Abstract
Autism spectrum disorder (ASD) is a developmental disorder with a rising prevalence and unknown etiology presenting with deficits in cognition and abnormal behavior. We hypothesized that the investigation of the synaptic component of prefrontal cortex may provide proteomic signatures that may identify the biological underpinnings of cognitive deficits in childhood ASD. Subcellular fractions of synaptosomes from prefrontal cortices of age-, brain area-, and postmortem-interval-matched samples from children and adults with idiopathic ASD vs. controls were subjected to HPLC-tandem mass spectrometry. Analysis of data revealed the enrichment of ASD risk genes that participate in slow maturation of the postsynaptic density (PSD) structure and function during early brain development. Proteomic analysis revealed down regulation of PSD-related proteins including AMPA and NMDA receptors, GRM3, DLG4, olfactomedins, Shank1-3, Homer1, CaMK2α, NRXN1, NLGN2, Drebrin1, ARHGAP32, and Dock9 in children with autism (FDR-adjusted P < 0.05). In contrast, PSD-related alterations were less severe or unchanged in adult individuals with ASD. Network analyses revealed glutamate receptor abnormalities. Overall, the proteomic data support the concept that idiopathic autism is a synaptopathy involving PSD-related ASD risk genes. Interruption in evolutionarily conserved slow maturation of the PSD complex in prefrontal cortex may lead to the development of ASD in a susceptible individual.
Keywords: autism, Brodmann area 9, childhood, proteomics, synaptopathy
Introduction
Autism is a severe neurodevelopmental disorder with a rising prevalence of 27.6 per 1,000 (1 in 36; Maenner et al. 2023). Autism is characterized by persistent deficits in social communication and social interaction across multiple contexts and restricted, repetitive patterns of behavior, interests, or activities (American Psychiatric Association 2013). Beyond core symptoms, people with autism display a number of comorbidities including seizure disorder, intellectual disability, and other cognitive impairments (Genovese and Butler 2023). Due to the heterogeneous nature of autism spectrum disorders (ASDs), it is not surprising that multiple gene families have been implicated in the pathology of autism. Hundreds of genes have been associated with core deficits of autism; however, they only account for 10–20% of ASD cases (Rylaarsdam and Guemez-Gamboa 2019). A number of neuropathologic abnormalities have been characterized in brains from individuals with autism including macrocephaly, volumetric and cellular abnormalities of the frontal cortex, parietal cortex, limbic, and cerebellar structures, and cortical minicolumnar disorganization (Casanova et al. 2002; Fatemi et al. 2012; Donovan and Basson 2017).
As ASD is an umbrella term that encompasses not only idiopathic autism (~85% of cases) but also syndromic forms such as fragile X syndrome or Angelman syndrome (~15% of cases) (Casanova et al. 2020), the search for consistent autism-specific biomarkers has been difficult. Nevertheless, much of the focus has been placed on investigating neurotransmitter systems including glutamatergic, gamma-aminobutyric acid (GABA)ergic, serotonergic, and cholinergic systems (Marotta et al. 2020). Imbalance between excitatory (glutamatergic) and inhibitory (GABAergic) signaling has been hypothesized to contribute significantly to core features of autism (Nelson and Valakh 2015). Furthermore, meta-analyses of blood and lymphocyte studies have also identified significant changes in methylation proteins (Guo, Ding et al. 2020), increased expression of brain-derived neurotrophic factor (Saghazadeh and Rezaei 2017), increased plasma glutamate levels (Zheng et al. 2016), and increased serotonin levels (Gabriele et al. 2014). The use of newer technologies such as proteomics has the potential to improve our understanding of brain protein expression in autism.
There are unique challenges for proteomic studies of autism including the etiological heterogeneity of autism, the presence of comorbid conditions, and how autism is defined (Szoko et al. 2017). A targeted selected reaction monitoring mass spectrometry proteomic study of Brodmann Area 10 (BA10) and cerebellum from subjects with autism vs. controls identified opposite directional changes for myelination, energy, and synaptic proteins in BA10 vs. cerebellum (Broek et al. 2014). More recently, Abraham et al. conducted a proteomic study of BA19 and posterior inferior cerebellum (Abraham, Szoko, Barnard, et al. 2019). In BA19, 27 proteins were significantly altered after adjusting for false discovery rate (FDR), while in the cerebellum, 38 proteins were significant (Abraham, Szoko, Barnard, et al. 2019). Upstream regulator analysis identified proteins associated with neurogeneration in both regions (Abraham, Szoko, Barnard, et al. 2019). While the studies by Abraham, Szoko, Barnard, et al. (2019) and Abraham, Szoko, and Natowicz (2019) are timely, there continues to be a dearth of proteomic data on children vs. adults with autism and no studies describing the proteome of synapse, necessitating the rationale behind the current study.
Methods and materials
Human brain procurement
All experimental procedures were approved by the Institutional Review Board of the University of Minnesota School of Medicine. Postmortem blocks of dorsolateral prefrontal cortex (Brodmann area 9 = BA9) (Table 1) were obtained from the Autism Research Foundation and their affiliated brain banks (NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland; the Harvard Brain Tissues Research Center; and the Autism Tissue Program). These samples, which have been used by our laboratory previously (Fatemi et al. 2013), are some of the most well-characterized and most-studied brain collections used by multiple groups (Palmen et al. 2004). Prior to freezing, donated brains were sectioned in half, dissected by anatomists, and stored at −80 °C until use. Consent from next of kin was given to the respective institutions. DSM-IV diagnoses were established prior to death by neurologists and psychiatrists using all clinical data from available medical records from family interviews. Samples were matched for age, brain area, and postmortem interval (PMI). All demographic information is listed in Table 1. None of the control samples had any history of neuropsychiatric disorders, seizure disorder, or intellectual disabilities. Each brain sample included both gray and white matter. The tissue samples were prepared for subcellular fractionation and proteomics as previously described and detailed below (Taha et al. 2014; Mueller et al. 2015; Folsom et al. 2019).
Table 1.
Clinical characteristics of BA9 brain tissues in children and adults with idiopathic ASD.
| Group | Children | Adults | ||
|---|---|---|---|---|
| Autism | Control | Autism | Control | |
| Total N | 5 | 5 | 5 | 5 |
| Ethnicity | 3C:1AA:1A | 3C:1AA:1 U | 4C:1AA | 2C:1AA:2 U |
| Age * | 10.6 ± 3.66 | 11.0 ± 5.0 | 29.4 ± 4.75 | 24.4 ± 6.65 |
| Sex | 4 M:1F | 5 M | 4 M:1F | 3 M:2F |
| PMI * | 16.2 ± 5.8 | 22.4 ± 8.6 | 26.4 ± 8.90 | 20.6 ± 7.06 |
*Comparison of autistic and control values were statistically nonsignificant using an unpaired t-test. A, Asian; AA, African American; C, Caucasian; F, Female; M, Male; U, Unknown.
Subcellular fractionation
Human BA9 samples (n = 5 per group, 20 per brain area) were subjected to homogenization and subcellular fractionation as previously described (Fatemi et al. 2017; Folsom et al. 2019), based upon established techniques (Taha et al. 2014; Mueller et al. 2015). BA9 samples were incubated for 4 min on ice in 1× isotonic extraction buffer (10-mM HEPES, 250-mM sucrose, 25-mM KCl, 1-mM EGTA, pH 7.8) plus protease inhibitors (PI) at a volume of 3× the weight of the sample. Subsequently, BA9 tissue was homogenized four times (30 s each) using a motorized pestle. Following homogenization, a 60-μL aliquot was saved as total homogenate. The remaining homogenate underwent a low-speed centrifugation (700× g) (5415D centrifuge, Eppendorf North America, Hauppauge, NY, USA) for 10 min at 4 °C to separate the intact nuclei and heavy membranes (pellet 1) from the supernatant (supernatant 1). Next, supernatant 1 was centrifuged at 15,000× g for 10 min at 4 °C, to separate the crude membrane fraction (pellet 2) from the supernatant (supernatant 2). Pellet 2 was reconstituted in sucrose homogenization buffer and added to ultracentrifuge tubes containing 3 mL of Triton X-100 buffer (10-mM Tris–HCl, 1-mM Na3PO4, 5-mM NaF, 1-mM EDTA, 1-mM EGTA, 0.5% v/v Triton X-100, pH 7.4, + PI). A 30,000× g centrifugation step for 20 min (Optima L-90 K Ultracentrifuge, Beckman-Coulter, Indianapolis, IN, USA) at 4 °C resulted in a Triton-insoluble pellet (pellet 3) constituting the synaptic fraction. The synaptic fraction was then reconstituted in 40 μL of PBS + PI. Protein levels for synaptic fraction were determined using a Bradford assay and samples were stored at −80 °C until proteomic analysis.
