Abstract
Internal ribosomal entry sites (IRESs) of certain plus-strand RNA viruses direct cap-independent initiation of protein synthesis both in vitro and in vivo, as can be shown with artificial dicistronic mRNAs or with chimeric viral genomes in which IRES elements were exchanged from one virus to another. Whereas IRESs of picornaviruses can be readily analyzed in the context of their cognate genome by genetics, the IRES of hepatitis C virus (HCV), a Hepacivirus belonging to Flaviviridae, cannot as yet be subjected to such analyses because of difficulties in propagating HCV in tissue culture or in experimental animals. This enigma has been overcome by constructing a poliovirus (PV) whose translation is controled by the HCV IRES. Within the PV/HCV chimera, the HCV IRES has been subjected to systematic 5′ deletion analyses to yield a virus (P/H710-d40) whose replication kinetics match that of the parental poliovirus type 1 (Mahoney). Genetic analyses of the HCV IRES in P/H710-d40 have confirmed that the 5′ border maps to domain II, thereby supporting the validity of the experimental approach applied here. Additional genetic experiments have provided evidence for a novel structural region within domain II. Arguments that the phenotypes observed with the mutant chimera relate solely to impaired genome replication rather than deficiencies in translation have been dispelled by constructing novel dicistronic poliovirus replicons with the gene order [PV]cloverleaf-[HCV]IRES-Δcore-R-Luc-[PV]IRES-F-Luc-P2,3-3′NTR, which have allowed the measurement of HCV IRES-dependent translation independently from the replication of the replicon RNA.
Higher-order structures of RNA can be deduced by a combination of different experimental approaches. These include (i) computer-aided folding, favorably under consideration of phylogenetic kinship of the RNAs, (ii) limited enzymatic and chemical degradation, and (iii) analysis by nuclear magnetic resonance spectroscopy or X-ray crystallography (11). Genetic analysis is another powerful tool, particularly if the RNA structure is a component of a self-replicating entity such as an RNA virus genome.
Internal ribosomal entry sites (IRESs) regulate translation in eukaryotic systems in a cap-independent manner. Present in genomes of some plus-strand RNA viruses and some cellular mRNAs, IRES elements belong to the most complex cis-acting signals known in the RNA world (52). In comparison to the Shine-Dalgarno signal (42), which determines internal entry of the ribosomal subunits into prokaryotic (multicistronic) mRNA, viral IRESs are colossal (up to 400 nucleotides), yet little if any of their sequence is dispensable (54).
IRES elements have been discovered in genomes of picornaviruses (20, 21, 33), of members of the genus Hepacivirus (hepatitis C virus [HCV] [47]) and Pestivirus (e.g. bovine viral diarrhea virus, [36]) of the family Flaviviridae, and of some insect RNA viruses (for example, Plautia stali intestine virus [41]). In animal viruses, IRES elements map to 5′ nontranslated regions (5′NTR) (53), whereas, remarkably, in some insect viruses, they are located several thousand nucleotides downstream from the 5′ end of the genome, separating different cistrons (41).
Early results of secondary-structure analyses of picornaviruses IRESs by Pilipenko et al. (35) were subsequently largely supported by biochemical analyses and genetics (reviewed by Stewart and Semler [46]). Genetic studies of the higher-order structure of the HCV IRES, however, cannot be carried out since no adequate tissue culture or animal system is available that allows efficient replication of HCV. Structural analyses of the HCV IRES therefore relied on computer folding, biochemical probing, and functional analysis of the IRES in vitro and in vivo in the context of expression vectors (reviewed by Lemon and Honda [24]).
We have recently shown that the IRES element of poliovirus (PV) can be exchanged with IRES elements of other viruses, resulting in viable chimeric viruses. The donor IRESs in these chimeras originated from different picornaviruses, such as encephalomyocarditis virus (2) and human rhinovirus 2 (12), or from HCV (see Fig. 1A) (27, 61). Such chimeric viruses replicate very well in tissue culture and thus have been useful for studies of genome replication or IRES function. We report here a genetic analysis of domain II of the HCV IRES in the context of the poliovirus genome. The results offer strong support for the notion that the 5′ border of the HCV IRES maps to the bottom of domain II (nucleotide [nt] 43), a result proposed previously (38). Importantly, the study has yielded a new structure for domain II of the HCV IRES.
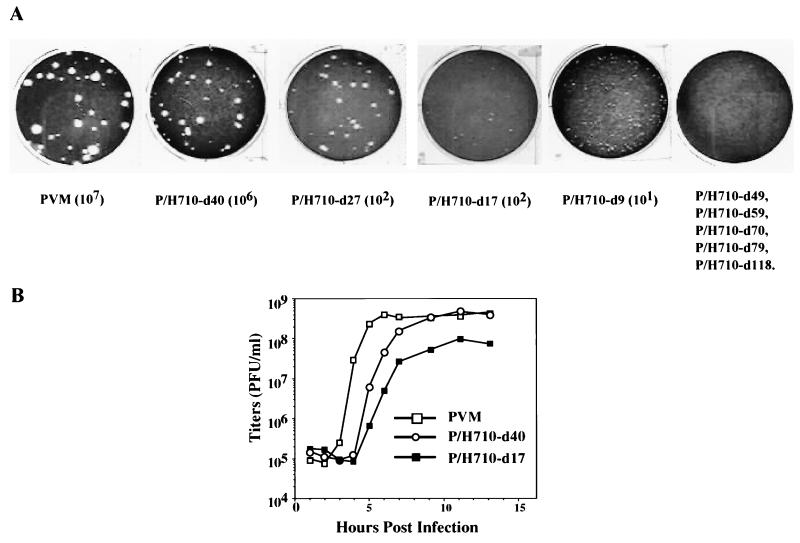
FIG. 1.
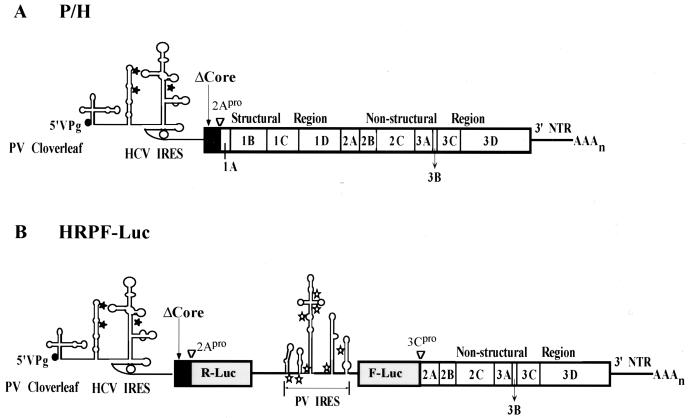
Schematic presentations of the P/H chimera and a dicistronic replicon, HRPF-Luc. (A) Diagram of the genomic organization of P/H chimeras. The cloverleaf-like RNA structure of PV, an essential cis-acting replication signal ending with the genome-linked protein VPg, is located at the 5′ end of the genome. Noninitiating AUG codons found in the HCV 5′ NTR are denoted by stars. The solid (HCV) and open (PV) boxes depict open reading frames encoding viral polypeptides; the position of the HCV core fragment (the first 123 amino acids) gene is denoted by ΔCore. Overall, the HCV-specific sequence in P/H710-d17 spans from nt 18 to 710 (27, 61). PV-encoded polypeptides within the polyprotein are indicated by 1A, 2B, etc. The PV-encoded proteinase 2Apro is responsible for the cleavage between ΔCore and capsid precursor P1. (B) Diagram of the genomic organization of HRPF-Luc, with the gene order [PV]cloverleaf-[HCV]IRES-Δcore-R-Luc-[PV]IRES-F-Luc-P2,3-3′NTR. The fusion between ΔCore and R-Luc and between F-Luc and 2A is cleaved by 2Apro. The HCV ΔCore contains 123 codons.
