low rates of genetic changes of CC factors and with most changes being point mutations in CC factor genes. Furthermore, individual patients harbored genetic changes in multiple components of the CC.
Genetic changes in bowel cancer types
Next, CC alterations in different types of bowel cancers was examined. The incidence of CC factor mutations was highest in mucinous adenocarcinomas of the colon and rectum (MACR) (>15%) followed by that in colon adenocarcinoma (CA) (Figure 1B). Mucinous adenocarcinomas are a unique clinicopathological subtype of colorectal cancer that are poorly differentiated, highly malignant, express large number of mucins and carry higher frequencies of BRAF and KRAS mutations, and lower frequency of TP53 mutations. 33 Thus, CC factor alterations appear at higher rates in a specific subtype of bowel cancer, MACR, and may be associated with specific driver genes such as BRAF or KRAS.
Survival data of patients with CC factor genetic alterations
Next, clinical features associated with genetic alterations of CC factors were examined. Interestingly, genetic alterations in CC factors positively affected patient survival. Colon cancer patients with alterations in CC factor genes have significantly better progression-free survival (q = 3.05e-8) as well as overall survival (q = 3.474e-3) (Figure 1C and D). Examination of individual CC factors indicated that genetic alterations in SF1, U2AF2, PRPF40B, or TCERG1 contributed to enhanced progression-free survival (Figure 1E to H). Patients with genetic alterations in PRPF40B also showed better overall survival rates (q = 9.326e-3) (data not shown). Defects in either U2AF1 or PRPF40A did not correlate with significant survival.
The Kaplan-Meier survival curves derived from cBioPortal often compare unequal number of patients in altered and unaltered groups. For example, survival curves in Figure 1C and D (progression-free survival and overall survival) compare 132 patients with CC gene alterations (altered group) and 1219 patients with no mutations in any CC factor (unaltered group) with q = 3.05e-8 and 3.474e-3, respectively. To demonstrate that the q-values of these survival curves are indeed significant in spite of unequal patient numbers, we performed further analysis where an Excel-based program randomly selected and compared survival of the 132 patients of the altered group with 132 randomly selected samples of patients from the unaltered group (Supplementary Method 1). Ten such survival curves were generated with different random subsets of 132 unaltered samples (Supplementary Figure 1). q-value of between 1.38e-09 and 2.95e-04 was observed for all survival curves using progression-free survival data. A similar analysis for overall survival found q between 1.79e-05 and 0.0187 for all except for one curve D, with q = 0.0963. This proves that although sample sizes may be unequal, 19 out of 20 times the q-values derived from cBioPortal are reliable indication that survival of altered and unaltered groups is significantly different.
Other clinical attributes are also significantly associated with specific CC factor mutations. For example, Supplementary Figure 2 shows selected clinical attributes significantly associated with SF1 mutations. Altered group (patients with SF1 mutations) and Unaltered group (patients with no mutations in any E-complex factor) are compared for MSI (microsatellite instability) profile (Supplementary Figure 2A and B), Mutation Count (Supplementary Figure 2A and C), Fraction Genome Altered (Supplementary Figure 2A and D), Diagnosis Age (Supplementary Figure 2A and E), and Race Category (Supplementary Figure 2F). Patients with SF1 mutations (Altered group) had lower mean levels of genome alterations and higher diagnosis age but different MSI profile. Furthermore, the demographic profile of patients with SF1 alterations is significantly different compared with patients without alterations in any CC factor (Unaltered group), as indicated in Supplementary Figure 2F. A number of factors could influence the demographic profiles including sex, socioeconomic factors, genetic susceptibility, diet, substance use, and so on.
Experimental data showed that deletion of 1 copy of Sf1 gene in mice resulted in decreased incidence of intestinal polyp or testicular tumor development.16,17 The SF1-deficient mice were not monitored for survival, but both experimental results in mice and human patient data from cBioPortal show that genetic loss of SF1 engenders better outcomes. Thus, to summarize, genetic alteration of CC components SF1, U2AF2, PRPF40B, or TCERG1 significantly enhanced bowel cancer patient survival compared with patients without genetic alterations in any CC component genes.
Mutation profile of CC components in bowel cancers
Examination of the type of mutations in the 6 different CC factors indicated some hotspots for missense and truncation mutations at or near specific motifs such as KH-1, zf-CCCH, RRM, or FF domains (Supplementary Figure 3). Truncation mutations would cause loss of functional protein while missense mutations could result in inefficient translation, structural changes of the encoded protein, or impaired functional activity of proteins. Alternatively, the mutations could also be gain-of-function and oncogenic. The biological significance of each mutation is not understood at present and remain to be experimentally determined. However, further analysis (described below) indicates that these missense mutations in individual CC factors likely result in loss or impaired function. Considering that the CC is a large complex, we speculate that small changes in the tertiary protein structure of individual CC factors, due to mutations, may adversely compromise protein-protein interactions, stability, or functional efficiency of the CC spliceosome.
