Abstract
Human cytomegalovirus (HCMV) stimulates arrested cells to enter the cell cycle by activating cyclin-dependent kinases (Cdks), notably Cdk2. Several mechanisms are involved in the activation of Cdk2. HCMV causes a substantial increase in the abundance of cyclin E and stimulates translocation of Cdk2 from the cytoplasm to the nucleus. Further, the abundance of the Cdk inhibitors (CKIs) p21cip1/waf1 (p21cip1) and p27kip1 is substantially reduced. The activity of cyclin E/Cdk2 increases as levels of CKIs, particularly p21cip1, fall. We have previously shown that these phenomena contribute to priming the cell for efficient replication of HCMV. In this study, the mechanisms responsible for the decrease in p21cip1 levels after HCMV infection were investigated by measuring p21cip1 RNA and protein levels in permissive human lung (LU) fibroblasts after HCMV infection. Northern blot analysis revealed that p21cip1 RNA levels increased briefly at 3 h after HCMV infection and then decreased to their nadir at 24 h; thereafter, RNA levels increased to about 60% of the preinfection level. Western blot analysis demonstrated that the relative abundance of p21cip1 protein roughly paralleled the observed changes in initial RNA levels; however, the final levels of protein were much lower than preinfection levels. After a transient increase at 3 h postinfection, p21cip1 abundance declined sharply over the next 24 h and remained at a very low level through 96 h postinfection. The disparity between p21cip1 RNA and protein levels suggested that the degradation of p21cip1 might be affected in HCMV-infected cells. Treatment of HCMV-infected cells with MG132, an inhibitor of proteasome-mediated proteolysis, provided substantial protection of p21cip1 in mock-infected cells, but MG132 was much less effective in protecting p21cip1 in HCMV-infected cells. The addition of E64d or Z-Leu-Leu-H, each an inhibitor of calpain activity, to HCMV-infected cells substantially increased the abundance of p21cip1 in a concentration-dependent manner. To verify that p21cip1 was a substrate for calpain, purified recombinant p21cip1 was incubated with either m-calpain or μ-calpain, which resulted in rapid proteolysis of p21cip1. E64d inhibited the proteolysis of p21cip1 catalyzed by either m-calpain or μ-calpain. Direct measurement of calpain activity in HCMV-infected LU cells indicated that HCMV infection induced a substantial and sustained increase in calpain activity, although there was no change in the abundance of either m- or μ-calpain or the endogenous calpain inhibitor calpastatin. The observed increase of calpain activity was consistent with the increases in intracellular free Ca2+ and phospholipid degradation in HCMV-infected LU cells reported previously from our laboratory. Considered together, these results suggest that the increase in calpain activity observed following HCMV infection contributes significantly to the reduction of p21cip1 levels and the resultant cell cycle progression.
Human cytomegalovirus (HCMV) infection is widespread among human populations, primarily as a subclinical persistent infection. Furthermore, HCMV infection is a majsor cause of morbidity and mortality in several well-studied risk groups. These include congenitally infected infants and individuals with compromised immune systems, particularly after human immunodeficiency virus infection or immunosuppressive therapy for tissue transplantation (for reviews, see references 8, 29, 68, and 72). The clinical management of these infections is still problematic. Although several agents with potent antiviral activity for HCMV infection both in vitro and in vivo have been identified, the toxicity associated with the long-term use of these drugs makes clinical management difficult, and drug-resistant strains of HCMV have emerged (for a review, see reference 61). Thus, there continues to be great interest in improving our understanding of the replication of HCMV with a view toward developing more effective approaches to control these infections.
HCMV replication is associated with extensive modifications of cellular metabolism (reviewed in references 4 and 5), leading to a number of physiologic changes and the activation of a large number of cellular genes (91). Initially, HCMV infection induces a series of cellular responses that resemble the immediate-early events observed following activation of serum-arrested cells by serum growth factors (4). These include hydrolysis of phosphatidylinositol 4,5-bisphosphate, yielding increased cellular levels of sn-1,2-diacylglycerol (DG) and inositol 1,4,5-trisphosphate (81); increased release of arachidonic acid and its metabolites (1, 2); changes in Ca2+ homeostasis, including Ca2+ influx, release of Ca2+ stores, and an increase in intracellular free Ca2+ (58); transcriptional activation of cellular oncogenes c-fos, c-jun, and c-myc (11, 12, 13); and increased activity of the DNA-binding proteins NFκB, AP-1, and CREB (14). The signaling cascade induced by HCMV infection induces a robust mitogenic response, as evidenced by the ability of HCMV to stimulate cell cycle entry by density-arrested cells, which are resistant to stimulation by serum growth factors (19). Recent results indicate that productive HCMV infection stimulates cell cycle progression in either serum- or density-arrested cells through late G1 phase to a point at or near the G1/S boundary (19, 30, 52). Closely associated with this limited traverse of the cell cycle is an increase in cyclin E/cyclin-dependent kinase 2 (Cdk2) activity, hereafter referred to as E kinase activity, and hyperphosphorylation of pRb, releasing E2F (45). Activation of E2F, together with Myc, leads to an increase in the cellular levels of a large number of genes involved in nucleotide biosynthesis which primes the infected cell for DNA synthesis (5, 7).
Three HCMV-induced events appear to be necessary for activation of E kinase activity: (i) transcriptional activation of cyclin E (17), (ii) translocation of Cdk2 from the cytoplasm to the nucleus (20), and (iii) a substantial decrease in the nuclear levels of the cyclin kinase inhibitors (CKIs) p21cip1/waf1 (hereafter p21cip1) and p27kip1 (19). Activation of E kinase appears to be critical for efficient HCMV replication, since drugs that interfere with the activity of Cdk2 substantially reduce infectious yields (18). The precise mechanisms through which these virus-induced cellular modifications are achieved are poorly understood. Our long-term goal is to better understand the events that prime the cell for HCMV replication. We are especially interested in the mechanism of reduction of cellular levels of p21cip1 during HCMV infection, since this event appears to be closely associated with the increase in E kinase activity (19).
p21cip1 is a potent inhibitor of Cdks (41, 42, 84) and is a critical p53-induced downstream effector in the growth-suppressive pathway (33). p21cip1 binds cyclin-Cdk complexes, thereby inhibiting the activity of Cdks, such as Cdk2, Cdk3, Cdk4, and Cdk6, and consequently inhibiting cell cycle progression. In addition, p21cip1 interacts with proliferating cell nuclear antigen (36) and gadd45 (48), affecting their function, e.g., interfering with DNA replication and repair (24, 36, 51, 60, 62, 82, 86). p21cip1 may also be involved in p53-mediated apoptosis (39) and in the control of cell senescence (57).
Despite significant advances in our understanding of how p21cip1 exerts its biological effects and is transcriptionally regulated, little is known about how the stability of p21cip1 is regulated under the physiologic conditions associated with disease and other forms of stress. Nonlysosomal cytoplasmic protease systems have been identified as important regulators of cell cycle progression (26, 28, 35, 59, 70, 71). Two prominent cytoplasmic protease pathways have been identified: the ubiquitin-proteasome pathway and the calpain pathway. Many of the cell cycle regulatory proteins that are degraded at specific points in the cell cycle, e.g., cyclins A, B, and E, are substrates of the ubiquitin-proteasome pathway. It has also been reported that p21cip1 is subject to proteolysis by ubiquitin-mediated proteasome degradation (10, 35, 38, 85). In this study, we undertook a series of experiments to investigate the proteolytic degradation of p21cip1 in cells productively infected with HCMV. Consistent with previous work, we observed that the ubiquitin-proteasome pathway is the predominant mechanism mediating the proteolysis of p21cip1 in mock-infected or noninfected human embryonic lung (LU) cells. In HCMV-infected LU cells, calpain is activated and is the predominant proteolytic mechanism mediating degradation of p21cip1. HCMV infection induced a substantial and prolonged increase in calpain activity when measured in intact cells using fluorogenic substrates or in lysates of infected cells by zymography. The time course for the HCMV-induced increase in calpain activity was consistent with the temporal pattern of p21cip1 degradation, and inhibitors of calpain-mediated proteolysis interfered with p21cip1 degradation, resulting in a substantial increase in its abundance. These findings provide an explanation for the sustained decrease in p21cip1 abundance observed in HCMV-infected cells and, accordingly, help to explain the prolonged activation of E kinase in HCMV infection.
MATERIALS AND METHODS
Cell culture and growth arrest.
LU cells (3) were propagated in Eagle's minimum essential medium containing 10% fetal bovine serum (FBS) and penicillin (100 U/ml)-streptomycin (100 μg/ml) in a 5% CO2 in air atmosphere. The cells were density arrested as described previously in detail (19).
Virus stocks and productive infection.
The AD169 strain of HCMV was propagated in LU cells as previously described in detail (6). The infectivities of virus stocks, determined by plaque assay (3), typically were between 8.0 × 106 and 4.0 × 107 PFU/ml. LU cells were infected with HCMV at a multiplicity of 5 PFU/cell to provide a uniform infection as described in detail by Bresnahan et al. (19). Virus stocks and cell cultures were routinely examined for mycoplasma and were free from detectable contamination.
RNA isolation.
RNA was extracted using Tri Reagent (Molecular Research Center, Inc., Cincinnati, Ohio), which contains phenol and guanidine thiocyanate, according to the manufacturer's protocol (25).
Preparation of probes.
A 32P-labeled DNA probe was prepared from plasmid pC-waf1-S (generously provided by B. Vogelstein [33]), which harbors a p21cip1 insert. A probe derived from plasmid p5B (16), which contains the cDNA for 18S rRNA, was used to ensure uniform loading of gels. Plasmids were introduced into competent Escherichia coli DH5α cells and amplified. DNA fragments comprising p21cip1 and 18S cDNA were excised from isolated plasmid DNA by using the the restriction endonucleases NotI and BamHI-EcoRI, respectively. The probes were labeled using the Megaprime DNA Labelling System (Amersham Pharmacia Biotech, Piscataway, N.J.).
Electrophoresis and Northern hybridization.
