Abstract
Atherosclerosis is a leading cause of cardiovascular diseases, characterized by endothelial dysfunction and lipid accumulation. Long non-coding RNAs (lncRNAs) are emerging as key regulators of endothelial cell behavior. This study aimed to investigate the role of lncRNA NORAD in endothelial cell proliferation and as a potential therapeutic target for atherosclerosis. A total of 75 CAD patients and 76 controls were recruited, and plasma NORAD levels were measured using qRT-PCR. HUVECs were transfected with si-NORAD to evaluate its effects on cell cycle, proliferation, migration, and apoptosis. Plasma NORAD levels were significantly elevated in CAD patients. The NORAD-miRNA-mRNA ceRNA regulatory network was constructed based on GEO database, and G1/S-specific cyclin-D1 (CCND1) was identified as one of the hub factors. NORAD deficiency suppressed cell migration and induced G1 cell cycle arrest in HUVECs by downregulating CCND1 in vitro. NORAD upregulated CCND1 in HUVECs via sponging miR-106a that inhibited cell migration. The dual-luciferase assay confirmed the direct targeting of miR-106a by NORAD, and overexpression of miR-106a inhibited HUVEC proliferation and migration. Si-NORAD transfection resulted in induced early apoptosis, increased intracellular ROS levels, decreased GSH levels, and reduced mitochondrial membrane potential. Additionally, si-NORAD decreased the expression of GPX4, FTH1, KEAP1, NCOA4, and Nrf2, while increasing Xct levels, confirming the involvement of ferroptosis. Our findings reveal that NORAD plays a critical role in endothelial cell proliferation, migration, and apoptosis, and its silencing induces ferroptosis. The regulatory network involving NORAD, miR-106a, and their target genes provides a potential therapeutic avenue for atherosclerosis.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76243-x.
Keywords: NORAD, Coronary artery disease, CCND1, miR-106a, Ferroptosis
Subject terms: Biochemistry, Cell biology, Chemical biology, Computational biology and bioinformatics, Developmental biology, Molecular biology, Biomarkers, Cardiology, Diseases, Medical research, Molecular medicine, Chemistry, Mathematics and computing
Introduction
The most common form of coronary artery disease (CAD) is characterized by the development of atherosclerotic plaques, consisting of lipid deposits and inflammatory lesions within large arteries. These plaques can lead to myocardial infarction (MI) and stroke. The high morbidity and mortality rates associated with CAD pose a significant public health challenge1,2. Research indicates that oxidative stress, inflammatory responses, lipid metabolism dysfunction, vascular cell autophagy, and apoptosis contribute to the onset and progression of atherosclerosis (AS)3. Endothelial cells can become dysfunctional due to damage from viruses, mechanical injury, immune complexes, and particularly by oxidized low-density lipoproteins, leading to the expression and release of active substances and adhesion molecules. This triggers excessive chronic inflammatory hyperplasia in local blood vessels, impairing the function of vascular endothelial cells and causing pathological changes, including reduced cell viability and altered cell cycle dynamics4–7. Damage to endothelial cells or alterations in their morphology, structure, and function compromise the vascular barrier, resulting in lipid and monocyte accumulation in the vascular endothelial space, which forms harmful foam cells and contributes to atherosclerosis8. These findings underscore the importance of preserving endothelial cell integrity and function for the prevention and treatment of atherosclerosis9.
Numerous studies have demonstrated that long non-coding RNAs (lncRNAs) are involved in the development of diseases, including atherosclerosis (AS). NORAD, a long non-coding RNA activated by DNA damage, is located on human chromosome 20q11.2310,11. It plays a role in maintaining chromosome stability and participating in mitosis. NORAD is primarily expressed in the cytoplasm, activated by DNA damage, evolutionarily conserved, and is highly expressed in human tissues and cell lines12. Gene microarray analysis has shown that NORAD is highly expressed in endothelial cells13. Research has shown that NORAD regulates cell proliferation, apoptosis, migration, invasion, and metabolism, thereby inhibiting tumor growth and metastasis. However, the specific mechanisms of NORAD involved in regulating endothelial cell function are unclear.
Circulating nucleic acids can modulate cellular phenotypes through targeted interference with gene expression14. Plasma lncRNAs exhibit high stability and detectability, with their levels potentially reflecting underlying disease mechanisms, offering a unique advantage for biomarker development15,16. Several studies investigated the diagnostic and prognostic role of circulating lncRNAs in cardiovascular disease. For example, plasma lncRNA-LIPCAR has been shown to predict cardiac remodeling and cardiovascular death in heart failure patients after MI17. Plasma lncRNA-CoroMarker has been found to assist in diagnosing CAD18. Additionally, changes in whole-blood lncRNA-ZFAS1 and CDR1AS have been associated with an increased risk of MI19. Elevated plasma lncRNA-DKFZP434I0714 levels have been linked to poor outcomes in uremic patients20.
This study was aimed to investigate the expression pattern of circulating NORAD in CAD patients with apparent atherosclerotic plaques and explore its potential as a biomarker for CAD. Bioinformatics analysis will be employed to investigate the underlying mechanisms of NORAD, and human umbilical vein endothelial cells (HUVECs) will be used to validate its effects on cell function and explore the possible mechanisms involved. Our findings may shed light on the role of NORAD in the pathogenesis of endothelial dysfunction and AS.
Methods
Patients and sample selection
From September 2021 to May 2022, we recruited participants (n = 151) from the Department of Cardiology, Zhongnan Hospital of Wuhan University, including CAD patients with coronary stenosis (n = 75) and controls (n = 76). Gensini scores estimated the degree of coronary stenosis after coronary computed tomography or coronary angiography21. At least two experienced cardiologists determined the diagnoses. Patients with a Gensini score of more than five points were enrolled in the CAD group. Based on physical examination and medical history, the control group had no other severe circulatory diseases or coronary stenosis (Gensini score < 5). We excluded patients with malignant tumors, malignant arrhythmia, severe heart failure, severe organic brain disease, concurrent infections, pregnancy, mental illness, or other significant diseases. The study was conducted in accordance with the Declaration of Helsinki, and approved by the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (protocol code 2021009).
RNA isolation and real-time quantitative PCR analysis
Plasma specimens were collected from each individual and stored at − 80 °C. Plasma RNA was extracted using a Biog cfRNA Easy Kit (Changzhou Bio-generating Biotechnology Corp, China). Total RNA was isolated using TRIzol reagent (Invitrogen, USA) and reverse transcribed into cDNA using a ReverTra Ace® qPCR RT Master Mix kit (TOYOBO, Osaka, Japan). Real-Time qPCR was performed on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) using SYBR ® Green Real-time PCR Master Mix (TOYOBO). The mRNA expression levels of human genes were normalized to 18 S ribosomal RNA. The sequences of PCR primers for amplifying human genes, including NORAD and CCND1, are presented in Supplementary Table S1. The relative mRNA expression level of each gene was calculated using the 2 −ΔΔCt method.
Data acquisition and preprocessing
The predicted miRNAs with NORAD binding site were obtained from starBase and miRcode, and compared with the miRNA microarray dataset GSE2885822 using Venn 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/). The target mRNAs were predicted using miRBD, miRTarBase, and miRWalk, consistent with GSE10092723. These datasets were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo). GSE28858 contains platelet miRNA profiles from CAD patients. The GSE100927 dataset contains transcriptome analysis of human peripheral arteries from carotid arteries with atherosclerotic and control tissue. The expression files were identified using the limma package in R with a criterion of P < 0.0524.
Regulatory network construction and analysis
The competing endogenous RNA (ceRNA) network of NORAD-miRNA-mRNA was constructed using Cytoscape software (3.8.0). Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis25,26 were performed using DAVID 6.8 and plotted using the ggplot2 package in R. The protein-protein interaction (PPI) network was constructed using STRING database software (https://string-db.org/) with a reliability threshold of a combined score of > 0.427. The top ten genes with maximum neighborhood component (MNC) were identified using the Cytoscape plug-in cytoHubba. The hub genes were analyzed using the Molecular Complex Detection (MCODE) algorithm. FunRich is introduced as a user-friendly, open-access bioinformatics tool that enables researchers to delve into the functional aspects and network interactions of genes and proteins, enhancing the understanding of complex biological data28. The study employed FunRich version 3.1.3 to conduct GO analysis focusing on biological process (BP), cellular component (CC), and molecular function (MF) categories of miRNAs. Statistical significance was determined by a P-value cut-off of < 0.05. The Molecular Signature Database (MSigDB) provided a comprehensive gene set, which, in conjunction with R package clusterProfiler and GSVA, facilitated gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA). The research highlights the five KEGG pathways with the most significant enrichment, based on a P-value threshold of < 0.05, to pinpoint key biological mechanisms potentially at play.