CC component expression in bowel cancers: TCGA PanCancer Atlas study
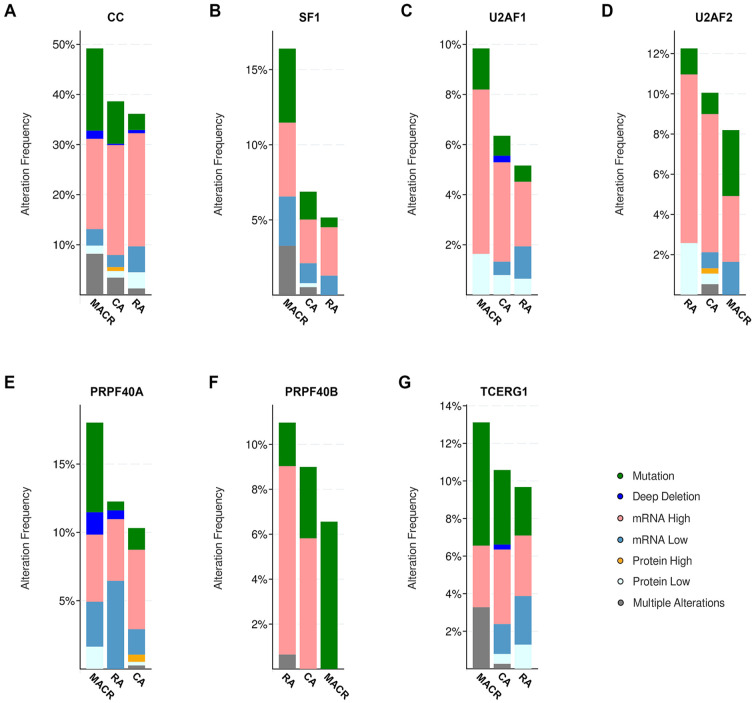
cBioPortal also reports changes in mRNA and protein levels in human cancer tissues in specific TCGA PanCancer Atlas studies. Thus, for each type of GI cancer, we separately examined their cognate TCGA PanCancer Atlas study. Each TCGA PanCancer Atlas study was simultaneously queried for alterations in genetic and expression level (mRNA and protein) changes in the 6 CC factors (see Supplementary Methods 2). Surprisingly, for each GI cancer type, the combined rate of alteration (genetic plus expression alteration frequency) of CC factors was much higher, between 30% and 39% (Table 1). Thus, although genetic changes of CC factors occur at a relatively low rate (<1%-13%), expression level changes of CC factors are present to a greater extent (>30%). The non-genetic changes predominantly involve changes in mRNA or protein levels or a small fraction carry a combination of alterations (Figure 2A).
Figure 2.
Analysis of TCGA, PanCancer Atlas dataset for incidence of alterations in CC factors in different types of bowel cancers (594 patients). TCGA, PanCancer Atlas dataset was queried for the 6 CC factors, selecting the search for mutations, structural variant, putative copy-number alterations from GISTIC, mRNA expression (mRNA expression z-scores relative to diploid samples; RNA Seq V2 RSEM), and protein/phosphoprotein level (protein expression z-scores; mass spectrometry by CPTAC). Graphs are results of cancer types detailed. Incidence of alterations of (A) all CC factors; (B) SF1; (C) U2AF1; (D) U2AF2; (E) PRPF40A; (F) PRPF40B; and (G) TCERG1 in different types of bowel cancers. Key: as in Figure 1B; pink represents mRNA high, medium blue represents mRNA low, orange represents protein high, and light blue represents protein low.
Survival rates in patients with high CC component mRNA levels
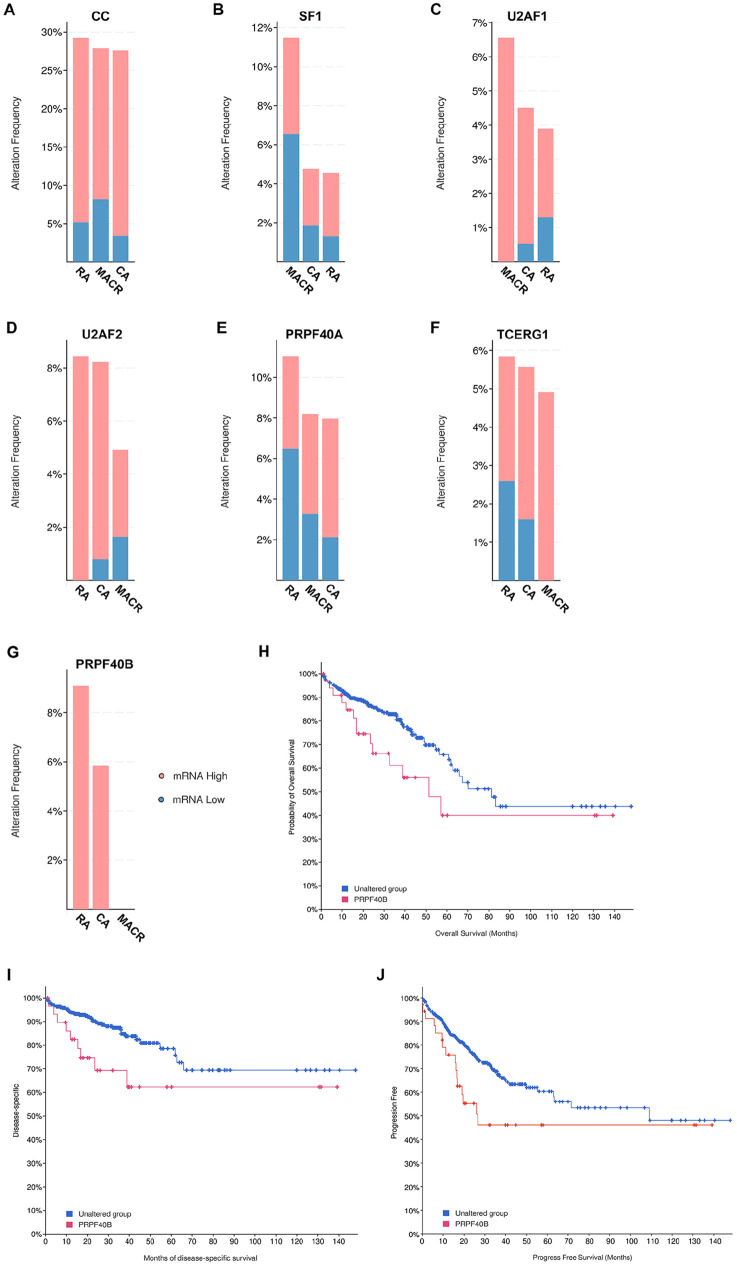
We examined bowel TCGA PanCancer Atlas dataset on how CC factor alterations contribute to patient survival but found no significant difference comparing patients with or without CC alterations (data not shown). We hypothesized that this may be because the TCGA PanCancer Atlas data includes a mix of all the different types of alterations such as high and low mRNA or protein levels as well as genetic mutations (Figure 2B to G). Each type of alteration could have distinct but contradictory effects that likely makes it difficult to observe a clear trend on patient survival outcome. To test this hypothesis, we queried the bowel TCGA PanCancer Atlas data for changes in mRNA expression only. The results of the query showed that for each CC factor, mRNA levels were either increased or decreased in different bowel cancer types (Figure 3A to G). However, in the case of PRPF40B, its mRNA levels were solely increased in all patient samples (Figure 3G). Examination of survival outcomes of the cohort of patients that expressed only high levels of PRPF40B mRNA found decreased overall survival (q = 0.047) (Figure 3H), disease-specific survival (q = 0.047) (Figure 3I), and progression-free survival (q = 0.047) (Figure 3J). Therefore, high mRNA levels correlate with negative outcomes for patient survival. This implies higher mRNA levels have oncogenic or gain-of-function effect. Conversely, we earlier described that mutation in CC factors correlated with enhanced survival rates (Figure 1C to H), thus indicating that CC mutations likely result in decreased mRNA or impaired CC factor function.
