Abstract
The human immunodeficiency virus type 1 (HIV-1) Nef protein has several independent functions that might contribute to efficient viral replication in vivo. Since HIV-1 adapts rapidly to its host environment, we investigated if different Nef properties are associated with disease progression. Functional analysis revealed that nef alleles obtained during late stages of infection did not efficiently downmodulate class I major histocompatibility complex but were highly active in the stimulation of viral replication. In comparison, functional activity in downregulation of CD4 and enhancement of HIV-1 infectivity were maintained or enhanced after AIDS progression. Our results demonstrate that various Nef activities are modulated during the course of HIV-1 infection to maintain high viral loads at different stages of disease progression. These findings suggest that all in vitro Nef functions investigated contribute to AIDS pathogenesis and indicate that nef variants with increased pathogenicity emerge in a significant number of HIV-1-infected individuals.
One factor that enables human immunodeficiency virus (HIV-1) to replicate efficiently and continuously despite a strong host antiviral immune response is the Nef protein. First, it has been shown that simian immunodeficiency virus (SIV) carrying a deletion in nef replicates inefficiently and does not cause disease in adult rhesus macaques (21). Subsequently, several long-term survivors of HIV-1 infection were identified, in whom only nef-deleted proviruses could be detected (10, 26). These individuals had low viral loads and unusually slow disease progression. Finally, the HIV-1 nef gene enhances the pathogenicity of SIV (1, 23).
A number of in vitro Nef activities which might contribute to the maintenance of high viral load and disease progression have been described (reviewed in references 9 and 12). Nef downregulates the cell surface expression of CD4, the primary receptor of HIV and SIV (14). A smaller number of CD4 molecules at the cell surface could promote the release of progeny virions, result in enhanced envelope incorporation into viral particles, prevent superinfection, alter T-cell receptor signaling, and impair immune functions of CD4+ helper T cells (2, 27, 34, 37). Nef also downregulates class I major histocompatibility complex (MHC) cell surface expression (35). This activity allows infected cells to resist killing by cytotoxic T lymphocytes (CTL) (6). Interestingly, class I haplotypes that protect the cells against lysis by natural killer cells are not affected by Nef (5). Selective downregulation of class I MHC proteins might contribute to immune evasion by HIV-1. In addition to inducing class I MHC and CD4 endocytosis, Nef increases the infectivity of viral particles derived from CD4-negative producer cells (28, 31) and enhances viral replication in peripheral blood mononuclear cells (PBMC) (4, 28, 31, 38).
All these Nef functions could potentially contribute to efficient viral replication and disease induction in vivo. Their physiologic relevance has been questioned, however, particularly since some effects were observed under artificial conditions or require very high Nef expression levels and since other HIV-1 proteins also downregulate CD4 (8, 42). Therefore, it is not entirely clear why Nef plays a key role in HIV-1 replication and progression to AIDS.
An interesting aspect of Nef function is that most in vitro activities seem to involve different interactions with the cellular signal transduction and endocytic machinery and are functionally separable (19, 28, 29, 32, 39). Several conserved activities of Nef are mediated by different regions of the protein, suggesting that they are independently selected in vivo. Since HIV-1 adapts very rapidly to its host environment (11), we speculated that different Nef functions may be selected for at different stages of infection.
To challenge this hypothesis, we investigated if the relative functional activities of Nef change during the course of HIV-1 infection. We found that nef alleles derived from asymptomatic individuals efficiently downregulated the cell surface expression of class I MHC. In contrast, nef alleles obtained after AIDS progression were impaired in class I MHC downmodulation but highly active in the stimulation of HIV-1 replication. Functional activity in CD4 downregulation and enhancement of infectivity was maintained or enhanced during the late stages of infection. In conclusion, in immunocompetent hosts, Nef properties are selected that allow the virus to efficiently escape the CTL response, whereas in AIDS patients the selective forces drive Nef variations that enhance viral spread in a more direct manner. Importantly, our results also suggest that all four in vitro Nef functions analyzed contribute to viral spread in vivo and that the pathogenic properties of HIV-1 change during progression to AIDS.
MATERIALS AND METHODS
Plasmid construction.
nef alleles predicting the five consensus Nef amino acid sequences were constructed using a combination of splice-overlap extension PCR mutagenesis and standard DNA techniques. The consensus nef alleles were generated based on the previous analysis of nef sequences derived from 91 HIV-1-infected individuals at different stages of disease (24). nef alleles were cloned into a full-length HIV-1 NL4-3 proviral clone essentially as described previously (3). Briefly, the env region of NL4-3 was amplified using primer pSi1 (5′-TAGTGAACGGATCCTTAGC-3′; 8572 to 8590) containing a BamHI restriction site (bold) and pSi3 (5′-TGACCACTTGCCACCCAT-3′; 8902 to 8919). The five consensus nef genes were amplified using primer pSi4 (5′-ATGGGTGGCAAGTGGTCA-3′; 8902 to 8919) and pSi2 (5′-TTCTTGTAGTACTCCGGATGC-3′; 9495 to 9515) containing an MroI site (bold). Numbers refer to the primer positions in the HIV-1 NL4-3 genome. The left- and right-half PCR products were gel purified, mixed at equimolar amounts, and subjected to a second PCR with primers pSi1 and pSi2. The PCR products were purified and inserted into a vector containing the env-nef-long terminal repeat region of NL4-3 by using the BamHI and MroI sites in the env and nef genes. Subsequently, the env-nef-long terminal repeat region was inserted into a modified pBR322 vector containing the full-length HIV-1 NL4-3 proviral DNA by using the unique BamHI and XbaI sites in the NL4-3 env and the vector sequences flanking the 3′ end of the provirus. For Nef expression in Jurkat T cells, the nef sequences were cloned into a bicistronic cytomegalovirus-based pCG expression vector, as described previously (29). All PCR-derived inserts were completely sequenced to confirm that they represented the desired nef gene sequences. Primary nef alleles were amplified by PCR as described previously (24). Amplification products were gel purified and used for a final round of amplification with primers that allowed cloning into the pCG expression vector and insertion into a nef-deleted NL4-3 clone (3), respectively. Cloning was performed essentially as described above, except that the patient-specific nef sequence variations were introduced into the primers used for PCR amplification. The digested PCR fragments were cloned as a pool, and the transformation and cloning efficiencies were determined essentially as described previously (23). Individual participants with earlier designations in the literature are as follows: LTNP4, HP (16, 17, 30); SP7, MB (24, 30); and P2, FA (24, 30).