It has been suggested that IRES elements may be involved in RNA replication (reviewed in reference 58). If so, changes introduced into an IRES element may affect viral macromolecular synthesis in addition to control of protein synthesis. To find whether the mutation engineered into the IRES of the PV/HCV (P/H) chimera influence translational control and RNA synthesis, we have separated the function of the HCV IRES from possible involvement in replication. We have constructed dicistronic replicons with the gene order [PV]clover leaf-[HCV]IRES-Δcore-R-Luc-[PV]IRES-F-Luc-P2,3-3′NTR (Fig. 1B). Mutations introduced into the HCV IRES produced a pronounced translation phenotype in Renilla luciferase (R-Luc) expression, independent of firefly luciferase (F-Luc) expression, an observation corroborating the results obtained with the P/H chimeric virus.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
HeLa R19 cell monolayers were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% bovine calf serum (BCS). PV type 1, strain Mahoney [PV1(M)] and its chimeric constructs were amplified in HeLa cells as described by Lu et al. (28). The titer of the virus stocks was determined by standard plaque assay on HeLa R19 monolayers (26). Briefly, HeLa cells were infected with cell lysates derived from transfection with the corresponding transcript RNA. Plasmid pT7PVM was a derivative of pT7PV1-5, a full-length cDNA clone of PV1(M) constructed in this laboratory (49). P/H710-d17 (61) contains nt 18 to 710 of the genome of HCV-1b, which includes domains II to IV of the HCV IRES and 123 codons of the core gene (Fig. 1A). Note that in previous studies, P/H710-d17 was designated a P/H701-2A because the chimeric genome contained the HCV-specific sequence up to position 710 of the HCV genome and the fusion peptide (Δcore-P1) is processed by the PV proteinase 2Apro (61). The numbering of the HCV 5′NTR in this paper conforms to the numbering of the full-length HCV 5′NTR (see, for example, reference 17).
Construction of 5′NTR mutants.
The region between EcoRI and SacI of the P/H710-d17 cDNA was removed to produce the cloning vector 710dES, lacking HCV IRES and core sequences. This cloning vector was used for the generation of a series of constructs shown in Tables 1 and 2. Basically, two strategies were used to generate the corresponding mutants. The templates used in the PCR for the generation of corresponding constructs are listed in Tables 1 and 2. First, oligonucleotide PVVP4 (5′-CGTTACTAGCTGAATCTCTATAATAATTAATGG-3′) was used as the universal minus-strand primer for the PCR mutagenesis reactions to generate the constructs by using the oligonucleotide in Table 1 as positive-strand primer. Second, oligonucleotide PV1-30 (5′-TTAAAACAGCTCTGGGGTTGTACCCACCCC-3′) and the negative-strand primer of each pair in Table 2 (negative sense, denoted by a minus sign after the name of the oligonucleotides) were used in the PCR A. PVVP4 and the positive-strand primer of each pair (positive sense, denoted by a plus sign after the name of the oligonucleotides) were used in PCR B. Gel-isolated PCR fragments from both PCR A and PCR B were used in PCR C with oligonucleotides PV1-30 and PVVP4 to produced the PCR fragment with the designed mutation. The mutated PCR fragment was digested with EcoRI and SacI and cloned into 710dES to yield the desired mutants, which was selected by restriction mapping and verified by sequencing.
TABLE 1.
Oligonucleotides and templates used for mutagenesis
| Construct | Template | Oligonucleotide | Sequence |
|---|---|---|---|
| P/H710-d9 | P/H710-d17 | ID9 | 5′-ACTTAGGAATTCCGATTGGGGGCGACACTCCACCATAGATCACTCCCCTGTGAGG-3′ |
| P/H710-d27 | P/H710-d17 | ID27 | 5′-ACTTAGGAATTCCACCATAGATCACTC CCCTGTGAGGAACTACTG-3′ |
| P/H710-d40 | P/H710-d17 | ID40 | 5′-ACTGAATTCTCCCCTGTGAGGAACTACTGTCTTCACG-3′ |
| P/H710-d49 | P/H710-d17 | ID49 | 5′-GAC TGAATTCAGGAACTACTGTCTTCACGCAGG-3′ |
| P/H710-d59 | P/H710-d17 | ID59 | 5′-GACTGAATTCGTCTTCACGCAGAAAGCGTCTAG-3′ |
| P/H710-d70 | P/H710-d17 | ID70 | 5′-GACTGAATTCGAAAGCGTCTAGCCATGGCG-3′ |
| P/H710-d79 | P/H710-d17 | ID79 | 5′-GACTGAATTCTAGCCATGGCGTTAGTATGAGTG-3′ |
| P/H710-d118 | P/H710-d17 | ID118 | 5′-GACTGAATTCACCCCCCCTCCCGGGAGAGCC-3′ |
| P/H710-d40-44–45 | P/H710-d40 | d40 44–45 | 5′-ACTTAGGAATTCTCCggTGTGAGGAACTACTGTC-3′ |
| P/H710-d40-49–51 | P/H710-d40 | d40 49–51 | 5′-ACTTAGGAATTCTCCCCTGTCTCGAACTACTGTCTTC-3′ |
| P/H710-d27 | P/H710-d17 | ID27 | 5′-ACTTAGGAATTCCACCATAGATCACTC CCCTGTGAGGAACTACTG-3′ |
| P/H710-d40 | P/H710-d17 | ID40 | 5′-ACTGAATTCTCCCCTGTGAGGAACTACTGTCTTCACG-3′ |
| P/H710-d49 | P/H710-d17 | ID49 | 5′-GAC TGAATTCAGGAACTACTGTCTTCACGCAGG-3′ |
| P/H710-d59 | P/H710-d17 | ID59 | 5′-GACTGAATTCGTCTTCACGCAGAAAGCGTCTAG-3′ |
| P/H710-d70 | P/H710-d17 | ID70 | 5′-GACTGAATTCGAAAGCGTCTAGCCATGGCG-3′ |
| P/H710-d79 | P/H710-d17 | ID79 | 5′-GACTGAATTCTAGCCATGGCGTTAGTATGAGTG-3′ |
| P/H710-d118 | P/H710-d17 | ID118 | 5′-GACTGAATTCACCCCCCCTCCCGGGAGAGCC-3′ |
| P/H710-d40-44–45 | P/H710-d40 | d40 44–45 | 5′-ACTTAGGAATTCTCCggTGTGAGGAACTACTGTC-3′ |
| P/H710-d40-49–51 | P/H710-d40 | d40 49–51 | 5′-ACTTAGGAATTCTCCCCTGTCTCGAACTACTGTCTTC-3′ |
| P/H710-d40-49–51/63–64 | P/H710-d40-49–51 | 63–64 | 5′-TTAGgaattcTCCCCTGTctcGAACTACTGTCgaCACGCAGAAAGCGTCTAGCC-3′ |
| P/H710-d40-49–51/63–65 | P/H710-d40-49–51 | 63–65 | 5′-CTTAgaattcTCCCCTGTctcGAACTACTGTCgagACGCAGAAAGCGTCTAGCC-3′ |
| P/H710-d40-56–57 | P/H710-d40 | 56–57 | 5′-CTTAggaattcTCCCCTGTGAGGAACgtCTGTCTTCACGCAGAAAGCG-3′ |
| P/H710-d40-58–59 | P/H710-d40 | 58–59 | 5′-CTTAggaattcTCCCCTGTGAGGAACTAgaGTCTTCACGCAGAAAGCG-3′ |
TABLE 2.
