Abstract
Cell entry of retroviruses is initiated by the recognition of cellular receptors and the subsequent membrane fusion between viral and cellular membranes. These two steps are mediated by the surface (SU) and transmembrane (TM) subunits of the retroviral envelope glycoprotein (Env), respectively. Determinants regulating membrane fusion have been described throughout SU and TM, but the processes coupling receptor recognition to fusion are still elusive. Here we establish that a critical interaction is formed between the receptor-binding domain (RBD) and the major disulfide loop of the carboxy-terminal domain (C domain) of the murine leukemia virus SU. Receptor binding causes an alteration of this interaction and, in turn, promotes further events of Env fusion activation. We characterize mutations which, by lowering this interaction and reducing the compatibility between the RBD and C domains of Env glycoprotein chimeras, affect both Env fusogenicity and sensitivity to receptor interference. Additionally, we demonstrate that suboptimal interactions in such mutant Env proteins can be compensated in trans by soluble RBDs in a manner that depends on their compatibility with the C domain. Our results therefore indicate that RBD/C domain interactions may occur in cis, via the proper RBD of the viral Env itself, or in trans, via a distinct RBD expressed by virion-free Env glycoproteins expressed endogenously by the infected cells or provided by neighboring Env trimers.
Membrane-enveloped viruses penetrate their host cells by fusing their membranes with those of the cells to which they have bound through interactions of their attachment glycoproteins with specific cell surface receptors. While in essence the fusion process always involves the activation of viral fusion proteins and their subsequent refolding into fusion-active conformations (17), two distinct pathways of fusion activation have been described. The fusogenicity of pH-dependent viruses, such as orthomyxoviruses, is activated by the acidic pH found in the endosomal vesicles into which the virions are routed following receptor binding (46). In contrast, the fusion activation of pH-independent membrane-enveloped viruses, such as paramyxoviruses (27) and most retroviruses (31), is induced by interaction with their receptors and is thought to occur at neutral pH at the cell surface.
For retroviruses, both the binding and fusion functions are carried by a single glycoprotein, named Env. The retroviral Env complex consists of trimers of two subunits derived from a single protein precursor: a surface subunit (SU), harboring the determinants of interaction with the cell surface receptor, and a transmembrane subunit (TM), whose functions include anchorage of the trimer complex in the viral membrane and membrane fusion (19). The manner by which interaction between the retrovirus receptor and the receptor-binding domain (RBD) of the SU is molecularly converted into a signal that activates the TM-located fusion machinery is currently being actively investigated since it represents a valuable target for antiretroviral therapeutic approaches (33). While the fusion determinants that result in merging of viral and cell membranes and subsequent formation of the fusion pore seem to reside into the TM subunit (23, 52), fusion activation determinants have been found spread within the whole of the SU subunit (2, 8, 28, 29, 34, 51). For type C mammalian retroviruses such as murine leukemia viruses (MLVs), the RBD is situated in the amino-terminal half of the SU (3–5, 10, 16, 32, 36, 49) and autonomously folds into a globular domain (12). This RBD, whose receptor-binding pocket is relatively well characterized (10, 50), is followed by a proline-rich region (PRR), which connects it to the SU carboxy-terminal domain (C domain) thought to interact with or to control activation of the TM subunit (38). The amino-terminal end of the RBD of type C mammalian retroviruses also harbors an essential fusion activation determinant containing a critical histidine located in a well-conserved PHQ motif (2, 29, 51). Recent results from our laboratory indicated that this determinant is activated on receptor binding, allowing the formation of a receptor-activated RBD, which is necessary to promote further events in Env fusion activation (29). Env mutants with a deletion of the amino-terminal histidine (delH mutants) fail to activate fusion despite undergoing efficient binding to their receptors. Interestingly, non-virion-associated SU or RBD-polypeptides are necessary and sufficient to compensate for in trans fusion-defective Env delH mutants, provided that these polypeptides are activated by their cell surface receptors (29).
In this study we investigated how the receptor-activated RBDs participate in the MLV Env fusion activation cascade and modulate in cis and in trans the postbinding events of retrovirus entry into cells. By using different types of RBD-containing polypeptides and Env mutants that carry modifications of selected fusion activation determinants, we first established that the RBD forms a critical contact with a median subregion of the Env C domain known to form a disulfide-bonded loop; alteration of this contact upon receptor binding is required to trigger Env fusogenicity. Next we showed that Env-derived RBD-containing polypeptides either expressed by the target cells or accompanying the incoming viral particles can block virus entry into cells at both the binding and postbinding steps before membrane fusion. Furthermore, in an apparent paradox, our results indicated that concomitantly with their blocking effect, non-virion-associated RBD-containing polypeptides may positively participate in trans in Env fusion activation and retrovirus entry. Thus, Env fusion triggering may proceed either in cis, via the proper RBD of the Env itself, or in trans, by using a distinct RBD present in the same Env trimer, in a neighboring trimer, or as a soluble form. Our data therefore shed light on the mechanisms that link receptor recognition to fusion. Moreover, they complement the current interpretation of receptor interference (18), which, by endogenous cell expression of retroviral envelope glycoproteins and interaction with receptors, leads to resistance to superinfection.
MATERIALS AND METHODS
Cell lines.
TELCeB6 cells (9), derived from TE671 human rhabdomyosarcoma cells (ATCC CRL8805), express Moloney MLV (MoMLV) Gag and Pol proteins and an nlsLacZ reporter MLV retroviral vector. Production of infectious retroviral particles by TELCeB6 cells depends on newly introduced Env expression vectors. Cear13 cells (26) were derived from CHO (Chinese hamster ovary) cells (ATCC CCL-61) and express both ecotropic and amphotropic receptors. Clones of CHO-PiT-2 cells expressing variable levels of PiT-2 amphotropic receptors were established by transfection of a PiT-2 expression vector (44) into CHO cells and subsequent characterization of PiT-2 expression levels in an amphotropic Env binding assay. NIH 3T3 mouse fibroblasts, XC rat sarcoma cells (ATCC CCL-165), TE671, TELCeB6, Cear13, CHO-PiT-2, and CHO cells were grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum.
Construction of envelope expression vectors.
Plasmids FBASALF and FBMOSALF, encoding the wild-type MLV-4070A amphotropic (denoted A) and MoMLV ecotropic (denoted MO) Env proteins, respectively, and carrying a phleomycin resistance gene, have been described elsewhere (28). The FBAdelHSALF plasmid (29), derived from FBASALF, was designed to produce a cell entry-defective form of the amphotropic Env by deleting codon 36 of the 4070A env gene (35). The resulting mutant envelope glycoprotein, in which residue 5 of the SU Env subunit was removed, was named AdelH (29). The FBMOdelHSALF expression plasmid encoding the fusion-defective MOdelH envelope glycoproteins (29), harboring the equivalent delH mutation, which was obtained by deleting residue 8 of the SU corresponding to codon 41 of the MoMLV env gene (41), was derived from FBMOSALF.
