Abstract
Background:
Biliary obstruction can be due to both malignant and benign pancreaticobiliary disease. Currently, there are no biomarkers that can accurately help make this distinction. MicroRNAs (miRNAs) are stable molecules in tissue and biofluids that are commonly deregulated in cancer. The MIRABILE study aimed to identify miRNAs in bile that can differentiate malignant from benign pancreaticobiliary disease.
Materials and methods:
There were 111 patients recruited prospectively at endoscopic retrograde cholangiopancreatography (ERCP) or percutaneous transhepatic cholangiography (PTC) for obstructive jaundice, and bile was aspirated for cell-free RNA (cfRNA) extraction and analysis. In a discovery cohort of 78 patients (27 with pancreatic ductal adenocarcinoma (PDAC), 14 cholangiocarcinoma (CCA), 37 benign disease), cfRNA was subjected to small-RNA sequencing. LASSO regression was used to define bile miRNA signatures, and NormFinder to identify endogenous controls. In a second cohort of 87 patients (34 PDAC, 14 CCA, 39 benign disease), RT-qPCR was used for validation.
Results:
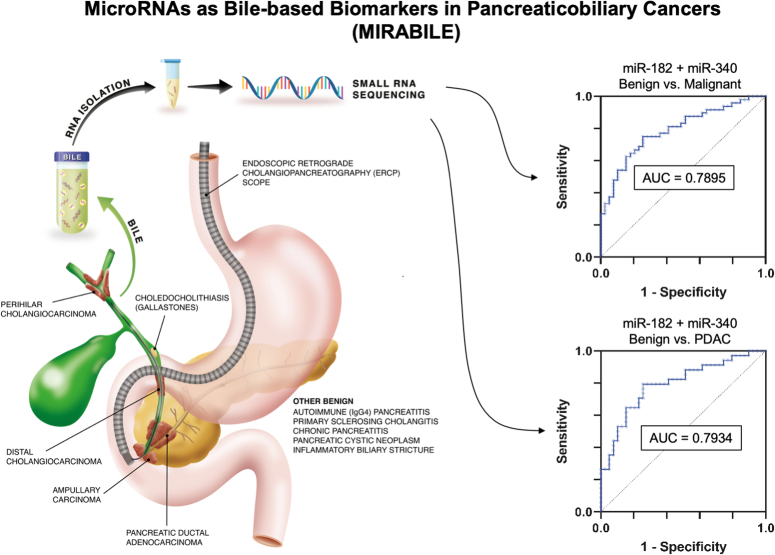
LASSO regression identified 14 differentially-expressed bile miRNAs of which 6 were selected for validation. When comparing malignant and benign pancreaticobiliary disease, bile miR-340 and miR-182 were validated and significantly differentially expressed (P<0.05 and P<0.001, respectively). This generated an AUC of 0.79 (95% CI: 0.70–0.88, sensitivity 65%; specificity 82%) in predicting malignant disease.
Conclusion:
Bile collected during biliary drainage contains miRNAs able to differentiate benign from malignant pancreaticobiliary diseases in patients with obstructive jaundice. These bile miRNAs have the potential to increase diagnostic accuracy.
Keywords: bile, biomarker, cholangiocarcinoma, microRNA, pancreatic ductal adenocarcinoma
Introduction
Highlights
This is the first study to assess bile miRNAs by small RNA-sequencing in a large number of patients presenting with obstructive jaundice.
This study demonstrated that bile cell-free miRNAs can be a source of biomarkers during a ‘window of opportunity’ prior to anticancer treatment to help stratify and personalise patient pathways.
Obstructive jaundice can be a clinical sign indicating occlusion or narrowing of bile ducts due to benign or malignant pancreaticobiliary disease. The most common malignant cause is pancreatic ductal adenocarcinoma (PDAC)1. Other malignant causes include cholangiocarcinoma (CCA) and ampullary cancer. Accurate diagnosis is important to prevent futile surgical resection in case of benign disease, such as chronic pancreatitis, and to treat malignant disease in a timely manner and avoid the chance of disease progression. However, obstructive jaundice with secondary biliary inflammation can make it challenging to recognise distinctive radiological features, or to obtain representative samples during endoscopic sampling1,2. Up to 20% of the biliary strictures may remain indeterminate, raising a therapeutic dilemma that is often decided by the maxim that a biliary stricture is malignant until proven otherwise3. Even when a biliary stricture is suspected malignant, it has been reported that 10% of these patients who undergo resection are postoperatively diagnosed with benign conditions3–5. To improve the diagnosis of biliary strictures causing obstructive jaundice, the MIRABILE study was designed to assess the feasibility and diagnostic value of cell-free RNAs (cfRNAs) in bile as diagnostic biomarkers to differentiate malignant from benign lesions.
Diagnostic work-up for a biliary stricture includes laboratory tests, imaging and endoscopic modalities. The diagnostic efficacy of blood-based biomarker carbohydrate antigen 19-9 (CA 19-9) has previously been assessed, but has been shown inadequate as a stand-alone biomarker to discriminate PDAC from benign pancreaticobiliary disease6. Serum CA 19-9 has a respective sensitivity and specificity of 78.2 and 82.8% when the threshold is set at 40 U/ml6. This is similar to computed tomography (CT), which has a respective sensitivity and specificity of 77% and 63% in the recognition of malignant versus benign biliary obstruction7. Contrast-enhanced MRI with magnetic resonance cholangiopancreatography (MRCP) has shown a sensitivity and specificity of ~85% for detecting malignant biliary strictures8–11. Furthermore, endoscopic retrograde cholangiopancreatography (ERCP) with brush cytology has a pooled sensitivity of 41–45%, while this is 48% for intraductal biopsy2,12. Endoscopic procedures remain the most optimal route to make a cytological diagnosis, enabling tissue or cells to be obtained. Exploring biomarkers from bile aspirated at this stage offers two benefits. Firstly, it is one of the initial stages in the patient’s diagnostic journey to manage biliary obstruction1,3. Secondly, bile is intimately related to the tumour site in terms proximity and anatomy, which enhances the potential for detecting molecular markers for pancreaticobiliary cancers13–16.