Oligonucleotides and templates used for mutagenesis
| Construct | Template | Oligonucleotide | Sequence |
|---|---|---|---|
| P/H710-d40-49–51/112–4 | P/H710-d40-49–51 | 112–4(+) | 5′-GTATGAGTGTCGTGCAGCgagCAGGACCCCCCCTCCC-3′ |
| P/H710-d40-49–51 | 112–4(−) | 5′-GGGAGGGGGGGTCCTGctcGCTGCACGACACTCATAC-3′ | |
| P/H710-d40-44–45/ 117–8 | P/H710-d40-44–45 | 117–118(+) | 5′-GTGCAGCCTCCACCACCCCCCCTCCCG-3′ |
| P/H710-d40-44–45 | 117–118(−) | 5′-CGGGAGGGGGGGTGGTGGAGGCTGCAC-3′ | |
| P/H710-d40-56–57/ 116–7 | P/H710-d40-56–57 | 116–117(+) | 5′-GCGTTAGTATGAGTGTCGatCAGCCTCCAGGACCC-3′ |
| P/H710-d40-56–57 | 116–117(−) | 5′-GGGTCCTGGAGGCTGatCGACACTCATACTAACGC-3′ | |
| P/H710-d40-58–59/ 109–110 | P/H710-d40-58–59 | 109–110(+) | 5′-CGTTAGTATGAGTGTCGTGCtcCCTCCAGGACCCCCCC-3′ |
| P/H710-d40-58–59 | 109–110(−) | 5′-GGGGGGGTCCTGGAGGgaGCACGACACTCATACTAACG-3′ |
Construction of dicistronic HRPF-Luc.
Oligonucleotides 5′-R-Luc-Sac I and 3′-R-Luc-Sac I (Table 3) were used to amplify the R-Luc gene from plasmid (Gtx 133-141)10(SI)9β/RPh (7). The PCR products were gel purified, digested with SacI, and ligated into SacI-linearized P/H710-d40. The construct 710-d40-R-Luc, with the correct orientation of the R-Luc gene, was selected by restriction analysis and verified by sequencing. Then a SalI site was introduced into PV-Luc (25) by PCR mutagenesis. Plasmid pT7PVM was used as the PCR template. Oligonucleotides PV1–30 and PV-SalI(−) were used in PCR A; oligonucleotides PV-SalI(+) and PVVP4 were used in PCR B. Gel-isolated PCR fragments from both PCR A and PCR B were used in PCR C with oligonucleotides PV1–30 and PVVP4 to produced the PCR fragment with the designed mutation. The PCR fragment was digested with AgeI and PmlI and cloned into PV-Luc-dAP, which was derived from PV-Luc by AgeI and PmlI digestion to release the small fragment. The resulting construct, PV-Luc-SalI, was selected by restriction analysis and verified by sequencing. To construct HRPF-Luc, oligonucleotides HCV-5NTR-SalI(+) and R-Luc-SalI(−) were used in a PCR to amplify the HCV 5′NTR, truncated core, and R-Luc gene from 710-d40-R-Luc, and then the PCR fragment was digested with SalI and cloned into SalI-linearized PV-Luc-Sal. HRPF-Luc was tested by transfecting HeLa cells and measuring the dual luciferases activities. HRPF-44–45, HRPF-44–45/117–8, HRPF-49–51, HRPF-49–51/112–4, HRPF-49–51/63–4, and HRPF-49–51/63–5 were constructed by replacing the PmlI-NheI fragment of HRPF-Luc with the corresponding fragment from P/H710-d40-44–45, P/H710-d40-44–45/117–8, P/H710-d40-49–51, P/H710-d40-49–51/112–4, P/H710-d40-49–51/63–4, and P/H710-d40-49–51/63–5, respectively.
TABLE 3.
Oligonucleotides used for the construction of HRPF-Luc
| Oligonucleotide | Sequence |
|---|---|
| 5′-R-Luc-SacI(+) | 5′-CCC ggg ctc aaa ggt ctc aca aca tat GGT ACT TCG AAA GTT TAT GAT CCA GAA CAA AGG-3′ |
| 3′-R-Luc-SacI(−) | 5′-CCC gag ctc TTA TTG TTC ATT TTT GAG AAC TCG CTC AAC GAA C-3′ |
| PV-SalI(+) | 5′-CCT TCC CGT AAC TTA gtc GAC GCA CAA AAC CAA G-3′ |
| PV-SalI(−) | 5′-CTT GGT TTT GTG CGT Cga cTA AGT TAC GGG AAG G-3′ |
| HCV-5NTR-SalI(+) | 5′-ACGC gtc gac TCC CCT GTG AGG AAC TAC TGT CTT CAC G-3′ |
| R-Luc-SalI(−) | 5′-ACGC gtc gac TTA TTG TTC ATT TTT GAG AAC TCG CTC AAC GAA C-3′ |
| HRPF-Luc-dSacI(+) | 5′-GGA AAG TCC AAA TTG Gaa CTC ACC ACA TAT GG-3′ |
| HRPF-Luc-dSacI(−) | 5′-CCA TAT GTG GTG Agt TCC AAT TTG GAC TTT CC-3′ |
HRPF-Luc-dSacI was generated by Quickmutagenesis (Stratagene) using oligonucleotides HRPF-Luc-dSacI(+) and HRPF-Luc-dSacI(−). By comparison with HRPF-Luc, deletion of the SacI site did not change the amino acid sequences and did not affect the dual luciferases activities (data not shown).
HRPF-56–57, HRPF-56–57/106–7, HRPF-58–59, and HRPF-58–59/109–110 were created by replacing the PmlI-SacI fragment of HRPF-Luc-dSacI with the corresponding fragment from P/H710-d40-56–57, P/H710-d40-56–57/106–7, P/H710-d40-58–59, and P/H710-d40-58–59/109–110, respectively.
Methods of molecular cloning and PCR mutagenesis.
Escherichia coli strain DH5α was used for plasmid transformation and propagation. PCR mutagenesis was performed by standard procedures (40). DNA cloning was done by the standard procedures. DNA fragments were ligated by use of a rapid-ligation kit (Roche Biochemicals).
Transcription, transfection, and translation.
For the production of infectious RNA transcripts in vitro, 1.0 μg of full-length cDNA of PV1(M) or P/H chimeric constructs was linearized at a unique restriction PvuI site downstream of the viral genome. RNAs were synthesized from linearized cDNA by T7 RNA polymerase in an in vitro system described previously (49). HeLa R19 monolayers maintained in 35-mm dishes were transfected by the DEAE-dextran method as described previously (26) and grown at 37°C for up to 5 days in 2 ml of DMEM containing 2% bovine calf serum (BCS). The virus yield from transfections was subjected to titer determination by plaque assay as described previously (26). Translation in vitro was performed at 30°C for up to 16 h or at 34°C for up to 7 h in a HeLa cell extract (30) supplemented with either PV1(M) transcript RNA or chimeric transcript RNAs. To label viral polypeptides in vivo, 35-mm dishes of HeLa cells were infected with lysates derived from cells transfected with PV1(M) or P/H transcript RNAs as described previously (61). In vitro- or in vivo-labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5 or 13.5% polyacrylamide) and newly synthesized proteins were visualized by autoradiography.
Characterization of viral growth phenotype.
The plaque phenotypes of wild-type PV and the P/H chimeras were measured by plaque assay on 35-mm dishes containing HeLa R19 cell monolayers. After incubation for 48 h at 37°C or longer, viral plaques were visualized by 1% crystal violet staining. One-step growth kinetic experiments were performed as described previously (61). Basically, aliquots of synchronously infected cells were harvested at the time points shown and the virus yield was determined by plaque assays.
Luciferase assays.