Expression vectors encoding Env chimeras in which polypeptides corresponding to the PRR, the SU C domain, or the TM subunit ectodomain (denoted TM) derived from the MoMLV Env were substituted individually or in combination (See Fig. 1) into the matching domains of the 4070A Env have been described previously (28). The resulting Env proteins were identified according to the substituted ecotropic domain(s) (Fig. 1). For amphotropic and ecotropic Env proteins, respectively, the boundaries of the various domains were defined as A32 to V237 and A34 to L262 for RBD, G238 to P297 and G263 to A308 for PRR, G298 to R458 and G309 to R469 for C, and E459 to P654 and E470 to P665 for TM. Residues are numbered starting from the initiation methionine deduced from the amino acid sequences of the 4070A MLV (35) and the MoMLV (41) Env proteins. The expression vector encoding the PRR-C2 amphotropic Env mutant, in which a 12-amino-acid peptide of the PRR was deleted (S284 to P295), was described previously (28). All subsequent constructs were generated by PCR-mediated and oligonucleotide site-directed mutagenesis (details and sequences are available upon request) and cloned in the FBASALF or FBAdelHSALF Env expression vectors.
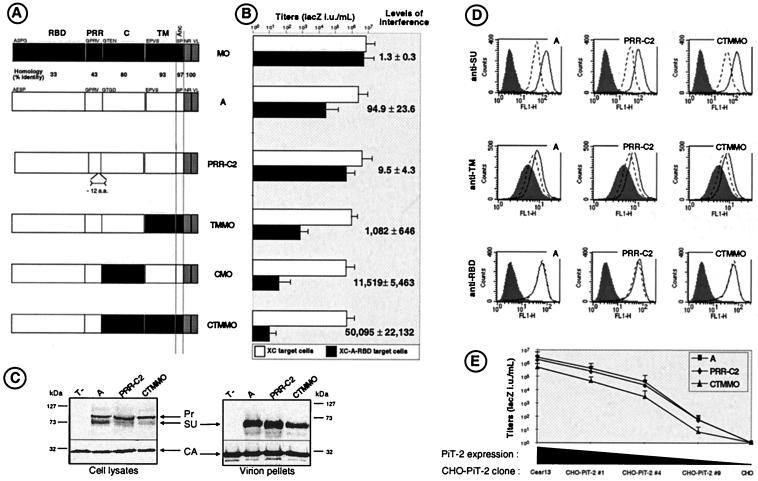
FIG. 1.
Schematic representation of Env chimeras and their properties. (A) Domain organization of parental Env proteins and chimeras. Open and solid boxes represent domains derived from ecotropic MoMLV Env (MO) and amphotropic MLV Env (A), respectively. The intracytoplasmic sequences are shown as gray boxes. Anc, anchor domain. The first amino-acids of each domain are indicated. The percentage of identical amino acids between each domain is indicated. (B) Titers (expressed as LacZ IU per milliliter) of retroviral vectors coated with the indicated Envs on XC target cells and on A-RBD-expressing XC cells. Error bars indicate standard deviations. The levels of interference were calculated according to the formula: titer[XC]/titer[XC-A-RBD]. (C) Detection of envelope glycoproteins in cell lysates and in pellets of retroviruses generated with the indicated Env proteins by immunoblotting with an anti-SU antiserum. Equivalent loading of viral samples was demonstrated by immunoblotting with an anti-capsid (CA) antiserum. Pr, Env precursor. (D) Binding assays of soluble (top histograms; probed with anti-SU antibodies) versus virion-associated (middle histograms; probed with anti-TM antibodies) wild-type amphotropic (A), PRR-C2, and CTMMO Env proteins on Cear13 cells (solid lines) and Cear13 cells expressing PiT-2 receptor-blocking A-RBD polypeptides (broken lines). The Env contents of the different samples were normalized by immunoblotting with viral supernatant or on viral pellet. No binding could be detected on CHO cells that lack PiT-2 receptors (data not shown). The background fluorescence (shaded histograms) was provided by incubating the cells with control supernatants containing viral particles devoid of Env. Detection of A-RBD polypeptides (bottom histograms; probed with anti-RGS-His tag antibodies) before (broken lines) and after (solid lines) the A-RBD-expressing Cear13 cells were incubated with retroviruses harboring the indicated Env proteins is shown. The background fluorescence (shaded histograms) was provided by using Cear13 cells not expressing A-RBD polypeptides. (E) Titers of retroviral vectors coated with the indicated Env proteins on CHO cell clones that express variable levels of PiT-2 amphotropic receptors, as determined in binding assays using A-SU. The levels of A-SU binding on cells of CHO-PiT-2 clone 1 and on cells of CHO-PiT-2 clone 4 were equivalent, respectively, to those detected on the parental XC and on the XC-A-RBD cells used as the target in panel B.
Expression vectors for the C1MO, C2MO, and C2MO or for the C1MOdelH, C2MOdelH, and C3MOdelH amphotropic Env chimeras were derived from FBASALF or FBAdelHSALF plasmids, respectively, by replacing DNA sequences encoding subregions of the amphotropic Env C domain, defined as G298 to N354, C355 to C409, and S410 to R458 in the 4070A MLV Env sequence, with the homologous subregions of the MoMLV Env, defined as G309 to N365, C366 to C420, and S421 to R469, respectively.
Plasmids encoding secreted RBDs were derived from FBASALF and FBMOSALF expression vectors. The carboxy-terminal ends of either amphotropic (A-RBD) and ecotropic (MO-RBD) RBDs, defined as A32 to G244 and A34 to G269, respectively, were fused in frame with a 9-amino-acid RGS-His tag (RGSHHHHHH) (21). Expression vectors encoding either A-RBDdelH or MO-RBDdelH were generated similarly by using the FBAdelHSALF or FBMOdelHSALF plasmids.
Production of retroviral particles.
Env expression plasmids were transfected into TELCeB6 cells as reported elsewhere (9). Transfected cells were selected with phleomycin, and phleomycin-resistant colonies were pooled. Virus-containing supernatants were collected after overnight production from confluent env-transfected cells, filtered through 0.45-μm pore-size membranes, and stored at 4°C.
Production of soluble SU or of soluble RBD fragments.
RBD or Env expression vectors were transfected in NIH 3T3, XC, or TE671 cells as reported elsewhere (29). Transfected cells were selected with phleomycin, and individual phleomycin-resistant colonies were isolated. The expression of SUs or of RBDs in each clone was analyzed by immunoblot analyses of cell lysates, using anti-SU or anti-RGS-His tag antibodies, respectively. Clones that expressed equivalent amounts of SU or RBD polypeptides were retained for production of soluble SU or RBD. SU-containing supernatants (i.e., supernatants containing SU that had accumulated as soluble material after dissociation from envelope complexes by shedding) or RBD-containing supernatants were collected after 48 h of production from confluent Env- or RBD-transfected cells, filtered through 0.45-μm pore-size membranes and stored at 4°C.
Standard infection assays.
Target cells were seeded in 24-well plates at a density of 5 × 104 cells per well and incubated overnight at 37°C. Unless otherwise indicated in the figure legends, 200 μl of viral supernatant dilutions containing 5 μg of Polybrene per ml was added to the cells after their supernatants were removed, and the cells were incubated for 5 to 6 h at 37°C. Cell supernatants were then removed, and the cells were incubated in regular medium for 48 h. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining and viral titer determination were performed as previously described and expressed as LacZ infectious units per milliliter of viral supernatants (9).
Immunoblots, binding assays, and antibodies.
Cell lysates and virus samples from Env-transfected TELCeB6 cells were prepared and analyzed in Western blots as previously described (28).
Binding assays of virions and of soluble Env-derived polypeptides were performed as described previously (29) by immunostaining using a fluorescence-activated cell sorter (FACSCalibur; Becton Dickinson) and anti-TM, anti-SU, or anti-RGS-His-tag antibodies.