Cell culture and transfection
HUVECs were acquired from the Classic Specimen Culture and Storage Center at Wuhan University (Wuhan, China). The cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 1% penicillin/streptomycin and 10% fetal bovine plasma (Gibco, Thermo Fisher, Waltham, MA, USA) in 5% CO2 at 37 °C in a humidified atmosphere. Cells (5 × 105 cells per well in six-well culture plates) were transfected with the si-NORAD or the control si-NC using the RNAiMAX reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. Cells were collected using RIPA or TRIzol reagent at the indicated times to measure the gene expression.
Western blot to determine protein level
HUVECs were harvested and lysed using RIPA (Beyotime Institute of Biotechnology, Shanghai, China) with 1% proteinase inhibitors (Roche, Basel, Switzerland). Sodium dodecyl sulfate-polyacrylamide electrophoresis and western blots were performed to determine the protein levels after quantifying the total protein concentration using a BCA protein assay kit (Bio-Rad, Hercules, CA, USA). The target proteins were detected using specific primary antibodies and horseradish peroxidase-conjugated secondary antibodies. The signal of the protein band was observed using an enhanced chemiluminescence kit (Advansta, San Jose, CA, USA). We used antibodies against human CCND1, peroxisome proliferator-activated receptor-gamma (PPARγ), forkhead box O3a (FOXO3a), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Invitrogen (Waltham, MA, USA), BCL2-Associated X (Bax) from Proteintech (Wuhan, China), β-Tubulin from Abmart (Shanghai, China), and glutathione peroxidase 4 (GPX4), ferritin heavy polypeptide 1 (FTH1), cysteinyl-aspartate specific protease 3 (Caspase 3) from cell signaling technology (CST, Boston, MA, USA).
Wound healing assay
The cells were overgrown. Three straight lines were drawn in each hole, and a horizontal line perpendicular to the straight line was drawn. The excess cells were washed out with a plasma-free medium and cultured in a low-plasma medium. The migration of each cell pore was photographed at 0 h, 6 h, and 12 h using an inverted microscope, and images were obtained at different positions of each pore simultaneously.
Cell cycle assay
Precooled anhydrous ethanol was used to fix the cells at 4 °C for 2 h. RNase A, stain buffer and propyl iodide stain were mixed to generate a propidium iodide staining solution. The HUVECs were incubated with the propidium iodide staining solution at 37 °C for 30 min. The fluorescence intensity was detected using a flow cytometer.
Dual-luciferase reporter assay
293T cells were seeded into 24-well plates and after 12 h incubation the confluence reaches to 70–80%. PMIR-NORAD/CCND1-RL WT and PMIR-NORADmut/CCND1mut-RL mutant reporter plasmids were constructed in advanced. According to the manufacturer’s instruction, 293T cells were transiently co-transfected with miR-106a-5p mimics together with PMIR-NORAD/CCND1-RL WT or PMIR-NORADmut/CCND1mut-RL mutant reporter plasmids using Lipofectamine 3000. After 48 h, The relative luciferase activities were measured using the Dual-luciferase Reporter Assay System (Beyotime, Shanghai, China) and normalized to Renilla activity by GloMax 20/20 Microplate Luminometer (Shengzhao, Shanghai, China).
Cell counting kit-8 (CCK-8) assay
HUVECs transfected with si-NORAD were seeded in 96-well plates and cell viability was estimated using a CCK-8 assay kit (Meilunbio, Dalian, China) following the manufacturer’s protocol. We added 10 µl of the working reagent to wells, and incubated them at 37 °C for 2 h. Each well’s absorbance at 450 nm was measured using a microplate reader.
Cell apoptosis detection
Cells in 6-well plates were harvested and stained with Annexin V-FITC and PI for 15 min at room temperature according to the manufacturer’s instructions (Elabscience, Wuhan, China). The cell suspensions were immediately analysed by flow cytometer (FACSAriaIII, Becton-Dickinson, San Jose, CA, USA) and the results were analyzed using Modifit software.
Intracellular Caspase 3 activity assay
HUVECs were harvested and the intracellular Caspase 3 activity was assessed by chemical colorimetric method. The absorbance was measured at 405 nm with a multifunctional microplate reader according to the Caspase 3 Activity Assay Kit (Beyotime Biotechnology, Shanghai, China).
Measurement of intracellular ROS generation
The intracellular levels of ROS were quantified using the fluorescent probe H2DCFDA (MCE, Shanghai, China), which reacts with intracellular free radicals and produces a fluorescent product, dichlorofluorescein (DCF). Hoechst 33,342 (MCE, Shanghai, China) was added to stain the cell nuclei for 10 min. Photographic recordings were made using the High Content Live Cell Imaging System (Operetta CLS, PerkinElmer, USA), and the fluorescence intensity of the cells was measured to verify the production of intracellular ROS.
Intracellular GSH quatification
HUVECs were harvested and the intracellular GSH level was assessed by chemical colorimetric method. The absorbance was measured at 420 nm with a multifunctional microplate reader according to the reduced Glutathione (GSH) Colorimetric Assay Kit (Elabscience, Wuhan, China).
Lipid peroxidation assessed by Liperfluo staining
Cells were stained with Liperfluo (Dojindo Molecular Technologies, Shanghai, China) for 30 min at 37 °C, collected by trypsinization, and analysed immediately using the 488 nm laser of a flow cytometer for excitation.
JC-1 staining
The mitochondrial membrane potential in cells was determined via a JC-1 fuorescent probe (Solarbio, Beijing, China). The cells in the 96-well plate were stained with JC-1 at 37 °C for 20 min in accordance with the manufacturer’s instructions, and the cell nuclei were stained with Hoechst 33342. The fluorescence was then detected, the mean fluorescence intensities quantified, and images captured by the High Content Live Cell Imaging System.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software 8.0 (GraphPad Software, Inc., La Jolla, CA, USA). The significant differences between groups were evaluated using unpaired two-tailed Student’s t-tests with Welch’s correction, or the chi-squared test. A two-way ANOVA followed by Newman-Keuls multiple comparison test was used to compare the control and multiple treatment groups. All data were expressed as means plus or minus the standard error of means. P-values < 0.05 were statistically significant.
Results
Plasma NORAD levels in the CAD and control groups
In this study, a total of 75 CAD patients and 76 controls were recruited to determine the plasma lncRNA NORAD levels using qRT-PCR. The baseline characteristics of patients with CAD and controls were presented in Table 1. The characteristic analysis revealed no significant differences in the characteristics of age, gender, smoking, and alcohol consumption. The prevalence of hypertension, diabetes history, as well as plasma total cholesterol (CHOL) and triglyceride (TG) levels were similar between the groups. Concerning the degree of stenosis, the Gensini score for the CAD group was 57.13 ± 42.21, whereas the score for the control group was nearly 0, making it a suitable control group for CAD patients with evident AS plaques.
Table 1.
General demographic characteristics of the research subjects.
| Demographic characteristics | Control group n (%) | CAD group n (%) | χ2/t | P | |
|---|---|---|---|---|---|
| Gender | Female | 40(52.63) | 28(37.33) | 3.569 | 0.0724 |
| Male | 36(47.37) | 47(62.67) | |||
| Age | < 65 | 49(64.47) | 48(64.00) | 0.0037 | > 0.9999 |
| ≥ 65 | 27(35.53) | 27(36.00) | |||
| Smoking | Yes | 15(19.74) | 25(33.33) | 3.584 | 0.0669 |
| No | 61(80.26) | 50(66.67) | |||
| Alcohol drinking | Yes | 10(13.16) | 10(13.33) | 0.0010 | > 0.9999 |
| No | 66(86.84) | 65(86.67) | |||
| Hypertension | Yes | 26(34.21) | 36(48.00) | 2.966 | 0.0993 |
| No | 50(65.79) | 39(52.00) | |||
| Diabetes | Yes | 8(10.53) | 14(18.67) | 2.010 | 0.1736 |
| No | 68(89.47) | 61(81.33) | |||
| CHOL | 4.44 ± 0.99 | 4.53 ± 0.86 | 0.5851 | 0.5594 | |
| TG | 1.38 ± 0.92 | 1.57 ± 0.65 | 1.483 | 0.1403 | |
| HDL-C | 1.26 ± 0.35 | 1.06 ± 0.22 | 4.284 | < 0.0001 | |
| LDL-C | 2.70 ± 0.84 | 2.96 ± 0.76 | 1.993 | 0.0481 | |
| sdLDL-C | 0.94 ± 0.39 | 1.02 ± 0.34 | 1.294 | 0.1977 | |
| LPa | 120.72 ± 97.56 | 146.02 ± 125.58 | 1.354 | 0.1778 | |
| NEFA | 410.26 ± 266.20 | 564.94 ± 317.93 | 3.213 | 0.0016 | |
| PLIP | 2.41 ± 0.46 | 2.34 ± 0.33 | 0.9797 | 0.3289 | |
| Gesini | 0.94 ± 1.49 | 57.13 ± 42.21 | 11.52 | < 0.0001 |
Data are expressed as means ± standard deviation (SD), median (25th–75th percentile), or counts (percentages). P < 0.05 was significant (t-tests with Welch’s correction, or chi-squared test). CHOL: total cholesterol; TG: triglyceride; HDL-C: high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; sdLDL-C: small dense low-density lipoprotein-cholesterol; LPa: lipoprotein(a); NEFA: non-esterified fatty acids; PLIP: phospholipid.