After transfection, HeLa cells were incubated at 37°C in DMEM medium supplemented with 10% BCS. After 12 h, the medium was removed from the 35-mm plate and the cells were washed gently with 2 ml of phosphate-buffered saline. The cells were incubated with 300 μl of passive lysis buffer (Promega). The culture plates were rocked at room temperature for 15 min before the lysate was transferred to a tube. A 50-μl volume of luciferase assay reagent II plus 10 μl of cell lysate were mixed, and the firefly luciferase activity was measured. After addition of 50 μl of Stop & Glow reagent, the R-Luc activity was measured.
RESULTS
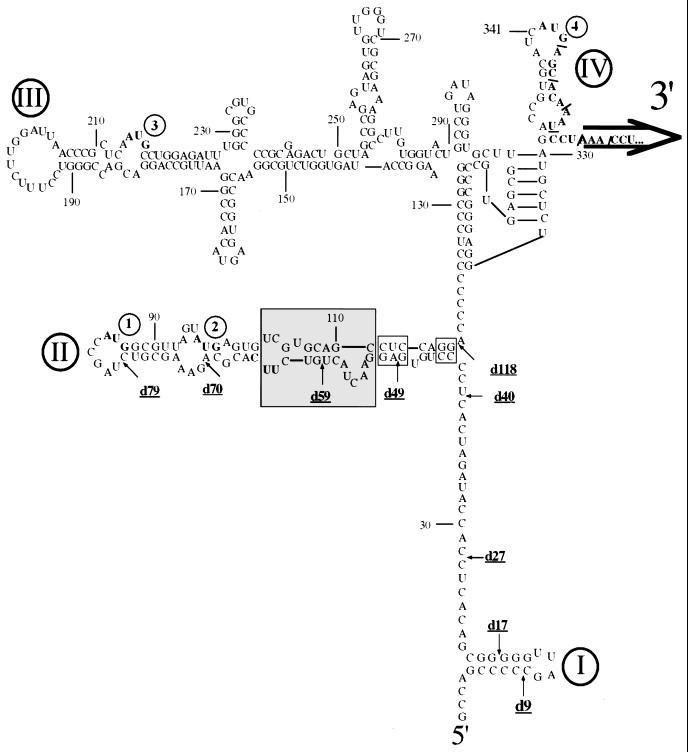
Depending on the strain, the 5′NTR of HCV is about 340 nt long. A variety of different experimental approaches have led to a putative higher-order structure that has been divided into four domains (Fig. 2) (6, 16, 17, 22–24). In contrast to picornavirus IRESs, the HCV IRES includes coding sequences of the HCV core protein (27, 38) although the minimal length of this core-specific sequence necessary for replication of HCV is not known. The HCV core protein itself or fragments thereof, however, are not required for the HCV IRES function (51, 61).
FIG. 2.
Proposed secondary structure of the HCV IRES (genotype 1b) based on evidence published previously and on data presented in this paper. Structural domains in the 5′NTR of the viral RNA are indicated by Roman numerals, and noninitiating AUGs are highlighted by bold type. Nucleotides involved in the genetic study presented here are boxed. The shaded area indicates a new folding pattern.
We have analyzed the relationship between the structure and function of the HCV IRES by two strategies. First, we have made use of the chimeric virus P/H710-d17, whose genome consists of the PV-specific 5′-terminal cloverleaf followed by HCV-specific sequences (nt 18 through 710) linked to the PV genomic sequence (Fig. 1A) (61). Specifically, a fragment (123 amino acids) of the open reading frame of the HCV core (Δcore) downstream of the HCV IRES is fused in frame to the open reading frame of the PV capsid precursor P1. During replication, the Δcore polypeptide is severed from the polyprotein by PV proteinase 2Apro. Except for the 693 nt of HCV replacing the PV IRES, the chimeric genome is built entirely from the PV1(M) sequence (61).
Second, we have constructed a novel dicistronic PV replicon harboring two reporter genes, R-Luc and F-Luc, whose expression is controlled by the IRESs of HCV and PV, respectively. The gene organization of this replicon is [PV]cloverleaf-[HCV]IRES-Δcore-R-Luc-[PV]IRES-F-Luc-P2,3-3′NTR (Fig. 1B). Assays for R-Luc and F-Luc activities in replicon-transfected cells have allowed us to assess independently the HCV and PV IRES functions in the context of a replicating genome.
Effect of 5′ deletions of the HCV IRES within P/H710-d17 on viral proliferation.
Deletion experiments have defined the 5′ border of the HCV IRES to map between nt 28 and 69 (18), between nt 28 and 45 (22) or to nt 40 (38). In contrast, Fukushi et al. (8) suggested that domain I (nt 5 to 20 [Fig. 2]) is also important for IRES function, whereas other groups reported that this structure was not only dispensable (27, 61) but also inhibitory in translation experiments (18, 39, 59). In one report, it was proposed that even the entire domain II (nt 44 to 118, [Fig. 2]) was dispensable (48). All these experiments were based on translations of mono- or dicistronic constructs either in vitro (using rabbit reticulocyte lysate) or in cultured cells transfected with expression plasmids. To test the effect of deletions in the context of the replicating viral chimera P/H710-d17, we have generated 5′ deletions within the HCV-specific sequence, designated d9, d17, d27, d40, d49, d59, d70, d79, and d118 (Fig. 2), and studied the phenotypes of the corresponding RNAs by replication in HeLa cells.
Briefly, the transcript RNAs harboring the deletions were transfected into HeLa cell monolayers as described in Materials and Methods. After evidence of cytopathic effects (CPE) was noted, the virus yield from each transfection was assayed in separate HeLa cell plaque assays. As shown in Fig. 3A, any of the deletions beyond nt 40 (d49, d59, d70, d79, and d118) conferred a lethal phenotype to viral proliferation. In these experiments, replication was assayed by two different methods. First, the transfected HeLa monolayers were inspected for CPE at various times posttransfection (p.t.), an assay indicative of intracellular replication (protein and RNA synthesis). Wild-type PV1(M) RNA induces CPE at 18 h p.t., a result also observed with P/H710-d40 RNA. RNAs of P/H710-d9, P/H710-d17, and P/H710-d27 yielded CPE only at about 48 h p.t. No CPE was observed with any of the other deletion mutant RNAs. Second, the cell lysates from the transfected monolayers were assayed for virus yield in a plaque assay. Because of significant differences in the appearance of CPE, the titers p.t. were determined 2 days [PV1(M) and P/H710-d40] or 3 days [P/H710-d9, P/H710-d17, and P/H710-d27] p.t. Only P/H710-d40 RNA produced virus yields and plaque sizes comparable to those of PV1(M); all other constructs expressed minute (P/H710-d9), small (P/H710-d17), or medium (P/H710-d27) plaque sizes (Fig. 3A). The higher yield with P/H710-d40 relative to d9, d17, and d27 was expected since we have reported recently that the HCV sequence up to nt 31 can engage in base pairing with the PV cloverleaf, resulting in impaired viral replication (60). Transfections with RNAs of P/H710-d49, P/H710-d59, P/H710-d70, P/H710-d79, and P/H710-d118 and assays for progeny virus after prolonged incubation were carried out several times, but no virus was found in any of these experiments. Since no CPE was found after transfection with any of these constructs, we suggest that these RNAs were unable to produce sufficient quantities of viral proteins to effect cell killing.
FIG. 3.
Characterization of 5′NTR deletion mutants of chimeric virus P/H710-d17. (A) Plaque phenotypes and yield of wt PV and of 5′ NTR deletion mutants of chimera P/H710-d17 after transfection of HeLa R 19 cell monolayers with transcript RNAs (see Materials and Methods). (B) One-step growth kinetics of PV, P/H710-d17, and P/H710-d40 on HeLa R19 cell monolayers.
