Abstract
Bacteria at different growth stages usually coordinate capsular polysaccharide (CPS) formation and may affect their susceptibility to phage. In this study, we evaluated the infection efficacy of phage vB_VspM_VS2 in V. splendidus AJ01 at different growth stages and explored the role of growth stage-related CPS translocon Wza in the susceptibility of V. splendidus to phage vB_VspM_VS2. The results showed that V. splendidus locked in the stationary growth stage (SGS) or early exponential stage (EES) infected with phage (EES-P) has a low susceptibility to phage vB_VspM_VS and exhibit a pronounced reduction in phage adsorption rate as compared to the EES bacteria. The expression of wza of CPS transport gene was significantly increased in EES bacteria compared to that bacteria locked in the SGS or EES-P. Bacteria with deleted wza (Δwza mutant) escaped phage adsorption due to absence of Wza mediated down-regulation of CPS expression, otherwise. Our results reveal that the Wza of V. splendidus can promotes phage to infect these bacteria via increasing the phage absorption, which provides important implications for using phages therapeutically target pathogenic bacteria in dynamics communities.
Subject terms: Phage biology, Bacteriophages
Vibrio splendidus with deleted wza (Δwza mutant) escaped phage adsorption due to absence of Wza mediated down-regulation of CPS expression.
Introduction
Phages are viruses that just infect microorganism including gram-negative (G−) bacteria and gram-positive (G+) bacteria, and represent one of the most abundant microorganism, with an estimated number as high as 1031 in the nature1. They have high host-specificity and do not infect animals or humans, which making them as ideal antibacterial biological agents2. With the issue of antibiotic-resistant bacteria getting worse, there is an urgent need for phages and phage-derived products (e.g., endolysins) to replace antibiotic drugs3,4. The infection cycle of phages is initiated by adsorption to the bacteria receptor, followed by injection of nucleic acid into the bacteria, DNA replication, biosynthesis of structural protein, assembly, and release5. Notably, the survival of phages depends on their ability to infect susceptible bacteria, and the critical step for phage lysing host bacteria is the stage of phage adsorption. This stage involves the binding of phages to the receptors on bacteria surface, which determines the host-specificity of phage. For example, Elhanan et al. reported that Bacillus subtilis can acquire phage sensitivity by exchanging the bacteria membrane protein with phage adsorption receptors of membrane vesicles6.
The cell-surface polysaccharide of G− bacteria is mainly composed of lipopolysaccharide (LPS, O antigen) and capsular polysaccharide (CPS, K antigen), which are encoded by lps and cps gene clusters, respectively7. CPS is located on the surface of G− bacteria cells and forms a vesicular structure covering on the cell surface8. In G− bacteria, the outer membrane size-exclusion barrier must be bypassed for translocation of CPS to the cell surface. ATP-binding cassette (ABC) transporter-dependent and Wzy-dependent pathways with different polymer biosynthesis strategies are involved in the assembly of most CPS9. Wza, the lipoprotein that forms a stable octamer is responsible for CPS translocon that from the periplasm to the cell surface in G− bacteria10. Compared with wild-type cells, Matanza et al. reported that Δwza mutants can be acapsulated in Photobacterium damselae11. Furthermore, Hao et al., demonstrated that the Δwza mutants can be converted to a nonmucoid phenotype and escape adsorption by phage SRD2021 in Klebsiella pneumoniae12.
Researchers have identified that CPS of G− bacteria serves as the adsorption receptor of many phages13,14. Before infecting G− bacteria, the tail filament protein of phages absorb to the CPS on the bacterial surface14. The adsorption sites on the bacterial surface may affect the infection efficacy and gene expression of phages, and even the bacterial survival15,16. Gong et al. reported that the CPS of Escherichia coli DE058 was the first irreversible binding receptor initially adsorbed by its phage PNJ1809-36 and further identified the tail protein ORF261 of the phage PNJ1809-36 as the receptor-binding protein17. In fact, Porter et al. utilized B. thetaiotaomicron strains expressing genetically distinct CPSs to isolate phages and found that the subsets of CPS determines the host tropism of phages18.
Bacteria in different growth stages have corresponding population densities19, and different growth stages usually coordinate individual behaviors, such as biomass, biochemical composition20, the AI-2 quorum sensing (QS) system19, and lipid membrane formation21. QS is a widely acceptable mechanism for regulating bacterial population behavior, universally acknowledged, G− bacteria commonly use N-acyl homoserine lactones as their autoinducers, and they are detected by LuxR-type receptors22, wherein LuxR family of transcription factors control QS gene expression in Vibrio23. Furthermore, phages can alter their lysis-lysogeny state based on host cell density, it can listen in on their host bacterium’s QS systems24. In addition, Xuan et al. reported that las QS mediates phage susceptibility, which depends on host bacterial populations25. Generally, bacterial populations influence the physiological state of cells to better protect them against phage attack. Nevertheless, in V. splendidus the effects of changes in the bacterial growth stage on phage sensitivity through the CPS translocon Wza are still unclear.
In the present study, we identified a potent anti-phage defense mechanism in V. splendidus and showed that different anti-phage strategies prevail among different growth stages and phage-infected state. Bacteria in stationary growth stage (SGS) or early exponential stage (EES) infected with phage (EES-P) exhibited down-regulated luxR and wza, resulting in low susceptibility to phage, and rendering individual cells almost unsusceptible to phage infection. Under the EES, in the contrary, Wza expression is unaffected, and individual cells are fully susceptible to infection. Our results reveal important roles for V. splendidus CPS translocon Wza and cells locked in the SGS or EES-P that allow V. splendidus to persist under phage predation, suggesting the presence of dynamic, temporary adaptations to phage infection pressure, and providing important implications for using phages therapeutically to target pathogenic bacteria in dynamic communities.
Results
Morphology, characteristics, and genome analysis of the isolated phage
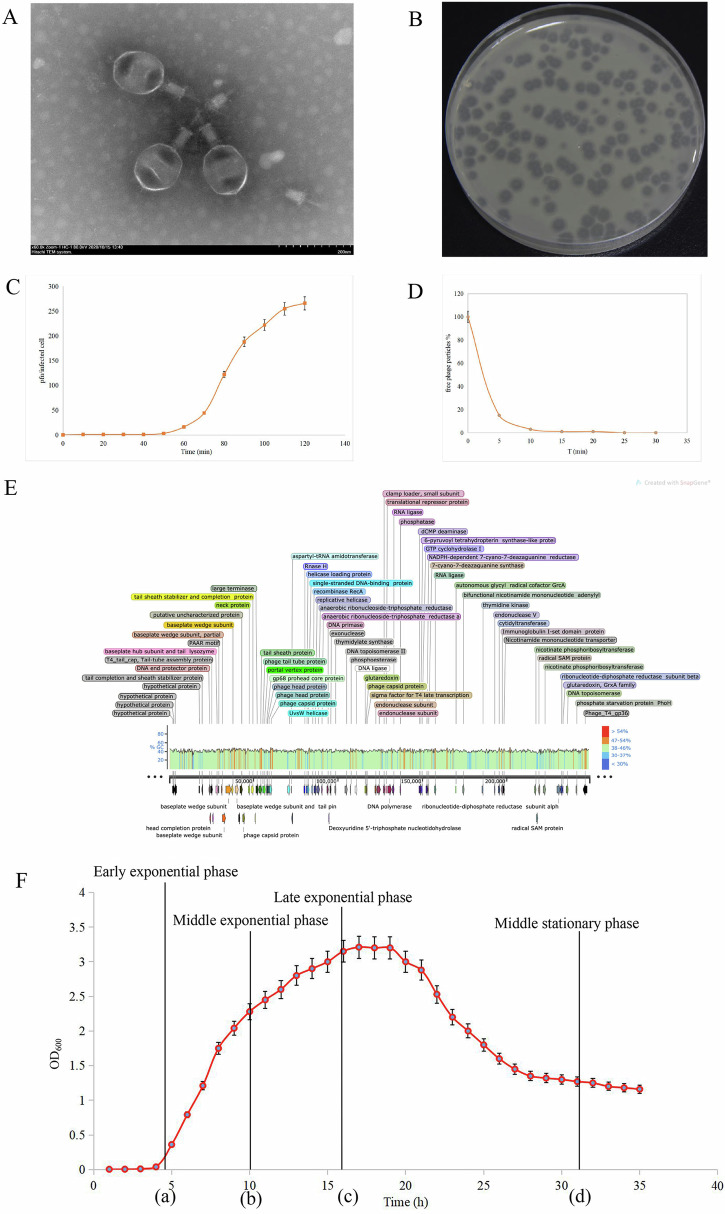
Virulent V. splendidus phage vB_VspM_VS2 was isolated from Apostichopus japonicus breeding pond silt in Dalian, China. TEM analysis revealed that the vB_VspM_VS2 virions had an icosahedral head of 122 × 140 ± 5 nm and a tail of 30 × 50 ± 5 nm in length (Fig. 1A), indicating it belongs to the family Straboviridae. The plaques of vB_VspM_VS2 were around 4 mm in diameter after overnight incubation at 28 °C (Fig. 1B). A one-step growth curve of vB_VspM_VS2 was obtained by inoculation of V. splendidus AJ01 at an multiplicity of infection (MOI) of 1 at 28 °C (Fig. 1C). The latent period of vB_VspM_VS2 was 50 min, and the titer of vB_VspM_VS2 reached the peak in 120 min. Additionally, the burst size of vB_VspM_VS2 was ~260 plaque-forming units (PFU)/cell. Regarding the adsorption rate of phage, after standing for 10 min at 28 °C, nearly 95% of the phage particles were adsorbed to the host bacterium V. splendidus AJ01. Moreover, after incubation for 25 min, almost all phages were adsorbed to the host bacterium V. splendidus AJ01 (Fig. 1D).
Fig. 1. Morphology, characteristics and genome organization of phage vB_VspM_VS2.
A Transmission electron micrograph showing that vB_VspM_VS2 belongs to the family Myoviridae and has a head of 122 × 140 ± 5 nm and a tail of 30 × 50 ± 5 nm. B Plaques formed by vB_VspM_VS2 on the host strain V. splendidus after overnight incubation at 28 °C. C, D Adsorption rate and population dynamics of vB_VspM_VS2 inoculated in V. splendidus culture. The values presented are means and standard deviations (SDs) of three independent biological repeats (n = 3). E Line map of the vB_VspM_VS2 genome. In the vB_VspM_VS2 genome track, the arrows represent the ORFs and point in the direction of transcription. The color intensity corresponds to G + C skew level. F Growth curve of V. splendidus. At the time of 4.5 h, the bacteria began to grow rapidly that enter the growth phase of exponential until 16 h. a: early exponential stage (EES, 4.5 h), b: middle exponential stage (MES, 10 h), c: late exponential stage (LES, 16 h), d: stationary growth stage (SGS, 31 h).
Phage vB_VspM_VS2 has a double-stranded molecule genome consisting of 250,514 bp with a G + C content of 42.51% (Fig. 1E). There are 398 protein-coding genes [open reading frames (ORFs)] were identified in the genome of vB_VspM_VS2. The complete genome sequence of phage vB_VspM_VS2 was deposited in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/nuccore) (Genebank Accession No. OQ722176.1). Bioinformatics analysis revealed that 398 ORFs can be categorized into five functional groups: DNA packaging and nucleic acid metabolism (such as DNA primase), cell lysis (such as lysozyme), structure proteins except tail fiber (such as phage head protein), tail-associated protein (such as tail completion and sheath stabilizer protein), and hypothetical protein. In addition, after evaluating the ability of the phage vB_VspM_VS2 to lyse V. splendidus AJ01, we found it cannot only lyse V. splendidus AJ01, but also produce more progeny phages, revealing that phage vB_VspM_VS2 is likely a virulent phage. Genes encoding putative lysis-associated proteins (particularly endolysin and tail-fiber/lysozyme) were found in the genome. Genes determining the activities of DNA replication and transcription, including a terminase, a replicative DNA helicase, a putative transcriptional regulator, a putative nucleoside triphosphate pyrophosphohydrolase, and a putative RNA polymerase, were also identified in the genome of vB_VspM_VS2 (Fig. 1E). No genes related to toxin production, temperate lifestyle, antibiotic resistance or virulence were identified.
