Abstract
Background
Inflammatory bowel disease (IBD) is a complex autoimmune disorder, although some medications are available for its treatment. However, the long-term efficacy of these drugs remains unsatisfactory. Therefore, there is a need to develop novel drug targets for IBD treatment.
Methods
We conducted two-sample Mendelian randomization (MR) analysis using Genome-Wide Association Study (GWAS) data to assess the causal relationships between plasma proteins and IBD and its subtypes. Subsequently, the presence of shared genetic variants between the identified plasma proteins and traits was explored using Bayesian co-localization. Phenome-wide MR was used to evaluate evaluated adverse effects, and drug target databases were examined for therapeutic potential.
Results
Using the Bonferroni correction (P < 3.56e-05), 17 protein-IBD pairs were identified. Notably, the genetic associations of IBD shared a common variant locus (PP.H4 > 0.7) with five proteins (MST1, IL12B, HGFAC, FCGR2A, and IL18R1). As a subtype of IBD, ulcerative colitis shares common variant loci with FCGR2A, IL12B, and MST1. In addition, we found that ANGPTL3, IL18R1, and MST1 share a common variant locus with Crohn’s disease. Furthermore, phenome-wide MR analysis revealed that except for ANGPTL3, no other proteins showed potential adverse effects. In the drug database, identified plasma proteins such as FCGR2A and IL18R1 were found to be potential drug targets for the treatment of IBD and its subtypes.
Conclusion
Six proteins (FCGR2A, IL18R1, MST1, HGFAC, IL12B, and ANGPTL3) were identified as potential drug targets for the treatment of IBD and its subtypes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00384-024-04744-2.
Keywords: Drug targets, Inflammatory bowel disease, Mendelian randomization
Introduction
Inflammatory bowel disease (IBD) is a prevalent global inflammatory disorder, comprising two main subtypes, ulcerative colitis (UC) and Crohn’s disease (CD). In the past three decades, IBD has had a significant impact on human health, with the global incidence rate increasing gradually [1] and a trend toward younger onset [2]. The precise etiology of IBD remains unclear, with researchers implicating a multifactorial interplay involving systemic immune responses, genetic factors, and disruptions in the intestinal microbiota [3–5]. There are a range of therapeutic approaches for managing IBD. For example, the application of traditional 5-aminosalicylic acid (5-ASA), corticosteroids, and immunosuppressants (e.g., thiopurines and methotrexate) are commonly used for treatment [6]. In recent years, several emerging drugs have been used to treat IBD, including biological agents (e.g., anti-TNF agents and cytokine inhibitors), anti-integrins (e.g., vedolizumab and natalizumab), and S1P inhibitor (ozanimod) [7–10]. However, this remains a vexing challenge as IBD is currently incurable. Furthermore, with prolonged treatment duration, a notable decline in the efficacy of certain medications has been observed. For instance, a study by Qiu et al. revealed that approximately one-third of IBD patients experience diminished drug responsiveness following prolonged usage of anti-TNF antibodies [11]. Therefore, the exploration of novel drug targets for IBD and the development of highly targeted, low-side effect therapies hold significant clinical significance.
Currently, most sequencing technologies focus on the DNA base sequences of tissues or cells or employ RNA-seq to study the impact of gene expression on disease biology [12, 13]. However, proteins are the primary executors of biological effects in the body, and there is substantial variability in the biological processes governing protein translation. Moreover, most drugs exert their pharmacological effects by modulating protein expression, especially in the case of plasma proteins [14]. In addition, many previous studies have explored the feasibility of plasma proteins as drug targets, such as type 2 diabetes [15], ischemic heart disease [16], and chronic kidney disease [17]. These proteins participate in a range of complex systemic biological processes through their circulation in the blood, making the development of therapeutic targets for IBD a promising avenue. Thus, plasma proteins were selected as the subjects of our study, in which we employed genetic variations related to proteins, known as protein quantitative trait loci (pQTL), in conjunction with IBD and its subtypes GWAS summary statistics as the foundation for a two-sample Mendelian randomization (MR) analysis [18–20]. This approach offers a fresh perspective on understanding the etiology and treatment of IBD since GWAS has identified numerous genetic loci associated with disease risk, thus providing evidence to elucidate the molecular pathways underlying these diseases [21].
In this study, we combined GWAS data with multiple large-scale proteomic studies to explore potential therapeutic targets for IBD and its subtypes. Firstly, we used MR to identify potential causal plasma proteins for IBD and its subtypes using GWAS data. Subsequently, Steiger filtering, Bayesian co-localization, and PheWAS analyses were implemented to consolidate the MR findings. Finally, we replicated the analysis using GWAS data from the FinnGen cohorts and plasma pQTL data from the study by Ferkingstad et al. as external validation to strengthen our conclusions. Additionally, we conducted a preliminary exploration of drugs related to potential target proteins and their respective mechanisms using a drug database.
Methods
Data sources
We obtained pQTL data from three separate studies on plasma proteins: Zheng et al. [22], which included data from five previously published GWAS studies [20, 23–26]; Pietzner et al. [19]; and Ferkingstad et al. [27]. Subsequently, we conducted preliminary screening of the pQTL data, with inclusion in the MR analysis contingent upon satisfying the following criteria: (a) selection of tier 1 or sentinel cis-pQTLs; (b) exclusion of the MHC region (GRCh38: chr6: from 29 to 33 Mb; GRCh37: chr6: from 26 to 34 Mb); (c) exclusion of proteins located on the sex chromosomes; (d) meeting linkage disequilibrium (LD) clumping criteria with r2 < 0.001; (e) meeting genome-wide significance levels with P < 5 × 10−8.
The GWAS summary statistics for the outcome variables (IBD, UC, and CD) were sourced from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). To enhance the reliability of our results, multiple external validations were conducted by utilizing IBD and its subtypes GWAS statistics from the FinnGen cohorts. In addition, to minimize potential bias from population diversity, we focused on individuals of European ancestry. These GWAS summary statistics were subsequently employed as outcome variables in the MR analysis. All research data were obtained through open data, and ethical approval was not required.