CfRNAs can be assessed as molecular markers in many bodily fluids, including blood17 and bile18. Of these cfRNAs, extracellular microRNAs (miRNAs) appear to be selectively protected from endogenous RNases found within the blood19. Furthermore, they are stable through freeze-thaw cycles and extremes of pH20,21. MiRNAs have a known role in the regulation of gene expression at a post-transcriptional level and are involved in many biological processes. They have shown unique expression profiles in tumours and have hitherto been shown to be differentially expressed markers in numerous biofluids22. MiRNAs reflect the current state of cells and tissues and, as such, form an attractive class of biomarkers for diagnostic purposes23,24.
MIRABILE (MIcroRNAs As BILE-based biomarkers in Pancreaticobiliary Cancers) was a prospective study that assessed cfRNAs in bile obtained from patients with pancreaticobiliary disease who presented at ERCP for obstructive jaundice. We identified a novel bile-based miRNA signature for the diagnosis of malignant pancreaticobiliary disease.
Materials and methods
Study design
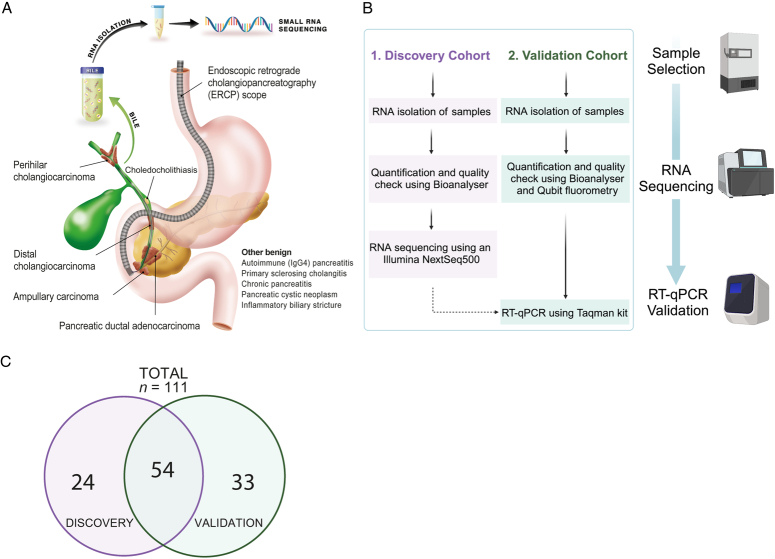
All study-related procedures and protocols were in accordance with the ethical guidelines of the Declaration of Helsinki and local ethical committees. Written informed consent was obtained from all individual subjects. The study is reported according to the Standars for the Reporting of Diagnostic accuracy studies (Supplemental Digital Content (SDC) 1, http://links.lww.com/JS9/D148, Figure S1; SDC 2, STARD Checklist, Supplemental Digital Content 2, http://links.lww.com/JS9/D149)25. We collected and analysed 111 bile samples from patients with benign and malignant pancreaticobiliary disease in this two-phase study (Fig. 1A-B), which consisted of biomarker discovery by small RNA sequencing and validation by RT-qPCR. Bile samples were allocated to the discovery or validation cohort based on sample availability Figure 1C.
Figure 1.
Study design of the MIRABILE study. (A) A graphical figure demonstrating the overall study design together with relevant pancreaticobiliary diseases. (B) Schematic diagram showing the workflow: RNA was isolated from bile samples, followed by quantification and then either RNA sequencing analysis or RT-qPCR. (C) Venn diagram showing the distribution of patients between the discovery and validation cohorts used in the analysis of bile cfRNA. RT-qPCR, quantitative reverse transcription polymerase chain reaction.
Sample collection and preparation
Bile was collected prospectively from consecutive patients attending the endoscopy unit between 2nd November 2017 and 11th March 2020. All patients were attending the endoscopic unit for evaluation of biliary obstruction. Inclusion criteria for patients recruited were age ≥18 years, able to provide written informed consent, scheduled for clinical reason to undergo an ERCP or a PTC, and WHO performance status <3. Exclusion criteria included patients with previous Roux-en-Y gastrojejunostomy, hepaticojejunostomy or Billroth II, pregnancy or those with WHO performance status 3 or 4. Definitions of benign and malignant, as well as procedures of sample collection and data collection are described in SDC 3, Materials and Methods (Supplemental Digital Content 3, http://links.lww.com/JS9/D150).
RNA isolation, analysis, library preparation, and quantification
Full detailed protocols for RNA isolation, library preparation and quantification are described in SDC 3, Materials and Methods (Supplemental Digital Content 3, http://links.lww.com/JS9/D150). Total RNA was isolated using TRIzol LS Reagent (Thermo Fisher Scientific) from 500 µl bile. RNA samples were prepared for sequencing using QIAseq small RNA Library Prep kit (Qiagen) and sequenced on a NextSeq 500 system (Illumina). Validation of candidate miRNAs was performed using Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) and target-specific stem–loop primer assays. CT values were calculated and normalised to endogenous miRNAs.
Statistical analysis
Small RNA-sequencing was used to identify differentially expressed miRNAs in bile. An adjusted P<0.05 was considered statistically significant. Receiver operating characteristic (ROC) curve analyses were performed for significantly upregulated miRNAs that showed the highest fold change (minimum log2(fold change) >2), leading to estimates of the area under the curve (AUC) with 95% CI. To prevent selection bias, all statistically significant miRNAs were included in Least Absolute Shrinkage and Selection Operator (LASSO) regression analysis. This type of regression analysis was chosen and performed in order to identify a parsimonious model26. It is a computational approach that uses L1 regularisation to combine potential candidates and reduce coefficients from a wide number of miRNA candidates to zero (i.e. adds penalty equivalent to the absolute value of the magnitude of coefficients)26. A selection of upregulated miRNAs (i.e. those that were considered upregulated and expressed in cancer may be more pragmatic to apply as cancer biomarker) was validated by RT-qPCR. Since strong collinearity between input variables (i.e. microRNAs) for logistic regression influences the accurate estimation of the miRNA model, Spearman’s Rank correlation analysis was performed for miRNAs and several laboratory markers. Next, multiple logistic regression analyses were performed to generate ROCs for potential diagnostic miRNAs and determine corresponding AUC values. Optimum cut-offs for sensitivity and specificity were determined from the ROC curve at the maximum Youden index.
MicroRNA functional analysis
To identify potential functional targets for the candidate miRNAs, gene set enrichment analysis (GSEA) tailored to miRNA was used to evaluate pathways, Gene Ontology terms, and targets27. Analyses were performed using the web-based application miRNA Enrichment Analysis and Annotation tool 2.1 (miEAA2.1). The input list of miRNAs was sorted in order of P-value from validation data. Validated miRNA-target interactions were obtained from miRWalk 2.0, HMDD2 and miRTarbase and converted to the current miRBase v21. P-values were computed by applying the Fisher’s exact test and adjusted for multiple comparisons (Bonferroni) with a standard significance threshold of P<0.05.