Figure 3.
Incidence of alterations in mRNA levels only from TCGA, PanCancer Atlas bowel cancer dataset (594 patients) of (A) all 6 CC genes (CC); (B) SF1; (C) U2AF1; (D) U2AF2; (E) PRPF40A; (F) TCERG1; and (G) PRPF40B. TCGA, PanCancer Atlas bowel cancer dataset was queried for the 6 CC factors and selected for mRNA expression only: mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM). Graph of cancer types detailed. Pink represents mRNA high and blue represents mRNA low. Key: as in Figure 1B. (H) Overall survival, (I) disease-specific survival, and (J) progression-free survival of patients with high PRPF40B mRNA. Blue line represents unaltered group (patients without alterations in mRNA levels of any CC factor) and red line represents patients with high mRNA levels of PRP40B. Overall survival using 459 patients (q = 0.047), disease-specific survival, 445 patients (q = 0.047) and progression free survival, 459 patients (q = 0.047).
Genetic and expression level of CC in other cancers
We also examined CC factor alteration rates in other cancer types and found expression level alterations of CC components to be always higher than genetic alteration rates (Supplementary Table 2). Thus, changes in the expression levels of the spliceosome CC factors are highly prevalent in most human cancers.
CC factor alterations associated with driver genes
In a previous study using genetically modified mouse strains, we observed that ApcMin/+;Sf1+/− mice develop significantly fewer intestinal polyps than ApcMin/+ mice, which indicated that SF1 deficiency reduces the strong polyp inducing potential of ApcMin driver gene. 17 In addition, the multi-step model of colorectal cancer development indicates that genetic alteration of driver gene APC occurs during early adenoma stage, KRAS alterations occur during intermediate to late adenoma stages and TP53 alterations occur in later adenocarcinoma stage.19-21 We therefore used cBioPortal database to investigate how changes in CC factors modulate the outcome of specific driver genes in colorectal cancer patients. Four cohorts of bowel cancer patients, each having genetic alterations in the driver genes APC, TRP53, KRAS, or BRAF, were isolated and examined for CC alterations (see also Supplementary Methods 3).
APC cohort
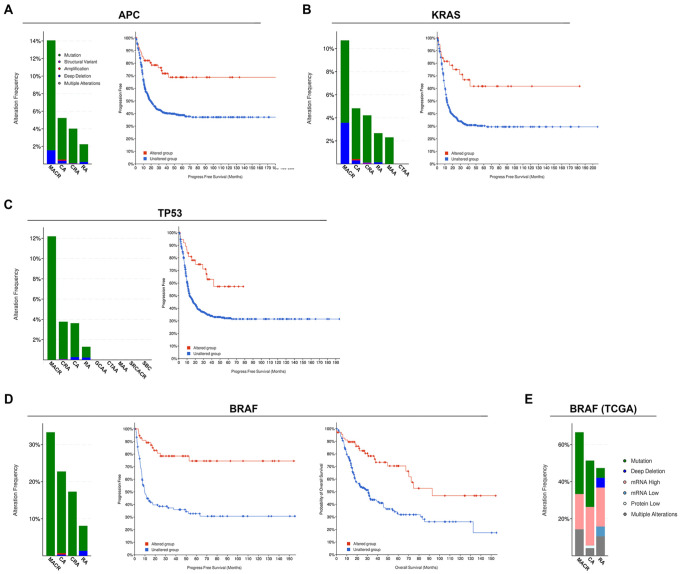
Sixty-eight percent of bowel cancer patients (or 4422 patients) had genetic changes in their APC gene (Table 2). Of these, a cohort of 4393 patients were selected that only carried APC gene mutations (excluding gene amplifications or deletions). CC factors were altered in 4% (177) of the patients carrying APC mutations. Progression-free survival was enhanced in patients with mutations in both APC and CC factors (APC/CC, indicated by red line) compared with that in patients with APC mutations alone (q = 1.160e-4) (Figure 4A).
Table 2.