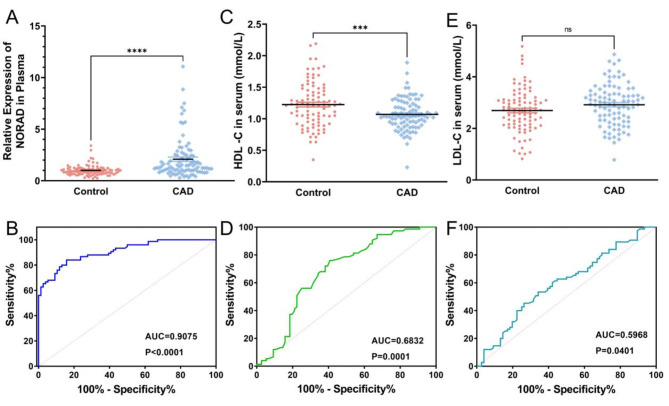
Plasma samples were obtained from both the CAD and control groups, followed by quantification of circulating NORAD levels using qRT-PCR (Fig. 1A). Plasma NORAD levels were significantly elevated in the CAD group compared to the control group which revealed that plasma NORAD exhibited remarkable discriminatory ability in identifying CAD patients from the control group (Fig. 1B). Our findings indicated lower HDL-C levels and higher LDL-C levels in CAD patients, yet the areas under the ROC curves were < 0.7, hence precluding the use of HDL-C and LDL-C as diagnostic indicators (Fig. 1C, D, E, F).
Fig. 1.
Circulating NORAD levels and serum lipid levels in the control and CAD groups. (A) Comparison of circulating NORAD between control and CAD group. (B) Receiver operating characteristic (ROC) curve analyses for NORAD. (C) Comparison of HCL-C between the control and CAD groups. (D) ROC curve analyses of HDL-C. (E) Comparison of circulating LDL-C between the control and CAD groups. (F) ROC curve analyses of LDL-C. ***P < 0.001, ****P < 0.0001, ns, not significant.
Analysis of the network regulation mechanism of NORAD
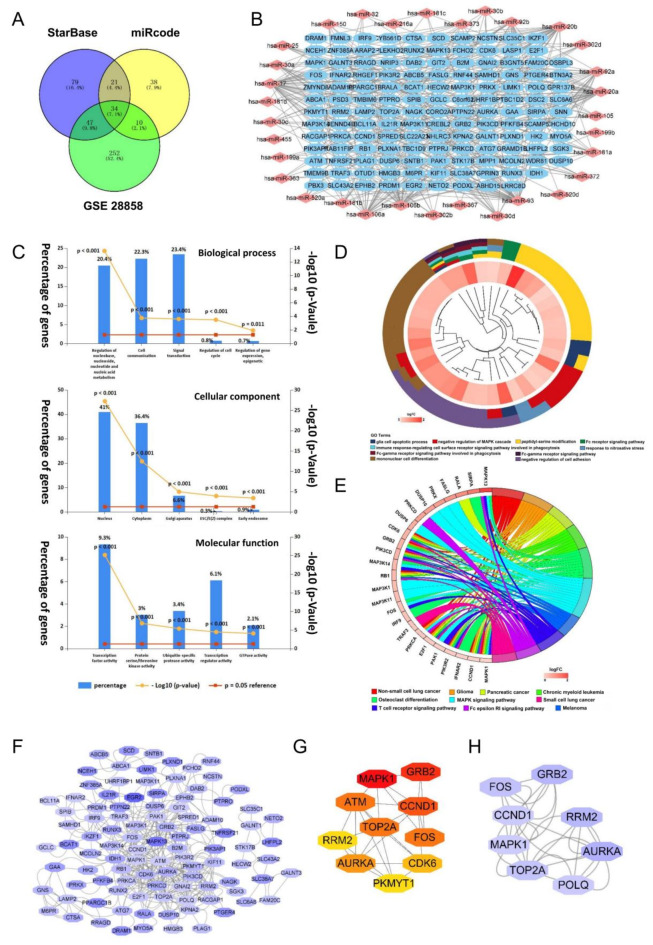
To investigate the regulation mechanism of NORAD in AS formation, we collected NORAD target miRNAs from starBase and miRcode. At the same time, we screened miRNAs with significantly reduced expression levels from the GSE28858 dataset. A Venn diagram was drawn to identify miRNAs overexpressed in all datasets, resulting in 34 miRNAs (Fig. 2A, Supplementary Table S2). The mRNAs predicted as targets of these miRNAs were obtained from miRBD, miRTarBase, and miRWalk database, and intersected with upregulated genes in GSE100927. Figure 2B shows the network of miRNAs and 138 target genes. We utilize FunRich 3.1.3 to perform GO enrichment analysis on the 34 significantly upregulated miRNAs targeting NORAD in CAD (Fig. 2C). In the Biological Process category, the target miRNAs are predominantly involved in regulating nucleobase, nucleoside, nucleotide, and nucleic acid metabolism, cell communication, and signal transduction. Within the Cellular Component category, the target miRNAs are primarily associated with the nucleus, cytoplasm, and Golgi apparatus. In the Molecular Function category, the target miRNAs are mainly linked to transcription factor activity, protein serine/threonine kinase activity, and ubiquitin-specific protease activity.
Fig. 2.
Construction of the ceRNA network of NORAD. (A) Venn diagram of miRNA from starBase database, miRcode database and GSE28858. (B) Network of miRNAs - mRNAs. (C) FunRich 3.1.3 to conduct GO enrichment analysis on the 34 significantly upregulated miRNAs targeting NORAD in CAD. (D) GO chord diagram of biological processes based on GO enrichment analysis of target mRNA. The inner circle indicates the mRNAs, and colors represent the log2 (fold change) change in expression. The different colors in the outer circle represent different GO terms. (E) KEGG circle diagram based on KEGG pathway analysis. The target mRNAs are shown on the left side, and distinct colored bands on the right-hand side symbolize different pathways. (F) The PPI network of targets miRNAs using STRING. The node color depth is proportional to the log2 change in expression. (G) The top ten genes subnetwork after CytoHubba filtration with MNC. (H) The significant module from the PPI network with an MCODE score of 4.286.
GO and KEGG pathway enrichment analyses were performed on target genes to explore the biological functions of NORAD. The GO chord diagram of biological processes revealed that NORAD was involved in apoptosis and negative regulation of the MAPK cascade (Fig. 2D). KEGG pathway enrichment analysis revealed that the targets of NORAD were enriched in cancer and the MAPK signaling pathway (Fig. 2E).
The 138 targets of NORAD were input into the STRING database to investigate the interrelation of the target proteins. The PPI network included 101 nodes and 448 edges (Fig. 2F). In cytoHubba, the first ten hub genes were identified using the MNC algorithm, including mitogen-activated protein kinase1 (MAPK1), growth factor receptor-bound protein 2 (GRB2), G1/S-specific cyclin-D1 (CCND1), among others (Fig. 2G). MCODE was used to conduct clustering analysis and the functional module was screened out from the PPI network (Fig. 2H). Based on the results of the bioinformatics analysis, we found that NORAD was involved in regulatory processes related to the cell cycle, and CCND1 may be a critical gene in the NORAD regulatory network.