We conclude that the 5′ boundary for efficient replication of the P/H chimeric viruses carrying the 5′NTR of HCV, genotype 1b, maps to approximately nt 40. This coincides with the currently accepted 5′ boundary of the HCV IRES obtained by studies with artificial dicistronic mRNAs (reviewed in reference 37). We feel confident, therefore, that the results reported are also a measure of the 5′ boundary of the HCV IRES.
For the genetic analyses described below, we have selected the chimera P/H710-d40 owing to its favorably high yield of virus after transfection. Indeed, a one-step growth experiment with HeLa cell monolayers revealed that the replication kinetics of this chimera was very similar to that of PV1(M) with the notable exception of a prolongation of the eclipse period (Fig. 3B). The extended eclipse period is also apparent with chimera P/H710-d17 (Fig. 3B), as reported previously (61). We assume that this phenomenon is related to a delay in uncoating and release of genomic RNA, but the precise mechanism remains to be determined.
It should be noted that the entire Δcore coding sequence of 369 nt was found to be retained in P/H710-d40 over six serial passages (data not shown). This conforms to previous results with P/H710-d17 (61). Thus, there was no apparent pressure on the part of the P/H710-d40 genome to delete any of the “foreign” core sequence, thereby generating variants with greater fitness to outcompete the parental genome. Considering the genetic plasticity of poliovirus genomes allowing for rapid deletion of foreign sequences fused to the P1 coding region of poliovirus (31), this observation can be interpreted to mean that the Δcore protein-encoding RNA sequence in P/H710-d40 is advantageous for efficient replication of P/H710-d40 RNA. Recent experiments have confirmed that the extension of the HCV-specific sequence downstream of the HCV 5′NTR is necessary for efficient HCV IRES function (W. D. Zhao, S. K. Jang, and E. Wimmer, unpublished data).
Genetic study of the stem-loop II structure of the IRES in P/H710-d40. Mutation at the bottom of domain II.
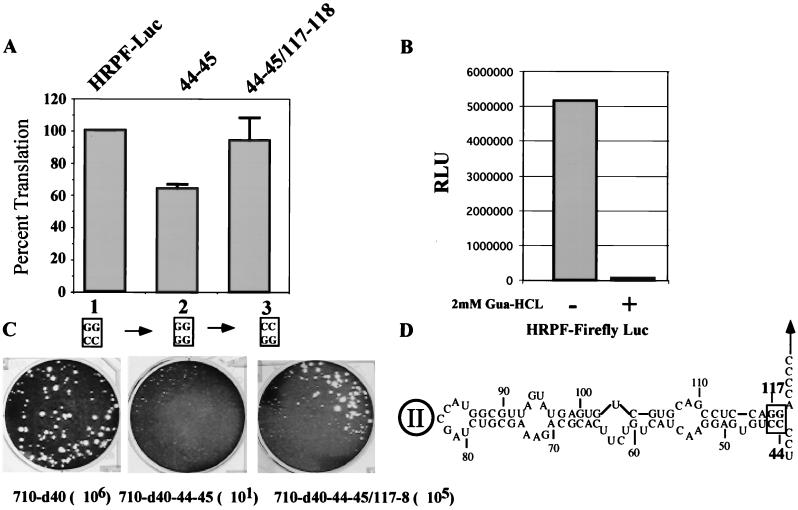
Using chimera P/H710-d40, we have embarked on a genetic analysis of the structure of domain II by introducing mutations disrupting stem regions followed by the appropriate compensatory mutations. We have tested this strategy first by mutating the base pair at the bottom of domain II (CC/GG: nt 44 to 45 pairing with nt 118 to 117 [Fig. 4D]). Remarkably, disrupting these base pairs at the bottom of domain II by changing nt 44 to 45 from CC to GG (construct P/H710-d40-44–45) severely impaired the proliferation of the virus, since the titer produced after transfection was drastically reduced (Fig. 4C). Compensatory mutation of nt 118 to 117 (construct P/H710-d40-44-45/117–118), on the other hand, which serves to restore base pairing at the bottom of domain II, led to the near-complete recovery of viral replication (Fig. 4C; titer, 105 dilution). These data could be interpreted to mean that the short helix at the bottom of domain II is important for the function of the HCV IRES in P/H710-d40 (Fig. 4C). Indeed, this has been concluded by Honda et al. (16), who performed a mutational analysis of this region of the HCV IRES and tested the effects by in vitro translation or expression in transfected cells. On the other hand, it could be argued that the rather small change in IRES structure in P/H710-d40-44–45 impaired genome replication rather than translational efficiency (see below).
FIG. 4.
Genetic analysis of the helix at the base of IRES domain II. The nucleotides changed by site-directed mutagenesis were nt 44 to 45 (CC) and 117 to 118 (GG; see the box at the bottom of domain II in panel D). (A) Mean relative IRES activities of mutants, measured as the ratio of R-Luc to F-Luc. The ratio of R-Luc to F-Luc obtained with HRPF-Luc was accorded a value of 100%. Error bars denote the standard deviation of values obtained in replicate experiments. (B) Detection of F-Luc activity with or without 2 mM guanidine HCl. Guanidine HCl was added to the DMEM medium after transfection. (C) Plaque phenotypes and yields of viruses derived from P/H710-d40, P/H710-d40-44–45, and P/H710-d40-44–45/117–118 transcript RNAs on HeLa R19 cell monolayers. The dilution factors for each construct are included in parentheses (D). Domain II structure of HCV 5′NTR (based reference 16). Nucleotides subjected to mutation analysis are boxed.
The effect of the mutation in the HCV IRES of P/H710-d40-44–45 was then analyzed within the context of a replicon in which replication was not dependent on the HCV function. For this purpose, dicistronic replicons were constructed in which the expression of the first reporter gene (R-Luc) was controlled by the HCV IRES while that of the second reporter gene (F-Luc) was controlled by the PV IRES (Fig. 1B; construct HRPF-Luc). Transfection of RNA harboring the d40-44–45 mutations into HeLa cells was followed by incubation of the cells for 12 h. Both R-Luc and F-Luc activities were then assayed as described in Materials and Methods. A mismatch of base pairs from CC/GG to GG/GG reduced the R-Luc signal relative to the L-Luc signal by 34% compared to the signal ratio in the parental replicon (Fig. 4A; compare lane 1 with lane 2). Restoring base pairing by compensatory mutation (GG/CC) in P/H710-d40-44–45/117–118 fully restored the observed R-Luc activity (lane 3). The detection of luciferase activity required RNA replication because, as shown in Fig. 4B, addition of 2 mM guanidine HCl reduced the signal to background level. 2 mM guanidine HCl is known to completely inhibit PV RNA replication (reference 34 and references therein).
The data obtained with the HRPF-LUC replicon indicate that the bottom of domain II carries a CC/GG base pair that contributes to the function of HCV IRES activity. However, the integrity of the short helix at the bottom of domain II does not appear to be absolutely essential for internal ribosomal entry within the dicistronic RNA, a conclusion drawn also by Honda et al. (16) using a different approach. The mechanism by which the CC/GG–to–GG/GG mutation in P/H710-d40-44-45 influences (inhibits) genome replication, as indicated by the drastic reduction of viral titer, remains to be determined. This issue is discussed below.
Analysis of the structure involving nt 49 to 65.
