Abstract
The viral ion channel protein M2 supports the transit of influenza virus and its glycoproteins through acidic compartments of the cell. M2 conducts endosomal protons into the virion to initiate uncoating and, by equilibrating the pH at trans-Golgi membranes, preserves the native conformation of acid-sensitive viral hemagglutinin. The exceptionally low conductance of the M2 channel thwarted resolution of single channels by electrophysiological techniques. Assays of liposome-reconstituted M2 yielded the average unitary channel current of the M2 tetramer—1.2 aA (1.2 × 10−18 A) at neutral pH and 2.7 to 4.1 aA at pH 5.7—which activates the channel. Extrapolation to physiological temperature predicts 4.8 and 40 aA, respectively, and a unitary conductance of 0.03 versus 0.4 fS. This minute activity, below previous estimates, appears sufficient for virus reproduction, but low enough to avert abortive cytotoxicity. The unitary permeability of M2 was within the range reported for other proton channels. To address the ion selectivity of M2, we exploited the coupling of ionic influx and efflux in sealed liposomes. Metal ion fluxes were monitored by proton counterflow, employing a pH probe 1,000 times more sensitive than available Na+ or K+ probes. Even low-pH-activated M2 did not conduct Na+ and K+. The proton selectivity of M2 was estimated to be at least 3 × 106 (over sodium or potassium ions), in agreement with electrophysiological studies. The stringent proton selectivity of M2 suggests that the cytopathology of influenza virus does not involve direct perturbation of cellular sodium or potassium gradients.
Viruses such as influenza virus (A, B, and C) and human immunodeficiency virus have evolved ion channel proteins that assist their invasion of the host cell or their egress from the biosynthetic machinery (for reviews, see references 14, 19, and 20). As the first viral ion channel protein to be discovered, as well as the target of the classic antivirals amantadine and rimantadine, the influenza A virus M2 protein has become the paradigm of this new class of viral proteins. For uncoating, the virus is dependent on the acidity of the endosome, but to protect the maturation of acid-sensitive hemagglutinin (HA [of the H7 and H5 subtypes]), it needs to avoid the low pH in the trans-Golgi network (TGN). The M2 protein fulfills both of these functions by equilibrating membrane pH gradients (14).
We developed procedures for the expression, isolation, and reconstitution of the M2 protein into liposomes, as well as a functional assay, demonstrating that M2 translocates protons in a rimantadine-sensitive manner (36). We now present quantitative data on single-channel conductance and ion selectivity determined in this system.
The initial report (28) on the electrophysiology of the M2 protein and several later studies represented M2 as an acid-activated sodium ion or unspecific monovalent cation channel (37, 41, 42). On the other hand, whole-cell recordings of M2-expressing MEL cells confirmed our observation of proton conductivity and, furthermore, showed that the channel was virtually impermeable to sodium ions (3, 27). Pinto and coworkers in a very recent electrophysiological study uncovered causes for these apparent discrepancies and came to the conclusion that plasma membrane-expressed M2 protein is proton selective as well in Xenopus oocytes and CV-1 cells (26).
The extremely low activity of the M2 ion channel has precluded the resolution of single proton channels by electrophysiological methods (3, 25, 27); however, the quantitative definition of the proteoliposome assay system (23, 36) has now enabled the determination of an average single-channel conductance for the M2 protein. We have also adapted our system to reveal the differences in selectivity of the isolated M2 protein for the physiological monovalent cations (protons, sodium, and potassium ions), avoiding interference by other proteins and cellular components present during whole-cell electrophysiological recordings. We found that the M2 protein did not conduct sodium or potassium ions either at neutral pH or at the weakly acidic pH that activates the proton channel.
MATERIALS AND METHODS
Expression isolation, and quantification of the M2 protein.
The M2 protein of influenza A/Germany/27 virus (H7N7 “Weybridge”) was expressed from a recombinant baculovirus in Trichoplusia ni insect cells and purified essentially as described previously (36), except that immunoaffinity chromatography was done by fast protein liquid chromatography (FPLC). The eluate was desalted, rebuffered into a mixture of 20 mM HEPES-buffered saline (pH 7.8) (HBS) and 40 mM β-octylglucoside (OG), and concentrated through Centriprep 30 or Centriplus 30 membrane (Amicon Millipore) at a relative centrifugal force of 1,500, and insoluble material was discarded. The purity of the M2 protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), staining with colloidal Coomassie (GELCODE Blue stain reagent; Pierce, Rockford, Ill.), and Western blotting. The preparation was checked for degradation products by developing Western blots with antibodies to the N terminus (K2) and C terminus (R54 or R66) of M2.
For native, horizontal agarose protein electrophoresis the REP automatic electrophoresis system (Helena Laboratories, Sunderland, United Kingdom), used in the diagnostics of human high- and low-density lipoprotein (HDL and LDL, respectively), was adapted. Custom-made 1% agarose gels in sodium barbital (pH 8.3) (HDL Plus Gel) were run at 4,000 V for 20°C for 5 min. The 1-μl samples contained 250 to 500 ng of M2 protein in HBS-OG. Where indicated, 0.05% sodium taurodeoxycholate or 0.34% Servablue (Coomassie blue; Serva) was included. The protein standard was human HDL-LDL (Helena Laboratories). Agarose gels were fixed in 10% acetic acid for 10 min at room temperature, washed with distilled water, stained with a cholesterol detection kit (REP HDL Plus reagent; Helena Laboratories), subsequently re-hydrated, washed in blotting buffer (25 mM Tris, 40 mM 6-amino-n-hexanoic acid, 20% methanol), and contact blotted onto nitrocellulose.
The concentration of M2 preparations was determined by UV spectroscopy from the absorbance maximum at 278 nm, the molar extinction coefficient (ɛtotal), and the molecular weight of the M2 tetramer, 45,560. The molecular weight was calculated from the sequence of Weybridge M2 (PubMed accession no. S07946, PID 77196) amino acids 2 to 97, including four phosphate and four palmitate groups: ɛtotal = no. of Trp × ɛTrp + no. of Tyr × ɛTyr + no. of Cys × ɛCys, where ɛTrp = 5,560, ɛTyr = 1,200, and ɛCys = 150 (2).
Reconstitution of M2 into liposomes.
