Abstract
A better understanding of the host and viral factors associated with human immunodeficiency virus (HIV) transmission is essential to developing effective strategies to curb the global HIV epidemic. Here we used the rhesus macaque-simian immunodeficiency virus (SIV) animal model of HIV infection to study the range of viral genotypes that are transmitted by different routes of inoculation and by different types of viral inocula. Analysis of transmitted variants was undertaken in outbred rhesus macaques inoculated intravenously (IV) or intravaginally (IVAG) with a genetically heterogeneous SIVmac251 stock derived from a well-characterized rhesus macaque viral isolate. In addition, we performed serial IV and IVAG passage experiments using plasma from SIV-infected macaques as the inoculum. We analyzed the V1-V2 region of the SIV envelope gene from virion-associated RNA in plasma from infected animals by the heteroduplex mobility assay (HMA) and by DNA sequence analysis. We found that a more diverse population of SIV genetic variants was present in the earliest virus-positive plasma samples from all five IV SIVmac251-inoculated monkeys and from two of five IVAG SIVmac251-inoculated monkeys. In contrast, we found a relatively homogeneous population of SIV envelope variants in three of five monkeys inoculated IVAG with SIVmac251 stock and in two monkeys infected after IVAG inoculation with plasma from an SIV-infected animal. In some IVAG-inoculated animals, the transmitted SIV variant was the most common variant in the inoculum. However, a specific viral variant in the SIVmac251 stock was not consistently transmitted by IVAG inoculation. Thus, it is likely that host factors or stochastic processes determine the specific viral variants that infect an animal after IVAG SIV exposure. In addition, our results clearly demonstrate that the route of inoculation is associated with the extent and breadth of the genetic complexity of the viral variant population in the earliest stages of systemic infection.
Human immunodeficiency virus (HIV) transmission by sexual, intravenous (IV), and perinatal routes is often associated with the acquisition of a limited distribution of genetic variants (45, 48–50). In many instances, the proviral DNA sequence of the transmitted variant represents a minor variant in the donor's virus population (45, 49, 50). Given the extent of genetic diversity between HIV isolates, these findings have been interpreted to suggest that HIV transmission may involve selective entry or selective amplification of specific viral variants (45, 48, 49). The inherent limitations of all studies using human samples include small sample sizes and uncertainty as to the genetic identity and the extent of genetic diversity of the virus population in the donor at the time of transmission. Thus, the mechanisms that underlie the sexual transmission of HIV variants are unclear, and they are difficult to assess because of the difficulties in establishing the precise time of infection and in obtaining samples before immune pressures affect viral variant populations (12).
The simian immunodeficiency virus (SIV) macaque model for intravaginal (IVAG) HIV transmission (30, 31) is particularly valuable for evaluating the role of viral selection during sexual transmission. The model allows access to information that is usually unattainable in human studies, such as the genotypic and phenotypic properties of the infecting virus, knowledge of the exact time of virus exposure, and the characteristics of viral variants in the infected host immediately after transmission. Previous studies of SIV transmission using rhesus macaques inoculated by the IV and IVAG routes have shown that the genetic complexity of the SIV populations detected in peripheral blood is associated with the method of challenge; greater diversity accompanied IV inoculation of SIV (10, 11, 36, 42). Some have concluded that these results mean that selective SIV transmission occurs after IVAG inoculation (11, 36); however, a specific SIV variant has not been detected in multiple individual outbred macaques after IVAG inoculation with a genetically diverse virus stock.
Here we extend previous investigations of SIV transmission in several ways. As in earlier SIV transmission studies, we IV- or IVAG-inoculated outbred rhesus macaques with the same genetically heterogeneous SIVmac virus stock and analyzed the extent of V1-V2 envelope diversity in plasma from each animal. In an effort to determine if a particular SIV variant or subset of variants would be consistently transmitted by serial mucosal passage, we also used virus in plasma from SIV-infected macaques to perform serial IV and IVAG inoculations. If viral variants possessing a unique ability to cross the vaginal mucosal surface and establish systemic infection exist in the SIV inoculum, then serial IVAG passage of plasma SIV variants should result in selection for a particular genetic variant(s). Further, it is likely that serial passage will select viral variants with increased pathogenic potential, as has been repeatedly demonstrated by serial parenteral passage of primate lentiviruses (9, 14, 16, 26, 40).
We found that both IV and IVAG inoculation resulted in transmission of genetically diverse populations of SIV V1-V2 variants from the stock inoculum to the animals. Compared to the complex SIV populations in the IV-inoculated animals, four of the five IVAG-inoculated animals were infected with SIV populations that had low genetic diversity in the env gene. However, we failed to identify a specific SIV envelope variant that was preferentially transmitted by either route of inoculation. We did find that the most common SIV V1-V2 variant in the genetically heterogeneous viral stock or plasma inoculum was transmitted to some IVAG-inoculated monkeys. These results contrast with the conclusion that homogeneous (21, 48–50) and rare donor (49, 50) viral variants are transmitted to HIV-infected men. However, our finding that genetically diverse SIV populations were transmitted to some IVAG-inoculated monkeys is consistent with the observation that a more genetically diverse population of viral variants is sexually transmitted from HIV-infected men to women (21, 39). Together, our results support the hypothesis that multiple viral and host factors determine which viral variants establish systemic infection following IVAG exposure to SIV. It is also likely that stochastic processes (e.g., multiple viral variants in equal frequency have equal probability of being involved in relatively rare virus-target cell interactions) play a role in determining the specific viral variants that infect an individual.
MATERIALS AND METHODS
Animal inoculations and virus stock.
All animals used in this study were colony-bred rhesus macaques from the California Regional Primate Research Center. All animals were seronegative for HIV type 2, SIV, type D retrovirus, and simian T-cell lymphotropic virus type 1 at the beginning of the study. The animals were housed in accordance with American Association for Accreditation of Laboratory Animal Care standards. We adhered to the Guide for the Care and Use of Laboratory Animals (1). When necessary, animals were anesthetized with ketamine hydrochloride (10 mg/kg of body weight; Parke-Davis, Morris Plains, N.J.) injected intramuscularly.
Rhesus macaques were inoculated with 1 ml of undiluted SIVmac251-8/95 virus stock containing 105 50% tissue culture infective doses (TCID50) per ml and 4 × 109 copies of RNA per ml. This SIVmac251 stock was produced by short-term expansion of a well-characterized, uncloned parent virus stock (kindly provided by R. Desrosiers [19]) on rhesus peripheral blood mononuclear cells (PBMC). The SIVmac251-8/95 virus stock has been used to successfully infect monkeys by the IVAG route (32). Adult male monkeys were inoculated by the IV route, and adult female monkeys were inoculated by the IVAG route, according to standard procedures (30, 32). For IVAG inoculation studies, multiparous, cycling female rhesus macaques were used. Blood samples were collected into tubes containing EDTA at 1, 2, 4, 6, and 8 weeks postinoculation (p.i.).
Virus isolation.
PBMC were isolated from whole blood by discontinuous Ficoll gradient separation (lymphocyte separation medium; Organon Teknika, Westchester, Pa.) and cocultured with CEMx 174 cells as previously described (20). Aliquots of the culture media were assayed regularly for the presence of SIV major core protein (p27) by antigen capture enzyme-linked immunosorbent assay (ELISA) (20, 27).
Detection of SIV RNA in plasma.
SIV RNA in plasma and the SIVmac251-8/95 stock were quantified using an SIV-specific branched DNA (bDNA) signal amplification assay (1). This assay is similar to the Quantiplex HIV RNA assay except that target probes were designed to hybridize with the pol region of the SIVmac group of strains including SIVmac251 and SIVmac239. SIV pol RNA in plasma samples was quantified by comparison with a standard curve produced with serial dilutions of cell-free SIV-infected tissue culture supernatant. The SIV RNA in tissue culture fluid was quantified by comparison with a standard curve of purified, DNA-free, in vitro-transcribed SIVmac239 pol RNA. SIV RNA associated with viral particles was measured after concentration from 1 ml of heparinized plasma by centrifugation (23,500 × g for 1 h at 4°C). The lower limit of detection of the assay was 1,500 copies of SIV RNA per ml of plasma.
RNA isolation and RT-PCR.
Plasma samples (1 ml) were centrifuged for 1 to 2 h at 4°C at 21,000 × g (Beckman Microfuge R centrifuge; Beckman Instruments, Fullerton, Calif.), and RNA was extracted with Trizol reagent (Gibco BRL Life Technologies, Grand Island, N.Y.) according to the manufacturer's protocol. RNA was then reverse transcribed (30 min at 42°C and then 5 min at 99°C) in a reaction mixture containing 1× PCR buffer II (Perkin-Elmer, Norwalk, Conn.), 5 mM MgCl2 (Perkin-Elmer), 2.5 μM random hexamer primers (Promega, Madison, Wis.), 1 mM deoxynucleoside triphosphate mix (Pharmacia, Piscataway, N.J.), 5 mM dithiothreitol (Gibco BRL), 20 U of RNasin (Promega), and 30 U of Superscript II reverse transcriptase (RT) (Gibco BRL). A 590-bp fragment encompassing the V1-V2 region of the SIV envelope gene was amplified from the resulting cDNAs in a nested PCR assay. Primer sequences (outer primers, 5′-TTATGGTGTACCAGCTTGGAGGAATGC-3′ and 5′-CCAAACCAAGTAGAAGTCTGTGTCTCCATC-3′; inner primers, 5′-CGATACTTGGGGAACAACTCAGTGCCTAC-3′ and 5′-CACATCTAAGCAAAGCATAACCTGGAGGTG-3′) were generously provided by R. Grant (University of California, San Francisco). For the first round of PCR, 70 μl of a mixture containing 1× PCR buffer, 0.6 μM each outer primer, and 2.5 U of Taq polymerase (Perkin-Elmer) was added to the entire RT reaction volume (30 μl) for each sample. Amplification was carried out in a Perkin-Elmer model 9600 thermal cycler under the following conditions: 3 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by 32 cycles of 94°C for 15 s, 55°C for 45 s, and 72°C for 1 min. The second round of PCR was carried out in a reaction mixture containing 1× PCR buffer, 1.5 mM MgCl2, 0.3 mM deoxynucleoside triphosphate mix, 0.4 μM each inner primer, and 2.5 U of Taq polymerase. Thermal cycling conditions for the second round of PCR were the same as those used for the first round of PCR. Specific precautions to avoid template and amplified product DNA carryover were in effect at all times (18).