Study populations.
Sequential samples were obtained from six individuals in the cohort monitored at the New England Hemophilia Center at the UMass/Memorial Health Care System, Worcester, Mass., since 1983. These individuals were selected by a CD4+ T-cell profile that showed an early rapid decline or a clear time point of inflection. They were further selected by the availability of samples from time points when CD4+ T-cell counts were normal and after declining to <100/μl. With the exception of P10 (1986), all individuals tested HIV positive in 1983. Currently, SP8 and P9 are alive with AIDS and the remaining individuals have died. All participants have given informed consent for these studies, with the approval of the institutional review board on the conduct of research on human subjects at the UMass Medical School.
Transfections and flow-cytometric analysis.
Transfection of Jurkat T cells was usually performed by electroporation as previously described (15, 19, 29). For the analysis of pooled primary nef alleles, DMRIE-C reagent (Gibco-BRL, Karlsruhe, Germany) was used as specified by the manufacturer. Flow cytometry analysis of CD4, class I MHC, and green fluorescent protein (GFP) reporter molecules in cells transfected with a bicistronic vector coexpressing Nef and GFP was measured as described previously (15, 29). The level of CD4 or class I MHC expression (red fluorescence) was measured from aliquots of the same transfection mixture as a function of GFP green fluorescence. For the quantitation of Nef-mediated CD4 or class I MHC downregulation, the mean channel numbers of red fluorescence were determined for cells expressing no (N), low (L), medium (M), or high (H) levels of GFP. The numbers obtained for cells transfected with vector expressing GFP only were divided by the corresponding numbers obtained for cells coexpressing Nef and GFP, to calculate the values for x-fold downmodulation.
Cell culture, infectivity, and viral replication assays.
Virus stocks were generated by transient transfection of 293T cells, and replication and infectivity assays were performed essentially as described previously (3). The effect of Nef on viral replication in human PBMC culture was to some extent dependent on the blood donor. Therefore, infections with the wild-type HIV-1 NL4-3 isolate and with a mutant containing a deletion of 260 -bp in the nef- unique region (3) were always performed in parallel with the analysis of primary or consensus nef alleles. Furthermore, the replicative capacity of all NL4-3 nef variants was determined in at least three independent experiments using different virus stocks and PBMC derived from different donors.
Statistical analysis.
Statistical analysis was performed using the InStat program version 3.0 (GraphPad Software, San Diego, Calif.).
RESULTS
Activity of progressor and nonprogressor consensus nef alleles.
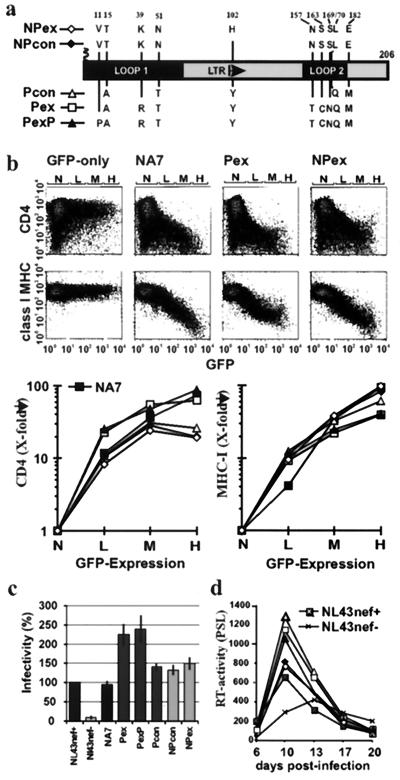
We have previously shown that certain amino acid variations in HIV-1 Nef are associated with different stages of disease (24). To assess the relevance of these variations for Nef function, we generated five nef alleles: the consensus alleles obtained from a large number of nonprogressors (NPcon) and from immunodeficient individuals (Pcon), which differ at only four amino acid positions (T15A, N51T, L170Q, and E182M); nef alleles that contained additional changes that were more commonly observed in nonprogressors (Y102H) (NPex), or in progressors (K39R, N157T, S163C, and S169N) (Pex); and an additional substitution (V11P), which creates an N-terminal PxxP sequence (PexP). The predicted Nef amino acid sequences differed only in the specific changes shown in Fig. 1a. An overview on the nef alleles analyzed in this study is given in Table 1.
FIG. 1.
Activity of nonprogressor and progressor consensus nef alleles. (a) Amino acid variations between NP and P consensus Nef sequences. Numbers refer to the corresponding position in the NL4-3 Nef amino acid sequence. (b) Flow cytometric analysis (upper panel) and quantitative effect of the indicated nef alleles on CD4 and class I MHC cell surface expression (lower panel). Flow cytometric analysis was performed, and CD4 and class I MHC expression was determined, as described in Materials and Methods. Ranges for green fluorescence on cells defined as expressing no (N), low (L), medium (M), or high (H) levels of GFP are indicated. (c) MAGI cells were infected in triplicate with aliquots of three different 293T cell-derived virus stocks containing 125 ng of p24 core antigen. Infectivity is shown relative to HIV-1 NL4-3 wild type. Error bars correspond to standard deviation. (d) Replication of the NL4-3 variants containing the indicated nef alleles (for symbols, see panel a) in PBMC. Cells were infected immediately after infection and stimulated with phytohemagglutinin 3 days later. All results were confirmed in at least three independent experiments. PSL, photostimulated light emission.
TABLE 1.