Anti-SU (ViroMed Biosafety Labs) was a goat antiserum raised against the Rauscher leukemia virus gp70 and was used as a 1/2,000 dilution for Western blots or 1/200 dilution for Env binding assays. Anti-CA (ViroMed Biosafety Labs) was a goat antiserum raised against the Rauscher leukemia virus p30 capsid protein (CA), used as a 1/10,000 dilution for Western blots. Anti-TM was a mouse monoclonal antibody 372 (ATCC CRL-1893) (7) cell culture supernatant against MLV TM used undiluted for fluorescence-activated cell sorter analysis. Anti-RGS-His-tag was a mouse monoclonal antibody (Qiagen) used as a 1/100 dilution for FACS analysis.
RESULTS
Env subdomain interactions modulate infection of receptor-interfering cells.
Previous results from our laboratory indicated that regions situated downstream of the MLV RBD modulate Env fusogenicity and sensitivity to receptor blocking (28). We therefore characterized in a receptor interference assay a series of amphotropic MLV Env chimeras (Fig. 1A) harboring modifications in the C and/or TM domains (CMO, TMMO, CTMMO), and compared them to a previously described amphotropic Env chimera (28) harboring a 12-amino-acid deletion at the carboxy-terminus of PRR (PRR-C2 Env). Retroviral vectors generated with the different Env chimeras were characterized in infection assays. All mutant Env proteins allowed infection of parental target cells, resulting in vector titers higher than 106 LacZ IU/ml, which were decreased by only up to 10-fold compared to those obtained with wild-type Env (Fig. 1B). A panel of different cell lines, including NIH 3T3 mouse fibroblasts, XC rat fibrosarcoma cells, and TE671 human rhabdomyosarcoma cells, were engineered to constitutively express or not express an amphotropic RBD polypeptide (A-RBD). This resulted both in partial blocking of PiT-2 amphotropic receptors (28) and in cell surface expression of RBDs (Fig. 1D). For each Env mutant, interference levels were determined as the ratio of infectious titer on parental target cells relative to that on A-RBD-expressing target cells (Fig. 1B). Compared to wild-type amphotropic Env, for which A-RBD expression decreased virus titers by about 100-fold, the Env chimeras fell into two groups of phenotypes. Consistent with our previous results (28), the Env chimera of the first group, PRR-C2, harboring a modified PRR, was more resistant to receptor interference than was parental amphotropic Env. In contrast, Env chimeras of the second group, harboring the heterologous C domain (CMO Env) or TM ectodomain (TMMO Env) derived from MoMLV ecotropic Env, had dramatically decreased infectivity on the PiT-2-blocked target cells. Among Env mutants of this group, the combination of both ecotropic C and TM domains (CTMMO Env) was found to further increase the sensitivity to receptor interference, resulting in close to zero infectious titers on A-RBD-expressing target cells and in interference levels higher than 50,000. Results identical to those shown in Fig. 1B were obtained by expressing into target cells either the entire A-SU polypeptides or the whole amphotropic Env, as a membrane-anchored form of SU and TM, rather than A-RBD polypeptides (data not shown). Moreover, the infectivity of retroviruses carrying Env chimeras of the different groups was similarly affected when the A-SU or A-RBD polypeptides were provided in trans rather than expressed by the target cells prior to infection (see Fig. 2 and 3A). Taken together, these results indicated that while a small deletion of the amphotropic Env PRR resulted in an increased resistance to PiT-2 receptor blocking, alterations of the Env domains located downstream of the PRR seemingly increased the sensitivity to receptor interference.
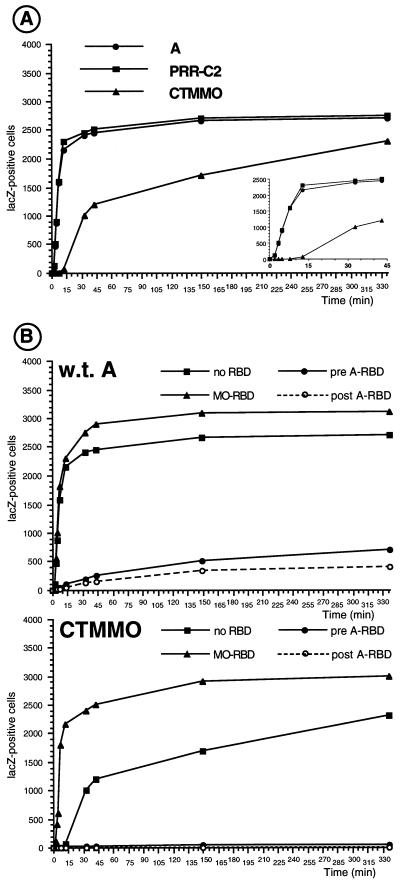
FIG. 2.
Virus-cell fusion kinetics of Env chimeras. The number of fusion events, reflected by LacZ-positive cells as a function of time, is shown. (A) Fusion kinetics on XC target cells for wild-type amphotropic, PRR-C2, and CTMMO Env proteins. (B) Fusion kinetics of wild-type amphotropic (w.t. A) (top) and CTMMO (bottom) Env proteins on XC target cells, on XC cells incubated with A-RBD before (pre A-RBD) or after (post A-RBD) virion binding at 4°C, or on XC cells preincubated with MO-RBD.
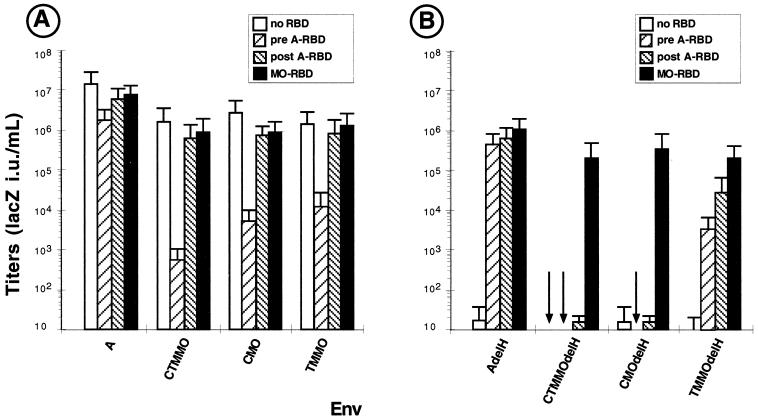
FIG. 3.
Effect of RBD fragments on infection assays with Env chimeras. Titers on XC target cells of retroviruses carrying Env chimeras (A) or H5del-mutated Env chimeras (B) in the presence of soluble RBD fragments are shown. A 200-μl volume of conditionned medium containing either MO-RBD or A-RBD was added to the target cells before (pre A-RBD) or after (post A-RBD) virion binding, carried out at 37°C.
