Abstract
All primate lentiviruses known to date contain one or two open reading frames with homology to the human immunodeficiency virus type 1 (HIV-1) vpr gene. HIV-1 vpr encodes a 96-amino-acid protein with multiple functions in the viral life cycle. These functions include modulation of the viral replication kinetics, transactivation of the long terminal repeat, participation in the nuclear import of preintegration complexes, induction of G2 arrest, and induction of apoptosis. The simian immunodeficiency virus (SIV) that infects African green monkeys (SIVagm) contains a vpr homologue, which encodes a 118-amino-acid protein. SIVagm vpr is structurally and functionally related to HIV-1 vpr. The present study focuses on how three specific functions (transactivation, induction of G2 arrest, and induction of apoptosis) are related to one another at a functional level, for HIV-1 and SIVagm vpr. While our study supports previous reports demonstrating a causal relationship between induction of G2 arrest and transactivation for HIV-1 vpr, we demonstrate that the same is not true for SIVagm vpr. Transactivation by SIVagm vpr is independent of cell cycle perturbation. In addition, we show that induction of G2 arrest is necessary for the induction of apoptosis by HIV-1 vpr but that the induction of apoptosis by SIVagm vpr is cell cycle independent. Finally, while SIVagm vpr retains its transactivation function in human cells, it is unable to induce G2 arrest or apoptosis in such cells, suggesting that the cytopathic effects of SIVagm vpr are species specific. Taken together, our results suggest that while the multiple functions of vpr are conserved between HIV-1 and SIVagm, the mechanisms leading to the execution of such functions are divergent.
The vpr gene from human immunodeficiency virus type 1 (HIV-1) encodes a 96-amino-acid protein with multiple functions in the viral life cycle. The first reported role for vpr was a moderate transactivation effect on the viral promoter, the long terminal repeat (LTR) (13). HIV-1 mutants with deletions in vpr replicate with slower kinetics than wild-type viruses do (5, 14). Vpr is encapsidated into virions in significant amounts (1, 12, 57). The presence of vpr in the viral particle facilitates efficient infection of macrophages and other nondividing cells (14, 17, 23) by mediating active nuclear import of preintegration complexes (16, 44). In addition, the presence of vpr enhances the transcriptional activity of the viral LTR in macrophages and T cells, allowing the production of a larger viral progeny (18, 52).
HIV-1 vpr contributes to the multiple cytopathic effects induced by HIV-1 by inducing cell cycle arrest in G2 (22, 27, 45, 46) and apoptosis (11, 49, 50, 55). The primate immunodeficiency virus strain, SIVagm, encodes an accessory gene, vprAGM, which bears sequence (31% amino acid identity) and functional conservation with vprHIV-1. At a functional level, vprAGM shares with vprHIV-1 its abilities to transactivate the viral LTR (39), cause cell cycle arrest in G2 (42, 51), and become incorporated in the virion particles for subsequent participation in nuclear import of preintegration complexes (1, 8).
Earlier studies have suggested that the HIV-1 and SIVagm vpr genes are functionally conserved in virion encapsidation, nuclear localization, cell cycle arrest, and effect on viral replication kinetics. However, noticeable differences have also been reported. Deletion of SIVagm vpr appears to have a more profound effect on viral replication in primary macrophages and peripheral blood mononuclear cells than does deletion of HIV-1 vpr. SIVagm vpr mutants are incapable of replication in nondividing cells, while HIV-1 vpr mutants are able to replicate at low level. To explain this difference, it was suggested by Campbell and Hirsch that HIV-1 contains redundant nuclear localization signals in addition to Vpr, such as the matrix protein, and perhaps that is not the case for SIVagm (8).
The cause-effect relationships among the different functions of vpr are currently being investigated. The present study focuses on how transactivation, G2 arrest, and apoptosis are related at a functional level, for both vprAGM and vprHIV-1.
MATERIALS AND METHODS
Cell lines.
The human breast cancer cell line SKBR3 was cultured in RPMI 1640 medium (BioWhittaker, Walkersville, Md.) containing 10% fetal calf serum (FCS) (Omega Scientific, Inc., Bedford, Ohio). African green monkey kidney Cos-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) supplemented with 10% FCS. The human embryonic 293T cells were propagated in Iscove's modified Dulbecco's medium (BioWhittaker) plus 10% FCS. The human cervical cancer cell line HeLa was maintained in DMEM with 5% FCS.
Plasmid constructs.