Overview of the selected HIV-1 Nef variants analyzed
| Group | nef allelea | Remarks |
|---|---|---|
| Group 1 (consensus nef alleles) | NPex | Nonprogressor, extreme |
| NPcon | Nonprogressor, consensus | |
| Pcon | Progressor, consensus | |
| Pex | Progressor, extreme | |
| PexP | Pex plus additional N-terminal proline | |
| Group 2 (progressor nef alleles) | P2-87 | Early allele from progressor P2; CD4+ cell count, 536/mm3 |
| P2-93 | Late nef allele from progressor P2; CD4+ cell count, 18/mm3 | |
| SP7-88 | Early nef allele from progressor SP7; CD4+ cell count, 1,264/mm3 | |
| SP7-91 | Late nef allele from progressor SP7; CD4+ cell count, 497/mm3 | |
| Group 3 (nonprogressor nef alleles) | LTNP4-91B1 | Clade 2 nef allele from LTNP4 |
| LTNP4-91(A,E) | Clade 3 nef allele from LTNP4 | |
| Group 4 (pooled sequential nef alleles) | SP8-84 | Pooled alleles derived from a PBMC sample drawn in 1984 |
| SP8-87 | Pooled alleles derived from a PBMC sample drawn in 1987 | |
| SP8-96 | Pooled alleles derived from a PBMC sample drawn in 1996 |
To investigate their effect on CD4 and class I MHC cell surface expression, the five consensus nef alleles were cloned in a bicistronic expression vector containing nef followed by GFP under the control of an internal ribosome entry site IRES element (29). Thus, the Nef proteins are expressed from a single bicistronic mRNA in a constant stoichiometry with the GFP reporter molecule. All five consensus nef alleles and the control NA7-Nef (15, 19) efficiently downregulated CD4 and class I MHC (Fig. 1b). The Pex and PexP nef alleles, however, showed two- to three-fold-higher activities in CD4 downregulation (L, 26.7 ± 4.4; M, 54 ± 9.6; H, 56 ± 17.4 [n = 6]) than did the NPcon and NPex alleles (L, 9.4 ± 2.1; M, 26.1 ± 3.1; H, 19.1 ± 3.7 [n = 6]). The values give x-fold downregulation for low, medium, and high GFP coexpression levels, respectively. In contrast, the NPcon and NPex nef alleles revealed two- to three-fold-higher maximum activity in class I MHC downregulation (Fig. 1b). These functional differences between the Pex and NP nef alleles were consistently observed in at least three experiments and were highly significant (P < 0.002). The results were not biased by different transfection efficiencies, because both CD4 and class I MHC were measured simultaneously from the same transfection. To assess their effect on virion infectivity and replication, the consensus nef alleles were cloned into the proviral HIV-1 NL4-3 genome. As shown in Fig. 1c, all nef alleles increased virion infectivity. Notably, the Pex and PexP alleles were about two fold more active than the Pcon, NPcon, NPex, NL4-3, and NA7 nef alleles. Similarly, all nef alleles stimulated viral replication in PBMC (Fig. 1d). The three progressor nef alleles showed consistently higher activity, however, than the two NP nef alleles. To confirm this result, we coinfected PBMC with these NL4-3 nef variants and passaged the virus four times in PBMC of different donors. Sequence analysis revealed that the progressor variants outgrew the nonprogressor nef variants in primary PBMC cultures (data not shown).
These results show that amino acid variations in Nef more frequently observed among nonprogressors were associated with high activity in downmodulation of class I MHC, a function which might allow the virus to escape antiviral CTL responses. In comparison, Nef proteins containing features commonly observed in immunodeficient patients were more active in downregulation of CD4 and enhancement of HIV-1 replication and infectivity.
Loss of efficient Nef-mediated class I MHC downregulation after progression to AIDS.
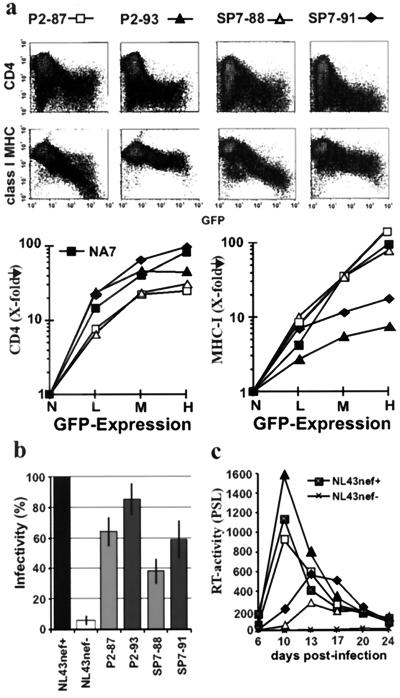
Next, we investigated if these differences in aspects of Nef function, observed using consensus nef alleles, are also obtained with primary nef genes. Previous analysis of sequential samples derived from two patients, P2 and SP7, revealed that several variations in Nef, which were more commonly observed in AIDS patients, were detected only during or after progression to immunodeficiency (24). To assess the effect of these alterations on Nef function, we analyzed two nef alleles derived from each patient. One was obtained early in infection (SP7-88 and P2-87), and the second was obtained during (SP7-91) or after (P2-93) progression to AIDS (Table 1). Notably, both nef alleles predicted amino acid sequences that were identical to >50% of the sequences amplified from the respective PBMC samples and thus were representative for these time points.
As shown in Fig. 2a the nef alleles obtained after the onset of immunodeficiency, P2-93 and SP7-91, were about two- to three-fold more active in downregulation of CD4 compared to the P2-87 and SP7-88 alleles. In comparison, the P2-87 and SP7-88 nef alleles, obtained during the asymptomatic stage of infection, downmodulated class I MHC with about 10- to 20-fold higher efficiency than did the late nef alleles (Fig. 2a). The P2-93 and SP7-91 nef alleles were slightly more active than the P2-87 and SP7-88 alleles in the enhancement of virion infectivity (Fig. 2b). The P2 nef alleles stimulated viral replication in PBMC more efficiently than the SP7 nef alleles did (Fig. 2c). For both patients, however, the late nef alleles were consistently more active than the nef genes amplified from the early samples (Fig. 2c). Coinfection experiments confirmed that the P2-93 variant outgrew the P2-87 variant (data not shown).
FIG. 2.
Inefficient Nef-mediated class I MHC downregulation after disease progression. (a) Flow cytometric analysis and quantitative presentation of CD4 and class I MHC downregulation by nef alleles obtained from patients P2 and SP7 prior (87 and 88 samples) and during or after disease progression (91 and 93 samples). (b and c) Viral infectivity (b) and enhancement of viral replication by P2 and SP7 nef alleles (c). Parameters were determined and reproduced in independent experiments, as described in the legend to Fig. 1.
The functional analysis of nef alleles derived from these two individuals with progressive disease extended the observations made with the consensus nef alleles and revealed a dramatic loss of Nef-mediated class I MHC downregulation after progression to AIDS. In contrast, other Nef functions that are probably associated with increased HIV-1 virulence (CD4 downmodulation, and enhancement of replication and infectivity) were increased during later stages of infection.
Nef alleles from a long-term nonprogressor are impaired in the stimulation of viral replication and CD4 downregulation.
The results obtained with the progressor samples suggested that Nef functions that allow the virus to evade the immune system are diminished after the onset of immunodeficiency whereas other activities are increased. In the next set of experiments we investigated which Nef functions are preserved in nonprogressive HIV-1 infection.