Summary of driver gene (APC, KRAS, TP53, or BRAF) and CC factor mutation rates in bowel cancer patient cohorts.
| Studies queried | Driver gene | Driver gene alteration rate | No. of patients in cohort a | CC alteration rate | TCERG1 alteration rate |
|---|---|---|---|---|---|
| All non-redundant bowel cancer studies | - | - | 6523 | 5% (310/6523) | 7% |
| APC | 68% (4422/6523) | 4393 a | 4% (177/4393) | 5% | |
| KRAS | 43% (2796/6523) | 2737 a | 4% (106/2737) | 5% | |
| TP53 | 66% (4302/6523) | 4275 a | 3% (123/4275) | 4% | |
| BRAF | 10% (670/6523) | 661 a | 19% (127/661) | 24% | |
| Colorectal adenocarcinoma (TCGA, PanCancer Atlas) | - | - | 594 | 39% (232/594) | 11% |
| BRAF | 19% (112/594) | 62 a | 61% (38/62) | 26% |
Incidence of TCERG1 alteration is shown.
Cohort comprises number of patients with gene mutation only, excluding other genetic changes.
Figure 4.
(A) (left) Incidence of CC factor alterations in different types of bowel cancers in cohort of patients with APC mutations (4393 patients). Key: as in Figure 1B. (right) Progression-free survival (905 patients; q = 1.160e-4) of patients with APC mutations. Blue line represents unaltered group (patients with APC mutations and without alterations in any CC factor) and red line represents patients with APC mutations and with at least 1 alteration in the 6 queried CC genes. (B) (left) Incidence of CC alterations in bowel cancers in patients with KRAS mutations (2737 patients). (right) Progression-free survival (555 patients, q = 2.191e-3) of patients with KRAS mutations. Blue line represents unaltered group (patients with KRAS mutations but without alterations in any CC factor) and red line represents patients with KRAS mutations and with at least 1 alteration in the 6 queried CC genes. (C) (left) Incidence of CC alterations in bowel cancers in patients with TP53 mutations (4275 patients). (right) Progression-free survival (775 patients, q = 5.970e-3) of patients with TP53 mutations. Blue line represents unaltered group (patients with TP53 mutations but without alterations in any CC factor) and red line represents patients with TP53 mutations and with at least 1 alteration in the 6 queried CC genes. (D) (left) Incidence of CC alterations in bowel cancers in patients with BRAF mutations (661 patients). (middle) Progression-free (185 patients, q = 1.319e-6) and (right) overall survival (310 patients, q = 1.204e-4) of patients with BRAF mutations. Blue line represents unaltered group (patients with BRAF mutations but without alterations in any CC factor) and red line represents patients with BRAF mutations and with at least 1 alteration in the 6 queried CC genes. (E) Incidence of CC alterations in bowel cancers of patients with BRAF mutations from PanCancer Atlas dataset (62 patients). Key: as in Figure 1B.
KRAS cohort
Forty-three percent (2796 patients) of bowel cancer patients had KRAS mutation (Table 2). Four percent (106 patients) of the KRAS cohort had mutations in CC factors. The KRAS/CC cohort had better progression-free survival rates (q = 2.191e-3) compared with patients with KRAS mutations alone (Figure 4B).
TP53 cohort
Sixty-six percent (4302 patients) with bowel cancers had TP53 mutations. Of these 3% (123 patients) also had CC factor mutations (Table 2). TP53/CC patients had enhanced progression-free survival (q = 5.970e-3) compared with patients with mutations in TP53 alone (Figure 4C). Overall, CC mutations occurred in 3% to 4% of patients harboring APC, KRAS, or TP53 mutations, but patients with specific driver gene mutations plus CC factor mutations, had better survival outcomes.
BRAF cohort
Unlike the other cohorts, BRAF gene was altered in a lower proportion, 10% (670 patients), of bowel cancer patients but of these, a higher fraction, 19% (127 patients), carried CC mutations (Table 2). Patients with both BRAF and CC factor mutations had better progression-free (q = 1.319e-6) and overall survival rates (q = 1.204e-4) compared with those with BRAF mutations alone (Figure 4D).
We also queried the TCGA PanCancer Atlas dataset (colorectal adenocarcinoma) for BRAF alterations (Table 2 and Figure 4E). In this study of 594 patients, BRAF was altered (with mutated BRAF plus expression level changes) in 19% (112 patients) of the patients. Of these, 62 patients carried only BRAF gene mutations and constituted the BRAF/TCGA cohort (Table 2). Surprisingly, a significantly large proportion, 61% (38 patients) of the BRAF/TCGA cohort, also had alterations in CC factors, with MACR patients having the highest alteration rates in CC genes (Figure 4E). There was no observable survival advantage for the BRAF/CC patients from the TCGA PanCancer Atlas study (data not shown) probably because of the small sample size (62 patients with BRAF mutations) and also because almost equal proportions of genetic mutations and other alterations of CC factors were present. Thus, patient outcomes due to CC factor genetic mutations cannot be differentiated from those due to expression level changes in CC factors.
Co-occurrence of multiple CC factor mutations
Interestingly, significant co-occurrence of multiple CC factor mutations occurred in the APC and KRAS but not in the TP53 or BRAF cohort (Supplementary Table 3). In the multi-step model of colorectal cancer development, APC is genetically altered in early adenoma stage, KRAS alterations occur during intermediate to late adenoma stages, whereas TP53 alterations occur in subsequent adenocarcinoma stage. 20 Our analysis indicates that APC driver gene is associated with a greater number of CC factor changes compared with driver genes activated at the later stages, indicating that mutations in multiple CC factors are favorable during the early adenoma stage but not selectively retained in the more advanced stages with activated TP53. This observation also supports the tumor suppressive role of genetically altered CC factors. Cancer cells at the early adenoma stage, with APC mutations, retain some tumor suppressive elements, in the way of CC factor genetic alterations. These CC factor genetic alterations are subsequently not retained in the later adenocarcinoma stage.