Orbitrap fusion liquid chromatography–mass spectrometry analysis
Following reconstitution of the dried peptide fractions in 97.9:2.0:0.1 H2O:acetonitrile (ACN):formic acid (FA), each sample was labeled with a specific label from the Thermo Scientific TMTpro™ 16plex Label Reagent Set, and samples were pooled and then analyzed by capillary LC–MS with a Thermo Fisher Scientific (Waltham, MA) Dionex UltiMate 3,000 system in-line with Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Peptides were loaded directly on-column in solvent A (99.9:0.1 H2O:FA) at maximum pressure (800 bar). Peptides were separated on a self-packed C18 column (Dr Maisch GmbH ReproSil-PUR) 1.9 μm, 120 A C18aq, 100 μm ID × 30-cm length at 55 °C with a biphasic gradient starting at 5% solvent B (99.9:0.1 ACN:FA) at a flow rate of 400 nl/min. The starting conditions were held for 2 min and then the gradient increased to 8% solvent B by 2.5 min. The flow was reduced to 315 nl/min and the gradient increased to 22% solvent B by 62 min and to 45% solvent B by 75 min. Finally, the gradient was increased to 90% solvent B by 77 min with a flowrate of 400 nl/min and held to 83 min followed by a return to starting conditions at 5% solvent B at 85 min and held to 92 min. Then, the utilization of a top 12 data dependent acquisition method with the following MS parameters: ESI voltage 2.1 kV, ion transfer tube 275 °C; Orbitrap MS1 scan 120-k resolution in profile mode from 380–1580 m/z with 100-ms maximum injection time, 100% (4E5) automatic gain control (AGC); MS2 triggered on the top 12 most abundant ions above 5E4 counts, 1.6-Da quadrupole isolation window, fixed HCD activation with 40% collision energy, Orbitrap detection with 60-K resolution at 200 m/z, first mass fixed at 122 m/z, 150-ms max injection time, 1E6 AGC, and 40-s dynamic exclusion duration with ±10 ppm mass tolerance.
Database search
Peptide tandem MS data were processed using Sequest [Thermo Fisher Scientific, San Jose, CA, in Proteome Discoverer (PD) 2.5]. The human (taxonID 9606) Universal Proteome (UP000005640) target protein sequence database was downloaded from UniProt (www.uniprot.org/) on 2019 July 12 and merged with a common lab contaminant protein database (http://www.thegpm.org/cRAP/index.html); the number of protein sequences was 74,234 sequences. Peptide search parameters specified trypsin digestion with a maximum of two missed cleavage sites, fragment ion mass tolerance 0.05 Da, and precursor tolerance 15 ppm. Variable modifications were set for the oxidation of methionine, pyroglutamic acid conversion from glutamine, deamidation of asparagine, acetyl and/or met-loss of the protein N-terminus, and TMT10plex of lysine and peptide N-terminus. Carbamidomethyl of cysteine was specified as a fixed modification.
Criteria for protein identification
One percent protein and peptide FDR filters were applied using the Percolator Algorithm (Käll et al. 2007) in PD.
Protein quantification
PD for TMT-based protein quantification was run with the following parameters: unique and razor peptides were included, shared peptides were excluded, impurity corrections were applied, co-isolation threshold maximum was 50%, normalization was performed on the total peptide amount; protein ratio calculations were performed using pairwise ratio-based mode; and hypothesis testing was performed using the background t-test approach. PD employed the Benamini–Hochberg FDR procedure to control for errors associated with multiple hypothesis tests (Benjamini and Hochberg 1995).
Statistical and Bioinformatic analyses of proteomic data
Peptide and protein quantification data were exported from PD software and imported into the R statistical programming language environment (R Core Team 2023). To improve sensitivity to biologically relevant changes, several complementary techniques were applied to analyze the data.
The peptide quantitations were used for a univariate approach; for each protein, a linear mixed model (LMM) was fitted to its quantitated, unique peptides; the LMM assumed correlation among those peptide levels and used the disease condition (autism) as a covariate. To account for multiple hypothesis testing, the Benjamini–Hochberg FDR control procedure was used to adjust the P-values obtained for the effect of disease status; these adjusted P-values were used as the criterion for assessments of significance of relative expression for individual proteins.
Pathway enrichment and coordinated expression of proteins was examined with multivariate techniques, using the inferred protein quantitations and pathway annotations from Reactome and WikiPathways exported from PD. First, the “predictive variable importance in projection” (VIP4p; Galindo-Prieto et al. 2014) metric from “orthogonal projection to latent structures discriminant analysis” (OPLS-DA) was computed for all proteins (without significance filtering). OPLS-DA separates the covariation among proteins that is correlated with the predictor (ASD vs. control) and from the covariation that is orthogonal to (i.e. uncorrelated with) the predictor; the VIP4p metric summarizes the impact of a protein on the former covariation, and variables with VIP4p > 1.0 are generally considered to have greater biological relevance with respect to the predictor (Mehmood et al. 2012). All proteins were ranked by VIP4p and subjected to “gene set enrichment analysis” (GSEA; Subramanian et al. 2005). As a complementary approach to increase sensitivity to enriched pathways, GSEA was also applied to the fold-changes of all protein quantitations inferred by PD, without reference to the LMM-based peptide quantitations or to any significance test. For these fold-changes, nominal P-values for t-tests of the inferred protein quantitations were considered when exploring possibly important proteins of biological significance to the etiology of autism but not for significance testing (Schwartzman and Lin 2011). When accounting for multiple hypothesis testing of GSEA enrichment scores, FDR-adjustment of P-values for these scores included correction for the fact that two ranking methods were used for GSEA. Gene sets enriched for Reactome and WikiPathways gene sets were identified using GSEA functions available through Tidyproteomics software (Jones et al. 2023). Enrichments of Reactome and WikiPathways gene sets were used to identify and explore relationships among significantly altered differentially expressed brain proteins.
For visual representation of protein quantitations using heatmaps, the hclust and dist functions of the R stats package were used for hierarchical clustering of both proteins and samples using Pearson correlation as the distance metric and the “ward.D2” agglomerating clustering method (Meunier et al. 2007). Finally, SynGo and Simon’s Foundation websites (https://syngoportal.org/ and https://gene.sfari.org/ respectively) were used for the identification of synaptic proteins and risk genes for autism.
Results
Proteomic analysis of brain synaptic fractions obtained from different groups (autistic vs. controls; children vs. adults) demonstrated a list of several thousand peptides per group. Quantitation of identified proteins and enumeration of differentially expressed proteins were based on three statistical methods. The statistical analytic techniques included an initial t-test leading to a differentially expressed list of up- or down-regulated proteins (nominal P < 0.05). We next used OPLS-DA which calculated VIP4p > 1 values to set out significant differential features between autistic and control groups, and lastly, peptide quantification approach was used to generate FDR-adjusted P-values for significantly differentially expressed proteins (Tables S1 and S2). The combination of these methods provided us with statistically reliable tests that discriminated between peptide and protein data ascribed to autistic pathology vs. control values.