Northern hybridization was performed as described previously (17). Total cellular RNA (20 μg/lane) was evaluated under denaturing conditions in formaldehyde-agarose gels, containing 1% agarose, 20 mM 3-(N-morpholino) propanesulfonic acid, 1 mM EDTA, 8 mM sodium acetate, and 2.2 M formaldehyde. After separation, the RNA was transferred to MSI nylon membranes (Micron Separations, Inc., Westborough, Mass.) for 18 h. The RNA was prehybridized in Rapid-Hyb buffer (Amersham Life Science, Arlington Heights, Ill.) containing denatured salmon sperm DNA (100 μg/ml) at 65°C for 1 h. 32P-labeled p21cip1 or 18S probe was added and hybridized for 2 h at 65°C. Membranes were washed twice with 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% sodium dodecyl sulfate (SDS) for 15 min at 65°C, once with 1× SSPE–0.1% SDS at 42°C, and twice with 0.1× SSPE–0.1% SDS at 42°C. The probe remaining on the filter was detected by autoradiography (Kodak [Rochester, N.Y.] X-Omat film for 1 to 16 h at −80°C).
Western blots.
Polyclonal and monoclonal antibodies for p21cip1 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Antibodies against m-calpain, μ-calpain, and calpastatin were generously provided by R. L. Mellgren (88). Cells were harvested as we described previously (19) by dislodging the cells with a cell lifter in phosphate-buffered saline (PBS). The cells were collected by sedimentation and lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.4] containing 150 mM NaCl and 0.5% NP-40, with 1 mM dithiothreitol [DTT], 1 mM NaVO3, 50 mM NaF, and protease inhibitors [1 mM phenylmethylsulfonyl fluoride {PMSF}, 25 μg of trypsin inhibitor/ml, 25 μg of aprotinin/ml, 1 mM benzamide, and 25 μg of pepstatin A/ml] added just before use). Cellular debris was removed by sedimentation, and the supernatant fluids were reserved. The protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). Whole-cell extracts (40 μg/lane) were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and the polypeptides were transferred to nitrocellulose membranes (Bio-Rad Laboratories, Hercules, Calif.) as described previously (19). Antigen-antibody reactions were detected by the enhanced chemiluminescence assay (Amersham Pharmacia Biotech) as recommended by the manufacturer.
Measurement of intracellular calpain activity.
Calpain activity was measured using the cell-permeant fluorogenic calpain substrate tert-butoxycarbonyl-l- leucyl-l-methionineamide-7-amino-4-chloromethylcoumarin (Boc-Leu-Met-CMAC; Molecular Probes, Eugene, Oreg.). HCMV- or mock-infected cells were loaded with Boc-Leu-Met-CMAC (10 μM) for 15 min at selected times after infection. Subsequently, the cells were dissociated by trypsinization and resuspended in PBS. After entering cells, Boc-Leu-Met-CMAC is conjugated to thiols, particularly glutathione, becoming impermeant for the plasmalemma. Cleavage of thiol-conjugated Boc-Leu-Met-CMAC by calpain releases thiol-conjugated 7-amino-4-methylcoumarin (AMC), unquenching the fluorescence. Thus, when cleavage is the rate-limiting step, the observed increase in fluorescence is proportional to the activity of calpain (67). Fluorescence intensities of Boc-Leu-Met-CMAC-loaded cells were measured with a SLM 4800S spectrofluorometer. The relative fluorescence intensities were determined by measuring the fluorescence emission values at intervals from 390 to 550 nm, with excitation set at 380 nm. The fluorescence signal was calculated by integrating the area under the peak at 460 nm. The time interval for loading the cells with Boc-Leu-Met-CMAC, harvesting the loaded cells, and measurement of fluorescence was constant for all samples.
Zymography.
Casein zymography was performed by the method of Zhao et al. (90). Cells were removed from culture dishes with a cell scraper into cold PBS, collected by sedimentation, and homogenized in freshly made homogenization buffer [20 mM Tris-HCl (pH 7.4) containing 5 mM EDTA, 5 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 10 μg of 4-(2-aminoethyl)benzenesulfonly fluoride (AEBSF)/ml, 5 μg of leupeptin/ml and 10 μg of pepstatin A/ml] at 4°C. Cellular debris was removed by sedimentation, and the supernatant fluids were collected, quick-frozen with liquid nitrogen, and maintained at −80°C until needed.
For electrophoretic separation of the target proteins, 100 μg of protein from each sample was mixed with the appropriate amount of 5× sample buffer (pH 6.8), containing 150 mM Tris-HCl, 20% glycerol, 2 mM 2-mercaptoethanol, and 0.004% bromophenol blue, to obtain a 1× sample buffer concentration. The samples were then fractionated in polyacrylamide gels. The separating gel contained 2% casein (wt/vol; sodium salt) copolymerized with 12% acrylamide, 0.32% (wt/vol) bisacrylamide, 375 mM Tris-HCl (pH 8.8), 0.04% ammonium persulfate, and 0.028% (vol/vol) N,N,N′,N′-tetramethylethylenediamine (TEMED). The stacking gel contained 4% acrylamide, 0.11% (wt/vol) bisacrylamide, 380 mM Tris-HCl (pH 6.8), 0.04% ammonium persulfate, and 0.028% (vol/vol) TEMED, without casein. The electrophoresis buffer contained 25 mM Tris base, 192 mM glycine, 1 mM EDTA, and 1 mM DTT (pH 8.3). After electrophoresis, the gel was removed and incubated in incubation buffer for 60 min at room temperature with slow agitation and with two changes of buffer. The incubation buffer contained 20 mM Tris-HCl, 10 mM DTT, 3 mM CaCl2 (pH 7.5 for μ-calpain; pH 7.3 for m-calpain). The gel was incubated for an additional 20 to 24 h and then stained with Coomassie brilliant blue.
Cleavage of p21cip1 by calpain.
Recombinant p21cip1 was a gift from J. Wade Harper (41) or Hengming Ke (53). We also prepared the recombinant p21cip1 by using the expression plasmid pET-p21, provided by J. Wade Harper (41), according to the published method (32). To evaluate the effect of calpains on p21cip1, 400 ng of purified recombinant p21cip1 was incubated with 0.004 U of either pure μ-calpain or m-calpain at 30°C for 30 min in 40 μl of cleavage buffer (25 mM Tris-HCl [pH 7.5] with 100 mM NaCl, 3 mM DTT, and 5 mM CaCl2) (50). EDTA was added to 10 mM to stop the reaction. An equal volume of 2× SDS-PAGE gel-loading buffer (19) was mixed with the digestion mixture, and the mixture was immediately boiled for 5 min. The digestion products were evaluated by Western blot analysis and SDS-PAGE. Antibody preparations used for the detection of p21cip1 (SC-397 and SC-6246) were obtained from Santa Cruz Biotechnology.
Chemicals.
The calpain inhibitors E64d [trans-epoxysuccinyl-l-leucylamido (4-guanidino)-butane] and Z-Leu-Leu-H (carbobenzoxy-l-leucyl-l-leucinal) were purchased from Peptides International, Inc. (Louisville, Ky.). Calpain II (m-calpain), AEBSF, casein, penicillin, streptomycin, diethyl pyrocarbonate, Tris, NaCl, NaVO3, NaF, PMSF, DTT, trypsin inhibitor, aprotinin, benzamide, and pepstatin A were purchased from Sigma (St. Louis, Mo.). Calpain I (μ-calpain) and NP-40 were obtained from Calbiochem (San Diego, Calif.). Boc-Leu-Met-CMAC and CMAC were purchased from Molecular Probes.
Image processing.
All experiments were repeated at least twice, and either data from a representative experiment or means of data from two or more experiments were selected for presentation. Chemiluminescent samples were exposed for intervals that ensured a linear response, as determined by standardization. All radiographic films were analyzed using an AlphaImager 2000 documentation and analysis system (Alpha Innotech Corp.) and the manufacturer's software (AlphaImager 2000, version 3.x). The images were quantified and recorded as tagged image format files, which were then used to prepare the graphic images presented in this report.
RESULTS
p21cip1 RNA and protein levels in HCMV-infected LU cells.
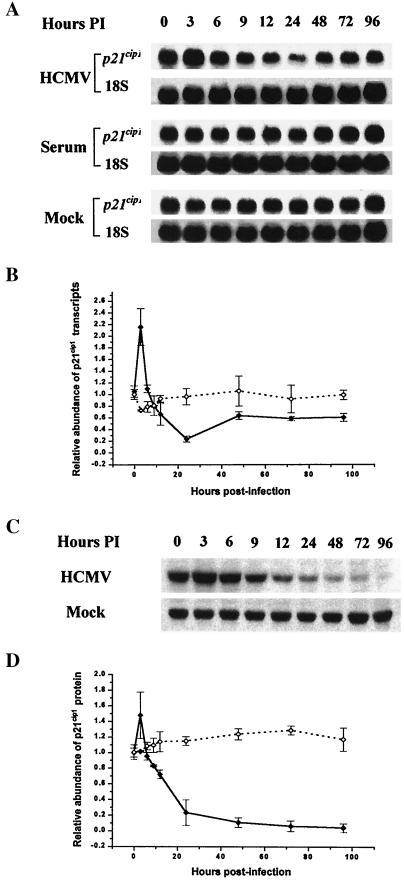
Previous studies from our laboratory demonstrated that HCMV infection resulted in a substantial decrease in the abundance of p21cip1 protein in LU cells (19). To investigate the mechanism responsible for this drop, p21cip1 transcript levels were measured by Northern blot analysis in density-arrested LU cells following stimulation with 10% FBS, HCMV (5 PFU/cell) infection, or mock infection. The results of a representative experiment are shown in Fig. 1A. In density-arrested cells, FBS treatment or mock infection had little effect on p21cip1 RNA levels. Following HCMV infection of density-arrested LU cells, p21cip1 RNA levels increased briefly at 3 h postinfection (p.i.) and then decreased over the next 21 h p.i., whereupon they began to recover, reaching 60% of their preinfection level at 96 h p.i. (determined by densitometry [Fig. 1B]). Western blot analysis (Fig. 1C) demonstrated that p21cip1 protein abundance was not correlated with the observed changes in RNA levels after 24 h p.i. After an early increase at 3 h p.i., p21cip1 protein levels fell dramatically from 3 to 24 h p.i., asymptotically approaching the limits of detection from 48 to 96 h p.i. (Fig. 1D). These findings provide clear evidence of a disparity between p21cip1 RNA and protein levels in the HCMV-infected cells after 24 h p.i.
FIG. 1.