Effects of growth stages and phage infection on the adsorption of phage to bacteria
To determine the relationship between the growth time and growth stage of V. splendidus, we conducted growth curve measurements and definitions. Parameters describing the growth stage of a (EES, ~4.5 h), b (middle exponential stage [MES] ~ 10 h), c (late exponential stage [LES] ~16 h) and d (SGS, ~31 h) were determined from the growth curves (Fig. 1F). During 0-3 h, the OD600 of V. splendidus was ~0, suggesting V. splendidus AJ01 was located at the growth stage lag phase. Subsequently, during 4.5–10 h, V. splendidus AJ01 was located at the early exponential growth phase, and among 10-16 h, V. splendidus AJ01 was located at the late exponential growth phase. Reaching late growth stage, V. splendidus AJ01 grew into a stable period 27 h later and persisted to 36 h (Fig. 1F).
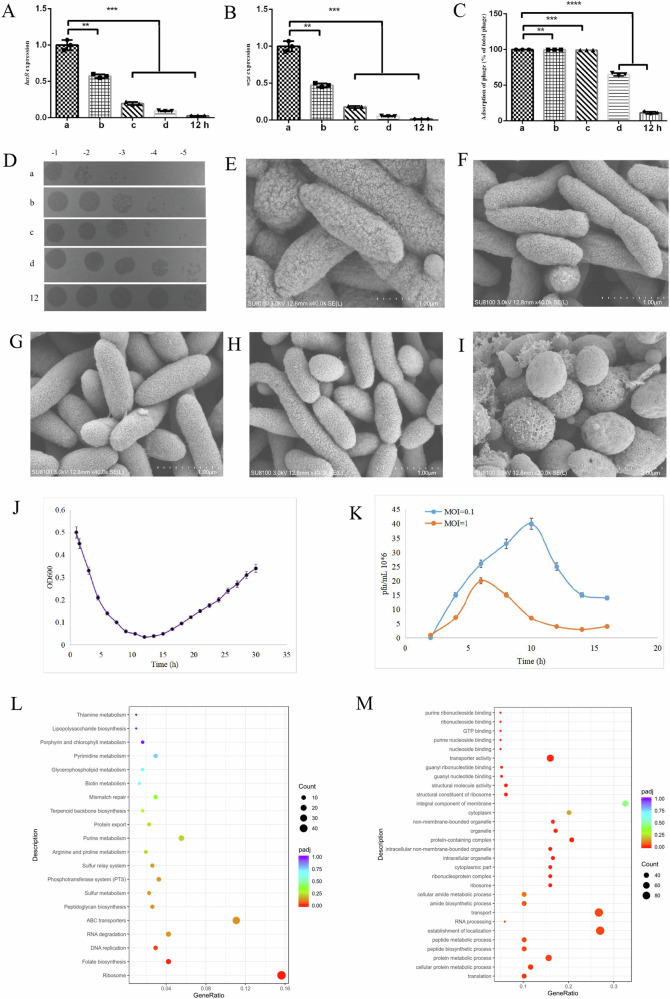
Wza mediates the translocon of phage adsorption target CPS. We performed RNA extraction, cDNA synthesis, and RT-qPCR detection among wild type, “growth stage a” with phage infected, and different “growth stages of a, b, c and d” to investigate the wza expression. Compared to the “growth stage a”, we found that cells in the “growth stage d” express low levels of luxR and wza genes (Fig. 2A, B), with a low susceptibility to phage due to few phages adsorbing on bacterial surface (Fig. 2C, D). In addition, we found that wza expression was significantly decreased after vB_VspM_VS2 infection and the adsorption of phage was significantly inhibited in V. splendidus at “growth stage d” and “growth stage a” infected with phage (Fig. 2B, C). These results show that the adsorption rate of vB_VspM_VS2 was consistent with the expression level of wza, and phage adsorption was most strongly inhibited after phage infection. Nevertheless, compared to bacteria in “growth stages b, c, and d”, bacteria in “growth stage a” are most easily adsorbed by phage.
Fig. 2. Effects of growth stages and phage on expression of wza and luxR regulating phage adsorption.
A, B The relative expression of AJ01 luxR and wza at different growth stages (a, b, c and d) and growth stage a infected with phage (EES-P), 16 s rRNA was the internal reference gene. C Adsorption rate of vB_VspM_VS2 by its host strain V. splendidus of different growth stages (a, b, c and d) and EES-P. D: Tenfold serial dilutions of 2 μL not adsorbed vB_VspM_VS2 plated on wild-type V. splendidus. E–I SEM images V. splendidus cells of different growth stages (a, b, c and d) and EES-P for 12 h. J Growth curves of AJ01 strain in 2216E medium. Optical densities (OD600) of cultures of AJ01 wild-type (WT) in the presence phage vB_VspM_VS2 at a multiplicity of infection (MOI) of 1. K Corresponding abundances of pfu/mL were quantified by plaque assay over a 16-h period of incubation in AJ01 cultures in the presence of phage vB_VspM_VS2 at a multiplicity of infection (MOI) of 0.1 and 1 were measured at different incubation times, respectively. Error bars represent standard deviations from all experiments carried out in duplicate. P < 0.05; **, P < 0.01; ***,P < 0.001; #, P > 0.05. L The abscissa in the figure is the ratio of the number of differential genes annotated on GO Term to the total number of differential genes. The ordinate is GO Term. The size of the dot represents the number of genes annotated on GO Term. The color from red to purple represents the significance of enrichment. The figure represented the comparison V. splendidus (V.s) between high-cell-density state and low-cell-density state. M The abscissa in the figure is the KEGG pathway, and the ordinate is the significance level of pathway enrichment. The higher the value is, the more significant it is. The figure represented the comparison V. splendidus (V.s) between high-cell-density state and low-cell-density state.
To further investigate the influence of growth stage and phage infection on cell morphology and expression profile, we used SEM to observe the morphology of V. splendidus AJ01 cells at the different growth stages (a, b, c and d) and growth stage a cells infected with phage. The images confirmed that growth stage a cells were rod-shaped and healthy with intact cell membranes and dense cytosol (Fig. 2E, F, G). Critically, the majority of the growth stage d cells were rod-shaped, spherical, and had smooth membranes (Fig. 2H). The results suggest that growth stage d cells undergo a morphological change. Vibrios morphological changes are often observed in phage infected cells, EES-P cells were more spherical, with ruptured and damaged membranes (Fig. 2I). Furthermore, phage vB_VspM_VS2 reduced cell density in the cultures of wild-type AJ01 within 12 h; however, wild-type AJ01 exhibited rapid regrowth after 12 h (Fig. 2J). To further explore this phenomenon we followed the kinetics of pfu/mL vB_VspM_VS2 in V. splendidus cultures at MOIs of 0.1 and 1, revealing a steep proliferation phase that gradually decreased (Fig. 2K). Additionally, ABC transports of GO and transport of KEGG were downregulated in growth stage d compared to the growth stage a (Fig. 2L, M).
Analysis of differentially expressed genes among V. splendidus in different stages under phage phage vB_VspM_VS2 infection
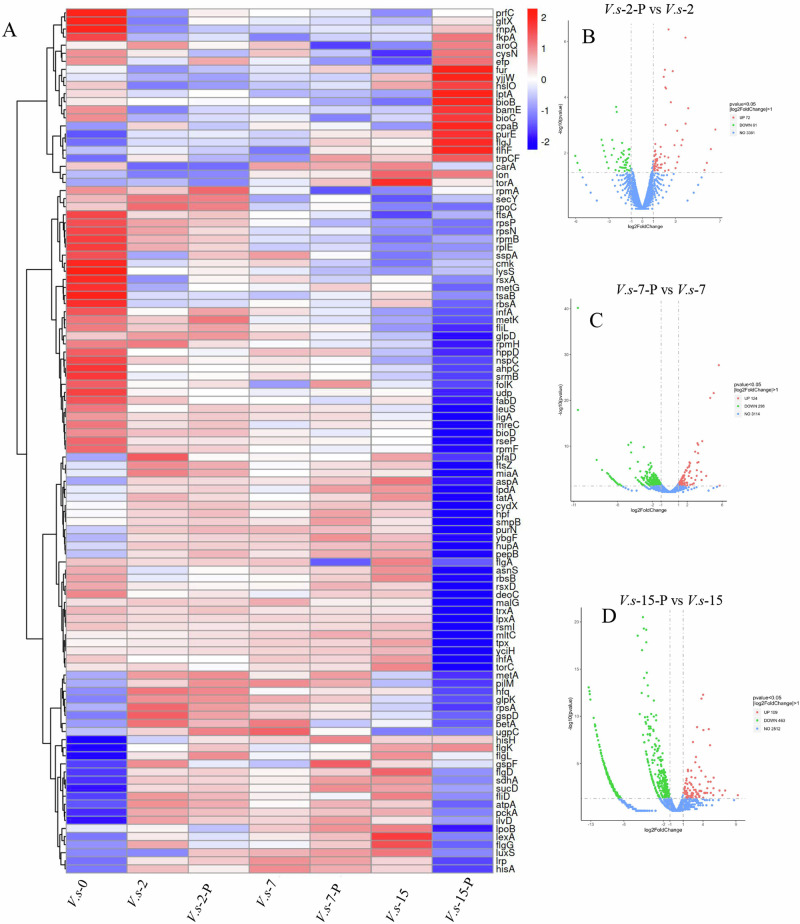
To profile global mRNA expression patterns, we analyzed the heat map of all differentially expressed mRNAs of the seven groups (V.s-0 [V. splendidus culture for 0 h], V.s-2 [V. splendidus culture for 2 h], V.s-2-P [V. splendidus infected with phage and culture for 2 h], V.s-7 [V. splendidus culture for 7 h], V.s-7-P [V. splendidus infected with phage and culture for 7 h], V.s-15 [V. splendidus culture for 15 h], V.s-15-P [V. splendidus infected with phage and culture for 15 h]). Here, we only included those that were statistically significantly different with a fold change ≥2 and a P < 0.05 (Fig. 3A). Compared to V.s-2 cells, 72 genes were up-regulated in cells of V.s-2-P, and 61 genes were down-regulated (Fig. 3B). Compared to V.s-7 cells, 124 genes were up-regulated in cells of V.s-7-P, and 298 genes were down-regulated (Fig. 3C). Compared to V.s-15 cells, 109 genes were up-regulated in cells of V.s-15-P, and 463 genes were down-regulated (Fig. 3D).
Fig. 3. Differential mRNA expression patterns in V. splendidus of the seven groups (V.s-0, V.s-2, V.s-2-P, V.s-7, V.s-7-P, V.s-15, V.s-15-P) were analyzed, V.s-2, V.s-2-P, V.s-7, V.s-7-P, V.s-15, V.s-15-P represented V. splendidus non-infected and phage infected at 2, 7, and 15 h, respectively.
V.s-0 represented V. splendidus non-infected at 0 h. A: Hierarchical cluster analysis of the differentially expressed mRNAs in V.s-0, V.s-2, V.s-2-P, V.s-7, V.s-7-P, V.s-15 and V.s-15-P groups. Significance was determined using a fold-change threshold of 2 and a p-value cutoff of 0.05. High expression of miRNAs is shown in red, and low expression is shown in blue. DEG distribution between V. splendidus and phage infected V. splendidus. V.s-2-P vs V.s-2 (B), V.s-7-P vs V.s-7 (C), and V.s-15-P vs V.s-15 (D) represented the comparison between V. splendidus (V.s) and phage infected V. splendidus at the growth of 2, 7, and 15 h, respectively. The significant level of the difference in gene expression between the V. splendidus and phage infected V. splendidus groups (−log10padj) versus the fold change of gene expression (log2Fold Change). Each dot represents one gene. Green and red dots represent down-regulated and up-regulated DEGs, respectively. Blue dots represent no DEGs.