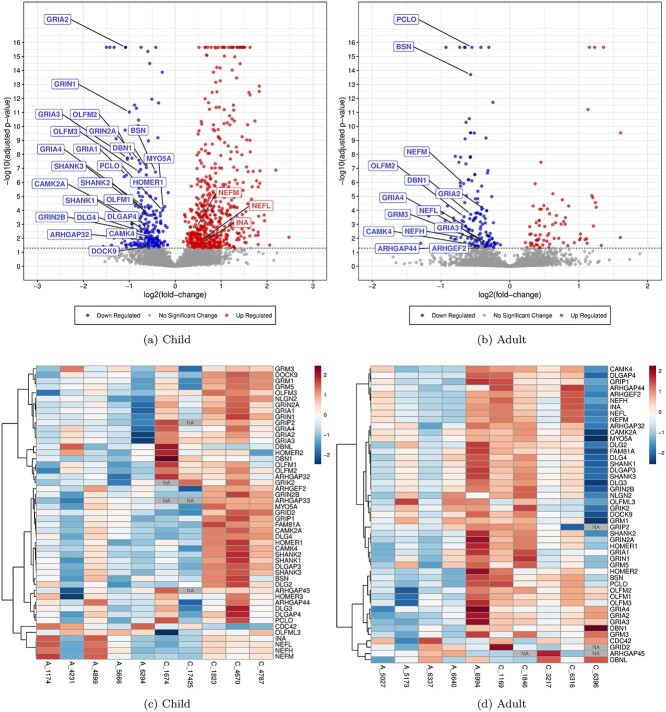
For BA9 of autistic children, the protein-based quantitation method identified 4,663 proteins, and 485 proteins were significant at the 5% level (nominal P < 0.05; 302 up-regulated and 183 down-regulated proteins). When an OPLS-DA analysis was performed on all proteins, 1,847 proteins were found to have VIP4p > 1.0 (1,015 up-regulated and 832 down-regulated) suggesting a biologically relevant covariation between the ASD-diagnostic status and these proteins. For the peptide-based LMM method, 77,452 peptides were detected; 4,677 proteins were identified based on the presence of at least two unique peptides or more per protein; this technique yielded 812 proteins significant at the 5% level (574 up-regulated and 238 down-regulated after FDR adjustment; Fig. 1a; Table S1).
Fig. 1.
Expression of selected synaptic proteins and their associated proteins. Panels (a) and (b), are plots for children and adults, respectively, of −log10 (adjusted P-value) vs. log2 (fold-change) for all proteins, computed for adjusted P-value of linear models of protein concentration based on peptide quantitation. Synaptic and associated proteins are labeled when above the threshold of significance. Panels (c) and (d) represent heatmaps of quantitations for synaptic and associated proteins. Quantities are centered and standard-scaled for each protein. Proteins are hierarchically clustered using Pearson correlation as the distance metric (longer lines perpendicular to the axis indicate lower correlation) and the “ward.D2” agglomerative clustering method. Undetected or unquantitated proteins are marked “NA.”
Proteomic data obtained from BA9 of adult subjects diagnosed with idiopathic autism identified 4,552 proteins, and 100 proteins were found significant at the 5% level (79 up-regulated and 21 down- regulated; nominal P < 0.05). Application of OPLS-DA analysis identified 1,629 proteins to have VIP4p > 1.0 (1,020 up-regulated and 609 down-regulated). Subsequent administration of the peptide analysis technique yielded 4,484 proteins with 226 proteins significant at the 5% level (FDR-adjusted P < 0.05; 73 up-regulated and 153 down-regulated; Fig. 1b; Table S2).
To further understand the significance of protein expression, we explored the enrichment of differentially expressed Reactome (Tables S3, S4, S7, and S8) and WikiPathways (Tables S5, S6, S9, and S10) gene sets using GSEA of proteins in BA9 of children and adults diagnosed with autism ranked by log2FC (Table 2; Figs. S1, S2, and S5–S11) and by VIP4p (Figs. S3 and S4). GSEA helped identify coordinated expression of proteins from several important pathways in BA9 of children including modulation of NMDA response, long-term potentiation, activation of AMPA receptors, neurexins, and neuroligins, glycolysis, Ras activation of calcium influx through NMDA receptor, and protein aggregation and folding (Fig. S1). Interestingly, WikiPathways analysis showed the enrichment of proteins involved in disruption of postsynaptic signaling by copy number variation (CNV), oxidative phosphorylation, electron transport chain, and mitochondria, glutathione metabolism, and glycolysis in senescence (Fig. S2). In contrast, enriched pathways in BA9 of adult subjects with autism involved cargo recognition for clathrin-mediated endocytosis and oxidative phosphorylation (Fig. S3; Table S10); cytoplasmic ribosomal proteins (Fig. S2); and peptide chain elongation, messenger RNA splicing, cytoskeletal components, and complement-related proteins (Fig. S1). Despite the differences in enriched pathways between childhood vs. adulthood in ASD values, several pathways overlapped in both age groups consisting of peptide chain elongation, eukaryotic translation termination, and selenocysteine synthesis (Fig. S1). For adults, GSEA analysis of Reactome gene sets showed an interesting pathway enrichment in regulation of complement cascade (Fig. S1; Tables S1, S2, and S4). Level of C3 factor was upregulated significantly in adult and child ASD subjects (respectively: log2FC = 0.8051, FDR-adjusted P < 0.0001; log2FC = 0.9933, FDR-adjusted P < 0.0001). GSEA did not score this enrichment for children, yet, in children, C4B was significantly upregulated (log2FC = 0.9575, FDR-adjusted P < 0.0001) and C1QBP was significantly downregulated (log2FC = −0.7569, FDR-adjusted P = 0.0304). Previous reports have indicated the involvement of the complement system in abnormal or excessive synaptic pruning or synaptic elimination in neurodegenerative diseases (Stephan et al. 2012; Presumey et al. 2017) or liability to the development of autism or schizophrenia (Sager et al. 2021). While a significant number of various brain specific proteins were deemed to be of importance in ASD, we focused our attention on changes in several glutamatergic and postsynaptic density (PSD)-related proteins because of their pathological involvement in neurodevelopmental disorders such as autism and schizophrenia (Hansen et al. 2021; Sabo et al. 2023; Wang et al. 2023).
Table 2.
GSEA enrichment of selected pathways in BA9 of children and adults with idiopathic ASD.
| Children | Adults | ||||
|---|---|---|---|---|---|
| Pathway | Adjusted P-value | Normalized enrichment score | Pathway | Adjusted P-value | Normalized enrichment score |
| NMDA activation (R) | <0.0001 | 2.3105 | Formation of the cornified envelope (R) | <0.0001 | 2.3246 |
| NMDA regulation (R) | 0.0006 | 2.1498 | Peptide chain elongation (R) | <0.0001 | 1.9071 |
| Long-term potentiation (R) | 0.0011 | 2.1379 | Common Pathway of Fibrin Clot Formation (R) | 0.0210 | 1.8545 |
| AMPA receptor activation (R) | 0.0035 | 1.9252 | mRNA processing (W) | 0.0003 | 1.7768 |
| Ras activation upon Ca influx through NMDA receptor (R) | 0.0499 | 1.862 | Oxidative phosphorylation (W) | 0.0117 | 1.7186 |
| Disruption of postsynaptic signaling by CNV (W) | 0.0064 | 1.8221 | mRNA splicing (R) | 0.002 | 1.6449 |
| Oxidative phosphorylation (W) | 0.0146 | 1.7258 | Clathrin-mediated endocytosis (R) | 0.0170 | 1.6138 |
| Peptide chain elongation (R) | 0.0037 | 1.7067 | Collagen chain trimerization (R) | 0.0123 | −2.2167 |
| Neurexin and Neuroligins (R) | 0.017 | 1.7048 | Regulation of complement cascade (R) | 0.0002 | −2.6783 |
| Electron transport chain OXPHOS mitochondria (W) | 0.0212 | 1.5995 | |||
| Inactivation of Cyclin B:Cdk1 by Chk1/Chk2(Cds1) (R) | 0.0374 | −2.0533 | |||
| Glycolysis (R) | 0.0406 | −2.0706 | |||
(W) WikiPathways,
(R) Reactome.
Children with ASD
Multiple glutamate receptor proteins were significantly downregulated in BA9 of children with autism (Table 3; Fig. 1 a and c). AMPA receptor proteins GRIA1 (log2FC = −1.0619, FDR-adjusted P < 0.0001), GRIA2 (log2FC = −1.0809, FDR-adjusted P < 0.0001), GRIA3 (log2FC = −1.0398, FDR-adjusted P < 0.0001), and GRIA4 (log2FC = −0.7505, FDR-adjusted P < 0.0001) were significantly decreased in ASD children. Kainate receptor GRIK2 was the only kainate receptor to show significant downregulation in ASD subjects (log2FC = −0.8421, nominal P = 0.0249). NMDA receptors GRIN1, GRIN2A, and GRIN2B exhibited a significant downregulation in BA9 of ASD children (log2FC = −0.9982, FDR-adjusted P < 0.0001; log2FC = −0.9411, FDR-adjusted P < 0.0001; log2FC = −0.6589, FDR-adjusted P = 0.0040, respectively, Table 3). GRID2 receptor showed a nonsignificant decrease in BA9 of ASD children (Table 3). Metabotropic glutamate receptors (mGRM 1 and 5) exhibited nonsignificant decreases. Glutamate receptor interacting protein 2 (GRIP2) showed a nonadjusted significant decrease in the same subjects (log2FC = −0.9485, nominal P = 0.0089). GRIP1 levels showed a nonsignificant decrease in expression in ASD BA9 of children (Table 3).
