Abstract
Migratory cells exist in the heart, such as immune cells, fibroblasts, endothelial cells, etc. During myocardium injury, such as ischemia–reperfusion (MIRI), cells migrate to the site of injury to perform repair functions. However, excessive aggregation of these cells may exacerbate damage to the structure and function of the heart, such as acute myocarditis and myocardial fibrosis. Myocardial injury releases exosomes, which are a type of vesicle with signal transduction function and the miRNA carried by exosomes can control cell migration function. Therefore, regulating this migratory cell population through cardiac-derived exosomal miRNA is crucial for protecting and maintaining cardiac function. Through whole transcriptome RNA sequencing, exosomal miRNA sequencing and single-cell dataset analysis, we (1) determined the potential molecular regulatory role of the lncRNA‒miRNA‒mRNA axis in MIRI, (2) screened four important exosomal miRNAs that could be released by cardiac tissue, and (3) screened seven genes related to cell locomotion that are regulated by four miRNAs, among which Tradd and Ephb6 may be specific for promoting migration of different cells of myocardial tissue in myocardial infarct. We generated a core miRNA‒mRNA network based on the functions of the target genes, which may be not only a target for cardiac repair but also a potential diagnostic marker for interactions between the heart and other tissues or organs. In conclusion, we elucidated the potential mechanism of MIRI in cardiac remodeling from the perspective of cell migration, and inhibition of cellular overmigration based on this network may provide new therapeutic targets for MIRI and to prevent MIRI from developing into other diseases.
Keywords: Myocardial ischemia‒reperfusion injury, Exosomal miRNA, Cell migration, Locomotion
Subject terms: Molecular biology, Physiology, Cardiology, Diseases, Pathogenesis, Risk factors
Introduction
Myocardial ischemia‒reperfusion injury (MIRI) refers to the occurrence of severe damage to the original ischemic myocardium after a short interruption in blood supply to the heart muscle and the restoration of blood supply within a certain period1. The mechanism by which MIRI occurs is complex and involves inflammatory reactions2, oxidative stress3, autophagy4, ferroptosis5 and pyroptosis6. MIRI is also associated with a variety of cardiac diseases, including heart failure, myocardial infarction, myocarditis, and myocardial fibrosis, and may be involved in the development of several cardiovascular diseases7–10. MIRI has become a major problem that affects the efficacy of cardiovascular surgery and is a bottleneck in clinical treatment. At present, there is a lack of effective prevention and treatment methods in clinical practice. Therefore, clarifying the pathogenesis of MIRI has become a difficult and hot topic in basic and clinical research to prevent and treat this disease.
Cell migration is the movement of living cells and is involved in processes such as angiogenesis11 and inflammatory responses12. After MIRI occurs, immune cells and fibroblasts are recruited and interact with other cardiac cells13,14, while endothelial cells migrate and proliferate to form a vascular network, forming granulation tissue and promoting cardiac repair15. This process aims to remove necrotic cells and promote healing. However, excessive cell migration and infiltration may also lead to excessive inflammatory response, exacerbate tissue injury, and even induce a fibrotic response9. Exosomes secreted by heart cells are involved in the pathological processes of heart disease, including myocardial injury16, cardiac fibrosis and cardiomyocyte hypertrophy17, inflammatory reactions18, and cardiomyocyte death19. Exosomes are released from the heart during MIRI as intercellular communication and may be involved in regulating cell migration. To further identify the important regulators of this process, we performed myocardial whole transcriptome analysis, circulating exosomal miRNA-seq analysis, and single-cell dataset analysis to identify possible cardiac-derived exosomes, which, in combination with cellular migratory functions, provide new insights into the mechanisms of MIRI and new therapeutic targets and strategies for MIRI-related cardiovascular diseases.
Materials and methods
Animals
Adult male SD rats (SPF grade, weighing 200–250 g) were provided by Chengdu Dashuo Experimental Animal Co., Ltd. (license number: SCXK Chuan-2020–030). The feeding environment consisted of 12 h of alternating light and dark, a temperature range of 16 °C to 28 °C, and a humidity range of 40% to 70%. Prior to the experiment, adaptive feeding took place for one week, and regular feeding of normal food and free access to water continued. The surroundings were familiar, the condition was good, and the activity was normal.
I/R model
By ligating the coronary arteries, the cardiac ischemia–reperfusion rat model was established. The anterior descending branch of the heart was exposed after a 2-cm incision was created in the third or fourth intercostal space. The incision was then closed and ligated with sutures. The living knot of the suture was opened to allow full reperfusion after the ventilator provided oxygen continuously for 30 min. The anterior descending branch of the coronary artery was threaded only without ligation in the sham-operated group, who were subjected to the same treatment as described above. The rats were sacrificed 7 days after reperfusion for testing.
TTC staining of the myocardial infarction area
The heart was removed and divided into 1 mm-thick cross-sections to measure infarct size. Areas at risk and infarct areas were identified using Evans blue/TTC double staining. ImageJ software v1.53 (https://imagej.en.softonic.com/mac) was used to analyse the Evans blue-stained area (blue), TTC-stained area (red; area at risk), and TTC-negative area (pale; infarct myocardium). The chest was directly closed and stitched, with the coronary artery left unstitched.
Hematoxylin–eosin (H&E) & masson staining
The rats were sacrificed, and the hearts were removed. In each group, six hearts were chosen for histological examination. By making a transverse slice between the apex and the atrioventricular sulcus, the heart was divided in half. The tissue was sliced into 5 µm-thick cross-sections, immersed in paraffin, and preserved in 4% paraformaldehyde. H&E staining was used to view the morphology of the cross-sections. Collagen deposition, which is depicted in blue in the cross-section of the heart, was identified using Masson’s trichrome staining.
Enzyme-linked immunosorbent assay (ELISA)
According to the instructions of the reagent kit (Wuhan Yilairuite Biotechnology Co., Ltd.), ELISA was used to determine the concentrations of lactate dehydrogenase (LDH), creatine kinase (CK), and CK-MB in rat serum.
Whole-transcriptome sequencing and RNA library construction
Personalbio (Shanghai, China) performed high-throughput sequencing analysis of rat myocardial tissues in the sham-operated and model groups to detect lncRNA, mRNA, and miRNA expression. Briefly, total RNA was first extracted by a TRIzol kit (Invitrogen Inc.), and the extraction efficiency was determined by agarose gel electrophoresis and a Nanodrop spectrophotometer. An Illumina HiSeq platform was used for RNA sequencing (RNA-Seq), and 2*150 bp paired-end reads were produced in accordance with the manufacturer’s protocol (Illumina, Inc.). The samples that contained miRNA were treated to extract total RNA using a serum miRNA purification kit (Norgen 57,600). Shanghai Personal Biotechnology Cp. Ltd. sequenced the small RNA libraries on the Illumina NovaSeq 6000 platform after they were prepared using the NEBNext Multiplex Small RNA Library Prep Set (New England Biolabs, Inc.) in compliance with the manufacturer’s instructions. Statistical thresholds of P-value < 0.05 and |log2FoldChange|> 1 were used to determine DEGs (lncRNAs and mRNAs). DEG miRNAs were identified using the statistical parameters |log2FoldChange|> 0.58 and P-value < 0.05. The fold change cutoff for dysregulated genes was calculated using a permutation-based test, as previously done20.