(A) Northern blot analysis of the effect of HCMV infection (HCMV), serum growth factors (Serum), or mock infection (Mock) on p21cip1 RNA transcripts in density-arrested LU cells. The density-arrested cells were infected with HCMV at a multiplicity of 5 PFU/cell, exposed to fresh FBS (10%), or mock infected. RNA was isolated at the times indicated, and 20 μg of RNA from each lysate was applied to the gel. 18s rRNA (18S) was used as a loading standard. (B) Results of the representative experiment illustrated in panel A and of two additional independent experiments, evaluated by densitometric analysis and plotted as the abundance relative to the mock-infected control at 0 h p.i. with standard deviation shown (⧫, p21cip1 RNA, HCMV-infected cells; ◊, p21cip1 RNA, mock-infected cells). (C) Western blot analysis of the abundance of p21cip1 after HCMV infection or mock infection. Parallel cultures of density-arrested LU cells were treated as in the legend to panel A. Whole-cell lysates were prepared at the indicated intervals, and 40 μg of protein from each was resolved by SDS-PAGE. The proteins were transferred to nitrocellulose membrane and probed with antibodies against p21cip1. The results of a representative experiment are illustrated. (D) The results of the experiment illustrated in panel C and two additional experiments, evaluated by densitometric analysis and plotted as the abundance relative to the mock-infected control at 0 h p.i. with standard deviation shown (⧫, p21cip1 protein, HCMV-infected cells; ◊, p21cip1 protein, mock-infected cells).
Differential effects of an inhibitor of the ubiquitin-proteasome proteolytic pathway on p21cip1 degradation during HCMV infection and mock infection.
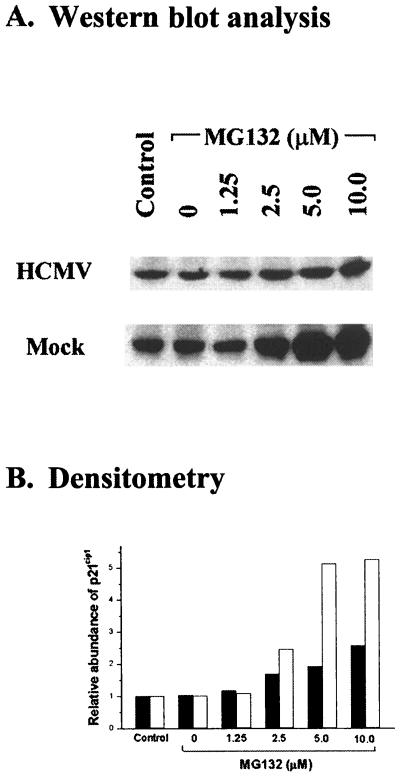
Since the ubiquitin-proteasome pathway is reported to be responsible for degrading many of the cell cycle regulatory proteins, we investigated whether or not the decrease of p21cip1 in HCMV-infected cells was due to a proteasome-mediated mechanism. HCMV (5 PFU/cell)-infected and mock-infected LU cells were treated with MG132, an inhibitor of proteasome-mediated proteolysis (56, 69), at 1 h p.i., and p21cip1 abundance was evaluated at 24 h p.i. The results of a representative experiment are shown in Fig. 2A. By 24 h p.i., p21cip1 levels were substantially lower in HCMV-infected LU cells (Fig. 2A) than in the control (mock-infected cells). MG132 stabilized p21cip1 in mock-infected, density-arrested LU cells but had only a limited effect on stabilization of p21cip1 in HCMV-infected cells (Fig. 2). Mock-infected, density-arrested LU cells treated with MG132 (0 to 10 μM) demonstrated a concentration-dependent increase in the abundance of p21cip1. A greater than fivefold increase in p21cip1 abundance relative to either the dimethyl sulfoxide (DMSO) control (lane 2) or cells in the absence of any chemical (lane 1) was observed at concentrations of 5 and 10 μM (Fig. 2). In HCMV-infected cells, smaller increases in p21cip1 abundance were observed at MG132 concentrations from 2.5 to 10 μM, reaching a maximum 2.5-fold increase at 10 μM. That the p21cip1 abundance was less responsive to protection by MG132 in HCMV-infected cells than in mock-infected cells is demonstrated by the significant difference (P < 0.01) in the slopes (determined by linear regression analysis) of the concentration effect of MG132 in HCMV-infected (0.800, P < 0.01) and mock-infected (1.964, P < 0.01) LU cells. These findings suggested that some additional MG132-insensitive pathway(s) may be involved in the proteolysis of p21cip1 in HCMV-infected cells. As the unidentified mechanism(s) seemed to be quantitatively more important in p21cip1 proteolysis in HCMV-infected cells than was proteasome-mediated degradation, we were interested in identifying the proteolytic mechanism(s) stimulated by HCMV infection.
FIG. 2.
Effect of MG132, an inhibitor of proteasome activity, on p21cip1 abundance in HCMV- or mock-infected, density-arrested LU cells. (A) Results of a representative experiment in which density-arrested LU cells were infected with HCMV (5 PFU/cell) or mock infected and then exposed to the indicated concentrations of MG132 beginning at 24 h p.i. Cell lysates were prepared at 30 h p.i. and 40 μg of protein from each lysate was resolved by SDS-PAGE. The proteins were transferred to nitrocellulose and probed with antibodies against p21cip1. Control, cells cultured in the absence of MG132 or its solvent, DMSO; 0, DMSO control, consisting of cells treated with the same concentration of DMSO (0.1%) present in the cultural fluids containing the highest concentration of MG132 (10 μM). (B) Change in abundance of p21cip1 after MG132 treatment, determined by densitometry of the results illustrated in panel A. The results for mock-infected cells (open bars) were calculated relative to the p21cip1 abundance in untreated mock-infected cells; the results for HCMV-infected cells (closed bars) were determined relative to the p21cip1 abundance in untreated HCMV-infected cells.
The calpain inhibitors E64d and Z-Leu-Leu-H protect p21cip1 from degradation during HCMV infection.
HCMV infection induces substantial increases in intracellular free Ca2+ (58) and in phospholipid degradation (1, 2, 81), raising the possibility that Ca2+-activated neutral proteases (calpains) might be activated by HCMV infection and participate in the proteolysis of p21cip1. Previous work has demonstrated that calpains are able to cleave cell cycle regulatory proteins, such as cyclin D1, cellular oncogene products (e.g., c-Mos, c-Jun, and c-Fos), and p53 (43, 59; for reviews, see references 21 and 28). The possibility that calpain-mediated proteolysis was involved in the degradation of p21cip1 in HCMV-infected cells was investigated first by examining the effect of the cell-permeable calpain inhibitors E64d (79) and Z-Leu-Leu-H (69) on p21cip1 levels following HCMV infection.
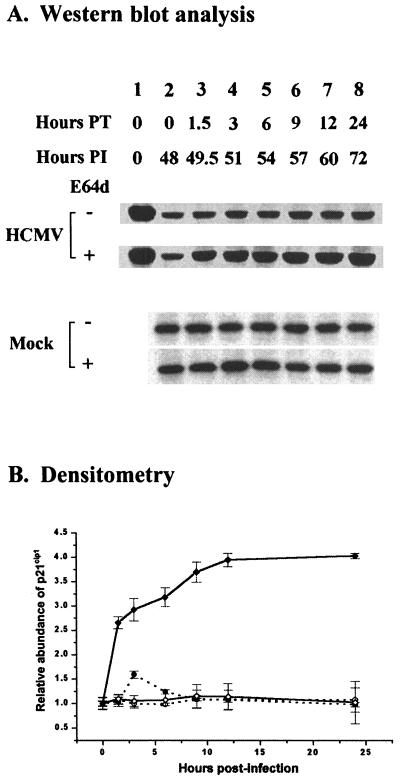
In initial experiments, we measured the effect of E64d (100 μM) on p21cip1 levels by Western blot analysis beginning at 48 h p.i., when p21cip1 was near its lowest level (Fig. 1) in HCMV-infected (5 PFU/cell), density-arrested LU cells. The abundance of p21cip1 was determined at selected intervals for 24 h following treatment with E64d. The results of a representative experiment are shown in Fig. 3. As noted previously, a substantial decrease in p21cip1 levels was observed at 48 h p.i. in HCMV-infected cells just prior to treatment with E64d (Fig. 3, compare lane 1 [lysate of mock-infected cells prepared at 0 h p.i.] to lane 2 [lysate of HCMV-infected cells in the absence of E64d prepared at 48 h p.i.]). Except at 3 h after treatment with E64d, when there was about a 1.5-fold increase in p21cip1 in the presence of E64d, the calpain inhibitor had little, if any, effect on the abundance of p21cip1 in mock-infected, density-arrested LU cells. In contrast, in HCMV-infected cells in the presence of E64d, a substantial accumulation of p21cip1 was observed, with a fourfold increase in the abundance of p21cip1 in HCMV-infected cells observed after 12 h of E64d treatment. In the absence of E64d, levels of p21cip1 in HCMV-infected cells remained at a constant low level. The maximum level of p21cip1 in the E64d-treated cells under these conditions was about 84% of that observed at 0 h. These data suggested that an E64d-sensitive proteolytic pathway was induced in HCMV-infected cells and was largely responsible for a substantial decrease in the abundance of p21cip1.
FIG. 3.
Cumulative effect of the calpain inhibitor E64d (100 μM) on p21cip1 protein levels in HCMV- or mock-infected, density-arrested LU cells. (A) LU cells arrested by contact inhibition as described in Materials and Methods were infected with HCMV (5 PFU/cell) or mock infected and then exposed to E64d beginning at 48 h p.i. At the times indicated, whole-cell lysates were prepared, and 40 μg of protein from each lysate was analyzed by SDS-PAGE. The proteins were transferred to nitrocellulose and probed with antibodies against p21cip1. The results of a representative experiment are shown. Hours PT (posttreatment) indicates the duration of E64d exposure. Hours PI indicates the time of harvest after infection. The exposure time for blots of lysates from HCMV-infected cells was about three times longer than that required for blots of lysates from mock-infected cells. Accordingly, to permit a quantitative comparison between the p21cip1 abundance at 0 h p.i. and at later times, lane 1 for the HCMV-infected cell lysates was loaded with a lysate from mock-infected cells not treated with E64d. (B) Accumulation of p21cip1 after E64d treatment, determined by densitometry of the data illustrated in panel A and from two additional experiments. The results for p21cip1 abundance in HCMV- and mock-infected cells with standard deviation shown are plotted relative to the p21cip1 abundance at 0 h in the absence of E64d in HCMV- and mock-infected cells, respectively (○, mock-infected cells; ●, mock-infected cells treated with E64d; ◊, HCMV-infected cells; ⧫, HCMV-infected cells treated with E64d).