Another observation was that the bridging factor, TCERG1, linking the 5′-ss factors with SF1 and other 3′-ss factors, was most frequently altered in all cohorts (ranging from 4% in TP53 cohort to 26% in the BRAF/TCGA, PanCancer Atlas cohort; Table 2 and Supplementary Figure 4). Thus, TCERG1 likely has additional functions in colorectal carcinomas. Studies have implicated TCERG1 as a colorectal carcinoma biomarker, 34 involved in transcription or transcriptional elongation.35,36
To summarize the driver gene cohort study findings, CC component mutations occurred in 3% to 4% of patients harboring APC, KRAS, or TP53 mutations, but patients with CC factor mutations had better survival outcomes. In contrast, 19% of patients with BRAF mutations also carried CC mutations. Further examination of TCGA dataset revealed 61% of patients with BRAF mutations had alterations (genetic or expression level changes) in CC factors and MACR patients had the highest CC alteration rates. Patients with both BRAF and CC factor mutations had better survival rates. Multiple CC factor mutations were simultaneously present in the APC and KRAS but not in the TP53 or BRAF cohort suggesting retention of tumor suppressor factors in early but not in the later stages of bowel cancer progression.
mRNA targets of CC factors
To understand the significance of CC factor alterations in cancer cells, it is important to know their RNA targets. Experimental studies using crosslinking and immunoprecipitation (CLIP) or RNA-Sequencing techniques have identified pre-mRNA targets and pathways targeted by SF1 from HeLa cells and from C elegans.2,18 In this study, we used the co-expression dataset in cBioPortal to obtain information regarding splicing targets of the macromolecular CC spliceosome.
We used the assumption that genes that are co-expressed with individual CC components and are common between multiple CC components could either be direct or indirect targets of the spliceosome CC or may regulate the CC. The bowel TCGA PanCancer Atlas database in cBioPortal was queried for mRNAs (RNA-sequencing data) and proteins (mass spectroscopy data) whose expression levels correlate with each CC factor (see Supplementary Methods 4). Co-expression gene lists were obtained for each CC factor: SF1, U2AF2, U2AF1, PRPF40A, PRPF40B, and TCERG1. Figure 5A shows one example of a gene, HCFC1, whose mRNA expression significantly and positively correlates with SF1 (q = 1.27e-60) expression in colorectal adenocarcinoma cells.
Figure 5.
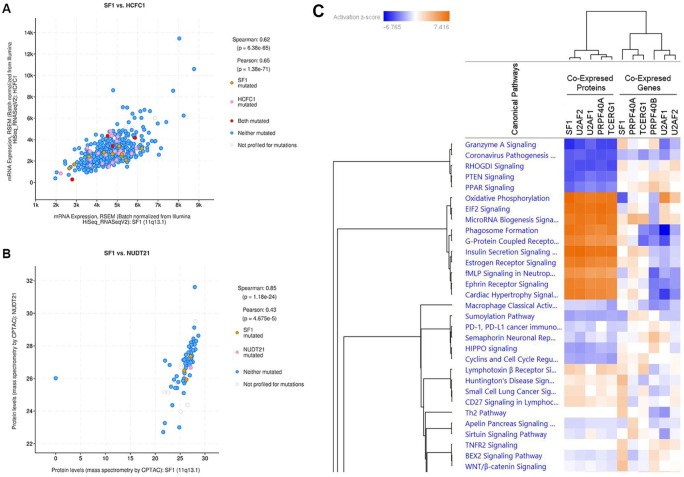
Co-expression and Ingenuity Pathway Analysis (IPA). (A) mRNA co-expression of HCFC1 with SF1 (q-value = 1.27e-60) in bowel cancer (TCGA PanCancer Atlas). mRNA expression is derived from RSEM (Batch normalized from Illumina HiSeq_RNASeqV2). Data are from 592 samples. (B) Protein co-expression of NUDT21 with SF1 (q-value = 6.53e-21) in bowel cancer (TCGA PanCancer Atlas). Protein co-expression is derived from mass spectrometry by CPTAC. Data are from 84 samples. (C) Heat map derived from QIAGEN Ingenuity Pathway Analysis using co-expressed proteins and mRNA associated with each of the 6 CC factors in bowel cancers. Left 5 lanes are derived from gene lists of proteins co-expressed with SF1, U2AF2, U2AF1, PRPF40A, and TCERG1. Right 6 lanes are derived from mRNA gene lists co-expressed with SF1, PRPF40A, TCERG1, PRPF40B, U2AF1, and U2AF2.
The colorectal adenocarcinoma (TCGA PanCancer Atlas) dataset also allowed for the query for proteins that are co-expressed with individual CC factors (example of NUDT21 co-expression with SF1, q = 6.53e-21, Figure 5B) (see Supplementary Methods 4). Protein co-expression data is available for SF1, U2AF1, U2AF2, PRPF40A, and TCERG1 but not for PRPF40B. We generated gene lists for protein co-expression for each of the CC factors.
We used the 5 lists of proteins co-expressed with each of the CC factors (SF1, U2AF1, U2AF2, PRPF40A, and TCERG1) together with the 6 lists of mRNAs co-expressed with each of the CC factors (SF1, U2AF1, U2AF2, PRPF40A, PRPF40B, and TCERG1) to directly perform QIAGEN Ingenuity Pathway Analysis (IPA). Surprisingly, when IPA was performed simultaneously on the co-expression protein and RNA data, the heat map shows that the protein and RNA data do not coincide (Figure 5C). However, the IPA heat map for protein data (Figure 5C, left lanes of heat map) has significant z-scores implying better reliability of the data. The likely explanation for this mismatch is that RNA expression levels usually cannot be directly correlated with protein expression levels in cells. Previous reports using experimental data found that for 90% of transcripts, transcription and translation are independent of each other and that it is translational control that regulates the cellular proteome.37,38 mRNA levels in cells have been found to correlate with approximately 40% of the total protein content in cells. Often, highly expressed transcripts are not translated or genes that are transcribed at low levels are translated efficiently. We recognize that this has significant implications in using and interpreting RNA expression data from experimental work and databases.