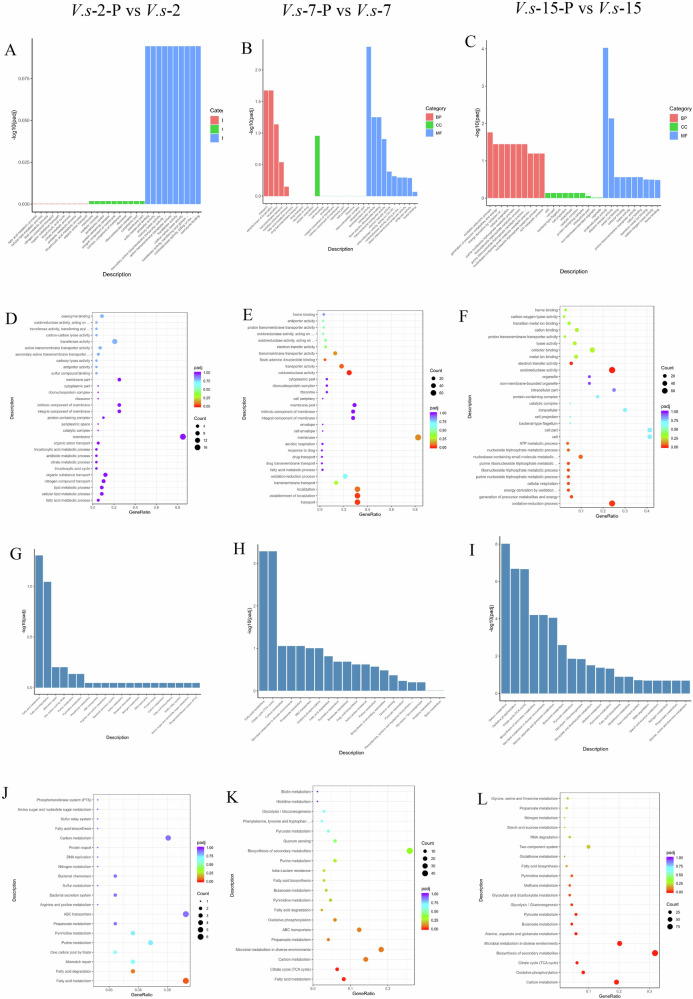
GO is a comprehensive database that can describe gene functions in three categories: biological process (BP), cellular component (CC), and molecular function (MF). To compare the functional categories of V. splendidus AJ01 under control and phage infection, we compared with V.s-2, V.s-7 and V.s-15; V.s-2-P, V.s-7-P and V.s-15-P; V. splendidus AJ01 infected with phage; V. splendidus AJ01 downregulated in transmembrane transport (GO:0055085) in BP, membrane (GO:0016020) in CC, and transmembrane transporter activity (GO:0022857) in MF (Fig. 4A–F). Furthermore, KEGG analysis showed that compared with V.s-2, V.s-7 and V.s-15, the differentially expressed genes were enriched mainly in the quorum sensing, ABC transporters, fatty acid biosynthesis and flagellar assembly in the cells at V.s-2-P, V.s-7-P and V.s-15-P groups (Fig. 4G–L). Overall, GO and KEGG analyses showed that phage infection played global roles in biosynthesis, ribosomes, metabolism, and secretion, which are pathways could associated with phage adsorption.
Fig. 4. Histogram and scatter plot based on the GO and KEGG analysis.
A–C The abscissa described the GO term, and the ordinate described the levels of enrichment expressed by −log10 (padj). V.s-2-P vs V.s-2 (A), V.s-7-P vs V.s-7 (B) and V.s-15-P vs V.s-15 (C) represented the comparison between V. splendidus (V.s) and phage infected V. splendidus at 2, 7, and 15 h, respectively. D–F: The abscissa in the figure is the ratio of the number of differential genes annotated on GO Term to the total number of differential genes. The ordinate is GO Term. The size of the dot represents the number of genes annotated on GO Term. The color from red to purple represents the significance of enrichment. V.s-2-P vs V.s-2 (D), V.s-7-P vs V.s-7 (E), and V.s-15-P vs V.s-15 (F) represented the comparison between V. splendidus (V.s) and phage infected V. splendidus at 2, 7, and 15 h, respectively. G–I The abscissa in the figure is the KEGG pathway, and the ordinate is the significance level of pathway enrichment. The higher the value is, the more significant it is. V.s-2-P vs V.s-2 (G), V.s-7-P vs V.s-7 (H) and V.s-15-P vs V.s-15 (I) represented the comparison between V. splendidus (V.s) and phage infected V. splendidus at 2, 7, and 15 h, respectively. J–L The abscissa represented the proportion of the number of differentially expressed genes of the specific KEGG pathway to the whole number of differentially expressed genes, and the ordinate represented the specific KEGG pathway. The percent of genes annotated to specific KEGG pathway is represented by the size of dots, and the color represents different padj value. V.s-2-P vs V.s-2 (J), V.s-7-P vs V.s-7 (K) and V.s-15-P vs V.s-15 (L) represented the comparison between V. splendidus (V.s) and phage infected V. splendidus at 2, 7 and 15 h, respectively.
Identification and characterization of V. splendidus gene knockout strains
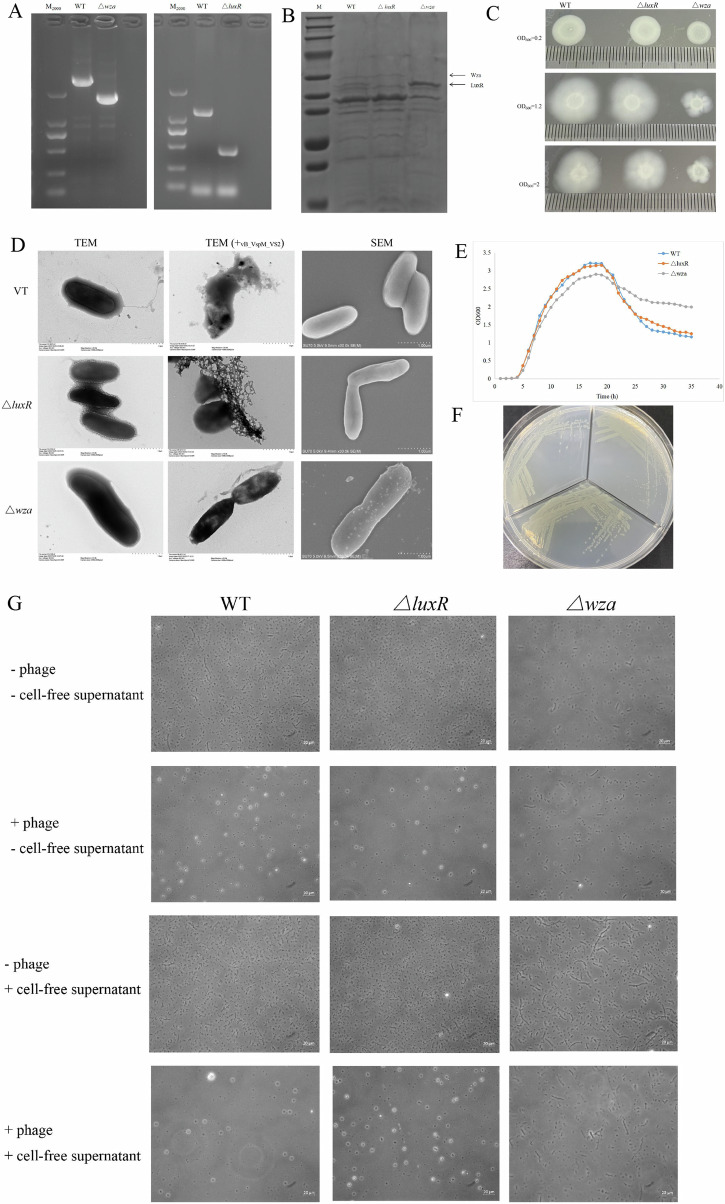
There were two genes annotated as luxR and wza in V. splendidus AJ01. The ORFs of the luxR and wza genes were 827 and 1137 bp, respectively, encoding LuxR family transcriptional regulator and CPS export protein. In this study, we obtained in-frame deletion of 1128 bp and 720 bp in the wza and luxR genes, respectively, through double homologous recombination (Fig. 5A). To compare the effects of knocking out genes on growth, we measured the growth of wild-type, ΔluxR and Δwza strains under the same culture conditions. When cultured in 2216E medium, the growth rate of the ΔluxR strain was almost the same as that of the wild-type strain, nevertheless, Δwza exhibited slight growth retardation compared to the wild-type strain at the logarithmic growth phase, and the cell density of Δwza decreased compared to that of the wild-type strain at the stationary phase. Interestingly, the cell density of Δwza was increased compared to that of the wild-type at the decay phase (Fig. 5E). In addition, the colony morphology of the Δwza strain was smaller than that of the wild-type strain under the same growth conditions (Fig. 5F). Purification and separation of outer membrane proteins using SDS-PAGE (Fig. 5B) among the wild-type, ΔluxR, and Δwza strains, with the ΔluxR and Δwza strains used as negative controls, confirmed the presence of the Wza and LuxR in the wild-type strains26. In agreement with the gene expression data, Wza appeared to be abundant in the ΔluxR and wild-type strains. SDS-PAGE outer membrane protein analysis also demonstrated that the CPS export protein Wza, which potentially functions in phage receptors (Fig. 5B).
Fig. 5. Identification and characterization gene knockout strains of V. splendidus.
A Agarose gel analysis of PCR product to detect the loss of V. splendidus wza and luxR genes. B SDS-PAGE analysis of the outer membrane protein with Coomassie blue staining. Lane M, protein markers; lanes 2–4, wild-type, ΔluxR and Δwza strains, respectively. C Swimming motility of wild-type, ΔluxR and Δwza strains at various cell densities. D TEM and SEM images of wild-type, ΔluxR and Δwza strains, and TEM images of wild-type, ΔluxR and Δwza strains infected with phage, and scale bar of SEM and TEM indicates 1 μm. E Growth curves of wild-type, ΔluxR and Δwza strains, wild-type, ΔluxR and Δwza were separately cultured in 2216E liquid medium, and OD600 was measured at 1.5 h intervals. Each sample was repeated three times. F Observation of wild-type (upper left), Δwza strain (upper right) and ΔluxR strain (below) using the naked eyes. G Visualization by phase-contrast microscopy of V. splendidus (wild type [WT], ΔluxR and Δwza mutants) in the presence or absence of vB_VspM_VS2 in either fresh 2216E broth or cell-free spent culture fluid obtained from a mid-log-phase culture of wild-type V. splendidus.
To verify whether the changes in CPS were consistent with swimming motility in V. splendidus AJ01, we determined the swimming motility of the wild-type, ΔluxR and Δwza strains in semisolid media. The colony diameter of Δwza was 9–10 mm less than the colony diameter of 16–17 mm for the wild-type and ΔluxR strains throughout all growth stages (Fig. 5C), indicating that Wza positively regulates swimming motility in V. splendidus AJ01.
SEM and TEM were used to explore the morphology of wild-type, ΔluxR and Δwza strains. The images confirmed that Δwza strains cells are larger and have smooth membranes (Fig. 5D). Critically, the majority of wild-type and ΔluxR cells are smaller, and they also have rough membrane (Fig. 5D). Phage-infected cells contained more spherical cells, which are either dense cytosol or empty cytosol types. These types are injured cells with damaged membranes (Fig. 5D).
Tan et al., demonstrated that the addition of phage to V. anguillarum resulted in increased biofilm formation and cell aggregation, providing protection against phage infection27,28. In this study, we tested whether an extracellular factor of V. splendidus might be involved in the regulation of host bacterial growth, phage tolerance, and the phage-induced aggregation phenotype. We experimentally examined in V. splendidus, the effect of adding a cell-free spent culture from the high-cell-density state of the V. splendidus strain to freshly inoculated cultures of the V. splendidus strain in the presence or absence of vB_VspM_VS2 with phase-contrast microscopy (Fig. 5G). Interestingly, the cell-free spent culture-induced aggregation phenotype of V. splendidus was completely inhibited by the presence of vB_VspM_VS2, suggesting that a QS signaling molecule may be involved in the regulation of phage defense in V. splendidus.
Wza is necessary for phage infection
Bacteria can defend against phage infection through various strategies, i.e., preventing phage entry, restriction-modification systems, abortive infection, and CRISPR-Cas systems29–32. In order to understand the defense mechanisms underlying phage-host interactions, we discovered that cells locked in the stationary growth state express low levels of the phage receptor CPS, resulting in a low susceptibility to phage due to growth stages mediated downregulation of wza expression. In addition, the luxR expression of V. splendidus was significantly decreased after phage infection, and phage adsorption was significantly inhibited. However, the molecular mechanism by how different growth stage effect the synthesis of CPS and influences phage adsorption remains unclear.