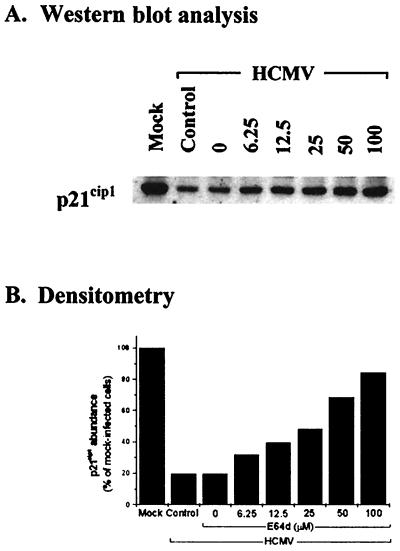
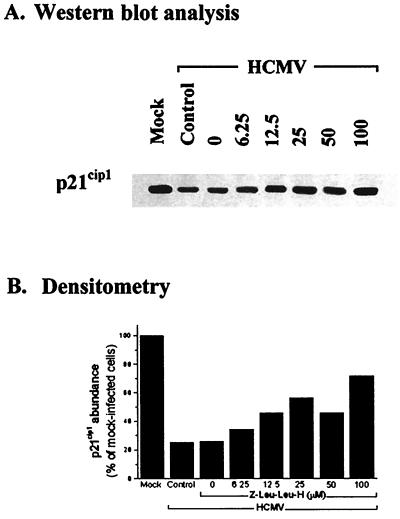
The concentration effect of E64d on p21cip1 abundance was examined in HCMV (5 PFU/cell)- and mock-infected, density-arrested LU cells by addition of selected concentrations of E64d at 48 h p.i. Whole-cell extracts were prepared 6 h later and evaluated by Western blot analysis. The results of a representative experiment are shown in Fig. 4. Since E64d had little, if any, effect on p21cip1 levels in mock-infected cells even at a concentration of 100 μM, as demonstrated in Fig. 3, the data for mock-infected cells are not illustrated in Fig. 4. In HCMV-infected cells, protection of p21cip1 was observed at all concentrations evaluated (6.25 to 100 μM), while the quantitative effect was directly dependent on the concentration of E64d. Cells treated with the highest concentration of E64d (100 μM) demonstrated about 85% of the p21cip1 abundance measured in mock-infected cells over the same time period (Fig. 4B, compare the first and last columns).
FIG. 4.
Effect of E64d concentration on p21cip1 abundance in HCMV-infected cells. (A) LU cells were density arrested as described in Materials and Methods, HCMV infected (5 PFU/cell) or mock infected, and at 48 h p.i. treated with the indicated concentrations of E64d. Whole-cell lysates were prepared 6 h later, and 40 μg of protein from each lysate was resolved by SDS-PAGE. The proteins were transferred to nitrocellulose and probed with antibodies to p21cip1. The results from a representative experiment are shown. Mock, mock-infected cells in the absence of drug; Control, untreated HCMV-infected cells; 0, DMSO control, consisting of HCMV-infected cells treated with the same concentration of DMSO (0.1%) contained in the highest concentration of E64d. (B) Densitometric analysis of the results shown in panel A.
Protection of p21cip1 by Z-Leu-Leu-H was evaluated by the same approach as described above. Z-Leu-Leu-H also protected p21cip1 in a concentration-dependent manner in HCMV (5 PFU/cell)-infected, density-arrested LU cells, with some protection provided by all concentrations of Z-Leu-Leu-H evaluated. The results of a representative experiment are shown in Fig. 5. The levels of protection provided by Z-Leu-Leu-H were similar to those observed with E64d (Fig. 4). A concentration of 100 μM of Z-Leu-Leu-H resulted in detection of about 72% of the mock-infected levels of p21cip1 (Fig. 5B, compare the first and last columns), compared to 85% for cells treated with E64d. Z-Leu-Leu-H had little, if any, effect on the abundance of p21cip1 in mock-infected cells (data not shown).
FIG. 5.
Effect of Z-Leu-Leu-H concentration on p21cip1 abundance in HCMV-infected cells. (A) LU cells were density arrested as described in Materials and Methods, infected with HCMV (5 PFU/cell) or mock infected, and at 48 h p.i. treated with selected concentrations of Z-Leu-Leu-H. Whole-cell lysates were prepared 6 h later, and 40 μg of protein from each lysate was resolved by SDS-PAGE. The proteins were transferred to nitrocellulose and probed with antibodies to p21cip1. The results illustrated are those from a representative experiment. Mock, Control, and 0 are defined in the legend to Fig. 4. (B) Densitometric analysis of the results illustrated in panel A.
Sensitivity of p21cip1 degradation to inhibition by E64d during the HCMV replication cycle.
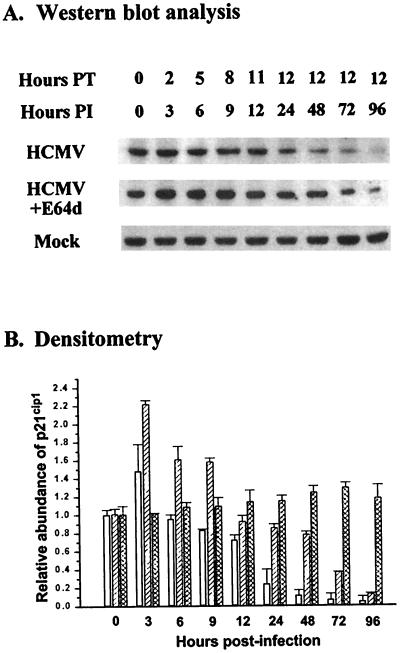
To determine the effect of E64d (100 μM) on p21cip1 levels throughout the time course of HCMV infection, density-arrested LU cell cultures were either infected with HCMV (5 PFU/cell) or mock infected and treated with E64d up to 12 h before harvest. For cells harvested at 12 h p.i. or before, the cells were treated with E64d from 1 h p.i. to the time of harvest (Fig. 6). E64d had little effect on p21cip1 levels in mock-infected cells, as previously noted (Fig. 3), and therefore the results for mock-infected cells are not shown. The results for HCMV-infected cells from a representative experiment are shown in Fig. 6A. In the absence of E64d, p21cip1 levels in HCMV-infected cells increased at 3 h and began to drop off by 6 h. By 72 and 96 h p.i., p21cip1 was only faintly detectable in the HCMV-infected cells in the absence of E64d. In the presence of E64d, p21cip1 levels in HCMV-infected cells were significantly (P < 0.05) greater (from 3 h to 96 h p.i., except at 12 and 96 h, when the difference, although greater, was not significant) than the levels observed in the absence of the calpain inhibitor (Fig. 6B). As a result of E64d treatment, p21cip1 levels remained at or above the preinfection levels through 9 h p.i. and within 85% of the preinfection levels through 24 h. After 48 h, p21cip1 levels in E64d-treated, HCMV-infected cells remained well above the levels in the absence of the calpain inhibitor, but the p21cip1 abundance dropped progressively below the preinfection levels. Considered together, these data suggested that calpain-mediated proteolysis contributed substantially to the degradation of p21cip1 in HCMV-infected cells from 3 h through the late phase of HCMV infection.
FIG. 6.
Time course for the effect of E64d on p21cip1 abundance in HCMV-infected, density-arrested cells. (A) LU cells density arrested as described in Materials and Methods were infected with HCMV (5 PFU/cell) or mock infected; at the intervals indicated, beginning 1 h p.i., subsets of the cells were treated with E64d (100 μM). Whole-cell lysates were prepared at the times indicated (up to 12 h posttreatment [PT]), and 40 μg of protein from each lysate was resolved by SDS-PAGE. The proteins were transferred to nitrocellulose and probed with antibodies to p21cip1. The results of a representative experiment are illustrated. (B) Densitometric analysis of the results shown in panel A and two additional experiments with standard deviation. Open bars, HCMV-infected cells; diagonally striped bars, HCMV-infected cells treated with E64d; cross-hatched bars, mock-infected cells.
Calpain is activated by HCMV infection.
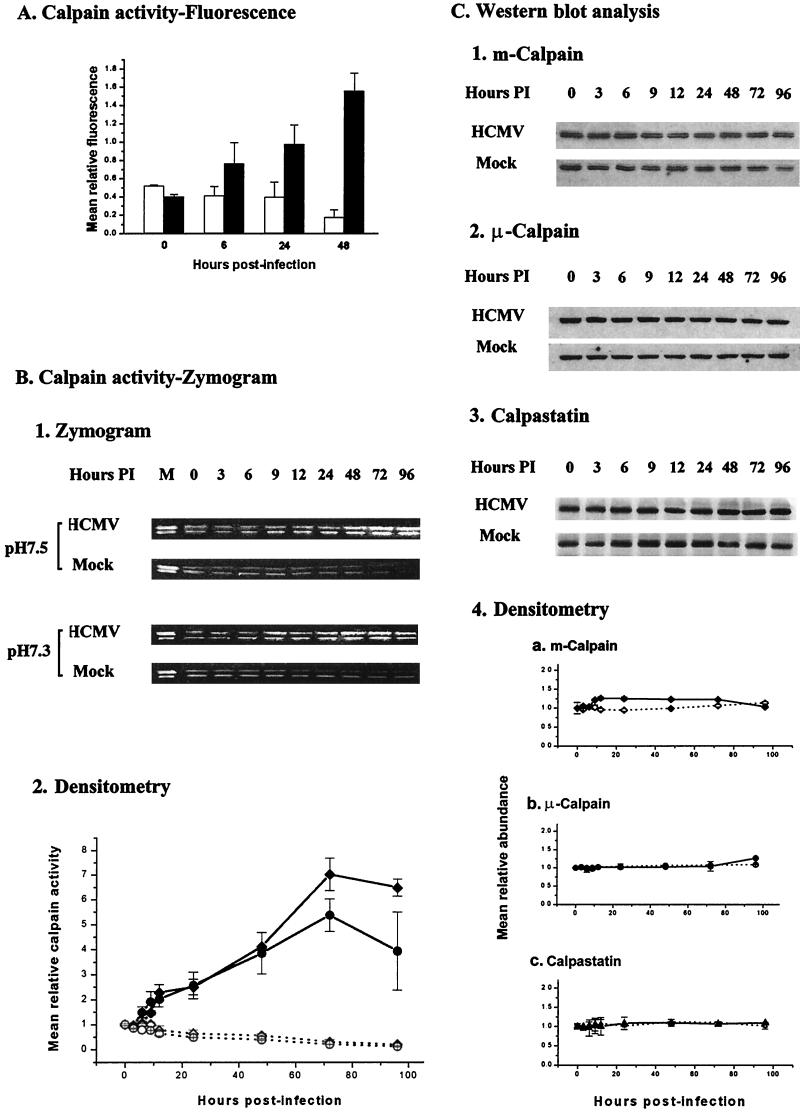
The level of calpain activity is normally very low in unstimulated cells due to the presence of the endogenous calpain inhibitor calpastatin (21). The findings that treatment of HCMV-infected cells with E64d or Z-Leu-Leu-H led to stabilization and accumulation of p21cip1 suggested that calpain is activated during HCMV infection. To investigate this possibility, we used the fluorogenic calpain substrate Boc-Leu-Met-CMAC to measure calpain activity in density-arrested LU cells (67). Density-arrested LU cells were infected with HCMV (5 PFU/cell) or mock infected and exposed to Boc-Leu-Met-CMAC (10 μM) 15 min before assay. The cells were harvested (see Materials and Methods), and the relative fluorescence was determined. The results (Fig. 7A) demonstrated that fluorescence from the product (AMC) of cleavage of the fluorogenic substrate fell gradually in mock-infected cells during the 48-h course of these experiments. HCMV infection, however, induced a substantial increase in calpain activity. An increase in AMC fluorescence was observed by 6 h p.i. in HCMV-infected cells. After 24 h, the AMC fluorescence in HCMV-infected cells increased to a value about 2.5-fold greater than that observed in mock-infected cells. The relative fluorescence observed in the HCMV-infected cells increased further by 48 h p.i., when it was about eightfold greater than the fluorescence in mock-infected cells. The increases in calpain activity from 24 and 48 h were significant (P < 0.05). These data closely reflect the time course for the changes in the abundance of p21cip1 following HCMV infection in our earlier work (19) and in this study. In these studies, a decrease in p21cip1 levels was first detected at 6 h p.i., and the abundance of p21cip1 continued to decrease through 48 h p.i.