Based on the z-scores from the heat map, we observe that pathways strongly upregulated are metabolic pathways needed for cell survival, nutrition, biosynthesis, autophagy, cellular movement (invasion), or immune surveillance, such as oxidative phosphorylation, 39 E1F2 signaling, 40 insulin secretion, 41 microRNA biogenesis, 42 growth factor receptor–mediated signaling, and phagosome formation. 43 Conversely, pathways strongly downregulated include tumor suppressor pathways that regulate cell proliferation and apoptosis, including PTEN, 44 RHGODI, 45 PPAR, 46 and Granzyme A signaling. 47 Thus, CC spliceosomes associate with pre-mRNA transcripts of genes in biosynthetic pathways that favor cell growth and proliferation.
CC targets in other GI cancers
Because protein co-expression data is unfortunately only available for bowel cancers but not for other GI cancers, we used the available RNA-Sequencing co-expression data from esophagus, stomach, liver, and pancreas to perform IPA (Supplementary Figure 5). IPA indicated that some of the pathways identified overlapped with that identified from bowel cancers (compare to Figure 5C) such as oxidative phosphorylation and E1F2 signaling. Other pathways such as DNA methylation, spliceosomal cycle, NER (nucleotide excision repair), mismatch repair, and BER (base excision repair) pathways overlapped with pathways detected with less significance in the bowel cancer IPA heat map, where both protein and RNA data were used (compare Supplementary Figures 5 and 6). Thus, pathway analysis using RNA-Sequencing data provides some overlap with protein co-expression data, but the significance of the pathways may be under or over-estimated.
In summary, using bowel cancer protein and RNA co-expression data allowed comparison of pathway analysis. Protein co-expression data yielded more reliable pathway data. Overall, we observe, similar pathways are targets of CC spliceosome in different GI tissues.
Limitation of study
One limitation of this in silico study using human cancer patient data is that there is genetic heterogeneity within the human population. Thus, in our analysis, as we isolate the group of individuals with CC component changes (altered cohort) compared with those without any changes (unaltered cohort), the genetic heterogeneity in both cohorts remains a confounding factor. How other genetic changes present in individuals of each cohort affect the clinical outcome cannot be controlled. Thus, the results are best referred to as correlations with statistical significance. Other confounding factors in analyzing human cancer patient data from cBioPortal include sex, socioeconomic factors, genetic susceptibility, diet, substance use, and so on. Contingent on available data, our future work will attempt to parse out the effects of these factors. Moreover, experimental studies using genetically defined animal model systems can be used in future studies to verify the in silico study results.
Discussion
Analysis of the TCGA PanCancer Atlas data of different cancer types found that overall there is high incidence of CC factor changes in human cancer cells, including GI cancers. Most cancer types did not carry high rates of genetic mutations in CC factor genes (genetic alteration ranging from <1% to 23%), but when changes in gene expression levels were included, the alteration frequencies increased to 15% to 59%.
Alterations in the CC factors are highly prevalent in a third of patients with GI cancers, with the most common alterations being acquired gene mutations or high expression of mRNA. Mucinous adenocarcinoma of the colon and rectum carrying BRAF mutations had very high rates of CC factor alterations. Bowel cancer patients with APC and KRAS mutations, but not those with TP53 and BRAF mutations, tended to harbor mutations in more than 1 CC factor. Interestingly, the bridging factor TCERG1 is the most frequently altered CC component, especially in BRAF associated mucinous adenocarcinomas. TCERG1, an RNA-binding protein, has been reported to interact with colorectal adenocarcinoma driver genes or may itself function as a driver gene. 34
Our results share similarities with a previously reported study where analysis of spliceosome genes was performed across 27 cancer types in 9070 patients derived from the TCGA database. 48 This study did not focus on different spliceosome complexes but screened across all the spliceosome genes. It reported that some spliceosome genes had high mutation rates in different cancer types and survival was better in patients with the mutations. The study also found that mutation in TCERG1 was significantly associated in patients with longer survival periods of >2 years. Moreover, some spliceosome genes were expressed at low levels in patients with all cancer types, and these patients had better survival outcomes.
A number of studies have also reported alterations in specific splicing factors in colon cancers. Examples include reported increases in SNRPB, 48 a factor found in multiple spliceosomal complexes; increased acetylation of PHF5a, a component of the U2 snRNP, which increases colorectal tumorigenesis 49 ; increased expression of alternative splicing factor PTBP1 which promotes colon tumorigenesis-triggering splicing isoforms 50 ; and changes in a number of splicing factor components that influence proliferation, apoptosis, angiogenesis, invasion and metastasis, and drug resistance in colon cancers. 51
The alterations in CC factors appear to be clinically significant because patients with CC factor mutations showed a consistent trend toward better overall or progression-free survival rates. In contrast, patients expressing high CC factor mRNA levels had lower survival rates. Thus, mutations in CC factor genes likely impair spliceosome function and may decrease production of splice variants that promote cancer cell survival. This is reflected in enhanced patient survival rates when CC factor genes are mutated. In contrast, higher mRNA expression of individual CC factor genes likely enhances CC assembly to favor increased splicing of oncogenic variants in the cell, as reflected by the lower survival rates of patients that express high CC factor mRNA.
Expression of CC factors correlated with greater activity of metabolic and synthetic pathways and inhibition of pathways that decrease cell proliferation and upregulate apoptosis. Thus, we surmise that patients carrying mutations in individual CC factors downregulate metabolic and cell proliferation pathways and upregulate apoptotic pathways, resulting in enhanced survival. The enhanced survival in human patients with CC factor mutations concur with our previous experimental observations that genetic loss of SF1 has tumor suppressive effects.16,17 However, it should be noted that the genetic profile of cells from human cancer patients in cBioPortal are complex and many factors, in addition to the CC factor mutations, likely contribute to the observed clinical outcomes.