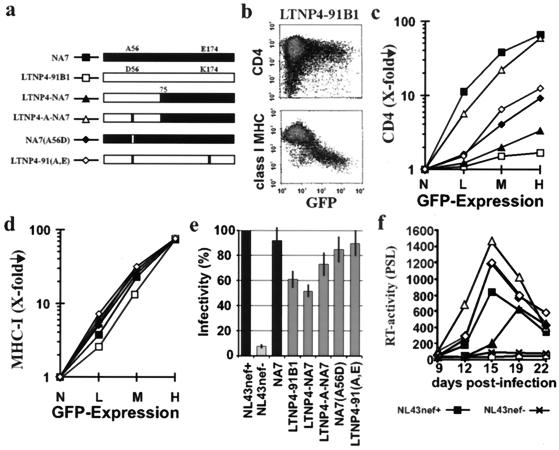
The nef alleles were derived from LTNP4, who shows no signs of immunodeficiency and has an undetectable viral RNA load despite more than 17 years of documented HIV-1 infection (16, 17). It has been previous shown that this long-term nonprogressor contains a high percentage of defective nef alleles (30): 32% of the reading frames were lacking the initiation codon or contained premature stop codons, and 37% predicted mutations of A56D and E174K that disrupted CD4 downregulation. The remaining 31% were functional in CD4 downregulation, but the activity was relatively low. Notably, these functional nef alleles, obtained over a 12-year period, were identical to each other and to an ancestral LTNP4 nef sequence, suggesting that they represent archival viral DNA rather than actively replicating virus (30).
To investigate if LTNP4 nef alleles were selectively impaired in CD4 downmodulation or also nonfunctional in other in vitro assays, we analyzed primary alleles and several recombinants with the functional NA7 nef allele (30) (Fig. 3a). The LTNP4-91B1 and LTNP4-91(A,E) nef alleles were representative for the majority of intact nef open reading frames amplified from LTNP4 over a 12-year period of PBMC sampling. In agreement with the previous study, the A56D and E174K substitutions resulted in inefficient CD4 downmodulation (Fig. 3b and c). However, even the LTNP4-91(A,E) Nef showed about a 10-fold reduced activity compared to the NA7 Nef (Fig. 3c). In contrast to the highly divergent effects of the different nef recombinants on CD4 cell surface expression, all forms efficiently downregulated class I MHC (Fig. 3d) and enhanced virion infectivity (Fig. 3e). The effects of these nef alleles on viral replication in PBMC were variable, depending on the donor. In two experiments, NL4-3 containing the LTNP4-A-NA7 and LTNP4-91(A,E) nef alleles replicated as efficiently as the NL4-3 and NA7 controls (an example is shown in Fig. 3f); and in three other infections, they showed an intermediate phenotype. Overall, the results from five independent experiments revealed that nef alleles that downregulated CD4 were also active in the stimulation of viral replication whereas the LTNP4-91B1 and LTNP4-NA7 Nef proteins were impaired in both assays. These data are consistent with reports that downregulation of CD4 and the enhancement of viral replication involve similar molecular interactions (27, 34).
FIG. 3.
nef alleles derived from a long-term nonprogressor are selectively impaired in CD4 downmodulation and stimulation of viral replication. (a) Structure of the chimeric and mutant Nef proteins that were analyzed (30). (b) Effect of the LTNP4-91-B1 nef allele on CD4 and class I MHC cell surface expression. (c to f) Functional activity of the indicated mutant and chimeric nef alleles in downregulation of CD4 (c), class I MHC downmodulation (d), enhancement of viral infectivity (e), and stimulation of HIV-1 replication in PBMC (f). Symbols are shown in panel a. For all four in vitro assay systems for Nef function, similar results were obtained in two to four independent experiments. Error bars shown in panel e represent standard deviation.
These results demonstrate that nef alleles derived from LTNP4 showed the opposite phenotype from nef alleles derived from AIDS patients: efficient class I MHC downregulation but inefficient downmodulation of CD4 and stimulation of viral replication. In agreement with a previous study (3), these results suggest that Nef functions which allow the virus to escape immune surveillance are maintained in nonprogressive HIV-1 infection whereas other Nef activities that probably contribute to the full pathogenic potential of HIV-1 might be impaired.
Functional activity of sequential nef alleles from progressors with HIV-1 infection.
The consensus Nef sequences were derived from a large number of patients (24), and representative nef alleles derived from patients P2, SP7, and LTNP4 were selected for functional analysis. Nonetheless, the number of alleles analyzed was limited, and the possibility that the results did not accurately reflect the course of primary HIV-1 infection could not be dismissed. Therefore, additional experiments with nef alleles derived from sequential patient samples were performed.
nef alleles derived from PBMC samples obtained from six individuals with progressive HIV-1 disease for between 6 and 14 years were analyzed. All study subjects showed a dramatic decrease in the number of CD4+ T cells and progressed to AIDS during the investigation period (Table 2). The amplified PCR products were cloned directly into the bicistronic pCG-Nef-IRES-GFP vector or used to introduce nef genes into the nef-deleted HIV-1 NL4-3 molecular clone by splice overlap extension-PCR. A complex mixture of primary nef alleles was cloned as a pool to ensure that the nef genes analyzed were representative of each patient and time point. Control experiments indicated that ≥95% of the expression vector and the proviral construct contained an insert of the expected size and that each plasmid preparation represented at least 250 independent transformants. Western blot analysis revealed that all plasmid populations resulted in efficient Nef protein expression (data not shown). A total of 80 colonies were randomly picked from the transformation of both the proviral (44 colonies) and the pCG-Nef-IRES-GFP (36 colonies) constructs. Agarose gel analysis confirmed that 43 (98%) and 34 (94%), respectively, of the clones contained an insert of the expected size, and sequence analysis showed that all positive clones contained nef alleles that were specific for the respective patient (data not shown).
TABLE 2.