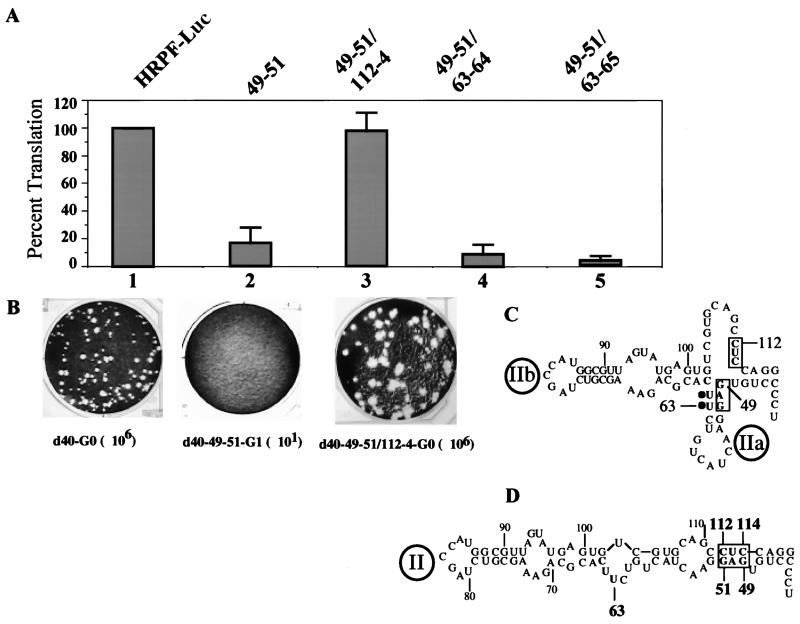
The original model of the HCV IRES predicted that domain II of the HCV IRES contained two subdomains, IIa and IIb (Fig. 5C) (6, 17). More recently, Honda et al. (16) suggested a modified structure (Fig. 5D) on the basis of phylogenetic considerations and mutational analyses. We have subjected this region of domain II to a genetic analysis very similar to that described for the bottom of domain II. Specifically, we have destabilized the putative helix of IIa (Fig. 5C) by changing the nucleotide sequence from GAG (nt 49 to 51) to CUC. Translational efficiency in a HeLa cell extract of variant RNA (d40-49–51) obtained by transcription with T7 RNA polymerase was reduced to 10% of that of the parental genotype, although all viral proteins were observed in the expected ratio (data not shown). On transfection of d40-49–51 RNA into HeLa cell monolayers, no virus was obtained in several independent experiments (Fig. 5B, d40-49–51-G1).
FIG. 5.
Genetic analysis of the structure of domain II. (A) Mean relative IRES activities of mutants, measured as the ratio of R-Luc to F-Luc. The ratio of R-Luc to F-Luc obtained with HRPF-Luc was given a value of 100%. Error bars denote the standard deviation of values obtained in replicate experiments. (B) Plaque phenotypes and yields of viruses derived from P/H710-d40, P/H710-d40-49–51, and P/H710-d40-49–51/112–4 transcript RNAs. The dilution factors for each construct are included in parentheses. (C) Domain II structure based on an older model (6, 17). (D) Structure of domain II based on data from the genetic analysis presented in this paper.
In an attempt to restore the viability of the d40-49–51 variant, we introduced three different (compensatory) mutations, of which two mapped to the complementary strand. Compensatory mutation at nt 63 to 64 (UU to GA [Fig. 5C]) yielded variant RNA d40-49–51/63–64, which was no more efficient in HeLa cell-free translation than was d40-49–51 RNA (data not shown). More importantly, d40-49–51/63–64 did not restore viral proliferation since no virus was retrieved in several different experiments (data not shown). Similarly, mutation of UUC to GAG at nt 63 to 65 also failed to restore proliferation in HeLa cells (data not shown). However, when mutations were introduced at nt 112 to 114 in d40-49–51 RNA from CUC to GAG (Fig. 5C, boxed trinucleotides 112 to 114), the resulting d40-49–51/112–4 RNA replicated efficiently in HeLa cells, yielding virus titers similar to that of d40 RNA (Fig. 5B). These data can be explained only if nucleotides GAGG (nt 49 to 52) of the HCV IRES base pair with nt CCUC (nt 112 to 114), as shown in Fig. 5D.
The mutations were then introduced into the dicistronic replicon HRPF-Luc (Fig. 1B), and their effect on HCV-controlled translation was measured in transfected HeLa cells as described above. Changing the nucleotide sequence from GAG (nt 49 to 51) to CUC reduced the signal of R-Luc relative to that of L-Luc to 25% (Fig. 5A, lane 2). Attempts to restore R-Luc activity by either one of the mutations at nt 63 to 64 or nt 63 to 65 further reduced the R-Luc signal to 14 and 7%, respectively (lanes 4 and 5). However, mutations at nt 112 to 114 (CUC to GAG) fully restored the R-Luc signal (lane 3).
These data can only be interpreted to mean that domain II of the HCV IRES has the structure shown in Fig. 5D, regardless of whether the assay was carried out using the P/H chimeric virus or the dicistronic replicon. Importantly, the strong inhibition and recovery of R-Luc expression by mutations and compensatory mutations, respectively, unambiguously relate HCV IRES structure to HCV IRES function on translation independently of genome replication. Our data support the results published by Honda et al. (16), who employed nonreplicating dicistronic mRNAs whose translational efficiency were assayed either in vitro or after transfection of corresponding plasmids into Huh-7 cells that were superinfected with a vaccinia virus vector expressing phage T7 RNA polymerase.
Evidence for a new structure within domain II of the HCV IRES.
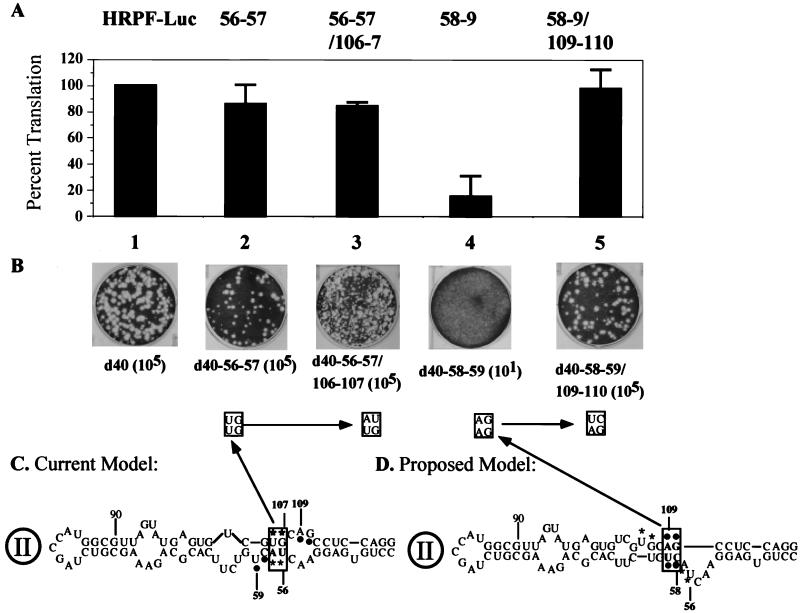
The data obtained so far suggest that our strategy to analyze the structure and function of domain II is correct. It has been suggested by Honda et al. (16) that nt 56 to 58 and nt 105 to 107 form a short stem (Fig. 6C). This region, however, can also be folded into an alternative structure (Fig. 6D) by using Zuker's mfold server (29, 62). To determine which of these structures is favored in P/H710-d40 RNA, the CU dinucleotide at nt 58 to 59 was changed to GA (Fig. 6C). Unexpectedly, these mutations (chimera d40-58–59) resulted in a lethal phenotype, an observation suggesting an important role for the CU dinucleotide (nt 58 to 59). In contrast, mutation of the UA at nt 56 to 57 (Fig. 6C) to GU, a change that should have interrupted the putative stem formed between 5′UA and 5′UG (boxed pair in Fig. 6C) did not have any significant effect on the virus yield (Fig. 6B). When 5′GU/5′UG (nt 106 to 107) was changed to 5′GU/5′AU, the resulting virus, d40-56–57/106–7 was viable, expressing a small plaque phenotype (Fig. 6B). These observations suggested to us that nt 106 to 107 are unlikely to base pair with nt 56 to 57.