Mammalian vectors directing the expression of thy-1 and a vpr gene, vprHIV-1, vprHIV-1-FS, or vprAGM, were described previously (42). A vector expressing thy-1 and vprAGM-FS was constructed by digesting the vprAGM expression vector with the restriction endonuclease BsrGI, treating it with Klenow to fill the ends, and religating. This treatment generated a frameshift at codon 85. The vector, pCMV-thy was described earlier (41).
(i) Vectors for luciferase expression.
LTRHIV-1-Luc was constructed by subcloning the 5′ LTR of HIV-1NL4-3 (2) into pBluescript II KS(+) (Invitrogen, Carlsbad, Calif.) using AvrII and HindIII. A fragment containing the luciferase gene followed by the simian virus 40 polyadenylation sequences was cloned downstream from the LTR using XhoI and XbaI. LTRAGM-Luc was constructed by replacing the HIV-1 LTR in the previous construct by that of pSIVagm677 (26), using HindIII and XbaI.
(ii) Plasmids for production of defective lentivirus vectors.
The envelope construct HCMV-VSVG (7) and the packaging construct pCMVΔR8.2-Δvpr (4) were described previously. D102 is a defective HIV-1-derived lentivirus vector that is described in detail elsewhere (Y. Zhu and V. Planelles, submitted for publication). Briefly, D102-mHSA was derived from the previously described vector HIV-GFP (49) as follows. A 484-bp PflMI-SalI fragment from HIV-GFP was replaced by a 630-bp fragment from pNL-r-HSAS (25) using the same restriction enzymes. This resulted in replacement of the vpr open reading frame with that of the murine heat-stable antigen (mHSA) and also in the inclusion of a unique restriction site, XbaI, for subsequent cloning of additional genes replacing the gene encoding mHSA. D102-vprH and D102-vprA were constructed by replacing mHSA with vprHIV-1 or vprAGM, respectively, using the newly introduced XbaI site and a natural site, EcoRI, within the vpr remnant.
(iii) Construction of green fluorescent protein (GFP) fusions.
pEGFP-N1 (Clontech, Palo Alto, Calif.) was digested with BglII and BamHI and religated to create pEGFP-bam/bgl-. A ligation was then performed to create pGFP-flex with the following three DNA fragments: an NheI-BsrGI fragment from pEGFP-bam/bgl-, a BamHI-NheI fragment from pcDNA-2, and a flexible linker, which was from pUC-flex (10), digested with BsrGI and BamHI. To construct a GFP-vprHIV-1 expression plasmid, an NcoI-XbaI fragment from BSVpr-Thy (42) was subcloned into pGFP-flex digested with XhoI and XbaI. In the previous subcloning, the BsrGI and XhoI sites were filled with Klenow. A GFP-vprAGM expression plasmid was made by subcloning the SIVagm vpr cDNA from the vector, BS-677X-Thy (42), using the same restriction sites and recipient plasmid as for its HIV-1 counterpart.
Transfections and luciferase assays.
Transient transfection of cells for luciferase measurement was performed using the TransFast reagent (Promega Corp., Madison, Wis.) as recommended by the manufacturer. Cells were plated at a density of 105/well in 12-well plates. One day later, 0.25 μg of LTRHIV-1-Luc or LTRAGM-Luc reporter plasmid and 0.25 μg of a vpr expression plasmid were mixed with 1.5 μl of TransFast reagent in 0.2 ml of serum-free RPMI 1640 (SKBR3 cells) or DMEM (Cos-7 cells), and the mixture was incubated at room temperature for 30 min. The mixture was then added to the cells and incubated at 37°C for 1 h, and then 2 ml of RPMI 1640 with 10% FCS was added. The control plasmid was pCMV-thy. After 72 h, the cells were lysed and assayed for luciferase activity with a commercially available luciferase assay kit (Promega Corp.), using a LumiCount microplate reader (Packard Instrument Corp., Meriden, Conn.). The luciferase assay was performed using the following settings: PMT, 1,100 V; gain level, 5.0; and read length, 0.5 s. The background of the luciferase reading was 25 ± 15 light units for six measurements of lysates from uninfected cells. Each experiment was performed in triplicate, and each measurement was the average of duplicate readings. Luciferase light units were normalized to 1 μg of protein content (Bio-Rad Laboratories, Hercules, Calif.) in cell lysates. Light units in experimental wells were converted to fold increase with respect to control wells.
Viral vector production and titer determination.