FIG. 7.
(A) The time course for calpain activity in HCMV (5 PFU/cell)-infected (solid bars) and mock-infected (open bars) density-arrested cells. Calpain activity was measured by exposing the cells to the cell-permeant fluorogenic calpain substrate Boc-Leu-Met-CMAC (18 μM) for 15 min before the fluorescence intensity was measured for equal numbers of HCMV- and mock-infected cells, using an SLM 4800S spectrofluorometer. Excitation was at 380 nm; emission was at 460 nm. The polarizers were set at 0° and 50°. The data plotted are the means (± standard deviation) of the findings from three independent experiments. (B) 1, representative casein zymogram. Density-arrested LU cells were either infected with HCMV (5 PFU/cell) or mock infected. The cells were harvested at the times indicated and processed for zymography as described in Materials and Methods. Since calpain activity is pH dependent, the gels were incubated in incubation buffer at either pH 7.5 (optimal for μ-calpain) or pH 7.3 (optimal for m-calpain). Purified μ- and m-calpain (0.1 U of each) were added to the sample in lane 1 as a positive control. The upper band in each lane corresponds to μ-calpain, whereas the lower band corresponds to m-calpain (89). 2, data for calpain activity based on the means (± standard deviation) of the intensity of bands at each time relative to the intensity of the corresponding band at 0 h p.i. for the zymograms shown above (⧫, μ-calpain, HCMV-infected cells; ◊, μ-calpain, mock-infected cells; ●, m-calpain, HCMV-infected cells; ○, m-calpain, mock-infected cells). (C) 1 to 3, abundances of m-calpain, μ-calpain, and calpastatin in parallel cell cultures as determined by Western blot analysis (results of a representative experiment); 4, densitometric analysis of the results shown in panel A and two additional experiments with standard deviation. Solid symbols (⧫, m-calpain; ●, μ-calpain, ▴, calpastatin) and line represent the means of the results for HCMV-infected cells; open symbols (◊, m-calpain; ○, μ-calpain; ▵, calpastatin) and dashed line represent the results for mock-infected cells. Error bars not shown are smaller than the symbol.
Zymography.
Since casein is a well-established target of calpain-mediated proteolysis (87), we used casein zymography to verify that calpain was activated in HCMV-infected cells, to determine which calpain isoenzymes were activated, and to extend the time course data for calpain activity. Density-arrested LU cells were infected with HCMV (5 PFU/cell) or mock infected, harvested at times from 0 to 96 h p.i., and evaluated by casein zymography. The results of casein zymography to evaluate cellular proteolysis are illustrated in Fig. 7B. When both of the ubiquitous calpains are activated, μ-calpain is observed as the upper band, while m-calpain is observed at a slightly lower position in the nondenaturing gels (89). The intensity of the bands was related linearly to the amount of protease activity. Since μ- and m-calpain have slightly different pH optima, duplicate gels were incubated at pH 7.5 or 7.3 to evaluate quantitative changes in isoenzyme activity. There was substantial activity of both ubiquitous calpains at each pH shown when both calpains were activated. The results for a representative experiment from this series of experiments (Fig. 7B) demonstrated that calpain activity gradually decreased in mock-infected cells during the 96-h time course of the experiment. In contrast, HCMV infection induced an increase in both μ- and m-calpain activities by 6 to 9 h p.i. (Fig. 7B, part 2). By 24 h, the calpain activity of HCMV-infected cells was about fourfold (μ-calpain) or fivefold (m-calpain) greater than that of mock-infected cells. The enzyme activity continued to increase and reached its maximum level at 72 h p.i. when it was about 23-fold (μ-calpain) or 25-fold (m-calpain) greater than that observed in mock-infected cells. The increases in calpain activities from 12 to 96 h were significant (P < 0.05) relative to the preinfection levels. Overall, the changes in calpain activity measured by casein zymography were remarkably similar to those obtained using the fluorogenic calpain substrate Boc-Leu-Met-CMAC in intact cells (Fig. 7A) and indicate that HCMV infection was activating both μ- and m-calpain.
To determine if the increased calpain activity observed in Fig. 7A and B was a result of increased expression of calpains or a decrease in the abundance of calpastatin, the abundance of these proteins was measured by Western blot analysis (Fig. 7C) in cells treated in parallel with those used to measure calpain activity (Fig. 7B). HCMV infection had little, if any, detectable effect on the abundance of μ-calpain or calpastatin through 96 h p.i. m-Calpain levels demonstrated a very slight increase from 6 h to 72 h p.i. over the abundance observed in mock-infected cells, and by 96 h, HCMV- and mock-infected cells demonstrated similar levels of m-calpain. Given the very small difference in the abundance of m-calpain in HCMV- and mock-infected cells, it was questionable if this difference had any significance. Mock infection had little, if any, effect on the cellular levels of μ-calpain, m-calpain, or calpastatin during the time course of this series of experiments. Thus, it is unlikely that the substantial increase in calpain activity observed in HCMV-infected cells was a result of a decrease in calpastatin abundance or of increases in the abundance of the ubiquitous calpains.
Calpain cleaves p21cip1.
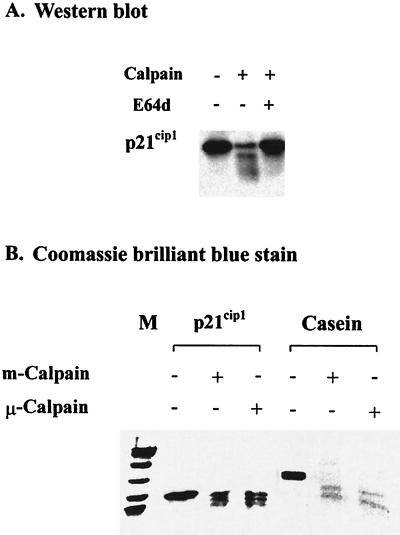
Although a number of proteins are now recognized as targets of calpain-mediated proteolysis (21), p21cip1 has previously not been included among them. Accordingly, we were interested in directly testing the sensitivity of p21cip1 to calpain-mediated proteolysis by incubating purified recombinant p21cip1, obtained from J. Wade Harper (41) or Hengming Ke (53), with either purified μ-calpain or m-calpain and evaluating the products by Western blot analysis. We also prepared recombinant p21cip1 by using the expression plasmid pET-p21, provided by J. Wade Harper (32, 41). Incubation of any of the purified p21cip1 preparations with purified μ-calpain or m-calpain resulted in the rapid cleavage of p21cip1, as illustrated with findings for μ-calpain in Fig. 8A. Two prominent p21cip1 fragments were observed after incubation with calpain, consistent with the results from earlier studies indicating that calpain typically cleaves its target proteins at a limited number of sites (for a review, see reference 78). Inclusion of E64d in the digestion inhibited the proteolysis of p21cip1 (Fig. 8A). To investigate if either of the ubiquitous calpains was removing or modifying the availability of the epitope that was the target of the anti-p21cip1 antibody used in the Western blot analysis, we incubated p21cip21 and casein separately with either μ-calpain or m-calpain and examined the proteolytic products by SDS-PAGE. The location of polypeptides in the gels was demonstrated by Coomassie brilliant blue staining. To minimize the loss of small peptides, electrophoresis of proteins applied to these gels was for a period of time shorter than used for Western blot analysis (Fig. 8A). A limited number of cleavage products was observed in the Coomassie blue-stained gels (Fig. 8B) for either casein or p21cip1 incubated with either of the ubiquitous calpains, as had been observed by Western blot analysis (Fig. 8A). Thus, p21cip1 appeared to be a target for proteolysis mediated by either μ- or m-calpain.
FIG. 8.
Cleavage of purified recombinant p21cip1 by purified calpain. (A) p21cip1 (0.4 μg) was incubated with μ-calpain (0.004 U) for 30 min at 30°C in the presence or absence of E64d (100 μM), and the resulting products were analyzed by Western blot analysis as described in Materials and Methods. The data illustrated are from a representative experiment. (B) p21cip1 (4 μg) or casein (3 μg) was incubated with 0.04 U of μ-calpain or m-calpain for 30 min at 30°C, and the products of the digestion were examined by SDS-PAGE and Coomassie brilliant blue staining. M, Rainbow colored protein molecular weight markers of 97.4, 66, 46, 30, 21.5, and 14.3 kDa (Amersham Life Science). The data illustrated are from a representative experiment.