Isolation and characterization of exosomes
We used the Keygenbio Exosome Extraction Kit (product no. KGPE003, JiangSu, China) to isolate exosomes from rat serum using gradient centrifugation and then purified the exosomes. We visualized the exosomes using a transmission electron microscope (HT-7700, Hitachi, Japan) and analyzed the size and concentration range of the exosomes with a particle size analyzer (N30E, NanoFCM, China), as described previously. We also determined the expression of the exosome surface proteins CD9 (555,371, BD, USA) and CD81 (551,108, BD, USA).
Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) analysis
The differentially expressed genes were analyzed by GO and KEGG21–23 analyses. The gene lists were submitted to DAVID v6.8 (http://david.abcc.ncifcrf.gov/), and GO enrichment and KEGG pathway analysis were performed. A P-value < 0.05 was used to determine whether the enrichment analysis was statistically significant.
Construction of the lncRNA‒miRNA‒mRNA network
To predict the lncRNA‒miRNA and mRNA‒miRNA relationships, the target gene prediction tool Miranda was used. Then, Pearson correlation analysis was used to determine the correlation between lncRNA and mRNA expression. Finally, hypergeometric tests were performed based on the shared miRNAs of lncRNAs and mRNAs to determine whether lncRNAs can act as ceRNAs. Then, the association between the lncRNAs, miRNAs, and mRNAs was graded for regulatory similarity and sensitivity. The lncRNA‒miRNA‒mRNA ceRNA regulatory network was created using Cytoscape v3.9.1 (https://cytoscape.org) for lncRNAs, miRNAs, and mRNAs with ceRNA interactions (screening results with sensitivity correlations > 0.3).
Protein‒protein interaction (PPI) network
We constructed PPI networks to identify interacting genes/proteins using the String database v11.0 (https://cn.string-db.org) and used Cytoscape v3.9.1 (https://cytoscape.org) for network graph visualization.
Western blotting (WB) & RT‒qPCR
The WB and RT-qPCR assays using samples independent of sequencing. The exosome-associated protein markers and 7 screened proteins were analyzed by WB. In brief, the protein homogenates were then transferred to PVDF membranes (Millipore) and divided into equal portions using SDS‒PAGE. The membranes were blocked in 5% nonfat milk and then incubated with primary antibodies for an entire night at 4 °C. After that, the membranes were incubated at room temperature for two hours with secondary antibodies. Finally, Pierce ECL Western blotting substrate was used to view the protein bands.
We extracted total RNA using a kit (Tiangen). Guangzhou RiboBio designed the Bulge-loop™ miRNA RT Primer and Forward Primer (one RT primer per group corresponds to one pair of qPCR primers) specifically for let-7i-5p, miR-149-5p, miR-29b-3p, miR-7a-5p and U6. After reverse transcription synthesis of cDNA, relative miRNA expression levels were normalized to U6.
Single-cell dataset analysis
We used the CELLxGENE database (https://cellxgene.cziscience.com/) to obtain the single-cell dataset of Kuppe et al.24 (the only single-cell sequencing dataset of myocardial tissues as of October 24, 2023) and analyzed the dataset with the gene expression function.
Statistical analysis
We used ANOVA or t tests as described in the results. A Tukey post hoc test was performed to compare means when ANOVA was used. A two-sided P-value < 0.05 was considered significant.
Ethics approval and consent to participate
The Chengdu University of Traditional Chinese Medicine’s Animal Welfare and Ethics Committee approved the experimental methods, which followed the ARRIVE guidelines (PLoS Bio 8 (6), e10004122010) set forth by the International Council for Experimental Animal Science. At the end of the experiment, the rats were sacrificed with high concentrations of carbon dioxide. All methods were carried out in accordance with relevant guideline and regulations.
Results
Phenotypic analysis of MIRI
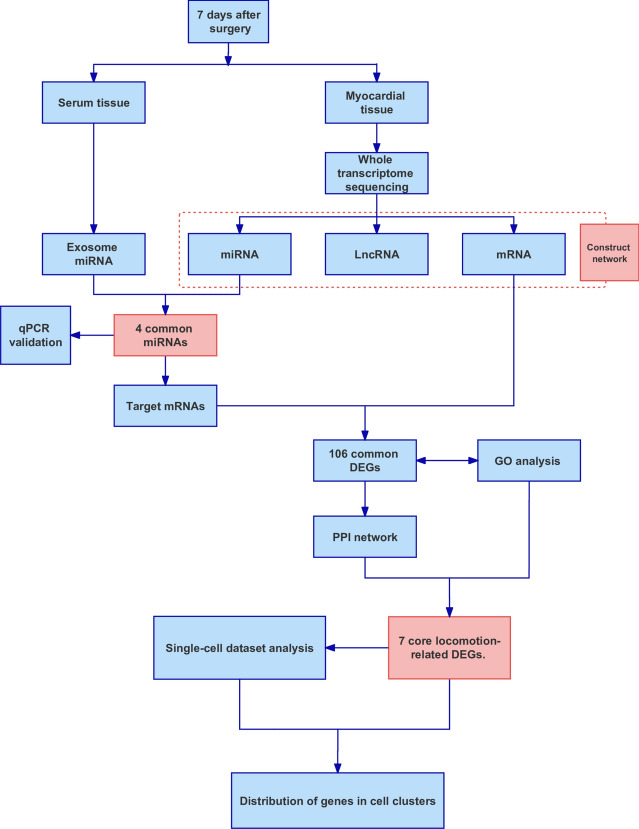
Figure 1 shows the flowchart of the experimental design. After successful modeling, whole transcriptome sequencing and exosomal miRNA sequencing were conducted, and the sequencing data were first analyzed individually. Then, myocardial tissue miRNAs and serum exosomal miRNAs were jointly analyzed, key intersecting miRNAs were screened out, and the functions of the target genes of the miRNAs were further analyzed. In addition, we performed RT‒qPCR validation of the screened key miRNAs and analyzed the single-cell dataset to further explore the function of the 7 mRNAs at the cellular level.
Fig. 1.
Flowchart of the experimental design.