Results
Patients
Patient details are summarised in Table 1 and further described in SDC 4, Supplementary Results (Supplemental Digital Content 4, http://links.lww.com/JS9/D151). A total of 111 samples were collected prospectively from patients presenting for biliary drainage. A total of 78 samples (27 PDAC, 14 CCA, and 37 benign) were selected for the discovery cohort. The validation cohort comprised 87 samples (34 PDAC, 14 CCA, and 39 benign).
Table 1.
Clinicopathological characteristics of patients in the discovery and validation cohorts.
| Total (n=111) | Discovery (n=78) | Validation (n=87) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDAC (n=38) | CCA (n=19) | Benign (n=54) | P | PDAC (n=27) | CCA (n=14) | Benign (n=37) | P | PDAC (n=34) | CCA (n=14) | Benign (n=39) | P | |
| Age, years | ||||||||||||
| <60 | 8 (21) | 4 (21) | 24 (44) | 8 (30) | 3 (21) | 16 (43) | 6 (18) | 3 (21) | 18 (46) | |||
| >60 | 30 (79) | 15 (79) | 30 (56) | 0.0143 | 19 (70) | 11 (79) | 21 (57) | 0.1565 | 28 (82) | 11 (79) | 21 (54) | 0.0098 |
| Mean (SD) | 69 (11) | 68 (9) | 62 (16) | 0.0054 | 68 (11) | 65 (7.8) | 63 (16) | 0.1260 | 70 (11) | 70 (8) | 61 (16) | 0.0029 |
| Sex (%) | ||||||||||||
| Female | 13 (34) | 6 (32) | 21 (39) | 12 (44) | 4 (29) | 15 (41) | 11 (32) | 4 (29) | 14 (36) | |||
| Male | 25 (66) | 13 (68) | 33 (61) | 0.5596 | 15 (56) | 10 (71) | 22 (59) | 0.8254 | 23 (68) | 10 (71) | 25 (64) | 0.6557 |
| History of PSC (%) | 0 (0) | 1 (5) | 4 (7) | 0.2017 | 0 (0) | 0 (0) | 3 (8) | 0.1105 | 0 (0) | 1 (7) | 4 (10) | 0.1688 |
| Chronic Pancreatitis (%) | 4 (11) | 0 (0) | 11 (20) | 0.0540 | 3 (11) | 0 (0) | 7 (19) | 0.1858 | 4 (12) | 0 (0) | 9 (23) | 0.0723 |
| Blood parameters | ||||||||||||
| CA19-9 (U/ml) | 9341 | 3186 | 278 | 0.0770 | 7825 | 555 | 347 | 0.1023 | 10158 | 4298 | 287 | 0.0823 |
| Bilirubin (ng/ml) | 248 | 148 | 42 | <0.0001 | 244 | 110 | 46 | <0.0001 | 254 | 149 | 48 | <0.0001 |
| CRP (μg/ml) | 50 | 57 | 48 | 0.9643 | 49 | 58 | 53 | 0.4931 | 52 | 47 | 38 | 0.7892 |
| Anatomical location | ||||||||||||
| Perihilar CCA | — | 10 (53) | — | — | 9 (64) | — | — | 7 (50) | — | |||
| Distal CCA | — | 7 (37) | — | — | 5 (36) | — | — | 5 (36) | — | |||
| Intrahepatic CCA | — | 2 (11) | — | — | 0 (0) | — | — | 2 (14) | — | |||
| Head of Pancreas PDAC | 31 (82) | — | — | 21 (78) | — | — | 27 (79) | — | — | |||
| Uncinate PDAC | 3 (8) | — | — | 3 (11) | — | — | 3 (9) | — | — | |||
| Mixed PDAC (HOP/uncinated) | 4 (11) | — | — | 3 (11) | — | — | 4 (12) | — | — | |||
| Tumour stage* (%) | ||||||||||||
| T1 | 0 (0) | 0 (0) | — | 0 (0) | 0 (0) | — | 0 (0) | 0 (0) | — | |||
| T2 | 7 (18) | 3 (17) | — | 6 (22) | 3 (21) | — | 7 (21) | 1 (7) | — | |||
| T3 | 10 (26) | 10 (53) | — | 7 (26) | 8 (58) | — | 8 (24) | 7 (50) | — | |||
| T4 | 19 (50) | 6 (33) | — | 14 (52) | 3 (21) | — | 19 (56) | 6 (43) | — | |||
| Nodal disease (%) | 23 (61) | 8 (42) | — | 18 (66) | 6 (42) | — | 23 (68) | 5 (36) | — | |||
| Metastatic disease (%) | 11 (29) | 1 (6) | — | 6 (22) | 0 (0) | — | 11 (32) | 1 (7) | — | |||
*In accordance with the eight edition AJCC Cancer Staging Manual.
P-values <0.05 are highlighted in bold.
CA19-9, carbohydrate antigen 19-9; CCA, cholangiocarcinoma; CRP, C-reactive protein; HOP, head of pancreas; PDAC, pancreatic ductal adenocarcinoma.