DISCUSSION
Involvement of p21cip1 in cell cycle progression in HCMV-infected cells.
Progression through the cell cycle is regulated by mechanisms that are beginning to be understood (31, 73, 74). Induction of cyclin E-associated kinase activity is essential for progression through the G1 phase of the cell cycle (for a review, see reference 49) and preparation of the cell for DNA synthesis. Data from this laboratory (19) and others (45) have demonstrated that HCMV infection substantially increases E kinase activity in arrested cells. In density-arrested cells, E kinase activity is first substantially increased at 24 h p.i., reaches its maximum level by 72 h, and is still detectable at 96 h (19), well into the late phase of HCMV replication.
Several mechanisms have been identified that regulate the activity of E kinase. For example, an increased cellular abundance of cyclin E is associated with activation of E kinase (reference 65 and references therein). HCMV infection has been shown to transcriptionally activate the expression of cyclin E, but the time course for the increase in E kinase activity is delayed relative to the changes in the cellular abundance of cyclin E (19). The cellular levels of cyclin E protein in density-arrested cells increase substantially by 12 h p.i., peak by 24 h, and are in decline by 48 h (19). Accordingly, not only does activation of E kinase lag behind the increase in cyclin E abundance, but E kinase remains activated longer than would be anticipated based solely on cyclin E abundance (19). For example, despite the decrease in cyclin E levels at 48 h and continuing reduction thereafter, E kinase activity increases remarkably at 48 h p.i., peaks at 72 h, and still remains high at 96 h p.i. The lack of congruity in the data for cyclin E abundance and E kinase activity suggest that other mechanisms are involved in the regulation of E kinase activity in HCMV-infected cells.
The activity of cyclin E-Cdk2 complexes is also regulated by CKIs, including p21cip1. In fact, it has been suggested that p21cip1 establishes a threshold in the nucleus (34, 44, 75, 86), which must be exceeded if E kinase activity is to develop from cyclin E-Cdk2 complexes. Increases in E kinase activity in HCMV-infected, density-arrested cells closely parallel a substantial reduction in the abundance of p21cip1 by 24 h p.i. (19). Our earlier results suggest that the delay in activation of E kinase activity after the increase in cyclin E abundance and the persistence of the cyclin E-associated kinase activity after cyclin E levels are sharply reduced are related to the effects of the CKIs p21cip1 and p27kip1 (19). Both p21cip1 and p27kip1 are abundant in quiescent LU cells, contributing to the observed delay between increasing cyclin E levels and activation of E kinase until cyclin E-Cdk2 complexes exceed the threshold established by p21cip1 and p27kip1. Later, as cyclin E levels are falling, E kinase remains active because HCMV infection stimulates reduction of both p21cip1 and p27kip1, particularly of p21cip1 (19). Thus, the initial phase of activation of E kinase activity is associated with a substantial increase in cylin E levels and decrease in p21cip1 levels, while a substantial and sustained decrease in p21cip1 is associated with the persistence of E kinase activity observed during HCMV infection (Fig. 1D).
Proteolysis of p21cip1 in mock- and HCMV-infected cells.
The abundance of each protein in a living cell depends on the balance between its synthesis and degradation, and therefore intracellular proteolysis is of critical importance in maintaining functional concentrations. In fact, many regulatory proteins are subject to proteolytic degradation, which allows cells to rapidly adjust their concentration. Reduction of p21cip1 levels in noninfected cells is reported to be mediated by ubiquitination and proteasome degradation (10, 35, 38, 85). Indeed, in mock-infected LU cells, proteasome degradation was the predominant pathway for p21cip1 proteolysis (Fig. 2). In contrast, the present results indicate that in HCMV-infected cells, the proteosome is responsible for little, whereas calpain activity is responsible for much, of the degradation of p21cip1 observed during the progression of the virus infection. The early decrease in p21cip1 abundance appears to be related in part to decreasing RNA levels (Fig. 1) and in part to increasing calpain-mediated degradation (Fig. 6). Since the half-life of p21cip1 is reported to be approximately 2 h (46), a decrease in RNA levels would predictably result in lower levels of the protein. When the activity of calpain was inhibited at these early times (e.g., 9 h p.i.), p21cip1 levels rose to 158% of the preinfection levels, compared with 83.1% in the absence of calpain inhibitors (Fig. 6). At 24 h p.i., when a substantial increase in E kinase activity is first detected, p21cip1 levels are at 23% of their preinfection levels, but when calpain activity is blocked, p21cip1 levels are at 85% of their preinfection levels. This level of p21cip1 is similar to the levels observed in absence of calpain inhibition at 9 and 12 h p.i. (83 and 72%, respectively), when E kinase activity is at a very low level (19). Thus, calpain-mediated mechanisms appear to be responsible for the degradation of a substantial portion of p21cip1, even at early times in HCMV replication. At later times (after 48 h), when RNA levels are relatively constant, calpain-mediated mechanisms are largely responsible for the continuing decrease in p21cip1 abundance. During this period (48 and 72 h p.i.), inhibition of calpain-catalyzed proteolysis produced cellular levels of p21cip1 that were from 77.5 to 36.4% of the preinfection levels, whereas in the absence of calpain inhibitors, the level of p21cip1 fell to 10.8 and 6.2%, respectively, of the preinfection level (Fig. 6). Thus, HCMV-induced activation of calpain is important at both early and late times in controlling the abundance of p21cip1 and, accordingly, the activity of the cyclin E-associated kinase. The calpain effect seems particularly noteworthy in extending the activation of E kinase well into the late phase of HCMV replication. Since E kinase activity has been shown to be necessary for high infectious yields of HCMV (18), it is tempting to speculate that the sustained calpain activity and persistent low levels of p21cip1 support durable activation of E kinase and production of high yields of infectious HCMV. Indeed, preliminary experiments indicate that a single exposure to Z-Leu-Leu-H (100 μM) is sufficient to reduce HCMV infectious yields at 96 h PI by 99.7% (P < 0.05).
Several experimental approaches support the view that calpain activity and p21cip1 levels are closely coupled in HCMV-infected cells. Calpain activity increases in a temporal pattern that is consistent with the time courses for the decrease in p21cip1 abundance and the increase in E kinase activity in HCMV-infected cells (compare Fig. 7 with Fig. 1; see also reference 19). The time course for E64d protection of p21cip1 (Fig. 3) closely parallels the decrease in p21cip1 (Fig. 1) and the increase in calpain activity (Fig. 7). The presence of inhibitors of calpain activity (E64d, Z-Leu-Leu-H) protects up to about 80% of p21cip1 (Fig. 4 and 5). The concentration effect of E64d in providing protection of p21cip1 in HCMV-infected LU cells is in the range (6.25 to 100 μM) observed in earlier studies to protect other targets of calpain-mediated proteolysis (54). In cell-free studies, purified calpain degrades purified recombinant p21cip1 (Fig. 8). Considered together, these results indicate that calpain activity is largely responsible for the substantial decrease in p21cip1 observed during HCMV infection.
Analysis of PEST sequences in p21cip1.
The degradation of cellular proteins frequently depends on some structural feature or consensus sequence. For example, PEST sequences, regions that are rich in proline (P), glutamic acid (E), serine (S), and threonine (T), flanked by clusters rich in basic amino acids, are routinely found in short-lived proteins (66; for a review, see reference 64). It has been demonstrated that many of the preferred substrates of calpain possess the PEST motif (64). Thus, the observation that p21cip1 is rich in PEST amino acids (33, 41) is of particular interest. Identification of putative PEST sites from these PEST amino acids can be facilitated by analysis with the PEST-FIND computer program (available at either http://www.icnet.uk/LRITu/projects/pest/index.html or http://emb1.bcc.univie.ac.at/embnet/tools/bio/PESTfind). Results of PEST-FIND analysis indicate that p21cip1 contains a PEST site, located between amino acids 122 and 140 (RSGEQAEGSPGGPGDSQGR), with a PEST-FIND score of +5.11, a mole fraction of PEDST of 33.47, and a hydrophobicity index of 26.59. Although p21cip1 also contains several other PEST amino acid-rich sequences, PEST-FIND analysis predicts that these motifs are poor target PEST sequences, because their PEST-FIND scores are less than 5 (−13.73, −5.23, +1.57 −21.61). PEST sequences are often conditional proteolytic signals that may be masked in certain situations but may be activated after binding to ligands or after posttranslational modification, such as protein phosphorylation (64). Although statistical analysis has shown a significant relationship between PEST motifs and rapid protein degradation (64, 66), the direct role of the sequence in calpain-mediated proteolysis is still not clear. For some proteins, PEST sequences are not required for calpain-mediated proteolysis (reference 22 and references therein).
Calpain-mediated cleavage of p21cip1.
Our results show that cleavage of purified recombinant p21cip1 resulted in two major protein fragments in cell-free experiments (Fig. 8), consistent with earlier work indicating that cleavage at a limited number of sites is a characteristic of calpain-mediated proteolysis (for a review, see reference 78). The absence of these calpain-mediated p21cip1 fragments in lysates of HCMV-infected LU cells suggests that these p21cip1 fragments are unstable in living cells. The products of calpain-mediated p21cip1 cleavage may be subject to further degradation by the ubiquitin-proteasome or other proteolytic pathways. This latter possibility is supported by the demonstration that calpain-dependent cleavage of p53 is necessary for subsequent ubiquitin-dependent degradation (50).
Calpain activity in HCMV infection.
These results are the first indication that calpain activity is involved in HCMV infection. Calpains (EC 3.4.22.17) are a family of cysteine proteases. The two major calpain isoforms, μ-calpain and m-calpain, which are ubiquitously distributed in cells, differ in the amount of Ca2+ required for activation in cell-free assays. They possess a calmodulin-like Ca2+-binding domain and are subject to intracellular Ca2+ regulation. Therefore, they are considered to participate in Ca2+-associated signal transduction pathways. Calpain activity is associated with the propagation of Ca2+ signaling (21), in some instances converting transient signals to more durable and amplified signals (92). It has been known for some time that HCMV infection induces a substantial increase in cytosolic Ca2+ activity (58). The timing of the intracellular free Ca2+ increase in HCMV-infected cells (58) is consistent with the increase in calpain activity illustrated in Fig. 7. Activation of calpains is also influenced by phospholipid degradation (9, 27), and HCMV infection has been previously shown to stimulate the breakdown of phospholipids. Valyi-Nagy et al. (81) reported that HCMV infection stimulated the hydrolysis of phosphatidylinositol 4,5-bisphosphate, yielding increases in the cellular levels of inositol 1,4,5-trisphosphate and DG. Increased cellular levels of DG are associated with activation of arachidonic acid metabolism. Indeed, in HCMV-infected cells, arachidonic acid metabolism is also stimulated, as revealed by the release of arachidonic acid and its metabolites (1, 2).