Statistical analysis
To include a broader range of circulating plasma proteins in our two-sample MR analysis, the filtered cis-pQTL data from Zheng et al. and Pietzner et al. were merged. However, multiple variant loci may appear in the same protein when we integrate protein data from different studies. To address this, we calculated the R-squared (R2) value for each variant locus using the formula [28], where EAF is the effect allele frequency (EAF) of the SNP and β is the estimated effect of SNP on trait. To determine the best instrumental variable for each circulating protein, the variant locus with the highest R2 value was selected as the sole instrumental variable [29]. Additionally, the impact of weak instrumental variables was eliminated through formulas: [30], because F > 10 suggested sufficient strength to ensure the validity of the SNPs. In contrast to cis-pQTL, trans-pQTL were located further away from the coding regions of the target protein genes. To avoid potential false-positive results, trans-pQTL was excluded from the MR analysis in this study [22].
MR analysis
MR, which follows the principle of random allocation of genetic variations, effectively avoids the influence of confounding factors and reverse causality when using genetic variations as instrumental variables to investigate potential causal relationships between exposure factors and traits, thus providing us with more definitive results [31, 32]. Therefore, we used plasma proteins as exposure factors and IBD and its subtypes as outcome variables. MR analysis was conducted using the “TwoSampleMR” package (Version 0.6.2) (https://github.com/MRCIEU/TwoSampleMR) in the R studio (Version 4.3.1). The MR approach was grounded on the following assumptions: (a) the genetic variants used as IVs were associated with plasma protein levels; (b) there were no other confounding factors influencing the relationship between plasma proteins and the outcomes; (c) the genetic variants exclusively affect the outcomes through changes in plasma protein levels [32].
In this study, we selected the best single SNP for each protein for MR analysis; the Wald ratio analysis method was employed for result analysis [29, 33]. For statistical correction of MR analysis results, we used Bonferroni correction, where we considered an instrument variable to have a causal effect on the disease if it met the threshold (P < 0.05/N, where N is the number of plasma proteins included in the final MR analysis). Additionally, it indicated that the protein increased the risk of the disease when the odds ratio (OR) value was greater than 1.
Reverse causality detection analysis
To ensure the correct direction of causality between the exposure protein and the outcomes of IBD, UC, and CD, Steiger filtering analysis was also performed by us. We used the same control criteria for external results validation with different plasma protein datasets as exposure conditions or with different IBD datasets as outcome variables. It was considered as evidence of the protein’s effect on the disease due to changes in the protein levels when P < 0.05.
Bayesian co-localization analysis
To assess whether two traits share a common variant locus, the “coloc” package (Version 5.2.3) (https://github.com/chr1swallace/coloc) was employed to conduct Bayesian co-localization analysis on candidate proteins (P1 = 1e-04, P2 = 1e-04, P12 = 1e-05). According to the five Bayesian hypothesis principles, no association with either trait (H0), association with trait 1, not with trait 2 (H1), association with trait 2, not with trait 1 (H2), association with trait 1 and trait 2, two independent SNPs (H3), association with trait 1 and trait 2, one shared SNP (H4). It was considered that there might be a correlation between two traits driven by the same causal variant locus when hypothesis H4 is met (PP.H4 > 0.7) [34]. To do this, the summary data studied by Ferkingstad et al. were obtained from the Decode database (https://www.decode.com/summarydata/). And all SNPs within ± 500 kb of the lead cis-pQTL variant for the target protein were selected for co-localization analysis. The target protein may directly mediate the disease risk associated with the variation, rather than being influenced by other biological processes when the co-localization analysis indicated a shared genetic variation between two traits.
Phenome-Wide Association Study (PheWAS)
To investigate whether these identified proteins have causal relationships with other phenotypes, we utilized the R package “ieugwasr” (Version 1.0.0) (https://mrcieu.github.io/ieugwasr/) and conducted a PheWAS using the phewas function. This analysis aimed to assess the relationships between the identified proteins and all phenotypes available in the UK Biobank, as provided by the IEU Open GWAS database (https://gwas.mrcieu.ac.uk/). The IEU Open GWAS database contains a comprehensive collection of 42,348 GWAS summary datasets, enabling a thorough exploration of potential causal relationships between the identified proteins and a wide range of human traits. In this study, the traits related to ICD-10 diagnosis codes from the UK Biobank database were specifically selected as outcome variables for the PheWAS analysis. And Bonferroni correction was implemented for quality control to account for multiple comparisons.
Drug target analysis
Based on the results from previous MR and Bayesian co-localization analyses, we selected plasma proteins that have causal relationships and co-localization with IBD and its subtypes for further potential drug target analysis. Next, we used Drugbank (https://go.drugbank.com/) and Therapeutic Target Database (http://db.idrblab.net/ttd/) to analyze drugs related to these potential drug targets and their respective mechanisms. These databases provide insights into the current state of research on drugs that target these proteins for the treatment of various diseases.
Results
Data overview
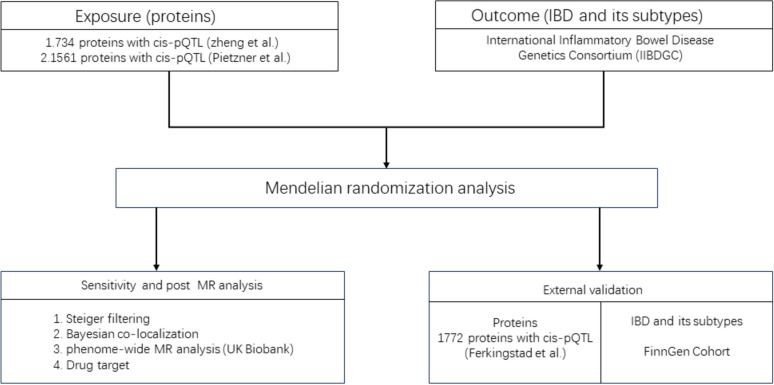
Following the filtering criteria described earlier, 734 plasma proteins from Zheng et al.’s study (Supplementary Table 1) and 1561 proteins from Pietzner et al.’s study (Supplementary Table 2) were obtained. To identify the most suitable single lead SNP for each plasma protein for MR analysis, we merged the proteins from both Zheng et al. and Pietzner et al., resulting in a final set of 1614 unique leader cis-pQTLs for MR analysis (Supplementary Table 3). As outcome data source, IIBDGC is a global collaboration comprising hundreds of researchers from over 20 countries across four continents, encompassing data from over 75,000 patients with IBD. In the IIBDGC database, IBD data included 12,882 cases and 21,770 controls, UC data included 6968 cases and 20,464 controls, and CD data included 5956 cases and 14,927 controls. As external validation data, the Ferkingstad et al. dataset contained 1772 plasma proteins. In FinnGen database, IBD data included 7625cases and 369,652 controls, UC data included 5034 cases and 371,530 controls, and CD data included 2007cases and 359,927 controls. In this study, the specific analysis flowchart was illustrated in Fig. 1.