Si-NORAD induces endothelial cell cycle arrest in the G0/G1 phase
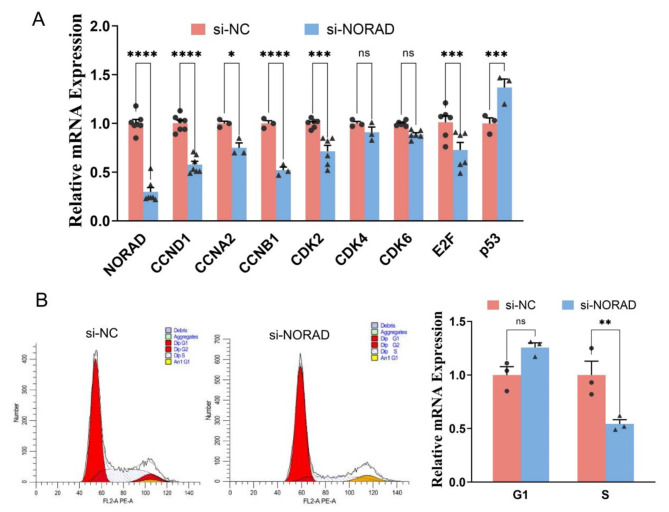
To determine the effect of NORAD on the cell cycle, we measured the expression levels of relevant genes using qPCR. As shown in Fig. 3A, the expression of NORAD was significantly reduced in HUVECs transfected with si-NORAD. Meanwhile CCND1, cell cycle protein A2 (CCNA2), G2/mitotic-specific cyclin-B1 (CCNB1), cyclin-dependent kinase 2 (CDK2), CDK6, and transcription factor 2 (E2F) were significantly decreased after NORAD knockdown in HUVECs. Then we examined endothelial cell proliferation cycles using flow cytometry, and we found that the number of endothelial cells in the G0/G1 phase increased significantly in the NORAD knockdown group. The proportion of cells in the S phase was significantly reduced (Fig. 3B). These results suggest that NORAD knockdown induces endothelial cell cycle arrest in the G0/G1 phase via downregulated cyclin D1 and cyclin-dependent kinases, then inhibits endothelial cell proliferation.
Fig. 3.
NORAD regulated the cell cycle. (A) The expression level of cell cycle-related genes in HUVECs. (B) Cell cycle determination using flow cytometry. N = 3–7. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
Si-NORAD inhibited HUVECs migration
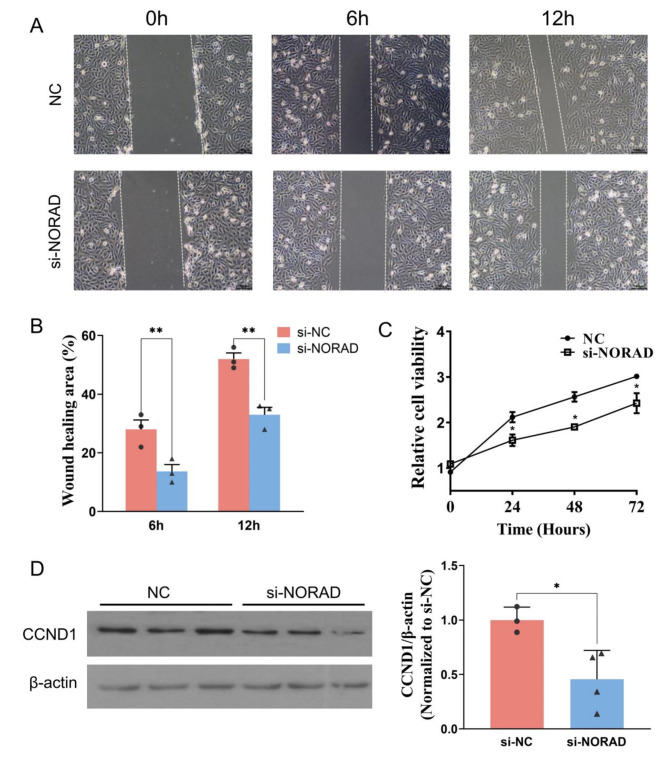
To investigate the effect of NORAD on the migration of HUVECs, we performed the wound-healing assay. As shown in Fig. 4A and B, the wound-healing areas at 6 and 12 h were estimated by a cell scratch test, and both significantly decreased in HUVECs with si-NORAD. Cell proliferations were measured at 0 h, 24 h, 48 h and 72 h using the CCK8 assay. The results suggest that si-NORAD significantly inhibited the proliferation of HUVECs compared with the control group (Fig. 4C). These findings suggest that si-NORAD inhibits HUVECs proliferation and migration. As indicated by the previous analysis, CCND1 may be the target of NORAD. We then detected the CCND1 protein levels using western blot. The result showed that CCND1 protein expression decreased in HUVECs transfected with si-NORAD (Fig. 4D).
Fig. 4.
Si-NORAD inhibited HUVECs migration. (A and B) Wound healing was estimated using a scratch assay in HUVECs at 0, 6, and 12 h. (C) Cell proliferation was determined using CCK8 analysis. (D) CCND1 protein levels were measured using western blot assay. N = 3 or 4. *P < 0.05, **P < 0.01.
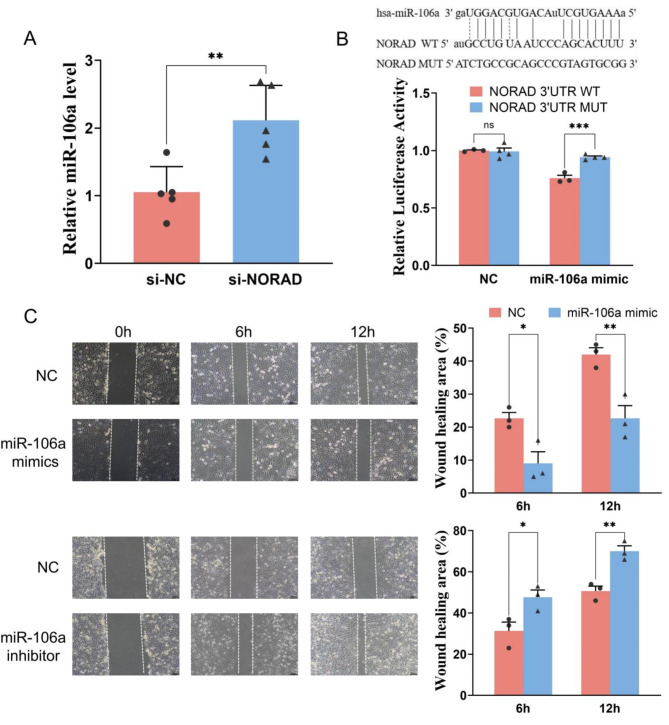
NORAD regulated the migration of HUVECs by targeting miR-106a
Based on miRNA targets prediction tool starBase database, miRcode database and combined with the data of GSE28858, we predicted NORAD could be targeted by miR-106a. And we suspected that miR-106a might be a sponge of NORAD involved in regulating CCND1 protein levels. To determine the targeting relationship between NORAD and miR-106a, we examined the expression of miR-106a in HUEVCs, and found elevated levels of miR-106 in HUVEC with si-NORAD (Fig. 5A). We predicted the potential binding site of the NORAD 3´UTR terminal with miR-106a (Fig. 5B). We verified the targeting relationship between them by a dual luciferase assay. The luciferase activity of the WT reporter was lower in NORAD-WT treated with miR-106a mimic compared to the NORAD-WT treated with miR-NC (Fig. 5B), but the repression was abrogated after mutating the putative binding sites of NORAD gene. The results supported the notion that miR-106a was directly targeted by NORAD. We also measured the effect of miR-106a on endothelial cells. After miR-106a was overexpressed in HUVECs, cell proliferation, and migration were detected using a cell scratch assay at 6 and 12 h. As shown in Fig. 5C, miR-106a mimic inhibited HUVEC proliferation and migration, while the miR-106a inhibitor promoted the proliferation and migration. These results suggest that NORAD may interact with miR-106a, potentially modulating its availability or activity, and thereby regulating the proliferation and migration of HUVECs.
Fig. 5.
NORAD acts as a sponge for miR-106a. (A) MiR-106a levels in HUVECs measured by qPCR. (B) A dual-luciferase reporter gene was used to verify the targeted relationship between NORAD and miR-106a. (C) Wound-healing was tested using a scratch assay in HUVECs treated with miR-106a mimic or inhibitor. N = 3–5. *P < 0.05, **P < 0.01.
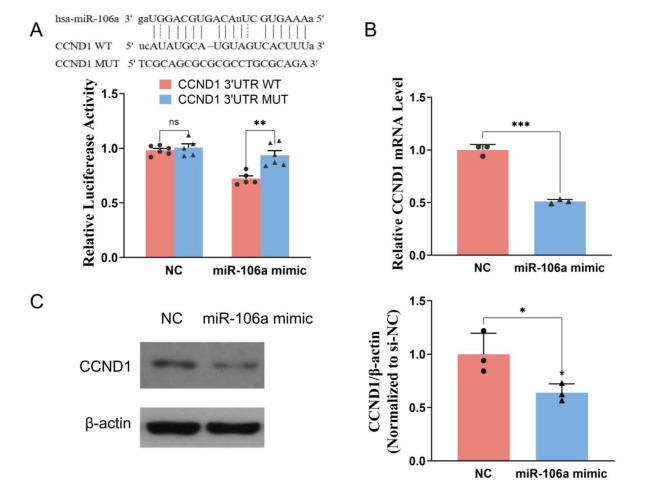
MiR-106a directly targets CCND1
In addition, miR-106a may target CCND1 directly, and the binding site was predicted using the starBase database. To confirm that miR-106a specifically binds to the 3’UTR of CCND1 mRNA to regulate the expression of CCND1, we performed a dual-luciferase reporter assay. The results showed that the luciferase activity in the CCND1-WT + miR-106a mimic co-transfected group was lower than that of the CCND1-WT + miR-NC co-transfected group, but there was no significant difference when miR-106a mimic or NC was co-transfected with CCND1-MUT (Fig. 6A). The result supported that miR-106a directly targeted the CCND1 3′UTR. To verify whether miR-106a affects the expression of CCND1 in HUVECs, we performed qPCR and western blot to measure the CCND1 expression. CCND1 mRNA and protein expression levels decreased (Fig. 6B and C) in HUVECs treated with miR-106a mimic. These results suggest that miR-106a directly targets CCND1, inhibiting the proliferation of endothelial cells.