Lentivirus vectors were produced by transient transfection of 293-T cells. D102-mHSA, D102-vprH, or D102-vprA was cotransfected with HCMV-VSVG and pCMV-ΔR8.2Δvpr using the calcium phosphate-mediated transfection. Virus was collected at 48, 72, 96 h posttransfection. The harvested supernatants were precleared by low-speed centrifugation at 2,000 rpm in a Sorvall RT7 with an RTH-750 rotor (Sorvall) and pelleted by ultracentrifugation at 25,000 rpm in a Discovery 100S centrifuge with a Surespin 630 rotor (Sorvall, Newton, Conn.). Virus pellets were resuspended in fresh tissue culture medium and frozen at −80°C. Vector titers were measured by infection of HeLa cells as described below, followed by flow cytometric analysis of cells positive for the reporter molecule, GFP. Vector titers were calculated as follows: titer = (F × C0 / V) × D, where F is the frequency of GFP-positive cells by flow cytometry, C0 is the total number of target cells at the time of infection; V is the volume of inoculum, and D is the virus dilution factor. The virus dilution factor (D) used for titer determinations was 10. The total number of target cells at the time of infection was 106.
Flow cytometry.
Cells were harvested and analyzed by direct immunofluorescence for GFP expression and with propidium iodide to analyze the DNA content. The cells were detached with 2 mM EDTA, washed in phosphate-buffered saline (PBS), fixed with 0.2% paraformaldehyde in PBS for 1 h, and stained with propidium iodide solution (20 μg of propidium iodide per ml, 0.2% Triton X-100, and 11.25 Kunitz units of RNase A per ml in PBS). The percentage of GFP-positive cells in G2/M was assessed and compared to that of untreated control cells. Flow cytometric analysis was performed in an Epics Elite ESP analyzer (Coulter Corp., Hialeah, Fla.). Gates for detection of GFP were established using mock-infected cells as background. Because electronic settings varied from experiment to experiment, gates were defined such that the percentage of false-positive events was not higher than 0.3% in the mock-infected population. Cell cycle analysis was performed using Multicycle AV software (Phoenix Flow Systems, San Diego, Calif.).
TdT-mediated dUTP-biotin nick end labeling (TUNEL).
To detect in situ apoptosis, the TdT-FragEL kit (Oncogene Research Products, Cambridge, Mass.) was used as specified by the manufacturer. Cells were seeded at 2 × 104 cells/well in chamber slides (Nalge Nunc International, Rochester, N.Y.). At 48 h following infection, cells were inspected for GFP expression to measure the level of infection. At 72 h postinfection, medium was removed and the cells were fixed with 4% formaldehyde (in PBS) for 10 min, washed with 80% ethanol, and stored overnight at −20°C. After being washed with Tris-buffered saline (TBS), the cells were permeabilized with proteinase K at room temperature for 5 min and washed again with TBS. The cells were then treated with 30% H2O2 in methanol for 5 min to inactivate endogenous peroxidases. Following application of equilibration buffer, the cells were incubated with a reaction mixture containing terminal deoxynucleotidyltransferase (TdT)- and biotin-labeled dUTP for 1.5 h in a humidified chamber at 37°C. The reaction was stopped by the supplied stop buffer. After the samples were washed with TBS, streptavidin-horseradish peroxidase conjugate was applied in a humidified chamber for 30 min. The cells were then counterstained with methyl green.
Drugs.
Caffeine was purchased from Sigma Chemical Co., and dissolved in water at 50 mM, and kept frozen at −80°C. Taxol (paclitaxel; Sigma Chemical Co.) was dissolved in dimethyl sulfoxide (Fisher Scientific, Fair Lawn, N.J.) at 7 mM and kept frozen at −80°C.
RESULTS
Induction of G2 arrest by vprAGM and vprHIV-1 differs in human and African green monkey cells.
We previously described a transient-transfection assay for the study of cell cycle effects by vprHIV-1 and related genes from primate lentiviruses (42). This method is based on the use of dual expression vectors which direct the expression of vpr and a reporter gene, murine thy-1 (Fig. 1A). Transfected cells are analyzed by direct immunofluorescence for Thy-1 surface expression and with propidium iodide for DNA content (see Materials and Methods) using flow cytometry. For simplicity, only the cell cycle profiles of Thy-1-positive cells (transfected) cells are shown (Fig. 2).
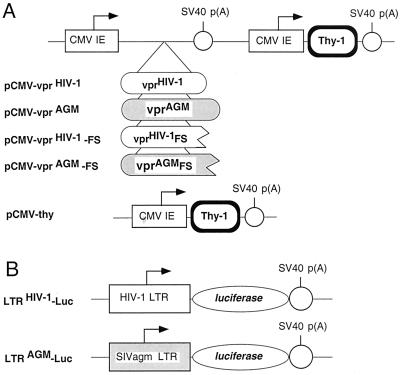
FIG. 1.