Analysis of SIV genetic complexity by HMA.
The extent of SIV genetic diversity in a sample was analyzed using several modifications of the heteroduplex mobility assay (HMA) (2). The HMA provides an estimate of nucleotide sequence variability based on differential electrophoretic mobility of PCR amplicons. Envelope V1-V2 fragments were generated by RT-PCR as described above, and the presence of sufficient product was confirmed on a 1.5% agarose gel. Ten-micro liter aliquots of the PCR products were then mixed with 1.5 μl of 10× annealing buffer (1 M NaCl, 100 mM Tris, 20 mM EDTA), denatured at 94°C for 2 min, and placed immediately on wet ice to promote heteroduplex formation. Samples were then run on nondenaturing 5% polyacrylamide gels and stained with ethidium bromide (0.5 μg/ml). Reverse images of the stained gels were photographed with a digital imaging system (Alpha Innotech Corporation, San Leandro, Calif.). The distribution and number of heteroduplex bands is a measure of SIV envelope diversity in a virus population (i.e., a high number of bands on a gel corresponds to a high number of V1-V2 variants in the sample). For DNA fragments of equal length, a minimum of 1 to 2% mismatches are required to detect heteroduplexes (3, 43).
We used two additional variations of the HMA to characterize SIV envelope variants. Like the heteroduplex tracking assay, both variations utilized mixtures of known viruses from different sources. The purpose of the first type of HMA mixture experiments was to accentuate nucleotide sequence differences between heteroduplex bands with apparently identical migration rates in the assay described previously (4, 5). To accomplish this, the V1-V2 fragment was amplified from SIVmac1A11 (25) plasmid DNA and mixed in equal volume (5 μl of each) with V1-V2 PCR products from each plasma sample, resulting in a relative excess of SIVmac1A11 fragments. Mixed samples were then analyzed on polyacrylamide gels as described above. This technique allowed comparison of V1-V2 variants among animals based on their relative sequence identity to the known SIVmac1A11 sequence.
Additionally, mixtures of PCR products from plasma of macaques or the SIVmac251-8/95 virus stock were analyzed by HMA to allow estimation of sequence similarity between homogeneous virus populations in two different samples (i.e., the most common variants in the inoculum and/or plasma from infected monkeys). This second type of mixture analysis was based on a HMA HIV subtyping protocol previously described (5). Briefly, 5-μl aliquots of PCR product amplified from each of two different samples were mixed and subjected to HMA analysis as described above. Using this technique, mixtures of PCR-amplified fragments that produce a single homoduplex band are 98 to 100% identical in nucleotide sequence (2). Mixtures that produce heteroduplexes comprise variant populations that differ by more than 1 to 2% in nucleotide sequence (2).
DNA sequencing of SIV V1-V2 region.
DNA sequencing was performed by standard PCR-based methods (PE Applied Biosystems, Norwalk, Conn.) using Big Dye terminator chemistry. Approximately 200 to 800 ng of PCR product DNA was amplified using the GeneAmp 9600 PCR system protocol as instructed by the manufacturer (PE Applied Biosystems). The PCR product DNA was precipitated using 100 μl of 72% ethanol, centrifuged at 2,500 × g for 30 min, inverted, and then dried by centrifuging the inverted tray at 700 × g for 1 min. Each DNA pellet was resuspended in 3 μl of formamide loading dye (Amersham-Pharmacia Biotech), and 1.2 μl was then loaded on a 2-mm by 48-cm 4% 6M polyacrylamide gel (Sooner Scientific). Electrophoresis was done on an ABI Prism 377 DNA sequencer according to protocols provided by the manufacturer and analyzed using Sequencing Analysis and AutoAssembler software (PE Biosystems, Norwalk, Conn.).
Quantification of p27 antigenemia.
Plasma levels of SIV major core antigen (p27) were determined using a commercial antigen capture ELISA kit (Coulter Immunology, Hialeah, Fla.) as instructed by the manufacturer. The lower limit of detection for this assay is 0.049 ng of SIV p27 per ml of plasma.
Detection of plasma antibodies.
Plasma samples at weeks 0, 4, 6, and 8 p.i. were tested for the presence of anti-SIV immunoglobulin G (IgG) antibodies; SIV IgG antibody titers were then determined for positive samples as described elsewhere (22).
Nucleotide sequence accession numbers.
The viral sequences have been submitted to GenBank under accession numbers AF303113 through AF303123. Accession numbers AF303113 through AF303122 are SIV env V1-V2 nucleotide sequences obtained from the plasma of animal 24762 at 2 weeks after IVAG inoculation with the SIVmac251-8/95 stock. GenBank accession number AF303123 is the SIV nucleotide sequence of the most common SIV env V1-V2 variant (viral stock endpoint variant [VSEV]) found in the SIVmac251-8/95 inoculum.
RESULTS
Characterization of viral variants in the SIVmac251-8/95 stock inoculum.
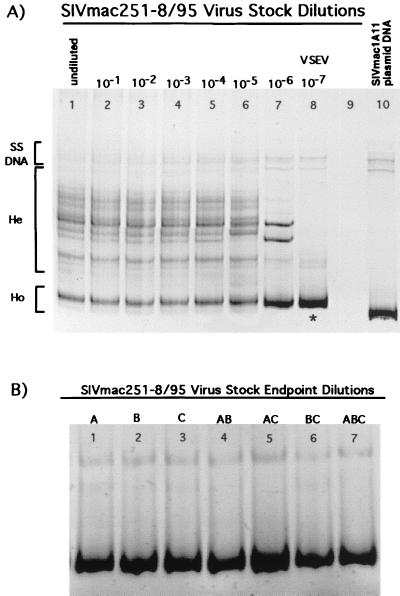
The SIVmac251-8/95 virus stock used to inoculate rhesus macaques contained ∼4 × 109 copies of total SIV RNA/ml as measured by bDNA assay. As indicated by the presence of numerous bands in HMA analyses, the undiluted virus stock was composed of a population of viral variants that were genetically diverse in the V1 to V2 region of the envelope gene (Fig. 1A, lane 1). To determine the most common SIV V1-V2 variant in the stock, we performed RT-PCR and HMA analyses on serial dilutions of viral RNA extracted from the virus stock. In three replicate experiments, a single variant was found at the highest dilution of viral RNA from the SIVmac251-8/95 stock (10−7) (Fig. 1A, lane 8) to yield a positive PCR result (endpoint dilution) with the V1-V2 PCR primers. Figure 1A shows representative results from one of those experiments, which demonstrate that multiple V1-V2 variants (indicated by multiple heteroduplex bands) were present in all dilutions of the stock from 10−1 to 10−5. However, by dilution 10−6, the number of variants (heteroduplex bands) decreased, and by dilution 10−7 only one V1-V2 variant (represented by a single homoduplex band) was detected. No SIV V1-V2 RT-PCR product was detected at a dilution of 10−8 in any of the three dilution experiments.
FIG. 1.
SIV V1-V2 variants present in serial dilutions of SIVmac251-8/95 virus stock. (A) Lanes 1 to 8, dilutions of virus stock from undiluted to 10−7. Heteroduplex (He) bands indicate the presence of multiple V1-V2 variants in the virus stock. The last dilution yielding a RT-PCR product (10−7) was comprised of a homogeneous variant population (i.e., a single variant) as depicted by the presence of a single homoduplex (Ho) band in lane 8. The V1-V2 sequence amplified from the 10−7 dilution (∗) was considered to be the most common envelope variant in the virus stock; this variant was designated VSEV. Lane 9, no sample loaded. Lane 10 contains the V1-V2 fragment of SIV amplified from SIVmac1A11 plasmid DNA. This clonal variant population is represented by a single homoduplex band on the gel and is included for comparison of the virus stock V1-V2 variants with a known reference variant. (B) SIV V1-V2 variants present in three separate endpoint dilutions (A to C) of SIVmac251-8/95 virus stock. Mixtures of the RT-PCR products from each of these three endpoint dilutions show they share the same DNA sequence in the V1-V2 envelope region and thus represent the same SIV variant. Lanes: 1, variant amplified from the endpoint of dilution A; 2, variant amplified from the endpoint of dilution B; 3, variant amplified from the endpoint of dilution C; 4, endpoint dilution variant A mixed with endpoint dilution variant B; 5, endpoint dilution variant A mixed with endpoint dilution variant C; 6, endpoint dilution variant B mixed with endpoint dilution variant C; 7, endpoint dilution variant A mixed with endpoint dilution variant B and endpoint dilution variant C.