Considered together, these findings indicate that HCMV infection induces an intracellular environment that is consistent with stimulation of calpain activity. Further, the data support the interpretation that the enhanced calpain activity plays a role in modulating p21cip1 abundance, facilitating initiation of cyclin E-associated kinase activity, and permitting continued E kinase activity well into the late phase of HCMV infection. We have postulated previously that the limited progression of the cell cycle observed with productive HCMV infection establishes a cellular environment conducive to efficient HCMV replication (5, 19). Activation of the ubiquitous calpains and degradation of p21cip1 appear to be important components in the cellular changes supporting limited cell cycle progression and HCMV replication.
A role for HCMV IE2-86 in cell cycle regulation has been reported. As noted previously, IE2-86 participates in activating transcription from the cyclin E gene (17). Other roles for IE2-86 in cell cycle regulation have also been proposed. IE2-86 interacts with p53 (15, 77, 80) and pRb (37, 40, 47, 63), which could help to facilitate HCMV-induced cell cycle traverse. IE2-86 has been reported to interact also with p21cip1 (76), which could potentially assist in releasing the p21cip1-mediated block of cell cycle progression. The observation that p21cip1 abundance substantially declines during productive HCMV infection (19) (Fig. 1) suggests that IE2-86 binding to p21cip1 may not be a quantitatively important mechanism for relief of the p21cip1 block in productively infected cells. It is possible that degradation of p21cip1 may have the peripheral effect of preventing a substantial reduction of the effective abundance of IE2-86 and a corresponding decrease in its transcriptional effect. It seems somewhat paradoxical that IE2-86 has also been reported to block cell cycle progression in the S phase (55, 76), since the published effects of IE2-86 on cyclin E expression and Cdk activities (through interaction with p21cip1) suggest that IE2-86 should promote cell cycle progression (23). A number of studies of HCMV infection in permissive arrested cells have noted that traverse of the cell cycle during productive infection is limited to a point at or near the G1/S boundary (19, 30, 52). Other studies of productive HCMV infection in which arrest is observed in the S phase (45, 55, 76) may be related to interpreting uncharacterized DNA synthesis as cellular, when it may be viral, or the effects of transfection and expression of IE2-86 in the absence of the entire repertoire of HCMV genes. Indeed, Wiebusch and Hagemeier (83) observed that IE2-86 blocked cell progression in G1. Although it is clear that IE2-86 participates in cell cycle activation by HCMV, additional work is needed to characterize its role and the role of cellular molecules in the limited cell cycle progression induced by productive HCMV infection.
ACKNOWLEDGMENTS
We thank the following researchers for reagents or constructs used in this study: B. Vogelstein for the WAF1 plasmid; J. W. Harper for plasmid pET-p21 and for recombinant p21cip1; H. Ke for recombinant p21cip1; R. L. Mellgren for antibodies with specificity to m-calpain, μ-calpain, or calpastatin; and B. Xu, X. Liang, and D. Coppenhaver for assistance with purification of recombinant p21cip1. Thanks also go to M. Rechsteiner, who provided the PEST-FIND computer program. The helpful advice of T. Wood is also much appreciated. We are grateful to W. R. Fleischmann, Jr., for helpful suggestions in preparing this report.
This work was supported by NIH grants DE11389 (T.A.), NS29261 (A.K.), and ES10018 (P. Muganda) and by NIEHS Center core grant ES06676.
REFERENCES
- 1.AbuBakar S, Boldogh I, Albrecht T. Stimulation of [3H] release from [3H]-arachidonic acid prelabeled cells. Arch Virol. 1990;113:255–266. doi: 10.1007/BF01316678. [DOI] [PubMed] [Google Scholar]
- 2.AbuBakar S, Boldogh I, Albrecht T. Human cytomegalovirus stimulates arachidonic acid metabolism through pathways that are affected by inhibitors of phospholipase A2 and protein kinase C. Biochem Biophys Res Commun. 1990;166:953–959. doi: 10.1016/0006-291x(90)90903-z. [DOI] [PubMed] [Google Scholar]
- 3.Albrecht T, Weller T H. Heterogeneous morphologic features of plaques induced by five strains of human cytomegalovirus. Am J Clin Pathol. 1980;73:648–654. doi: 10.1093/ajcp/73.5.648. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht T, Boldogh I, Fons M P. Receptor-initiated activation of cells and their oncogenes by herpes-family viruses. J Investig Dermatol. 1992;98(Suppl.):29S–35S. doi: 10.1111/1523-1747.ep12462169. [DOI] [PubMed] [Google Scholar]
- 5.Albrecht T, Boldogh I, Fons M, Lee C H, AbuBakar S, Russell J M, Au W W. Cell-activation responses to cytomegalovirus infection: relationship to the phasing of CMV replication and to the induction of cellular damage. Subcell Biochem. 1989;15:157–202. [PubMed] [Google Scholar]
- 6.Albrecht T, Cavallo T, Cole N L, Graves K. Cytomegalovirus: development and progression of cytopathic effects in human cell culture. Lab Investig. 1980;42:1–7. [PubMed] [Google Scholar]
- 7.Albrecht T, Nachtigal M, St. Jeor S C, Rapp F. Induction of cellular DNA synthesis and increased mitotic activity in Syrian hamster embryo cells abortively infected with human cytomegalovirus. J Gen Virol. 1976;30:167–177. doi: 10.1099/0022-1317-30-2-167. [DOI] [PubMed] [Google Scholar]
- 8.Alford C A, Stagno S, Pass R F, Britt W J. Congenital and perinatal cytomegalovirus infections. Rev Infect Dis. 1990;12(Suppl.):S745–S753. doi: 10.1093/clinids/12.supplement_7.s745. [DOI] [PubMed] [Google Scholar]
- 9.Arora A S, de Groen P, Emori Y, Gores G J. A cascade of degradative hydrolase activity contributes to hepatocyte necrosis during anoxia. Am J Physiol. 1996;270:G238–G245. doi: 10.1152/ajpgi.1996.270.2.G238. [DOI] [PubMed] [Google Scholar]
- 10.Blagosklonny M V, Wu G S, Omura S, el-Deiry W S. Proteasome-dependent regulation of p21WAF1/CIP1 expression. Biochem Biophys Res Commun. 1996;227:564–569. doi: 10.1006/bbrc.1996.1546. [DOI] [PubMed] [Google Scholar]
- 11.Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 12.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldogh I, AbuBakar S, Millinoff D, Deng C Z, Albrecht T. Cellular oncogene activation by human cytomegalovirus: lack of correlation with virus infectivity and IE gene expression. Arch Virol. 1991;118:163–177. doi: 10.1007/BF01314027. [DOI] [PubMed] [Google Scholar]
- 14.Boldogh I, Fons M P, Albrecht T. Increased activity of DNA-binding proteins in nuclei of human cytomegalovirus-infected MRC-5 cells. Biochem Biophys Res Commun. 1993;197:1505–1510. doi: 10.1006/bbrc.1993.2647. [DOI] [PubMed] [Google Scholar]
- 15.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman L H, Rabin B, Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981;9:4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 18.Bresnahan W A, Boldogh I, Chi P, Thompson E A, Albrecht T. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology. 1997;231:239–247. doi: 10.1006/viro.1997.8489. [DOI] [PubMed] [Google Scholar]
- 19.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 20.Bresnahan W A, Thompson E A, Albrecht T. Human cytomegalovirus infection results in altered Cdk2 subcellular localization. J Gen Virol. 1997;78:1993–1997. doi: 10.1099/0022-1317-78-8-1993. [DOI] [PubMed] [Google Scholar]
- 21.Carafoli E, Molinari M. Calpain: a protease in search of a function? Biochem Biophys Res Commun. 1998;247:193–203. doi: 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- 22.Carillo S, Pariat M, Steff A M, Jariel-Encontre L, Poulat F, Berta P, Piechaczyk M. PEST motifs are not required for rapid calpain-mediated proteolysis of c-fos protein. Biochem J. 1996;313:245–251. doi: 10.1042/bj3130245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castillo J P, Yurochko A D, Kowalik T F. Role of human cytomegalovirus immediate-early proteins in cell growth control. J Virol. 2000;74:8028–8037. doi: 10.1128/jvi.74.17.8028-8037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen I-T, Smith M L, O'Connor P M, Fornace A J., Jr Direct interaction of Gadd45 with PCNA and evidence for competitive interaction of Gadd45 and p21Waf1/Cip1 with PCNA. Oncogene. 1995;11:1931–1937. [PubMed] [Google Scholar]
- 25.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15:532–534. [PubMed] [Google Scholar]
- 26.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 27.Coolican S A, Hathaway D R. Effect of l-α-phosphatidylinositol on a vascular smooth muscle Ca2+-dependent protease. Reduction of the Ca2+ requirement for autolysis. J Biol Chem. 1984;259:11627–11630. [PubMed] [Google Scholar]
- 28.Croall D E, DeMartino G N. Calcium-activated neutral protease (calpain) system: structure, function, and regulation. Physiol Rev. 1991;71:813–847. doi: 10.1152/physrev.1991.71.3.813. [DOI] [PubMed] [Google Scholar]
- 29.de Jong M D, Galasso G J, Gazzard B, Griffiths P D, Jabs D A, Kern E R, Spector S A. Summary of the 2nd International Symposium on Cytomegalovirus. Antiviral Res. 1998;39:141–162. doi: 10.1016/s0166-3542(98)00044-8. [DOI] [PubMed] [Google Scholar]
- 30.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Draetta G F. Mammalian G1 cyclins. Curr Opin Cell Biol. 1994;6:842–846. doi: 10.1016/0955-0674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 32.Dynlacht B D, Ngwu C, Winston J, Swindell E C, Elledge S J, Harlow E, Harper J W. Purification and analysis of CIP/KIP proteins. Methods Enzymol. 1997;283:230–244. doi: 10.1016/s0076-6879(97)83019-4. [DOI] [PubMed] [Google Scholar]