Small RNA sequencing of bile cfRNA revealed differentially expressed bile miRNAs
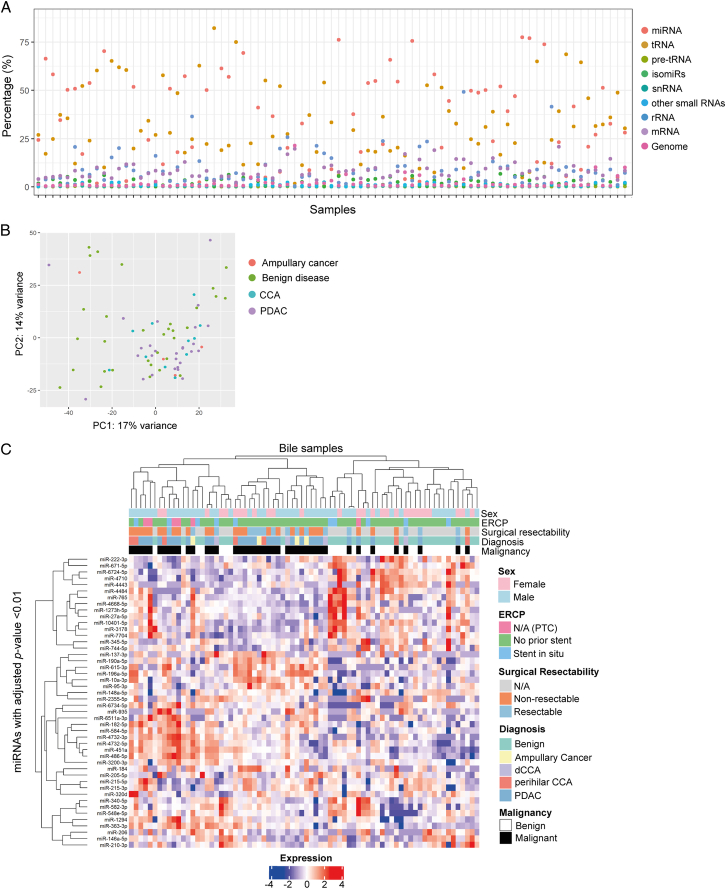
The composition of small noncoding RNAs that were present in bile samples of the discovery cohort is shown in Figure 2A. MiRNAs were a highly expressed species of RNA in human bile (mean proportion of reads was 36%). After filtering for reads, a total of 457 miRNAs were identified for differential expression analysis. Correlation of different samples according to miRNA expression values is shown in a principal component analysis (PCA) plot (Fig. 2B). For miRNAs with adjusted P-values <0.01, a heatmap was generated with unsupervised hierarchical clustering according to similar expression in samples (Fig. 2C).
Figure 2.
Bile cell-free RNA (cfRNA) demonstrated differences in RNA quantity and expression. (A) Relative distribution of small RNA reads for each bile sample from the discovery cohort (n=78). (B) Principal component analysis (PCA) plot of bile miRNA expression showing clustering of samples. (C) Heatmap with unsupervised hierarchical clustering showing malignant samples grouping to the left and including all miRNAs with adjusted P-values <0.01. CCA, cholangiocarcinoma; dCCA, distal CCA; ERCP, endoscopic retrograde cholangiopancreatography; mRNA, messenger RNA; N/A, not applicable; PC, principal component; PDAC, pancreatic ductal adenocarcinoma; PTC, percutaneous transhepatic cholangiography; rRNA, ribosomal RNA; snRNA, small nuclear RNA; tRNA, transfer RNA.
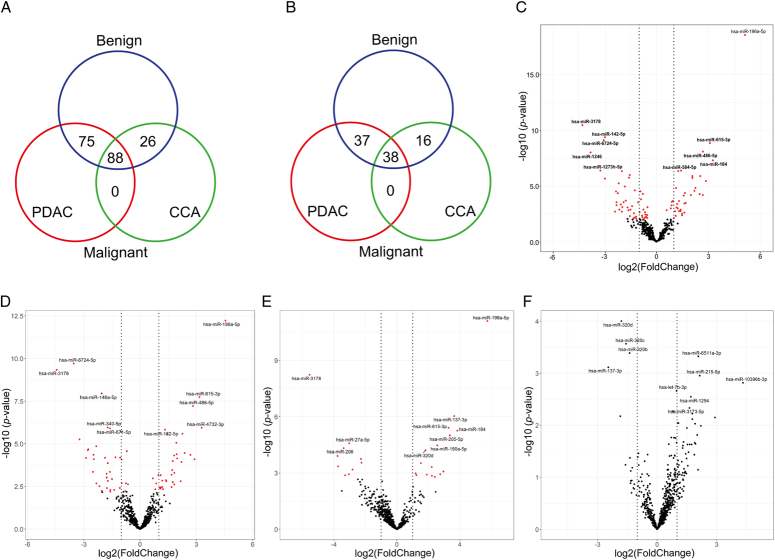
The numbers of statistically significantly (adjusted P<0.05) differentially expressed miRNAs for all pairwise comparisons are displayed in a Venn diagram in Figure 3A. From these dysregulated miRNAs, upregulated miRNAs may be to more pragmatic as clinical diagnostic biomarkers (i.e. considered upregulated and identifiable in samples). Upregulated miRNAs are hence shown in a second Venn diagram (Fig. 3B). When comparing malignant and benign disease, 88 miRNAs were significantly differentially expressed (Fig. 3C). The top 20 dysregulated miRNAs with highest fold change are shown in SDC 5, Table S1 (Supplemental Digital Content 5, http://links.lww.com/JS9/D152). Furthermore, 75 miRNAs showed differential expression for the pairwise comparison PDAC vs. benign disease (Fig. 3D; SDC 5, Table S2, Supplemental Digital Content 5, http://links.lww.com/JS9/D152), 26 for CCA vs. benign disease (Fig. 3E; SDC 5, Table S3, Supplemental Digital Content 5, http://links.lww.com/JS9/D152), and 0 for CCA vs. PDAC (Fig. 3F). Although analysis of transfer RNA fragment (tRF) expression was not the aim of this study, small RNA sequencing revealed that tRFs were highly expressed in our bile cfRNA samples and on average made up 37% of the small RNA composition in bile (Fig. 2A). In order to evaluate these for future investigations, differential expression analysis was performed using the available small RNA sequencing data (SDC 4, Supplementary Results, Supplementary Results, Supplemental Digital Content 4, http://links.lww.com/JS9/D151).
Figure 3.
Differential expression analysis identified differentially expressed miRNAs in bile samples. Venn diagrams for (A) the total number of differentially expressed miRNAs (adjusted P-value <0.05), and (B) the total number of upregulated miRNAs only when comparing PDAC, CCA, and benign cohorts. Volcano plots demonstrate differentially expressed miRNAs with the top 10 most statistically significant miRNAs annotated (upregulated and downregulated) for the pairwise comparisons: (C) malignant (PDAC and CCA) vs. benign disease; (D) PDAC vs. benign; (E) CCA vs. benign and (F) PDAC vs. CCA. Red illustrates miRNAs with FDR<0.05. Vertical dotted lines indicate log2(fold change)=+/− 1. CCA, cholangiocarcinoma; hsa, homo sapiens; PDAC, pancreatic ductal adenocarcinoma.