Fig. 6.
MiR-106a directly targets CCND1. (A) The putative binding sites and corresponding mutant region for miR-106a within CCND1 and dual-luciferase reporter assay to verify the targeted relationship. (B) The effect of miR-106a on the mRNA level of CCND1 was determined using qPCR. (C) The effect of miR-106a on the protein expression of CCND1 was determined using western blot. N = 3–6. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
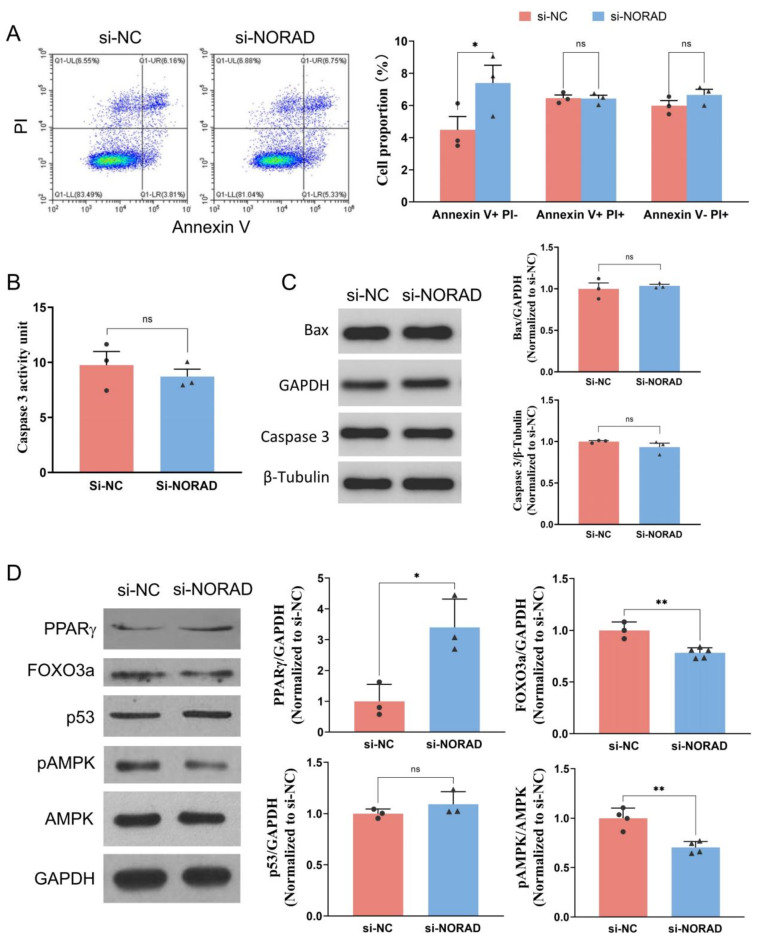
Si-NORAD regulated cell apoptosis
The apoptosis of HUVEC induced by NORAD was detected by PI and Annexin V-FITC staining. As shown in Fig. 7A, compared with HUVEC transfected with NC, HUVEC transfected with si-NORAD exhibited high proportions (7.4% vs. 4.5%, > 20%) of Annexin V + PI− (early apoptotic), but there was no significant difference in the proportions of Annexin V + PI+ (late apoptotic) and Annexin V- PI+ (necrotic) cells between those transfected with NC or si-NORAD. These results collectively suggest that si-NORAD induced HUVEC early apoptotic.
Fig. 7.
Si-NORAD induced early apoptosis. (A) Cell apoptosis analysis of HUVEC transfected with si-NORAD by flow cytometry. (B) Intracellular Caspase 3 activity assay. (C) and (D) The protein levels were measured using western blot. GAPDH or β-Tubulin was used as the protein loading control. N = 3–5. *P < 0.05, **P < 0.01, ns, not significant.
Additionally, the expression levels of apoptotic key molecules were examined. The enzyme activity assay revealed no statistically significant difference in Caspase 3 activity between the two groups (Fig. 7B). Furthermore, Western blot analysis demonstrated that following transfection with si-NORAD, there was no remarkable alteration in the protein expression levels of Caspase 3 and Bax in HUVECs, as compared to the NC group (Fig. 7C). Based on the above results, it may sugguest that si-NORAD dose not activate the Caspase 3-mediated apoptotic pathway, thereby not proceeding to the terminal stage of apoptosis.
Protein electrophoresis results showed that compared with the control group, the protein levels of PPARγ increased, but FOXO3a protein levels were down-regulated in HUVEC with NORAD knockdown. Besides, the protein level of p53 did not increase, but the phosphorylated AMPK decreased, as induced by si-NORAD in HUVECs (Fig. 7D).
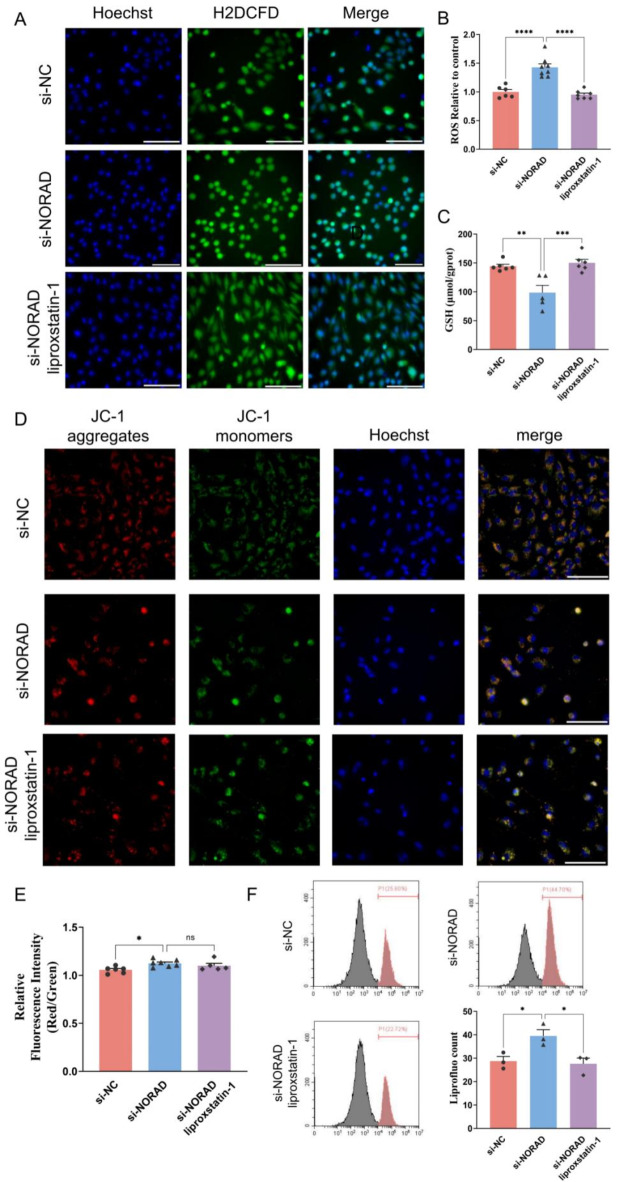
Modulation of endothelial redox state by si-NORAD
The impact of si-NORAD transfection on endothelial ROS levels and redox state was investigated using H2DCFD staining and high-content analysis. Transfection of HUVECs with si-NORAD resulted in a significant increase in intracellular ROS levels. However, the addition of liproxstatin-1 attenuated this increase (Fig. 8A and B). Additionally, si-NORAD transfection led to a decrease in intracellular glutathione (GSH) levels, which were restored upon the addition of liproxstatin-1, indicating a restorative effect of liproxstatin-1 on the cellular redox balance (Fig. 8C). Further analysis of mitochondrial membrane potential using the JC-1 probe revealed that si-NORAD transfection induced a decrease in mitochondrial membrane potential. Although liproxstatin-1 was administered, it did not significantly affect the mitochondrial membrane potential, suggesting that the protective effects of liproxstatin-1 may not be mediated through this particular mitochondrial pathway (Fig. 8D and E). To assess lipid peroxidation, the liprofluo fluorescent probe was employed, which revealed that si-NORAD transfection significantly increased the level of oxidized lipids in HUVECs. However, this increase was reversible with the application of liproxstatin-1, indicating that liproxstatin-1 can effectively reduce the oxidative stress induced by si-NORAD (Fig. 8F). Collectively, these results suggest that si-NORAD induces oxidative stress in HUVECs.