Depending on the type of liposome to be prepared, the isolated protein was rebuffered into buffer containing a single metal ion; KPS (12 mM K2HPO4, 50 mM K2SO4 [pH 7.4]) or NaPS (12 mM Na2HPO4, 50 mM Na2SO4 [pH 7.4]). Complex liposomes (45) were composed of phosphatidylcholine (P7763; all lipids from Sigma), sphingomyelin (S0756), phosphatidylethanolamine (P8704), phosphatidylserine (P6641), phosphatidylinositol (P0639), gangliosides (G9886), and cholesterol (C8667) (molar ratio, 10:3:3:1:0.5:0.32:14), and simple liposomes (6) contained l-α-dimyristoylphosphatidylcholine (DMPC)-phosphatidylserine (PS) (85:15). M2 and control vesicles were prepared as described previously (23, 36) with 0.2 mol% valinomycin, except for ion selectivity and inhibitor preincubation experiments, in which ionophores were omitted. The lipid film was carefully taken up in 10 μl of 400 mM OG, followed immediately by 90 μl of buffer (NaPS or KPS) and 50 μg of M2 in 100 μl of the same buffer containing 40 mM OG at 37°C. Liposomes were formed in a dialysis cassette (Slide-a-lyzer; Pierce) by dialysis against three changes (every 4 h and overnight) of 3 vol of the same buffer, followed by three changes of 10 vol, and finally 2 changes of 5 liters for 12 h in the presence of Amberlite XAD-2 (Sigma). All buffers except the last contained 0.04% sodium azide. The fluorescent pH indicator pyranine (2 mM; Molecular Probes), present during the first two steps of dialysis, became entrapped in the liposomes. The integrity of liposome-inserted M2 was checked by PAGE and Western blots. Control liposomes were prepared in parallel without M2. The internal pHs of M2 and control vesicles are often not identical (23).
Determination of the size and buffer capacity of the liposomes.
The size of the liposomes was determined by photon correlation spectroscopy (12, 22) with a Coulter model N4MD Sub-micro Particle Analyzer with multiple scattering angle detection and size distribution processor analysis (30). Assays were run in duplicate or triplicate at 18°C. The buffer capacity of the liposome lumen was calculated as described by Dencher et al. (6) from the decay kinetics of a pH gradient by using liposomes made up in 12, 24, or 36 mM potassium phosphate buffer and 100 mM KCl.
Analysis of the orientation of the M2 protein to the liposome membrane.
M2 vesicles (100 μl) were digested with trypsin (20 μg) for 30 min at 37°C and immediately diluted into ice-cold 100 mM Tris-HCl (pH 7.4)–1% Triton X-100. To assess N- and C-terminal accessibility to trypsin, 1-μl aliquots were spotted onto two nitrocellulose sheets subsequently developed with rabbit sera specific to the N terminus and the C terminus of M2. Digests were also analyzed by PAGE and Western blots.
Cation translocation assay.
Reagents were equilibrated and reactions were usually recorded at 18°C. M2 or control vesicles (5 to 10 μl) were injected into 2 ml of incubation buffer, NaPS, or KPS with a syringe. Where indicated, ionophores (monensin, 5 nM; or valinomycin, 50 nM) were added. The sample was stirred continuously. Pyranine emission at 510 nm at two excitation wavelengths (410 and 460 nm) was recorded essentially as described previously (36) at 1-s intervals with an SLM AB 2 fluorimeter (Aminco-Bowman). The fluorescence ratio, calibrated with standard buffers (KPS or NaPS at pH 5.0 to 9.0 in increments of 0.1 to 0.2 pH units), is proportional to the internal pH of the liposome (6). Plots were generated as the average of three to five recordings. Inhibitor studies were performed by preincubating vesicles in the presence of rimantadine under incubation conditions and triggering proton translocation by adding ionophore.
Derivation of single-channel permeability.
Under constant field conditions (1) at the reversal potential (Erev), the single-channel permeability (p) for an ion (X+) is related to the single-channel conductance γ = pZ′[X+]out[X+]in([X+]out − [X+]in)−1, with Z′ = Erevz3F3(RT)−2. F is the Faraday constant, R is the gas constant, T is the absolute temperature, and z is the charge. When calculating p from γ, a factor of 1,000 is introduced to transform the units of volume from liters to cubic centimeters. As demonstrated by Ogden et al. (3, 27), the Erev of M2 is close to the proton equilibrium potential. Therefore Z′ = 2.303ΔpH × F2(RT)−1 = Z × ΔpH × T−1. For [H+]in << [H+]out, the equation reduces to γ = pZ × ΔpH × T−1 [H+]in. For other ion channels, the unitary permeability was calculated from published conductance and relative permeability (α) data by using the formula γ = pZ′([K+]out − [K+]in + α[H+]out)−1([K+]out + α[H+]out)[K+]in (1), the Goldman-Hodgkin-Katz equation. In the absence of a concentration gradient, the proton permeability of a symmetric ion channel is expressed by the relation p = γRTz−2 F−2 [H+]−1 (35).
RESULTS AND DISCUSSION
Characterization of the purified M2 protein and its insertion into liposomes.
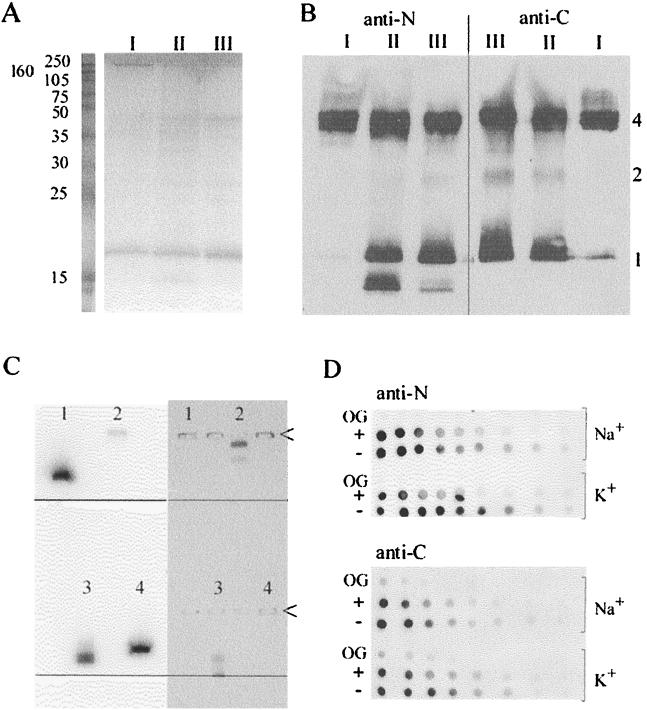
We have shown previously that the purification scheme yields M2 free of other proteins (36). The original procedure was scaled up, and immunoaffinity chromatography was performed by FPLC, yielding 0.5 to 1 mg of M2 per run. Purity was checked by SDS-PAGE, with about 1 μg of the M2 protein loaded per lane and with Coomassie blue staining (Fig. 1A). Because M2 stains very poorly with Coomassie blue (36), the M2 load is very high and protein contaminants are likely to be visualized. The concentration of purified M2 protein was determined by UV spectroscopy. The presence of degradation products was assessed by developing Western blots with antibodies to both termini of the protein. Figure 1A and B show three M2 preparations, two of which (II and III) contain a C-terminal degradation product. M2 forms a tetramer (15, 39), but SDS-PAGE resolves higher oligomers >200 kDa that are partially refractive to boiling with SDS and reducing agents (15, 36, 39). Such a high-molecular-mass complex predominates in the more concentrated preparation I, with hardly any tetramer stained by Coomassie blue (Fig. 1A), although visible in the blot (Fig. 1B). Large M2 complexes >200 kDa transfer inefficiently from polyacrylamide gels to blots (36).