FIG. 6.
Analysis of two possible structures for domain II of the HCV IRES. (A) Mean relative IRES activies of mutants, measured as the ratio of R-Luc to F-Luc. The ratio of R-Luc to F-Luc obtained with HRPF-Luc was given a value of 100%. Error bars denote the standard deviation of values obtained in replicate experiments. (B) Plaque phenotypes and yields of viruses derived from d40-56–57, d40-56–57/106–107, d40-58–59, and d40-58–59/109–110 transcript RNAs on HeLa R19 cell monolayers. The dilution factors for each construct are included in parentheses. (C) Structure proposed by Honda et al. (16) and supported by data described here. Nucleotides involved in this analysis are boxed. (D) Proposed domain II structure, by subjecting the sequence to computer folding (29) and to the genetic analysis presented here.
When mutations were introduced into the nonviable construct d40-58–59 at nt 109 to 110 (AG to UC) (Fig. 6C) to yield d40-58–59/109–110, the RNA replicated highly efficiently in HeLa cells, yielding virus titers similar to that of d40 RNA. These genetic data strongly favor the structure of domain II shown in Fig. 6D.
These mutations were then engineered into the dicistronic replicon. The mutations at nt 56 to 57 (d40-56–57) had only a small effect on the translational efficiency of the HCV IRES (Fig. 6A, lane 2). The mutations at nt 56 to 57 and 106 to 107 reduced the expression of R-Luc further, indicating some detrimental effect on structure (lane 3). However, mutations at nt 58 to 59 profoundly inhibited expression of R-Luc (lane 4), an effect that could be reversed by the compensatory mutations at nt 109 to 110 (lane 5). These data are compatible only with the structure in this region of domain II as depicted in Fig. 6D. It should be noted that this model is also supported by limited RNase T1 digestion studies recently published by Kieft et al. (23), who failed to find cleavage at G110 as would be predicted if this residue resided in the internal loop of the published models (Fig. 6C). They are also supported by phylogenetic considerations (see Discussion).
DISCUSSION
Viral IRES elements arguably belong to the most complex cis-acting signals in any RNA viral genome. Their huge size appears to contain much more information than seems necessary to promote internal ribosomal entry, particularly if one compares the IRES with the Shine-Dalgarno signal (42) operating in multicistronic mRNAs of prokaryotic cells (55). Elucidation of IRES structure and function is therefore a major challenge.
The structure of the HCV IRES has been probed by various methods (reviewed in reference 24), to which we have now added viral genetics and assays in the context of a P/H replicon. Genetic analysis of the HCV IRES within the context of its cognate viral genome is not feasible at present because of the poor replication properties of HCV. However, the exchange of the PV IRES with the HCV IRES has yielded a rapidly growing variant, P/H710-d40, that has allowed us to assess the effect of mutations in the HCV IRES-specific segment of genomic RNA on viral proliferation. Based on the data, a firm prediction can be made about the IRES structure necessary for translation. The question has been raised, however, whether any effect of altering the HCV IRES can be solely equated with internal ribosomal entry and control of translation.
Previously, it has been suggested that viral IRESs serve as cis-acting elements not only in translation but also in RNA synthesis (reviewed in references 5 and 58). If so, changes introduced into the viral IRES may affect viral RNA synthesis in addition to or independently of the control of protein synthesis. In some cases, such changes may be apparent only as host range phenotypes (19, 43). On the other hand, exchanges of the PV IRES with those of different picornaviruses, specifically encephalomyocarditis virus, human rhinoviruses 2 and 14, coxsackievirus A9, coxsackievirus A24v, and coxsackievirus B4 (references 13 and references therein), would have been expected to yield replication phenotypes. If assayed in HeLa cells under standard conditions, however, these chimeric viruses did not yield growth phenotypes. As shown here, the P/H chimera P/H710-d40 carrying an IRES quite different in structure from that of the cognate PV IRES expressed replication kinetics mimicking that of PV1(M).
If IRES elements function in cis in RNA synthesis, the PV replication proteins (replication machinery) in the IRES chimera must be able to recognize cis-acting signals inherent to all the different IRES elements tested. This is difficult to comprehend in view of the apparent difference in IRES structure, a consideration particularly striking when the HCV IRES is compared with that of PV. Why should the HCV IRES carry a replication signal recognized by the replication machinery of a virus belonging to a different family? On the other hand, a single cellular factor, X, capable of binding all viral IRESs could be recognized by the PV replication machinery, thereby mediating the identification of a “generic” cis-acting signal. It is known that viral IRES elements can form complexes with several different cellular RNA binding proteins, such as polypyrimidine binding protein (PTB), polycytidylic acid binding protein (PCBP), unr (upstream of N-ras), or La auto antigen (reviewed in reference 9). Of these proteins, a dual role in translation and in replication has been suggested only for PCBP. PCBP has affinity to the 5′-terminal cloverleaf of PV, a cis-acting signal in replication, as well as to the cognate IRES (10, 32). We have argued previously, however, that PCBP is not necessarily linked to PV RNA replication since PCBP can be replaced in all in vitro binding experiments to the cloverleaf by viral protein 3AB (15, 56, 57). Nevertheless, PCBP2 also interacts with the IRES of EMCV, even though this interaction is not required for EMCV IRES-dependent translation (50). Similarly, PCBP binds to the HCV IRES (45). Could PCBP2 be a hypothetical “host factor X” playing a dual role in IRES-dependent translation and replication of all IRES-containing picornaviruses? Until the mechanism by which cellular RNA binding proteins may function in picornavirus and HCV replication has been elucidated, a putative role of polypeptides such as PCBP, PTB, or La as universal “host factors” in viral RNA synthesis will remain elusive.
To circumvent the problem of the putative dual IRES function in genome replication, we have constructed chimeric dicistronic replicons (1, 2) carrying two luciferase genes. The first luciferase gene (R-Luc) was controlled by the HCV IRES, and the second (F-Luc) was controlled by the PV IRES. After transfection into HeLa cells, a significant luciferase signal was detectable only if the RNA replicated, as shown by the inhibitory effect of 2 mM guanidine HCl. Mutations studied in P/H710-d40 were then constructed into the replicon, and the effect on either of the luciferase genes was tested. The results of the experiments unambiguously showed that some of the mutations introduced into domain II of the HCV IRES produced a negative effect on translation of its reporter gene while being ignored by the PV replication machinery. Thus, the results obtained with the replicons corroborated the results obtained with P/H710-d40.
The remarkable sensitivity of P/H710-d40 replication to the mutation at nt 44 to 45, however, remains a mystery, since the data obtained with the replicon carrying the same mutations showed only a 34% reduction of translational activity of the HCV IRES (Fig. 4). In this case, we consider it possible that these mutations may have a negative effect not only on translation but also on RNA synthesis, although the mechanism is unknown. However, both analyses employing either P/H710-d40 or the replicon support previous data suggesting the importance of the CC/GG base pair at the bottom of domain II (reference 16 and references therein).
Similarly, the analysis of base pairing of nt 49 to 51 has led to an unambiguous designation of the structure shown in Fig. 5D. Here, the low translational activities of the R-Luc signal observed with mutated replicons, combined with the effect of compensatory mutations, also strongly support the structure of domain II proposed by Honda et al. (16). We suggest that the inability of these P/H710-d40 variants to replicate is due to insufficient amounts of viral proteins that were synthesized under the control of the mutated HCV IRES.
Following the same strategy, we have analyzed the possibility of a structural arrangement in domain II that differs from that in previous reports. Mutations of nt 58 to 59 and compensatory mutations unambiguously support the domain II structure shown in Fig. 6D, regardless of whether the analysis was carried out with the replicon or with P/H710-d40.