The V1-V2 homoduplex band detected in the highest RT-PCR positive dilution of the SIVmac251-8/95 stock (i.e., 10−7 dilution) was designated VSEV (Fig. 1A, lane 8). Each of the three serial dilution experiments resulted in amplification of a single SIV V1-V2 fragment with the same gel mobility from the endpoint dilution. HMA mixture analysis of all pairwise combinations of the PCR product from the three 10−7 dilutions of SIVmac251-8/95 stock indicated that these fragments were from the same SIV envelope variant (nucleotide identity of 98 to 100%) (Fig. 1B). The ability to detect a single variant by multiple endpoint dilution experiments confirms that VSEV is the most common V1-V2 variant present in the SIVmac251-8/95 stock. DNA sequence analysis of the bulk-amplified product DNA (GenBank accession number AF303123) confirmed the homogeneity of the virus population at the 10−7 dilution of the original virus stock. These results eliminate the possibility that several SIV V1-V2 variants are present at high frequencies in the SIVmac251-8/95 stock. Based on this endpoint dilution-HMA analysis, we estimate that this stock contains ∼107 VSEV virions per ml or ∼2 × 107 copies of VSEV RNA per ml.
IV inoculation of rhesus macaques with the SIVmac251-8/95 virus stock.
Five monkeys were inoculated IV with 1 ml of the SIVmac251-8/95 virus stock to evaluate the diversity of viral variants capable of establishing systemic infection in rhesus macaques (Fig. 2A). Because there is no physical barrier to infection following IV inoculation, we reasoned that animals inoculated by this route would likely be infected with the greatest number of SIV V1-V2 variants possible with this virus stock. At 1 week p.i., all five monkeys inoculated IV with SIVmac251 were SIV positive by virus culture of PBMC and/or RT-PCR of plasma (Table 1) and remained positive by virus culture and/or RT-PCR throughout the 8-week study period.
FIG. 2.
Design for the initial inoculation with SIVmac251 (105 TCID50 per ml) and the serial IV (A) and serial IVAG (B) passage experiments. For passage 1, plasma was collected from one IV-inoculated (22673) and one IVAG-inoculated (21743) monkey at 2 weeks p.i., then introduced IV or IVAG to one or two naive monkeys. This process was repeated for passage 2. Symbols (+) and (−) indicate whether each monkey became infected with SIV, as determined by virus isolation.
TABLE 1.
Detection of SIVmac251-8/95 in IV- and IVAG-inoculated monkeys after inoculation
| Animal | Virus isolation
|
RT-PCR
|
p27 antigen (ng/ml)
|
Serum SIV-specific IgG titera
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1b | 2 | 4 | 6 | 8 | 1 | 2 | 4 | 6 | 8 | 1 | 2 | 4 | 4 | 6 | 8 | |
| IV | ||||||||||||||||
| 22673 | + | + | +c | + | + | + | + | +c | − | − | 0.39 | 0.06 | —f | 128,000c | 128,000 | 128,000 |
| 22801 | + | + | + | + | + | + | + | + | + | + | — | 0.24 | — | 4,000 | 2,000 | 1,000 |
| 25299 | + | + | + | + | + | + | + | + | + | + | 1.14 | 0.09 | — | 64,000 | 1,024,000 | 2,048,000 |
| 25388 | + | + | + | + | + | + | + | + | + | + | 0.66 | 0.91 | 0.83 | 32,000 | 128,000 | 256,000 |
| 27606 | + | + | + | + | + | + | + | + | + | + | 0.15 | 0.20 | 64,000 | 64,000 | 64,000 | |
| IVAG | ||||||||||||||||
| 23224 | − | + | + | + | + | − | + | − | + | + | — | — | — | 8,000 | 32,000 | 128,000 |
| 24483 | + | + | + | + | + | + | + | + | − | + | — | 0.28 | — | 512,000 | 256,000 | 256,000 |
| 25562 | + | + | + | + | + | + | + | + | + | + | — | 1.48 | — | 128,000 | 512,000 | 1,024,000 |
| 21743 | NDd | + | + | ND | + | + | + | + | ND | + | ND | 0.76 | — | 16,000 | ND | 256,000 |
| 24762 | ND | + | + | ND | +e | − | + | + | ND | +e | ND | — | — | 64,000 | ND | 512,000e |
Reciprocal of inverse titer.
Week p.i.
Week 5 p.i.
ND, not done.
Week 7 p.i. (necropsy).
—, p27 < 0.49 ng/ml (below assay cutoff).
Peak levels of SIV RNA in plasma occurred at 1 week p.i. in the IV-inoculated monkeys and were between 107 and 108 copies per ml (Fig. 3A). In four animals, viral RNA remained between 106 and 108 copies per ml for the 8-week study period. Plasma SIV RNA in the fifth monkey (22673) dropped from a peak level at 1 week p.i. to low levels by 5 weeks p.i. and remained at low levels for the duration of the study. By 4 weeks p.i., all five IV-inoculated animals also had detectable plasma anti-SIV IgG; peak anti-SIV IgG titers ranged from 4 × 103 to 106 and occurred at 4 to 8 weeks p.i. (Table 1).
FIG. 3.
Plasma RNA levels as measured by bDNA assay. (A) Animals inoculated IV with SIVmac251-8/95; (B) animals inoculated IVAG with SIVmac251-8/95.
Viral sequence diversity in peripheral blood from rhesus macaques inoculated IV with the SIVmac251-8/95 virus stock.
HMA was performed on the earliest plasma samples from which SIV RNA was detected by RT-PCR. Multiple heteroduplex bands with different electrophoretic mobilities were observed in plasma samples from all five IV-inoculated monkeys by 1 week p.i., indicating infection with multiple SIV V1-V2 variants (Fig. 4, lanes 2 to 6). However, the HMA banding patterns of the V1-V2 region amplified from plasma from all IV-inoculated monkeys were very similar to the HMA banding patterns of V1-V2 region amplified from the SIVmac251-8/95 virus stock. This indicates that IV inoculation results in infection with all SIV variants in the virus stock used for the inoculum (Fig. 4, lane 1). Furthermore, the HMA banding patterns were similar among all IV-inoculated animals, suggesting that IV-inoculated animals were infected with a similar population of SIV V1-V2 variants. Thus, host factors appear to have little influence on the earliest population of viral variants in animals inoculated IV with a high dose of genetically diverse SIV.
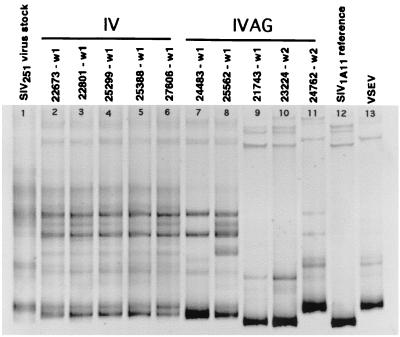
FIG. 4.
Plasma envelope variants in monkeys inoculated IV or IVAG with SIVmac251-8/95. Variants were analyzed in plasma collected at the first SIV-positive time point for each animal (w1, week 1 p.i.; w2, week 2 p.i.). Lane 1, SIVmac251 virus stock; lanes 2 to 6, IV-inoculated monkeys; lanes 7 to 11, IVAG inoculated monkeys; lane 12, SIVmac1A11 V1-V2 reference variant; lane 13, VSEV.
IVAG inoculation of rhesus macaques with the SIVmac251-8/95 virus stock.
We next sought to determine the extent of viral diversity after IVAG inoculation and to assess whether or not infection by this route uniformly results in transmission of a specific subset of genetically distinct SIV variants. We inoculated five monkeys IVAG with 1 ml of SIVmac251-8/95 (Fig. 2B) and analyzed the SIV V1-V2 banding patterns obtained by HMA analysis of the first SIV-positive plasma sample from each animal. At 1 week p.i., three of the five IVAG-inoculated monkeys were SIV positive by virus culture and/or RT-PCR (Table 1). By 2 weeks p.i., all five IVAG-inoculated animals were positive for SIV. All five of these monkeys remained positive by virus culture and/or RT-PCR throughout the 8-week study period.
Peak levels of SIV RNA documented in plasma from the IVAG-inoculated animals occurred at 2 weeks p.i. for two (24483 and 25562) of three monkeys with samples available at all study time points (Fig. 3B). The peak concentration of SIV RNA was not seen in the plasma of the third monkey (23224) until week 4. At 1 week p.i., peripheral blood samples were not available for the remaining two monkeys (21743 and 24762). Monkey 21743 had SIV RNA levels in plasma of 4.0 × 107 copies per ml at 2 weeks p.i., a level comparable to that of the IV-inoculated animals; SIV RNA levels in plasma of monkey 21743 dropped only slightly for the remaining 6 weeks of the study. In contrast, monkey 24762 had 100-fold-lower plasma SIV RNA levels (1.6 × 105 copies per ml) at 2 weeks p.i. that persisted to week 8 p.i. Plasma SIV RNA in three of the five IVAG-inoculated animals (21743, 24483, and 25562) reached peak levels that were similar to those of IV-inoculated animals (107 to 108 copies per ml). However, unlike most IV-inoculated animals, the SIV RNA levels in plasma from IVAG-inoculated monkeys 24483 and 25562 dropped below 106 copies per ml by 4 weeks p.i. Viral RNA levels in plasma from two IVAG-inoculated monkeys (23224 and 24762) never exceeded 2.4 × 105 copies per ml; this pattern of low SIV RNA in plasma was not observed for any of the five IV-inoculated macaques.