FIG. 1.
Characterization of isolated and liposome-reconstituted M2 protein. (A) SDS-PAGE (12.5% polyacrylamide) of M2 preparations (I, II, and III) stained with Coomassie blue. Left lane: Amersham RPN800 molecular mass markers (in kilodaltons). (B) Aligned Western blots of the same gel, developed with antiserum to the M2 N terminus (left panel) or C terminus (right panel). 1, monomer; 2, dimer; 4, tetramer. (C) Native 1% agarose gel. Lane 1, 500 ng of M2 plus 40 mM OG; lane 2, protein standard HDL-LDL; lane 3, 250 ng of M2 plus 0.34% Coomassie blue; lane 4, 250 ng of M2, 0.05% taurodeoxycholate, and 40 mM OG. In the right panel, the gel was stained with a cholesterol detection kit to visualize the protein standard in lane 2 (LDL in the upper band and HDL in the lower band). Lane 3 shows prestained M2 and unbound Coomassie blue at the front indicated by a line. The left panel shows an aligned Western blot, developed with anti-M2 rabbit serum. <, loading pockets. (The HDL-LDL standard contained a nonspecifically reacting band migrating to the cathode.) (D) Orientation of liposome-reconstituted M2. Serial twofold dilutions of M2 vesicles, prepared with NaPS (Na+) or KPS (K+), digested with trypsin in the absence (+) or presence of 40 mM OG (OG), and untreated controls (−) were dot blotted and developed with antiserum to the M2 N terminus (upper panel) or C terminus (lower panel).
Native gel electrophoresis was employed to investigate the homogeneity of M2 preparations. Polyacrylamide gradient gels at different pHs in the presence of neutral detergents like Triton X-100 or OG caused M2 protein to smear. When the anionic dye Coomassie blue (“blue native electrophoresis” in references 33 and 34) or the nondenaturing anionic detergent taurodeoxycholate were incorporated into sample and running buffers to complex the protein, the laddered band structure was similar to that resolved by SDS-PAGE (data not shown). However, electrophoresis through 1% agarose, which separates proteins on the basis of charge and shape, proved a suitable medium for native gel electrophoresis. Here M2 migrated as a single band in the presence of nonionic or anionic detergents (OG or taurodeoxycholate) or Coomassie blue (Fig. 1C), suggesting that its tendency to form a spectrum of oligomers on polyacrylamide gels, sucrose gradients, and Superose 12 gel filtration (data not shown) is influenced by experimental conditions. The mobility of the M2 band was somewhat greater in the presence of Coomassie blue, which confers a greater negative charge to the protein (Fig. 1C).
Since the permeabilities of channel proteins differ in the inward and outward directions according to their physiological function, the polarity of the M2 protein to the liposome membrane had to be determined. The orientation of the protein was probed by digesting M2 vesicles with trypsin. Digests were analyzed by PAGE, Western blotting (not shown), and quantitative dot blotting (Fig. 1D) with antisera specific to the N and the C termini. M2 sequences within the liposome were masked from proteolysis, but permeabilization of vesicles with 40 mM OG allowed total degradation of M2 during trypsinization. Liposomal M2 was degraded from both termini with equal efficiency, leaving half of the material intact, thus demonstrating random membrane insertion. Therefore, only half of the liposomal M2 population is engaged in proton translocation, depending on the direction of proton flux (into or out of the vesicles).
Stringent proton selectivity of the M2 channel.
The objective of this study was a comparison of the permeation of protons and sodium and potassium ions through the M2 ion channel. Available fluorescent Na+ and K+ probes detect physiological concentrations of these ions in the 1 to 150 mM range (13). Considering that M2 is active at 104- to 105-fold-lower proton concentrations, 0.04 to 10 μM (pH 5 to 7.4) and, as detailed below, proton flux is minute, it was necessary to monitor Na+ and K+ fluxes with the same sensitivity as the pH. This was achieved by an inversion of the fluorimetric assay of proton translocation (36) as follows. M2 vesicles prepared in a single-cation buffer were exposed to a medium containing the other metal ion. In a closed liposome system, any ion flux is coupled to a counterflow of ions compensating for the loss or accumulation of charge in the liposome (7). It is therefore straightforward to examine whether metal ion fluxes are mediated by the same molecular species as proton flux, i.e., by the M2 protein, or require the introduction of other carriers. Coupling allows Na+ or K+ fluxes to be sensitively monitored via the internal pH, as reported by pyranine fluorescence.
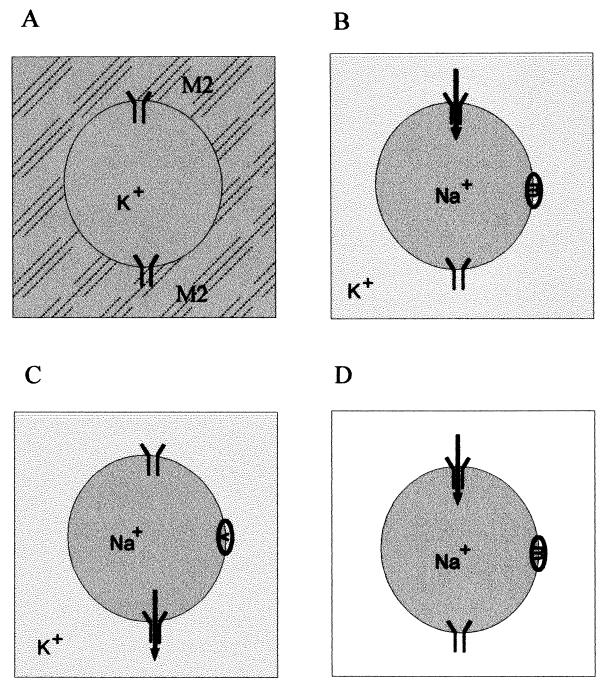
The setup of experiments investigating ion selectivity is illustrated in Fig. 2. Figure 2A shows an M2 vesicle immersed in incubation buffer. Ionic conditions and/or pH differ on either side of the membrane. Cation flux in both directions was examined with respect to the N-C polarity of the M2 protein by preparing vesicles containing either sodium or potassium ions (the buffers NaPS or KPS) and assaying them in buffers containing the other metal ion or impermeant cations and anions (N-methyl-d-glucamine HEPES [NMDGH]). The experiment illustrated in Fig. 2B and C involves vesicles prepared in NaPS, which were introduced into KPS of the same pH, 7.4. Protein-free control vesicles did not exhibit internal pH changes, indicating that their membrane (of a complex lipid composition similar to plasma membrane) (45) was impermeable to cations, as required for these experiments (Fig. 3A). As expected, exposure of these M2 vesicles to NaPS did not elicit any pH change. Moreover, exposure to KPS also caused no pH change, demonstrating that M2 allowed neither an influx of K+ ions nor an efflux of Na+ ions, either of which could have been compensated for by proton counterflow. In contrast, introduction of the sodium ionophore monensin (Fig. 2B) caused an immediate pH decrease (proton influx), and the potassium carrier valinomycin (Fig. 2C) elicited an immediate pH increase (proton efflux) (Fig. 3A). Clearly, the ionophores enabled the metal ion counterflow necessary to drive proton flux through M2.