In order to examine the effects of different growth stages exploit Wza to modify the interaction between phage vB_VspM_VS2 and V. splendidus, two mutants were constructed which represent cell behavior at the growth stage d high (Δwza) or the growth stage a (ΔluxR), respectively. By contrast, a ΔluxR mutant has lost the ability to regulate QS-associated functions and is therefore locked in growth stage a phenotype. Use of the two extreme growth stages phenotypes of Δwza or ΔluxR mutants in the current study allowed us to explore the role of growth stage-mediated phage protection in V. splendidus.
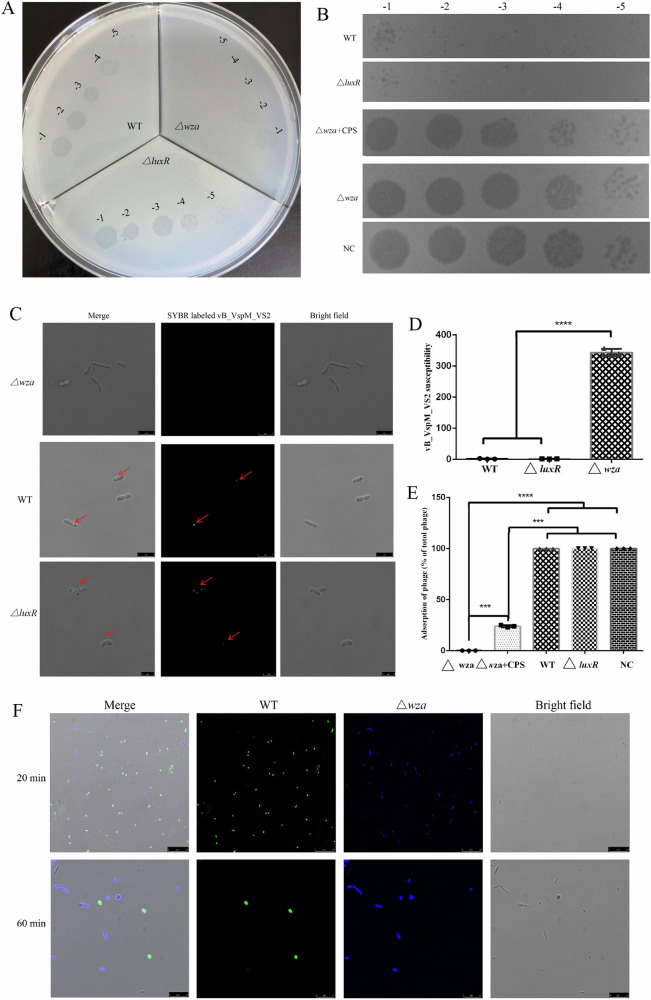
To better understand how the CPS translocon Wza modifies phage susceptibility in V. splendidus strains, we investigated the phage resistance of V. splendidus. Compared with the wild-type and ΔluxR mutant strains, the Δwza mutant strain was found to be resistant to phage vB_VspM_VS2 infection (Fig. 6A). However, the deletion of luxR did not affect the transparency of the plaques and phage adsorption, suggesting that luxR deletion did not affect the resistance of the bacteria to this phage (Fig. 6A, B, D). Nevertheless, both the wild-type and the ΔluxR mutant strains, can be adsorbed and lysed by phages and their sensitivity to phages does not differ (Fig. 6A, D). These results strongly suggesting that Wza, but not the LuxR, positively regulates phage sensitivity in V. splendidus. Indeed, these findings not only support previous observations for that CPS specific for phage adsorption, but also provide additional evidence for the role of Wza of V. splendidus in phage host interactions.
Fig. 6. Wza expression confers phage infection.
A Phage sensitivity assay. Tenfold serial dilutions of phage vB_VspM_VS2 plated on wild-type, ΔluxR and Δwza strains. B Phage adsorption assay of spot test assay, tenfold serial dilutions of 2 μL not adsorbed vB_VspM_VS2 plated on wild-type V. splendidus. Unadsorbed free phages were determined as a ratio of free phage at the time point divided by the total phage added at the beginning of the assay. Results are representative of three independent experimental assays (n = 3). NC negative control. C Visualization of vB_VspM_VS2 adsorption on wild-type, ΔluxR and Δwza strains by SYBR gold-labeled vB_VspM_VS2 under a phase-contrast epifluorescence microscope. D Bar chart comparison of phage sensitivity. E Identification of CPS as an important receptor for vB_VspM_VS2 infection. Δwza mutant significantly reduced the adsorption of phage vB_VspM_VS2. Extracted CPS was used for adsorption assays. The adsorption rate was increased in the CPS-treated group compared to that in the control group. Data are averages of three samples with standard deviations (error bars); Adsorption rate of phage vB_VspM_VS2 by its host strain wild-type, ΔluxR and Δwza mutants. Data are averages of six samples with standard deviations (error bars). **, P < 0.01 (Student’s paired t test). F Wild-type (green) and Δwza (blue) cells were mixed, infected with vB_VspM_VS2 at 5:1 (phages:bacteria) MOI, and followed by time-lapse fluorescence microscopy. Shown are overlay images from blue, green, and phase contrast (gray), captured at the indicated time points postinfection. Scale bar, 1 μm.
To further investigate the molecular mechanisms associated with the altered susceptibility of V. splendidus strains to vB_VspM_VS2 infections, we examined the adsorption rate of phage vB_VspM_VS2 by different strains of wild-type, ΔluxR and Δwza. The Δwza mutant strain exhibited a pronounced reduction in phage adsorption rates compared to the wild-type strain. The ΔluxR mutant, however, exhibited no differences in adsorption rate compared to that of the wild-type strain (Fig. 6B). Thus, Wza positively regulates phage susceptibility by increasing the phage adsorption receptor CPS. Additionally, the extracted CPS was used for adsorption assays, Δwza mutant strain revealed an increase in the adsorption rate when CPS was added to the reaction system (Fig. 6B, E), confirming CPS as a receptor for V. splendidus phage vB_VspM_VS2. Besides, Wza is involved in V. splendidus CPS core synthesis8. The expression of wza is dependent on the growth stage, with its expression at growth stage a being higher than that at growth stage d or phage-infected state (Fig. 2B). Also, growth stage d should lead to lower phage susceptibility due to the synthesis of fewer CPS receptors. As expected, the adsorption rate of stationary stage cells was significantly lower than that of exponential stage cells (Fig. 2C).
To further examine the mechanisms of tolerance or resistance of vB_VspM_VS2 in the wild-type, ΔluxR, and Δwza strains, we incubated the wild-type, ΔluxR and Δwza strains with SYBR gold-labeled vB_VspM_VS2. Fluorescently labeled phage vB_VspM_VS2 attached to the sensitive cells of the wild-type and ΔluxR strains (Fig. 6C), while no phage attachment was observed in the phage-resistant mutants of Δwza (Fig. 6C). In addition, wild-type (green) and Δwza (blue) cells were mixed, infected with vB_VspM_VS2 at 5:1 (phages:bacteria) MOI, and followed by time-lapse fluorescence microscopy. Overlay images from blue, green, and phase contrast (gray) captured at the indicated time points post-infection are shown (Fig. 6F). The scale bar is 1 μm. To assay Wza activity in bacteria, we added vB_VspM_VS2 to wild-type (green) and Δwza (blue) cells. Time-lapse microscopy revealed repositioning and massive nucleoid deployment in the resistant bacteria (Fig. 6F). The green fluorescence signal of wild-type cells dropped 60 min post-infection (Fig. 6F).
wza alter the sensitivity of phages in liquid culture
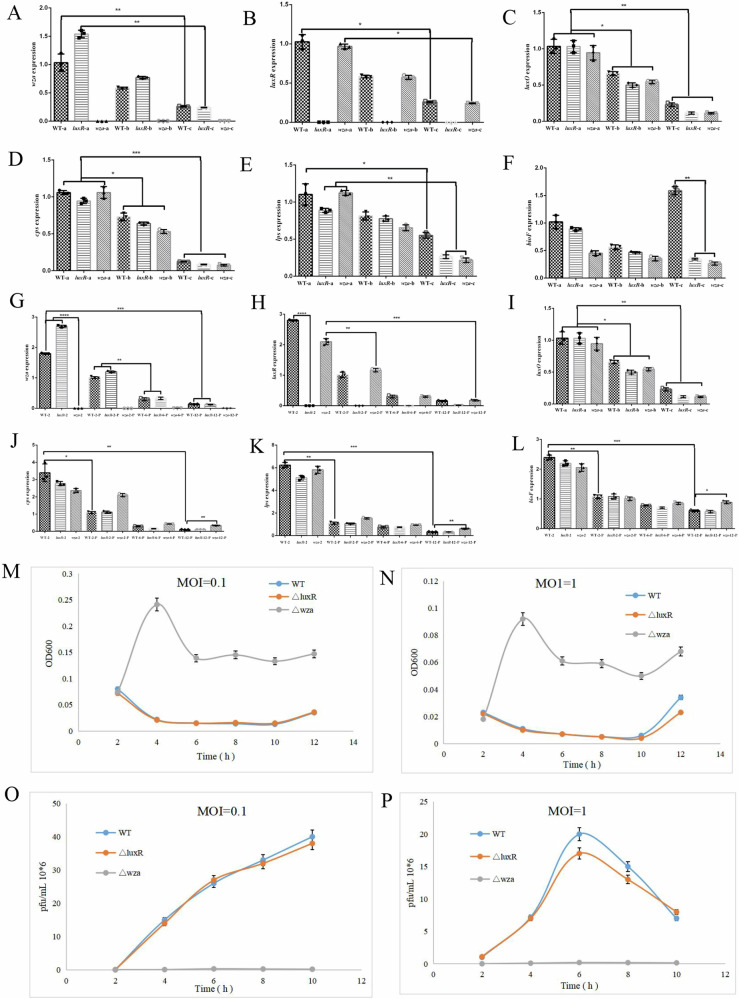
To further examine the molecular mechanism underlying the differences in adsorption rates among wild-type, ΔluxR and Δwza and growth stage-mediated strains and to link these differences to growth stage regulation, we quantified the expression of phage vB_VspM_VS2 receptor related cps, wza, luxR, luxO, lps and bioF in 9 different cultures (wild type, ΔluxR and Δwza, exponential state, stationary growth state, phage infection 2 h, phage infection 6 h, and phage infected 12 h) by quantitative real-time PCR (qPCR). The relative wza, luxR, luxO, cps and lps expression levels of wild-type and ΔluxR strains in early exponential state were approximately four times higher than the relative expression in stationary growth state and phage infected cells, suggesting a downregulation of Wza when the cells were infected by phage in the regulatory state mimicking stationary growth state (Fig. 7). In addition, the expression of wza and luxR genes could not be detected in ΔluxR and Δwza mutants (Fig. 7).
Fig. 7. wza alter the sensitivity of phage in liquid culture.
A–F Relative wza, luxR, luxO, cps, lps and bioF expression measured by RT-qPCR in V. splendidus cells at low and high cell densities (OD600, 0.1, 0.8, and 2.5, respectively). The reference gene was 16 s rRNA. P < 0.05; **, P < 0.01; ***, P < 0.001; #, P > 0.05. G–L Relative wza, luxR, luxO, cps, lps and bioF expression at phage infection of 2, 6, and 12 h in wild-type, ΔluxR and Δwza strains. Data are averages of three samples with standard deviations. P < 0.05; **, P < 0.01; ***, P < 0.001; #, P > 0.05. M, N Growth curves of wild-type, ΔluxR and Δwza strains in 2216E medium. Optical densities (OD600) of cultures of wild-type, ΔluxR and Δwza mutants in the presence of phage vB_VspM_VS2 at a multiplicity of infection (MOI) of 0.1 and 1 were measured at different incubation times. Data are averages of six samples with standard deviations. P < 0.05; **, P < 0.01; ***, P < 0.001; #, P > 0.05. O, P Corresponding abundances of pfu/ mL were quantified by plaque assay over a 10 h period of incubation in wild-type, ΔluxR and Δwza mutants cultures in the presence of phage vB_VspM_VS2 at a multiplicity of infection (MOI) of 0.1 and 1 were measured at different incubation times, respectively. Error bars represent standard deviations from all experiments carried out in duplicate. P < 0.05; **, P < 0.01; ***, P < 0.001; #, P > 0.05.
To further investigate the sensitivity of wild type, ΔluxR and Δwza strains to phages in liquid culture medium, we found that Δwza mutant exhibited a slower reduction in cell density within 12 h than wild type and ΔluxR mutant at MOI of 0.1 and 1 (Fig. 7M, N). In addition, we observed that phage vB_VspM_VS2 propagation in the wild-type andΔluxR were ~150-fold more than Δwza mutants cultures at a multiplicity of infection (MOI) of 0.1 and 1 (Fig. 7O, P).