Fig. 8.
Si-NORAD induced HUVEC oxidation. (A) and (B) The images of HUVEC stained with H2DCFD for ROS and Hoechst (blue fluorescent) for nuclei by the high-content live cell imaging system. (C) The GSH level in HUVEC transfected with si-NORAD. (D) HUVEC stained with JC-1 for mitochondria and Hoechst (blue fluorescent) for nuclei. JC-1 forms aggregates (red fluorescent) under high mitochondrial potential condition and becomes monomers (green fluorescent) under low mitochondrial potential condition. Bar = 100 μm. (E) The ratios of red fluorescent intensity to green fluorescent intensity were shown. (F) Representative flow cytometric profiles are shown to demonstrate lipid peroxides with Liperfluo signals. N = 3–8. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
Regulatory roles of NORAD in ferroptosis
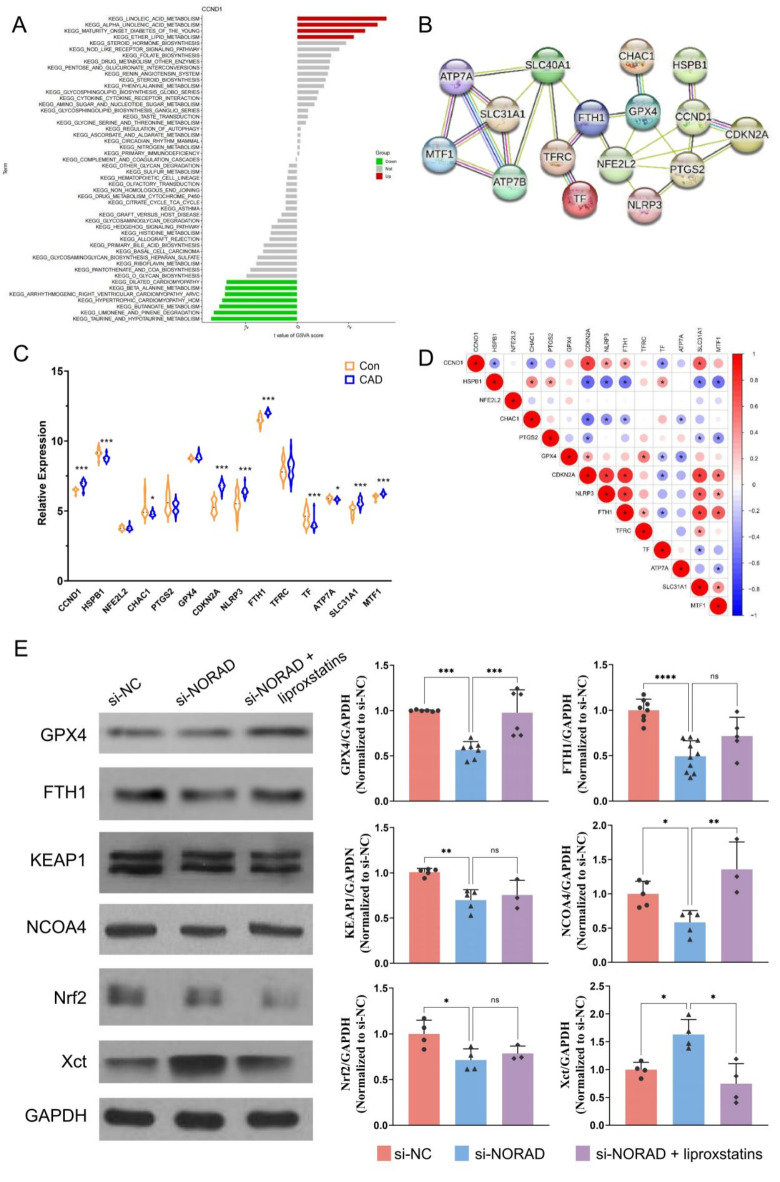
Through the application of Gene Set Variation Analysis (GSVA), our investigation into the biological processes and potential mechanisms of CCND1 expression in atherosclerosis (AS) revealed distinct pathway enrichments. High expression of CCND1 was associated with the significant enrichment of metabolic pathways such as linoleic acid metabolism, α-linolenic acid metabolism, and ether lipid metabolism. Conversely, low expression of CCND1 correlated with the enrichment of pathways including butanoate metabolism, limonene and pinene degradation, and taurine and hypotaurine metabolism (Fig. 9A).
Fig. 9.
Si-NORAD regulated cell ferroptosis-related genes. (A) GSVA analysis on CCND1. (B) The PPI network of CCND1 and ferroptosis-related proteins. (C) Relative expression of CCND1 and ferroptosis-related genes in GSE100927. (D) Corrplot of CCND1 and ferroptosis-related genes in GSE100927. (E) Protein levels were measured using western blot. N = 3–10. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
Besides, our research demonstrated that the silencing of NORAD via si-NORAD led to a marked increase in cellular oxidative stress levels. This oxidative stress response was found to be reversible by the administration of liproxstatin-1, a ferroptosis inhibitor, suggesting a potential role for NORAD in the regulation of ferroptosis, a form of cell death linked to iron metabolism. Utilizing the String database, we conducted an analysis to elucidate the potential interactions between CCND1 and ferroptosis-related factors (Fig. 9B). Analysis of the GSE100927 dataset revealed a positive correlation between the expression levels of CCND1 and ferroptosis inhibitors such as GPX4 and FTH1 in AS plaques (Fig. 9C and D). This analysis provided a framework for understanding the molecular connections that may underlie the observed correlations between CCND1 expression and ferroptosis in AS.
To further delineate the effects of NORAD silencing on ferroptosis-related proteins, we transfected HUVECs with si-NORAD and assessed the expression levels of key proteins. Our results indicated that NORAD silencing led to a decrease in the expression of GPX4, FTH1, KEAP1, NCOA4, and Nrf2, while Xct levels were increased (Fig. 9E). These alterations in protein expression were found to be reversible by the treatment with liproxstatin-1, confirming the involvement of ferroptosis in the cellular response to NORAD silencing.
Discussion
Atherosclerotic lesions are slow-progressing diseases characterized by the accumulation of lipids, inflammatory cells, and necrotic cell debris, transforming into atheromatous plaques in the intimal space beneath the endothelial cell layer. This process often leads to significant reductions in blood flow, resulting in conditions such as angina, and can cause life-threatening events like myocardial infarction (MI) or stroke if clots block blood flow to the heart or brain. These mechanisms are significant causes of death and disability, particularly in the elderly, highlighting the critical need for sensitive and specific biomarkers for early atherosclerosis (AS) detection and treatment1.
Plasma NORAD has been identified as a potential biomarker for various conditions, including pulmonary tuberculosis, gestational hypertension, and neonatal sepsis29–31. Zhang et al. studied peripheral blood mononuclear cells NORAD levels linked to blood lipids and an increased risk of CAD32. Given the importance of lipid metabolism, and inflammation in the pathology and progression of CAD, NORAD emerges as a candidate biomarker for CAD patients. The current study provides insights into the role of plasma NORAD expression levels in coronary artery disease (CAD) diagnosis and prognosis. Our findings demonstrate that plasma NORAD levels are significantly elevated in CAD patients compared to healthy controls, indicating its potential as a novel biomarker for CAD. NORAD levels were significantly higher in CAD patients with Gesini scores greater than 5, whereas HDL-C levels were significantly lower. Reduced plasma HDL-C levels are associated with the CAD development, but this study found that the ROC score of HDL-C was lower than NORAD suggesting that NORAD could be used as a diagnostic marker for CAD. The ROC curve analysis further supports the diagnostic value of NORAD in CAD, with the ability to differentiate CAD patients from healthy controls with high accuracy.
Plasma free lncRNAs can influence the function of vascular wall cells via several mechanisms. Targeting binding plasma free miRNAs can reduce miRNA entry and regulate the function of vascular wall cells. Circulating lncRNAs and miRNAs together may also lead to advances in the diagnosis and outcomes of CAD14,33. MiR-106a is a multifunctional miRNA that regulates cell proliferation, apoptosis, and other cell signaling pathways that influence the development of diseases such as cancer. A study showed that miR-106a promoted cell proliferation and inhibited apoptosis in human vascular smooth muscle cells34. MiR-106a regulated proliferation and apoptosis in colon cancer cells by targeting the PTEN/PI3K/AKT signaling pathway35. Our findings demonstrate that NORAD upregulates CCND1 in HUVECs via sponging miR-106a, which inhibits cell migration. CCND1 is a critical protein regulating the G1 phase of the cell cycle and is crucial to the regulation of the cell proliferation cycle. After NORAD knockdown, CCND1 protein and mRNA expression levels were significantly reduced, suggesting that NORAD acts as a sponge for miR-106a and affects CCND1 levels to arrest cells in the G0/G1 phase, affecting endothelial cell proliferation.