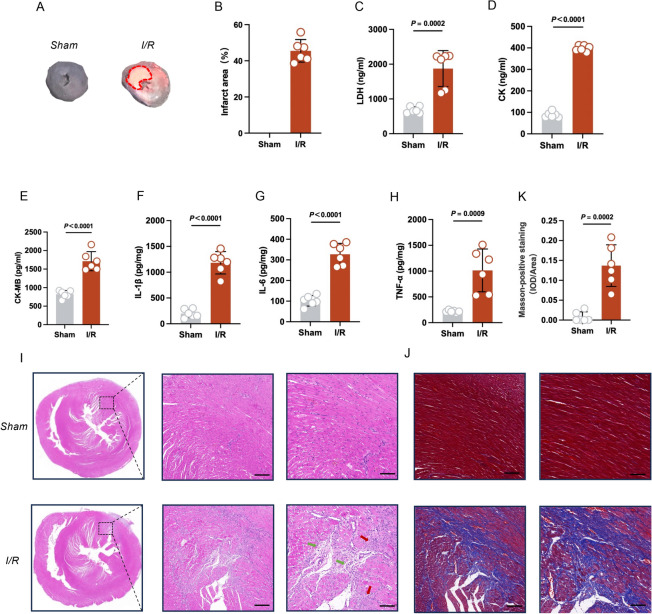
After 7 days of reperfusion, the infarct area was measured by TTC staining. There was a significant difference in infarct size between sham and I/R rats (Fig. 2A,B). Serum myocardial-specific enzyme levels were measured after 7 days of reperfusion, including LDH, CK, and CK-Mb. After I/R, LDH, CK, and CK-Mb levels all markedly increased (Fig. 2C–E). Inflammatory factors, including IL-1β, IL-6, and TNF-α, were significantly elevated in myocardial tissue (Fig. 2F–H), suggesting significant myocardial injury and inflammatory infiltration. H&E staining suggested the granulation tissue contained large amounts of neovascularization and inflammatory cells infiltration in the I/R group (Fig. 2I). Masson staining showed that the collagen fiber content was significantly higher in the I/R group than the sham group, suggesting significant fibrosis in the I/R group (Fig. 2J,K).Overall, myocardial ischemic rats showed significant damage to myocardial tissue after 7 days of reperfusion, with a large amount of neovascularization and inflammatory cell infiltration in the granulation tissue, and myocardial fibrosis.
Fig. 2.
Phenotypic analysis of MIRI (n = 6 per group). (A) TTC staining. The red dotted line is the area of infarction. (B) The quantitative results of TTC staining. (C–E) Serum levels of lactate dehydrogenase (LDH), creatine kinase (CK), and CK-MB. (F–H) Tissue levels of IL-1β, IL-6, and TNF-α. (I) H&E staining of myocardial tissue. Green arrows indicate neovascularization, and yellow arrows indicate inflammatory infiltration. Scale bar: 200 μm (left); Scale bar: 100 μm (right). (J) Masson’s trichrome of myocardial tissue. Scale bar: 200 μm (left); Scale bar: 100 μm (right). (K) The quantitative results of Masson staining.
Isolation, identification, and sequencing analysis of serum exosomes
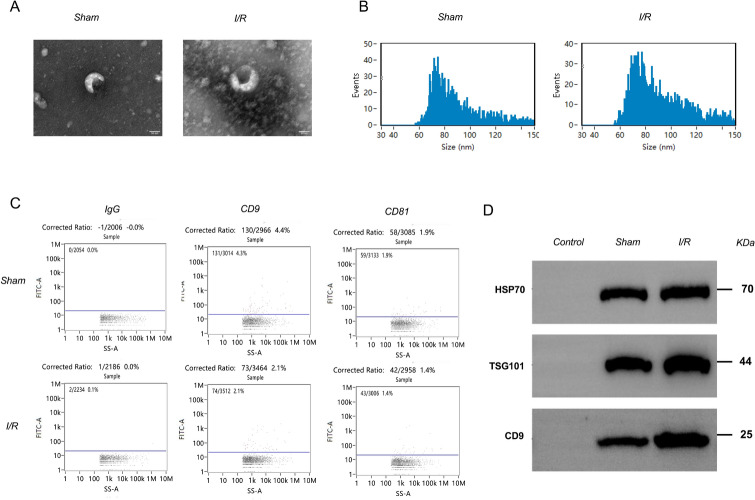
Extraction, isolation, identification, and sequencing analysis have shown that serum exosomes can originate from stem cells, macrophages, and serum. To investigate the effect of exosomes on myocardial injury induced by I/R, exosomes were isolated from the serum of rats in the sham surgery group and I/R group. Transmission electron microscopy showed that exosomes exhibited a membranous structure (Fig. 3A). The particle size of exosomes was analyzed by NTA. Each sample was examined three times, and the results represent the average of the three NTA analyses. The exosome diameter was approximately 100 nm, with an average of 89.57 nm in the sham surgery group and 90.13 nm in the I/R group (Fig. 3B). Fluorescence analysis (Fig. 3C) and Western blotting (Fig. 3D) showed that exosomes in the sham-operated group and I/R group expressed CD9, TSG101, and HSP70. In summary, these results indicate that exosomes were successfully isolated.
Fig. 3.
Identification of exosomes in the sham surgery group and model group by (A) transmission electron microscopy, (B) particle size analysis, (C) fluorescence analysis of the exosome markers CD9 and CD81 and IgG control, and (D) western blot analysis of the exosome markers HSP70, TSG101, and CD9 in the sham and I/R groups. The blots cut prior to hybridization with antibodies and the original blots/gels were presented in supplementary Fig. 1. (n = 3 per group).
Differential expression of serum exosomal miRNAs in MIRI
Subsequently, serum exosomal miRNAs were sequenced, and hierarchical clustering analysis revealed significant changes in miRNA expression in the I/R group compared to the sham group. A total of 51 miRNAs were differentially expressed (Fig. 4A,B): 41.2% were upregulated, while 58.8% were downregulated. The top five upregulated and downregulated exosomal miRNAs are listed in Table 1. The top twenty molecular functions related to the differentially expressed genes were examined by GO pathway analysis and included “positive regulation of cellular process” and “positive regulation of biological process”. (Fig. 4C). Moreover, KEGG analysis of differentially expressed exosomal miRNAs is shown in Fig. 4D. The complete results of DEG miRNA, GO and KEGG analysis were listed in Table S1.
Fig. 4.
Analysis of differentially expressed miRNAs in the serum exosomal (n = 3 per group). (A) Volcano plot of DEG miRNAs. (B) Heatmap of DEG miRNAs. (C) GO analysis of DEG miRNAs. (D) KEGG analysis of DEG miRNAs. Count: the number of enriched genes; rich: the ratio of the number of differential genes under the metabolic pathway to the number of all genes annotated to the pathway, with larger values indicating greater enrichment.
Table 1.
Top 5 significantly up- and downregulated exosomal miRNAs.
| Gene ID | Log2 fold change (I-R/sham) | P-value | Q-value |
|---|---|---|---|
| rno-miR-26a-5p | 1.25 | 0.0001 | 0.0104 |
| rno-let-7c-5p | 1.29 | 0.0002 | 0.0129 |
| rno-let-7a-5p | 1.15 | 0.0002 | 0.0129 |
| rno-miR-186-5p | 1.50 | 0.0002 | 0.0129 |
| rno-let-7f.-5p | 1.01 | 0.0007 | 0.0265 |
| rno-miR-199a-3p | −1.93 | < 0.0001 | 0.0002 |
| rno-miR-22-3p | −1.19 | 0.0003 | 0.0129 |
| rno-miR-10b-5p | −1.21 | 0.0003 | 0.0129 |
| rno-miR-27b-3p | −1.06 | 0.0022 | 0.0529 |
| rno-miR-146a-5p | −1.04 | 0.0023 | 0.0529 |
A adjusted p-value was represented as q-value.