Plasma anti-SIV IgG antibody responses were found in three animals inoculated IVAG with SIVmac251 by 4 weeks p.i. (Table 1), and peak anti-SIV IgG antibody titers ranged from 105 to greater than 106. As observed for IV-inoculated monkeys, SIV antibody levels in all five monkeys inoculated IVAG with SIVmac251 were high and sustained, regardless of the levels of SIV RNA in plasma.
Viral sequence diversity in plasma from rhesus macaques inoculated IVAG with the SIVmac251-8/95 virus stock.
HMA was performed on the earliest plasma samples from which SIV RNA was detected by RT-PCR. Variant analysis of SIV in plasma from two (24483 and 25562) of five monkeys inoculated IVAG with SIVmac251-8/95 revealed the presence of multiple heteroduplex bands at week 1 p.i. (Fig. 4, lanes 7 and 8); this observation indicates infection with multiple SIV V1-V2 variants. One of these monkeys, 25562 (Fig. 4, lane 8), had an HMA banding pattern that was similar in complexity to the patterns found in the IV-inoculated animals. Each of the other four IVAG-infected monkeys (24483, 21743, 23224, and 24762) (Fig. 4, lanes 7 and 9 to 11) had an HMA pattern with fewer heteroduplex bands than the HMA patterns of the IV-infected monkeys. These simpler HMA patterns indicate that these four IVAG-inoculated animals were infected with a less genetically diverse population of SIV V1-V2 variants than were the IV-inoculated animals. HMA analysis of the first RT-PCR-positive plasma sample from three IVAG-inoculated animals (21743 at 1 week p.i., 23224 at 2 weeks p.i., and 24762 at 2 weeks p.i.) revealed one dominant homoduplex and one or two minor but detectable heteroduplex bands. This HMA pattern is consistent with infection by a very simple SIV variant population.
One IVAG-infected animal, 24483, had an HMA pattern less complex than that of IV-inoculated animals but more complex than that of three IVAG-inoculated animals (21743, 23224, and 24762). At the conclusion of the 8-week observation period, all of the animals were necropsied and the genital tracts were examined. Of the five IVAG-inoculated monkeys, only 24483 had cervical ectopy. In this anatomic condition, the simple columnar epithelium of the endocervix is directly exposed to the contents of the vaginal lumen (13). Apparently, this simple epithelium does not provide as stringent a barrier to transmission of multiple SIV variants as the intact ectocervical mucosa. This interpretation is consistent with epidemiological data for humans indicating that cervical ectopy is associated with increased risk of HIV infection through sexual contact (33).
There was not a consistent HMA banding pattern shared by all of the monkeys inoculated by the IVAG route. This contrasts with the similar plasma HMA patterns observed among all monkeys inoculated by the IV route. Two of the vaginally infected animals had homoduplex bands with similar electrophoretic mobilities (Fig. 4, 21743 and 23224, lanes 9 and 10). In addition, the plasma HMA pattern from the week 1 p.i. sample of one IVAG animal (24762) showed a homoduplex band (Fig. 4, lane 11) with the same electrophoretic mobility as the homoduplex band of the most common V1-V2 from the SIVmac251-8/95 inoculum (VSEV) (Fig. 5, lane 13). To determine if the nucleotide sequences of these V1-V2 variants with similar electrophoretic mobilities were identical, we performed two variations of the HMA (see below).
FIG. 5.
Plasma envelope variants from monkeys inoculated with SIVmac251-8/95 (Fig. 8) mixed with SIVmac1A11 reference variant (V1-V2 region amplified from SIVmac1A11 plasmid DNA). Lane 1, SIVmac251-8/95 virus stock; lanes 2 to 6, IV-inoculated monkeys; lanes 7 to 11, IVAG-inoculated monkeys; lane 12, SIVmac1A11 reference variant. w1 and w2, week 1 and week 2 p.i.
Comparison of distinct genetic variants in plasma with the SIVmac1A11 reference variant.
Similarities and differences among the nucleotide sequences of SIV V1-V2 regions from different infected animals can be enhanced by mixing variant populations from the plasma samples to be compared with a known standard homogeneous virus isolate (4, 5). To clarify nucleotide sequence relationships between heteroduplex bands with apparently identical migration rates, HMA analysis was performed on the mixed PCR-amplified products from the V1-V2 region of SIVmac1A11 and the SIV RNA from plasma samples shown in Fig. 4. The resulting heteroduplex patterns are shown in Fig. 5. Similar to the standard HMA analysis (Fig. 4), this reference-variant mixture HMA analysis demonstrates the high V1-V2 diversity (reflected by the presence of multiple heteroduplex bands) in plasma samples from all five of the IV-inoculated animals. This analysis confirmed that the viral variant populations in the IV-inoculated animals were indistinguishable from each other. The consistent nature of the viral variant pool supports the conclusion that after IV inoculation, host factors do not significantly influence which viral variants infect animals and replicate in vivo.
The HMA SIVmac1A11 mixture analysis provided more evidence that two of the five IVAG-inoculated animals (24483 and 25562) were infected with complex mixtures of V1-V2 viral variants in the first few days after IVAG SIV inoculation. Thus, this analysis supports the conclusion that IVAG SIV inoculation produced one of three distinct outcomes: infection with either a very simple viral variant population, a moderately complex viral population, or a highly complex variant pool. The HMA mixture analysis also provided direct evidence of the genetic similarity of SIV V1-V2 variants in earliest plasma samples from monkeys 21743 and 23224. The same pattern of heteroduplexes was formed when V1-V2 PCR products from either of these two animals were mixed with the SIVmac1A11 V1-V2 reference PCR product (Fig. 5, lanes 9 and 10). Thus, these two animals were infected with very similar viral variant pools. In sum, the reference variant mixture analysis confirmed that compared to the uniformity of the viral variant populations among five of five IV-inoculated monkeys, similar viral variant populations are rare among animals IVAG-inoculated with the identical inoculum. Thus, individual host factors or stochastic processes, and not viral determinants, are likely to be most important in determining which viral variant(s) infects an individual after IVAG inoculation.
Genetic similarity of SIV variants transmitted to IVAG-inoculated animals.
As noted above, two of five IVAG-inoculated animals (21743 and 23224) seemed to be infected with the same variant populations. To further evaluate the degree of viral sequence similarity between the V1-V2 variants in monkey 21743 (week 1 p.i. plasma.) and monkey 23224 (week 2 p.i. plasma), additional HMA analyses of SIV variant mixtures were conducted (Fig. 6, lanes 1 to 6). When V1-V2 PCR products from the plasma samples of monkeys 21743 and 23224 were mixed together and analyzed by HMA, the banding pattern showed a strong homoduplex band. The presence of a single homoduplex band indicates that the SIV V1-V2 envelope region nucleotide sequences of these viral variants are 98 to 100% identical (2); the faint heteroduplex band indicates that a minimum of two SIV V1-V2 variants infected both animals (Fig. 6, lane 6). Thus, these two IVAG-inoculated animals were infected with the same SIV V1-V2 variants. The fact that a second homoduplex band was not visualized for the mixture of PCR product from plasma of monkeys 21743 and 23224 could be explained if the level of the second V1-V2 variant was below the limit of detection of the HMA.
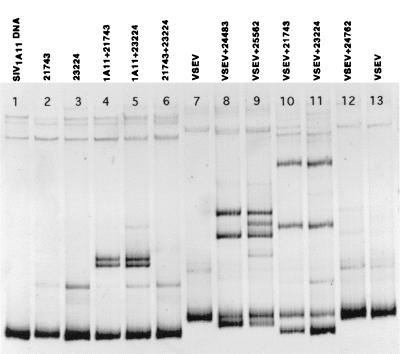
FIG. 6.
Plasma variants from IVAG-inoculated monkeys (Fig. 8) mixed with the SIVmac1A11 reference variant (lane 1) or the endpoint dilution of the virus stock (VSEV; lanes 7 and 13). The 21743 and 23224 variants are shown alone (lanes 2 and 3), mixed with the 1A11 variant (lanes 4 and 5), and mixed together (lane 6). Monkeys 21743 and 23224 were infected with the same envelope variant, based on the formation of a homoduplex when the two variants were mixed (lane 6). Mixtures of VSEV with plasma variants (lanes 8 to 12) indicate that monkey 24762 was infected with the most common variant in the stock (lane 12), but that multiple V1-V2 variants were transmitted to four other monkeys (lanes 8 to 11).
To determine whether the most common virus stock variant was transmitted to any of the IVAG-inoculated monkeys, we performed HMA analysis on mixtures of VSEV and the SIV V1-V2 PCR product amplified from the plasma of each IVAG-inoculated animal (Fig. 2B). Figure 6 (lanes 8 to 12) shows that one of the five IVAG-inoculated monkeys (24762) was infected with VSEV (Fig. 6, lane 12).