Table 3.
Selected synaptic proteins, and their partners in BA9 of children and adults with idiopathic ASD: Regulation, assignment to postsynaptic density complex, and identification of ASD risk genes.
| Class | Protein | Children | Adults | Risk Gene | PSD |
|---|---|---|---|---|---|
| AMPA receptor | GRIA1 GRIA2 GRIA3 GRIA4 |
↓ ↓ ↓ ↓ |
NC ↓ ↓ ↓ |
+ + + − |
+ + + + |
| NMDA receptor | GRIN1 GRIN2A GRIN2B |
↓ ↓ ↓ |
NC NC NC |
+ + + |
+ + + |
| metabotropic glutamate receptor | GRM3 | ? | ↓ | − | + |
| membrane associated guanylate kinase (MAGUK) | DLG4 DLGAP4 |
↓ ↓ |
NC NC |
+ − |
+ + |
| Olfactomedin | OLFM1 OLFM2 OLFM3 |
↓ ↓ ↓ |
Trend ↓ ↓ Trend ↓ |
− − − |
+ + − |
| scaffolding protein | SHANK1 SHANK2 SHANK3 HOMER1 |
↓ ↓ ↓ ↓ |
NC Trend ↓ NC NC |
+ + + + |
+ + + + |
| presynapse protein | BSN PCLO |
↓ ? |
↓ ↓ |
− + |
− − |
| serine/threonine protein kinase | CAMK2A CAMK4 |
↓ ↓ |
NC ↓ |
+ + |
+ + |
| synaptic adhesion molecule | NRXN1 NLGN2 |
↓ Trend ↓ |
Trend ↓ NC |
+ + |
+ + |
| rho GTPase | ARHGAP 32 ARHGAP 44 ARHGEF 2 ARHGEF 3 CDC 42 DBN DOCK 9 MYO5A |
↓ NC NC ↑ ↑ ↓ ↓ ↓ |
NC ↓ ↓ NC NC ↓ NC NC |
+ − + − + − − + |
+ + + − + + + + |
| PSD protein | FAM81A | ↓ | NC | − | + |
| neurofilament protein | NEFL NEFM NEFH INA |
↑ ↑ Trend ↑ ↑ |
↓ ↓ ↓ ↓ |
− − − − |
+ + + + |
| ↓ decrease in protein level, FDR-adjusted P < 0.05 ↑ increase in protein level, FDR-adjusted P < 0.05 NC, no change in protein level Trend, near significant change in protein level + assignment established − no assignment established |
+ ASD risk gene listed on SFARI web site | + postsynaptic density protein listed on Syngo web site | |||
We next evaluated alterations in the expression of several partners of glutamate receptors including scaffolding genes, kinases, Rho GTPases, and ion channels. Analysis of the “disks large homolog family of proteins” (DLG) in BA9 of ASD children revealed coordinated significant downregulation of DLG4 (log2FC = −0.7379, FDR-adjusted P = 0.0032, Table 3) and of DLG4 associated protein 4 (DLGAP4, log2FC = −0.6134, FDR-adjusted P = 0.0026). No alterations were observed in levels of DLG2, DLG3, or DLGAP3. Another set of important scaffolding partners for glutamate receptors include the shank family of proteins (Shi et al. 2017); levels of shank 1, shank 2, and shank 3 proteins were reduced significantly in BA9 of autistic children (SHANK1, log2FC = −0.6396, FDR-adjusted P = 0.0029; SHANK2, log2FC = −0.6733, FDR-adjusted P = 0.0001; SHANK3, log2FC = −0.7989, FDR-adjusted P < 0.0001, Table 3). Levels of other scaffolding protein family members homer 1 and homer 3 were reduced in BA9 of children with ASD (HOMER1 log2FC = −.06333, FDR-adjusted P < 0.0001; HOMER3, log2FC = −0.6830, FDR-adjusted P = 0.0694).
A family of anchoring proteins that control dynamics and functions of AMPA receptors, noelins/olfactomedins, exhibited a significant downregulation in the BA9 of both children and adults with autism. Levels of olfactomedins 1, 2, and 3 were reduced significantly in BA9 of ASD children (OLFM1, log2FC = −0.5602, FDR-adjusted P < 0.0001; OLFM2, log2FC = −0.7902, FDR < 0.0001; OLFM3, log2FC = −0.7895, FDR-adjusted P < 0.0001, Table 3).
The rho family of GTPases plays major roles in neuronal morphology and function. Levels of several members of this group of proteins were reduced in BA9 of children with Autism. ARHGAP32 is a neuron-associated protein that modulates dendritic spine structure and strength impacting postnatal remodeling and fine tuning of neural circuits (Akshoomoff et al. 2015). Levels of ARHGAP32, 33, and 45 were decreased in BA9 of children with autism (ARHGAP32, log2FC = −0.5838, FDR-adjusted P = 0.0141; ARHGAP33, log2FC = −1.9058, nominal P = 0.0048; ARHGAP45, log2FC = −1.0883, nominal P = 0.0248). The maintenance of normal levels of ARHGAP32 is necessary for early development of neuronal circuits. We did not observe any alterations in the levels of ARHGAP32 or 45 in BA9 of adult subjects with ASD. Lastly, level of DOCK9, an atypical guanine nucleotide exchange factor (Zizimin1) with roles in dendrite development and involvement in autism, schizophrenia, and bipolar disorder, was reduced significantly in ASD children (Shi 2013; log2FC = −0.4379, FDR-adjusted P = 0.0340).
The neurofilament family of proteins is involved in maintenance of neuronal morphology and function and in correct modulation of synaptic plasticity via regulation of NMDA and dopamine activity. Levels of NEFL, NEFM and α-internexin were elevated in BA9 of children with ASD (NEFL, log2FC = 0.5945, FDR-adjusted P = 0.0119; NEFM, log2FC = 0.4659, FDR-adjusted P = 0.0016; α-internexin, log2FC = 0.5948, FDR-adjusted P = 0.0205). Level of NEFH was elevated nonsignificantly in autistic children. Upregulation of NEFL has been observed in CSF of subjects with Alzheimer’s disease (van der Ende et al. 2023).
Adults with ASD
The main glutamatergic receptors exhibiting significant alterations in the BA9 of autistic adults included AMPA receptors GRIA2, GRIA3, and GRIA4 (Table 3) and metabotropic glutamate receptor 3 (GRM3). The former four receptors were significantly downregulated (GRIA2, log2FC = −0.3377, FDR-adjusted P < 0.0001; GRIA3, log2FC = −0.3875, FDR-adjusted P = 0.0183; GRIA4, log2FC = −0.3301, FDR-adjusted P = 0.0203; GRM3, log2FC = −0.3372, FDR-adjusted P = 0.0244) in BA9 of adult subjects with autism. There were no notable alterations in other inotropic and metabotropic glutamate receptors. Evaluation of the scaffolding proteins showed the downregulation of shank 2 protein at a near significant level (log2FC = −0.3241, FDR-adjusted P = 0.0619). In contrast, levels of other important scaffolding proteins shank 1, shank 3, homer 1, and homer 2 did not change statistically.
Levels of noelins/olfactomedins (important AMPA-receptor partners) exhibited near statistically significant reductions in olfactomedin 1 (log2FC = −0.7065, nominal P = 0.0662) and olfactomedin 3 (log2FC = −0.5256, nominal P = 0.0672). The level of olfactomedin 2 was significantly decreased in BA9 of adult ASD subjects (log2FC = −0.6660, FDR-adjusted P < 0.0001). Interestingly, OLFML3, which exhibited a nearly significant increase (log2FC = 1.1652, nominal P = 0.0594), plays an important role in placental and embryonic development (Zeng et al. 2004) and in enrichment of microglia cells (Chiu et al. 2013) during neuroinflammation (Grassivaro et al. 2021).