Validation of candidate miRNAs from LASSO regression analysis by RT-qPCR
All miRNAs with adjusted P<0.05 were used as input for LASSO regression analysis. Further details and results of LASSO regression analysis are described in SDC 4, Supplementary Results, Supplementary Results (Supplemental Digital Content 4, http://links.lww.com/JS9/D151). Following LASSO regression analysis, six upregulated miRNAs (i.e. those that were considered upregulated and expressed in cancer may be easier to apply as cancer biomarker) were selected as candidates for RT-qPCR validation: miR-196a-5p, miR-199a-5p, miR-335-5p, miR-182-5p, miR-340-5p and miR-424-5p. These six miRNAs were selected because a) they were all included in the 14-miRNA LASSO model that discriminated malignant from benign disease with an AUC of 0.93 (95% CI: 0.84–1.00), and b) the first four miRNAs were also included in the LASSO regression model that discriminated PDAC from benign disease. Endogenous miRNAs with stable expression were used to calculate the relative expression of candidate miRNAs. These normalisers were identified by applying the NormFinder algorithm to small RNA sequencing data (SDC 5, Table S4, Supplemental Digital Content 5, http://links.lww.com/JS9/D152)28. As a result, miR-26a and miR-let-7b were used as normalisers in subsequent RT-qPCR analyses.
During validation, two of the six miRNAs (miR-199a-5p and miR-424-5p) did not show adequate expression (>50% of samples showing no expression) and were not further assessed. Expression levels of the other four candidate miRNAs (miR-196a-5p, miR-182-5p, miR-335-5p, and miR-340-5p) were considered adequate for further analysis. Correlation analysis of these miRNAs is described in SDC 4, Supplementary Results, Supplementary Results (Supplemental Digital Content 4, http://links.lww.com/JS9/D151).
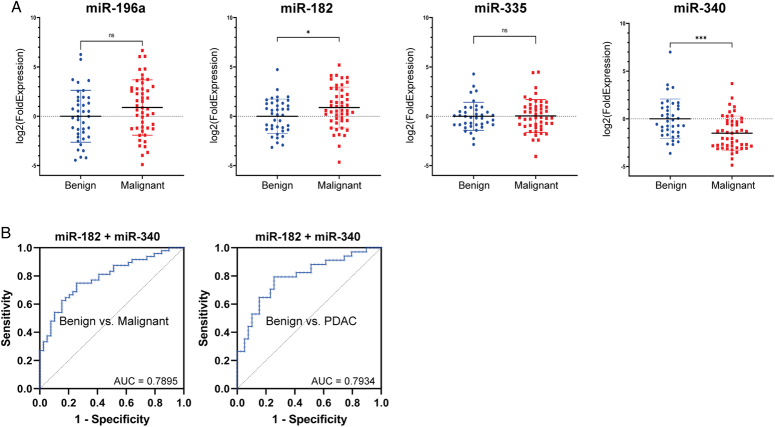
Differential expression of the four final candidate miRNAs were evaluated in the pairwise comparison of malignant (i.e. PDAC and CCA) vs. benign disease is shown in Figure 4A. MiR-182-5p was significantly upregulated in malignant samples (P=0.0179), while miR-340-5p was significantly downregulated in malignant samples (P=0.0002). MiR-196a-5p showed a trend towards upregulation in malignancy (P=0.0671) and miR-335-5p showed no significant difference between malignant and benign samples (P=0.4607). When PDAC and CCA were compared to benign disease individually, no statistically significant difference was shown for miR-182-5p, miR-196a-5p, and miR-335-5p. However, miR-340-5p was statistically significantly differentially expressed in both PDAC and CCA comparisons with benign disease (P=0.0275 and P=0.0007, respectively).
Figure 4.
RT-qPCR validation of candidate miRNAs identified by LASSO regression analysis. (A) Expression of candidate miRNAs in benign and malignant pancreaticobiliary disease (left to right): miR-196a, miR-182, miR-335 and miR-340. *P<0.05, **P<0.005, ***P<0.0005. (B) ROC curve with corresponding AUC value for the 2-miRNA signature (miR-182 and miR-340) to predict malignant disease vs. benign disease (left), or to predict PDAC vs. benign disease (right). AUC, area under the curve; PDAC, pancreatic ductal adenocarcinoma.
A bile miRNA signature with diagnostic potential to discriminate malignant from benign disease
Within the validation cohort, serum CA 19-9 values were missing for 25 out of 39 (64%) patients with benign disease, 4 out of 48 (8%) patients with malignant disease, and 2 out of 34 (6%) patients with PDAC. Logistic regression removes patients with missing values for CA 19-9, resulting in a smaller sample size and potential bias in effect estimates. ROC and AUCs were calculated for CA 19-9 only (SDC 1, Figure S2, http://links.lww.com/JS9/D148), but in order to generate the most reliable miRNA signature CA 19-9 was not included in logistic regression analysis with candidate miRNAs.
Multiple logistic regression was performed for the miRNA combination miR-182-5p and miR-340-5p, generating a ROC curve which showed an AUC of 0.79 (95% CI: 0.70–0.88) in predicting malignant vs. benign disease (Fig. 4B, left). At the optimum cut-off, determined by maximum Youden Index, sensitivity and specificity were 64.6 and 82.1%, respectively. In addition, the negative predictive value (NPV) was 65.3% and the positive predictive value (PPV) 81.6%. In discriminating PDAC from benign disease, the miRNA signature showed an AUC of 0.79 (95% CI: 0.69–0.90) with a sensitivity of 82% and specificity of 59% (Fig. 4B, right). Moreover, NPV and PPV were 79 and 64%, respectively.
Functional analysis of candidate miRNAs to identify common pathways
Fourty functional pathways were identified for all six candidates with an adjusted P-value <0.05. The top 10 is shown in SDC 5, Table S5 (Supplemental Digital Content 5, http://links.lww.com/JS9/D152). This analysis highlighted pathways including bone morphogenetic proteins (BMP) signalling, Toll-like receptor (TLR) pathways and involvement of the endosome. BMPs regulate developmental epithelial-to-mesenchymal transition (EMT) and there is evidence that there is a role for BMP signalling in promoting the metastatic cascade29.