In vitro activities of pooled nef alleles obtained at different stages of HIV-1 infection
| Patient | Yr of PBMC sampling | No. of CD4+ cells/mm3 | Gag-specifica CTL activity | MHC downregulationb
|
CD4 downregulationb
|
Infectivityc | Replicationc | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L | M | H | L | M | H | ||||||
| SP8 | 1984 | 616 | NDd | 2.8 | 5.2 | 28.4 | 2.9 | 8.7 | 67.3 | 10.8 ± 1.5 | 5.0 ± 1.7 |
| 1987 | 601 | ND | 2.7 | 5.2 | 28.9 | 2.8 | 9.1 | 75.6 | 11.2 ± 1.2 | 3.4 ± 0.6 | |
| 1996 | 39 | ND | 2.1 | 2.5 | 2.2 | 3.2 | 9.9 | 87.4 | 14.9 ± 1.8 | 10.8 ± 2.3 | |
| P5 | 1983 | 616 | +1,987 | 2.4 | 5.4 | 45.1 | 2.1 | 3.3 | 45.3 | 13.2 ± 1.4 | 3.2 ± 0.2 |
| 1991 | 121 | +1,992 | 2.3 | 3.6 | 6.7 | 2.4 | 4.0 | 54.9 | 9.9 ± 0.5 | 5.7 ± 1.8 | |
| 1995 | 5 | +1,994 | 2.2 | 3.3 | 5.3 | 2.6 | 5.1 | 59.5 | 9.1 ± 1.2 | 11.0 ± 3.1 | |
| P7 | 1982 | ND | ND | 2.8 | 7.4 | 38.5 | 2.5 | 7.5 | 76.8 | 8.9 ± 1.5 | 4.5 ± 2.4 |
| 1985 | 822 | ND | 2.5 | 4.9 | 24.0 | 2.9 | 10.3 | 78.0 | 19.6 ± 3.9 | 4.8 ± 0.7 | |
| 1993 | 7 | ND | 2.0 | 2.9 | 8.7 | 2.9 | 8.4 | 70.3 | 26.2 ± 5.1 | 7.2 ± 2.0 | |
| P8 | 1983 | 526 | +1987 | 2.9 | 5.0 | 10.3 | 2.7 | 6.5 | 60.8 | 7.8 ± 1.2 | 4.0 ± 1.0 |
| 1987 | 126 | ND | 3.0 | 4.6 | 9.9 | 3.3 | 11.4 | 78.7 | 9.1 ± 1.5 | 11.3 ± 3.5 | |
| 1989 | 12 | ND | 3.3 | 4.9 | 9.0 | 3.2 | 14.6 | 89.3 | 11.1 ± 0.5 | 9.0 ± 2.3 | |
| P9 | 1984 | 636 | ND | 4.8 | 15.4 | 142.7 | 1.6 | 2.3 | 19.1 | 2.3 ± 0.4 | 2.0 ± 0.8 |
| 1989 | 118 | ND | 3.3 | 5.3 | 6.5 | 3.8 | 17.1 | 76.8 | 6.6 ± 1.3 | 3.4 ± 2.1 | |
| 1998 | 95 | ND | 1.6 | 3.2 | 7.0 | 2.5 | 9.3 | 69.3 | 6.9 ± 0.4 | 9.2 ± 1.1 | |
| P10 | 1984 | 1,021 | ND | 2.8 | 6.8 | 44.8 | 3.0 | 6.5 | 58.0 | 8.8 ± 2.0 | 2.0 ± 1.0 |
| 1986 | 1,357 | -1993 | 3.9 | 13.3 | 120.2 | 3.0 | 6.7 | 60.0 | 10.8 ± 2.3 | 7.3 ± 2.4 | |
| 1996 | 9 | ND | 2.7 | 4.4 | 7.2 | 6.9 | 30.8 | 89.3 | 12.4 ± 1.8 | 13.9 ± 5.1 | |
| n = 8e | >600 | 3.1 ± 0.8 | 8.0 ± 4.0 | 59.1 ± 45.7 | 2.6 ± 0.5 | 6.8 ± 2.9 | 60.0 ± 19.9 | 10.7 ± 4.8 | 4.0 ± 1.8 | ||
| n = 4e | 100–600 | 2.9 ± 0.4 | 4.6 ± 0.7 | 8.4 ± 2.0 | 3.1 ± 0.6 | 9.8 ± 5.7 | 67.8 ± 11.7 | 8.4 ± 1.5 | 6.1± 3.6 | ||
| n = 6e | <100 | 2.3 ± 0.6 | 3.5 ± 0.9 | 6.6 ± 2.5 | 3.5 ± 1.6 | 13.0 ± 9.2 | 77.5 ± 12.8 | 13.4 ± 6.8 | 10.2 ± 2.2 | ||
CD4 and class I MHC expression levels were determined for cells expressing low (L), medium (M), and high (H) levels of GFP, as described in Materials and Methods. Results were confirmed in two additional experiments.
Average values and standard deviations for the enhancement of viral infectivity, compared to nef-deleted NL4-3 infection. Enhancement of replication in PBMC culture is indicated for 9 to 13 days postinfection, when peak activities where observed for wild-type NL4-3 infection. Numbers show x-fold enhancement of reverse transcriptase activity relative to nef-deleted infection. Values given for infectivity and replication were derived from three or four experiments.
ND, not determined.
e Number of samples with a CD4+ cell count >600/mm3.
As indicated in Table 2, primary nef alleles derived from all 18 samples downmodulated the cell surface expression of CD4. In three of the six patients (SP8, P5, and P7) the efficiencies remained relatively constant over the course of infection. For the remaining individuals, P8, P9, and P10, a significantly (P < 0.01) increased activity was observed after progression to AIDS. At medium expression levels, nef alleles obtained during late stages of infection were about two-fold more active than nef alleles obtained during the asymptomatic stage (Table 2). The functional differences in class I MHC downmodulation were more dramatic. nef alleles amplified early in infection from five of the six study subjects showed activities that were comparable to that of the strong NA7 control nef allele. Importantly, in all five cases, nef alleles amplified after AIDS progression showed strongly diminished activity (P < 0.01). In contrast, nef alleles amplified from P8 always showed low activity in class I MHC downregulation, even for the sample drawn prior to progression to AIDS (Table 2). The results obtained for Nef function were consistent with the sequencing data, showing that only in P8 no specific sequence variations in Nef were observed during progression to AIDS (data not shown). On average, the maximum levels of Nef-mediated class I MHC downregulation dropped about 10-fold after progression. To assess the effect on infectivity and replication, virus stocks were generated by transient transfection of 293T cells with the NL4-3 proviral clones carrying the primary HIV-1 nef alleles. Infection of MAGI cells revealed that in four of six patients the ability of nef to increase virion infectivity remained constant or increased only slightly throughout the course of infection. nef alleles amplified from P7 and P9 showed about a three-fold increase in activity after progression to AIDS (P < 0.01). Similar to the results for infectivity, all inserted nef pools were able to stimulate HIV-1 replication in human PBMC cultures. Notably, nef alleles amplified from late PBMC samples consistently showed two- to three-fold-higher activity than did nef alleles amplified from samples drawn early in infection (Table 2). This increase in the ability of Nef to stimulate HIV-1 replication in PBMC culture was highly significant (P < 0.001).