Endothelial dysfunction, caused by inflammatory factors, oxidized lipids, and other stimuli, is one of the first significant events in the development of AS36. NORAD regulates cellular function via multiple pathways, involves in the development and progression of cardiovascular disease. NORAD inhibits VEGF gene transcription by increasing H3K9 deacetylation, promoting vascular endothelial cell injury and AS37. NORAD Knockdown reduces Krüppel-like factor 5 by increasing miR-495-3P, thus exerting an atherosclerotic inhibitory effect38. NORAD sponges miR-590-3p and promotes angiogenesis induced by ox-LDL39. NORAD knockdown inhibited ox-LDL induced endothelial cell injury by regulating miR-30c-5p/Wnt7b/β-catenin40. Conflicting findings have also been reported, investigators found that NORAD knockdown increased ox-LDL induced reactive oxygen species, they also demonstrated that NORAD knockdown increased lipid disorders and atherosclerotic lesions in apoE-/- mice via NF-κB and p53-p21 signaling pathways41. To elucidate the regulatory role of NORAD, we constructed a ceRNA regulatory network of NORAD using GSE28858 and GSE100927 and demonstrated that NORAD is primarily involved in cell proliferation, death, and other processes affecting disease development. Our founding suggested that NORAD acts as a sponge for miR-106a and affects CCND1 levels to arrest cells in the G0/G1 phase, affecting endothelial cell proliferation. CCND1 is acritical protein regulating the G1 phase of the cell cycle, and is crucial to the regulation of the cell proliferation cycle. After NORAD knockdown, CCND1 protein and mRNA expression levels were significantly reduced. Studies showed that CCND1 forms a complex with CDK4 or CDK6 and functions as a regulatory subunit, whose activity is required for G1/S transition in the cell cycle42. CDK2 promote cell-cycle progression in G1, S, or G2 phases by binding with cyclin E or cyclin A43. CDK2/6 and transcription factor E2F are essential in cell cycle regulation. Our finding suggest that NORAD knockdown reduced intracellular mRNA levels of CDK6, but the changes in CDK4 mRNA levels were insignificant. Expression levels of cell cycle regulation-related factors CDK2, CCNA2, and transcription factor E2F were reduced, resulting in a G0/G1 phase block.
HUVECs transfected with si-NORAD exhibited increased proportions of early apoptotic cells, indicating that NORAD silencing may induce cell death. Si-NORAD may not directly affect the protein levels of Caspase 3 and Bax or the enzymatic activity of Caspase 3 in HUVECs. This observation aligns with our previous results, which showed that si-NORAD induced early apoptosis in HUVECs, as evidenced by increased Annexin V + PI- cells, without significant changes in late apoptosis. This may indicate that si-NORAD does not activate the Caspase 3-mediated apoptotic pathway, thereby not proceeding to the terminal stage of apoptosis. Protein electrophoresis results revealed that NORAD knockdown led to an increase in PPARγ protein levels and a decrease in FOXO3a protein levels, suggesting that NORAD may modulate the expression of these proteins, which are known to be involved in various cellular processes, including cell cycle regulation and apoptosis. PPARγ is a nuclear hormone receptor shown to cause cell arrest in the G0/G1 phase44. PPARγ is involved in the regulation of cellular differentiation, while FOXO3a is a transcription factor that promotes cell cycle arrest and apoptosis. The increase in PPARγ and decrease in FOXO3a protein levels in HUVECs with NORAD knockdown suggest NORAD may also modulate cellular responses to apoptosis and oxidative stress. The decrease in phosphorylated AMPK indicates a potential reduction in cellular energy sensing and response, which is reasonable given the role of AMPK in maintaining cellular energy homeostasis.
The observed alterations in apoptosis, reactive oxygen species (ROS) levels, and mitochondrial function in response to si-NORAD transfection are intricately linked to the process of ferroptosis, a form of regulated cell death associated with iron metabolism and lipid peroxidation45. The increase in early apoptotic cells, rather than late apoptotic cells, following si-NORAD transfection suggests that NORAD may play a role in the initiation phase of apoptosis. While distinct from ferroptosis, the induction of early apoptosis could potentially prime cells for ferroptosis by compromising cellular integrity and increasing susceptibility to oxidative stress46. ROS are critical signaling molecules and also potent inducers of oxidative stress. The significant rise in intracellular ROS levels post si-NORAD transfection aligns with the characteristics of ferroptosis, where ROS, particularly those generated via iron-catalyzed reactions, drive lipid peroxidation leading to cell death47. Thus, the elevation of ROS in response to si-NORAD may facilitate the onset of ferroptosis. Mitochondria are central to cellular energy metabolism and are also a major source of ROS. The decrease in mitochondrial membrane potential observed following si-NORAD transfection reflects mitochondrial dysfunction, which is consistent with the mitochondrial damage observed in ferroptosis. Impaired mitochondrial function can lead to the disruption of the electron transport chain, increasing ROS production and thereby promoting ferroptosis48.
Lipid disorder-mediated ferroptosis effects on AS are multifaceted and an essential intermediate between initial and advanced AS. A study found that cell cycle-related factors regulate ferroptosis, p53 is a well-established critical cell cycle regulator and contributed to ferroptosis, they found E2F suppresses ferroptosis49. GPX4 scavenges oxidized lipids from cells, reduces levels of oxidative stress, blocks ferroptosis and attenuates AS damage50. FTH1 is a ferritin complex that stores iron and prevents its oxidation by ROS, thereby inhibiting ferroptosis51. FTH1 binds to nuclear receptor cofactor 4, a cargo receptor that degrades ferritin and sends autophagosomes to lysosomes, where ferritin is degraded to increase intracellular iron content52. The downregulation of proteins such as GPX4, FTH1, KEAP1, NCOA4, and Nrf2, and the upregulation of Xct in response to si-NORAD transfection, are all indicative of a shift in the cellular environment that could favor ferroptosis. These proteins are known to play crucial roles in iron homeostasis and the cellular response to oxidative stress53–55. Their altered expression may impair the cell’s defences against ferroptosis. In the GSE100927 dataset, we discovered higher levels of GPX4 and FTH1 expression in AS plaques in CAD patients. Together with our experimental findings, we hypothesize that elevated plasma NORAD levels in CAD patients may promote endothenial cell growth via inhibiting ferroptosis, prevent AS. Our results are consistent with earlier observations that NORAD knockdown enhanced AS plaques in apoE-/- mice39. The decrease in the expression of GPX4, FTH1, KEAP1, NCOA4, and Nrf2, and the increase in Xct levels following NORAD silencing, all point towards a disruption in the ferroptosis regulatory pathway. This is a reasonable finding as these proteins are known to be involved in the cellular response to oxidative stress and iron metabolism, both of which are central to ferroptosis. However, our experiments did not provide direct evidence that NORAD inhibits ferroptosis and AS. Thereby, the mechanism of action of ferroptosis-related factors and the mechanism of action on AS require further investigation.
Conclusion
In summary, plasma free NORAD levels are elevated in patients with significant AS plaques in CAD. NORAD influences endothelial cell proliferation by regulating the endothelial cell cycle via the miR-106a/CCND1 axis. The si-NORAD-induced changes in apoptosis, ROS levels, and mitochondrial function are consistent with the molecular mechanisms underlying ferroptosis. These observations suggest a model where NORAD deficiency leads to increased oxidative stress and cellular damage, culminating in the promotion of ferroptosis. The interplay between these cellular processes and the dysregulation of iron metabolism and redox balance are critical areas for further research, as they may offer novel therapeutic targets for conditions where ferroptosis contributes to pathology.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to extend our sincere appreciation to our colleagues for their invaluable efforts and insightful comments on this paper.
Author contributions
T.H collected the plasma from patients and analysis the sample. J.P. analyzed the data, performed the experiments; H.G carried out qPCR; T.L. and X.L. participated in cell experiments; Y.Z. and J.C. contributed to Western blot; H.Y. guided experiments; Z.L and F.D. conceived and designed the experiments. F.D. wrote the paper. All authors reviewed the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81670412), in part by the grant from the Natural Science Foundation of Hubei Province, China (grant numbers 2021CFB487, 2020CFB738) and by Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (grant number ZNLH201907).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no conflict of interest. The corresponding author is responsible for submitting a competing interests statement on behalf of all authors of the paper.