Fig. 1.
Schematic diagram of specific experimental design for MR analysis
Potential drug targets for the treatment of IBD
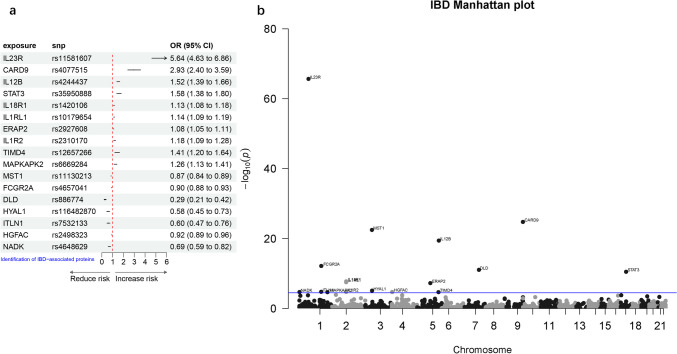
To investigate the causal relationships between 1614 plasma proteins (unique leader cis-pQTLs) and IBD. Upon harmonizing the data through the utilization of the harmonize_data function within the TwoSampleMR package, a total of 1403 proteins were obtained for two-sample MR analysis. After Bonferroni correction (P < 3.56e-05, 0.05/1403), we identified 17 proteins with causal relationships with IBD. Among them, ten proteins (IL23R, CARD9, IL12B, STAT3, IL18R1, IL1RL1, ERAP2, IL1R2, TIMD4, MAPKAPK2) were found to increase the risk of IBD. Our careful analysis found that the interleukin family accounted for 50% of the ten risk proteins, which is consistent with previous studies showing that the interleukin family plays an important role in IBD disease progression [35]. STAT3, a member of the JAK/STAT pathway, is also a potential risk protein [36]. However, seven proteins (MST1, FCGR2A, DLD, HYAL1, ITLN1, HGFAC, NADK) were associated with a decreased risk of IBD (Table 1, Fig. 2). Furthermore, Steiger filtering analysis indicated that there was no reverse causality between our exposure and outcome.
Table 1.
The causal relationship between plasma proteins and IBD was analyzed by MR analysis
| Exposure | Outcome | snp | pval | OR (95%CI) | R2 | F | steiger_dir | steiger_pval | Sources |
|---|---|---|---|---|---|---|---|---|---|
| IL23R | IBD | rs11581607 | 2.28E-66 | 5.64 (4.63–6.86) | 0.022054 | 73.46939 | TRUE | 1.98E-03 | Zheng et al |
| CARD9 | IBD | rs4077515 | 1.80E-25 | 2.93 (2.40–3.60) | 0.013607 | 154.761 | TRUE | 7.37E-09 | Pietzner et al |
| IL12B | IBD | rs4244437 | 3.97E-20 | 1.52(1.39–1.66) | 0.102585 | 1308.832 | TRUE | 2.84E-155 | Pietzner et al |
| STAT3 | IBD | rs35950888 | 3.31E-11 | 1.58(1.38–1.80) | 0.031953 | 415.219 | TRUE | 1.74E-47 | Pietzner et al |
| IL18R1 | IBD | rs1420106 | 1.81E-08 | 1.13(1.08–1.18) | 0.287255 | 1222.309 | TRUE | 2.90E-197 | Zheng et al |
| IL1RL1 | IBD | rs10179654 | 3.10E-08 | 1.14(1.09–1.19) | 0.268339 | 5168.482 | TRUE | 0 | Pietzner et al |
| ERAP2 | IBD | rs2927608 | 5.75E-08 | 1.08(1.05–1.11) | 0.666989 | 22,832.39 | TRUE | 0 | Pietzner et al |
| IL1R2 | IBD | rs2310170 | 1.91E-05 | 1.18(1.09–1.28) | 0.091001 | 1162.917 | TRUE | 4.48E-163 | Pietzner et al |
| TIMD4 | IBD | rs12657266 | 2.22E-05 | 1.41(1.20–1.64) | 0.021831 | 243.7193 | TRUE | 8.99E-31 | Pietzner et al |
| MAPKAPK2 | IBD | rs6669284 | 2.29E-05 | 1.26(1.13–1.41) | 0.048292 | 54.89463 | TRUE | 6.62E-11 | Zheng et al |
| MST1 | IBD | rs11130213 | 3.58E-23 | 0.87(0.84–0.89) | 0.627728 | 18,526.23 | TRUE | 0 | Pietzner et al |
| FCGR2A | IBD | rs4657041 | 7.31E-13 | 0.90(0.88–0.93) | 0.741726 | 29,463 | TRUE | 0 | Pietzner et al |
| DLD | IBD | rs886774 | 9.24E-12 | 0.29(0.21–0.42) | 0.004341 | 48.28206 | TRUE | 5.83E-03 | Pietzner et al |
| HYAL1 | IBD | rs116482870 | 8.37E-06 | 0.58(0.45–0.73) | 0.00959 | 107.5644 | TRUE | 5.77E-12 | Pietzner et al |
| ITLN1 | IBD | rs7532133 | 1.78E-05 | 0.60(0.47–0.76) | 0.009995 | 124.2569 | TRUE | 2.18E-14 | Pietzner et al |
| HGFAC | IBD | rs2498323 | 1.98E-05 | 0.92(0.89–0.96) | 0.409896 | 7502.529 | TRUE | 0 | Pietzner et al |
| NADK | IBD | rs4648629 | 2.12E-05 | 0.69(0.59–0.82) | 0.021121 | 236.2483 | TRUE | 1.05E-29 | Pietzner et al |
Fig. 2.