Differential expression of miRNAs,lncRNAs and mRNAs in MIRI
Using RNA-seq, hierarchical clustering analysis revealed that compared to the Sham group, there was a significant change in cardiac expression of miRNAs in the myocardial I/R group. A total of 44 miRNAs were differentially expressed (Figure S2A,B): 54.5% were upregulated. while 45.5% were downregulated. The top five upregulated and downregulated miRNAs are listed in Table S2. GO and pathway analyses were performed (Figure S2C,D).
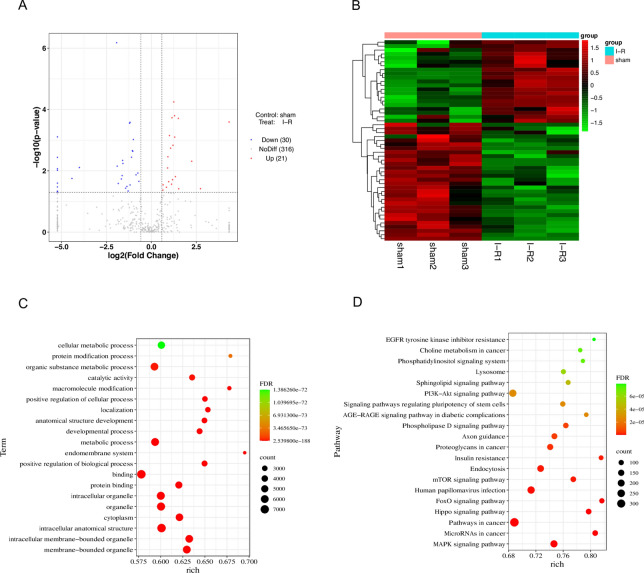
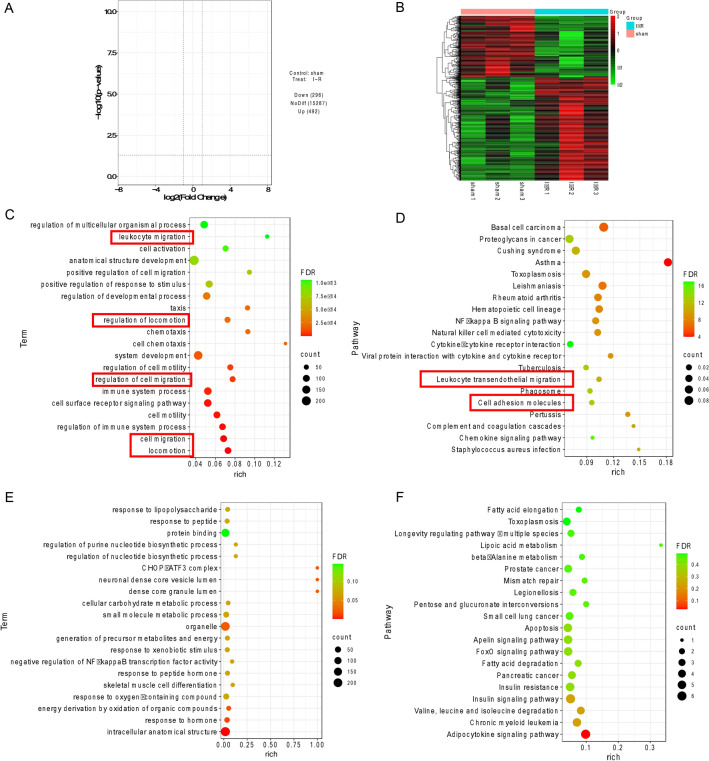
Differentially expressed mRNAs were identified in the myocardial I/R group compared to the sham group (|log2FoldChange|> 1 and P-value < 0.05). Among them, 62.4% of mRNAs were upregulated, and 37.6% of mRNAs were downregulated (Fig. 5A,B). In addition, lncRNAs were upregulated (33.9%) and downregulated (66.1%) (Figure S3A). The top five upregulated and downregulated mRNAs are listed in Table 2. GO analysis showed that the molecular functions associated with the differentially expressed up-mRNAs included “leukocyte migration”, “regulation of locomotion”, “regulation of cell migration”, “locomotion” and “cell migration” (Fig. 5C). The KEGG pathways also showed the functions of these mRNAs mostly involved “leukocyte transendothelial migration” and “cell adhesion molecules” (Fig. 5D). The functions in red boxes are related to cell migration and locomotion. And Fig. 5E showed the GO analysis of the down-DEG mRNAs. Figure S3C shows the functions of the differentially expressed lncRNAs, and the KEGG pathways of the differentially expressed down-mRNAs and lncRNAs are shown in Figs. 5F and S3D. The complete results of DEG mRNA, GO and KEGG analysis were listed in Table S1.
Fig. 5.
Analysis of differentially expressed mRNAs in myocardial tissue (n = 3 per group). (A) Volcano plot of DEG mRNAs. (B) Heatmaps of DEG mRNAs.(C) GO analysis of up-DEG mRNAs. (D) KEGG analysis of up-DEG mRNAs. (E) GO analysis of down-DEG mRNAs. (F) KEGG analysis of down-DEG mRNAs. Count: the number of enriched genes; rich: the ratio of the number of differential genes under the metabolic pathway to the number of all genes annotated to the pathway, with larger values indicating greater enrichment.
Table 2.
Top 5 significantly up- and downregulated mRNAs.
| Ensembl ID | Gene symbol | Log2 fold change (I-R/sham) | P-value | Q-value |
|---|---|---|---|---|
| ENSRNOG00000031801 | Ephb3 | 2.75 | < 0.0001 | 0.0004 |
| ENSRNOG00000028896 | A2m | 3.46 | < 0.0001 | 0.0017 |
| ENSRNOG00000064521 | Hist2h3c2 | 1.94 | < 0.0001 | 0.0065 |
| ENSRNOG00000007907 | Tmem178a | 2.70 | < 0.0001 | 0.0067 |
| ENSRNOG00000009263 | Ifi27 | 1.43 | < 0.0001 | 0.0077 |
| ENSRNOG00000003537 | Spta1 | − 2.24 | < 0.0001 | < 0.0001 |
| ENSRNOG00000007999 | Abra | −2.61 | < 0.0001 | < 0.0001 |
| ENSRNOG00000019719 | Kcna5 | −2.00 | < 0.0001 | 0.0002 |
| ENSRNOG00000033261 | Fam107a | −1.91 | < 0.0001 | 0.0009 |
| ENSRNOG00000003300 | Btg2 | −1.55 | < 0.0001 | 0.0022 |
A adjusted p-value was represented as q-value.
A network diagram was constructed using Cytoscape to display the lncRNA‒miRNA‒mRNA ceRNA regulatory network, which included 18 lncRNAs, 39 miRNAs, and 36 mRNAs (Figure S4A). We found that rno-miR-30c-1-3p could sponge the lncRNAs MSTRG.577221.1, MSTRG.87625.2, MSTRG.495.2, and MSTRG.51713.3 to regulate mRNAs (e.g., Nfam1, Sdc1, and P2ry6) (Figure S4C).