Global analysis of mRNA expression patterns among wild-type (WT), ΔluxR and Δwza strains cells
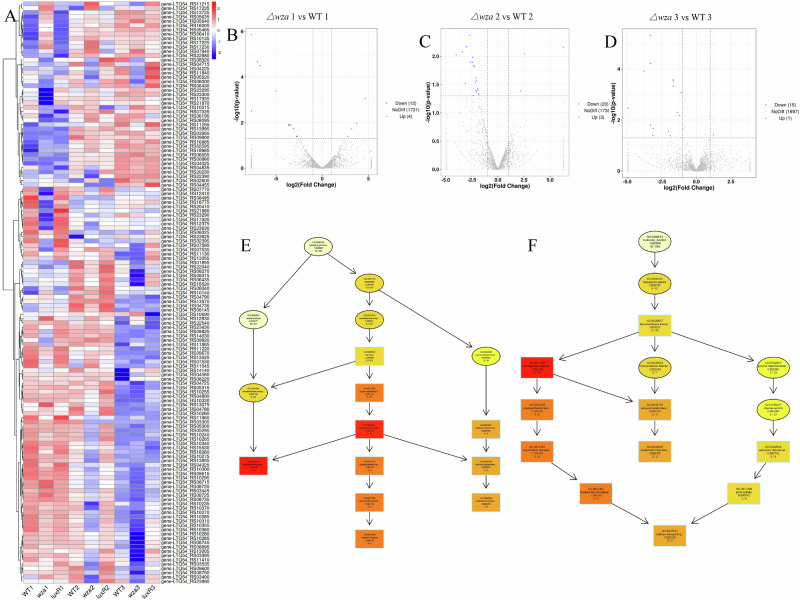
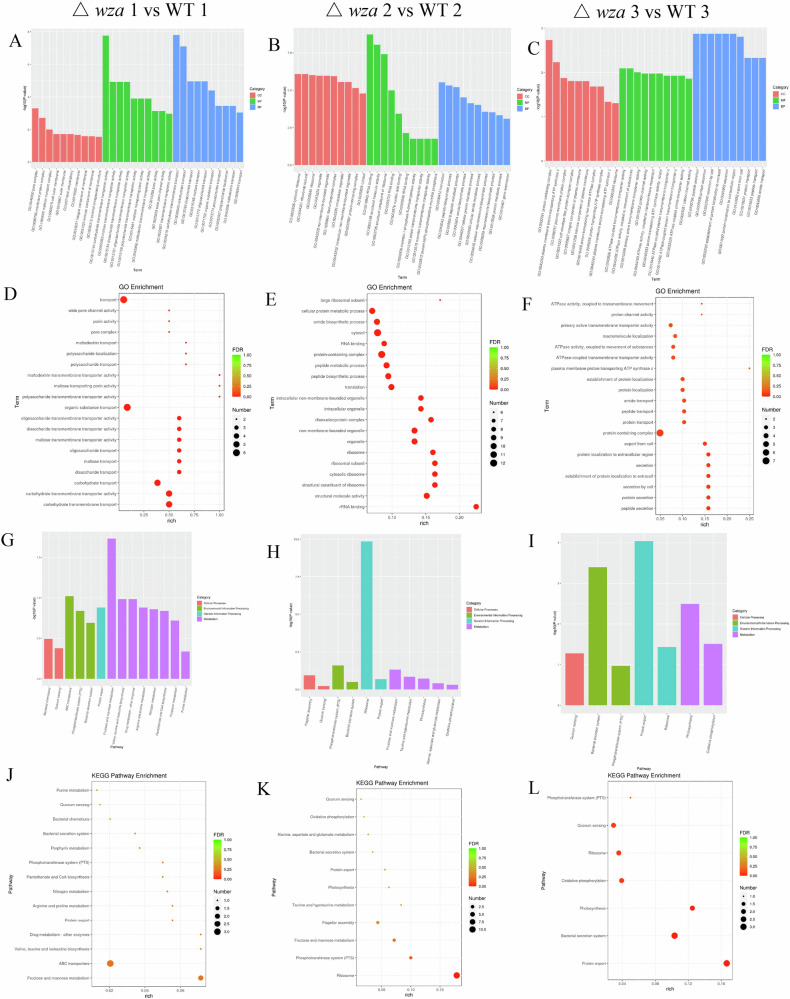
We next to analyze the global mRNA expression patterns, the heat map of all differentially expressed mRNAs of the nine groups [WT1 (OD600 = 0.2), wza1 (OD600 = 0.2), luxR1 (OD600 = 0.2), WT2 (OD600 = 1.2), wza2 (OD600 = 1.2), luxR2 (OD600 = 1.2), WT3 (OD600 = 1.9), wza3 (OD600 = 1.9), and luxR3 (OD600 = 1.9)] (Fig. 8A, Supplementary Data 1). In the cells at OD600 of 0.2, 1.2, and 1.9, there were 16, 23, and 16 differentially expressed genes in Δwza compared to the WT strain, respectively. Indeed, the volcano map visually displayed the distribution of differentially expressed genes in WT and Δwza at the same cell density (Fig. 8B–D). In conclusion, These differentially expressed genes indicated that Wza had a global regulatory effect on the gene expression of V. splendidus AJ01. Compared to the WT1 cells, 4 genes were up-regulated in cells of Δwza1, and 12 genes were down-regulated in cells of Δwza1 (Fig. 8B). Notably, in Directed Acyclic Graph (DAG), polysaccharide and carbohydrate transport were the most highly represented genes in BP and MF in the Δwza strain cells compared with WT1 (Fig. 8-E).
Fig. 8. Differential mRNA expression patterns in V. splendidus of the nine groups (WT1, wza1, luxR1, WT2, wza2, luxR2, WT3, wza3 and luxR3) were analyzed.
A Hierarchical cluster analysis of the differentially expressed mRNAs in WT1, wza1, luxR1, WT2, wza2, luxR2, WT3, wza3 and luxR3 groups. Significance was determined using a fold-change threshold of 2 and a p-value cutoff of 0.05. High expression of miRNAs is shown in red, and low expression is shown in blue. DEG Distribution of the differentially expressed genes in WT and Δwza at different growth stages. Δwza1 vs WT1 (B), Δwza2 vs WT2 (C) and Δwza3 vs WT3 (D) represented the comparison between WT and Δwza at OD600 = 0.2, 1.2, and 1.9, respectively. The significant level of the difference in gene expression between the WT and Δwza groups (−log10padj) versus the fold change of gene expression (log2FoldChange). The red dots indicated the genes that are upregulated in the absence of Δwza strain, and the green dots indicated the genes that are downregulated in the absence of Δwza strain. E, F In the figure, the branch represents the inclusion relationship, and the function range defined from top to bottom is getting smaller and smaller. Select the GO Term with the top 5 most significant GO enrichment results of each difference comparison combination as the main node of the directed acyclic graph and display the associated GO Term together through the inclusion relationship, and the depth of color represents the enrichment degree.
As mentioned earlier, GO is a comprehensive database that can describe gene functions in three categories: BP, CC, and MF. Compared with WT1, polysaccharide transmembrane transporter activity (GO:0015159) and polysaccharide transport (GO:0015774) were downregulated in BP, transmembrane transporter activity (GO:0022857) was downregulated in MF in Δwza1 group. In addition, KEGG analysis showed that compared with WT1, the differentially expressed genes were enriched mainly in the carbohydrate metabolism, membrane transport and bacterial secretion system in Δwza1 group, which were related to phage adsorption (Fig. 9). Overall, GO and KEGG analyses showed that Wza played global roles in biosynthesis, transport, energy metabolism, QS and secretion that were associated with phage adsorption through various pathways.
Fig. 9. Histogram and scatter plot based on the GO and KEGG analysis.
The abscissa described the GO term, and the ordinate described the levels of enrichment expressed by - log10 (padj). Δwza1 vs WT1 (A), Δwza2 vs WT2 (B), and Δwza3 vs WT3 (C) represented the comparison between wild-type (WT) and Δwza at OD600 = 0.2, 1.2, and 1.9, respectively. A–F The abscissa in the figure is the ratio of the number of differential genes annotated on GO Term to the total number of differential genes. The ordinate is GO Term. The size of the dot represents the number of genes annotated on GO Term. The color from green to purple represents the significance of enrichment. Δwza1 vs WT1 (D), Δwza2 vs WT2 (E), and Δwza3 vs WT3 (F) represented the comparison between wild-type (WT) and Δwza at OD600 = 0.2, 1.2, and 1.9, respectively. G–L The abscissa in the figure is the KEGG pathway, and the ordinate is the significance level of pathway enrichment. The higher the value is, the more significant it is. Δwza1 vs WT1 (G), Δwza2 vs WT2 (H) and Δwza3 vs WT3 (I) represented the comparison between wild-type (WT) and Δwza at OD600 = 0.2, 1.2, and 1.9, respectively. The abscissa represented the proportion of the number of differentially expressed genes of the specific KEGG pathway to the whole number of differentially expressed genes, and the ordinate represented the specific KEGG pathway. The percent of genes annotated to specific KEGG pathway is represented by the size of dots, and the color represents different padj value. Δwza1 vs WT1 (J), Δwza2 vs WT2 (K) and Δwza3 vs WT3 (L)represented the comparison between wild-type (WT) and Δwza at OD600 = 0.2, 1.2, and 1.9, respectively.
CPS alters bacterial phage susceptibility
To investigate the molecular mechanisms associated with the altered susceptibility of V. splendidus strains to phage vB_VspM_VS2 infections, we examined the adsorption rate of phage vB_VspM_VS2 on different cell-density state and phage infected state of Vibrio. As mentioned above, the Δwza mutant exhibited a significant reduction in phage adsorption rates compared to the wild-type strain, while the ΔluxR mutant showed no differences in adsorption rate compared to the wild-type strain (Fig. 6). Therefore, Wza positively regulated phage susceptibility by increasing the phage adsorption rate. Furthermore, after phage infection, the optical density of stationary stage cells decreased slower than that of the exponential stage cells (Fig. 10A). Interestingly, these results suggest that Wza had a high correlation with the phage vB_VspM_VS2 adsorption rate, indicating that Wza corresponded with the adsorption rate of phage vB_VspM_VS2 (Fig. 10). Additionally, phage vB_VspM_VS2 do not have the ability to lyse Vibrio harveyi and Vibrio antiquarius (Fig. S1). Furthermore, the strains of Staphylococcus aureus, Salmonella bongori, Acinetobacter lwoffii, Escherichia marmotae, V. harveyi and V. antiquarius showed a significant reduction in phage adsorption rates compared to the V. splendidus (Fig. S1). Compared to other bacterial species, phage vB_VspM_VS2 has a certain adsorption capacity for other Vibrio, but whether it can lyse bacteria depends crucially on whether the phage genome can enter the bacteria and whether the expressed lytic enzymes can lyse the cell wall of the bacterium.