FIG. 2.
Experimental setup for analysis of M2 activity and ion selectivity. (A) M2 vesicles containing either potassium or sodium ions and a fluorescent pH indicator are introduced into assay buffers, which impose metal ion or pH gradients. The M2 protein is present in both orientations. Metal ion fluxes coupled to proton counterflow are monitored via internal pH. (B) M2 vesicles containing sodium ions are introduced into a buffer containing potassium ions. If no pH change is observed, an ionophore specific for the internal metal ion is added to elicit proton flux (arrow). Addition of monensin (m) supports escape of Na+ ions, enabling proton influx through M2. (C) Addition of an ionophore specific for the external metal ion, valinomyin (v), supports K+ ion influx, enabling proton efflux through M2. (D) Introduction of M2 vesicles into a buffer lacking both metal ions.
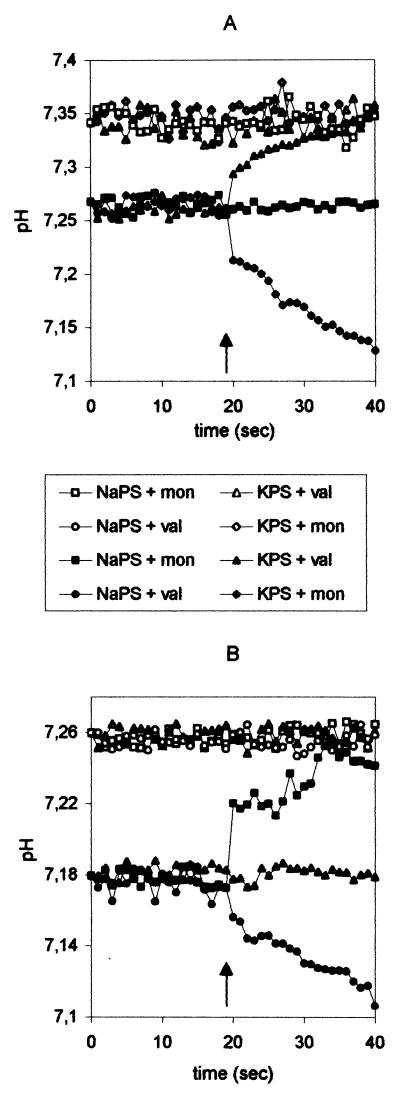
FIG. 3.
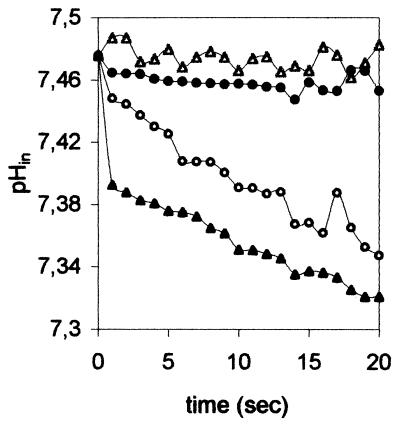
Cation selectivity of the M2 ion channel in the presence of Na+ and K+ ions. (A) M2 (solid symbols) or control vesicles (open symbols) containing NaPS were introduced into Na+- or K+-containing buffer. (B) M2 (solid symbols) or control vesicles (open symbols) containing KPS were introduced into Na+ or K+ buffer. Fluorimetrically monitored internal pH is plotted against time; the initial pH is 7.4 on the outside of the membrane. The experimental setup is explained in the legend to Fig. 2C and D. Addition of ionophores is indicated by an arrow. Incubation buffers and added ionophores are displayed in the box. Val, valinomycin; mon, monensin.
In the “mirror” experiment, M2 vesicles were made up in K+ buffer and exposed to Na+ buffer. Again, there was no reaction until an ionophore was provided. Valinomycin triggered a pH decrease, while monensin caused a pH rise (Fig. 3B). In both series of experiments (Fig. 3A and B), proton fluxes into or out of the M2 vesicles were in the reverse direction to ionophore-mediated metal ion flux. The ionophores are highly selective for either Na+ or K+ ions (8, 29). Obviously, the direction of Na+ or K+ flux depended both on the preset cation gradient and the specificity of the ionophore, which either carried the liposome-trapped metal ion outwards or moved metal ions from the external buffer into the liposome.
In the experiments described above, both Na+ and K+ were present. In the following experiment, illustrated schematically in Fig. 2D, M2 and control vesicles prepared in NaPS (Fig. 4A) or KPS (Fig. 4B) were introduced into the metal ion-free buffer NMDGH. For clarity, the KPS and NaPS controls are omitted, because they were included in Fig. 3. Again, no pH change occurred unless the appropriate ionophore was added. Thus, M2 was incapable of translocating potassium or sodium ions into a metal ion-free medium.
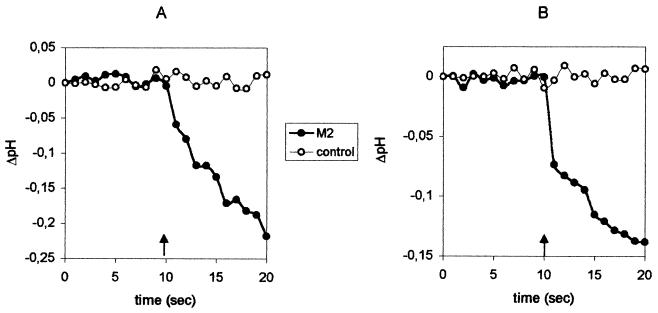
FIG. 4.
Cation selectivity of the M2 ion channel in the presence of a single metal ion. M2 or control vesicles containing NaPS (A) or KPS (B) were introduced into metal ion-free NMDGH buffer, as shown schematically in Fig. 2D. Monensin (A) or valinomycin (B) was added after 10 s (arrow). ΔpH = pHin − pHin (t = 0 s). The initial pH is 7.4 on both sides of the membrane. Other symbols are as introduced in the legend to Fig. 3.
Since the M2 ion channel is activated at weakly acidic pH (3, 28), it was conceivable that the activated channel also becomes more permeable to other ions. When M2 vesicles prepared in NaPS at neutral pH were introduced into K+ or Na+ buffer at pH 5.7, no ion fluxes were induced (Fig. 5). Hence, a higher protonation state of the channel did not increase its permeability to K+ or Na+ ions. Addition of valinomycin had no effect, because an influx of potassium ions could not be balanced by an efflux of protons against the pH gradient. In both K+ and Na+ buffer, only monensin elicited proton influx through M2 by mediating the efflux of Na+.