Two markers of Rho GTPase family of proteins, namely ARHGAP44 (log2FC = −0.5033, FDR-adjusted P = 0.0356) and ARHGEF2 (log2FC = −0.3234, FDR-adjusted P = 0.0477), exhibited significant downregulation in adults with ASD (Table 3). Recent data implicate an important role for ARHGEF2 in intellectual disability and midbrain–hindbrain malformations (Ravindran et al. 2022). Finally, levels of neurofilament family of proteins in BA9 of ASD adults showed a significant downregulation in almost all members of this group (NEFL, log2FC = −0.6389, FDR-adjusted P = 0.0014; NEFM, log2FC = −0.6722, FDR-adjusted P < 0.0001; NEFH, log2FC = −0.7125, FDR-adjusted P = 0.0087; INA, log2FC = −0.4704, FDR-adjusted P = 0.0562) in contrast to elevations in the same proteins observed in children with ASD (Table 3).
Three other presynaptic and synaptic proteins showed significant downregulation in BA9 of adult subjects with autism (Table 3). Proteins bassoon (BSN) and piccolo (PCLO) were significantly reduced in BA9 of adults with ASD (Bassoon, log2FC = −0.5702, FDR-adjusted P < 0.0001; Piccolo, log2FC = −0.5516, FDR-adjusted P < 0.0001). Drebrin (DBN1), a major cytoskeletal binding protein involved in regulation of cortical neuron axon branch lamination (Dorskind et al. 2023), was reduced significantly (log2FC = −0.5681, FDR-adjusted < 0.0001) in adult patients with Autism. Drebrin and drebrin-like (DBNL) proteins are involved in PSD maturation process (Wang et al. 2023). Drebrin loss of function may result in ectopic collateral axon branching (Dorskind et al. 2023), an important event that may lead to abnormal axonal circuit formation and neuronal mis-wiring in autistic disorder.
Discussion
The current proteomic study has investigated, identified, and compared the presence of autism risk genes from 812 differentially expressed proteins (FDR-adjusted P < 0.05; 571 upregulated and 241 downregulated) from children with ASD vs. 226 differentially expressed proteins (FDR-adjusted P < 0.05; 73 upregulated and 153 downregulated) from adults with ASD. Evaluation of the identified proteins in BA9 of children with ASD revealed a complex set of proteins enriched in autism risk genes that impact PSD structure and functioning during early brain development.
PSD is a biologically important protein complex composed of more than 1,000 proteins encompassing excitatory synapses (AMPA and NMDA receptors), scaffold proteins (DLG4, shank, homer), olfactomedins, and Rho GTPases (Sheng and Kim 2011; Kaizuka and Takumi 2018; Kaizuka et al. 2022; Wang et al. 2023). By the same token, prefrontal cortex (PFC), a site involved in cognition, executive functions, working memory, and attention (Webster et al. 2011), is one of the last brain areas to undergo maturation during brain development (Somel et al. 2009; Wang et al. 2023). This expected, evolutionarily conserved delay in human brain development (neoteny) throughout childhood gives way to rapid change in early adolescence (~10 years of age; Somel et al. 2009) that coincides with early acquisition of cognitive abilities and extension of neuronal plasticity associated with the process of learning (Somel et al. 2009; Wang et al. 2023). Moreover, synaptic growth predominates with increases in synaptic markers such as complexin 2 and PSD-95 (DLG4) reaching a steady state level in adolescence (Webster et al. 2011). Additional supportive evidence indicates that neoteny of synaptic spines in human PFC may continue beyond adolescence and into the third decade of life in humans (Petanjek et al. 2011).
Thus, as the maturation of PSD domain in human brain is a slow process (compared with mice and macaques; Wang et al. 2023), alterations in the structure or functioning of PSD can have major implications on normal development of the human brain. Abnormalities in genes coding for proteins that act as important partners for PSD proteins, such as NMDA receptors (GRIN2B or GRIN2A), AMPA receptors (GRIA1, GRIA2, GRIA3, GRIA4), and DLG3 or DLG4, may result in the development of autism, schizophrenia, or other neurodevelopmental disorders (Coley and Gao 2018; Hansen et al. 2021; Tumer et al. 2023). Based on the consensus among protein levels for several components of the PSD complex in BA9 of children and adults with ASD, we propose that idiopathic autism is a synaptopathy involving genes that support the processes of cognition and synaptic plasticity, especially impacting the growth of the brain during childhood.
We identified significant downregulation in the AMPA receptors (GRIA1, GRIA2, GRIA3, and GRIA4) in BA9 of children with autism (Table 3). While GRIA1 level did not change in BA9 of adults with autism, levels of GRIA2, GRIA3, and GRIA4 were downregulated significantly in ASD adults (Table 3). AMPA receptors are expressed early in brain development and may be involved in neuronal cell migration and neurite growth (Hansen et al. 2021). Additionally, GRIA1, GRIA2, and GRIA3 are participants in dendritic growth, and these three plus GRIA4 participate in synaptogenesis, synapse maturation, postsynaptic function, and synaptic plasticity (Hansen et al. 2021). Moreover, because of rapid kinetics of AMPA receptor activation, its contribution to synaptic plasticity is significant. Thus, AMPA receptors contribute to synaptic signaling and interact with multiple secreted proteins including olfactomedins. There is ample evidence for the presence of GRIA2 variants in autism (Salpietro et al. 2019). Furthermore, mutations in synaptic partners of AMPA receptors such as shank 1 or shank 2, CaM kinase α2, and neuroligin can lead to altered AMPA receptor function (Etherton et al. 2011; Shi et al. 2017; Stephanson et al. 2017). Rescue of AMPA receptor function leads to improvement in abnormal behavior in an animal model for ASD (Kim et al. 2019). Recent evidence linking AMPA receptor trafficking abnormalities to autism (Niescier and Lin 2021) is supported by our current and novel proteomic data. Furthermore, several recent reports link GRIA1 and shank 3 (Ross and Aizenman 2023), GRIA2 (Salpietro et al. 2019; Latsko et al. 2022), and GRIA3 proteins to etiology of schizophrenia and autism (Singh et al. 2022). Our observation of GRIA4 receptor downregulation in children and adult ASD individuals is novel (Table 3).
NMDA receptors GRIN1, GRIN2A, and GRIN2B levels were significantly reduced in BA9 of children with autism (Table 3). In contrast, the levels of the same receptors did not change in autistic adults (Table 3). NMDA receptors are localized to pre-, post-, and extra-synaptic compartments and regulate many functions including neurogenesis, neurite development, synaptic function, transmission, excitability, and plasticity (Hansen et al. 2021). Levels of NMDA receptors GRIN2A and GRIN2B follow a strict timetable which coincides with critical periods in the development of various brain areas. In general, GRIN2B levels have higher expression prenatally with a drop at postnatal stages. In contrast, GRIN2A levels are low at prenatal stages and increase at birth (Myers et al. 2022). By the same token, the molar ratio of GRIN2B to GRIN2A remains at 4:1 in all brain areas except for cerebellum where GRIN2A predominates GRIN2B levels (Frank et al. 2016). Thus, alterations in levels of these receptors during early brain development impact the normal schedule of brain development adversely (Wang et al. 2023). Reduction in levels of GRIN1, GRIN2A, and GRIN2B in BA9 of children with autism is supported by previous reports demonstrating the presence of variants in GRIN2A and 2B in developmental encephalopathies and autism (Hansen et al. 2021; Gandal et al. 2022; Wang et al. 2022; Vollweiter et al. 2023).