Integrative analysis of the serum exosomal miRNAs and myocardial miRNAs
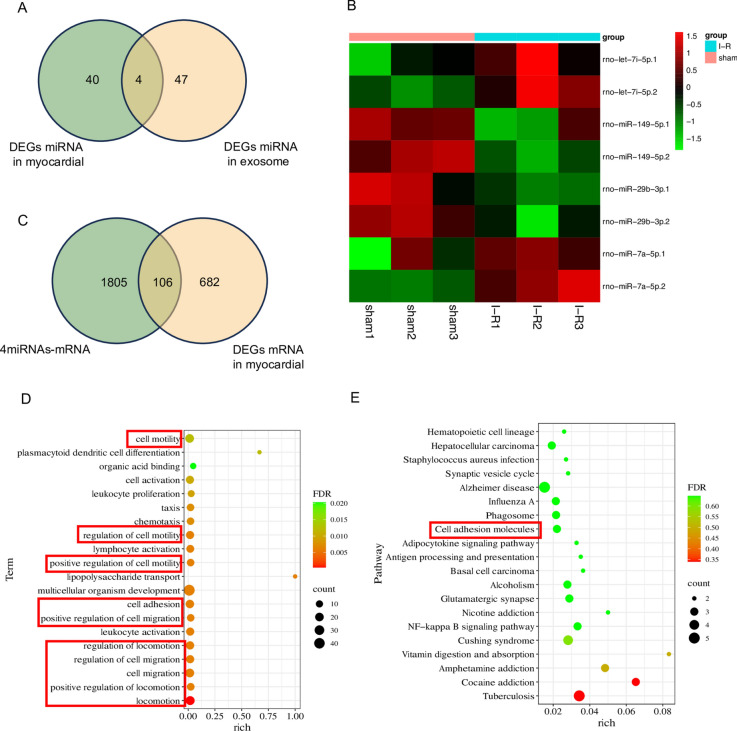
After sequencing myocardial miRNAs and serum exosomal miRNAs, we screened miRNAs that were differentially expressed in both samples. Four miRNAs intersected in the two samples (Fig. 6A): rno-let-7i-5p, rno-miR-149-5p, rno-miR-29b-3p, and rno-miR-7a-5p. Interestingly, we found that the changes in the expression of these four miRNAs were consistent in myocardial tissue and serum (Fig. 6B). This finding indicates that there may be a strong correlation between these four miRNAs in serum exosomes and cardiac tissues. However, using these four miRNAs, we also constructed IncRNA-miRNA‒mRNA networks (Figure S5) based only on the binding relationship between them.
Fig. 6.
Integrative analysis of serum exosomal miRNAs and myocardial miRNAs (n = 3 per group). (A) Venn plot of intersecting miRNAs in myocardial tissue and exosomes. (B) Heatmap of intersecting miRNAs in myocardial tissue and exosomes; the number “1” behind the gene symbol of miRNA represents myocardial tissue, and “2” represents serum. (C) Venn plot of the intersection of the target mRNAs of miRNAs and differentially expressed mRNAs in myocardial tissue. (D) GO analysis of intersecting mRNAs. (E) KEGG analysis of intersecting mRNAs. Count: the number of enriched genes; rich: the ratio of the number of differential genes under the metabolic pathway to the number of all genes annotated to the pathway, with larger values indicating greater enrichment.
In addition, we tried to intersect the target genes (screening based on miRNA expression, e.g. if miRNA expression is up-regulated in the model, the target gene expression is down-regulated) of these 4 intersecting miRNAs with the previously identified differentially expressed mRNAs in myocardial tissue and found 106 common mRNAs (Fig. 6C). Similarly, we tried to intersect lncRNAs; however, due to the relatively limited annotation of rat lncRNA genes in the database, we did not analyze this factor, and we provide the IDs of the intersecting lncRNAs in the Supplementary material (Figure S6 and table S3). Further GO and KEGG analysis of common mRNAs showed that the functions of these mRNAs mostly involved aspects of cell migration and locomotion (Fig. 6D,E). The functions in red boxes are related to cell migration and locomotion. The complete results of GO and KEGG analysis were listed in Table S1.
The functional analysis of target genes by intersecting 4 miRNAs
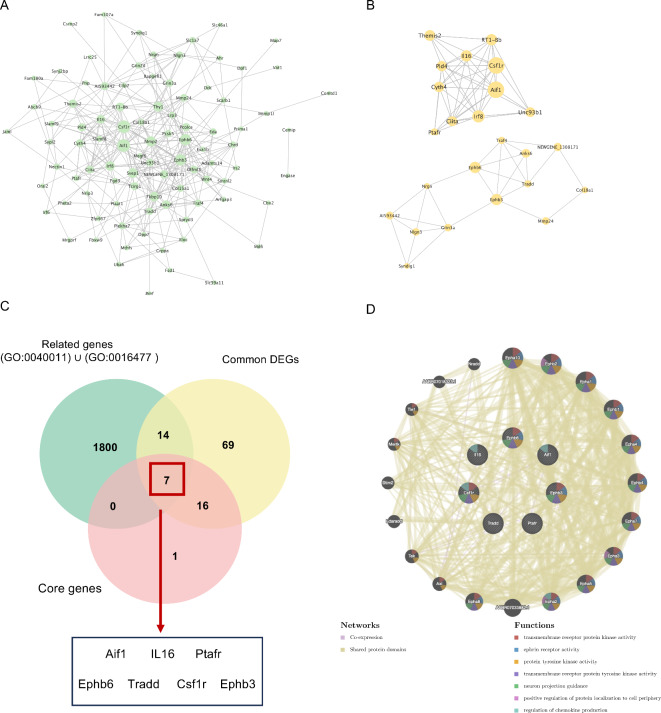
To determine the interactions among common genes, we performed PPI network construction on the 106 common mRNAs (Fig. 7A), and according to the MCODE plugin, we obtained two core protein clusters (Fig. 7B). Then, we extracted all the genes related to locomotion (GO: 0040011) and cell migration (GO:0016477) from the GO database using R software v4.3.1(https://cloud.r-project.org), and all the gene symbols are provided in Table S4. We intersected the extracted genes with common DEGs and core protein clusters, and finally, we identified 7 genes (Fig. 7C). In the PPI network diagram, we showed the interaction of these seven genes with other proteins and their main functions (Fig. 7D).
Fig. 7.
The analysis of target genes by intersecting 4 miRNAs. (A) PPI network interaction diagram of the intersecting mRNAs. (B) Two core protein groups in the PPI network. (C) Related genes, common DEGs and core protein groups in the Venn diagram of intersections. (D) PPI network interaction diagram of the 7 screened proteins.