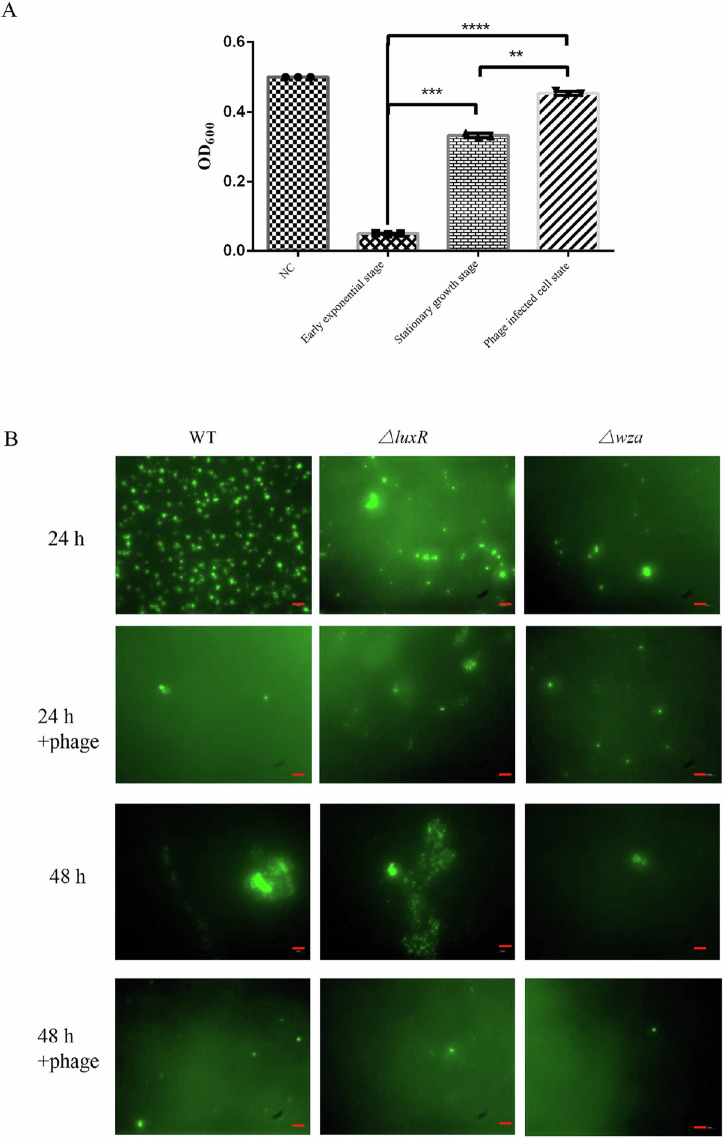
Fig. 10. Effects of phage on biofilm formation.
A Optical densities (OD600) of cultures of V. splendidus high-cell-density state, low-cell-density state and phage infected cell-state in the presence of phage vB_VspM_VS2 at a multiplicity of infection (MOI) of 1 were measured in 20 mL of each culture using a microplate reader at 10 h. Data are averages of six samples with standard deviations (error bars). **, P < 0.01 (two-way analysis of variance [ANOVA]). B Fluorescence microscopic images of microcolony formation of strain wild-type (left columns, WT), strain ΔluxR (middle columns) and strain Δwza (right columns) at different time points in the presence and absence of phage vB_VspM_VS2, respectively. Samples were stained with 0.5% SYBR gold for 15 min. Scale bar, 100 μm.
Effects of phages on biofilm formation
To investigate the effect of phage on establishment and growth of biofilm produced by the three strains, wild-type, ΔluxR, and Δwza, we examined over a long-term experiment in parallel to an experiment performed with control cultures without phage. The Δwza strain formed simple, single-layered microcolonies where individual cells could easily be identified, whereas the wild-type and ΔluxR strains formed complex, 3-dimensional volcano-shaped structures (Fig. 10B). In addition, phage vB_VspM_VS2 efficiently controlled wild-type and ΔluxR microcolonies according to microscope observations, reducing the total colony area after 24 and 48 h of incubation and leaving mainly single cells on the filters following phage exposure. Nevertheless, for the Δwza strain, the individual colonies were unaffected by phage exposure, whereas single cells were lysed by phage vB_VspM_VS2, leaving mainly colonies on the filters at 24 h post addition (Fig. 10B). These data suggest that the wza mediates the formation of biofilm, and that phage can effectively inhibit biofilm formation and growth.
Discussion
Phages are the most abundant and diverse biological entities on our planet33,34. In nature, phage-bacteria relationships are complex and extend beyond microorganisms they parasitize, encompassing dynamics during health and disease35–37. Recently, some researchers found that populations in G− bacteria is involved in the antiphage process by downregulation the phage receptor OmpK38 and LPS39. In bacteria, changes in growth stages correspond with population densities19, however, it remains unclear how different stages regulates the synthesis of CPS and influences phage adsorption, and what is the molecular mechanism in V. splendens. Through knockout of the luxR and wza genes, we demonstrated that growth stage regulates CPS synthesis and increases resistance to phage susceptibility. In addition, using RNA-seq technology, we confirmed that the regulatory network of CPS synthesis is enriched mainly in the membrane and ABC transports, which are commonly related to phage adsorption. In the present study, we expected to uncover the molecular mechanism of CPS regulated by growth stage according to population density, and to build a foundation for understanding bacteria–phage interactions in different growth states and other complex ecosystems.
The outer membrane exclusion-size barrier must be bypassed for translocation of CPS to the cell surface, and this is achieved by outer membrane export proteins, with E. coli Wza as the prototype9,40. The wza was a highly conserved gene in the cps cluster and Δ wza mutant leading non-capsulated strain41. Furthermore, the Δ wza mutant strain was more able to auto-aggregate, more hydrophobic and increased biofilm formation in Riemerella anatipestifer42. Compared with wild-type strains, Δ wza mutant strains displayed lower antiserum complement killing ability, lower adhesion to A549 cells, lower phage adsorption capacity, and lower mortality of mice12,43.
There are several ways in which CPSs could prevent or promote phage infection44. First, CPSs may stereoscopically mask surface receptors to block phage binding. Additionally, some CPSs could increase the affinity and adherence of a phage for the bacterial cell surface or serve as obligate receptors14,45,46. The host tropism of phages is dictated by subsets CPSs in Bacteroides thetaiotaomicron strains and the expression of non-permissive CPS variants strains enables survival under phage predation, in the absence of CPSs, the alteration of expression of eight distinct phase-variable lipoproteins allows escape from phage predation in B. thetaiotaomicron18. Here, we found that the adsorption rate of phage vB_VspM_VS2 differed among Vibrio spp, S. aureus, S. bongori, A. lwoffii and E. marmotae strains. In addition, phylogenetic tree analysis indicated that the adsorption rate corresponded with the evolutionary relationship (Fig. 10).
Growth stage could regulate phage receptor expression according to the density of the bacterial population47. The cas gene expression was activated at high cell density, which protects the Pseudomonas aeruginosa PA14 against phage infection using the cell-cell communication process48,49. We aimed at explore the role of growth stage in regulating susceptibility to phage vB_VspM_VS2 in V. splendidus strain AJ01 through downregulation of the phage receptor CPS. The susceptibility of phage vB_VspM_VS2 in V. splendidus strain AJ01 was reduced in the growth stage d and phage-infected state. This conclusion was further confirmed by subsequent studies of phage adsorption and sensitivity in the constructed V. splendidus mutant, the Δwza strain. These results showed that phage adsorption and susceptibility were reduced in the Δwza strain, phage infected state, and growth stage d, nevertheless enhanced in the growth stage a of the wild type, directly confirming that growth stage and Wza played a key role in the protection against vB_VspM_VS2 infection (Fig. S2). Additionally, there were 5-fold bacterial density and 300- fold lower phage production in cultures of the Δwza strain relative to the wild type strain after phage addition emphasized that Wza mediated an efficient promotion phage infection. On the opposite aside, Xuan et al., reported that las QS, instead of rhl QS, upregulated the expression of galU for phage receptor lipopolysaccharide synthesis, thus increasing phage adsorption and infection in Pseudomonas aeruginosa PAO125. Previously, bacteria population determines the choice of antiphage defense in other Vibrio species, and reported that V. anguillarum cells locked in the growth stage d were almost completely unsusceptible due to growth stage-mediated downregulation of phage receptor OmpK expression28.
Additionally, our results also showed that downregulation of wza and luxR expression for defense against phage adsorption was not the only growth stage-regulated mechanism in V. splendidus. Cell formation of a biofilm, motility and aggregation in response to phage addition in the wild-type and Δwza strains suggested that the biofilm was also a way of protection against phage infection28. The exact mechanism by which phages and growth stage induce cell morphological change, however, remains to be discovered. While the membrane vesicles and OmpK of the host mediated phage sensitivity have been suggested previously6,38, here, we suggest that this mechanism is growth stage- and phage-controlled in V. splendidus. Addition of phage vB_VspM_VS2 strongly changed cell morphological in the wild type and growth stage d. Moreover, morphologically, cells exhibited the following characteristics: smaller size, smoother surface, and closer to a circular shape (Fig. 2). Furthermore, the expression trends of wza, luxR, luxO, cps, lps and bioF also showed similar changes between high-cell-density or phage-infected state cells (Fig. 7). Hence, the data suggest that biofilm formation, cell morphology, and cell aggregation are important phage defense mechanisms.
Global transcriptomic analysis of the interactions between phage vB_VspM_VS2 and V. splendidus showed that transport (GO:0006810), transmembrane transport (GO:0055085), transporter activity (GO:0005215), ABC transporters (KEGG: vsp02010) and quorum sensing (KEGG: vsp02024) were significant different between the wild type and phage-infected type. In comparison, Yang et al., reported that the expression of virulence factors was significantly changed after phage infection, but the drug resistance genes were mainly activated in Acinetobacter baumannii50. Also, Mojardín et al., reported that DNA replication, translation, ribose and inositol were significantly changed after phage infection in Bacillus subtilis51. Similar, Kortright et al., queried those genes encoding membrane proteins for candidate receptors of phage T6 through transposon insertion sequencing52. Taken together, our data on phage infection in V. splendidus, collectively reveal the general overview of the impact of phages on host bacteria.
Here, we point out the existence of complex relationships between phages sensitivity and bacteria growth stage through through CPS translocon Wza in the liquid culture environment. Our findings provide strong evidence implicating CPS as a receptor for phage vB_VspM_VS2 in V. splendens. An interesting phenomenon we discovered is that cells locked in the growth stage d express low levels of wza, resulting in a low susceptibility to phages due to growth stage-mediated downregulation of CPS. In addition, wza expression was significantly decreased after phage vB_VspM_VS2 infection, and phage adsorption was significantly inhibited in V. splendidus. Interestingly, similar results were found in cell morphological changes, with cells becoming smaller, rounder, and smoother in both the growth stage and phage-infected state. Given these considerations, our results reveal important roles for V. splendidus CPS translocon Wza and cells locked in the growth stage d or phage-infected state, which allow V. splendidus to persist under phage predation, and hold important implications for using phages therapeutically to target pathogenic bacteria in dynamic communities.
Materials and methods
Bacterial strains, plasmids and growth conditions
V. splendidus AJ01 strain in our laboratory was isolated from a diseased sea cucumbers with skin ulcer syndrome53. The phage and bacterial strains used in the present study are listed in Supplementary Table S1. All primers used in the study are listed in Supplementary Table S2. V. splendidus was cultured in 2216E medium (5 g tryptone and 1 g yeast extract in 1 L seawater, pH 7.6–8.0) at 28 °C with shaking at 180 rpm. Plasmid constructions were performed in E. coli DH5α using standard methods, and the cells were cultured in LB broth medium (10 g tryptone, 5 g yeast extract and 10 g NaCl in 1 L water, pH 7.6–8.0) at 37 °Cwith shaking at 180 rpm.
Phage isolation and purification
V. splendidus phage was isolated from an Apostichopus japonicus breeding pond silt in Dalian, China. The propagation method was modified from Park54. Briefly, 2 g of a breeding pond silt sample was mixed with 5 mL phosphate buffer solution buffer in a 15 mL centrifuge tube and incubated with shaking at 180 rpm for 1 h at room temperature. Then, the sewage sample was centrifuged at 6500 × g for 4 min, and the supernatants were filtered with a 0.22 μm filter membrane. After filtration, 2 mL of each filtrate was inoculated onto log phase-grown V. splendidus in 15 mL of 2216E culture broth and incubated for 24 h. Then, the culture was centrifuged at 6500 × g for 4 min, and the supernatant was filtered with a 0.22 μm filter membrane. The filtrate was serially diluted 10 times, mixed with 5 mL molten 0.45% 2216E soft agar containing exponential phase V. splendidus (2 × 108 cfu/mL), and immediately added to a 2216E plate containing 1.8% agar. The growth of overnight cultures and plaque formation were observed after 12 h. A single phage plaque was selected for phage purification, and the process was repeated three times.
One-step growth curve, adsorption and lysis activity of the isolated phage
To measure the adsorption rate, the co-culture of phage vB_VspM_VS2 (1 × 108 pfu/mL) and exponential phase V. splendidus was mixed at an multiple of infection (MOI) of 1 and cultured at 28 °C. The phage titer was measured after 0, 5, 10, 15, 20, 25, and 30 min. The one-step growth curve of phage vB_VspM_VS2 was carried out as follows. Briefly, 20 mL of exponential phase V. splendidus culture was harvested by centrifugation (6500 g, 4 min, 4 °C), and the pellet was resuspended in 20 mL of fresh 2216E to obtain an OD600 of 1.0. Next, 20 mL of phage vB_VspM_VS2 was added to reach an MOI of 1 and allowed to adsorb for 15 min at 28 °C. The mixture was centrifuged at 6500 × g for 4 min at 4 °C, and the pellet was resuspended in 50 mL of fresh 2216E. Samples were taken every 10 min for 120 min, then the supernatants were plated on 2216E agar plate to determine the phage titer.