Ethics approval and consent to participate
This work complies with the Declaration of Helsinki and was performed under the oversight and regulations of the ethics committee of the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (protocol code 2021009).
Informed Consent Statement
In this study, we have strictly adhered to ethical and legal standards to ensure that all human participants, or their legal guardians, have been fully informed about the nature, purpose, procedures, potential risks, benefits, and privacy measures of the research. All participants have voluntarily signed an Informed Consent Form prior to their involvement in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tao He and Junxing Pu.
Contributor Information
Zhibing Lu, Email: luzhibing222@163.com.
Fen Du, Email: fen.du@whu.edu.cn.
References
- 1.Bjorkegren, J. & Lusis, A. J. Atherosclerosis: Recent developments. Cell. 185(10), 1630–1645 (2022). [DOI] [PMC free article] [PubMed]
- 2.Kobiyama, K. & Ley, K. Atherosclerosis Circ. Res. 123(10), 1118–1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo, F. X. et al. Shear Stress in Autophagy and its possible mechanisms in the process of atherosclerosis. DNA Cell. Biol. 36 (5), 335–346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gimbrone, M. J. & Garcia-Cardena, G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 118 (4), 620–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu, W. N. et al. The suppression of ox-LDL-induced inflammatory response and apoptosis of HUVEC by lncRNA XIAT knockdown via regulating miR-30c-5p/PTEN axis. Eur. Rev. Med. Pharmacol. Sci. 23 (17), 7628–7638 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Laslett, L. J. et al. The worldwide environment of cardiovascular disease: prevalence, diagnosis, therapy, and policy issues: a report from the American College of Cardiology. J. Am. Coll. Cardiol. 60 (25 Suppl), S1–49 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Fowkes, F. G. et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet. 382 (9901), 1329–1340 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Qin, B. et al. MicroRNA-142-3p induces atherosclerosis-Associated endothelial cell apoptosis by directly targeting Rictor. Cell. Physiol. Biochem. 47 (4), 1589–1603 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Pal, P. B. et al. Aldose reductase regulates hyperglycemia-induced HUVEC death via SIRT1/AMPK-alpha1/mTOR pathway. J. Mol. Endocrinol. 63(1), 11–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munschauer, M. et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 561 (7721), 132–136 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Lee, S. et al. Noncoding RNA NORAD regulates genomic Stability by Sequestering PUMILIO proteins. Cell. 164 (1–2), 69–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tichon, A. et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 7, 12209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalik, K. M. et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 114 (9), 1389–1397 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Anfossi, S. et al. Clinical utility of circulating non-coding RNAs - an update. Nat. Rev. Clin. Oncol. 15 (9), 541–563 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Bayoumi, A. S. et al. Circular noncoding RNAs as potential therapies and circulating biomarkers for ardiovascular diseases. Acta Pharmacol. Sin. 39 (7), 1100–1109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busch, A., Eken, S. M. & Maegdefessel, L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann. Transl Med. 4 (12), 236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumarswamy, R. et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114 (10), 1569–1575 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Yang, Y. et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. (Lond). 129 (8), 675–685 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y. et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict Acute myocardial infarction. Sci. Rep. 6, 22384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, C. F. et al. Circulating long noncoding RNA DKFZP434I0714 predicts adverse cardiovascular outcomes in patients with end-stage renal disease. Int. J. Cardiol. 277, 212–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rampidis, G. P. et al. A guide for Gensini score calculation. Atherosclerosis. 287, 181–183 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Sondermeijer, B. M. et al. Platelets in patients with premature coronary artery disease exhibit upregulation of miRNA340* and miRNA624*. PLoS One 6(10), e25946 (2011). [DOI] [PMC free article] [PubMed]
- 23.Steenman, M. et al. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci. Rep. 8 (1), 3940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa, M. et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44 (D1), D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45 (D1), D362–D368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseka, P. et al. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 433 (11), 166747 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Sun, W. et al. Diagnostic value of lncRNA NORAD in pulmonary tuberculosis and its regulatory role in Mycobacterium tuberculosis infection of macrophages. Microbiol. Immunol. 66 (9), 433–441 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Liang, Z. & Wang, L. Expression and clinical significance of lncRNA NORAD in patients with gestational hypertension. Ginekol. Pol. 94 (6), 429–434 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Zhang, H. et al. Clinical significance of the serum lncRNA NORAD expression in patients with neonatal Sepsis and its association with miR-410-3p. J. Inflamm. Res. 14, 4181–4188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, X. et al. Increased long non-coding RNA NORAD reflects serious cardiovascular stenosis, aggravated inflammation status, and higher lipid level in coronary heart disease. J. Clin. Lab. Anal. 36 (11), e24717 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Gonzalo-Calvo, D. et al. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur. Heart J. 40 (20), 1643–1650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y. et al. Exosome-mediated miR-106a-3p derived from ox-LDL exposed macrophages accelerated cell proliferation and repressed cell apoptosis of human vascular smooth muscle cells. Eur. Rev. Med. Pharmacol. Sci. 24 (12), 7039–7050 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Qin, Y. et al. mir-106a regulates cell proliferation and apoptosis of colon cancer cells through targeting the PTEN/PI3K/AKT signaling pathway. Oncol. Lett. 15 (3), 3197–3201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabas, I., Garcia-Cardena, G. & Owens, G. K. Recent insights into the cellular biology of atherosclerosis. J. Cell. Biol. 209 (1), 13–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kai, H. et al. LncRNA NORAD promotes vascular endothelial cell Injury and Atherosclerosis through suppressing VEGF gene transcription via enhancing H3K9 deacetylation by recruiting HDAC6. Front. Cell. Dev. Biol. 9, 701628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu, D. N. et al. Silenced long non-coding RNA activated by DNA damage elevates microRNA-495-3p to suppress atherosclerotic plaque formation via reducing Kruppel-like factor 5. Exp. Cell. Res. 401 (2), 112519 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Bao, M. H. et al. Long noncoding RNA LINC00657 acting as a mir-590-3p sponge to Facilitate Low Concentration Oxidized Low-Density Lipoprotein-Induced Angiogenesis. Mol. Pharmacol. 93 (4), 368–375 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Wu, H., Liu, T. & Hou, H. Knockdown of LINC00657 inhibits ox-LDL-induced endothelial cell injury by regulating miR-30c-5p/Wnt7b/beta-catenin. Mol. Cell. Biochem. 472 (1–2), 145–155 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Bian, W. et al. Downregulation of LncRNA NORAD promotes Ox-LDL-induced vascular endothelial cell injury and atherosclerosis. Aging (Albany NY). 12 (7), 6385–6400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qie, S. & Diehl, J. A. Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. (Berl). 94 (12), 1313–1326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudsen, E. S. et al. CDK/cyclin dependencies define extreme cancer cell-cycle heterogeneity and collateral vulnerabilities. Cell. Rep. 38 (9), 110448 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng, J. et al. WLIP derived from Lasiosphaera Fenzlii Reich exhibits anti-tumor activity and induces cell cycle arrest through PPAR-gamma associated pathways. Int. Immunopharmacol. 19 (1), 37–44 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 149 (5), 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, P. et al. Organelle-Specific Mechanisms in Crosstalk between Apoptosis and Ferroptosis. Oxid Med Cell Longev. 3400147 (2023). (2023). [DOI] [PMC free article] [PubMed]
- 47.Endale, H. T., Tesfaye, W. & Mengstie, T. A. ROS induced lipid peroxidation and their role in ferroptosis. Front. Cell. Dev. Biol. 11, 1226044 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, X. et al. The mechanisms of ferroptosis and its role in atherosclerosis. Biomed. Pharmacother. 171, 116112 (2024). [DOI] [PubMed] [Google Scholar]
- 49.Kuganesan, N. et al. Tumor suppressor p53 promotes ferroptosis in oxidative stress conditions independent of modulation of ferroptosis by p21, CDKs, RB, and E2F. J. Biol. Chem. 297 (6), 101365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouyang, S. et al. Ferroptosis: the potential value target in atherosclerosis. Cell. Death Dis. 12 (8), 782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie, Y. et al. Ferroptosis: process and function. Cell. Death Differ. 23 (3), 369–379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin, L. et al. Autophagy, Pyroptosis, and ferroptosis: New Regulatory mechanisms for atherosclerosis. Front. Cell. Dev. Biol. 9, 809955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ingold, I. et al. Selenium utilization by GPX4 is required to Prevent Hydroperoxide-Induced ferroptosis. Cell. 172 (3), 409–422 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Cao, J. Y. et al. A genome-wide haploid genetic screen identifies regulators of glutathione abundance and ferroptosis sensitivity. Cell. Rep. 26 (6), 1544–1556 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, X. et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 63 (1), 173–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.