The PRR-C2 and the CTMMO Env chimeras, representative of the two groups of phenotypes, were retained for further analysis. Western blot analysis of their pattern of expression revealed a less efficient intracellular processing than that of the wild-type amphotropic Env proteins (Fig. 1C). This led to a slightly decreased formation of mature SU glycoproteins for these two chimeras. However, no significant differences between the PRR-C2, CTMMO, and wild-type amphotropic Env proteins could be found in their incorporation into viral particles (Fig. 1C), as assessed by Western blot analysis of viral pellets, thus indicating that all of them were stably expressed on retroviruses. Moreover, binding assays performed with the SUs and with the viral particles corresponding to the three different Env glycoproteins demonstrated identical capacities to specifically attach to cells expressing PiT-2 amphotropic receptors (Fig. 1D). Furthermore, virions carrying the three different Env glycoproteins exhibited a similar reduction of binding on target cells for which the PiT-2 receptors were partially blocked with A-RBD (Fig. 1D). Finally, none of the cell-bound virions carrying these different Env glycoproteins could remove the A-RBD PiT-2-blocking polypeptides from the cell surface (Fig. 1D), suggesting that viral particles and A-RBD polypeptides were colocalized on the plasma membrane of target cells at the onset of infection. Collectively, these results demonstrated that retroviruses harboring the PRR-C2, CTMMO, and wild-type amphotropic Env proteins had similar expression patterns and binding characteristics on receptor-interfering target cells. This strongly contrasted with the marked differences of resistance and sensitivity to PiT-2 receptor blocking observed for retroviruses carrying the PRR-C2 and CTMMO Env chimeras compared to those harboring the wild-type amphotropic Env (Fig. 1B).
A possible explanation of these findings is that the modifications introduced in the Env domains located downstream of the RBD could affect the viral fusion activation thresholds. Hence, we sought to directly address the possibility that retroviruses harboring the PRR-C2 and the CTMMO Env chimeras might require fewer and more SU-receptor interactions, respectively, to trigger their fusogenicity compared to those carrying wild-type amphotropic Env. We therefore generated a panel of CHO-derived cell lines that expressed variable numbers of PiT-2 receptors. The infectivity of retroviral vectors carrying the PRR-C2, CTMMO, or wild-type amphotropic Env proteins was evaluated on these PiT-2-transfected CHO cells (Fig. 1E). When the number of PiT-2 receptors on the CHO target cells was reduced, as assessed by the reduced binding capacity of amphotropic Env (data not shown), a progressive reduction of infectivity was observed. Unexpectedly, the same reduction of infectious titers was detected for viruses carrying each of the three Env types, although, from the data in Fig. 1B, one might have expected less and more decreased infectivity for PRR-C2 and CTMMO retroviruses, respectively, compared to wild-type retroviruses when PiT-2 receptor expression was lessened. Thus, dramatic differences in behavior were found when target CHO cells expressing low levels of PiT-2 receptors were compared with cells displaying a similar low level of available binding sites achieved by using PiT-2-blocking polypeptides (Fig. 1B and E). These data therefore indicated that in terms of available binding sites on the target cell surface, there is no functional equivalence between low receptor expression and partial receptor blocking using an Env-derived polypeptide. Thus, the lack of correlation between low receptor expression and receptor blocking suggested that the RBD-containing polypeptides bound on the target cells could modulate infectivity by mechanisms other than their receptor-blocking capacity. We therefore investigated the possibility that non-virion-associated SU or RBD, localized at the surface of the infected cells, may modulate virus entry not only by limiting receptor accessibility but also by directly interfering, positively or negatively, with the postbinding entry events.
Virion-free SU or RBD modulate fusion kinetics in a receptor-dependent manner.
Our results provide evidence that subtle modifications of the conformation of the Env complex can induce important variations in the modulation of infectivity exerted by an RBD-containing polypeptide bound on target cells, most probably at a postbinding step. Therefore we thought that further characterization of the properties of the Env chimeras that exhibited different levels of resistance to receptor interference could be valuable to analyze the effect of RBD polypeptides bound to receptors at the time of infection. Thus, the kinetics of infection of lacZ retroviral vectors carrying the wild-type amphotropic Env, the PRR-C2 Env, or the CTMMO Env were determined (Fig. 2). After an initial stage of virus-cell binding at 4°C followed by two washing steps to remove the unbound virions, membrane fusion was allowed to proceed for various periods by shifting the cell temperature to 37°C. The viral particles which had remained at the cell surface without undergoing internalization and/or membrane fusion were then inactivated by a 1-min acidic shock at pH 3 (25). The levels of infection, reflecting virus-cell fusion, were assessed 2 days later by X-Gal staining. The kinetics of infection were found to be the same for retroviruses carrying either wild-type or PRR-C2 envelope glycoproteins and reached a plateau after 45 min of incubation at 37°C (Fig. 2A). In comparison, despite their similar binding capacity (Fig. 1D), the kinetics of infection of retroviral vectors carrying the CTMMO Env chimera was delayed by about 15 to 20 min in all cell types tested, including XC (Fig. 2A), NIH 3T3, Cear13, and TE671 (data not shown). However, when fusion events were allowed to proceed for longer periods (about 5 to 6 h), the infectivity of CTMMO retroviruses was found to be equivalent to or slightly lower than that of vectors carrying wild-type Env (Fig. 1B and 2A). A similar retardation of the fusion kinetics was observed in cell-cell fusion (syncytium) assays for CTMMO Env compared to wild-type Env (data not shown). Hence, the delayed kinetics of infection measured with the CTMMO retroviruses indicated that the CTMMO Env harbored a defect which affected an early stage of virus entry into the cell, after binding and before or during fusion, rather than a step that follows membrane fusion.
The presence of A-RBD or A-SU during infection of the target cells was not found to greatly affect the fusion kinetics of retroviruses generated with the PRR-C2 Env (data not shown), consistent with its reduced sensitivity to receptor interference (Fig. 1B). In contrast, either of these polypeptides, if present before infection of the target cells, could extensively decrease the fusion kinetics of viruses carrying wild-type and CTMMO Env proteins (Fig. 2B), with a particularly marked effect for the latter chimera. Strikingly, the negative effect exerted by A-RBD-containing polypeptides on membrane fusion mediated by the CTMMO Env or by the wild-type Env was also observed when A-RBD was added to the target cells after the phase of virus binding at 4°C (Fig. 2B). As demonstrated in Fig. 1D, it should be noted that similar numbers of viral particles harboring the PRR-C2, CTMMO, and wild-type Env proteins were bound on target cells before membrane fusion was initiated by elevating the temperature to 37°C. Therefore, the results of the fusion kinetics experiment confirmed that in addition to reducing the number of receptor-binding sites, the A-RBD-containing polypeptides could negatively modulate infection at a postbinding step. These data also indicated that the magnitude of the negative effect exerted by the A-RBD polypeptide on Env fusion kinetics depends on the structure of the carboxy-terminal domains of Env and is correlated with the results of interference assays (Fig. 1B). Thus, it is likely that the strong sensitivity to receptor interference of the CTMMO retroviruses is a reflection of a particularly potent negative effect of A-RBD polypeptide on this Env chimera.
Conformational rearrangements of the Env complex induced by receptor binding are necessary for triggering membrane fusion, and they involve multistep interactions between the different Env subdomains (28). Therefore, we thought that the slow kinetics of infection determined for the CTMMO Env chimera could be due to nonoptimal interactions between its RBD, of amphotropic MLV origin, and its carboxy-terminal domains, derived from ecotropic MoMLV. Since RBD fragments rescue some fusion-defective MLV Env proteins in trans (29), we sought to investigate the effect on the CTMMO Env fusion kinetics of polypeptides harboring an ecotropic RBD (MO-RBD), also derived from MoMLV SU (MO-SU). The presence of either MO-RBD or MO-SU was not found to alter the kinetics of infection of retroviruses carrying PRR-C2 (data not shown) or wild-type amphotropic Env (Fig. 2B). In contrast, either MO-RBD (Fig. 2B) or MO-SU (data not shown) could accelerate the fusion kinetics of CTMMO Env and allowed infection to proceed at wild-type rates. Additionally, slightly increased titers could reproducibly be obtained for all three Env proteins at the plateau stage of infection compared to the maximal titers obtained in the absence of RBD (Fig. 2B). Since a similar stimulating effect achieved by MO-RBD on CTMMO Env could also be demonstrated in cell-cell fusion assays (data not shown), these results indicated that MO-RBD or MO-SU could positively modulate cell entry of CTMMO retroviruses at a step situated before or during membrane fusion. Additionally, enhancement of both the membrane fusion kinetics and the overall infectivity of the CTMMO retroviruses in the presence of MO-RBD or MO-SU could be detected on various target cell types, such as XC rat cells, NIH 3T3 mouse cells, and Cear13 hamster cells, which express ecotropic receptors. Importantly however, no increase of the CTMMO Env fusion kinetics by MO-RBD was observed on human cells, such as TE671 cells, that lack the MLV ecotropic receptor (data not shown). Taken together, these data suggested that non-virion-associated ecotropic MO-SU or MO-RBD could stimulate the postbinding events of cell entry of CTMMO retroviruses in a receptor-dependent manner.