FIG. 5.
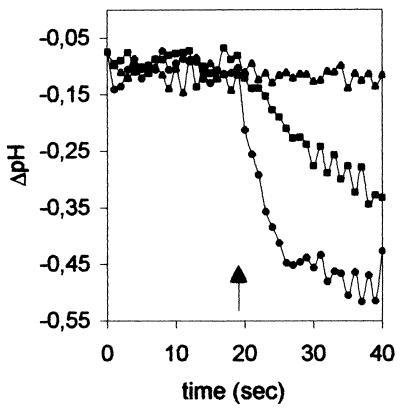
Effect of acidic pH on cation selectivity of the M2 ion channel protein. Vesicles prepared in NaPS (pH 7.4) were introduced into NaPS or KPS at pH 5.7. The data are presented as plots of differences between recordings on M2 vesicles and control (c) vesicles: ΔpH = pHin(M2) − pHin(c). Ionophores were added at 20 s (arrow). Incubation conditions: ▴, KPS, pH 5.7 (plus valinomycin); ■, NaPS, pH 5.7 (plus monensin); ●, KPS, pH 5.7 (plus monensin).
Our experiments provide an estimate of the proton selectivity of the M2 ion channel as the ratio of the highest sodium or potassium ion concentration (120 mM) at which metal ion flux was not detectable and the lowest proton concentration (40 nM [pH 7.4]) at which proton flux was recorded. Therefore, the proton selectivity of M2 with respect to sodium and potassium ions is at least 3 × 106. This is consistent with the value of 1.7 × 106 previously determined by whole-cell recordings with Weybridge M2-expressing MEL cells (3, 27) and with the reevaluated proton selectivities of 1.8 × 106 and 1.5 × 106 determined by patch clamping of CV-1 cells and Xenopus oocytes expressing the M2 protein of influenza A/Udorn/72 virus (26).
Determination of single-channel parameters unitary current, conductance, and permeability.
Because of the low channel activity of the M2 protein (27), it was imperative to ensure that no other ion channel was present, since even tiny amounts of a foreign activity could influence the conductance of the proteoliposomes. Therefore, the susceptibility to the selective M2 inhibitor rimantadine (reviewed in reference 14) was tested. Figure 6 shows a complete block of proton translocation after a 5-min preincubation with 1 μM rimantadine, confirming the identity of the channel and the absence of interfering activities. Moreover, the vesicular pH did not rise during the preincubation period, proving that rimantadine did not permeate the vesicles. The complex lipid composition employed here forms an effective seal against rimantadine permeation, in contrast to vesicles composed only of DMPC-PS (23). In agreement with observations in whole-cell patch-clamp studies (3, 27, 28, 44), preincubation is essential for total inhibition of M2 by amantadine and rimantadine. The inhibitor strengths of rimantadine were similar in KPS and NaPS and somewhat reduced at a channel-activating lower pHout (data not shown).
FIG. 6.
Inhibition of the M2 proton channel activity by rimantadine. Vesicles prepared in KPS (pH 7.4) were introduced into NaPS (pH 7.4), with 1 μM rimantadine, at 18°C and incubated for 5 min (●) before addition of valinomycin to initiate proton translocation. Inhibition without preincubation (○) was observed by introducing vesicles into NaPS containing valinomycin and rimantadine. The uninhibited reaction (▴) was recorded in NaPS (pH 7.4), and the background (▵) was recorded in KPS (pH 7.4).
Proton fluxes through the M2 ion channel were calculated from the internal pH on the basis of the volume and buffer capacity of the liposome. Two types of liposome differing in composition and size were analyzed (Table 1). Vesicles of complex lipid composition had a significantly, 10-fold-larger volume than simple DMPC-PS vesicles, as determined by photon correlation spectroscopy. Protein-free control vesicles were generally 40% smaller than M2 proteoliposomes. The buffer capacities which depend also on the phospholipid head groups (6), were similar in both systems (Table 2). Complex M2 vesicles contained 500 M2 tetramers, and the simple vesicles contained 100 M2 tetramers in both orientations to the membrane (Fig. 1D).
TABLE 1.
M2 and control vesicle parameters
| Parameter | Result fora:
|
|||
|---|---|---|---|---|
| DMPC-PS vesicles
|
Complex vesicles
|
|||
| + M2 | Control | + M2 | Control | |
| Diam (nm) | 115 ± 38 | 95 ± 17 | 256 ± 97 | 221 ± 87 |
| Surface (S [nm2]) | 4.13 × 104 | 2.83 × 104 | 2.05 × 105 | 1.53 × 105 |
| Lipid content (2 × S/SPL)b | 1.18 × 105 | 0.81 × 105 | 5.87 × 105 | 4.37 × 105 |
| Total liposomes (1 mg)c | 6.64 × 1012 | 9.96 × 1012 | 1.33 × 1012 | 1.79 × 1012 |
| Liposome vol (nm3)d | 7.89 × 105 | 4.48 × 105 | 8.75 × 106 | 5.62 × 106 |
| Total liposome vol (μl) with 1 mg of liposomes | 5.24 | 4.33 | 11.7 | 10.1 |
There were 100 M2 tetramers per liposome for DMPC-PS vesicles (50 in either orientation) and 500 M2 tetromers per liposome for complex vesicles (250 in either orientation).
Average lipid surface area (SPL) = 0.7 nm2.
One milligram of liposomes contains 7.83 × 1017 lipid molecules and 50 μg of M2 (1.1 nmol = 6.65 × 1014 tetramers).
TABLE 2.
Derivation of M2 single-channel parameters
| Parameter | Result for:
|
|||
|---|---|---|---|---|
| DMPC/PS vesicles
|
Complex vesicles
|
|||
| 1st determination | 2nd determination | 1st determination | 2nd determination | |
| pHout | 7.4 | 5.7 | 7.4 | 5.7 |
| Δφ (mV)a | 150 | −94 | 150 | −94 |
| ΔpHin (initial pH change/s) | −0.1134 ± 0.0362 | −0.2664 ± 0.0175 | −0.0514 ± 0.0210 | −0.1701 ± 0.0616 |
| Initial proton translocation rate (dH+/dt [pmol/s])b | 40.5 ± 12.9 | 95 ± 6 | 42.9 ± 17.5 | 142 ± 51 |
| H+/s/M2 tetramerc | 7.3 ± 2.3 | 17.2 ± 1.1 | 7.7 ± 3.2 | 25.7 ± 9.3 |
| Unitary current at an average of 18°C (aA)d | 1.2 | 2.7 | 1.2 | 4.1 |
| Unitary current extrapolated to 37°C (aA)e | 5 | 40 | NDf | ND |
| Unitary conductance at 18°C (aS)g | 8 | 29 | 8 | 44 |
| Unitary conductance extrapolated to 37°Ce | 33 aS | 0.4 fS | ND | ND |
| Unitary permeability at 18°C (cm3 s−1)h | ND | 5.4 × 10−12 | ND | 8.2 × 10−12 |
| Unitary permeability at 37°C [cm3 s−1] | ND | 74 × 10−12 | ND | ND |
Transmembrane potential.
dH+/dt = ΔpHin × B × V, where V is absolute liposome volume per assay (10 μg of liposomes, see Table 1) A and B is buffer capacity (6.8 mM for DMPC-PS vesicles and 7.13 mM for complex vesicles).