The new structure (shaded area in Fig. 2) is also supported by phylogenetic considerations. Of 124 sequences analyzed (http://s2as02.genes.nig.ac.jp), 83 folded as proposed in Fig. 2. In 16 sequences, G107 was changed to A107, yielding a preferred UA base pair in this position; in 5 cases, the C58/G110 base pair was changed to an U58/A110 base pair. Neither of these changes had an apparent effect on the overall structure. Of interest are three published sequences in which an A57 has been replaced by a U57. This change has no apparent effect on the structure proposed here but would disrupt a short (3-bp) helix in the previously published structure (Fig. 6C).
The HCV IRES can form complexes with PTB (3), La (4), hnRNPL (14), eIF3 (44), and PCBP (45). To what extent the binding of these proteins, either alone or in complex with each other, influences IRES function in vivo is unknown. It is likely that disruptions of the domain II structure interfere with protein binding, thereby influencing IRES function.
ACKNOWLEDGMENTS
We are indebted to Sung Key Jang and Ann Jacobson for valuable discussions and suggestions. We thank A. Nomoto for his gift of HCV cDNA subclones of HCV. The critical reading of the manuscript by A. Paul is highly appreciated. We thank F. Maggiore for excellent technical assistance.
This work was supported in part by NIH grants NIAID 2R01AI151 and 5R01AI321.
REFERENCES
- 1.Alexander L, Lu H H, Gromeier M, Wimmer E. Dicistronic polioviruses as expression vectors for foreign genes. AIDS Res Hum Retroviruses. 1994;10:S57–S60. [PubMed] [Google Scholar]
- 2.Alexander L, Lu H H, Wimmer E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman A M, Deliat F G, Kean K M. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown E A, Zhang H, Ping L H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chappell S A, Edelman G M, Mauro V P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 9.Gale M, Jr, Tan S L, Katze M G. Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamarnik A V, Andino R. Interactions of viral protein 3CD and Poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gesteland R F, Atkins J F. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 12.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci USA. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gromeier M, Mueller S, Solecki D, Bossert B, Bernhardt G, Wimmer E. Determinants of poliovirus neurovirulence. J Neurovirol. 1997;3(Suppl. 1):S35–S38. [PubMed] [Google Scholar]
- 14.Hahm B, Cho O H, Kim J E, Kim Y K, Kim J H, Oh Y L, Jang S K. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- 15.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 16.Honda M, Beard M R, Ping L H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 18.Honda M, Ping L H, Rijnbrand R C, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 19.Ishii T, Shiroki K, Iwai A, Nomoto A. Identification of a new element for RNA replication within the internal ribosome entry site of poliovirus RNA. J Gen Virol. 1999;80:917–920. doi: 10.1099/0022-1317-80-4-917. [DOI] [PubMed] [Google Scholar]
- 20.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamoshita N, Tsukiyama-Kohara K, Kohara M, Nomoto A. Genetic analysis of internal ribosomal entry site on hepatitis C virus RNA: implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology. 1997;233:9–18. doi: 10.1006/viro.1997.8600. [DOI] [PubMed] [Google Scholar]
- 23.Kieft J S, Zhou K, Jubin R, Murray M G, Lau J Y, Doudna J A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 24.Lemon S, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 25.Li X, Lu H-H, Mueller S, Wimmer E. The C-terminal residues of poliovirus proteinase 2Apro are critical for viral RNA replication but not for cis- or trans-proteoloytic cleavages. J Gen Virol. 2001;82:397–408. doi: 10.1099/0022-1317-82-2-397. [DOI] [PubMed] [Google Scholar]
- 26.Lu H H, Alexander L, Wimmer E. Construction and genetic analysis of dicistronic polioviruses containing open reading frames for epitopes of human immunodeficiency virus type 1 gp120. J Virol. 1995;69:4797–806. doi: 10.1128/jvi.69.8.4797-4806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu H H, Yang C F, Murdin A D, Klein M H, Harber J J, Kew O M, Wimmer E. Mouse neurovirulence determinants of poliovirus type 1 strain LS-a map to the coding regions of capsid protein VP1 and proteinase 2Apro. J Virol. 1994;68:7507–7515. doi: 10.1128/jvi.68.11.7507-7515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves predict of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 30.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 31.Mueller S, Wimmer E. Expression of foreign proteins by poliovirus polyprotein fusion: analysis of genetic stability reveals rapid deletions and formation of cardioviruslike open reading frames. J Virol. 1998;72:20–31. doi: 10.1128/jvi.72.1.20-31.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 34.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 35.Pilipenko E V, Blinov V M, Romanova L I, Sinyakov A N, Maslova S V, Agol V I. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- 36.Poole T L, Wang C, Popp R A, Potgieter L N, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 37.Reed K E, Rice C M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–26. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 43.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sizova D V, Kolupaeva V G, Pestova T V, Shatsky I N, Hellen C U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spangberg K, Schwartz S. Poly(C)-binding protein interacts with the hepatitis C virus 5′ untranslated region. J Gen Virol. 1999;80:1371–1376. doi: 10.1099/0022-1317-80-6-1371. [DOI] [PubMed] [Google Scholar]
- 46.Stewart S R, Semler B L. RNA determinants of picornavirus cap-independent translation initiaon. Semin Virol. 1997;8:242–254. [Google Scholar]
- 47.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urabe M, Hasumi Y, Ogasawara Y, Matsushita T, Kamoshita N, Nomoto A, Colosi P, Kurtzman G J, Tobita K, Ozawa K. A novel dicistronic AAV vector using a short IRES segment derived from hepatitis C virus genome. Gene. 1997;200:157–162. doi: 10.1016/s0378-1119(97)00412-5. [DOI] [PubMed] [Google Scholar]
- 49.van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter B L, Nguyen J H, Ehrenfeld E, Semler B L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T H, Rijnbrand R C, Lemon S M. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J Virol. 2000;74:11347–11358. doi: 10.1128/jvi.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wimmer E. In Recognition signals on RNA genomes. Introduction. Semin Virol. 1997;8:151–152. [Google Scholar]
- 53.Wimmer E, Hellen C U, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 54.Witherell G, Wimmer E. Cap-independent translation of picornavirus mRNAs. In: Doerfler P B W, editor. Virus strategies. Weinheim, Germany: Verlag Chemie; 1993. pp. 237–247. [Google Scholar]
- 55.Witherell G W, Gil A, Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- 56.Xiang W, Cuconati A, Paul A V, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 57.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang W, Paul A, Wimmer E. RNA Signals in entero- and rhinovirus genome replication. Semin Virol. 1997;8:256–273. [Google Scholar]
- 59.Yoo B J, Spaete R R, Geballe A P, Selby M, Houghton M, Han J H. 5′ end-dependent translation initiation of hepatitis C viral RNA and the presence of putative positive and negative translational control elements within the 5′ untranslated region. Virology. 1992;191:889–899. doi: 10.1016/0042-6822(92)90264-p. [DOI] [PubMed] [Google Scholar]
- 60.Zhao W D, Lahser F C, Wimmer E. Genetic analysis of a poliovirus/hepatitis C virus (HCV) chimera: interaction between the poliovirus cloverleaf and a sequence in the HCV 5′ nontranslated region results in a replication phenotype. J Virol. 2000;74:6223–6226. doi: 10.1128/jvi.74.13.6223-6226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao W D, Wimmer E, Lahser F C. Poliovirus/hepatitis C virus (internal ribosomal entry site-core) chimeric viruses: improved growth properties through modification of a proteolytic cleavage site and requirement for core RNA sequences but not for core-related polypeptides. J Virol. 1999;73:1546–1554. doi: 10.1128/jvi.73.2.1546-1554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure predictioń: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Hingham, Mass: NATO ASI Series, Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]