Migratory function of miRNA‒mRNA analysis by single-cell sequencing data
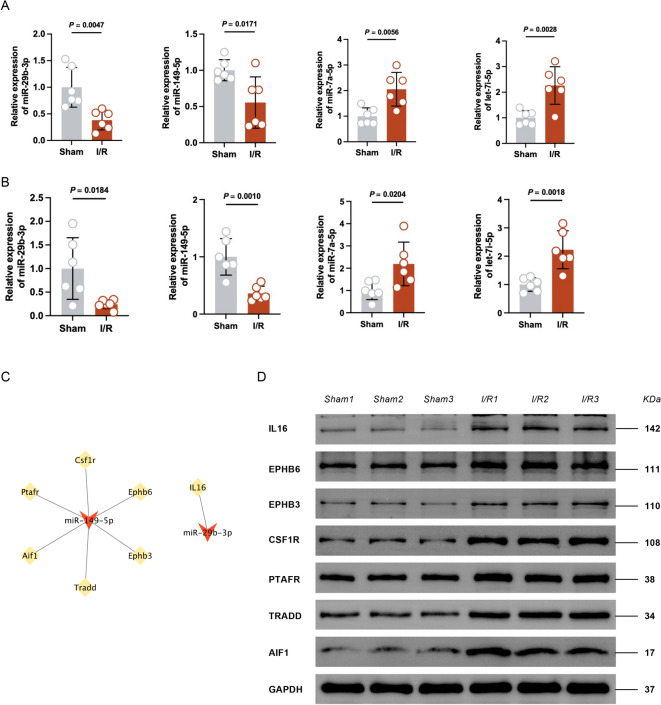
We next verified the expression of these four miRNAs by qPCR. Compared with those of the sham group, the levels of miR-29b-3p and miR-149-5p in myocardial tissues (Fig. 8A) and plasma exosomes (Fig. 8B) in the I/R group were significantly lower, and the levels of let-7i-5p and miR-7a-5p were significantly higher, which was consistent with our sequencing results.
Fig. 8.
Validating the potential function of miRNA‒mRNA. (A) The relative expression of myocardial intersecting miRNAs by RT‒qPCR. n = 6 per group. (B) The relative expression of serum exosomal intersecting miRNAs by RT‒qPCR. n = 6 per group. (C) The miRNA‒mRNA ceRNA regulatory network. (D) western blot analysis of the 7 screened proteins in the sham and I/R groups. The blots cut prior to hybridization with antibodies and the original blots/gels were presented in supplementary Fig. 7. (n = 3 per group).
Then, we tried to construct a miRNA-mRNA network (Fig. 8C), and the expression of both miR-149-5p and miR-29b-3p decreased after modeling. We verified the expression of the seven target genes by western-blot, and the expression of all seven genes was elevated in the I/R group compared with the sham group, consistent with the sequencing results (Fig. 8D).
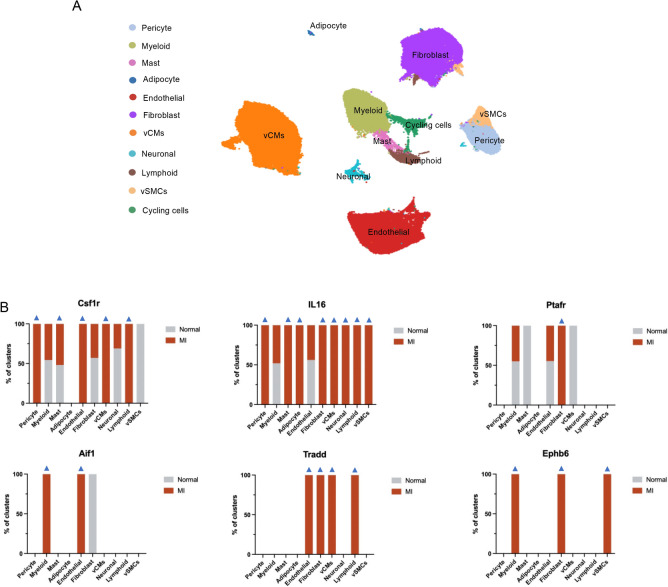
Finally, to explore the migratory function of these 7 genes at the cellular level, we obtained the single-cell dataset of Kuppe et al24 from the CELLxGENE database and analyzed it with CELLxGENE. These samples were derived from the left ventricles of 23 individuals and included 4 nontransplanted donor hearts as controls in addition to samples from patients with acute myocardial infarction (MI), and the information about these patients was detailed in Table S5. We referenced the original article24 to annotate the cells (Fig. 9A), which contained a total of 11 cell types. We then analyzed the expression of the above 7 genes in different cell clusters (Fig. 9B), except for the cycling cells, which may mix multiple cell subpopulations. We found that Csf1r expression was increased in pericytes, mast cells, endothelial cells, ventricular cardiomyocytes (vCMs), and lymphoid cells in the MI group compared to the normal group; IL16 expression was increased except in both myeloid and endothelial cells; Ptafr was highly expressed in fibroblasts; and Aif1 was highly expressed in myeloid and endothelial cells. Notably, Tradd and Ephb6 were barely expressed in the normal group, but Tradd was highly expressed in endothelial cells, fibroblasts, vCMs, and lymphoid cells in the MI group; Ephb6 was highly expressed in myeloid cells, fibroblasts and vascular smooth muscle cells (vSMCs). Overall, based on analysis of a single-cell database, these seven genes regulated by 4 miRNAs were significantly expressed in clusters of cells with migratory functions.
Fig. 9.
Analyzing the potential function of miRNA‒mRNA by single-cell sequencing data. (A) UMAP of snRNA-seq data (n = 191,795). vCMs, ventricular cardiomyocytes. vSMCs, vascular smooth muscle cells. (B) The relative expression of the 6 screened proteins in different cell clusters by snRNA-seq analysis in the normal and MI groups. Blue arrows indicate the gene changing trends in various clusters of cells consistent with gene changes in the RNA-sequencing results of the MI group.
Discussion
In this study, we conducted sequencing analysis of serum exosomal miRNAs, and full transcriptome sequencing of myocardial tissue, including miRNA, mRNA, and lncRNA sequencing. We found 4 intersecting miRNAs by cross-tabulation analysis of differentially expressed miRNAs and exosomal miRNAs in myocardial tissue, and interestingly, the changes in expression were consistent in myocardial tissues and serum. We hypothesized that these four miRNAs have important regulatory functions during MIRI and might be exosomal miRNAs released by the heart. Then, we analyzed the target genes of these 4 miRNAs and found 7 target genes that were functionally related to cell migration. Among the 7 target genes, our analysis of the single-cell dataset showed that 6 target genes were highly expressed in different cell populations in the MI group compared to the normal group. In particular, some cell clusters with high gene expression, such as endothelial cells, myeloid, and fibroblasts, exhibited significant migratory functions. Based on these findings, we surmised that a miRNA-mRNA regulatory network of regulation of cell migration function exists within cardiac tissues when MIRI occurs. In conclusion, we elucidated the potential mechanism of MIRI in cardiac remodeling from the perspective of cell migration, and inhibition of cellular overmigration based on this network may provide new therapeutic targets for MIRI and to prevent MIRI from developing into other cardiovascular diseases.
The 4 miRNAs we identified play important roles in the heart, and previous studies have shown that they not only affect disease progression in MIRI, but also other cardiovascular diseases, playing an important role in myocardial inflammation and fibrosis. A previous study showed that miR-29b-3p is a novel regulator of cardiac development that inhibits cardiomyocyte proliferation through NOTCH2 and causes cardiac dysfunction25, and it is also closely related to fibrosis, with its expression level inversely correlating with plaque fibrosis26. MiR-7a-5p is involved in cardiomyocyte injury induced during myocardial ischemia/reperfusion, and the downregulation of miR-7a-5p reduces cardiomyocyte apoptosis27,28. The lncRNA HFRL/miR-149-5p axis plays an important role in regulating cardiac inflammation, proliferation, and fibrosis through synergistic effects29. Notably, ptafr30 and ephb331 are involved in the regulation of myocardial fibrosis and their high expression can exacerbate fibrosis. Besides, our sequencing results revealed that they are highly expressed during MIRI and have a targeting relationship with miR-149-5p. Let-7i-5p may be a hypoxia-induced exosomal miRNA with potential antiapoptotic effects32.