Discussion
Differential diagnosis between malignant and benign disease in patients with obstructive jaundice is challenging. As bile is in close proximity to the tumour and can be easily obtained during ERCP, we have identified bile miRNAs as compelling diagnostic biomarkers in the differentiation of patients with malignant and benign biliary strictures. This study is the largest study to date to identify miRNAs from bile using small RNA sequencing (see SDC 5, Table S6, Supplemental Digital Content 5, http://links.lww.com/JS9/D152, for an overview of previous studies on bile miRNAs). In total, 111 unique patient samples were used in this study, with a sequencing strategy to determine miRNA (and tRF candidates) from a discovery cohort of 78 samples (27 PDAC, 14 CCA, and 37 Benign). We were able to investigate all 2656 mature known human miRNAs (miRBase v22) and all 26 531 known tRNA fragments (MINTbase v2.0) in bile, indicating that bile is an ample source for potential RNA markers. A total of 457 miRNAs were identified with sufficient expression for analysis. After multiple hypotheses correction, 88 differentially expressed miRNAs were identified when comparing malignant and benign disease, 75 when comparing PDAC and benign disease and 26 when comparing CCA and benign disease. Using LASSO regression analysis, a sparse model was generated which included 14 miRNAs that could discriminate benign from malignant disease with an AUC of 0.933. Six miRNAs were chosen for validation RT-qPCR analysis, which confirmed a significant upregulation of miR-182-5p and downregulation of miR-340-5p in malignant samples. In combination, they showed an AUC value of 0.79 (95% CI: 0.70–0.88, P<0.0001) for predicting malignancy, with a sensitivity of 64.6% and of specificity 82.1%, and an AUC of 0.79 (95% CI: 0.69–0.90) for predicting PDAC, with a sensitivity of 82% and specificity of 59%. Compared to the CancerSEEK test, a blood test that assesses circulating proteins and mutations in cell-free DNA for early cancer detection, our miRNA signature showed a higher sensitivity for detecting PDAC (82 vs. 70%)30. A previous study already showed that miRNAs can also be assessed in the bile obtained from the gallbladder by surgical aspiration18. The study used nCounter Nanostring profiling to identify candidate miRNAs from surgically aspirated bile, and concluded that bile-based miR-148a-3p, miR-125b-5p, and miR-194-5p could be used to distinguish pancreaticobiliary disease. Compared to this previous study, we performed discovery by small RNA sequencing of a wider range of miRNAs (all known miRNAs according to miRbase v22, which includes 2656 mature miRNAs), and collected bile directly from the bile duct in a preoperative setting during ERCP. Therefore, we were able to obtain candidates that were different, but with greater predictive ability for pancreaticobiliary malignancy (AUC of 0.752 using miR-148a-3p compared with 0.796 using miR-340 and miR-182). No significantly differentially expressed miRs were identified comparing PDAC to CCA, which may be due to the small samples size for CCA (n=14), which contributes to the probability of a type II error.
Functional analysis of the candidate miRNAs indicated involvement in BMP and TLR pathways. Several BMP family members, in particular BMP 2 and 4, have been shown to induce EMT through increased matrix metalloproteinase (MMP)-2 and suppression of Smad1 in Panc-1 cells29. Furthermore, TLRs are known to be upregulated in PDAC and play a role in the antitumour immune response and inflammation through the recognition of pathogen and danger-associated molecular patterns31. Indeed, TGF-β induced miR-182 has been shown to modulate the TLR4/NF-κB signalling pathways, indicating its role in tumour progression32,33. Upregulation of our validated miR-182 has previously also been described in PDAC compared to chronic pancreatitis in blood, and in CCA compared to chronic cholangitis in cells from bile cytology34,35. Similarly, miR-340 has previously been shown to be downregulated in PDAC, and upregulation can be shown to inhibit tumour growth by influencing the tumour immune microenvironment in murine models36.
Surprisingly, small RNA sequencing also revealed that tRNA-derived fragments (tRFs) were highly expressed in bile and on average tRNAs composed 37% of the assessed bile samples. tRNA biology is complex with over 270 different tRNA sequences present among approximately 450 tRNA human genes37. It undergoes several chemical modifications which can alter its fragmentation and function38. Using high-throughput sequencing, several studies have reported on differential expression of tRFs in cancer, but also emphasised that further extensive investigations are warranted to understand the roles and underlying mechanisms of these RNA fragments38. Although this study focused on miRNAs, our preliminary findings may contribute to future studies into the potential roles of tRFs in pancreaticobiliary cancer.
Our study contained several limitations. Firstly, an independent validation in a separate cohort would provide stronger evidence for our model and would strengthen implementation in clinical practice. Secondly, a prediagnostic cohort of patients presenting with biliary obstruction was investigated, which meant that patients presented to our study with advanced disease. Furthermore, LASSO regression is a useful tool to identify biomarker signatures, but it does not take into account functional relationships. Correlation analysis did, however, show low correlation between candidate miRNAs and current laboratory markers, indicating a potential cancer-related role distinct from inflammation or obstruction. Our methodology also had a number of notable strengths. Firstly, we used an NGS approach for biomarker discovery. NGS is the preferred standard for miRNA biomarker discovery because it does not cause a priori selection bias39. Secondly, a relatively large cohort of patients was sequenced (n=78), after which results were also validated in a second cohort by RT-qPCR. Moreover, this is the first study to analyse tRFs in bile for diagnosis of PDAC and CCA38.
This study was designed for the hypothesis that bile-based microRNAs could act as an adjunct to help clinical decision making. Future studies could assess the diagnostic performance of a model including routinely measured biomarkers (e.g. CA 19-9 and bilirubin), bile-based miRNAs, and other novel markers (e.g. radiomics-base machine learning models, circulating tumour DNA, and microbiome)40–44. Furthermore, RNA sequencing in patients with PDAC has previously been used to develop promising tissue based transcriptomic predictive signatures for response to gemcitabine (GemCore) or FOLFIRINOX (single sample classifier PurIST)45,46. Possibly, bile-based miRNAs could contribute to such predictive signatures. Therefore, in our ongoing prospective multicentric clinical trial, RESECTA-BILE, we are studying bile miRNAs to validate their diagnostic potential, but also to investigate their potential roles as prognostic and predictive biomarkers for patients with resectable or borderline resectable PDAC.
Conclusion
In conclusion, there is differential expression of miRNAs in bile associated with malignant pancreaticobiliary disease. This is the first study to assess bile miRNAs by small RNA-sequencing in a large number of patients presenting with obstructive jaundice. This study indicates that bile miRNAs could be clinically useful biomarkers for confirming malignant pancreaticobiliary disease. This opens the possibility that these bile miRNAs could be used during a ‘window of opportunity’ prior to anticancer treatment to help stratify and personalise patient pathways. The final diagnostic model with miR-182-5p and miR-340-5p generated an AUC of 0.79 for the detection of malignancy, and 0.79 when used for the prediction of PDAC only. Functional target analysis showed that these two miRNAs target BMP signalling, indicating that they could be associated with EMT and tumour progression. Thus, our findings demonstrated that bile cell-free miRNAs can be a source of biomarkers and potential adjuncts for the detection of PDAC and CCA.