We examined the extent of genetic complexity of the SIV population in the week 2 p.i. plasma from animal 24762 by DNA sequence analysis. Within this animal, the range of viral sequence homology in 10 plasmid clones was 97.6 to 99.8% (GenBank accession numbers AF303113 through AF303122). The range of nucleotide homology between animal 24762 and the VSEV (GenBank accession number AF303123) was 97.6 to 100%, thus confirming that this animal was infected with a single, predominant V1-V2 viral variant and validating our HMA results. The DNA sequence results are also consistent with the expected resolution limits for HMA to detect nucleotide sequences differing by more than 1 to 2% (2).
The pattern of heteroduplexes in lanes 8 to 11 in Fig. 6 indicates that each of the other four IVAG-inoculated monkeys was infected with SIV variants that contained V1-V2 sequences that differed from the predominant cDNA sequence of VSEV. However, this HMA mixture analysis cannot exclude the possibility that VSEV was also present in the plasma of the other four monkeys at low levels but was masked by other V1-V2 variants that were present at higher frequency. The fact that VSEV, a variant in high frequency in the stock, was rarely transmitted to IVAG-inoculated animals is consistent with the conclusion that factors other than the multiplicity of infection (MOI) of a specific variant in a complex mixture determine which variant establishes systemic infection after IVAG inoculation.
SIVmac251 variants transmitted by serial IV passage.
To assess transmission of SIV variants by IV serial passage, we used plasma collected from one of the five IV-inoculated animals (22673) at 2 weeks p.i. to IV inoculate an SIV naive monkey (27666, passage 1-[Fig. 2A]). A second naive animal (26108) was then IV inoculated with plasma drawn from 27666 at 2 weeks p.i. (Fig. 2A, passage 2). Each monkey inoculated IV with plasma from the SIV-infected donor became infected with SIV by week 1 p.i., as determined by isolation of virus in culture (Fig. 2A) and by detection of SIV RNA in plasma (Fig. 7A).
FIG. 7.
Plasma viral load in monkeys serially inoculated by the IV (A) or IVAG (B) route, as measured by bDNA assay. Viral load was not measured in IVAG-inoculated monkeys after week 2 p.i. (A) Monkey 22673 was inoculated IV with SIVmac251-8/95; monkey 27666 was inoculated with plasma from 22673; monkey 26108 was inoculated with plasma from 27666. (B) Monkey 21743 was inoculated IVAG with SIVmac251-8/95; monkeys 30446 and 30469 were inoculated with plasma from 21743; monkeys 30450 and 30472 were inoculated with plasma from 30469. p1 and p2, passages 1 and 2; ∗, plasma sample used for serial IVAG passage.
SIV RNA levels in the plasma of the three animals involved in the IV passage were similar during the first 2 weeks after inoculation (Fig. 7A). Unlike the initial donor animal, 22673, SIV RNA levels in the two recipient monkeys (27666 and 26108) remained elevated for the duration of the 8-week study period. This result is consistent with the conclusion that serial IV passage produced a variant pool in the plasma with increased in vivo replication capacity (fitness) relative to the SIVmac251-8/95 stock inoculum.
HMA analysis of virus in plasma samples from the two serial IV passage animals at 1 week p.i. demonstrated multiple heteroduplex bands that migrated similarly (Fig. 8, lanes 11 and 13). Thus, 1 week after IV inoculation, similar, genetically heterogeneous populations of SIV V1-V2 variants were present in the plasma of the serial IV passage animals. Thus, no obvious changes in the genetic diversity of the SIV V1-V2 variant populations resulted from serial IV passage of virions in plasma.
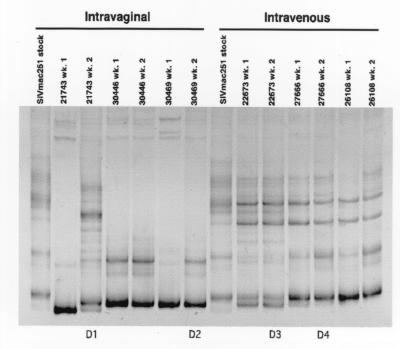
FIG. 8.
Plasma variants present in monkeys infected in a serial IVAG and serial IV passage. After inoculation with SIVmac251, plasma collected at week 2 p.i. was inoculated into monkeys IVAG or IV. Plasma donors 1 and 2 for the IVAG passages (D1 and D2) and donors 3 and 4 for the IV passages (D3 and D4) are indicated. Lanes 1 and 8, SIVmac251-8/95 virus stock; lanes 2 and 3, variants at weeks 1 and week 2 (wk. 1 and wk. 2) p.i. from monkey 21743 inoculated IVAG with SIVmac251; lanes 4 to 7, variants at weeks 1 and 2 p.i. in monkeys inoculated IVAG with plasma from 21743; lanes 9 and 10, variants at weeks 1 and 2 p.i. from a monkey (22673) inoculated IV with SIVmac251-8/95; lanes 11 and 12, variants at weeks 1 and 2 p.i. in a monkey inoculated IV with plasma from 22673; lanes 13 and 14, variants at weeks 1 and 2 p.i. in a monkey inoculated IV with plasma from 22766.
SIVmac251 variants transmitted by serial IVAG passage.
To determine whether specific SIV V1-V2 variants in plasma would be transmitted after serial IVAG passage, plasma collected at 2 weeks p.i. from one IVAG-inoculated monkey (21743) was used to IVAG inoculate two naive animals (Fig. 2B, 30446 and 30469, passage 1). The choice of monkey 21743 as a donor was based on a relatively diverse variant population at 2 weeks p.i. (Fig. 8, lane 3) and high plasma SIV RNA levels, the second highest among the IVAG monkeys (Fig. 3). Both monkeys inoculated with plasma containing SIV in IVAG passage 1 were SIV positive by 1 week p.i. (Table 1). IVAG passage of SIV in plasma was repeated once more (passage 2) using plasma from the passage 1 monkey, 30469 (Fig. 8, lane 7, D2), with the higher plasma SIV RNA levels at 2 weeks p.i. No infectious virus was isolated from either of the two monkeys (Fig. 2B, 30450 and 30472) inoculated with plasma-associated SIV in vaginal passage 2.
Plasma SIV RNA was also measured in the IVAG passage animals. At week 1 or 2 p.i., the plasma SIV RNA concentration was highest in the first donor animal (21743), slightly lower in the passage 1 animals (30446 and 30469) and at the limit of detection in one of the two passage 2 animals (30472) at 1 week p.i. only (Fig. 7B). Because the number of SIV RNA copies detected in plasma from animal 30472 was at the lower limit of the assay and no infectious virus was isolated from PBMC, we concluded that 30472 was not SIV infected.
Low or undetectable plasma SIV RNA levels in the IVAG passage 2 animals are consistent with the conclusion that serial IVAG passage produced a variant pool in the plasma with decreased in vivo replication capacity (fitness) relative to the SIVmac251 stock. It is important to note that while the concentrations of viral RNA in the plasma used for IVAG passages 1 and 2 were very similar, infectious viral titers of the plasma inocula were not determined. Thus, the MOIs of the plasma samples used for IVAG passages 1 and 2 cannot be directly compared. The results of serial IVAG passage of plasma are in sharp contrast to the results for the serial IV plasma passage experiment. In the IV passages, the SIV RNA levels were as high and more sustained in the IV passage 1 and IV passage 2 monkeys than in all of the SIVmac251 stock IV-inoculated animals. Thus, serial IV passage of SIVmac251 seems to select for more fit variants, while serial IVAG passage seems to produce less fit variants.
We analyzed the diversity of the SIV V1-V2 variants in plasma collected from the serial IVAG passage animals 1 week after IVAG inoculation (Fig. 8). The HMA banding pattern showed a homoduplex with several faint heteroduplexes for the initial donor monkey 21743 at week 1 p.i. indicating that there was a relatively homogeneous SIV V1-V2 variant population at this time. However by 2 weeks p.i., a complex banding pattern of multiple homoduplex and heteroduplex bands was found in 21743 plasma, demonstrating the presence of a diverse population of SIV V1-V2 variants (Fig. 8, lanes 2 and 3). There are two possible explanations for this rapid development of viral diversity. Either a series of new mutations in the SIV variant pool arose de novo after infection, or viral variants below the level of detection in our PCR-based assay at week 1 p.i. were detectable by the assay at week 2 p.i. Although the explanations are not mutually exclusive, the latter explanation probably accounts for most of the new SIV variants seen in plasma from 21743 at week 2 p.i.
When the heterogeneous SIV variant population in the 21743 week 2 p.i. plasma was inoculated IVAG into two monkeys (30446 and 30469), both became infected with a relatively homogeneous virus (Fig. 8, lanes 4 and 6). The same predominant variant appeared to be transmitted to monkeys 30446 and 30469; this variant was also present at high frequency in the plasma of donor 21743 (Fig. 9 and 10). For the second IVAG passage, plasma from monkey 30469 at 2 weeks p.i. was used to IVAG inoculate two additional monkeys (30450 and 30472, passage 2) (Fig. 2B). We were unable to detect convincing evidence of systemic SIV infection in either IVAG passage 2 animal. Thus, although the IVAG passage 1 recipient animals were infected with the same SIV variant population, this variant population was not capable of infecting either of the passage 2 IVAG-inoculated animals. The inability to transmit in IVAG passage 2 may be due to decreased viral dose or the lower genetic diversity of SIV in the plasma of donor 2 (30469) compared to the plasma of donor 1 (21743) or both. However, regardless of the reason for our inability to infect the passage 2 recipients, it is clear that serial IVAG passage did not select SIVmac251 variants adapted to IVAG transmission.