M2 with the ectodomain at the vesicle surface.
1 A = 6.25 × 1018 charges per s. 1 aA = 10−18 A.
See Fig. 7.
ND, not done.
Unitary conductance (γ) is the quotient of unitary current and transmembrane potential, 1 S = 1 A V−1.
Unitary permeability (p) was derived from conductance as described in the text.
Single-channel currents were determined from the initial (1 s) proton translocation rate into vesicles containing valinomycin to support cation counterflow (Fig. 2). The variation in activity between independent M2 preparations and recordings did not exceed 40%. Two pHout were compared: pH 7.4, where proton translocation is driven by the potassium ion concentration gradient and the channel is in its ground state, and pH 5.7, which activates the channel (3, 27, 28). The vesicles contained KPS (pH 7.4) with valinomycin incorporated in the membrane; the incubation buffer was NaPS (pH 7.4) or KPS (pH 5.7). Table 2 runs through the calculation of single-channel currents. The average proton translocation rates were similar for both types of vesicles: 7.3 protons per s per tetramer in DMPC-S and 7.7 protons per s per tetramer in complex vesicles (Table 2). At a pHout of 5.7, the flux increased to 17 and 26 protons per second, respectively. The lipid composition therefore had no significant influence on single-channel conductance.
These proton currents are equivalent to 1.2 to 4.1 aA, 4 orders of magnitude below the noise level (<10 fA) of whole-cell patch clamp recordings previously defined as the upper boundary of M2 unitary currents (27). Recently, Mould et al. (25) offered three estimates of maximal M2 single-channel currents (influenza A/Udorn/72 virus): first, ≈10 fA, on the basis of the dissociation rate of protonated histidine, given that His37 is the activation site of the channel (43); second, ≈1 fA, from the proton diffusion rate and the minimal buffer concentration supporting M2 currents; and third, 0.5 fA, from the total current recorded in M2-expressing oocytes and the estimated number of M2 channels at the cell surface. The latter approach is most similar to our own and was therefore scrutinized. Repeating the calculation, we obtained a result 10 times lower than the reported 0.5 fA. Per oocyte, 3 ng of M2 was expressed, generating a current of 0.7 μA at pH 6.2 and a potential of −130 mV (25), in agreement with previous findings of these authors, which were 0.16 and 0.26 μA per ng of M2 in oocytes and CV-1 cells, respectively (16, 42). Three nanograms of M2 is equivalent to 66 fmol or 4 × 1010 tetramers (molecular mass of 45.6 kDa, predicted from the sequence and modifications of M2) and translates into a single-channel current of 17 aA. Following the assumption of Mould et al. that only half of the M2 is expressed on the oocyte plasma membrane (25), the value is 34 aA, and based on the somewhat higher molecular weight of 60,000 used by these authors, the single-channel current becomes 47 aA. This is still an order of magnitude above the experimentally determined unitary current (Table 2); however, the M2 proteins were from different virus strains. Despite the purity and homogeneity of the isolated M2 protein (Fig. 1), it is impossible to rule out or to quantify inactive M2 in the preparation, which would cause an underestimation of M2 activity. The average M2 single-channel parameters determined here will translate into higher values if open and closed states of M2 exist, which up to now have proved impossible to resolve (27).
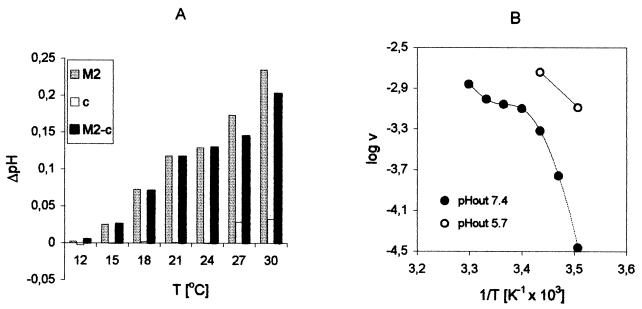
Proton translocation assays were run at 18°C, a standard temperature for fluorimetric and electrophysiological ion channel recordings (27). In order to obtain an estimate of channel activity at physiological temperature, we monitored the temperature dependence of M2 activity (Fig. 7). The assay temperature was limited by the permeability increase of the liposome membrane (Fig. 7A) near the lipid-phase transition temperature (6). The relation of M2 activity to 1/T was approximately linear up to 21°C and then leveled off (Fig. 7B). Single-channel currents were extrapolated to 37°C by linear extension of the plots, yielding a maximum of ≈ 40 aA for the low-pH-activated state of the channel.
FIG. 7.
Temperature dependence of the proton translocation rate. (A) Proton translocation into M2 and control (c) vesicles. ΔpH = pHin (t = 0) − pHin (t = 1 s). (B) Arrhenius plot. The initial proton translocation rate (v [M s−1]) was calculated from ΔpHin as described in Table 1; log v was plotted against 1/T.
Also of interest is the single-channel conductance, the quotient of the current and the transmembrane potential. Assays were performed at 150 mV (K+ gradient) and −94 mV (proton gradient); the resting potential of cells is usually around −70 mV (24). The unitary conductance of M2 was between 8 and 44 aS (Table 2). Low-pH activation enhanced the conductance 3.5- to 5-fold, a difference which extrapolated to >10-fold (0.4 fS) at physiological temperature (Fig. 6 and Table 2). Previously, whole-cell recordings on M2-expressing MEL cells could not resolve single-channel conductance below 0.1 pS (3, 27). The unitary current and conductance of the M2 protein are, to our knowledge, the lowest ever reported for an ion channel. Before the single-channel parameters of the M2 protein were known, its designation as an ion channel remained tentative. Compared to sodium or potassium channels with ion transport rates of 107 to 108 per s, M2 seems to be an exceedingly slow channel. M2 proton translocation rates of 7 to 26 per s (Table 2) are closer to figures for transporters (102 to 104 molecules per second) and pumps (1 to 1,000 ions per s) (reviewed in reference 24).