The causal relationship between plasma proteins and IBD analyzed by MR analysis (a) Forest plot, (b) Manhattan plot
To investigate whether the association of the variant between proteins used as IVs and IBD outcomes are shared. Bayesian co-localization analysis was conducted on the 17 proteins that were identified to have a causal relationship with IBD. We found that five of these proteins (FCGR2A, IL18R1, MST1, HGFAC, IL12B) may share variant loci with IBD (PP.H4 > 70%). Among them, FCGR2A (PP.H4 = 75.2%) and IL18R1 (PP.H4 = 71.7%) showed moderate co-localization with IBD. Notably, MST1 (PP.H4 = 98%), HGFAC (PP.H4 = 93.6%), and IL12B (PP.H4 = 95.2%) exhibited strong co-localization with IBD, indicating a robust shared variant locus (Supplementary Table 4).
To comprehensively assess whether the identified proteins with shared co-localization exhibit any side effects, the PheWAS analysis was conducted. The results were corrected through Bonferroni correction. It indicated that there were no potential causal relationships between the five identified proteins (MST1, IL12B, FCGR2A, IL18R1, HGFAC) and other traits in the UK Biobank diagnoses ICD10 dataset (Supplementary Table 5–9). This indirectly suggested that these five proteins have the potential to be used as treatments for IBD with minimal side effects and significant targeting potential.
In addition, we explored proteins causally related to IBD as potential drug targets. Our analysis of the drug dataset showed that drugs targeting FCGR2A, MST1, and IL12B have been approved for the treatment of certain diseases. For example, the drug Daclizumab, which targets FCGR2A, can be used to treat multiple sclerosis [37]. Interestingly, the drugs Ustekinumab, which targets IL12B, or Briakinumab, can be used for the treatment of IBD [38, 39], which is in line with our study. However, the targeting potential of IL18R1 and HGFAC has yet to be developed, suggesting potential areas for further research (Supplementary Table 10).
Potential drug targets for the treatment of UC
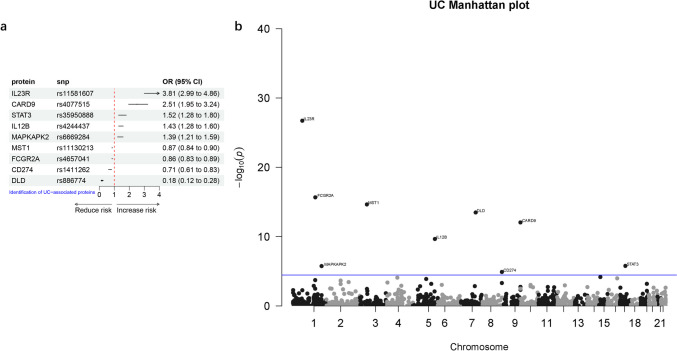
Next, for a more precise study of IBD, we conducted an analysis focusing on one of its subtypes, UC. We conducted a two-sample MR analysis using a total of 1437 proteins obtained by harmonizing the plasma protein cis-pQTLs and UC GWAS data. After correcting for multiple testing using Bonferroni correction (P < 3.48e-05, 0.05/1437), we identified nine proteins with causal relationships with UC (Table 2, Fig. 3). Among them, five proteins (IL23R, CARD9, IL12B, STAT3, MAPKAPK2) were found to increase the risk of UC. In contrast, three proteins, MST1, FCGR2A and DLD, were associated with a reduced risk of UC. It is worth noting that the Steiger filtering analysis indicated that CD274 (P = 0.16) was excluded from further analysis due to the presence of reverse causality, while the remaining proteins showed no reverse causality with the outcome (Table 2).
Table 2.
The causal relationship between plasma proteins and UC was analyzed by MR analysis
| Exposure | Outcome | snp | pval | OR (95%CI) | R2 | F | steiger_dir | steiger_pval | Sources |
|---|---|---|---|---|---|---|---|---|---|
| IL23R | UC | rs11581607 | 1.92E-27 | 3.81 (2.99–4.86) | 0.022054 | 73.46939 | TRUE | 6.35E-06 | Zheng et al |
| CARD9 | UC | rs4077515 | 9.07E-13 | 2.51 (1.95–3.24) | 0.013607 | 154.761 | TRUE | 1.57E-11 | Pietzner et al |
| STAT3 | UC | rs35950888 | 1.72E-06 | 1.52 (1.28–1.80) | 0.031953 | 415.219 | TRUE | 1.63E-48 | Pietzner et al |
| IL12B | UC | rs4244437 | 2.22E-10 | 1.43 (1.28–1.60) | 0.102585 | 1308.832 | TRUE | 2.28E-157 | Pietzner et al |
| MAPKAPK2 | UC | rs6669284 | 1.81E-06 | 1.39 (1.21–1.59) | 0.048292 | 54.89463 | TRUE | 2.66E-10 | Zheng et al |
| MST1 | UC | rs11130213 | 2.31E-15 | 0.87 (0.84–0.90) | 0.627728 | 18,526.23 | TRUE | 0 | Pietzner et al |
| FCGR2A | UC | rs4657041 | 2.18E-16 | 0.86 (0.83–0.89) | 0.741726 | 29,463 | TRUE | 0 | Pietzner et al |
| CD274 | UC | rs1411262 | 1.29E-05 | 0.71 (0.61–0.83) | 0.030452 | 345.5225 | FALSE | 0.16395 | Pietzner et al |
| DLD | UC | rs886774 | 3.35E-14 | 0.18 (0.12–0.28) | 0.004341 | 48.28206 | TRUE | 0.061251 | Pietzner et al |
Fig. 3.
The causal relationship between plasma proteins and UC analyzed by MR analysis (a) Forest plot, (b) Manhattan plot
To investigate whether the association of the variant between proteins used as IVs and UC outcomes is shared, Bayesian co-localization analysis revealed that three proteins (FCGR2A, MST1, IL12B) exhibited very strong co-localization with UC (Supplementary Table 4), with MST1 (PP.H4 = 98.2%), IL12B (PP.H4 = 99.3%), and FCGR2A (PP.H4 = 100%) showing robust shared variant loci.
Similarly, we used the same variable control strategy for the PheWAS analysis of shared variant site proteins. PheWAS analysis demonstrated that these three proteins (FCGR2A, MST1, IL12B) had no associations with other traits (Supplementary Table 5–7). In drug datasets, drugs that target IL12B have been shown to be useful for the treatment of UC [40]. Focusing on FCGR2A and MST1 as potential drug targets could significantly advance drug development for UC (Supplementary Table 10).