FIG. 9.
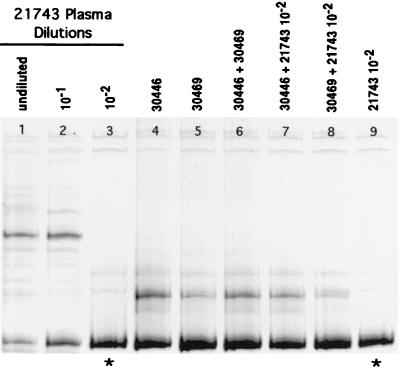
(A) SIV V1-V2 variants present in three replicate serial dilutions of plasma from donor monkey 21743 (week 2 p.i.). Lanes 1 to 4, dilution series A (undiluted to 10−3); lanes 5 to 7, dilution series B (undiluted to 10−2); lanes 8 to 11, dilution series C (undiluted to 10−3). The highest dilutions yielding a RT-PCR product (10−2 and/or 10−3) comprised homogeneous variant populations (i.e., single variants) as depicted by the presence of a single homoduplex band (lanes 4, 7, 10, and 11). These V1-V2 sequences (∗) were considered to be the most common envelope variants in the plasma of 21743 at 2 weeks p.i. The variants depicted in lanes 7 and 11 are the same as determined by the formation of a homoduplex when the two variants were mixed together (B) Therefore, there were three major variants present at similar frequencies in the plasma of 21743 at week 2 p.i. (B) HMA gel showing mixtures of the endpoint variants from three dilutions (A to C) of donor 21743 (week 2) RNA (see also panel A). Lane 9 shows that the endpoint variant from dilution B was the same as one of the endpoint variants from dilution C (10−3).
FIG. 10.
SIV V1-V2 variants present in dilution series B (Fig. 9) of plasma from donor monkey 21743 at 2 weeks p.i. Lanes 1 to 3, dilutions of plasma from undiluted to 10−2 (same as lanes 5 to 7 in Fig. 9). The V1-V2 sequence amplified from the 10−2 dilution (∗; lanes 3 and 9) was considered to be one of the three most common envelope variants in the plasma of 21743 (see text and Fig. 9). Lanes 4 to 8 show plasma variants at week 1 p.i. from the two monkeys inoculated in serial vaginal passage 1 (Fig. 3), separately and mixed together (lanes 4 to 6) and mixed with one of the most common V1-V2 variants (lane 3) in the plasma of the donor animal (lanes 7 and 8). Monkeys 30446 and 30469 were infected with the same predominant variant (lane 6), and this variant was the same as one of the most common variants in the plasma of donor animal 21743 (lanes 7 and 8), as indicated by the formation of homoduplexes when variants from two different animals were mixed.
DNA sequence similarity of the most common SIV genetic variant in plasma and the predominant virus transmitted by IVAG inoculation.
To characterize the V1-V2 variant population in the first serial IVAG donor animal (21743 at 2 weeks p.i.), we used a serial dilution of SIV cDNA prepared from the plasma inoculum. The experiment was performed three separate times in the same manner as described for the virus stock dilution (Fig. 9A). In contrast to the results obtained from the virus stock dilution experiments, homoduplex bands with different migration rates were detected in each replicate. Thus, distinct SIV V1-V2 variants were amplified from the three endpoint dilutions of 21743 plasma (Fig. 9A, lanes 4, 7, and 11). HMA of the V1-V2 fragments amplified from the 10−2B dilution (Fig. 9A, lane 7) and the 10−3C dilution (Fig. 9A, lane 11) confirmed DNA sequence identity of these two fragments. Further, these fragments were distinct from the V1-V2 fragments amplified from the 10−3 A (Fig. 9A, lane 4) and the 10−2C dilutions (Fig. 9A, lane 10), as shown by the HMA patterns when these products were mixed together (Fig. 9B). These results indicate that a minimum of three SIV variants were present at approximately similar levels in the plasma from 21743 at 2 weeks p.i. Additional mixture experiments confirmed that none of the three most common variants in plasma of 21743 was the most common variant in the SIVmac251-8/95 virus stock (VSEV [data not shown]).
We next evaluated whether the SIV V1-V2 variants transmitted to monkeys 30446 and 30469 in serial vaginal passage 1 were the same by HMA and whether these variants matched the viral sequence of one of the most common variants in the plasma from donor animal 21743. When mixed together, the predominant V1-V2 variants from 30446 and 30469 produced a homoduplex (Fig. 10, lane 6), confirming 98 to 100% identical V1-V2 nucleotide sequences. One of the endpoint variants from donor 21743 (Fig. 10, lane 3) formed a homoduplex when mixed with the variant transmitted to 30446 and 30469 (Fig. 10, lanes 7 and 8). This result indicates that both passage 1 recipients became infected with one of the three most common SIV V1-V2 variants present in the plasma of the donor 1 animal (21743).
DISCUSSION
In this study, we examined the extent of genetic diversity of the SIV env variant pool in the plasma of outbred rhesus macaques infected with a genetically complex SIVmac251 inoculum by either IVAG or IV inoculation. Further, viral variant populations were analyzed in plasma of animals during either serial IVAG or IV passage of the virus stock. IV-inoculated monkeys had a complex population of V1-V2 env variants, and the variant populations in all IV-inoculated animals were similar. Like others (11, 42), we found a tendency for lower SIV genetic diversity in the venous blood of IVAG-inoculated monkeys compared to IV-inoculated monkeys. However, this finding was not uniform among the IVAG-inoculated animals. One of the five IVAG-inoculated animals had a plasma virus population with a level of genetic diversity that was similar to that of the animals infected by the IV route of inoculation. One animal became infected with a moderately complex viral variant population, and three animals had a relatively simple population of SIV variants in plasma. The variability in the outcome of vaginal SIV inoculation sharply contrasts with the remarkable consistency of the variant population infecting IV-inoculated monkeys. This result could be explained if nonviral (host) factors, or stochastic processes, are the important determinants of which SIV env variants infect IVAG-inoculated monkeys. The result is also consistent with the conclusion that host factors do not significantly affect the composition of viral variant populations in the first days after IV inoculation. Last, despite the more limited level of viral genetic diversity in animals infected by the IVAG route, we did not find in the stock a specific set of STV V1-V2 variants that was consistently transmitted by IVAG SIV inoculation.
Transmission of a genetically diverse population of SIV variants to two of the five macaques infected by the IVAG route appears to contradict some findings in HIV-infected humans. Specifically, people recently infected with HIV after sexual exposure harbor relatively homogeneous virus populations, even when the virus donor is infected with a genetically diverse population of viral variants (3, 38, 44, 48–50). However, in agreement with our results, a recent study found that greater viral diversity was present in the PBMC of women compared to men infected with HIV by sexual contact (21). It seems likely that any discrepancies between our findings and HIV transmission studies can be explained by the differences in experimental design. In human studies, it is difficult to know with certainty when HIV infection occurred relative to the time when blood samples were obtained. In the SIV/rhesus macaque model studies, blood samples were obtained in the earliest stages after viral transmission. Furthermore, studies in human are limited by the fact that paired samples from the HIV-infected donor(s) and the uninfected partner(s) at the time of HIV transmission cannot be obtained reliably.
Our results are consistent with the conclusion that there is a subset of genetic variants in the SIVmac251-8/95 stock that is capable of establishing systemic infection after IVAG inoculation. This variant subset is smaller than the variant population that infects all monkeys after IV inoculation. We have previously demonstrated that the ability of molecularly cloned (i.e., relatively genetically homogeneous) SIV and SIV/HIV chimeric (SHIV) variants to establish systemic infection in rhesus macaques after IVAG inoculation is directly related to ability of the clone to replicate in rhesus macaques following IV infection. Molecularly cloned viruses that replicate persistently, but at low levels, or produce transient plasma viremia in rhesus macaques are not reliably transmitted by IVAG inoculation (32). Furthermore, tissue culture assessments of SIV or SHIV cell tropism do not predict IVAG transmission (32). The capacity for high, persistent in vivo replication is likely to be one common characteristic of all SIV V1-V2 variants detected in IVAG-inoculated animals. Thus, any genetic changes that increase the in vivo replication capacity of a particular SIV variant are likely to enhance the ability of that variant to be transmitted by the IVAG route.
Others have observed that serial IV passage of SIV (9) and SHIV (16, 24, 26, 40) in macaques consistently results in increased viral virulence and high concentrations of virus in plasma. In this study we also found that the concentrations of viral RNA (Fig. 7A) and p27 antigen (data not shown) in plasma increased, and remained high longer, after each successive serial IV monkey passage of SIV. The plasma inoculum used in the IV inoculations contained large viral populations, >107 SIV RNA copies. Thus, serial IV virus passage of large viral variant populations results in increased fitness of the SIV variant pool. In addition, monkeys infected with SIV after serial IV inoculation consistently had a genetically diverse population of V1-V2 variants in their plasma, similar in complexity to the variant population found in the SIVmac251-8/95 stock. A similar trend toward increased viral population fitness after serial passage of large viral populations has been documented in carefully controlled tissue culture serial passage studies of RNA viruses. Serial passage of large populations (>106 infectious virions) of RNA viruses reproducibly gained genetic diversity and increased in fitness, while serial passage of relatively small and genetically homogeneous viral populations was associated with reduced fitness (reviewed in reference 34).