Compatibility between RDB and the major loop of the C domain is required for efficient fusion.
Results shown in Fig. 2B led us to hypothesize that fusion activation may require an interaction between the RBD and the carboxy-terminal domains of Env. Indeed because they are derived from relatively different retroviruses, it is possible that the RBD of the CTMMO Env itself is not fully compatible with the carboxy-terminal domains of this chimera. This may impair interaction(s) between these Env domains during the fusion activation process. MO-RBD might restore in trans the fusion defect of the CTMMO chimera by interacting with the C or/and TM domains of the latter Env. This interaction is likely to be optimal because the MO-RBD polypeptide and the carboxy-terminal domains of the CTMMO Env are both derived from the same glycoprotein. Nevertheless, as supported by the fact that the CTMMO retroviruses are infectious in the absence of MO-RBD (Fig. 1 and 2), the RBD of the CTMMO chimera itself also acts as a partner in cis of the fusion reaction, despite a putatively reduced compatibility with its C and/or TM domains. Hence to better investigate the interaction between the RBD and the other Env domains, we sought to distinguish the effect mediated either by the “endogenous” RBD of the CTMMO Env itself or by the “exogenous” RBD, provided in trans during virus-cell fusion. This was achieved by knocking out the capacity of either RBD to activate fusion.
The amino-terminal end of the RBDs of several type C mammalian retroviruses contains an essential fusion activation determinant (2, 29, 51). Its disruption by the delH point mutation (H5del for amphotropic Env and H8del for ecotropic Env, respectively referred to as the AdelH and MOdelH mutant Env proteins) can be fully compensated in trans by RBD polypeptides harboring an intact amino-terminal end (29). The effectiveness of this complementation is dose dependent, although it is inhibited at high RBD polypeptide concentrations by receptor blocking (29). Thus, to address the role played by the exogenous RBD during CTMMO Env fusion activation and to investigate which Env carboxy-terminal domain it might interact with, the delH mutation was first introduced into the CMO, CTMMO, and TMMO Env chimeras (Fig. 1A). As expected, similarly to virions carrying delH-mutated amphotropic Env, none of the retroviruses generated with these delH-mutated Env proteins could infect target cells in the absence of RBD polypeptides (Fig. 3B). The rescue of the fusion activity of these delH mutants was then investigated in an infection assay in the presence of A-RBD or MO-RBD polypeptides. A-RBD was found to efficiently rescue the infectivity of the AdelH retroviruses, consistent with our previous results (29), and of the TMMOdelH Env chimera (Fig 3B). In contrast, no rescue of virus-cell fusion could be detected for the delH-mutated versions of the CTMMO and CMO Env chimeras, harboring a non A-RBD-matching C domain (Fig. 3B), regardless of the A-RBD concentration used in the complementation assays. These results indicated that the carboxy-terminal domains of the two latter Env chimeras were not compatible with the A-RBD polypeptide and suggested that the RBD could interact with the Env C domain rather than with the TM ectodomain. In contrast to A-RBD polypeptides, MO-RBD could efficiently restore the fusogenicity of the CTMMOdelH and CMOdelH Env chimeras and, more surprisingly, of the AdelH and TMMOdelH Env proteins. Taken together, these results indicated that optimal interactions in the Env domains during fusion activation require compatible RBD and C domains. They also confirm that suboptimal interactions of these two domains, as in the case of the CTMMO and CMO Env proteins, can be overcome in trans by a C domain-compatible RBD polypeptide. However, the molecular basis for this compatibility is elusive since, in contrast to A-RBD, MO-RBD could restore the infectivity of all the delH-mutated Env proteins, whether they carried an amphotropic or an ecotropic C domain.
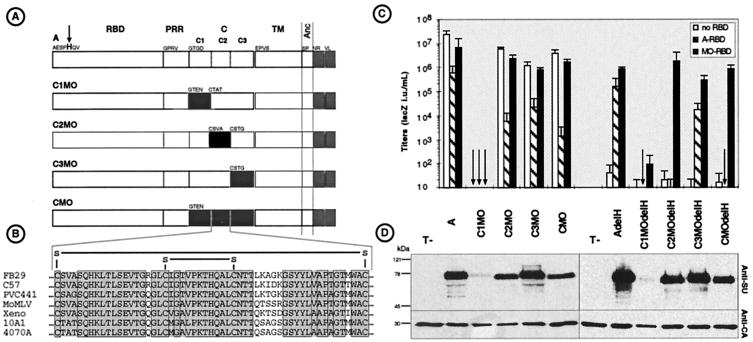
To localize the determinants of the C domain which may be responsible for interaction with the receptor-activated RBD, the MLV Env C domain was subdivided into three regions, named C1, C2, and C3 (Fig. 4). Each of these subdomains of the amphotropic Env was replaced by the corresponding C1, C2, and C3 regions derived from MoMLV Env (Fig. 4A). Viral particles generated with the C1MO chimera were not found to incorporate envelope glycoproteins and were not infectious (Fig. 4C and D). In contrast, retroviruses generated with the C2MO and C3MO Env chimeras incorporated normal Env levels and were as infectious as viruses carrying the parental CMO Env. Receptor interference assays indicated that the infectivity of the C3MO retroviruses on PiT-2-blocked target cells was inhibited by approximately the same order of magnitude than that of retroviruses carrying wild-type amphotropic Env proteins (Fig. 4C). In contrast, compared to CMO, the C2MO retroviruses exhibited the same phenotype of sensitivity to receptor interference, resulting in a very strong reduction of infectivity on target cells expressing A-RBD (Fig. 4C) or A-SU polypeptides. These results therefore suggested that the C2 region of the Env C domain, harboring a conserved disulfide-bonded loop (30) (Fig. 4B), contains determinants which account for the sensitivity to receptor interference and for the compatibility with the RBD.
FIG. 4.
Representation and properties of SU carboxy-terminal chimeras. (A) Domain organization of parental and chimeric Env proteins. Open and solid boxes represent domains derived from amphotropic MLV-4070A and ecotropic MoMLV Envs, respectively. The H5del mutation (arrow) was also introduced into each of the indicated Env proteins. (B) Sequence alignement of the C2 regions of several MLVs, showing the conserved disulfide bonds. (C) Titers (expressed as LacZ IU per milliliters) of retroviral vectors coated with the indicated Env proteins on XC target cells expressing the indicated RBDs. (D) Western blot analysis of pellets of retroviruses expressing the indicated Env proteins using anti-SU and anti-CA antisera.