Unlike the unitary conductance and current of an ion channel, which are positively correlated to the concentration of the permeant, the single-channel permeability, p (reviewed in reference 1), as an intrinsic property is potentially more informative regarding the nature of a transport process. Since p is not directly accessible to measurement and the equations by which p is calculated only hold under restricted conditions, this quantity is rarely tabled. Knowledge of the unitary conductance γ of M2 allowed the derivation of p for one of the two experimental conditions, the pH gradient (Table 2). As detailed in Materials and Methods, γ = p × Z × ΔpH × T−1[H+]in. At 18°C, p = 5.4 × 10−12 and 8.2 × 10−12 cm3 s−1 (for DMPC-PS and complex vesicles), and extrapolated to 37°C, p = 74 × 10−12 cm3 s−1. Unitary proton permeabilities of other ion channels were estimated from published data (17, 32) as described in Materials and Methods. The relative permeability of a potassium-activated cation channel (17; S. S. Kolesnikov, personal communication) for H+ and K+, 3,600:1 and γ(K+) of 0.3 pS ([K+]out = [K+]in = 10 mM; pHin = pHout = 7.2; Erev = 7.8 mV) yielded a unitary proton conductance of 0.9 fS and a unitary proton permeability of 10−11 cm3 s−1, close to that of the M2 protein. Desformylgramicidin, a peptide forming a symmetric cation channel, has a proton conductance of 17 pS at pH 2.5 (32). The simple relation p = γRTF−2 [H+]−1 which holds in this case of a nondirectional channel (35; P. Pohl, personal communication) yields p = 1.4 × 10−12 cm3 s−1, which is somewhat below the proton permeability of M2. Published K+ permeabilities of potassium channels are about an order of magnitude lower: 1 × 10−13 to 2.6 × 10−13 cm3 s−1 (1, 9, 38). In summary, the single-channel current of M2 appears especially minute because it is limited by the low physiological proton concentration, but its unitary proton permeability is within the range of other proton-conducting channels, supporting the classification of M2 as an ion channel, rather than another type of transporter.
The following considerations indicate that the activity of M2 is sufficient to acidify the virus interior within less than a minute. The initial pH decrease in DMPC-PS vesicles was 0.26 pH units per s (Table 1). Extrapolated to 37°C, the rate increases ≈15-fold. Since the virion has approximately the same size as a DMPC-PS vesicle, but contains 4- to 10-fold-less M2 protein (46), initial acidification is expected to be of the order of 0.4 to 1 pH unit per s. The acidification of the virion is driven by the membrane potential and the pH gradient and is obviously dependent on the buffer capacity and the solute volume within the virion, which are not speculated upon here. The function of M2 to equilibrate trans-Golgi pH (4, 5, 11), and thus to avoid the low pH-induced irreversible conformational change of HA (40), is apparently achieved by virtue of its high level of expression on the membranes of the TGN (21, 31).
We have shown that the M2 ion channel is impermeable to Na+ and K+ ions in both flux directions, in its low-pH-activated state as well as in its ground state at neutral pH. Our data prove that stringent proton selectivity and low single-channel conductance are inherent to the M2 protein (i.e., independent of other proteins or gating factors present in whole-cell experiments). Both the low single-channel conductance and the strict proton selectivity of the M2 protein allow it to function with minimal perturbation of ionic conditions during virus replication. However, hyperexpression of wild-type M2 causes significant cytotoxicity in cells cultivated in low-pH media, such as insect (36) and yeast (18) cells. Influenza virus strains with particularly acid-labile HA, like the Rostock strain, encode for M2 proteins more active than Weybridge M2 (11), and certain amantadine-resistant M2 variants are more active than the wild-type proteins (10, 16, 28, 42, 43). Variant M2 proteins with enhanced activity and reduced ion selectivity may become as disruptive to the host cell as hyperexpressed wild-type M2 and could even be abortive to infection. In this context, it is relevant to investigate the cytopathic potential of the M2 protein during generalized infection, elicited by virulent and pandemic strains or by current influenza virus strains in immunocompromised hosts. Here, M2 expression in nonpermissive cells may interfere with physiological gradients and currents.
ACKNOWLEDGMENTS
We gratefully acknowledge support by the Deutsche Forschungsgemeinschaft (grants Schr 554/2-1 and -2-2) and grants from the Humboldt University Medical School (Charité).
We thank Stephen A. Wharton (National Institute for Medical Research [NIMR], London, United Kingdom) for critical discussion of our work and Andreas Herrmann (Institute of Biophysics, Humboldt University) for generous access to fluorimeters. We are grateful to Brigitte Brux for introduction to the REP electrophoresis system, to Alan J. Hay (NIMR) for samples of antisera, and to Laurence H. Pinto (Howard Hughes Medical Institute, Northwestern University, Evanston, Ill.) for communicating results from his paper in press. We are obliged to Stanislav Kolesnikov (Institute of Cell Biophysics, Russian Academy of Sciences, Pushchino, Russia) and Peter Pohl (Institut für Medizinische Physik und Biophysik, Martin-Luther Universität Halle, Halle, Germany) for information on the calculation of single-channel permeabilities from their published data. We are obliged to Harald Heider for help with M2 expression and purification and Kathlen Schröder for expert technical assistance.