DLG4 (PSD-95) levels were significantly reduced in the BA9 of children with autism. Additionally, levels of DLGAP4 protein, which interacts with DLG4, were also reduced significantly in BA9 of ASD children (Table 3). In contrast, levels of DLG4 and DLGAP4 were not altered in BA9 of adult subjects with ASD. DLG4 is a member of an important family of scaffolding proteins that is involved in synaptic signaling, strength, and plasticity through its interactions with members of the PSD in mammalian brain (Levy et al. 2022). DLG4 is a major gene which impacts both intellectual/cognitive processes and is considered a risk gene for developmental encephalopathies as well as autism (Lelieveld et al. 2016; Tumer et al. 2023). Mouse DLG4 (PSD-95) mutants exhibit repetitive behaviors, impaired motor coordination, and increased anxiety-related behaviors which are relevant to autism and William’s syndrome symptomology (Feyder et al. 2010). Dysfunction of DLG4 has also been associated with early-onset psychosis and catatonia (Yang et al. 2023) which is responsive to treatment with clozapine. Furthermore, mutations in DLGAP4, a membrane-associated guanylate kinase that interacts with DLG4 (PSD-95) through its guanylate kinase domain, affect cognition and cause ASD-like behaviors such as impaired vocal communication and social interaction abnormalities in SAPAP-4 (DLGAP4)-deficient mice (Schob et al. 2019). Deficits of DLG4 and DLGAP4 in ASD children are consistent with previous reports indicating age- and brain site-dependent consequences of PSD-95 deficiency in medial prefrontal cortex, an area of the brain that exhibits a prolongated postnatal maturation period (Gao and Mack 2021). Thus, abnormalities in levels of DLG4 during the postnatal life impact development of cognition and synaptic circuit formation adversely (Kirov et al. 2012; Grant 2015; Wang et al. 2023).
An important partner in PSD function is the family of olfactomedins (Tomarev and Nakaya 2009). Proteomic results showed a significant decrease in levels of olfactomedins 1, 2, and 3 in BA9 of children with autism (Table 3). In adult BA9, level of OLFM2 was reduced significantly, with trends for reduction in OLFM1and OLFM3 (Table 3). OLFM4 levels did not change in adults with ASD and OLFM4 levels were not detected in children with ASD. The olfactomedin family of PSD-related proteins have significant roles in brain development and neural crest cell production (Barembaum et al. 2000). Recent evidence indicates that olfactomedins interact with AMPA receptors and play a major role in synaptic plasticity (Boudkkazi et al. 2023). Olfactomedins stabilize AMPA receptors at the surface of synapses and dendrites (Boudkkazi et al. 2023). Alterations in interaction between olfactomedins and AMPA receptors lead to decreases in AMPA receptor density and impacted long-term synaptic plasticity adversely (Boudkkazi et al. 2023). Reduction in levels of olfactomedins 1–3 in BA9 of children and in olfactomedin 2 in BA9 of adults with ASD may play a role in cognitive deficits observed in subjects with autism.
Level of CaMK2α, a major serine/threonine kinase with roles in synaptic plasticity and cognition (Hell 2014; Lee et al. 2021; Lintas et al. 2023), was reduced in BA9 of children with ASD. De novo mutation in CaMK2α in a subject with autism led to reduction in binding of this kinase to Shank3 and NMDA receptors (Stephanson et al. 2017). CaMK2α mutant mice exhibit diverse synaptic and behavioral abnormalities such as deficits in cognition during pre-adolescence period (Gustin et al. 2011), scaffolding disruption in synaptic signaling, and reduction in ratio of CaMK2α to CaMK2/3 during childhood (Gustin et al. 2011). Interestingly, levels of this kinase could not be detected in BA9 of adult subjects with ASD (Table 3).
Scaffolding proteins shank 1–3 and homer 1 and 3 were reduced significantly in BA9 of children with ASD. In contrast with children, levels of these proteins in adult ASD subjects did not change significantly (Table 3). Mutations in shank 1–3 have been detected in subjects with autism (Jung and Park 2022) and participation of variants in Shank proteins have been associated with neurodevelopmental disorders that result in intellectual disability (Sala et al. 2015). Homer1 variants have also been associated with autism and schizophrenia (Soler et al. 2018; Meliskova et al. 2021).
Lastly, levels of several members of Rho GTPases were altered significantly in BA9 of children and adults with autism. While ARHGAP32 and DOCK9 levels were reduced in children, levels of ARHGAP 44 and ARHGEF2 decreased in BA9 of adults with autism. ARHGAP32 is considered to be a risk gene for autism (Guo et al. 2020). Wang et al. (2023) determined that increased synaptic Rho GTPase activity helps to delay early maturation of synapse in line with the slow maturation of synaptic structures in human brain. Thus, reductions in ARHGAP32 in autistic BA9 of children disrupt the normal timetable required for normal developmental sequence of synaptic structure and function in autism. Furthermore, alterations in levels of ARHGAP44 and ARHGEF2 in adult subjects with autism support the notion that synaptic dysmaturation is a continual pathological process in autistic brain which is observed in both children and adults with autism.
In conclusion, proteomic investigation of the frontal cortex in children with idiopathic autism demonstrates global down regulation of PSD members (AMPA and NMDA receptors, DLG4, olfactomedins, Serine/Threonine kinases, and Rho GTPases), affecting synaptic development and plasticity adversely and leading to deficits in cognition, a hallmark of autism. These novel results point to etiopathogenesis of autism as a developmental synaptopathy and offer potential targets for pharmacotherapeutic interventions.
Supplementary Material
Acknowledgments
Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders. The Autism Tissue Program is also gratefully acknowledged. Technical support by Dr LeeAnn Higgins and Dr Kevin Murray from the Center for Mass Spectrometry, Proteomics, and Metabolomics at the University of Minnesota is greatly appreciated.
Contributor Information
S Hossein Fatemi, Department of Psychiatry and Behavioral Sciences, University of Minnesota Medical School, 420 Delaware Street SE, Minneapolis, MN 55455, USA.
Arthur Eschenlauer, Minnesota Supercomputing Institute, 599 Walter Library, 117 Pleasant Street, Minneapolis, MN 55455, USA.
Justin Aman, Department of Psychiatry and Behavioral Sciences, University of Minnesota Medical School, 420 Delaware Street SE, Minneapolis, MN 55455, USA.
Timothy D Folsom, Department of Pediatrics, University of Minnesota Medical School, Minneapolis, MN 55455, USA; Masonic Cancer Center, University of Minnesota Medical School, Minneapolis, MN 55455, USA; Center for Genome Engineering, University of Minnesota Medical School, Minneapolis, MN 55455, USA; Stem Cell Institute, University of Minnesota Medical School, Minneapolis, MN 55455, USA.
Thierry Chekouo, University of Minnesota School of Public Health, Minneapolis, MN 55455, USA.
Author contributions
S. Hossein Fatemi (Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing—original draft, Writing—review & editing), Arthur Eschenlauer (Data curation, Formal analysis, Software, Visualization, Writing—review & editing), Justin Aman (Data curation, Resources, Writing—review & editing), Timothy D. Folsom (Investigation, Resources, Writing—review & editing), and Thierry Chekouo (Formal analysis, Software, Writing—review & editing)
Funding
Grant support by Autism Research Institute to S.H.F. is gratefully acknowledged.