For phage adsorption and lysis activity experiments, stationary phase V. splendidus cells were resuspended in 2216E medium at the desired concentration and cultured at 28 °C for 12 h. At the same time, phage was added to the culture at a MOI of 0.1 and 1. Samples were taken every 1.5 h (0–9 h), and the optical density (OD600) was measured using a spectrophotometer. Phages titers in the filtrates were determined by titration on the double-layer agar. The phage adsorption rate was calculated as follows: adsorption rate = [(initial titer of phage − titer in the supernatant of phage)/(initial titer of phage)] × 100 (%). The lysis activity of phage was evaluated on the basis of their ability to form plaques on bacterial lawns. Lawns of V. splendidus (positive control), S. aureus, S. bongori, A. lwoffii and E. marmotae (negative control), V. harveyi and V. antiquarius were prepared as described above using the double layer agar plate method. Two microliter of phage suspension was spotted onto the pre-solidified bacterial lawn. The plates were incubated at 28 °C overnight. The formation lysis zone was evaluated.
TEM analyses of the isolated phage
The morphology of the phage vB_VspM_VS2 particles was analyzed using transmission electron microscopy (TEM) mainly with the steps described below. Dilutions of the phage vB_VspM_VS2 stock (~5 × 109 pfu/mL) were deposited on carbon film and stained with 2% uranyl acetate. Phage vB_VspM_VS2 samples were observed using a Philips EM 300 electron microscope operated at an acceleration voltage of 120 kV at Ningbo University (Ningbo, China). Phage vB_VspM_VS2 was identified and classified according to the International Committee on Taxonomy of Viruses.
Sequencing and analysis
Purified phage vB_VspM_VS2 was concentrated through a 10 kDa filter and treated with DNase and RNase at 37 °C for 1 h. A Takara Minibest Viral RNA/DNA Extraction Kit (Cat#9766) was used to extract phage vB_VspM_VS2 genomic DNA. Sequencing of the phage vB_VspM_VS2 genomic DNA was carried out using the Illumina HiSeq platform (Sangon Biotech, China) and assembled using SPAdes and FastQC assembler software. NCBI BLAST was used to compare sequences from multiple databases and open reading frames (ORFs), including TrEMBL, KOG, COG, CDD, NT, PFAM, SwissProt and NR, to obtain functional annotation information for the phage vB_VspM_VS2 gene protein sequences.
Total RNA extraction and sequencing
A total volume of 15 mL V. splendidus culture (with an optical density at 600 nm of 0.5) was infected with phage vB_VspM_VS2 at an MOI of 2, while the same volume of V. splendidus culture served as the control group. Seven samples for RNA isolation were taken from the non-infected/infected culture at three time points post-infection (2, 7, and 15 h). RNA extraction was performed using the Bacterial RNA extraction kit (Omega, China). RNA quality and quantity were checked using the Agilent 2100 bioanalyzer.
RNA sequencing analyses
Total RNA from all samples (300 ng) were depleted of rRNA using the Ribo-zero Kit. The cDNA libraries were sequenced and constructed on an Illumina HiSeq/ MiSeq sequencing platform (Illumina, Novogene, Beijing, China). RNA sequence reads were aligned to the V. atlanticus strain LGP32 (FM954973.2) sequences using Bowtie2. RNAseq data analysis was performed using HTSeq software. Gene expression values were determined using HTSeq software. Statistical analysis of gene expression was performed to calculate differentially expressed genes between non-infected and infected at 2, 7, and 15 h growth stages. Genes with a fold change value of less than 0.05 were considered differentially expressed genes. The cluster Profiler software was used to perform GO (gene ontology) term enrichment and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis. To further investigate the mechanism of phage infection regulated by Wza, 1% overnight culture of wild type (WT), Δwza or ΔluxR strains were reinoculated into 200 mL of 2216E liquid medium, respectively. The cells were grown to OD600 values of 0.2, 1.2, and 1.9, respectively, and the cell pellets were collected. Bacterial RNA extraction, RNA integrity detection, library construction, sequencing, GO and KEGG analysis methods were consistent with phage-infected methods.
Construction of mutant strains
To construct chromosomal deletions in V. splendidus, the flanking regions of the wza and luxR genes were cloned into pDM4. The plasmid was transferred into V. splendidus via conjugation with E. coli S17λpir harboring the recombinant plasmid. Positive clones were selected on TCI plates (3.9% Columbia blood agar base, 0.5% sodium thiosulphate, 0.5% sodium chloride, and 4.6% potassium iodide) plates containing 5 µg/mL chloramphenicol or Thiosulfate Citrate bile salts sucrose (TCBS, Becton Dickinson, NJ, USA), followed by selection for plasmid loss on LB plates containing 5% sucrose. For Δwza or ΔluxR mutant construction, the pDM4wza and pDM4 luxR plasmids were constructed as previously described38,55, using the primers listed in Supplementary Table S2.
Effects of cell-free supernatant enrichment on vB_VspM_VS2 phage infection
Cell free supernatant was prepared from mid-exponential phase of V. splendidus cultures in 2216E meida. The supernatant was obtained by centrifugation (12,000 g for 5 min), sterile filteration through a 0.22 μm membrane, and added to freshly inoculated cultures of wild type (WT), Δwza or ΔluxR strains, with and without phage vB_VspM_VS2, to examine the potential effects of autoinducer molecules produced by V. splendidus on the lytic effects of the phage vB_VspM_VS2. The cells were incubated for 24 h, and images were taken.
Outer membrane preparation and SDS-PAGE analysis
The outer membrane protein preparation was made as following methods56. Briefly, cells were pelleted from 50 mL overnight cultures of wild type (WT), Δwza or ΔluxR strains, resuspended in 5 mL of water, and sonicated on ice (amplitude of 120, 5 min). To solubilize the V. splendidus cytoplasmic membranes, N-lauroylsarcosine was added to a final concentration of 2% (w/w) and incubated at 25 °C for 30 min. To pellet the V. splendidus outer membranes, the mixture was ultracentrifuged (40,000 rpm for 1.5 h at 4 °C; SW41 Ti rotor). The pellet was washed with precooled water and ultracentrifuged again (40,000 rpm for 1 h at 4 °C; SW41 Ti rotor). The pellet was then resuspended in 100 μL 2% SDS and 100 mM Tris-HCl (pH 8) buffer and separated on a 12% SDS-polyacrylamide gel and stained with heating Coomassie blue.
Motility analysis
The bacterial motility analysis was measured as previously described by Wang et al. 57. The wild type, Δwza or ΔluxR strains were separately cultured overnight and diluted to 1:200 in fresh 2216E culture. Five microliters of wild type, Δwza or ΔluxR cultures (OD600 = 0.2, 1.2, and 2) were dropped onto 2216E plates containing 0.4% agar at 28 °Cfor 24 h. The colony size of motility circles of wild type, Δwza or ΔluxR strains on the agar plate was measured, respectively. All the experiments were repeated for three times.
TEM and SEM analyses of V. splendidus
The morphology of all wild type, Δwza strain, ΔluxR strain, EES-P, and different growth stages of a, b, c, and d of wild type strains were analyzed using TEM with the following steps. Dilutions of wild type, Δwza, ΔluxR, EES-P, and different growth stages (~5 × 107 cfu/mL) were deposited on carbon film and stained with 2% uranyl acetate. Cells samples were observed using a Philips EM 300 electron microscope operated at an acceleration voltage of 120 kV at Ningbo University (Ningbo, China). Next, the recovered cultures were washed twice with PBS buffer and fixed with 2.5% precooling glutaraldehyde at room temperature for 2 h in the dark. The cells were washed twice with PBS buffer and dehydrated with an increasing ethyl alcohol gradient (15%, 30%, 40%, 50%, 70%, 100% v v−1) for 15 min for each step. Afterwards, the cells were dried overnight, and the results were obtained through scanning electron microscopy (SEM) (SU-70) with an accelerating voltage of 20 kV.
Growth curve of wild type, Δwza and ΔluxR strains
Wild type, Δwza and ΔluxR strains seed were kept in −80 °C refrigerator and revived in 2216E medium broth with 4‰ (V/V), cultured at 28 °C for 12 h, then the same vaccination rate 4‰ (V/V) inoculated in fresh 2216E medium, also static cultured at 28 °C. Sampling the culture of every 1.5 h (0–36 h) and using spectrophotometer to determination optical density (OD600). The co-culture of phage vB_VspM_VS2 (1 × 108 pfu/mL) and logarithmic phase V. splendidus were mixed at an MOI of 0.1 and 1, cultured at 28 °C and measured optical density (OD600), meanwhile.
Effect of extracellular CPS on phage adsorption
Wild type CPS was extracted using an CPS extraction kit (iNtron Biotechnology, China). The concentration of CPS was determined using the phenol-sulfuric acid method. For CPS adsorption assays, 100 μL of extracted CPS (1 mg/mL) was added to 1 mL 2216E broth and mixed with the phage (108 pfu/mL) at 28 °C for 20 min to allow adsorption. Control samples were transferred into 2216E broth with 100 μL of phosphate-buffered saline (PBS) before mixing with phages. Samples were centrifuged at 10,000 g at 4 °C for 5 min, and then their titers were determined.
Phage sensitivity assay
Overnight cultures of wild type, Δwza and ΔluxR strains were inoculated in fresh 2216E medium for 16 h until they reached the late exponential growth phase (OD600, 1.5). Then, 200 mL of the culture was mixed with 5 mL 0.45% melted 2216E agar to prepare double-layered agar plates. In control samples, an equivalent volume of DMSO was added as a solvent control. The phage vB_VspM_VS2 were then subjected to a 10-fold gradient dilution in 2216E buffer, and 2-μL aliquots were spotted onto a plate and incubated at 28 °C for 16 h. Phage vB_VspM_VS2 was mixed with wild type strains at growth stage a, b, c, d, and the growth stage a infected with phage, according to the ratio of MOI = 1. The concentration of bacterial solution was adjusted to OD600 = 2, and the mixture was adsorbed in 5 mL of 2216E for 20 min, followed by contrifugation at 10,000 g for 2 min. Immediately after, 0.22 μm membrane filtration was carried out, the pfu value of phage in the supernatant was measured using the double-layer plate method, and the adsorption rate of host bacteria to phage under different growth conditions was calculated.
Quantification of mRNA expression
For qRT-PCR analyses, specific primers for V. splendidus were employed (Table S2). The total RNA of wild type, Δwza mutant, ΔluxR mutant, phage infected at MOI = 1 at 2, 6, 12 h states, as well as at different growth stages of EES, MES and LES of wild type strains were collected from triplicate cultures with varying OD600 values. The total RNA of cells was extracted with the Trizol reagent (Takara, Japan), and cDNA was prepared using gDNA Eraser Kit PrimeScript RT reagent (Takara, Japan). Real-time polymerase chain reaction (RT-PCR) was performed using a one-step relative real-time qRT-PCR in an ABI 7500 RT-PCR detection system (Applied Biosystems) with a total volume of 20 μL (0.8 μL of each primer [10 mM], 8 μL of cDNA, 0.4 μL of Dye-II [ROX], and 10 μL of TB GREENTM Premix Ex Taq II), followed by 40 cycles of 95 °C for 15 s, 60 °C for 20 s, 72 °C for 20 s. The relative expression level of genes was analyzed using the comparative critical threshold (2−ΔΔCT) method.
Effects of vB_VspM_VS2 on microcolony formation and initial bacterial attachment
To quantify the effect of vB_VspM_VS2 on initial bacterial attachment and microcolony formation on the wild type, Δwza mutant, ΔluxR mutant strains, 5 μL of diluted log-phase cultures (108 cfu/mL) was filtered onto 30_x0005_ + replicate 20-mm-diameter, 0.22 μm-pore-size polycarbonate filters and incubated on 2216E agar in 6 well plates58. Phage stock (108 pfu/mL, 2 mL) was added, and controls were prepared with the addition of 2 mL SM buffer (0.01% gelatin, 100 mM NaCl, 50 mM Tris-Cl [pH 7.5], 8 mM MgSO4). Duplicate filters were collected every 12 h for 24 h, transferred to a 20 mm diameter filtration manifold, and stained with 0.5% SYBR gold for 15 min (Invitrogen, USA), followed by 3 rinses with PBS buffer. The filters were then quantification by epifluorescence microscopy.