To directly determine whether the C2 loop harbored components that participate in interaction with the RBD, the delH mutation was introduced into the C2MO and C3MO Env chimeras. The rescue of the fusogenicity of the resulting C2MOdelH and C3ModelH Env chimeras was then attempted in an infection assay in the presence of either A-RBD or MO-RBD polypeptides (Fig. 4C). In a manner similar to that of retroviruses carrying the parental CMOdelH Env chimera, the infectivity of retroviruses generated with the C2MOdelH Env could be restored with MO-RBD but not with A-RBD polypeptides. In contrast, the fusogenicity of the C3MOdelH Env chimera could be rescued with both the A-RBD and the MO-RBD fragments (Fig. 4C). These results therefore indicated that the C2 disulfide loop harbored a determinant responsible for the compatibility with the RBD domain with which it might directly interact.
A-RBD or A-SU both positively and negatively affect cell entry.
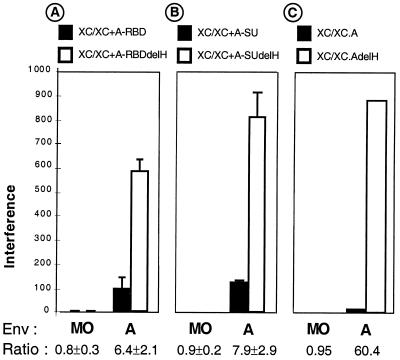
For MLVs, the SU is not tightly attached to the TM subunit and tends to dissociate from Env complexes (13). Shed SU therefore accompanies the viral particles as soluble components, and it is likely that virion-free and virion-associated SUs colocalize when they reach the cell surface during the early events of infection. Although the soluble SU may compete with the viral particles for receptor occupancy at high concentrations, the results presented in this report raise the possibility that non-virion-associated SU polypeptides themselves may also positively participate in trans in Env fusion activation (Fig. 2 to 4). This might be achieved by direct interaction of the RBD of the receptor-bound virion-free SU with the C domain of the viral Env. Therefore, we sought to decipher the antagonistic properties of non-virion-associated SU during infection, i.e., receptor blocking versus fusion activation. Since the delH mutation leads to binding-competent but fusion activation-deficient RBDs (29), this alteration was introduced into A-SU or A-RBD polypeptides. The different polypeptides were then compared for their capacity to inhibit the infection of retroviruses carrying wild-type amphotropic envelope glycoproteins in a receptor interference assay. As shown previously (29), each pair of polypeptides (A-RBD plus A-RBDdelH and A-SU plus A-SUdelH) exhibited identical binding levels on the target cells and could similarly reduce binding of the viral particles (data not shown). Interference levels, calculated for each type of polypeptides (A-RBD versus A-RBDdelH (Fig. 5A) and A-SU vs. A-SUdelH (Fig 5B), were considerably augmented when the receptor-interfering polypeptides harbored the delH mutation. Similar results were obtained by expressing in target cells the whole amphotropic Env proteins as membrane-anchored forms of SU and TM rather than using the entire A-SU or the A-RBD polypeptides (Fig 5C). These data indicated that despite identical expression and receptor-binding capacities, the A-RBDdelH-containing polypeptides were more potent than their parental counterparts for blocking virus-cell fusion, most probably because of the disruption of their amino-terminal fusion activation determinant. From these data, we could deduce that in addition to their receptor-blocking capacity that negatively affects virus entry, virion-free A-RBD-containing polypeptides, expressed by the target cells or provided in trans, may positively participate in activation of amphotropic Env fusogenicity upon receptor binding.
FIG. 5.
Comparative interference assays. Interference levels calculated for retroviruses coated with the indicated Env proteins and used to infect XC cells preincubated with A-RBD polypeptides (A), preincubated with A-SU polypeptides (B), or expressing complete SU/TM amphotropic Env glycoproteins (C) harboring or not harboring the delH mutation are shown. For each Env type, the ratio of interference levels calculated for the delH-mutated A-RBD-containing fragments relative to that of the parental A-RBD-containing polypeptides is indicated.
DISCUSSION
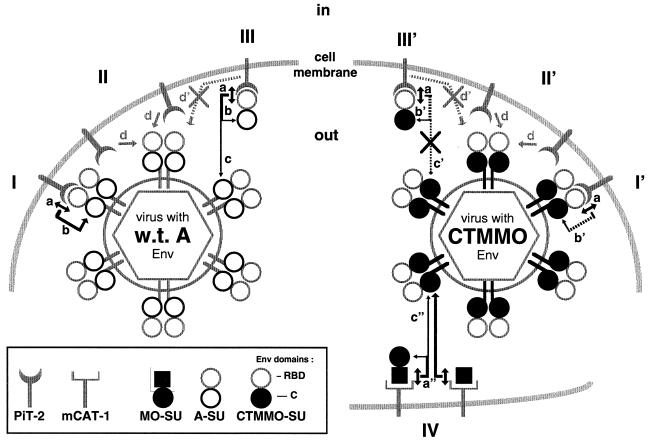
Acting at a preliminary step of retrovirus entry into cells, receptor interference, leading to resistance to superinfection, has initially been described as the process by which the endogenous expression of an Env glycoprotein, by interacting with a given retrovirus receptor, can inhibit infection by an exogenous retrovirus that uses the same receptor (18). Thus, Env-derived polypeptides containing an RBD, either expressed endogenously by the target cells or provided in trans by neighboring cells, can block infection by impairing virion accessibility to receptors, most probably through their masking and/or downregulation (22, 26). In addition to these mechanisms, the results reported here indicate that RBD-containing polypeptides, expressed by or provided in trans to the target cells, can block infection not only by impairing virion binding but also by negatively modulating virion penetration at a postbinding step before membrane fusion. These results therefore imply that abundant expression of Env in receptor-interfering cells is not necessarily required to efficiently block infection by exogenous retroviruses. They are consistent with the findings that the concentration of A-RBD polypeptide required to inhibit virus entry was lower than that required to inhibit the binding of virus particles at the cell surface (5). Additionally, our data suggest that expression of Env at the surface of infected cells may positively modulate the fusion activation of cell surface-bound retroviruses. Although this stimulation may not be effective in the context of receptor-interfering cells, where the blocking effect of endogenously expressed envelope glycoproteins is likely to be dominant, it might provide some insights in the mechanisms evolved by MLVs to optimize infection of normal cells. Indeed, since SU can easily dissociate from the MLV Env complex and accompanies virions as soluble materials, our data have several implications for our understanding of how both virion-associated and virion-free SU participate in Env fusion activation and modulate the post-binding events of retrovirus entry into cells. The availability of different types of RBD-containing polypeptides which can be expressed by the target cells or, alternatively, provided in trans at various stages of the infection process, allowed us to highlight different events that may modulate virus entry in vivo, including virus entry under conditions of receptor interference. These mechanisms are summarized in Fig. 6.
FIG. 6.