Processes such as angiogenesis, atherosclerosis, and cancer metastasis all involve cell migration. Endothelial cell migration promotes new blood vessel formation or angiogenesis33; fibroblast migration promotes the repair of damaged tissue34; and mast cells not only promote endothelial cell migration35, but their migration may affect the movement of tumor-derived peptides36. Our study showed that the 4 exosomal miRNAs are important signals for cell locomotion and may promote the migration of different cells in heart muscle tissue by targeting seven key genes. Tradd and Ephb6 may be specific genes that promote cell locomotion during MI. In particular, Ephb6, which positively or negatively regulates cell adhesion and migration37, was highly expressed in myeloid cells, fibroblasts, and vSMs in our single-cell dataset analysis. Notably, aberrant release of exosomal miRNAs is involved in malignant tumorigenesis and plays an important role in the regulation of the tumor microenvironment38–40. The downregulation of miR-149-5p promotes hnRNPA1 expression, which is responsible for the loading of miR-let-7a-5p into exosomes, whereas the expression of exosomal miR-let-7a-5p is increased in the blood of breast cancer patients41. The upregulation of exosomal miR-149-5p exerts therapeutic effects by inhibiting the growth and migration of pituitary adenoma cells, as well as tube formation in endothelial cells42. Interestingly, Ephb6 could affect the formation of cancer stem-like cells, drug resistance and metastasis43, and its mutation could induce paclitaxel resistance in cancer patients and aggravate cancer44. Recent a study has shown that cardiac remodeling and heart failure can significantly accelerate tumor progression via exosomal miRNAs, suggesting that exosomal miRNAs released after myocardial injury or entering the circulation are involved in the tumor microcirculation to influence tumorigenesis45. Therefore, these four exosomal miRNAs that may be released by the heart and the 7 locomotion-related target genes identified in this study are likely to be not only targets for cardiac repair but also potential diagnostic markers for interactions between the heart and other tissues or organs.
However, Epbh3 was not detected in the nucleus because it was sequenced in the nucleus, and Epbh3 is likely to be expressed in the cytoplasm after post-transcriptional transport. Only two (miR-149-5p and miR-29b-3p) of the four miRNAs screened were involved in the construction of the miRNA-mRNA network, and the other two miRNAs (let-7i-5p and miR-7a-5p) were not included probably due to the database version. Secondly, due to the lack of annotation of rat lncRNA data, this study only provided gene IDs for transcriptome-wide differentially expressed lncRNAs and constructed a differentially expressed ceRNA network in myocardial tissue without detailed analysis. Finally, although the functional roles of four miRNAs and seven target proteins were explored in this study by enrichment analysis and single-cell dataset analysis, their targeting and function were not validated in this study, and further experiments are still needed to validate their targeting relationships as well as their ability to migrate cells in the future.
Conclusions
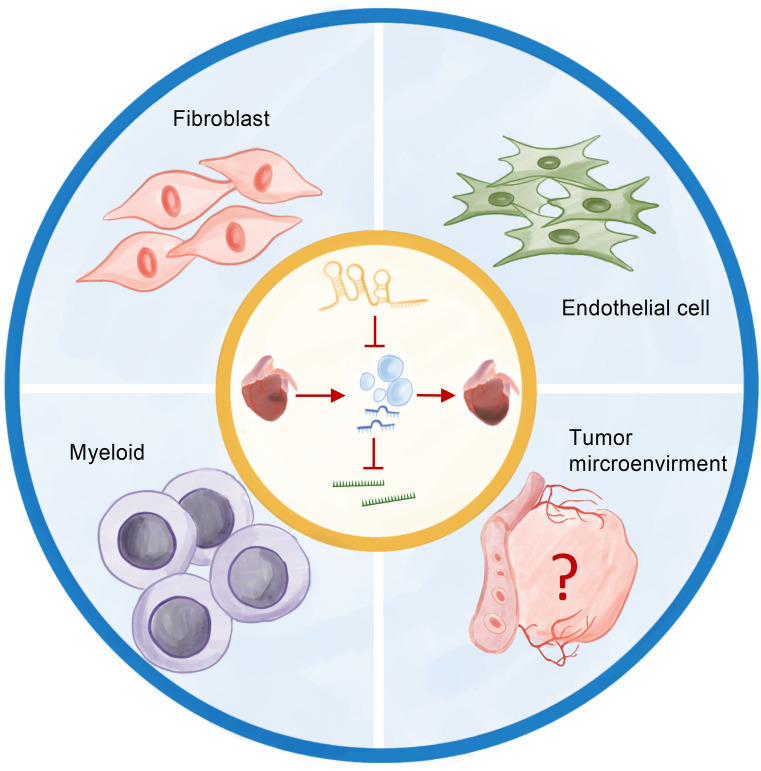
Exosomal miRNAs play an important role in disease progression and can stimulate angiogenesis and promote cancer metastasis.We have identified four key miRNAs that may be derived from exosomal of the heart. Our study also identified a miRNA‒mRNA network that promotes cell migration. Notably, this network may be not only a target for cardiac repair but also a potential diagnostic marker for interactions between the heart and other tissues or organs (Fig. 10). In conclusion, we elucidated the potential mechanism of MIRI in cardiac remodeling from the perspective of cell migration, and inhibition of cellular overmigration based on this network may provide new therapeutic targets for MIRI and to prevent MIRI from developing into other diseases.
Fig. 10.
Graphic summary of the function for potential cardiac-derived exosomal miRNAs involved in cardiac healing and remodeling after MIRI.
Supplementary Information
Author contributions
Y.L. and J.C.: Conceptualization, Formal analysis, Writing—original draft preparation. J.X., J.H., and L.Y.: Methodology, Data curation. Y.S., Y.W., Y.Z., and X.L.: Validation. Q.Z.: Supervision. W.Q.:Methodology, Writing—review and editing, Funding acquisition. F.L.: Writing—review and editing, Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (No. 82074556 and No. 82205286). The regional cooperation program of the National Natural Science Foundation of China (No. U21A20404). Natural Science Foundation of Sichuan Province (No. 2023NSFSC1819 and No. 2023NSFSC0697).
Data availability
The datasets generated and/or analysed during the current study have been uploaded to the GEO database (GSE253760), and go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253760 (enter token ozatioyqxzobbgx into the box) to review.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu Liu and Jiao Chen.
Contributor Information
Wen-Chuan Qi, Email: qiwenchuan710@126.com.
Fan-Rong Liang, Email: acuresearch@126.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75517-8.