Potential drug targets for the treatment of CD
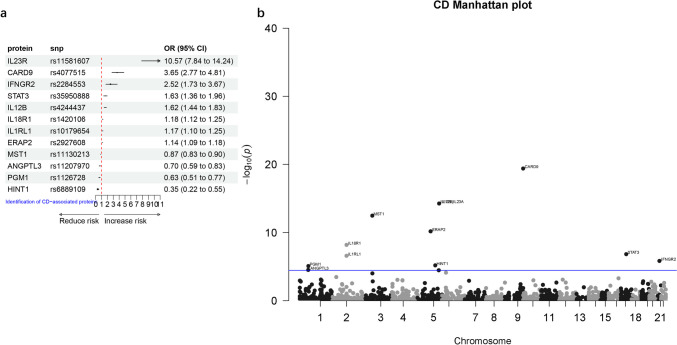
Finally, we conducted a preliminary study on potential drug targets for another subtype of IBD, CD. After harmonizing the data, a total of 1437 proteins were obtained for the two-sample MR analysis. The results revealed twelve proteins with causal relationships with CD. Among them, eight proteins (IL23R, CARD9, IFNGR2, STAT3, IL12B, IL18R1, IL1RL1, ERAP2) were found to increase the risk of CD. Conversely, four proteins (MST1, ANGPTL3, PGM1, HINT1) were associated with a reduced risk of CD (Table 3, Fig. 4). To avoid reverse causality, it was indicated that there was no reverse causality between the twelve proteins and CD according to Steiger filtering analysis.
Table 3.
The causal relationship between plasma proteins and CD was analyzed by MR analysis
| Exposure | Outcome | snp | pval | OR (95% CI) | R2 | F | steiger_dir | steiger_pval | Sources |
|---|---|---|---|---|---|---|---|---|---|
| IL23R | CD | rs11581607 | 3.61E-54 | 10.57 (7.84–14.24) | 0.022054 | 73.46939 | TRUE | 0.02626242 | Zheng et al |
| CARD9 | CD | rs4077515 | 4.03E-20 | 3.65 (2.77–4.81) | 0.013607 | 154.761 | TRUE | 2.08E-06 | Pietzner et al |
| IFNGR2 | CD | rs2284553 | 1.49E-06 | 2.52 (1.73–3.67) | 0.007207 | 79.82183 | TRUE | 8.45E-06 | Pietzner et al |
| STAT3 | CD | rs35950888 | 1.51E-07 | 1.63 (1.36–1.96) | 0.031953 | 415.219 | TRUE | 5.62E-41 | Pietzner et al |
| IL12B | CD | rs4244437 | 5.52E-15 | 1.62 (1.44–1.83) | 0.102585 | 1308.832 | TRUE | 2.06E-130 | Pietzner et al |
| IL18R1 | CD | rs1420106 | 6.57E-09 | 1.18 (1.12–1.25) | 0.287255 | 1222.309 | TRUE | 4.75E-180 | Zheng et al |
| IL1RL1 | CD | rs10179654 | 2.62E-07 | 1.17 (1.10–1.25) | 0.268339 | 5168.482 | TRUE | 0 | Pietzner et al |
| ERAP2 | CD | rs2927608 | 6.75E-11 | 1.14 (1.09–1.18) | 0.666989 | 22,832.39 | TRUE | 0 | Pietzner et al |
| MST1 | CD | rs11130213 | 3.39E-13 | 0.87 (0.83–0.90) | 0.627728 | 18,526.23 | TRUE | 0 | Pietzner et al |
| ANGPTL3 | CD | rs11207970 | 3.11E-05 | 0.70 (0.59–0.83) | 0.035909 | 435.4178 | TRUE | 3.47E-47 | Pietzner et al |
| PGM1 | CD | rs1126728 | 8.38E-06 | 0.63 (0.51–0.77) | 0.028997 | 97.94174 | TRUE | 6.12E-14 | Zheng et al |
| HINT1 | CD | rs6889109 | 6.68E-06 | 0.35 (0.22–0.55) | 0.004849 | 58.64065 | TRUE | 0.0003197 | Pietzner et al |
Fig. 4.
The causal relationship between plasma proteins and CD analyzed by MR analysis (a) Forest plot, (b) Manhattan plot
Similarly, Bayesian co-localization analysis was conducted for the proteins that were identified to have a causal relationship with CD. The results showed that ANGPTL3, IL18R1, and MST1 shared variant loci with CD, with MST1 (PP.H4 = 94.2%), IL18R1 (PP.H4 = 96.5%), and ANGPTL3 (PP.H4 = 72.3%) exhibiting strong co-localization (Supplementary Table 4).
Subsequently, the PheWAS analysis revealed that MST1 and IL18R1 had no associations with other traits (Supplementary Table 5, 8). However, ANGPTL3 was correlated with hypercholesterolemia and hyperlipidemia, indicating the need for special attention to the potential side effects related to ANGPTL3 (Supplementary Table 11). In the drug database, MST1, IL18R1, and ANGPTL3 are candidate drug targets for CD, and there is currently no corresponding drug research (Supplementary Table 10). Therefore, research targeting these targets may provide promising research directions.
Bidirectional external validation of the causal relationship between plasma proteins and IBD and its subtypes
Multiple validations were conducted to enhance the credibility of our research results. First, we kept the outcome variable data unchanged and conducted external validation by selecting other plasma protein data as exposure factors. A total of 1772 proteins obtained from the study by Ferkingstad et al. were used as exposure factors. IBD and its subtypes from the IIBDGC database were used as outcome variables. We strictly followed the filtering criteria used in the previous analysis for external validation. After harmonization, a total of 1132 proteins were included in the two-sample MR analysis with IBD as the outcome variable. The results showed that twelve plasma proteins (MST1, IL12B, FCGR2A, STAT3, TNFSF15, IL1RL1, ERAP2, HYAL1, HGFAC, MAPKAPK2, NADK, and TIMD4) were causally related to IBD. Among them, except for TNFSF15, the other eleven plasma proteins were consistent with our research findings. When CD was used as the outcome variable, a total of 1129 proteins were included in the two-sample MR analysis. The results showed that eight plasma proteins (MST1, IL12B, ERAP2, IL1RL1, TNFSF15, STAT3, ATF6B, and TIMD4) had causal relationships with CD, and MST1, IL12B, ERAP2, IL1RL1, and STAT3 were consistent with our previous analysis results. Lastly, when UC was used as the outcome variable, seven plasma proteins (MST1, IL12B, FCGR2A, MAPKAPK2, STAT3, CD274, and NQ01) were found to have causal relationships with UC, and all except NQO1 were consistent with our previous research results (Supplementary Tables 12–14).