Model of retroviral fusion activation and receptor interference. (Left) Following interaction with PiT-2 receptors (a), activated RBD activates the C domain (b) through a specific interaction with the C2 subdomain, leading to fusion triggering of the Env complex (I). The interaction between activated RBD and C2 can occur in cis (b) or in trans (c). Receptor recruitment (d) by untriggered Env proteins continues long after initial virus attachment (II), supporting a model of accrual of a critical number of triggered complexes to result in membrane fusion (15). Receptor interference (III) is predominantly due to receptor blockade and reduction of available free receptors (d′). However, it is mitigated by the ability of receptor-activated A-SU molecules to trans activate fusion by interaction with the C domains of virion Env (c). w.t. A, wild-type amphotropic. (Right) Due to low compatibility between receptor-activated RBD and C domain (b′), fusion activation of CTMMO Env is impaired (I′) and is critically dependent on recruitment of additional PiT-2 receptors (II′). Blocking of PiT-2 receptors by A-SU (III′) is therefore a strong inhibitor of CTMMO Env fusogenicity because of the critical dependence of CTMMO Env for free PiT-2 receptors and because the low compatibility of A-SU with CTMMO Env C domain prevents fusion trans activation (c′). However, full activation of CTMMO Env can be restored in trans by mCAT-1-activated MO-SU (c") or, more efficiently, by MO-RBD molecules (a") through intermolecular contacts with the CTMMO Env C domain (IV).
Multiple Env-receptor interactions are required for retrovirus fusion.
Our results are consistent with a model of MLV entry which involves numerous Env-receptor interactions that must occur in a cooperative manner to induce the formation of a sufficient number of fusion-activated envelope glycoproteins required for achieving virus-cell membrane fusion. Indeed, A-RBD fragments added after a phase of virion binding could inhibit infection and slow the fusion kinetic of amphotropic retroviruses. An inference of this result is that the masking of PiT-2 amphotropic receptors by the A-RBD polypeptides on virion-bound cells inhibits the recruitment of free receptors that are required to promote further events of viral fusion activation. This indicates that the initial binding of viral particles, to a limited number of receptors on target cells, may not be sufficient for triggering virus-cell membrane fusion and that additional interactions between unbound virion-associated Env and other free receptors are necessary for virus penetration into cells (15). However, successful virus penetration may occur at later stages upon dissociation of A-RBD–receptor complexes or when free receptors, newly expressed by the target cell, reach the cell surface to assist membrane fusion of the cell surface-bound viral particle. Such a recruitment of additional receptors is consistent with the observation of receptor clustering during infection (42, 45). It is also compatible with the notion that different subpopulations of the same receptor coexist on the cell surface and may differentially influence virus binding and entry (5, 40). Receptor recruitment is expected to be dependent on cofactors (42, 45) and on drugs (37) that modulate the mobility of molecules along the plasma membrane. Variations between cell types in the quantity of such cofactors may explain differences in virus fusion kinetics and syncytium formation. Of note, compared to NIH 3T3 mouse fibroblasts, more rapid fusion kinetics were observed on XC and CHO-PiT-2 cells (data not shown), which are particularly permissive to retrovirus-induced cell-cell fusion (42). Likewise, mouse fibroblasts can be rendered permissive to MoMLV-induced syncytium formation after treatment with amphotericin B (37), a drug that increases membrane fluidity (24, 48), or following their oncogenic transformation (47), a process known to alter the structure and function of the plasma membrane.
Fusion activation requires optimal RBD/C domains interaction.
Our results indicate that the inhibition of virus entry by A-RBD-containing polypeptides depends on the intrinsic capacity of Env fusogenicity to be triggered on receptor binding and reflects the requirement for recruitment of free receptors necessary to initiate viral/cell membrane fusion (Fig. 6). Fusogenicity of MLV Env is thought to involve a cascade of conformational changes within the Env complex, which are initiated by the binding of the RBD to the receptor and which end with the folding of the TM subunit into its fusogenic conformation (18). Two key events of this pathway involve, on the one hand, a receptor-mediated modification of the RBD (29) and, on the other hand, an interaction between the receptor-activated RBD and the SU C domain, as shown here. Compared to wild-type Env, activation of the fusogenicity of the CTMMO Env chimera is less readily achieved on receptor binding, as a result of a low compatibility between the RBD and C domains of its SU. Membrane fusion mediated by retroviruses carrying this mutant Env would therefore require more time to occur and/or more Env-receptor interactions. Thus, CTMMO-mediated fusion is critically dependent on the recruitment of additional free receptors, and this probably explains its greater sensitivity to receptor blocking mediated by A-RBD-containing polypeptides. In contrast, interactions between the RBD and C domains of the PRR-C2 envelope glycoprotein occur more easily than for wild-type Env (D. Lavillette and F.-L. Cosset, unpublished data), explaining the lower sensitivity of the former Env to receptor interference. Importantly, the analysis of Env chimeras such as those described here, which carry RBD and C domains of different retroviruses, allowed us to delineate a subregion of the MLV C domain that might interact with the RBD during the fusion activation cascade. This subregion (Fig. 4B), which is known to form a disulfide-bonded loop (30), encompasses a stretch of 4 or 5 amino acids whose sequence is highly variable despite its conserved position in the C domains of MLVs and of other type C mammalian retroviruses, such as feline leukemia viruses (FeLVs). This motif might therefore represent a critical determinant of the interaction between RBDs and C domains. Moreover, the sequence of this motif also varies within the different strains in a given group of type C mammalian retroviruses that includes pathogenic and nonpathogenic strains. In fact, the evolution of nonpathogenic strains to cytopathic ones is associated with the acquisition of mutations in the SU and, more particularly, in this motif for both ecotropic MLVs (Fig. 4B) and FeLVs (11). We propose that variation of this motif allows a fine-tuning of the RBD-C domain interaction and might reflect the ease with which receptor-activated RBD activates the Env fusion cascade and Env cytotoxicity.
Receptor blocking and fusion activation are overlapping mechanisms.
In an apparent contradiction of their negative role in virus entry, our results suggest that non-virion-associated RBD-containing polypeptides may concomitantly participate in Env fusion activation (Fig. 6). However, such a positive effect is difficult to assess directly since it is intrinsically associated with receptor blocking (Fig. 5), an effect which is detected predominantly in receptor interference assays. Nevertheless, a beneficial effect of virion-free RBD-containing polypeptides could be revealed for retroviruses harboring fusion-defective delH-mutated Env proteins (Fig. 3 and 4). Interestingly, infection of T cells by the feline FAIDS virus, FeLV-T, requires a T-cell-secreted cofactor, named FeLIX, whose sequence is closely related to that of FeLV-B RBD (1). Moreover the FeLV-T envelope glycoprotein is fusion-defective as it lacks the critical histidine at position 6 of its own RBD (11). Thus, in agreement with the finding that RBD fragments can restore delH-mutated fusion-defective type C mammalian retrovirus Env proteins (29), FeLIX-dependent replication of FeLV-T may represent a natural situation of a fusion-helper function provided by a non-virion-associated RBD fragment. Several endogenous retrovirus loci express Env-derived polypeptides in vertebrates (6, 20, 39). Whether their expression confers protection against exogenous retrovirus infection or whether they may provide fusion helper functions to as yet unknown retroviruses remains to be determined.
For more classical retroviruses, the importance of non-virion-associated Env-derived polypeptides in cell entry is questionable. In MLVs, the proteolytically processed SU and TM subunits are held together via a labile disulfide bond whose disruption in reducing environments is likely to reduce the stability of the Env complex (38). Thus MLV SU has the propensity to dissociate from the Env complex (13) by a process known as shedding. Virion-free SU is therefore an abundant contaminant of infectious retroviral particles and, when expressed at excessive levels, may act as a competitor for retrovirus infection (43). Nevertheless, the results of this study raise the possibility that non-virion-associated SU, whether it accompanies the viral particles or whether its dissociation from virions is induced when they approach the target cell surface, could also play a positive role during the early events of the infection process before receptor interference is established.