Ingenuity Pathway Analysis allowed for comparison of outcomes when protein or RNA data is used. Our IPA results support the evidence that transcription and translation are independent of each other and not always tightly correlated.37,38 There is, however, a paucity of protein co-expression data and thus most pathway analysis will have to currently rely on RNA-Sequencing data.
By using the cBioPortal co-expression database, we have been able to identify the likely targets and pathways of a large and transient CC (E-complex). This would be difficult to perform experimentally. Usually, experimental methods assess RNA targets of each splicing factor individually but it is difficult to assess the targets of a complex. For example, previous experimental work identified the targets of SF1 using ectopically expressed SF1 in HeLa and C elegans.2,18 RNA that experimentally bound to SF1 were coprecipitated, isolated, identified, and subsequently used for pathway analysis. The studies indicated that a number of metabolic pathways are regulated by SF1 and especially the nutrition sensing TORC1 pathway that regulates cell growth and development. 2 Our studies, using a slightly different approach in considering the CC macromolecule and using protein data, have also identified a number of metabolic pathways as targets of the CC. However, it is to be expected that the targets and pathways regulated by SF1 and CC are unlikely to be exactly the same in different cell types.
It also raises the question as to how increasing the complexity of the spliceosome complex could affect target specificity, that is, whether the mRNA targets that bind individually to specific splicing factors are the same as that of the spliceosome complex. These questions remain to be answered. In this study, we also sought to reduce the complexity in our analysis by excluding U1 snRNP which is comprised of at least 10 proteins.25,26 Future work will examine the contributions of U1 snRNP.
Conclusions
Our results found surprisingly high incidence of CC component alterations in GI cancers. Patients with mutations or low expression of CC genes exhibited a consistent trend of favorable survival rates. This is likely related to the fact that the CC regulates specific metabolic and tumor suppressor pathways. The findings from our in silico study imply that therapeutic lowering of expression levels of CC factors in colon cancer patients may have positive effects on patient survival, especially for patients with mucinous adenocarcinoma. Our results will serve to guide further experimental work to evaluate the function and clinical significance of alterations of the CC in human cancers.
Supplemental Material
Supplemental material, sj-pdf-1-bbi-10.1177_11779322241287115 for Commitment Complex Splicing Factors in Cancers of the Gastrointestinal Tract—An In Silico Study by Yun Zhang, Alexandria Carrasquillo Simko, Uzondu Okoro, Deja Jamese Sibert, Jin Hyung Moon, Bin Liu and Angabin Matin in Bioinformatics and Biology Insights
Footnotes
Author contributions: AM and YZ conceived the idea. ACS, UO, DJS, and JHM assisted in data gathering and analysis. YZ performed the IPA analysis and BL contributed to statistical analysis. AM and YZ contributed to writing and editing the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: A.C.S., D.J.S., and J.H.M. were supported by the 2022 Summer Scholars Program at MUSM. Y.Z. is supported by NIGMS (1SC2GM135111 and 1R16GM149425), NIH RCMI (U54MD007605), and CPRIT (RP180748).
Data availability: Data and analytic methods will be made available to other researchers upon request.
ORCID iD: Angabin Matin  https://orcid.org/0000-0002-7592-3438
https://orcid.org/0000-0002-7592-3438
Supplemental material: Supplemental material for this article is available online.
References
- 1. Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13-19. doi: 10.1038/ng0102-13 [DOI] [PubMed] [Google Scholar]
- 2. Heintz C, Doktor T, Lanjuin A, et al. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature. 2017;541:102-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhadra M, Howell P, Dutta S, Heintz C, Mair WB. Alternative splicing in aging and longevity. Hum Genet. 2020;139:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradley RK, Anczuków O. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer. 2023;23:135-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer—implications for care. Nat Rev Clin Oncol. 2020;17:457-474. [DOI] [PubMed] [Google Scholar]
- 6. Sveen A, Kilpinen S, Ruusulehto A, et al. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2015;35:2413-2427. [DOI] [PubMed] [Google Scholar]
- 7. Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311-5318. [DOI] [PubMed] [Google Scholar]
- 8. Will C, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larson J, Hoskins A. Dynamics and consequences of spliceosome E complex formation. Elife. 2017;6:e27592. doi: 10.7554/eLife.27592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borao S, Ayté J, Hümmer S. Evolution of the early spliceosomal complex—from constitutive to regulated splicing. Int J Mol Sci. 2021;22:12444. doi: 10.3390/ijms222212444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berglund JA, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rino J, Desterro JM, Pacheco TR, Gadella TW, Jr, Carmo-Fonseca M. Splicing factors SF1 and U2AF associate in extraspliceosomal complexes. Mol Cell Biol. 2008;28:3045-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Selenko P, Gregorovic G, Sprangers R, et al. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11:965-976. [DOI] [PubMed] [Google Scholar]
- 14. Kent OA, Ritchie DB, Macmillan AM. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol Cell Biol. 2005;25:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kielkopf C, Rodionova N, Green M, et al. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595-605. [DOI] [PubMed] [Google Scholar]