Fluorescence microscopy
For fluorescence microscopy, wild type and Δwza mutant cells (3 mL, OD600, 0.5) were stained with FITC (Sigma; 1 mg/ mL) and 4-6-diamidino-2-phenylindole, followed, centrifuged and suspended in 10 mL of 2216E. Infection experiments were carried out at an MOI = 5 (phage:bacteria). Samples were photographed using a laser scanning spectral confocal microscope (TCS SP2; Leica, Germany).
Adsorption experiments with SYBR gold labeled phages
Phage adsorption to selected wild type and Δwza mutant cells was tested by adding SYBR gold-labeled phage vB_VspM_VS2 to cultures followed by visual inspection of phage binding to the cells using laser scanning spectral confocal microscope59. Aliquots of 1 mL phage vB_VspM_VS2 were digested by addition of DNase I (Takara, Japan) at 28 °C for 1.5 h, and then stained with SYBR gold (final concentration, 5×, Invitrogen) overnight at 4 °C. Ten microliters of chloroform were then added to inactivate the DNase I. The above solution was filtered through a 50 kDa ultrafiltrationspin device at 1000 g for 1 h to remove the free SYBR gold. Labeled phage vB_VspM_VS2 was added to the log phase wild type and Δwza mutant cells, respectively, at an MOI of 5 and incubated at room temperature for 15 min. Solution were pelleted at 8000 g for 4 min, and the pellet was resuspended in a small volume of SM buffer, mixed with 0.45% (m/v) agarose preheated to around 40 °C, and transferred to a gel coated slide glass for a laser scanning spectral confocal microscope (TCS SP2; Leica, Germany).
Statistics and reproducibility
For all the statistical analyses and production of the graphs GraphPad Prism version 8 was used. The data are presented as the means ± SDs (n = 3) relative to the untreated group and shown in bar graphs. Error bar represent SDs. Data points on graphs represent three biological replicates. Statistical significance was performed using Dunnett’s test. P values in the present study is considered statistically significant denoted as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ23C190005, the National Natural Science Foundation of China (42206093, 32325050), Ningbo Yongjiang Talent Introduction Programme (No. 2021B-029-C), and the K.C. Wong Magna Fund at Ningbo University.
Author contributions
Conceived and designed the experiments: L.M.J., C.H.L., and J.S.W. Performed the experiments: D.M.T., C.H.L. Analyzed the data: C.H.L., L.M.J., D.M.T., J.S.X., and J.Q.L. Contributed reagents/materials/analysis tools: L.M.J. and C.H.L. Wrote the paper: L.M.J. All authors read and approved the final manuscript.
Peer review
Peer review information
Communications Biology thanks Francisco Rodriguez-Valera and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Tobias Goris.
Data availability
The data that support the findings of this study are available in the methods and/or Supplementary Material of this article. NCBI Accession: V. s-0: SAMN33923098; V. s-2: SAMN33923099; V. s-7: SAMN33923101; V. s-15: SAMN33923103; V. s-2-P: SAMN33923100; V. s-7-P: SAMN33923102; V. s-15-P: SAMN33923104. The complete genome sequence of phage vB_VspM_VS2 was deposited in the NCBI GenBank database with accession number of OQ722176.1. The Supplementary Materials-corresponding explanations for genes in Fig. 8 are provided in Supplementary Data 1. All statistical source data that underlie the graphs in figures are provided in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Liming Jiang, Jinsheng Wen.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07038-z.
References
- 1.Hatfull, G. F. Bacteriophage genomics. Curr. Opin. Microbiol.11, 447–453 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katharios, P., Kalatzis, P. G., Kokkari, C., Sarropoulou, E. & Middelboe, M. Isolation and characterization of a N4-like lytic bacteriophage infecting Vibrio splendidus, a pathogen of fish and bivalves. PLoS ONE12, e0190083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra, A. K. et al. Expression and lytic efficacy assessment of the Staphylococcus aureus phage SA4 lysin gene. J. Vet. Sci.14, 37–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutateladze, M. & Adamia, R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol.28, 591–595 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Aksyuk, A. A. & Rossmann, M. G. Bacteriophage assembly. Viruses3, 172–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzipilevich, E., Habusha, M. & Ben-Yehuda, S. Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell168, 186–199.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem.75, 39–68 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Whitfield, C., Wear, S. S. & Sande, C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu. Rev. Microbiol.74, 521–543 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Cuthbertson, L., Mainprize, I. L., Naismith, J. H. & Whitfield, C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev.73, 155–177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, C. et al. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature444, 226–229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matanza, X. M., López-Suárez, L., do Vale, A. & Osorio, C. R. The two-component system RstAB regulates production of a polysaccharide capsule with a role in virulence in the marine pathogen Photobacterium damselae subsp. damselae. Environ. Microbiol.23, 4859–4880 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Hao, G. et al. Bacteriophage SRD2021 recognizing capsular polysaccharide shows therapeutic potential in serotype K47 Klebsiella pneumoniae Infections. Antibiotics10, 894 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertozzi Silva, J., Storms, Z. & Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett.363, fnw002 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Cai, R. et al. Three capsular polysaccharide synthesis-related glucosyltransferases, GT-1, GT-2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae. Front Microbiol.10, 1189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, K., Young, R. & Zeng, L. Bacteriophage P1 does not show spatial preference when infecting Escherichia coli. Virology542, 1–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letarov, A. V. & Kulikov, E. E. Adsorption of bacteriophages on bacterial cells. Biochemistry82, 1632–1658 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Gong, Q. et al. Novel host recognition mechanism of the K1 capsule-specific phage of Escherichia coli: capsular polysaccharide as the first receptor and lipopolysaccharide as the secondary receptor. J. Virol.95, e0092021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter, N. T. et al. Phase-variable capsular polysaccharides and lipoproteins modify bacteriophage susceptibility in Bacteroides thetaiotaomicron. Nat. Microbiol5, 1170–1181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, L. et al. LuxS quorum sensing system mediating Lactobacillus plantarum probiotic characteristics. Arch. Microbiol.203, 4141–4148 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Abdullahi, Z. H., Marselin, F. N., Khaironizam, N. I. A., Fauzi, N. F. A. & Wan Maznah, W. O. Growth stage-related biomass, pigments, and biochemical composition of Stichococcus bacillaris, Synechococcus sp., and Trentepohlia aurea isolated from Gua Tempurung, a cave in Malaysia. Plant Physiol. Biochem.197, 107633 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Lee, T. H. et al. The impact of antibacterial peptides on bacterial lipid membranes depends on stage of growth. Faraday Discuss232, 399–418 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Silpe, J. E. & Bassler, B. L. Phage-encoded LuxR-type receptors responsive to host-produced bacterial quorum-sensing autoinducers. mBio10, e00638-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman, J. D. & van Kessel, J. C. Purification of the Vibrio quorum-sensing transcription factors LuxR, HapR, and SmcR. Methods Mol. Biol.2346, 173–182 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Silpe, J. E. & Bassler, B. L. A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell176, 268–280.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xuan, G., Lin, H., Tan, L., Zhao, G. & Wang, J. Quorum sensing promotes phage infection in Pseudomonas aeruginosa PAO1. mBio13, e0317421 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue, T., Matsuzaki, S. & Tanaka, S. A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett.125, 101–105 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Tan, D., Gram, L. & Middelboe, M. Vibriophages and their interactions with the fish pathogen Vibrio anguillarum. Appl. Environ. Microbiol.80, 3128–3140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, D., Dahl, A. & Middelboe, M. Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Appl. Environ. Microbiol.81, 4489–4497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Bertani, G. & Weigle, J. J. Host controlled variation in bacterial viruses. J. Bacteriol.65, 113–121 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luria, S. E. & Human, M. L. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol.64, 557–569 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, M. J., Stierhof, Y. D. & Henning, U. Location and unusual membrane topology of the immunity protein of the Escherichia coli phage T4. J. Virol.67, 4905–4913 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Díaz-Muñoz, S. L. & Koskella, B. Bacteria-phage interactions in natural environments. Adv. Appl. Microbiol.89, 135–183 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Olszak, T., Latka, A., Roszniowski, B., Valvano, M. A. & Drulis-Kawa, Z. Phage life cycles behind bacterial biodiversity. Curr. Med. Chem.24, 3987–4001 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Duerkop, B. A. et al. Murine colitis reveals a disease-associated bacteriophage community. Nat. Microbiol.3, 1023–1031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manrique, P. et al. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA113, 10400–10405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell160, 447–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, D., Svenningsen, S. L. & Middelboe, M. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio6, e00627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoque, M. M. et al. Quorum regulated resistance of Vibrio cholerae against environmental bacteriophages. Sci. Rep.6, 37956 (2016). Published 2016 Nov 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickerson, N. N. et al. Trapped translocation intermediates establish the route for export of capsular polysaccharides across Escherichia coli outer membranes. Proc. Natl. Acad. Sci. USA111, 8203–8208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, D., Yuminaga, Y., Shi, J. & Hao, J. Non-capsulated mutants of a chemical-producing Klebsiella pneumoniae strain. Biotechnol. Lett.40, 679–687 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Yi, H. et al. Identification of a wza-like gene involved in capsule biosynthesis, pathogenicity and biofilm formation in Riemerella anatipestifer. Micro. Pathog.107, 442–450 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Niu, T. et al. Wza gene knockout decreases Acinetobacter baumannii virulence and affects Wzy-dependent capsular polysaccharide synthesis. Virulence11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Sordi, L., Lourenço, M. & Debarbieux, L. The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract. Cell Host Microbe25, 210–218 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA110, 10771–10776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns, S. M. & Hull, S. I. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun.66, 4244–4253 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Høyland-Kroghsbo, N. M., Maerkedahl, R. B. & Svenningsen, S. L. A quorum-sensing-induced bacteriophage defense mechanism. mBio4, e00362-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Høyland-Kroghsbo, N. M. et al. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. USA114, 131–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patterson, A. G. et al. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol. Cell64, 1102–1108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, Z. et al. Global transcriptomic analysis of the interactions between phage φAbp1 and extensively drug-resistant Acinetobacter baumannii. mSystems4, e00068-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mojardín, L. & Salas, M. Global transcriptional analysis of virus-host interactions between phage ϕ29 and Bacillus subtilis. J. Virol.90, 9293–9304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kortright, K. E., Chan, B. K. & Turner, P. E. High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proc. Natl. Acad. Sci. USA117, 18670–18679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, W., Liang, W. & Li, C. Inhibition of marine Vibrio sp. by pyoverdine from Pseudomonas aeruginosa PA1. J. Hazard. Mater.302, 217–224 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Park, M. et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol.78, 58–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milton, D. L., O’Toole, R., Horstedt, P. & Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol.178, 1310–1319 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, S. Y., Lauritz, J., Jass, J. & Milton, D. L. Role for the major outer-membrane protein from Vibrio anguillarum in bile resistance and biofilm formation. Microbiology149, 1061–1071 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z. et al. Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Front. Microbiol.4, 379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Højberg, O., Binnerup, S. J. & Sørensen, J. Growth of silicone-immobilized bacteria on polycarbonate membrane filters, a technique to study microcolony formation under anaerobic conditions. Appl. Environ. Microbiol.63, 2920–2924 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunisaki, H. & Tanji, Y. Intercrossing of phage genomes in a phage cocktail and stable coexistence with Escherichia coli O157:H7 in anaerobic continuous culture. Appl. Microbiol. Biotechnol.85, 1533–1540 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that support the findings of this study are available in the methods and/or Supplementary Material of this article. NCBI Accession: V. s-0: SAMN33923098; V. s-2: SAMN33923099; V. s-7: SAMN33923101; V. s-15: SAMN33923103; V. s-2-P: SAMN33923100; V. s-7-P: SAMN33923102; V. s-15-P: SAMN33923104. The complete genome sequence of phage vB_VspM_VS2 was deposited in the NCBI GenBank database with accession number of OQ722176.1. The Supplementary Materials-corresponding explanations for genes in Fig. 8 are provided in Supplementary Data 1. All statistical source data that underlie the graphs in figures are provided in Supplementary Data 1.