Next, we kept the exposure data unchanged and conducted external validation by selecting other IBD and its subtype data as outcome variables. The unique proteins obtained from the combined data of Zheng et al. and Pietzner et al. were used as exposures. The IBD and its subtypes GWAS summary statistics from the FinnGen database were used as outcome variables for two-sample MR analysis. Using the same filtering criteria as previously described, the results showed that six plasma proteins (MST1, IL23R, HLA-DQA2, FCGR2A, FCGR3A, and FCGR3B) had causal relationships with IBD, among which MST1, IL23R, and FCGR2A were consistent with our previous analysis results. Interestingly, when UC was used as an outcome variable, the results showed that the same six plasma proteins as IBD were causally associated with UC. Among them, MST1, IL23R, and FCGR2A were consistent with our previous UC analysis results (Supplementary Tables 15–16). In addition, under the same conditions, no external validation of CD as an outcome variable was performed because there was no independent CD data in the FinnGen dataset.
Discussion
In this study, two-sample MR analysis was conducted to investigate the causal relationships between a large number of plasma proteins and IBD and its subtypes. A total of 22 proteins that might have causal associations with IBD and its subtypes were identified. Through Bayesian co-localization and PheWAS analyses, we explored the potential of these identified proteins as therapeutic targets for IBD and its subtypes. Ultimately, six proteins (FCGR2A, IL18R1, MST1, HGFAC, IL12B, ANGPTL3) were identified as potential drug targets for the treatment of IBD and its subtypes. Among them, MST1 serves as a shared potential target for IBD and its subtypes, whereas IL18R1 is a potential target for both IBD and CD. In addition, FCGR2A and IL12B are potential targets for IBD and UC. Interestingly, HGFAC is the only potential target for the treatment of IBD and ANGPTL3 has been identified as the sole potential target for the treatment of CD.
Currently, several drugs targeting IL12 inhibitors have been developed, such as Ustekinumab, which has been approved for the treatment of IBD and UC [38, 40]. Mechanistically, it primarily aims to interfere with Th1/Th17-mediated adaptive immune responses by targeting the P40 subunit shared by IL12 and IL23. In our study, we also confirmed that IL12B (rs424437) is a viable drug target for IBD and UC. However, we did not identify IL12B as a potential drug target for CD. Surprisingly, studies have shown that the IL12B inhibitor Ustekinumab can also inhibit the progression of CD [41]. This discrepancy may be due to incomplete data, and we plan to analyze additional data in the future. It should also be noted that age is an independent risk factor for the use of new biological agents in the elderly. Therefore, the use of biological agents in the elderly should be comprehensively evaluated for their risks and used with caution [42].
Previous research has demonstrated that IL18 is a significant factor in IBD progression [43]. IL18R1 acts as the downstream target of IL18 signaling, blocking the binding of IL18 to IL18R1 and reducing the risk of inflammatory and some autoimmune diseases [44]. The rationale behind this is that intestinal epithelial cells can secrete IL18, which acts on CD4 T cells expressing IL18R1 to limit Th17 cell differentiation, thereby maintaining barrier function in the gut [43]. Moreover, Nowarski et al. showed that blocking IL18RL inhibited the progression of inflammatory bowel disease in a mouse model of colitis [45]. Therefore, it is theoretically feasible to study drugs that target IL18R1 for the treatment of IBD and its subtypes. Our study also found that IL18R1 is a potential drug target for IBD and CD, indicating a significant research potential for drugs targeting IL18R1.
The identification of genetic variants in the 3p21-22 region as a high-risk factor for IBD is intriguing, especially because MST1 is located in this region [46]. Some polymorphic loci of MST1 have been confirmed to play important roles in the biological processes of IBD and its subtypes progression [47]. Our study identified MST1 as a potential target for IBD, UC, and CD with strong evidence. This result is consistent with a study by Lee et al., which found that MST1 can negatively regulate TNFα-induced NF-KB signaling by targeting LUBAC, thereby inhibiting inflammation [48]. Interestingly, no studies have targeted MST1 for the treatment of IBD and its subtypes, and our findings support the feasibility of targeting MST1 for treatment.
Fcγ receptors refer to a family of receptors located on the cell surface. It is expressed by various innate and adaptive immune cells and mediates inflammatory responses by binding to the Fc portion of immunoglobulin G (IgG) [49]. Some studies have found that FCGR2A plays a crucial role in inflammation and autoimmune diseases such as sepsis, systemic lupus erythematosus, Kawasaki disease, and UC [50–53]. Our results indicate that FCGR2A is a potential drug target for the treatment of IBD and UC, consistent with the findings of McGovern et al. [53]. The mechanism by which FCGR2A is involved in immunity and inflammation is thought to play a central role in antigen–antibody complex recognition, and FCGR2A can be regulated by multiple proximal and distal genomic regions [54]. Currently, drugs targeting FCGR2A with known pharmacological effects include antagonists (human immunoglobulin G), agonists (Catumaxomab), and the FCGR2A-targeting drug SM-101, which has been used to treat idiopathic thrombocytopenic purpura and systemic lupus erythematosus. Therefore, the development of drugs targeting FCGR2A for the treatment of IBD and UC holds significant research value.
In addition, we found that ANGPTL3 is a potential drug target for the treatment of CD. Regarding the mechanism of ANGPTL3 in inflammatory diseases, Zhang et al. found that ANGPTL3 can interact with IL1R1 and IL1RAP through its intracellular C-terminal fibrinogen-like domain, disrupting the assembly of IL1R1-related complexes, and thereby inhibiting the activation of the NF-KB signaling pathway to prevent inflammation progression [55]. Although ANGPTL3 has shown great potential in the treatment of hypercholesterolemia, there are currently no reports of drugs targeting ANGPTL3 in IBD and its subtypes. Interestingly, we identified a new target, HGFAC, in our study. However, the feasibility of targeting HGFAC as a drug target was mainly based on literature reports [56], and specific target drugs have not been thoroughly researched.