The relative in vitro activities of Nef change during the course of HIV-1 infection.
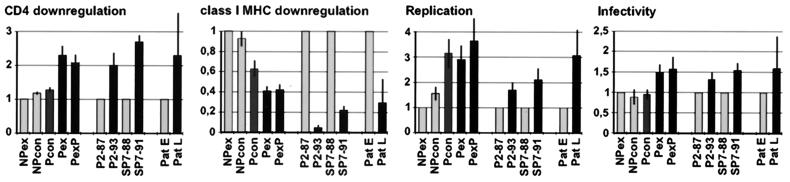
The relative functional differences observed between the various consensus Nefs proteins and between primary nef alleles obtained before and after disease progression are summarized in Fig. 4. Consensus Nef proteins containing features that were more frequently found in progressors (Pex and PexP) showed slightly to moderately higher activity in three in vitro assay systems, (i) CD4 downregulation, (ii) enhancement of virion infectivity, and (iii) stimulation of HIV-1 replication in PBMC, than did proteins from NPcon and NPex. The NP nef alleles were about 2.5-fold more effective, however, in downmodulation of class I MHC cell surface expression (Fig. 4). Despite these functional differences, all consensus nef alleles were highly active compared to patient-derived nef alleles. The consensus progressor and nonprogressor nef alleles predicted the predominant amino acid residue observed in a large number of patient-derived samples at each position (24). Therefore, they might express optimized Nef proteins.
FIG. 4.
Functional differences between nef alleles representing different stages of HIV-1 infection. Functional activity of a consensus nonprogressor nef allele (NPex), primary alleles obtained during the asymptomatic stage of infection (P2-87 and SP7-88), and the average activities obtained from the first samples drawn from the six patients in Fig. 4 (Pat E) were assigned a value of unity. The functional activity of other nef alleles is shown relative to these forms. Values for CD4 (at medium GFP expression levels) and class I MHC (at high GFP expression levels) downregulation were obtained from three to six independent experiments. Values obtained for the enhancement of viral replication were derived from four to six infections and represent relative reverse transcriptase activities measured at the peak activity of the NL4-3 wild-type strain, which was observed between 9 and 13 days postinfection. Infectivity values represent at least nine measurements with three independent virus stocks. Error bars give the standard deviations.
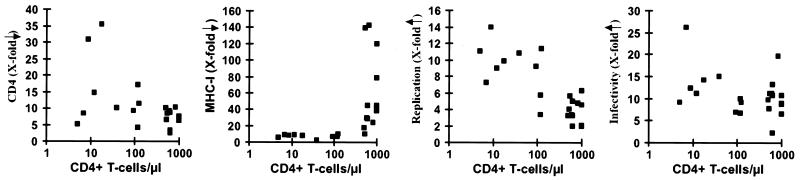
The observation that functional differences in Nef are selected during or after AIDS progression was confirmed using primary patient-derived nef alleles. The functional activities of primary nef alleles obtained at different stages of infection are indicated in Fig. 5. nef alleles derived from AIDS patients generally showed very low activity in class I MHC downmodulation (<100 CD4+ cells/μl, 6.7 ± 2.1, [n = 7]; >500 CD4+ cells/μl, 63.6 ± 46.2, [n = 11]) (Fig. 5). However, these late nef alleles were frequently more active than nef alleles derived from asymptomatic individuals with high CD4+ T cell counts in other in vitro assays for Nef function, particularly in the stimulation of viral replication (<100 CD4+ cells/μl, 10.1 ± 1.8 [n = 7]; >500 CD4+ cells/μl, 3.9 ± 1.4, [n = 11]) (Fig. 5). The results obtained with these primary nef alleles confirmed that (i) efficient Nef-mediated class I MHC downregulation is lost after progression to AIDS, (ii) late nef alleles frequently show higher activity in stimulation of viral replication in PBMC cultures, and (iii) functional activity in downmodulation of CD4 and enhancement of virion infectivity are usually maintained and sometimes increased during late stages of infection (Fig. 4 and 5; Table 2).
FIG. 5.
Functional activity of primary nef alleles obtained at different stages of infection. The x-axis gives the number of CD4+ cells per microliter at the time of PBMC sampling. A total of 20 samples derived from eight patients (SP7, SP8, P2, P5, P7, P8, P9, and P10 (Tables 1 and 2) were analyzed. Average values (■) for downregulation of CD4 (at medium GFP expression levels) and class I MHC (at high GFP expression levels), as well as enhancement of HIV-1 replication and infectivity, are indicated. Values were determined as described in Materials and Methods and in Table 2.
DISCUSSION
This study demonstrates that nef alleles found during asymptomatic infection are more active in class I MHC downregulation whereas Nef features typically found after progression to AIDS are associated with higher activity in stimulation of viral replication and frequently also with more effective downmodulation of CD4 and enhancement of virion infectivity. Our results indicate a link between Nef function and the immune status of the infected individual and are in agreement with recent reports suggesting that Nef performs multiple separable in vitro activities that might be independently selected for in vivo (19, 20, 28, 29, 32, 39). Obviously, a selective advantage might also result from other factors, e.g., escape from neutralizing-antibody and CTL responses or an expanded cell or coreceptor tropism (11). Therefore, suboptimal HIV-1 Nef variants might become predominant if the viruses contain mutations elsewhere in the genome that result in a significant growth advantage. Nonetheless, our findings suggest that more virulent Nef variants are selected in a significant number of infected individuals.
The observed changes in Nef function probably reflect an adaptation of HIV-1 to its host environment (Fig. 6). Presumably, the selective pressure for efficient Nef-mediated class I MHC downregulation is high during the asymptomatic phase of infection, particularly in patients who show effective CTL responses. In contrast, HIV-1 Nef variants that induce efficient class I MHC downregulation would have no selective advantage after progression to AIDS, when the immune system is destroyed, and in rapid progressors who are unable to mount an efficient antiviral CTL response. This might explain the low activity of nef alleles derived from P8, who progressed rapidly to AIDS. The alterations in the ability of Nef to downmodulate class I MHC at different stages of disease strongly suggest that this in vitro activity of Nef does play a role in immune evasion and efficient viral persistence in the infected host.