References
- 1.Li, Q. et al. Indole-3-Carbinol (I3c) protects the heart from ischemia/reperfusion injury by inhibiting oxidative stress, inflammation, and cellular apoptosis in mice. Front. Pharmacol. 13, 924174 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu, J., Wang, H. & Li, J. Inflammation and inflammatory cells in myocardial infarction and reperfusion injury: a double-edged sword. Clin. Med. Insights Cardiol. 10, 79–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang, M. et al. Role of oxidative stress in reperfusion following myocardial ischemia and its treatments. Oxid. Med. Cell. Longev. 2021, 6614009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsui, Y. et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 100, 914–922 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Fang, X., Ardehali, H., Min, J. & Wang, F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat. Rev. Cardiol. 20, 7–23 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, S., Duanmu, X., Xu, L., Zhu, L. & Wu, Z. Ozone pretreatment alleviates ischemiareperfusion injury-induced myocardial ferroptosis by activating the Nrf2/Slc7a11/Gpx4 axis. Biomed. Pharmacother. 165, 115185 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Ovize, M., Thibault, H. & Przyklenk, K. Myocardial conditioning. Circ. Res. 113, 439–450 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Heusch, G. Cardioprotection: Chances and challenges of its translation to the clinic. Lancet 381, 166–175 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Frangogiannis, N. G. Regulation of the inflammatory response in cardiac repair. Circ. Res. 110, 159–173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferdinandy, P., Schulz, R. & Baxter, G. F. Interaction of cardiovascular risk factors with myocardial ischemia/reperfusion injury, preconditioning, and postconditioning. Pharmacol. Rev. 59, 418–458 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Shang, J., Gao, Z.-Y., Zhang, L.-Y. & Wang, C.-Y. Over-expression of JAZF1 promotes cardiac microvascular endothelial cell proliferation and angiogenesis via activation of the Akt signaling pathway in rats with myocardial ischemia-reperfusion. Cell. Cycle 18, 1619–1634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, Y. et al. Research on the biological mechanism and potential application of CEMIP. Front. Immunol. 14, 1222425 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Algoet, M. et al. Myocardial ischemia-reperfusion injury and the influence of inflammation. Trends Cardiovasc. Med. 33, 357–366 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Rurik, J. G., Aghajanian, H. & Epstein, J. A. Immune cells and immunotherapy for cardiac injury and repair. Circ. Res. 128, 1766–1779 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, C. et al. M2b macrophages stimulate lymphangiogenesis to reduce myocardial fibrosis after myocardial ischaemia/reperfusion injury. Pharm. Biol. 60, 384–393 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta, S. & Knowlton, A. A. HSP60 trafficking in adult cardiac myocytes: role of the exosomal pathway. Am. J. Physiol. Heart Circ. Physiol. 292, H3052-3056 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Bang, C. et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 124, 2136–2146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H. et al. Exosomes derived from dendritic cells improve cardiac function via activation of CD4(+) T lymphocytes after myocardial infarction. J. Mol. Cell. Cardiol. 91, 123–133 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y., Wang, M., Yu, Y., Li, C. & Zhang, C. Advances in the study of exosomes derived from mesenchymal stem cells and cardiac cells for the treatment of myocardial infarction. Cell. Commun. Signal 21, 202 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montero-Calle, A. et al. In-depth quantitative proteomics analysis revealed C1GALT1 depletion in ECC-1 cells mimics an aggressive endometrial cancer phenotype observed in cancer patients with low C1GALT1 expression. Cell Oncol. 46, 697–715 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, Q. et al. Aberrant expression of miR-29b-3p influences heart development and cardiomyocyte proliferation by targeting NOTCH2. Cell Prolif. 53, e12764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leistner, D. M. et al. Transcoronary gradients of vascular miRNAs and coronary atherosclerotic plaque characteristics. Eur. Heart J. 37, 1738–1749 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Liang, D. et al. Down-regulation of Xist and Mir-7a-5p improves LPS-induced myocardial injury. Int. J. Med. Sci. 17, 2570–2577 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, H. et al. Butyrate suppresses atherosclerotic inflammation by regulating macrophages and polarization via GPR43/HDAC-miRNAs axis in ApoE-/- mice. PLoS ONE 18, e0282685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, X. et al. Enhancement of LncRNA-HFRL expression induces cardiomyocyte inflammation, proliferation, and fibrosis via the sequestering of miR-149-5p-mediated collagen 22A inhibition. Ann. Transl. Med. 10, 523 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leisegang, M. S. LET’s sponge: How the lncRNA PFL promotes cardiac fibrosis. Theranostics 8, 874–877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, M. et al. Gene expression analysis to identify mechanisms underlying improvement of myocardial fibrosis by finerenone in SHR. Biochem. Pharmacol. 220, 115975 (2024). [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J. et al. Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 cells apoptosis. Int. J. Mol. Sci. 18, 711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glaser, B. M., D’Amore, P. A., Seppa, H., Seppa, S. & Schiffmann, E. Adult tissues contain chemoattractants for vascular endothelial cells. Nature 288, 483–484 (1980). [DOI] [PubMed] [Google Scholar]
- 34.Ross, R. & Odland, G. Human wound repair. II. Inflammatory cells, epithelial-mesenchymal interrelations, and fibrogenesis. J. Cell Biol.39, 152–168 (1968). [DOI] [PMC free article] [PubMed]
- 35.Azizkhan, R. G., Azizkhan, J. C., Zetter, B. R. & Folkman, J. Mast cell heparin stimulates migration of capillary endothelial cells in vitro. J. Exp. Med. 152, 931–944 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole, T. J. & Zetter, B. R. Stimulation of rat peritoneal mast cell migration by tumor-derived peptides. Cancer Res. 43, 5857–5861 (1983). [PubMed] [Google Scholar]
- 37.Matsuoka, H., Obama, H., Kelly, M. L., Matsui, T. & Nakamoto, M. Biphasic functions of the kinase-defective Ephb6 receptor in cell adhesion and migration. J. Biol. Chem. 280, 29355–29363 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Sun, Z. et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 17, 147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, J. et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genom. Proteom. Bioinform. 13, 17–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, F. et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol. Cancer 16, 148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drula, R. et al. 17β-estradiol promotes extracellular vesicle release and selective miRNA loading in ERα-positive breast cancer. Proc. Natl. Acad. Sci. U. S. A. 120, e2122053120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, P. et al. Up-regulation of the expressions of MiR-149-5p and MiR-99a-3p in exosome inhibits the progress of pituitary adenomas. Cell. Biol. Toxicol. 37, 633–651 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Wu, J.-E. et al. DNA methylation maintains the CLDN1-EPHB6-SLUG axis to enhance chemotherapeutic efficacy and inhibit lung cancer progression. Theranostics 10, 8903–8923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon, S. et al. EPHB6 mutation induces cell adhesion-mediated paclitaxel resistance via EPHA2 and CDH11 expression. Exp. Mol. Med. 51, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, Y. et al. Exosomes secreted from cardiomyocytes suppress the sensitivity of tumor ferroptosis in ischemic heart failure. Signal Trans. Target Ther. 8, 121 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study have been uploaded to the GEO database (GSE253760), and go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253760 (enter token ozatioyqxzobbgx into the box) to review.