Ethical approval
All study-related procedures and protocols were in accordance with the ethical guidelines of the Declaration of Helsinki. Ethical approval was obtained as part of the Imperial College Healthcare Tissue Bank (ICHTB). ICHTB was approved on July 25th 2017 by Wales REC3 to release human material for research (ICHTB HTA licence 12275; REC Wales approval 17/WA/0161).
Consent
Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
Source of funding
This research was funded by The Jon Moulton Charity Trust, UK (AEF); Action Against Cancer, UK (DKSL, MMP, ER, BP, LRJ, JK AEF); the Royal College of Surgeons of Edinburgh, UK (AEF); the Royal College of Surgeons of England, UK (AEF, DSKL); the Mason Medical Research Foundation, UK (AEF); the S.A.L. Charitable Fund, UK (AEF); the Bennink Foundation, the Netherlands (JRP, GK, EG); Italian Association for Cancer Research AIRC, Italy (EG), Fondazione Pisana Per La Scienza, Italy (EG); and the Dutch Cancer Society KWF (RJS, EG, GK). These sponsors did not play a role in the collection, analysis or interpretation of data; nor in the writing of the manuscript; nor in the decision to submit the manuscript for publication.
Author contribution
D.G., G.C., N.P., C.W., P.V., J.P., S.S., B.R.D., L.R.J., J.K., and A.E.F.: sample provision; D.S.K.L. and A.E.F.: clinical data collection; D.S.K.L., J.R.P., A.E.F., and E.G.: original draft preparation; A.E.F., J.K., E.G., and G.K.: funding acquisition. All authors contributed in concept and design, read and approved the final manuscript, and statistical analysis and interpretation, and critical revision and editing of the manuscript.
Conflicts of interest disclosure
The authors have no disclosures for this work.
Research registration unique identifying number (UIN)
The unique identifying number that was issued by ClinicalTrials.gov is NCT06258824.
Guarantor
All authors accept full responsibility for the work and/or the conduct of the study. Adam Frampton and Elisa Giovannetti controlled the decision to publish. Adam Frampton, Dr. Daniel Liu and Dr. Jisce Puik had access to the data.
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Provenance and peer review
Not applicable.
Supplementary Material
Acknowledgements
Assistance with the study: none.
Footnotes
Daniel S.K. Liu and Jisce R. Puik contributed equally to this work as co-first authors.
Jonathan Krell and Adam E. Frampton contributed equally to this work as co-senior authors.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.lww.com/international-journal-of-surgery.
Published online 23 July 2024
Contributor Information
Daniel S.K. Liu, Email: Daniel.liu08@imperial.ac.uk.
Jisce R. Puik, Email: jiscepuik@gmail.com;j.puik@amsterdamumc.nl.
Morten T. Venø, Email: morten.veno@omiics.com.
Mireia Mato Prado, Email: m.prado@ucl.ac.uk.
Eleanor Rees, Email: Erees0103@gmail.com.
Bhavik Y. Patel, Email: bhavik.patel@surrey.ac.uk.
Nabeel Merali, Email: nabeel.merali@gmail.com.
Daniel Galloway, Email: Daniel.galloway1@nhs.net.
Grace Chan, Email: gracepschan@gmail.com.
Natalie Phillips, Email: Natalie.phillips2@nhs.net.
Christopher Wadsworth, Email: Christopher.wadsworth@nhs.net.
Panagiotis Vlavianos, Email: Panagiotis.vlavianos@nhs.net.
Jonathan Potts, Email: jonathan.potts@nhs.net.
Shivan Sivakumar, Email: S.sivakumar@bham.ac.uk.
Brian R. Davidson, Email: b.davidson@ucl.ac.uk.
Marc G. Besselink, Email: m.g.besselink@amsterdamumc.nl.
Rutger-Jan Swijnenburg, Email: r.j.swijnenburg@amsterdamumc.nl.
Long R. Jiao, Email: l.jiao@imperial.ac.uk.
Geert Kazemier, Email: g.kazemier@amsterdamumc.nl.
Elisa Giovannetti, Email: elisa.giovannetti@gmail.com.
Jonathan Krell, Email: j.krell@imperial.ac.uk.
Adam E. Frampton, Email: adam.frampton@surrey.ac.uk.