REFERENCES
- 1.Aidley D J, Stanfield P R. Ion channels: molecules in action. Cambridge, United Kingdom: Cambridge University Press; 1996. [Google Scholar]
- 2.Beaven G H, Holiday E R. Ultraviolet absorption spectra of proteins and amino acids. Adv Protein Chem. 1952;7:319–368. doi: 10.1016/s0065-3233(08)60022-4. [DOI] [PubMed] [Google Scholar]
- 3.Chizhmakov I V, Geraghty F M, Ogden D C, Hayhurst A, Antoniou M, Hay A J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciampor F, Bayley P M, Nermut M V, Hirst E M A, Sugrue R J, Hay A J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus haemagglutinin to the low pH conformation occurs in an acidic trans-Golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- 5.Ciampor F, Thompson C A, Hay A J. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22:247–258. doi: 10.1016/0168-1702(92)90056-f. [DOI] [PubMed] [Google Scholar]
- 6.Dencher N A, Burghaus P A, Grzesiek S. Determination of the net proton-hydroxide ion permeability across vesicular lipid bilayers and membrane proteins by optical probes. Methods Enzymol. 1986;127:746–760. doi: 10.1016/0076-6879(86)27059-7. [DOI] [PubMed] [Google Scholar]
- 7.Garcia M L, Kitada M, Eisenstein H C, Krulwich T A. Voltage-dependent proton fluxes in liposomes. Biochim Biophys Acta. 1984;766:109–115. doi: 10.1016/0005-2728(84)90222-6. [DOI] [PubMed] [Google Scholar]
- 8.Gennis R B. Biomembranes: molecular structure and function. New York, N.Y: Springer-Verlag; 1989. [Google Scholar]
- 9.Gogelein H, Greger R. Properties of single K+ channels in the basolateral membrane of rabbit proximal straight tubules. Pflueg Arch. 1987;410:288–295. doi: 10.1007/BF00580279. [DOI] [PubMed] [Google Scholar]
- 10.Grambas S, Bennet M S, Hay A J. Influence of amantadine-resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 11.Grambas S, Hay A J. Maturation of influenza A virus haemagglutinin —estimates of the pH encountered during transport and its regulation by the M2 protein. Virology. 1992;190:11–18. doi: 10.1016/0042-6822(92)91187-y. [DOI] [PubMed] [Google Scholar]
- 12.Green D J, Sattelle D B, Westhead E W, Langley K H. Relative size and dispersity of isolated chromaffin granules. In: Pike E R, Cummis H Z, editors. Photon correlation spectroscopy and velocimetry. New York, N.Y: Plenum Publishing; 1977. pp. 477–481. [Google Scholar]
- 13.Haugland R P. Handbook of fluorescent probes and research chemicals. 7th ed. Eugene, Oreg: Molecular Probes, Inc.; 1999. Fluorescent Na+ and K+ indicators. . (CD ROM.), [Google Scholar]
- 14.Hay A J. The action of adamantanamines against influenza A viruses: inhibition of the M2 ion channel protein. Semin Virol. 1992;3:21–30. [Google Scholar]
- 15.Holsinger L J, Lamb R A. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulphide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 16.Holsinger L J, Nichani D, Pinto L H, Lamb R A. Influenza A virus M2 ion channel protein: a structure-function analysis. J Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolesnikov S S, Margolskee R F. Extracellular K+ activates a K+- and H+-permeable conductance in frog taste receptor cells. J Physiol. 1998;502:415–432. doi: 10.1111/j.1469-7793.1998.415bt.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtz S, Luo G, Hahnenberger K M, Brooks C, Gecha O, Ingalls K, Numata K-I, Krystal M. Growth impairment resulting from expression of influenza virus M2 protein in Saccharomyces cerevisiae: identification of a novel inhibitor of influenza virus. Antimicrob Agents Chemother. 1995;39:2204–2209. doi: 10.1128/aac.39.10.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb R A, Holsinger L J, Pinto L H. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Cellular receptors of animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 303–321. [Google Scholar]
- 20.Lamb R A, Pinto L H. Do Vpu and Vpr of human immunodeficiency virus type 1 and NB of influenza B virus have ion channel activities in the viral life cycles? Virology. 1997;229:1–11. doi: 10.1006/viro.1997.8451. [DOI] [PubMed] [Google Scholar]
- 21.Lamb R A, Zebedee S L, Richardson C D. Influenza virus M2 protein is an integral membrane protein expressed on the infected cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 22.Langley K H, Newton S A, Ford N C, Sattelle D B. Photon correlation analysis of protoplasmic streaming in the slime mold Physarum polycephalum. In: Pike E R, Cummis H Z, editors. Photon correlation spectroscopy and velocimetry. New York, N.Y: Plenum Publishing; 1977. pp. 519–525. [Google Scholar]
- 23.Lin T, Heider H, Schroeder C. Different modes of inhibition by adamantane amine derivatives and natural polyamines of the functionally reconstituted influenza virus M2 proton channel protein. J Gen Virol. 1997;78:767–774. doi: 10.1099/0022-1317-78-4-767. [DOI] [PubMed] [Google Scholar]
- 24.Lodish H, Beerk H, Matsudaira P, Baltimore D, Zipurski L, Darnell J. Molecular cell biology 3.0. New York, N.Y: W. H. Freeman and Company; 1996. . (CD-ROM.) [Google Scholar]
- 25.Mould J A, Li H-C, Dudlak C S, Lear J D, Pekosz E, Lamb R A, Pinto L H. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 26.Mould J A, Drury J E, Frings S M, Kaupp U B, Pekosz E, Lamb R A, Pinto L H. Permeation and activation of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:31038–31050. doi: 10.1074/jbc.M003663200. [DOI] [PubMed] [Google Scholar]
- 27.Ogden D, Chizhmakov I V, Geraghty F M, Hay A J. Virus ion channels. Methods Enzymol. 1999;294:490–506. doi: 10.1016/s0076-6879(99)94029-6. [DOI] [PubMed] [Google Scholar]
- 28.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 29.Pressman B C. Biological applications of ionophores. Annu Rev Biochem. 1976;45:501–530. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- 30.Provencher S W, Hendrix J, DeMaeyer L J. Direct determination of molecular weight distribution of polystyrene in cyclohexane with photon correlation spectroscopy. Chem Phys. 1978;69:4273–4275. [Google Scholar]
- 31.Sakaguchi T, Leser G P, Lamb R A. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J Cell Biol. 1996;133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saparov S M, Antonenko Y N, Koeppe R E, Pohl P. Desformylgramicidin: a model channel with an extremely high water permeability. Biophys J. 2000;79:2526–2534. doi: 10.1016/S0006-3495(00)76493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 34.Schägger H, Cramer W A, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional blue native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt R F, Thews G. Lehrbuch der Physiologie des Menschen. Berlin, Germany: Springer-Verlag; 1990. [Google Scholar]
- 36.Schroeder C, Ford C F, Wharton S A, Hay A J. Functional reconstitutition in lipid vesicles of influenza virus M2 protein expressed by baculovirus: evidence for proton transfer activity. J Gen Virol. 1994;75:3477–3484. doi: 10.1099/0022-1317-75-12-3477. [DOI] [PubMed] [Google Scholar]
- 37.Shimbo K, Brassard D L, Lamb R A, Pinto L H. Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spruce A E, Standen N B, Stanfield P R. Studies on the unitary properties of adenosine-5′-triphosphate-regulated potassium channels of frog skeletal muscle. J Physiol. 1987;382:13–36. doi: 10.1113/jphysiol.1987.sp016364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugrue R J, Hay A J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugrue R J, Bahadur G, Zambon M C, Hall-Smith M, Douglas A R, Hay A J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tosteson M T, Pinto L H, Holsinger L J, Lamb R A. Reconstitution of the influenza M2 ion channel in lipid bilayers. J Membr Biol. 1994;142:117–126. doi: 10.1007/BF00233389. [DOI] [PubMed] [Google Scholar]
- 42.Wang C K, Lamb R A, Pinto L H. Direct measurement of the influenza A virus M2 protein ion channel activity in mammalian cells. Virology. 1994;205:133–140. doi: 10.1006/viro.1994.1628. [DOI] [PubMed] [Google Scholar]
- 43.Wang C K, Lamb R A, Pinto L H. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C K, Takeuchi K, Pinto L H, Lamb R A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wharton S A, Skehel J J, Wiley D C. Studies of influenza haemagglutinin-mediated membrane fusion. Virology. 1986;149:27–35. doi: 10.1016/0042-6822(86)90083-8. [DOI] [PubMed] [Google Scholar]
- 46.Zebedee S L, Lamb R A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]