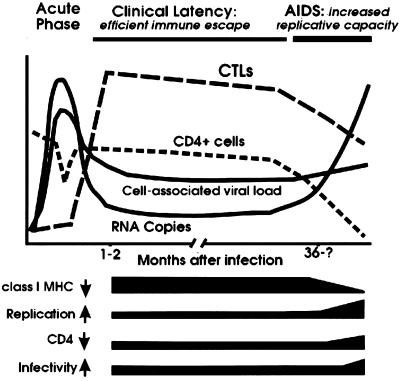
FIG. 6.
Schematic model for the modulation of independent Nef functions during the course of HIV-1 infection. The black bars indicate the changes in Nef function observed during progression to AIDS. In immunocompetent hosts, Nef efficiently downmodulates class I MHC to prevent CTL lysis of infected cells. After the breakdown of the immune system, the selective forces drive variations in Nef that are associated with an increased activity in the stimulation of HIV-1 replication and reduced functional activity in class I MHC downregulation. The ability of Nef to downregulate CD4 and to enhance virion infectivity is enhanced in some AIDS patients and is likely to optimize viral spread in a host environment where uninfected CD4+ target cells become limited. The acute phase of infection has not been investigated in this study. Symbols: ↓, downmodulation; ↑, enhancement.
Despite the mechanisms developed by HIV-1 to escape the antiviral immune response, infected cells have a short half-life and are efficiently eliminated in immunocompetent individuals (18, 41). Highly aggressive SIV nef variants, which cause strong T-cell activation, can evolve in immunodeficient macaques (13, 25). However, the mutations in Nef which are associated with high virulence revert after the onset of the cellular immune response (36). The functional differences observed in our study were more subtle. Nonetheless, these observations suggest that immunodeficiency viruses containing nef alleles that stimulate viral gene expression and replication have only a moderate a selective advantage over variants that cause stronger T-cell activation in immunocompetent hosts. Notably, we have previously found that nef alleles derived from a long-term survivor of HIV-1 infection did not stimulate viral replication in primary PBMC culture (3).
In agreement with the observation that downregulation of CD4 and class I MHC are independent nef functions (15, 39), late nef alleles were highly active in CD4 downmodulation. In all progressing individuals investigated, functional activity remained stable, suggesting that CD4 downregulation plays a role in viral replication throughout the course of infection. However, some primary nef alleles obtained after progression to AIDS and the Pex nef alleles were particularly active. Possibly some consequences of CD4 downmodulation, e.g., prevention of superinfection and enhancement of virion production, might be more important for optimal viral spread during late stages of disease, when a high percentage of CD4+ target cells are already infected. Interestingly, nef alleles derived from two long-term nonprogressors were selectively impaired in CD4 downmodulation (3, 30) (Fig. 3). CD4 is crucial for helper T-cell signaling, and the lack of efficient virus-specific proliferative responses might be of major importance for progression to AIDS (33). To understand how the virus can be controlled by the immune system, it will be important to clarify if nonprogressors frequently harbor HIV variants that are defective in this aspect of Nef function and are able to maintain strong HIV-1-specific T-helper-cell responses.
The enhancement of virion infectivity was preserved among all nef alleles investigated. Thus, although infection of MAGI cells represents a relatively artificial system, it might reflect a Nef activity that plays an important role in vivo. Obviously, enhancement of virion infectivity should always be advantageous for viral spread. However, the selective pressure might be particularly high late in infection, when the number of uninfected CD4+ target cells is greatly reduced. Under these conditions, HIV-1 variants that show higher infectivity, or an expanded cell and coreceptor tropism (7), might have a substantial selective advantage.
Do the observed variations in Nef cause progression to AIDS, or are they just a phenomenon that might be unrelated to disease? We feel that cause and effect cannot be separated in HIV-1 infection. Clearly, the ability of HIV-1 to persist and replicate at high levels in infected individuals is linked to progression. In a continuous evolutionary process, HIV-1 variants that are better adapted to spread efficiently in their respective host environment are constantly selected. New variants predominate because they have some selective advantage, either by replicating better or by being less efficiently eliminated by the antiviral immune response. Thus, while more aggressive Nef variants might occur only when the immune system is already damaged, it is obvious that these forms do emerge because they have some advantage over the existing viral quasispecies. As a consequence, HIV-1 nef alleles that are functionally optimized to the host environment should lead to a higher viral load and therefore should accelerate progression to AIDS.
Recent studies in the SIV-macaque model demonstrated that SIV becomes more virulent during disease progression (22). Our results suggest that variations in Nef that evolve during progression to AIDS might contribute to the increased virulence of HIV-1. Late-stage HIV-1 Nef variants, which are more active in stimulation of viral replication in T lymphocytes, should lead to higher levels of viremia and more rapid progression to AIDS. In support of an increased virulence of late HIV-1 isolates, it has been shown that a low maternal CD4+ cell count was associated with rapid progression to AIDS in vertically infected children (40). Additional studies with well-characterized patient cohorts and with pathogenic Nef-SHIVs might lead to the elucidation of the relevance of different HIV-1 Nef properties for virulence.
In conclusion, our results suggest that the selective forces on different Nef functions depend on the clinical stage of HIV-1 disease. The adaptive evolution of HIV to its host environment results in particularly high activity in Nef functions that promote the escape of HIV-1 from immune surveillance during the asymptomatic stage. In comparison, the in vivo selective forces drive Nef properties that enhance viral spread in a more direct manner after progression to AIDS. Importantly, our findings provide strong evidence that these independent in vitro Nef functions all contribute to viral spread in vivo. Therefore, pharmaceutical agents should disrupt molecular interactions of Nef that are critical for all these in vitro functions, e.g., membrane association. Finally, our results suggest that HIV-1 nef variants with increased pathogenicity emerge in a significant number of infected individuals and contribute to the progression to AIDS.
ACKNOWLEDGMENTS
We thank Bernhard Fleckenstein for support and encouragement. We also thank Joachim Hauber and Ronald C. Desrosiers for critical reading of the manuscript. We acknowledge Rod Daniels and Philippa Easterbrook for providing patient samples and PCR-amplified nef alleles in the early phase of this study. We are also indebted to all the patients who participated in the study.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), the Wilhelm-Sander-Stiftung, and the U.S. Public Health Service and by Cold Spring Harbor Laboratory funds.