Conflict of interest statement: The authors declare no conflicts of interest.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD048017 and doi: 10.6019/PXD048017: https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD048017
References
- Abraham JR, Szoko N, Barnard J, Rubin RA, Schlatzer D, Lundberg K, Li X, Natowicz MR. Proteomic investigations of autism brain identify known and novel pathogenic processes. Sci Rep. 2019:9(1):13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham JR, Szoko N, Natowicz MR. Proteomic investigations of autism spectrum disorder: past findings, current challenges, and future prospects. Adv Exp Med Biol. 2019:1118:235–252. [DOI] [PubMed] [Google Scholar]
- Akshoomoff N, Mattson SN, Grossfeld PD. Evidence for autism spectrum disorder in Jacobsen syndrome: identification of a candidate gene in distal 11q. Genet Med. 2015:17(2):143–148. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 5th ed. Washington (DC): American Psychiatric Association; 2013 [Google Scholar]
- Barembaum M, Moreno TA, LaBonne C, Sechrist J, Bronner-Fraser M. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol. 2000:2(4):219–225. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple hypothesis testing. JR Stat Soc B. 1995:57(1):289–300. [Google Scholar]
- Boudkkazi S, Schwenk J, Nakaya N, Brechet A, Kollewe A, Harada H, Bildl W, Kulk A, Dong L, Sultana A, et al. A noelin-organized extracellular network of proteins required for constitutive and context-dependent anchoring of AMPA-receptors. Neuron. 2023:111(16):2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek JA, Guest PC, Rahmoune H, Bahn S. Proteomic analysis of post mortem brain tissue from autism patients: evidence for opposite changes in prefrontal cortex and cerebellum in synaptic connectivity-related proteins. Mol Autism. 2014:5(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002:58(3):428–432. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Casanova EL, Frye RE, Baeza-Velasco C, LaSalle JM, Hagerman RJ, Scherer SW, Natowicz M. Editorial: secondary versus idiopathic autism. Front Psych. 2020:11:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ETA, Goodarzi H, Liao JT, O’Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports (Cambridge). 2013:4(2):385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley AA, Gao W-J. PSD95: A synaptic protein implicated in schizophrenia or autism? Prog Neuro-Psychopharmacol Biol Psychiatry. 2018:82:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan AP, Basson MA. The neuroanatomy of autism – a development perspective. J Anat. 2017:230(1):4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorskind JM, Sudarsanam S, Hand RA, Ziak J, Amoah-Dankwah M, Guzman-Clavel L, Soto-Vargas JL, Kolodkin AL. Drebrin regulates collateral axon branching in cortical layer II/III somatosensory neurons. J Neurosci. 2023:43(46):7745–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Tabuchi K, Sharma M, Ko J, Sudhof TC. An autism-associated point mutation in neuroligin cytoplasmic tail selectively impairs AMPA receptor-mediated synaptic transmission in hippocampus. EMBO J. 2011:30(14):2908–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CO, Blatt GJ, Chauhan A, Chauhan V, Dagler SR, Dickson PE, et al. Consensus paper: pathologic role of the cerebellum in autism. Cerebellum. 2012:11(3):777–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Kneeland RE, Yousefi MK, Liesch SB, Thuras PD. Impairment of fragile X mental retardation protein-metabotropic glutamate receptor 5 signaling and its downstream cognates ras-related C3 botulinum toxin substrate 1, amyloid beta A4 precursor protein, striatal-enriched protein tyrosine phosphatase, and homer 1, in autism: a postmortem study in cerebellar vermis and superior frontal cortex. Mol Autism. 2013:4(1):21–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Thuras PD. Altered subcellular localization of fragile X mental retardation signaling partners and targets in superior frontal cortex of individuals with schizophrenia. Neuroreport. 2017:28(16):1066–1070. [DOI] [PubMed] [Google Scholar]
- Feyder M, Karlsson RM, Mathur P, Lyman M, Bock R, Momenam R, Munasinghe J, Scattoni ML, Ihne J, Camp M, et al. Association of mouse DLG4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorder and Williams’ syndrome. Am J Psychiatry. 2010:167(12):1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom TD, Higgins L, Markowski TW, Griffin TJ, Fatemi SH. Quantitative proteomics of forebrain subcellular fractions in fragile X mental retardation 1 knockout mice following acute treatment with 2-methyl-6-(phenylethynyl)pyridine: relevance to developmental study of schizophrenia. Synapse. 2019:73(1):e22069. [DOI] [PubMed] [Google Scholar]
- Frank RAW, Komiyama NH, Ryan TJ, Zhu F, O’Dell TJ, Grant SGN. NMDA receptors are selectively partitioned into complexes and super complexes during synapse maturation. Nat Commun. 2016:7(1):11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele S, Sacco R, Persico AM. Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. Eur Neuropsychopharmacol. 2014:24(6):919–929. [DOI] [PubMed] [Google Scholar]
- Galindo-Prieto B, Eriksson L, Trygg J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J Chemom. 2014:28(8):623–632. [Google Scholar]
- Gandal MJ, Haney J, Wamsley B, Yap CH, Parhami S, Emani PS, Chang N, Chen GT, Hoftman GD, de Alba D, et al. Broad transcriptomic dysregulation occurs across the cerebral cortex in ASD. Nature. 2022:611(7936):532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W-J, Mack NR. From Hyposociability to Hypersociability—the effects of PSD-95 deficiency on the dysfunctional development of social behavior. Front Behav Neurosci. 2021:15:618397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese A, Butler M. The autism spectrum: behavioral psychiatric and genetic associations. Genes. 2023:14(3):677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SGN. The molecular evolution of the vertebrate behavioral repertoire. Philos Trans R Soc B. 2015:371(1685):20150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassivaro F, Martino G, Farina C. The phenotypic convergence between microglia and peripheral macrophages during development and neuroinflammation paves the way for new therapeutic perspectives. Neural Regen Res. 2021:16(4):635–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BQ, Ding SB, Li HB. Blood biomarker levels of methylation capacity in autism spectrum disorder: a systematic review. Acta Psychiatr Scand. 2020:141(6):492–509. [DOI] [PubMed] [Google Scholar]
- Guo D, Yang X, Shi L. Rho GTPase regulators and effectors in autism spectrum disorders: animal models and insights for therapeutics. Cells. 2020:9(4):835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin RM, Shonesy BC, Robinson SL, Rentz TJ, Bauum AJ II, Jalan-Sakrikar N, Winder DG, Stanwood GD, Colbran RJ. Loss of Thr286 phosphorylation disrupts synaptic CaMKIIα targeting, NMDAR activity and behavior in pre-adolescent mice. Mol Cell Neurosci. 2011:47(4):286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti F, Sobolevsky AI, Swanson CT, Swanger SA, Greger IH, Nakagawa T, et al. Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol Rev. 2021:73(4):1469–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell JW. CaMKII: claiming center stage in postsynaptic function and organization. Neuron. 2014:81(2):249–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, MacKrell EJ, Wang TY, Lomenick B, Roukes ML, Chou TF, Chou TF. Tidyproteomics: an open-source R package and data object for quantitative proteomics post analysis and visualization. BMC Bioinformatics. 2023:24(1):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Park M. Shank postsynaptic scaffolding proteins in autism spectrum disorder: mouse models and their dysfunctions in behaviors, synapses, and molecules. Pharmacol Res. 2022:182:106340. [DOI] [PubMed] [Google Scholar]
- Kaizuka T, Takumi T. Postsynaptic density proteins and their involvement in neurodevelopmental disorders. J Biochem. 2018:163(6):447–455. [DOI] [PubMed] [Google Scholar]
- Kaizuka T, Suzuki T, Kishi N, Kilimann MW, Ueyama T, Watanabe M, Okano H, Dohme N, Takumi T. Developmental dynamics of the postsynaptic proteome to understand synaptic maturation and dysmaturation. BioRxiv. 2022. 10.1101/2022.05.05.490828. [DOI] [Google Scholar]
- Käll L, Canterbury J, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007:4(11):923–925. [DOI] [PubMed] [Google Scholar]
- Kim J-W, Park K, Kang RJ, Gonzales EL, Kim DG, Oh HA, Seung H, Ko MJ, Kwon KJ, Kim KC, et al. Pharmacological modulation of AMPA receptor rescues social impairments in animal models of autism. Neuropsychopharmacology. 2019:44(2):314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Halmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signaling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012:17(2):142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latsko MS, Koboldt DC, Franklin SJ, Hickey SE, Williamson RK, Garner S, Ostendorf AP, Lee K, White P, Wilson RK. De novo missense variant in GRIA2 in a patient with global developmental delay, autism spectrum disorder, and epileptic encephalopathy. Cold Spring Harbor Molecular Case Studies. 2022:8(4):a006172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LC, Su MT, Huang HY, Cho YC, Yeh TK, Chang CY. Association of CaMK2A and MeCP2 signaling pathways with cognitive ability in adolescents. Mol Brain. 2021:14(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelieveld SH, Reijnders MRF, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, de Vries BBA, Willemsen MH, Kleefstra T, Lohner K, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016:19(9):1194–1196. [DOI] [PubMed] [Google Scholar]