In this study, some plasma proteins were identified as potential drug targets for the treatment of IBD and its subtypes. However, our study has some limitations. Firstly, the research results lack support from basic experiments. Secondly, the research relies on data derived from plasma proteins rather than directly from tissues or organs. Consequently, drug development targeting plasma proteins may exhibit unpredictability due to tissue specificity. Finally, this study primarily focused on European populations. Therefore, our findings should be interpreted considering these limitations.
Supplementary Information
Below is the link to the electronic supplementary material.
Authors’ contributions
Ji-Chang Fan conceived, designed, and write the manuscript. Yuan Lu analyzed the data and generated the figures and tables. Jin-Heng Gan helped to search for some relevant papers for this research. Hao Lu guided the research process and review the manuscript. All authors reviewed the manuscript.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ji-Chang Fan and Yuan Lu contributed equally to this work.
References
- 1.Cao F, He YS, Wang Y, Zha CK, Lu JM, Tao LM et al (2023) Global burden and cross-country inequalities in autoimmune diseases from 1990 to 2019. Autoimmun Rev 22(6):103326. 10.1016/j.autrev.2023.103326 [DOI] [PubMed] [Google Scholar]
- 2.Zhang ZM, Lin ZL, He BX, Yan WT, Zhang XY, Zhang ZH et al (2023) Epidemiological analysis reveals a surge in inflammatory bowel disease among children and adolescents: a global, regional, and national perspective from 1990 to 2019 - insights from the China study. J Glob Health 13:04174. 10.7189/jogh.13.04174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan GG, Windsor JW (2021) The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 18(1):56–66. 10.1038/s41575-020-00360-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramos GP, Papadakis KA (2019) Mechanisms of disease: inflammatory bowel diseases. Mayo Clin Proc 94(1):155–165. 10.1016/j.mayocp.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336(6086):1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awan H, Fatima U, Eaw R, Knox N, Alrubaiy L (2023) The efficacy of currently licensed biologics for treatment of ulcerative colitis: a literature review. Cureus 15(4):e37609. 10.7759/cureus.37609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jefremow A, Neurath MF (2023) Novel small molecules in IBD: current state and future perspectives. Cells 12(13):1730. 10.3390/cells12131730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu X, Biao Y, Liu C, Zhang Y, Liu C, Ma JZ et al (2023) Network meta-analysis on efficacy and safety of different biologics for ulcerative colitis. BMC Gastroenterol 23(1):346. 10.1186/s12876-023-02938-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wangchuk P, Yeshi K, Loukas A (2024) Ulcerative colitis: clinical biomarkers, therapeutic targets, and emerging treatments. Trends Pharmacol Sci. 10.1016/j.tips.2024.08.003.10.1016/j.tips.2024.08.003 [DOI] [PubMed] [Google Scholar]
- 10.Yeshi K, Jamtsho T, Wangchuk P (2024) Current treatments, emerging therapeutics, and natural remedies for inflammatory bowel disease. Molecules 29(16).10.3390/molecules29163954 [DOI] [PMC free article] [PubMed]
- 11.Qiu Y, Chen BL, Mao R, Zhang SH, He Y, Zeng ZR et al (2017) Systematic review with meta-analysis: loss of response and requirement of anti-TNFα dose intensification in Crohn’s disease. J Gastroenterol 52(5):535–554. 10.1007/s00535-017-1324-3 [DOI] [PubMed] [Google Scholar]
- 12.Xiang L, Rao J, Yuan J, Xie T, Yan H (2024) Single-cell RNA-sequencing: opening new horizons for breast cancer research. Int J Mol Sci 25(17):9482. 10.3390/ijms25179482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Yu B, Wang F, Yang J (2024) Single-cell RNA sequencing to map tumor heterogeneity in gastric carcinogenesis paving roads to individualized therapy. Cancer Immunol Immunother 73(11):233. 10.1007/s00262-024-03820-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, Richardson TG et al (2023) Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622(7982):329–338. 10.1038/s41586-023-06592-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao P, Iona A, Pozarickij A, Said S, Wright N, Lin K et al (2024) Proteomic analyses in diverse populations improved risk prediction and identified new drug targets for type 2 diabetes. Diabetes Care 47(6):1012–1019. 10.2337/dc23-2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazidi M, Wright N, Yao P, Kartsonaki C, Millwood IY, Fry H et al (2023) Plasma proteomics to identify drug targets for ischemic heart disease. J Am Coll Cardiol 82(20):1906–1920. 10.1016/j.jacc.2023.09.804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Si S, Liu H, Xu L, Zhan S (2024) Identification of novel therapeutic targets for chronic kidney disease and kidney function by integrating multi-omics proteome with transcriptome. Genome Med 16(1):84. 10.1186/s13073-024-01356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T et al (2018) Author Correction: Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun 9(1):3853. 10.1038/s41467-018-06231-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietzner M, Wheeler E, Carrasco-Zanini J, Cortes A, Koprulu M, Wörheide MA et al (2021) Mapping the proteo-genomic convergence of human diseases. Science 374(6569):eabj1541. 10.1126/science.abj1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R et al (2018) Co-regulatory networks of human serum proteins link genetics to disease. Science 361(6404):769–773. 10.1126/science.aaq1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz T, Lam K, Chen Y, Xia Y, Liu C (2019) A decade in psychiatric GWAS research. Mol Psychiatry 24(3):378–389. 10.1038/s41380-018-0055-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J, Haberland V, Baird D, Walker V, Haycock PC, Hurle MR et al (2020) Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet 52(10):1122–1131. 10.1038/s41588-020-0682-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J et al (2018) Genomic atlas of the human plasma proteome. Nature 558(7708):73–79. 10.1038/s41586-018-0175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J et al (2017) Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 8:14357. 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T et al (2018) Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun 9(1):3268. 10.1038/s41467-018-05512-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folkersen L, Fauman E, Sabater-Lleal M, Strawbridge RJ, Frånberg M, Sennblad B et al (2017) Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet 13(4):e1006706. 10.1371/journal.pgen.1006706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL et al (2021) Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 53(12):1712–1721. 10.1038/s41588-021-00978-w [DOI] [PubMed] [Google Scholar]