REFERENCES
- 1.Alexander L, Du Z, Howe A Y, Czajak S, Desrosiers R C. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J Virol. 1999;73:5814–5825. doi: 10.1128/jvi.73.7.5814-5825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson R E, Sanfridson A, Ottinger J S, Doyle C, Cullen B R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral superinfection. J Exp Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carl S, Daniels R, Iafrate A J, Easterbrook P, Greenough T C, Skowronski J, Kirchhoff F. Partial “repair” of defective nef genes in a long-term nonprogressor with human immunodeficiency virus type 1 infection. J Infect Dis. 2000;181:132–140. doi: 10.1086/315187. [DOI] [PubMed] [Google Scholar]
- 4.Chowers M Y, Spina C A, Kwoh T J, Fitch N J, Richman D D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 6.Collins K L, Chen B K, Kalams S A, Walker B D, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 7.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crise B, Buonocore L, Rose J K. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–669. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 10.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, Mc Phee D A, Greenway A L, Ellett A, Chatfield C. Genomic structure of an attenuate quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 11.Desrosiers R C. Strategies used by human immunodeficiency virus that allow persistent viral replication. Nat Med. 1999;5:723–725. doi: 10.1038/10439. [DOI] [PubMed] [Google Scholar]
- 12.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 13.Fultz P N, McClure H M, Anderson D C, Switzer W M. Identification and biologic characterization of an acutely lethal variant of simian immunodeficiency virus from sooty mangabeys (SIV/SMM) AIDS Res Hum Retroviruses. 1989;5:397–409. doi: 10.1089/aid.1989.5.397. [DOI] [PubMed] [Google Scholar]
- 14.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg M E, Iafrate A J, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenough T C, Brettler D B, Kirchhoff F, Alexander L, Desrosiers R C, O'Brien S J, Somasundaran M, Luzuriaga K, Sullivan J L. Long-term nonprogressive infection with human immunodeficiency virus type 1 in a hemophilia cohort. J Infect Dis. 1999;180:1790–802. doi: 10.1086/315128. [DOI] [PubMed] [Google Scholar]
- 17.Greenough T C, Somasundaran M, Brettler D B, Hesselton R M, Alimenti A, Kirchhoff F, Panicali D, Sullivan J L. Normal immune function and inability to isolate virus in culture in an individual with long term human immunodeficiency virus infection. AIDS Res Hum Retroviruses. 1994;10:395–403. doi: 10.1089/aid.1994.10.395. [DOI] [PubMed] [Google Scholar]
- 18.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 19.Iafrate A J, Bronson S, Skowronski J. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 1997;16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iafrate A J, Carl S, Bronson S, Stahl-Hennig C, Hunsmann G, Skowronski J, Kirchhoff F. The ability of Nef to downregulate CD4 and to enhance virion infectivity are important for the replication of the simian immunodeficiency virus in rhesus macaques. J Virol. 2000;74:9836–9844. doi: 10.1128/jvi.74.21.9836-9844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 22.Kimata J T, Kuller L, Anderson D B, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 23.Kirchhoff F, Münch J, Carl S, Stolte N, Mätz-Rensing K, Fuchs D, Ten Haaft P, Heeney J L, Swigut T, Skowronski J, Stahl-Hennig C. The human immunodeficiency virus type 1 nef gene can to a large extent substitute for the simian immunodeficiency virus nef in vivo. J Virol. 1999;73:8371–8383. doi: 10.1128/jvi.73.10.8371-8383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhoff F, Easterbrook P J, Douglas N, Troop M, Greenough T C, Weber J, Carl S, Sullivan J L, Daniels R S. Sequence variations in human immunodeficiency virus type 1 Nef are associated with different stages of disease. J Virol. 1999;73:5497–5508. doi: 10.1128/jvi.73.7.5497-5508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhoff F, Carl S, Kuhn E M, Nißlein T, Hunsmann G, Stahl-Hennig C. Emergence of the acute pathogenic SIVmac239 Nef R16Y variant in a rapidly progressing macaque with exceedingly high levels of plasma viremia. Virology. 1999;257:61–70. [Google Scholar]
- 26.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term, nonprogressing survivor of HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 27.Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 28.Lang S M, Iafrate A J, Stahl-Hennig C, Kuhn E M, Niβlein T, Haupt M, Hunsmann G, Skowronski J, Kirchhoff F. Association of simian immunodeficiency virus Nef with cellular serine/threonine kinases is dispensable for the development of AIDS in rhesus macaques. Nat Med. 1997;3:860–865. doi: 10.1038/nm0897-860. [DOI] [PubMed] [Google Scholar]
- 29.Lock M, Greenberg M E, Iafrate A J, Swigut T, Münch J, Kirchhoff F, Shohdy N, Skowronski J. Two elements target SIV Nef to the AP-2 clathrin adaptor complex, but only one is required for the induction of CD4 endocytosis. EMBO J. 1999;18:2722–2733. doi: 10.1093/emboj/18.10.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–114. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piguet V, Schwartz O, Le Gall S, Trono D. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168:51–63. doi: 10.1111/j.1600-065x.1999.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 34.Ross T M, Oran A E, Cullen B R. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol. 1999;9:613–621. doi: 10.1016/s0960-9822(99)80283-8. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 36.Schwiebert R S, Tao B, Fultz P N. Loss of the SIVsmmPBj14 phenotype and nef genotype during long-term survival of macaques infected by mucosal routes. Virology. 1997;230:82–92. doi: 10.1006/viro.1997.8469. [DOI] [PubMed] [Google Scholar]
- 37.Skowronski J, Parks D, Mariani R. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J. 1993;12:703–713. doi: 10.1002/j.1460-2075.1993.tb05704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swigut T, Iafrate A J, Münch J, Kirchhoff F, Skowronski J. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility antigen expression. J Virol. 2000;74:5691–5701. doi: 10.1128/jvi.74.12.5691-5701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tovo P A, de Martino M, Gabiano C, Galli L, Tibaldi C, Vierucci A, Veglia F. AIDS appearance in children is associated with the velocity of disease progression in their mothers. J Infect Dis. 1994;170:1000–1002. doi: 10.1093/infdis/170.4.1000. [DOI] [PubMed] [Google Scholar]
- 41.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 42.Willey R L, Maldarelli F, Martin M A, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]