There have been no previous reports of serial IVAG passage of lentiviruses. Based on the outcome for serial IV passage of SIV in animals, we expected similar results for serial IVAG passage of SIV. Instead, we found that serial IVAG passage of SIV-infected plasma resulted in successively lower plasma levels of SIV RNA and lower viral genetic diversity. The results of the serial IVAG passage experiment are best understood in the context of general features of RNA virus population biology. HIV, and all RNA viruses, has a high degree of mutation due to the error-prone copying activity of RNA replicases and RTs (7). In the case of HIV, it is estimated that one point mutation is introduced into the proviral genome at every round of reverse transcription (6). Because of the high frequency of mutant genomes, HIV or SIV populations replicating in an individual are extremely dynamic mutant swarms termed viral quasispecies. Thus, viral genomes cannot be described as a defined unique structure but as a weighted average of a large number of different and transient sequences. When a mutant variant, or a set of variants which are more suited to replicate in an altered environment are present in the quasispecies, the virus population gains fitness through selective amplification of this subset of genomes. The basic biology of HIV and SIV replication has profound implications for transmission of these viruses.
Population (genetic) bottlenecks act as founder events as a result of strong reductions in the genetic diversity of RNA virus populations and amplification of progeny virus from only one or a few founder virions. In vitro experiments using RNA viruses have demonstrated that serial passage of a small virus population with limited genetic diversity results in average fitness losses in the passaged populations relative to the parental population (reviewed in reference 34). The term “fitness” describes the replicative capacity of a virus in a particular environment. Fitness loss accompanying repeated bottleneck events is the expected result of accumulation of deleterious mutations. HIV and SIV do not possess mechanisms that can efficiently compensate for the accumulation of deleterious mutations. Muller first proposed that the accumulation of irreversible mutational lesions would lead to genetic deterioration, and the process is termed “Muller's ratchet” (35). In vitro studies confirm that after a relatively few serial passages of small HIV populations (103 to 104 infectious particles), the fitness of the resulting HIV population is severely decreased compared to the starting population. In fact, HIV was more susceptible to Muller's ratchet after of serial bottleneck passages than any other RNA virus tested (47).
The probability that a transmitted viral population will have lower fitness than the parental population from which it is taken depends on a number of factors, including the dose of the inoculum and the fitness of the parental population. Thus, the size of the genetic bottleneck that leads to fitness losses depends on the mean initial fitness of the parent viral population (37). To maintain fitness, parental populations of high initial fitness require that a relatively large number of genetic variants be transferred at each serial passage. In contrast, parental populations of low initial fitness maintain fitness with a serial passage of a relatively small number of genetic variants. This is because the high fitness population contains a higher proportion of low fitness clones and a bottleneck transmission will include only progeny of lower fitness than the parental virus population (8).
The SIVmac251 stock used for our studies contains 105 infectious virions per ml and is a complex virus variant population. This stock replicates to very high levels in monkeys after IV inoculation, and thus the stock consists of a highly fit virus population. We have previously shown that IVAG inoculation with this stock results in infection of approximately 103 to 104 target cells in the vaginal mucosa (15). Even if one assumes that each of these cells is infected with a distinct genetic variant, a strong genetic bottleneck exists in the experimental vaginal transmission of this highly fit SIVmac251 virus population. Based on the above considerations, the variant pool that infects an animal following IVAG inoculation with the stock is most likely to include only progeny of lower fitness than the parental virus population. We selected plasma from an animal with a genetically homogeneous virus population but a high viral load. Thus, the plasma inoculum used for IVAG passage was composed of a relatively homogeneous population of highly fit env variants, making the transmitted viral population vulnerable to the effects of Muller's ratchet. The animals in the first vaginal passage did become infected but had lower viral loads than the donor animal. This outcome is consistent with fitness losses in the virus population transmitted in passage 1. As this effect occurred in both animals, host factors are not likely involved. The plasma used for the second vaginal passage consisted of a population of variants more homogeneous than the SIV population used for the first passage, and systemic infection was not detected in animals exposed to this inoculum. Because the plasma used for the first and second IVAG passages contained similar SIV RNA concentrations, the failure to transmit SIV in the second vaginal passage cannot be explained by a decreased MOI in the passage 2 inoculum.
Although we do not believe that the outcome of the IVAG passage experiment is due primarily to the SIV dose in the plasma inoculum, we have shown that virus dose plays a role in the outcome of IVAG SIV stock inoculation. Serial dilutions of two SIVmac251 stocks that are related to the stock used in the studies described here were made and used for vaginal transmission studies (30). A large proportion of the animals that became infected after IVAG inoculation with the lower doses of the stocks had unusual virologic outcomes characterized by a brief transient viremia, failure to mount anti-SIV antibody responses, and persistent SIV-specific CD4+ T-cell proliferative responses. These animals had a persistent infection with low-level replication in systemic lymphoid tissues, but virus was undetectable in peripheral blood (28, 30). This occult SIV infection of monkeys is similar to the condition of some HIV-exposed women who have anti-HIV cellular immune responses but no evidence of HIV infection in blood (41). A feature common to these monkeys and such HIV-resistant women is vaginal exposure to a simple variant population in a low-dose lentivirus inoculum. We have shown that repeated IVAG inoculation of low-dose inocula increases the efficiency of IVAG lentivirus transmission (23, 29, 31). The assumption was that many exposures eventually resulted in infection of enough target cells to establish an infection. However, it is possible that as viral genomes accumulate in long-lived antigen-presenting cells or memory T cells in the vaginal mucosa and draining lymph node, recombination occurs between two weak but persistent SIV genomes, producing a more fit variant population that produces a classic systemic SIV infection. In vivo recombination between SIV genomes can occur rapidly after systemic coinfection with large virus variant populations (46). This virus variant population-based explanation of IVAG variant transmission is consistent with recent reports of emerging systemic infections in the previously apparently resistant commercial sex workers (17).
We have shown that the route of SIV inoculation determines the genetic diversity, but not the identity, of viral variant populations that infect rhesus macaques. Even for a well-characterized SIV inoculum, the specific viral variant(s) that will infect a particular outbred macaque after IVAG inoculation with SIV cannot be predicted. These results suggest that the route of HIV exposure may determine the extent of genetic diversity, but not the specific variants, in the HIV population that infects a person. Moreover, the results from these SIV/macaque studies support the position that a vaccine to prevent heterosexual HIV transmission should be designed to protect against multiple, rather than specific, HIV variants.
ACKNOWLEDGMENTS
We are grateful for the expert technical support of Steven Joye, Yichuan Wang, Judith Torten, Kristen Bost, Stephen Dillard-Telm, Linda Hirst, David Bennet, and Wilhelm Von Morgenland, whose assistance was crucial to the successful completion of these studies.