- 28.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ et al (2012) Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res 21(3):223–242. 10.1177/0962280210394459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce BL, Ahsan H, Vanderweele TJ (2011) Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40(3):740–752. 10.1093/ije/dyq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S, Thompson SG (2011) Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 40(3):755–764. 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 31.Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM et al (2019) Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res 4:186. 10.12688/wellcomeopenres.15555.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boef AG, Dekkers OM, le Cessie S (2015) Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol 44(2):496–511. 10.1093/ije/dyv071 [DOI] [PubMed] [Google Scholar]
- 33.Davey Smith G, Hemani G (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23:R89-98. 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10(5):e1004383. 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourgonje AR, Ungaro RC, Mehandru S, Colombel JF (2024) Targeting the interleukin 23 pathway in inflammatory bowel disease. Gastroenterology. 10.1053/j.gastro.2024.05.036.10.1053/j.gastro.2024.05.036 [DOI] [PubMed] [Google Scholar]
- 36.Tian Z, Zhao Q, Teng X (2024) Anti-IL23/12 agents and JAK inhibitors for inflammatory bowel disease. Front Immunol 15:1393463. 10.3389/fimmu.2024.1393463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gold R, Radue EW, Giovannoni G, Selmaj K, Havrdova EK, Montalban X et al (2020) Long-term safety and efficacy of daclizumab beta in relapsing-remitting multiple sclerosis: 6-year results from the SELECTED open-label extension study. J Neurol 267(10):2851–2864. 10.1007/s00415-020-09835-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang S, Zhang S, Zhang C, Wang L (2023) Effectiveness and safety of ustekinumab for pediatric inflammatory bowel disease: a systematic review. Paediatr Drugs 25(5):499–513. 10.1007/s40272-023-00586-7 [DOI] [PubMed] [Google Scholar]
- 39.Panaccione R, Sandborn WJ, Gordon GL, Lee SD, Safdi A, Sedghi S et al (2015) Briakinumab for treatment of Crohn’s disease: results of a randomized trial. Inflamm Bowel Dis 21(6):1329–1340. 10.1097/mib.0000000000000366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowan CR, Boland K, Harewood GC (2020) Ustekinumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med 382(1):91. 10.1056/NEJMc1915042 [DOI] [PubMed] [Google Scholar]
- 41.Globig AM, Sommer NP, Wild K, Schardey J, Zoldan K, Thomann AK et al (2021) Ustekinumab inhibits T follicular helper cell differentiation in patients with Crohn’s disease. Cell Mol Gastroenterol Hepatol 11(1):1–12. 10.1016/j.jcmgh.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cottone M, Kohn A, Daperno M, Armuzzi A, Guidi L, D’Inca R et al (2011) Advanced age is an independent risk factor for severe infections and mortality in patients given anti-tumor necrosis factor therapy for inflammatory bowel disease. Clin Gastroenterol Hepatol 9(1):30–35. 10.1016/j.cgh.2010.09.026 [DOI] [PubMed] [Google Scholar]
- 43.Harrison OJ, Srinivasan N, Pott J, Schiering C, Krausgruber T, Ilott NE, Maloy KJ (2015) Epithelial-derived IL-18 regulates Th17 cell differentiation and Foxp3+ Treg cell function in the intestine. Mucosal Immunol 8(6):1226–1236. 10.1038/mi.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinarello CA, Kaplanski G (2005) Interleukin-18 treatment options for inflammatory diseases. Expert Rev Clin Immunol 1(4):619–632. 10.1586/1744666x.1.4.619 [DOI] [PubMed] [Google Scholar]
- 45.Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W et al (2015) Epithelial IL-18 equilibrium controls barrier function in colitis. Cell 163(6):1444–1456. 10.1016/j.cell.2015.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latiano A, Palmieri O, Corritore G, Valvano MR, Bossa F, Cucchiara S et al (2010) Variants at the 3p21 locus influence susceptibility and phenotype both in adults and early-onset patients with inflammatory bowel disease. Inflamm Bowel Dis 16(7):1108–1117. 10.1002/ibd.21176 [DOI] [PubMed] [Google Scholar]
- 47.Goyette P, Lefebvre C, Ng A, Brant SR, Cho JH, Duerr RH et al (2008) Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunol 1(2):131–138. 10.1038/mi.2007.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee IY, Lim JM, Cho H, Kim E, Kim Y, Oh HK et al (2019) MST1 negatively regulates TNFα-induced NF-κB signaling through modulating LUBAC activity. Mol Cell 73(6):1138-1149.e6. 10.1016/j.molcel.2019.01.022 [DOI] [PubMed] [Google Scholar]
- 49.Nimmerjahn F, Ravetch JV (2008) Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 8(1):34–47. 10.1038/nri2206 [DOI] [PubMed] [Google Scholar]
- 50.Beppler J, Koehler-Santos P, Pasqualim G, Matte U, Alho CS, Dias FS et al (2016) Fc gamma receptor IIA (CD32A) R131 polymorphism as a marker of genetic susceptibility to sepsis. Inflammation 39(2):518–525. 10.1007/s10753-015-0275-1 [DOI] [PubMed] [Google Scholar]
- 51.Duits AJ, Bootsma H, Derksen RH, Spronk PE, Kater L, Kallenberg CG et al (1995) Skewed distribution of IgG Fc receptor IIa (CD32) polymorphism is associated with renal disease in systemic lupus erythematosus patients. Arthritis Rheum 38(12):1832–1836. 10.1002/art.1780381217 [DOI] [PubMed] [Google Scholar]
- 52.Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C, Wright VJ et al (2011) Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet 43(12):1241–1246. 10.1038/ng.981 [DOI] [PubMed] [Google Scholar]
- 53.McGovern DP, Gardet A, Törkvist L, Goyette P, Essers J, Taylor KD et al (2010) Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 42(4):332–337. 10.1038/ng.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahlqvist J, Fulco CP, Ray JP, Liechti T, de Boer CG, Lieb DJ et al (2022) Systematic identification of genomic elements that regulate FCGR2A expression and harbor variants linked with autoimmune disease. Hum Mol Genet 31(12):1946–1961. 10.1093/hmg/ddab372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Zhang ZT, Wan SY, Yang J, Wei YJ, Chen HJ et al (2023) ANGPTL3 negatively regulates IL-1β-induced NF-κB activation by inhibiting the IL1R1-associated signaling complex assembly. J Mol Cell Biol 15:mjad053. 10.1093/jmcb/mjad053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J, Xu F, Ruan X, Sun J, Zhang Y, Zhang H et al (2023) Therapeutic targets for inflammatory bowel disease: proteome-wide Mendelian randomization and colocalization analyses. EBioMedicine 89:104494. 10.1016/j.ebiom.2023.104494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.