- Levy AM, Gomez-Puertas P, Tumer Z. Neurodevelopmental disorders associated with PSD-95 and its interaction partners. Int J Mol Sci. 2022:23(8):4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintas C, Facchiano A, Azzarà A, Cassano I, Tabolacci C, Galasso C, Gurrieri F. Deletion of a single lysine residue at position 292 of CAMK2A disrupts protein function, causing severe epileptic encephalopathy and intellectual disability. Genes. 2023:14(7):1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner MJ, Warren Z, Williams AR, Amoakohene E, Bakian AV, Bilder DA, Durkin MS, Fitzgerald RT, Furnier SM, Hughes MM, et al. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill Summ. 2023:72(2):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta R, Risoleo MC, Messina G, Parisi L, Carotenuto M, Vetri L, Roccella M. The neurochemistry of autism. Brain Sci. 2020:10(3):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood T, Warringer J, Snipen L, Saebo S. Improving stability and understandability of genotype-phenotype mapping in saccharomyces using regularized variable selection in L-PLS regression. BMC Bioinformatics. 2012:13(1):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meliskova V, Havranek T, Bacova Z, Bakos J. The role of selected postsynaptic scaffolding proteins at glutamatergic synapses in autism-related animal models. J Integr Neurosci. 2021:20(4):1047–1057. [DOI] [PubMed] [Google Scholar]
- Meunier B, Dumas E, Piec I, Bechet D, Hebraud M, Hocquette J-F. Assessment of hierarchical clustering methodologies for proteomic data mining. J Proteome Res. 2007:6(1):358–366. [DOI] [PubMed] [Google Scholar]
- Mueller TM, Remedies CE, Haroutunian V, Meador-Woodruff JH. Abnormal subcellular localization of GABAA receptor subunits in schizophrenia brain. Transl Psychiatry. 2015:5(8):e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SJ, Yuan H, Kang J-Q, Tan FCK, Traynelis SF, Low C-M. Distinct roles of GRIN2A and GRIN2B variants in neurological conditions. F1000Res. 2022:8:1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Valakh V. Excitatory/inhibitory balance and circuit homeostasis in autism spectrum disorders. Neuron. 2015:87(4):684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niescier RF, Lin Y-C. The potential role of AMPA receptor trafficking in autism and other neurodevelopmental conditions. Neuroscience. 2021:479:180–191. [DOI] [PubMed] [Google Scholar]
- Palmen SJMC, van Engeland H, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004:127(12):2572–2583. [DOI] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HBM, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. PNAS. 2011:108(32):13281–13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presumey J, Bialas AR, Carroll MC. Complement system in neural synapse elimination in development and disease. Adv Immunol. 2017:135:53–79. [DOI] [PubMed] [Google Scholar]
- R Core Team (2023). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/. [Google Scholar]
- Ravindran E, Ullah N, Mani S, Chew EGY, Tandiono M, Foo JN, Khor CC, Kaindl AM, Siddiqi S. Case report: expanding the phenotype of ARHGEF17 mutations from increased intracranial aneurysm risk to a neurodevelopmental disease. Front Neurol. 2022:13:1017654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MM, Aizenman E. GluA1-Shank3 interaction decreases in response to chronic neuronal depolarization. Neurosci Lett. 2023:809:137305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylaarsdam L, Guemez-Gamboa A. Genetic causes and modifiers of autism Spectrum disorder. Front Cell Neurosci. 2019:13:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo SL, Lahr JM, Offer M, Weekes ALA, Sceniak MP. GRIN2B-related neurodevelopmental disorder: current understanding of pathophysiological mechanisms. Front Synaptic Neurosci. 2023:14:1090865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager REH, Walker AK, Middleton F, Robinson K, Webster MJ, Weickert CS. Trajectory of change in brain complement factors from neonatal to young adult humans. J Neurochem. 2021:157(3):479–493. [DOI] [PubMed] [Google Scholar]
- Saghazadeh A, Rezaei N. Brain-derived neurotrophic factor levels in autism: a systematic review and meta-analysis. J Autism Dev Disord. 2017:47(4):1018–1029. [DOI] [PubMed] [Google Scholar]
- Sala C, Vicidomini C, Bigi I, Mossa A, Verpelli C. Shank synaptic scaffold proteins: keys to understanding the pathogenesis of autism and other synaptic disorders. J Neurochem. 2015:135(5):849–858. [DOI] [PubMed] [Google Scholar]
- Salpietro V, Dixon CL, Guo H, Bello OD, Vandrovcova J, Efthymiou S, Maroofian R, Heimer G, Burglen L, Valence S, et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat Commun. 2019:10(1):3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schob C, Morellini F, Ohana O, Bakota L, Hrynchak MV, Brandt R, Brockmann MD, Cichon N, Hartung H, Hanganu-Opatz IL, et al. Cognitive impairment and autistic-like behavior in SAPAP4-deficient mice. Transl Psychiatry. 2019:9(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman A, Lin X. The effect of correlation in false discovery rate estimation. Biometrika. 2011:98(1):199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol. 2011:3(12):a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L. Dock protein family in brain development and neurological disease. Commun Integr Biol. 2013:6(6):e26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Redman P, Ghose D, Hwang H, Liu Y, Ren X, Ding LJ, Liu M, Jones KJ, Xu W. Shank proteins differentially regulate synaptic transmission. eNeuro. 2017:4(6):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Poterba T, Curtis D, Akil H, Al Eissa MA, Barchas JD, Bass N, Bigdeli TB, Breen G, Bromet EJ, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022:604(7906):509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler J, Fananas L, Parellada M, Krebs M-O, Rouleua GA, Fatjo-Vilas M. Genetic variability in scaffolding proteins and risk for schizophrenia and autism-spectrum disorders: a system review. J Psychiatry Neurosci. 2018:43(4):223–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. Transcriptional neoteny in the human brain. PNAS. 2009:106(14):5743–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012:35(1):369–389. [DOI] [PubMed] [Google Scholar]
- Stephanson JR, Wang X, Perfitt TL, Parrish WP, Shonesy BC, Marks CR, Mortlock DP, Nakagawa T, Sutcliffe JS, Colbran RJ. A novel human CamK2A mutation disrupts dendritic morphology and synaptic transmission, and causes ASD-related behaviors. J Neurosci. 2017:37(8):2216–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005:102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szoko N, McShane AJ, Natowicz MR. Proteomic explorations of autism spectrum disorder. Autism Res. 2017:10(9):1460–1469. [DOI] [PubMed] [Google Scholar]
- Taha MS, Nouri K, Milroy LG, Moll JM, Herrmann C, Brunsveld L, Piekorz RP, Ahmadian MR. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein FMRP with nucleolin. PLoS One. 2014:9(3):e91465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009:40(2):122–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumer Z, Dye TJ, Prada C, White-Brown AM, Mackenzie A, Levy AM. DLG4-related synaptopathy. In: Adam MP, et al., editors. GeneReviews. Seattle (WA): University of Washington; 2023. pp. 1993–2023.
- van der Ende EL, in ‘t Veld SGJG, Hanskamp I, van der Lee S, Dijkstra JIR, Hok-A-Hin YS, Blujdea ER, van Swieten JC, Irwin DJ, Chen-Plotkin A, et al. CSF proteomics in autosomal dominant Alzheimer’s disease highlights parallels with sporadic disease. Brain. 2023:146(11):4495–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollweiter D, Shergill JK, Hilse A, Kochlamazashvili G, Koch SP, Mueller S, Boehm-Sturm P, Haucke V, Maritzen T. Intersectin deficiency impairs cortico-striatal neurotransmission and causes obsessive-compulsive behaviors in mice. PNAS USA. 2023:120(35):e2304323120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo Z, Mei D, Zhang Y, Zhao S, Hu S, Luo S, Wang Q, Gao C. The GluN2B-Trp373 NMDA receptor variant is associated with autism-, epilepsy-related phenotypes and reduces NMDA receptor currents in rats. Neurochem Res. 2022:47(6):1588–1597. [DOI] [PubMed] [Google Scholar]
- Wang L, Pang K, Zhou L, Cebrián-Silla A, González-Granero S, Wang S, Bi Q, White ML, Ho B, Li J, et al. A cross-species proteomic map reveals neoteny of human synapse development. Nature. 2023:622(7981):112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MJ, Elashoff M, Weickert CS. Molecular evidence that cortical synaptic growth predominates during the first decade of life in humans. Int J Dev Neurosci. 2011:29(3):225–236. [DOI] [PubMed] [Google Scholar]
- Yang M, Rubin A, Wondimu R, Grebe T, Ritfeld G. Significant improvement of psychotic symptoms in treatment-resistant schizophrenia with clozapine in an adolescent with SHINE syndrome: a case report. BMC Psychiatry. 2023:23(1):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L-C, Liu F, Zhang X, Zhu Z-D, Wang Z-Q, Han ZG, Ma WJ. hOLF44, a secreted glycoprotein with distinct expression pattern, belongs to an uncharacterized olfactomedin-like subfamily newly identified by phylogenetic analysis. FEBS Lett. 2004:571(1–3):74–80. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Zhu T, Qu Y, Mu D. Blood glutamate levels in autism spectrum disorder: a systematic review and meta-analysis. PLoS One. 2016:11(7):e0158688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD048017 and doi: 10.6019/PXD048017: https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD048017