This work was supported by Public Health Service grants RR00169 from the National Center for Research Resources, AI39109 (M.L.M.), AI39435 (C.J.M.), and AI35545 (C.J.M.) from the National Institute of Allergy and Infectious Diseases, and HD37356 (S.M.W.) from the National Institute of Child Health and Human Development and by Elizabeth Glaser Scientist award 8-97 (M.L.M.) from the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Committee on Care and Use of Laboratory Animals. Guide for the care and use of laboratory animals. Washington, D.C.: Institute of Laboratory Animal Resources, National Research Council; 1996. [Google Scholar]
- 1a.Dailey P J, Zamroud M, Kelso R, Kolberg J, Urdea M. Quantitation of simian immunodeficiency virus (SIV) RNA in plasma of acute and chronically infected rhesus macaques using a branched DNA (bDNA) signal amplification assay. 1995. p. 180. . 13th Annual Symposium on Nonhuman Primate Models of AIDS, Monterey, Calif. [Google Scholar]
- 2.Delwart E, Shaper E, McCutchan F, Louwagie J, Grez M, Rubsamen-Waigmann H, Mullins J. Genetic relationships determined by a heteroduplex mobility assay: analysis of HIV env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 3.Delwart E, Sheppard H, Walker B, Goudsmit J, Mullins J. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delwart E L, Gordon C J. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods Companion Methods Enzymol. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 5.Delwart E L, Herring B, Rodrigo A G, Mullins J I. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl. 1995;4:S202–S216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 6.Drake J. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake J, Holland J. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duarte E A, Novella I S, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. Subclonal components of consensus fitness in an RNA virus clone. J Virol. 1994;68:4295–4301. doi: 10.1128/jvi.68.7.4295-4301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmonson P, Murphey-Corb M, Martin L N, Delahunty C, Heeney J, Kornfeld H, Donahue P R, Learn G H, Hood L, Mullins J I. Evolution of a simian immunodeficiency virus pathogen. J Virol. 1998;72:405–414. doi: 10.1128/jvi.72.1.405-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enose Y, Kentaro I, Toshihide S, Ui M, Hayami M. Genomic analysis of the viral population in genital secretions early after infection of simian immunodeficiency viruses in macaque monkeys. Microbiol Immunol. 1998;42:715–722. doi: 10.1111/j.1348-0421.1998.tb02344.x. [DOI] [PubMed] [Google Scholar]
- 11.Enose Y, Okada M, Sata T, Ma W, Igarashi T, Ibuki K, Ido E, Hayami M. Restriction of viral populations by intravaginal infection of simian immunodeficiency viruses in macaque monkeys. Arch Virol. 1997;142:37–51. doi: 10.1007/s007050050057. [DOI] [PubMed] [Google Scholar]
- 12.Evans D T, Knapp L A, Jing P C, Mitchen J L, Dykhuizen M, Montefiori D C, Pauza D C, Watkins D I. Rapid and slow progressors differ by a single MHC class I haplotype in a family of MHC-defined rhesus macaques infected with SIV. Immunol Lett. 1999;66:53–59. doi: 10.1016/s0165-2478(98)00151-5. [DOI] [PubMed] [Google Scholar]
- 13.Ferenczy A, Winkler B. Anatomy and histology of the cervix. In: Kurman R J, editor. Blaustein's pathology of the female genital tract. 3rd ed. New York, N.Y: Springer-Verlag Inc.; 1987. pp. 141–157. [Google Scholar]
- 14.Harouse J M, Gettie A, Tan R C H, Blanchard J, Cheng-Mayer C. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science. 1999;284:816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- 15.Hu J, Gardner M B, Miller C J. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J Virol. 2000;74:6087–6095. doi: 10.1128/jvi.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L J, Adany I, Pinson D M, McClure H M, Narayan O. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaul R, Kimani J, Dong T, Yang H, Bwayo J, Macdonald K, Plummer F A, Rowland-Jones S. Proceedings of the Seventh Conference on Retroviruses and Opportunistic Infection. Calif: San Francisco; 2000. Late seroconversion in HIV-resistant Nairobi prostitutes is associated with a preceding decrease in HIV exposure. [Google Scholar]
- 18.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. . (Erratum, 339:490.) [DOI] [PubMed] [Google Scholar]
- 19.Lewis M, Bellah S, McKinnon K, Yalley-Ogunro J, Zack P, Elkins W, Desrosiers R, Eddy G. Titration and characterization of two rhesus-derived SIVmac challenge stocks. AIDS Res Hum Retroviruses. 1994;10:213–292. doi: 10.1089/aid.1994.10.213. [DOI] [PubMed] [Google Scholar]
- 20.Lohman B, Higgins J, Marthas M, Marx P, Pedersen N. Development of simian immunodeficiency virus isolation, titration, and neutralization assays which use whole blood from rhesus monkeys and an antigen capture enzyme-linked immunosorbent assay. J Clin Microbiol. 1991;29:2187–2192. doi: 10.1128/jcm.29.10.2187-2192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long E M, Martin J, L. H, Kreiss J K, Rainwater S M J, Lavreys L, Jackson D J, Rakwar J, Mandaliya K, Overbaugh J. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6:71–75. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, Kiyono H, Lu D, Kawabata S, Torten J, Srinivasan S, Dailey P, McGhee J, Lehner T, Miller C. Targeted lymph-node immunization with whole inactivated simian immunodeficiency virus (SIV) or envelope and core subunit antigen vaccines does not reliably protect rhesus macaques from vaginal challenge with SIVmac251. AIDS. 1998;12:1–10. doi: 10.1097/00002030-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, Brosio P, Lafaile M, Li J, Collman R G, Sodroski J, Miller C J. Vaginal transmission of chimeric simian/human imunodeficiency viruses in rhesus macaques. J Virol. 1996;70:3045–3050. doi: 10.1128/jvi.70.5.3045-3050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y, Pauza C D, Lü X, Montefiori D C, Miller C J. Rhesus macaques that become systemically infected with pathogenic SHIV 89.6-PD after intravenous, rectal, or vaginal inoculation and fail to make an antiviral antibody response rapidly develop AIDS. J Acquir Immune Defic Syndr Hum Retroviral. 1998;19:6–18. doi: 10.1097/00042560-199809010-00002. [DOI] [PubMed] [Google Scholar]
- 25.Luciw P, Shaw K, Unger R, Planelles V, Stout M, Leung N, Banapour B, Marthas M. Genetic and biologic comparisons of pathogenic and non-pathogenic molecular clones of simian immunodeficiency virus (SIVmac) AIDS Res Hum Retroviruses. 1992;8:395–402. doi: 10.1089/aid.1992.8.395. [DOI] [PubMed] [Google Scholar]
- 26.Luciw P A, Mandell C P, Li J, Lowe T A, Schmidt K A, Shaw K E S, Cheng-Mayer C. Fatal immunopathogenesis by SIV/HIV-1 (SHIV) containing a variant form of the HIV-1SF33 env gene in juvenile and newborn rhesus macques. Virology. 1999;263:112–127. doi: 10.1006/viro.1999.9908. [DOI] [PubMed] [Google Scholar]
- 27.Marthas M, Ramos R, Lohman B, Van Rompay K, Unger R, Miller C, Banapour B, Pedersen N, Luciw P. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed using attenuated and pathogenic molecular clones of SIVmac. J Virol. 1993;67:6047–6055. doi: 10.1128/jvi.67.10.6047-6055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McChesney M, Collins J, Lu D, Lu X, Torten J, Ashley R, Cloyd M, Miller C. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4+ T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J Virol. 1998;72:10029–10035. doi: 10.1128/jvi.72.12.10029-10035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller C, Alexander N, Sutjipto S, Joye S, Hendrickx A, Jennings M, Marx P. Effect of virus dose and Nonoxynol-9 on the genital transmission of SIV in rhesus macaques. J Med Primatol. 1990;19:401–409. [PubMed] [Google Scholar]
- 30.Miller C, Marthas M, Torten J, Alexander N, Moore J, Doncel G, Hendrickx A. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J Virol. 1994;68:6391–6400. doi: 10.1128/jvi.68.10.6391-6400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller C J, Alexander N J, Sutjipto S, Lackner A A, Hendrickx A G, Gettie A, Lowenstine L J, Jennings M, Marx P A. Genital mucosal transmission of simian immunodeficiency virus: animal model for heterosexual transmission of human immunodeficiency virus. J Virol. 1989;63:4277–4284. doi: 10.1128/jvi.63.10.4277-4284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C J, Marthas M, Greenier J, Lu D, Dailey P, Lu Y. In vivo replication capacity rather than in vitro macrophage tropism predicts efficiency of vaginal transmission of simian immunodeficiency virus or simian/human immunodeficiency virus in rhesus macaques. J Virol. 1998;72:3248–3258. doi: 10.1128/jvi.72.4.3248-3258.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss G B, Clemetson D, D'Costa L, Plummer F A, Ndinya-Achola J O, Reilly M, Holmes K K, Piot P, Maitha G M, Hillier S L, Kiviat N C, Cameron C W, Wamola I A, Kreiss J K. Association of cervical ectopy with heterosexual transmission of human immunodeficiency virus: results of a study of couples in Nairobi, Kenya. J Infect Dis. 1991;164:588–591. doi: 10.1093/infdis/164.3.588. [DOI] [PubMed] [Google Scholar]
- 34.Moya A, Elena S F, Bracho A, Miralles R, Barrio E. The evolution of RNA viruses: a population genetics view. Proc Natl Acad Sci USA. 2000;97:6967–6973. doi: 10.1073/pnas.97.13.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller H J. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 36.Neildez O, Le Grand R, Caufour P, Vaslin B, Cheret A, Matheux F, Theodor F, Roques P, Dormont D. Selective quasispecies transmission after systemic or mucosal exposure of macaques to simian immunodeficiency virus. Virology. 1998;243:12–20. doi: 10.1006/viro.1997.9026. [DOI] [PubMed] [Google Scholar]
- 37.Novella I, Elena S F, Moya A, Domingo E, Holland J J. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang S, Shlesinger Y, Daar E S, Moudgil T, Ho D D, Chen I S. Rapid generation of sequence variation during primary HIV-1 infection. AIDS. 1992;6:453–460. doi: 10.1097/00002030-199205000-00003. . (Erratum, AIDS 6:following 606.) [DOI] [PubMed] [Google Scholar]
- 39.Poss M, Martin H, Kreiss J, Granville L, Chohan B, Nyange P, Mandaliya K, Overbaugh J. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reimann K A, Li J T, Veazey R, Halloran M, Park I W, Karlsson G B, Sodroski J, Letvin N L. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland-Jones S L, Dang T, Fowke K R, Kimani K, Krausa P, Newell H, Blanchard T, Ariyoshi K, Oyugi J, Ngugi E, Bwayo J, MacDonald K S, McMichael A J, Plummer F A. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Investig. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodora D L, Lee F, Dailey P J, Marx P A. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res Hum Retroviruses. 1998;14:171–181. doi: 10.1089/aid.1998.14.171. [DOI] [PubMed] [Google Scholar]
- 43.Upchurch D A, Shankarappa R, Mullins J I. Position and degree of mismatches and the mobility of DNA heteroduplexes. Nucleic Acids Res. 2000;28:e69. doi: 10.1093/nar/28.12.e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfs T F, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 45.Wolinsky S M, Wike C M, Korber B, Hutto C, Parks W P, Rosenblum L L, Kunstman K J, Furtado M R, Muñoz J L. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 46.Wooley D, Smith R, Czajak S, Desrosiers R. Direct demonstration of retroviral recombination in a rhesus monkey. J Virol. 1997;71:9650–9653. doi: 10.1128/jvi.71.12.9650-9653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuste E, Sanchez-Palomino S, Casado C, Domingo E, Lopez-Galindez C. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J Virol. 1999;73:2745–2751. doi: 10.1128/jvi.73.4.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 50.Zhu T, Wang N, Carr A, Nam D, Moor-Jankowski R, Cooper D, Ho D. Genetic characterization of human immunodeficiency virus type 1 in blood and gential secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]