- 33.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 34.Elledge S J, Harper J W. Cdk inhibitors: on the threshold of checkpoints and development. Curr Opin Cell Biol. 1994;6:847–852. doi: 10.1016/0955-0674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 35.Elledge S J, Harper J W. The role of protein stability in the cell cycle and cancer. Biochim Biophys Acta. 1998;1377:M61–M70. doi: 10.1016/s0304-419x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 36.Flores-Rozas H, Kelman Z, Dean F B, Pan Z Q, Harper J W, Elledge S J, O'Donnell M, Hurwitz J. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase δ holoenzyme. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortunato E A, Sommer M H, Yoder K, Spector D H. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71:8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukuchi K, Tomoyasu S, Nakamaki T, Tsuruoka N, Gomi K. DNA damage induces p21 protein expression by inhibiting ubiquitination in ML-1 cells. Biochim Biophys Acta. 1998;1404:405–411. doi: 10.1016/s0167-4889(98)00089-5. [DOI] [PubMed] [Google Scholar]
- 39.Gervais J L, Seth P, Zhang H. Cleavage of CDK inhibitor p21Cip1/Waf1 by caspases is an early event during DNA damage-induced apoptosis. J Biol Chem. 1998;273:19207–19212. doi: 10.1074/jbc.273.30.19207. [DOI] [PubMed] [Google Scholar]
- 40.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 42.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirai S, Kawasaki H, Yaniv M, Suzuki K. Degradation of transcription factors, c-Jun and c-Fos, by calpain. FEBS Lett. 1991;287:57–61. doi: 10.1016/0014-5793(91)80015-u. [DOI] [PubMed] [Google Scholar]
- 44.Hunter T, Pines J. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 45.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johannesesn L E, Knardal S L, Madshus I H. Epidermal growth factor increases the level of cyclin-dependent kinase (CDK) inhibitor p21/CIP1 (CDK-interacting protein 1) in A431 cells by increasing the half-lives of the p21/CIP1 transcript and the p21/CIP1 protein. Biochem J. 1999;337:599–606. [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson R A, Yurochko A D, Poma E E, Zhu L, Huang E S. Domain mapping of the human cytomegalovirus IE1–72 and cellular p107 protein-protein interaction and the possible functional consequences. J Gen Virol. 1999;80:1293–1303. doi: 10.1099/0022-1317-80-5-1293. [DOI] [PubMed] [Google Scholar]
- 48.Kearsey J M, Coates P J, Prescott A R, Warbrick E, Hall P A. Gadd45 is a nuclear cell cycle regulated protein which interacts with p21Cip1. Oncogene. 1995;11:1675–1683. [PubMed] [Google Scholar]
- 49.Keyomarsi K, Herliczek T W. The role of cyclin E in cell proliferation, development and cancer. Prog Cell Cycle Res. 1997;3:171–191. doi: 10.1007/978-1-4615-5371-7_14. [DOI] [PubMed] [Google Scholar]
- 50.Kubbutat M H, Vousden K H. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol Cell Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li R, Waga S, Hannon G J, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 52.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayrose D R, Nichols M A, Xiong Y, Ke H. Purification and crystallization of cyclin-dependent kinase inhibitor p21. Protein Sci. 1996;5:1928–1930. doi: 10.1002/pro.5560050920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mellgren R L. Evidence for participation of a calpain-like cysteine protease in cell cycle progression through late G1 phase. Biochem Biophys Res Commun. 1997;236:555–558. doi: 10.1006/bbrc.1997.7003. [DOI] [PubMed] [Google Scholar]
- 55.Murphy E A, Streblow D N, Nelson J A, Stinski M. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74:7198–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray E J, Bentley G V, Grisanti M S, Murray S S. The ubiquitin-proteasome system and cellular proliferation and regulation in osteoblastic cells. Exp Cell Res. 1998;242:460–469. doi: 10.1006/excr.1998.4090. [DOI] [PubMed] [Google Scholar]
- 57.Noda A, Ning Y, Venable S F, Pereira-Smith O M, Smith J R. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 58.Nokta M, Eaton D, Steinsland O S, Albrecht T. Ca2+ responses in cytomegalovirus-infected fibroblasts of human origin. Virology. 1987;157:259–267. doi: 10.1016/0042-6822(87)90268-6. [DOI] [PubMed] [Google Scholar]
- 59.Pahl H L, Baeuerle P A. Control of gene expression by proteolysis. Curr Opin Cell Biol. 1996;8:340–347. doi: 10.1016/s0955-0674(96)80007-x. [DOI] [PubMed] [Google Scholar]
- 60.Pan Z Q, Reardon J T, Li L, Flores-Rozas H, Legerski R, Sancar A, Hurwitz J. Inhibition of nucleotide excision repair by the cyclin-dependent kinase inhibitor p21. J Biol Chem. 1995;270:22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- 61.Perez J L. Resistance to antivirals in human cytomegalovirus: mechanisms and clinical significance. Microbiologia. 1997;13:343–352. [PubMed] [Google Scholar]
- 62.Podust V N, Podust L M, Goubin F, Ducommun B, Hubscher U. Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry. 1995;34:8869–8875. doi: 10.1021/bi00027a039. [DOI] [PubMed] [Google Scholar]
- 63.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E S. The human cytomegalovirus IE1–72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 65.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 67.Rosser B G, Powers S P, Gores G J. Calpain activity increases in hepatocytes following addition of ATP. Demonstration of a novel fluorescent approach. J Biol Chem. 1993;268:23593–23600. [PubMed] [Google Scholar]
- 68.Rubin R H. Impact of cytomegalovirus infection on organ transplant recipients. Rev Infect Dis. 1990;12(Suppl.):S754–S766. doi: 10.1093/clinids/12.supplement_7.s754. [DOI] [PubMed] [Google Scholar]
- 69.Saito Y, Tsubuki S, Ito H, Kawashima S. The structure-function relationship between peptide aldehyde derivatives on initiation of neurite outgrowth in PC12h cells. Neurosci Lett. 1990;120:1–4. doi: 10.1016/0304-3940(90)90153-z. [DOI] [PubMed] [Google Scholar]
- 70.Santella L. The role of calcium in the cell cycle: facts and hypotheses. Biochem Biophys Res Commun. 1998;244:317–324. doi: 10.1006/bbrc.1998.8086. [DOI] [PubMed] [Google Scholar]
- 71.Santella L, Kyozuka K, De Riso L, Carafoli E. Calcium, protease action, and the regulation of the cell cycle. Cell Calcium. 1998;23:123–130. doi: 10.1016/s0143-4160(98)90110-5. [DOI] [PubMed] [Google Scholar]
- 72.Schooley R T. Cytomegalovirus in the setting of infection with human immunodeficiency virus. Rev Infect Dis. 1990;12(Suppl.):S811–S819. doi: 10.1093/clinids/12.supplement_7.s811. [DOI] [PubMed] [Google Scholar]
- 73.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 74.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 75.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 76.Sinclair J, Baillie J, Bryant L, Caswell R. Human cytomegalovirus mediates cell cycle progression through G1 into early S phase in terminally differentiated cells. J Gen Virol. 2000;81:1553–1565. doi: 10.1099/0022-1317-81-6-1553. [DOI] [PubMed] [Google Scholar]
- 77.Speir E, Huang E S, Modali R, Leon M B, Shawl F, Finkel T, Epstein S E. Interaction of human cytomegalovirus with p53: possible role in coronary restenosis. Scand J Infect Dis Suppl. 1995;99:78–81. [PubMed] [Google Scholar]
- 78.Suzuki K, Sorimachi H. A novel aspect of calpain activation. FEBS Lett. 1998;433:1–4. doi: 10.1016/s0014-5793(98)00856-4. [DOI] [PubMed] [Google Scholar]
- 79.Tamai M, Matsumoto K, Omura S, Koyama L, Ozawa Y, Hanada K. In vitro and in vivo inhibition of cysteine proteinases by EST, a new analog of E-64. J Pharmacobiodyn. 1986;9:672–677. doi: 10.1248/bpb1978.9.672. [DOI] [PubMed] [Google Scholar]
- 80.Tsai H L, Kou G H, Chen S C, Wu C W, Lin Y S. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem. 1996;271:3534–3540. [PubMed] [Google Scholar]
- 81.Valyi-Nagy T, Bandi Z, Boldogh I, Albrecht T. Hydrolysis of inositol lipids: an early signal of human cytomegalovirus infection. Arch Virol. 1988;101:199–207. doi: 10.1007/BF01311001. [DOI] [PubMed] [Google Scholar]
- 82.Waga S, Hannon G J, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 83.Wiebusch L, Hagemeier C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 85.Yu Z K, Gervais J L, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W, Mellgren R L. Calpain subunits remain associated during catalysis. Biochem Biophys Res Commun. 1996;227:891–896. [PubMed] [Google Scholar]
- 88.Zhang W, Lane R D, Mellgren R L. The major calpain isozymes are long-lived proteins. Design of an antisense strategy for calpain depletion in cultured cells. J Biol Chem. 1996;271:18825–18830. doi: 10.1074/jbc.271.31.18825. [DOI] [PubMed] [Google Scholar]
- 89.Zhao X, Newcomb J K, Posmantur R M, Wang K K, Pike B R, Hayes R L. pH dependency of mu-calpain and m-calpain activity assayed by casein zymography following traumatic brain injury in the rat. Neurosci Lett. 1998;247:53–57. doi: 10.1016/s0304-3940(98)00219-5. [DOI] [PubMed] [Google Scholar]
- 90.Zhao X, Posmantur R, Kampfl A, Liu S J, Wang K K, Newcombm J K, Pike B R, Clifton G L, Hayes R L. Subcellular localization and duration of mu-calpain and m-calpain activity after traumatic brain injury in the rat: a casein zymography study. J Cereb Blood Flow Metab. 1998;18:161–167. doi: 10.1097/00004647-199802000-00006. [DOI] [PubMed] [Google Scholar]
- 91.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zimmerman U J, Wang M, Nelson J B, Ekwunife F S, Liu L. Secretagogue-induced proteolysis of cAMP-dependent protein kinase in intact rat alveolar epithelial type II cells. Biochim Biophys Acta. 1996;1311:117–123. doi: 10.1016/0167-4889(95)00181-6. [DOI] [PubMed] [Google Scholar]