References
- 1. Singh A, Gelrud A, Agarwal B. Biliary strictures: diagnostic considerations and approach. Gastroenterol Rep (Oxf) 2015;3:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burnett AS, Calvert TJ, Chokshi RJ. Sensitivity of endoscopic retrograde cholangiopancreatography standard cytology: 10-y review of the literature. J Surg Res 2013;184:304–311. [DOI] [PubMed] [Google Scholar]
- 3. Bowlus CL, Olson KA, Gershwin ME. Evaluation of indeterminate biliary strictures. Nat Rev Gastroenterol Hepatol 2016;13:28–37. [DOI] [PubMed] [Google Scholar]
- 4. Gerritsen A, Molenaar IQ, Bollen TL, et al. Preoperative characteristics of patients with presumed pancreatic cancer but ultimately benign disease: a multicenter series of 344 pancreatoduodenectomies. Ann Surg Oncol 2014;21:3999–4006. [DOI] [PubMed] [Google Scholar]
- 5. Abraham SC, Wilentz RE, Yeo CJ, et al. Pancreaticoduodenectomy (Whipple Resections) in patients without malignancy: are they all ‘Chronic Pancreatitis’? Am J Surg Pathol 2003;27:110–120. [DOI] [PubMed] [Google Scholar]
- 6. Poruk KE, Gay DZ, Brown K, et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med 2013;13:340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rösch T, Meining A, Frühmorgen S, et al. A prospective comparison of the diagnostic accuracy of ERCP, MRCP, CT, and EUS in biliary strictures. Gastrointest Endosc 2002;55:870–876. [DOI] [PubMed] [Google Scholar]
- 8. Yu XR, Huang WY, Zhang BY, et al. Differentiation of infiltrative cholangiocarcinoma from benign common bile duct stricture using three-dimensional dynamic contrast-enhanced MRI with MRCP. Clin Radiol 2014;69:567–573. [DOI] [PubMed] [Google Scholar]
- 9. Suthar M, Purohit S, Bhargav V, et al. Role of MRCP in differentiation of benign and malignant causes of biliary obstruction. J Clin Diagn Res 2015;9:Tc08–Tc12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim JY, Lee JM, Han JK, et al. Contrast-enhanced MRI combined with MR cholangiopancreatography for the evaluation of patients with biliary strictures: Differentiation of malignant from benign bile duct strictures. J Magn Reson Imaging 2007;26:304–312. [DOI] [PubMed] [Google Scholar]
- 11. Kim MJ, Mitchell DG, Ito K, et al. Biliary dilatation: differentiation of benign from malignant causes--value of adding conventional MR imaging to MR cholangiopancreatography. Radiology 2000;214:173–181. [DOI] [PubMed] [Google Scholar]
- 12. Navaneethan U, Njei B, Lourdusamy V, et al. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: a systematic review and meta-analysis. Gastrointest Endosc 2015;81:168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li YC, Li KS, Liu ZL, et al. Research progress of bile biomarkers and their immunoregulatory role in biliary tract cancers. Front Immunol 2022;13:1049812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voigtländer T, Metzger J, Husi H, et al. Bile and urine peptide marker profiles: access keys to molecular pathways and biological processes in cholangiocarcinoma. J Biomed Sci 2020;27:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li L, Masica D, Ishida M, et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology 2014;60:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu F, Hao X, Liu B, et al. Bile liquid biopsy in biliary tract cancer. Clin Chim Acta 2023;551:117593. [DOI] [PubMed] [Google Scholar]
- 17. Meijer LL, Puik JR, Le Large TYS, et al. Unravelling the diagnostic dilemma: a MicroRNA panel of circulating MiR-16 and MiR-877 as a diagnostic classifier for distal bile duct tumors. Cancers (Basel) 2019;11:1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mato Prado M, Puik JR, Castellano L, et al. A bile-based microRNA signature for differentiating malignant from benign pancreaticobiliary disease. Exp Hematol Oncol 2023;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
- 21. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sohail AM, Khawar MB, Afzal A, et al. Multifaceted roles of extracellular RNAs in different diseases. Mil Med Res 2022;9:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaporozhchenko IA, Ponomaryova AA, Rykova EY, et al. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn 2018;18:133–145. [DOI] [PubMed] [Google Scholar]
- 24. Verhoeven CJ, Farid WR, de Ruiter PE, et al. MicroRNA profiles in graft preservation solution are predictive of ischemic-type biliary lesions after liver transplantation. J Hepatol 2013;59:1231–1238. [DOI] [PubMed] [Google Scholar]
- 25. Bossuyt, Reitsma PM, Bruns DE JB, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tibshirani R. Regression shrinkage and selection via the Lasso. J Royal Statist Soc Series B-Methodological 1996;58:267–288. [Google Scholar]
- 27. miEEA 2.1. Accessed 8 July 2023, https://ccb-compute2.cs.uni-saarland.de/mieaa/
- 28. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004;64:5245–5250. [DOI] [PubMed] [Google Scholar]
- 29. Gordon KJ, Kirkbride KC, How T, et al. Bone morphogenetic proteins induce pancreatic cancer cell invasiveness through a Smad1-dependent mechanism that involves matrix metalloproteinase-2. Carcinogenesis 2009;30:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaz J, Andersson R. Intervention on toll-like receptors in pancreatic cancer. World J Gastroenterol 2014;20:5808–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma C, He D, Tian P, et al. miR-182 targeting reprograms tumor-associated macrophages and limits breast cancer progression. Proc Natl Acad Sci USA 2022;119:e2114006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu M, Li Y, Sun K. MicroRNA-182-5p inhibits inflammation in LPS-treated RAW264.7 cells by mediating the TLR4/NF-κB signaling pathway. Int J Clin Exp Pathol 2018;11:5725–5734. [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Q, Yang L, Xiao Y, et al. Circulating microRNA-182 in plasma and its potential diagnostic and prognostic value for pancreatic cancer. Med Oncol 2014;31:225. [DOI] [PubMed] [Google Scholar]
- 35. Uchihata Y, Arihiro K, Kaneko Y, et al. Analysis of MicroRNA in Bile cytologic samples is useful for detection and diagnosis of extrahepatic cholangiocarcinoma. Am J Clin Pathol 2022;158:122–131. [DOI] [PubMed] [Google Scholar]
- 36. Xi Q, Zhang J, Yang G, et al. Restoration of miR-340 controls pancreatic cancer cell CD47 expression to promote macrophage phagocytosis and enhance antitumor immunity. J Immunother Cancer 2020;8:e000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 2009;37(Database issue):D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fu M, Gu J, Wang M, et al. Emerging roles of tRNA-derived fragments in cancer. Mol Cancer 2023;22:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coenen-Stass AML, Magen I, Brooks T, et al. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol 2018;15:1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boyd LNC, Ali M, Comandatore A, et al. Prediction model for early-stage pancreatic cancer using routinely measured blood biomarkers. JAMA Network Open 2023;6:e2331197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol 2020;17:53–64. [DOI] [PubMed] [Google Scholar]
- 42. Groot VP, Mosier S, Javed AA, et al. Circulating tumor DNA as a clinical test in resected pancreatic cancer. Clin Cancer Res 2019;25:4973–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mukherjee S, Patra A, Khasawneh H, et al. Radiomics-based machine-learning models can detect pancreatic cancer on prediagnostic computed tomography scans at a substantial lead time before clinical diagnosis. Gastroenterology 2022;163:1435–1446.e3. [DOI] [PubMed] [Google Scholar]
- 44. Merali N, Chouari T, Terroire J, et al. Bile microbiome signatures associated with pancreatic ductal adenocarcinoma compared to benign disease: a UK pilot study. Int J Mol Sci 2023;24:16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fraunhoffer N, Chanez B, Teyssedou C, et al. A Transcriptomic-Based Tool to Predict Gemcitabine Sensitivity in Advanced Pancreatic Adenocarcinoma. Gastroenterology 2023;164:476–480.e4. [DOI] [PubMed] [Google Scholar]
- 46. Rashid NU, Peng XL, Jin C, et al. Purity independent subtyping of tumors (PurIST), a clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clin Cancer Res 2020;26:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.