ACKNOWLEDGMENTS
We are grateful to François Mallet and Thomas Schulz for helpful comments on the manuscript.
This work was supported by Agence Nationale pour la Recherche contre le SIDA (ANRS), AFIRST, Association Française contre les Myopathies (AFM), Association pour la Recherche contre le Cancer (ARC), Centre National de la Recherche Scientifique (CNRS), the European Community, and Institut National de la Santé Et de la Recherche Médicale (INSERM).
REFERENCES
- 1.Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 2.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini J L, Rodrigues P, Müller R, Danos O, Heard J-M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blond J-L, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset F-L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro B, Wehrly K, Cloyd M, Britt W, Portis J, Collins J, Nishio J. Characterization of mouse monoclonal antibodies specific for Friend murine leukemia virus-induced erythroleukemia cells: friend-specific and FMR-specific antigens. Virology. 1981;112:131–144. doi: 10.1016/0042-6822(81)90619-x. [DOI] [PubMed] [Google Scholar]
- 8.Chung M, Kizhatil K, Albritton L M, Gaulton G N. Induction of syncytia by neuropathogenic murine leukemia viruses depends on receptor density, host cell determinants, and the intrinsic fusion potential of envelope protein. J Virol. 1999;73:9377–9385. doi: 10.1128/jvi.73.11.9377-9385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosset F-L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995b;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey R A, Zuo Y, Cunningham J M. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 13.Gliniak B C, Kozak S L, Jones R T, Kabat D. Disulfide bonding controls the processing of retroviral envelope glycoproteins. J Biol Chem. 1991;266:22991–22997. [PubMed] [Google Scholar]
- 14.Haywood A. Characteristics of Sendai virus receptors in a model membrane. J Mol Biol. 1974;83:427–436. doi: 10.1016/0022-2836(74)90504-x. [DOI] [PubMed] [Google Scholar]
- 15.Haywood A. Virus receptors: Binding, adhesion strengthening, and changes in viral structure. J Virol. 1994;68:1–5. doi: 10.1128/jvi.68.1.1-5.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heard J-M, Danos O. An amino-terminal fragment of the friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 18.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 19.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda H, Sugimura H. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 1989;63:5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janknecht R, Martynoff G D, Lou J, Hipskind R A, Nordheim A, Stunnenberg H G. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jobbagy Z, Garfield S, Baptiste L, Eiden M V, Anderson W B. Subcellular redistribution of Pit-2 Pi transporter/amphotropic leukemia virus (A-MuLV) receptor in A-MuLV-infected NIH 3T3 fibroblasts: involvement in superinfection interference. J Virol. 2000;74:2847–2854. doi: 10.1128/jvi.74.6.2847-2854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kataoka T, Koprowski H. Lipids and cell fusion in vitro: effect of amphotericin B. Proc Soc Exp Biol Med. 1975;149:447–451. doi: 10.3181/00379727-149-38825. [DOI] [PubMed] [Google Scholar]
- 25.Kizhatil K, Albritton L M. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virol. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak S L, Siess D C, Kavanaugh M P, Miller A D, Kabat D. The envelope glycoprotein of an amphotropic murine retrovirus binds specifically to the cellular receptor/phosphate transporter of susceptible species. J Virol. 1995;69:3433–3440. doi: 10.1128/jvi.69.6.3433-3440.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb R A. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 28.Lavillette D, Maurice M, Roche C, Russell S J, Sitbon M, Cosset F-L. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol. 1998;72:9955–9965. doi: 10.1128/jvi.72.12.9955-9965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavillette D, Ruggieri A, Russell S J, Cosset F-L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J Virol. 2000;74:295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure M O, Sommerfelt M A, Marsh M, Weiss R A. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 32.Morgan R A, Nussbaum O, Muenchau D D, Shu L, Couture L, Anderson W F. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:4712–4721. doi: 10.1128/jvi.67.8.4712-4721.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunberg J H, Follis K E, Trahey M, LaCasse R A. Turning a corner on HIV neutralization? Microbes Infect. 2000;2:213–221. doi: 10.1016/s1286-4579(00)00267-7. [DOI] [PubMed] [Google Scholar]
- 34.Nussbaum O, Roop A, Anderson W F. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:7402–7405. doi: 10.1128/jvi.67.12.7402-7405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia virus: close relationship to mink cell focus-forming viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinter A, Chen T-E, Lowy A, Cortez N G, Siligari S. Ecotropic murine leukemia virus-induced fusion of murine cells. J Virol. 1986;57:1048–1054. doi: 10.1128/jvi.57.3.1048-1054.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson H L, Lamoreux W F. Expression of endogenous ALV antigens and susceptibility to subgroup E ALV in three strains of chickens (endogenous avian C-type virus) Virology. 1976;69:50–62. doi: 10.1016/0042-6822(76)90193-8. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues P, Heard J M. Modulation of phosphate uptake and amphotropic murine leukemia virus entry by posttranslational modifications of PIT-2. J Virol. 1999;73:3789–3799. doi: 10.1128/jvi.73.5.3789-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukemia virus. Nature (London) 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 42.Siess D C, Kozak S L, Kabat D. Exceptional fusogenicity of chinese hamster ovary cells with murine retroviruses suggests roles for cellular factor(s) and receptor clusters in the membrane fusion process. J Virol. 1996;70:3432–3439. doi: 10.1128/jvi.70.6.3432-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slingsby J H, Baban D, Sutton J, Esapa M, Price T, Kingsman S M, Kingsman A J, Slade A. Analysis of 4070A envelope levels in retroviral preparations and effect on target cell transduction efficiency. Hum Gene Ther. 2000;11:1439–1451. doi: 10.1089/10430340050057512. [DOI] [PubMed] [Google Scholar]
- 44.VanZeijl M, Johann S V, Cross E, Cunningham J, Eddy R, Shows T B, O'Hara B. An amphotropic virus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Paul R, Burgeson R, Keene D, Kabat D. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J Virol. 1991b;65:6468–6477. doi: 10.1128/jvi.65.12.6468-6477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 47.Wilson C A, Marsh J W, Eiden M V. The requirements for viral entry differ from those for virally induced syncytium formation in NIH 3T3/DTras cells exposed to Moloney murine leukemia virus. J Virol. 1992;66:7262–7269. doi: 10.1128/jvi.66.12.7262-7269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshimura A, Kobayashi T, Hidaka K, Kuwano M, Ohnishi S. Altered interaction between Sendai virus and a Chinese hamster cell mutant with defective cholesterol synthesis. Biochim Biophys Acta. 1987;904:159–164. doi: 10.1016/0005-2736(87)90099-x. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, Soong N, Anderson W F. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J Virol. 1995;69:6557–6562. doi: 10.1128/jvi.69.10.6557-6562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zavorotinskaya T, Albritton L M. A hydrophobic patch in ecotropic murine leukemia virus envelope protein is the putative binding site for a critical tyrosine residue on the cellular receptor. J Virol. 1999;73:10164–10172. doi: 10.1128/jvi.73.12.10164-10172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zavorotinskaya T, Albritton L M. Suppression of a fusion defect by second-site mutations in the ecotropic murine leukemia virus surface protein. J Virol. 1999;73:5034–5042. doi: 10.1128/jvi.73.6.5034-5042.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu N L, Cannon P M, Chen D, Anderson W F. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J Virol. 1998;72:1632–1639. doi: 10.1128/jvi.72.2.1632-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]