- 16. Zhu R, Heaney J, Nadeau J, et al. Deficiency of Splicing Factor 1 (SF1) suppresses occurrence of testicular germ cell tumors. Cancer Res. 2010;70:7264-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Godavarthi J, Polk S, Nunez L, et al. Deficiency of Splicing Factor 1 (SF1) reduces intestinal polyp incidence in ApcMin/+ Mice. Biology. 2020;9:398. doi: 10.3390/biology9110398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corioni M, Antih N, Tanackovic G, et al. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2010;39:1868-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vogelstein B, Fearon E, Hamilton S, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [DOI] [PubMed] [Google Scholar]
- 20. Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159-170. [DOI] [PubMed] [Google Scholar]
- 21. Tanaka T. Colorectal carcinogenesis: review of human and experimental animal studies. J Carcinog. 2009;8:5. doi: 10.4103/1477-3163.49014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao J, Aksoy B, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Bruijn I, Kundra R, Mastrogiacomo B, et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR project GENIE biopharma collaborative in cBioPortal. Cancer Res. 2023;83:3861-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilkinson M, Charenton C, Nagai K. RNA splicing by the spliceosome. Annu Rev Biochem. 2020;89:359-388. [DOI] [PubMed] [Google Scholar]
- 26. Campagne S, de Vries T, Malard F, et al. An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands. Nucleic Acids Res. 2021;49:e63. doi: 10.1093/nar/gkab135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin KT, Lu RM, Tarn WY. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol Cell Biol. 2004;24:9176-9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Makarov EM, Owen N, Bottrill A, Makarova OV. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res. 2012;40:2639-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Becerra S, Montes M, Hernández-Munain C, Suñé C. Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo. RNA. 2015;21:438-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suñé C, Hayashi T, Liu Y, Lane WS, Young RA, Garcia-Blanco MA. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol. 1997;17:6029-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bedford MT, Leder P. The FF domain: a novel motif that often accompanies WW domains. Trends Biochem Sci. 1999;24:264-265. [DOI] [PubMed] [Google Scholar]
- 32. Becerra S, Andrés-León E, Prieto-Sánchez S, et al. Prp40 and early events in splice site definition. Wiley Interdiscip Rev RNA. 2015;7:17-32. [DOI] [PubMed] [Google Scholar]
- 33. Huang A, Yang Y, Shi J, et al. Mucinous adenocarcinoma: a unique clinicopathological subtype in colorectal cancer. World J Gastrointest Surg. 2021;13:1567-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. García-Cárdenas JM, Armendáriz-Castillo I, García-Cárdenas N, et al. Data mining identifies novel RNA-binding proteins involved in colon and rectal carcinomas. Front Cell Dev Biol. 2023;11:1088057. doi: 10.3389/fcell.2023.1088057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Banman S, McFie P, Wilson H, et al. Nuclear redistribution of TCERG1 is required for its ability to inhibit the transcriptional and anti-proliferative activities of C/EBPalpha. J Cell Biochem. 2010;109:140-151. [DOI] [PubMed] [Google Scholar]
- 36. Sánchez-Hernández N, Boireau S, Schmidt U, et al. The in vivo dynamics of TCERG1, a factor that couples transcriptional elongation with splicing. RNA. 2016;22:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337-342. [DOI] [PubMed] [Google Scholar]
- 38. Tebaldi T, Re A, Viero G, et al. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012;13:220. doi: 10.1186/1471-2164-13-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ashton T, McKenna W, Kunz-Schughart L, et al. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24:2482-2490. [DOI] [PubMed] [Google Scholar]
- 40. García-Jiménez C, Goding C. Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metab. 2019;29:254-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Giovannucci E. Insulin, insulin-like growth factors and colon cancer: a review of the evidence. J Nutr. 2001;131:3109S-3120S. doi: 10.1093/jn/131.11.3109S [DOI] [PubMed] [Google Scholar]
- 42. Blahna MT, Hata A. Regulation of miRNA biogenesis as an integrated component of growth factor signaling. Curr Opin Cell Biol. 2013;25:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang T, Song X, Yang Y, et al. Autophagy and hallmarks of cancer. Crit Rev Oncog. 2018;23:247-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641-669. [DOI] [PubMed] [Google Scholar]
- 45. Harding MA, Theodorescu D. RhoGDI signaling provides targets for cancer therapy. Eur J Cancer. 2010;46:1252-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo Y, Xie C, Brocker CN, et al. Intestinal PPARα protects against colon carcinogenesis via regulation of methyltransferases DNMT1 and PRMT6. Gastroenterology. 2019;157:744-759. doi: 10.1053/j.gastro.2019.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Legrand F, Driss V, Delbeke M, et al. Human eosinophils exert TNF-α and granzyme A-mediated tumoricidal activity toward colon carcinoma cells. J Immunol. 2010;185:7443-7451. [DOI] [PubMed] [Google Scholar]
- 48. Ye Z, Bing A, Zhao S, Yi S, Zhan X. Comprehensive analysis of spliceosome genes and their mutants across 27 cancer types in 9070 patients: clinically relevant outcomes in the context of 3P medicine. EPMA J. 2022;13:335-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Z, Yang X, Liu C, et al. Acetylation of PHF5A modulates stress responses and colorectal carcinogenesis through alternative splicing-mediated upregulation of KDM3A. Mol Cell. 2019;74:1250-1263. [DOI] [PubMed] [Google Scholar]
- 50. Hollander D, Donyo M, Atias N, et al. A network-based analysis of colon cancer splicing changes reveals a tumorigenesis-favoring regulatory pathway emanating from ELK1. Genome Res. 2016;26:541-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Y, Huang M, Liu X, et al. Alternative splicing of mRNA in colorectal cancer: new strategies for tumor diagnosis and treatment. Cell Death Dis. 2021;12:752. doi: 10.1038/s41419-021-04031-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-bbi-10.1177_11779322241287115 for Commitment Complex Splicing Factors in Cancers of the Gastrointestinal Tract—An In Silico Study by Yun Zhang, Alexandria Carrasquillo Simko, Uzondu Okoro, Deja Jamese Sibert, Jin Hyung Moon, Bin Liu and Angabin Matin in Bioinformatics and Biology Insights