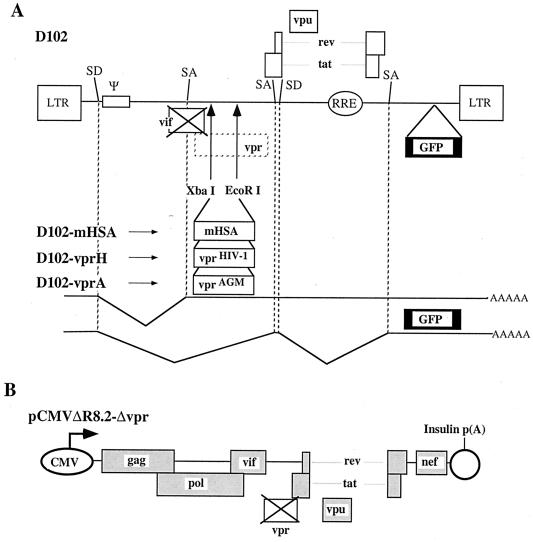
Expression vectors used for analysis of cell cycle distribution and transactivation. (A) Dual expression vectors for vpr variants and the reporter gene, thy-1. Arrows denote promoters. SV40 p(A), simian virus 40 polyadenylation signal; CMV IE, human cytomegalovirus immediate-early promoter; vprHIV-1-FS and vprAGM-FS, carboxy-terminal truncations of vprHIV-1 and vprAGM, generated by introducing frameshift mutations at codons 64 and 85, respectively. (B) Luciferase reporter vectors in which the viral LTR serve as a basal promoters.
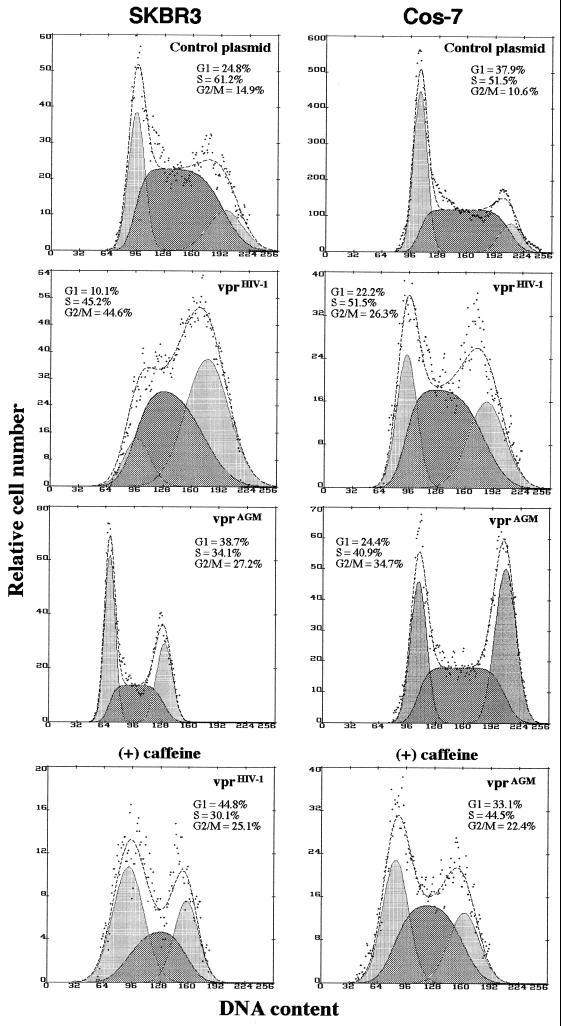
FIG. 2.
Flow cytometric analysis of vpr-induced G2 arrest. Cells were transfected with control plasmid (pCMV-thy) or dual expression vectors encoding the murine thy-1 gene and the indicated vpr version (top of each histogram). At 48 h after transfection, the cells were analyzed for thy-1 expression and DNA content. Where indicated, the cells were treated by adding 2 mM caffeine to the medium 1 h posttransfection. Transfected cells were distinguished from untransfected ones by Thy-1 expression and were electronically gated. Histograms depict the cell cycle profile of gated Thy-1-positive cells only. The left peaks constitute cells in G1, and the right peaks constitute cells in G2/M; cells in S phase are between the G1 and G2 peaks. The frequencies of cells in different stages of the cell cycle were calculated using Multicycle AV software (Phoenix Flow Systems, San Diego, Calif.).
We used parallel constructs encoding vpr mutants, vprHIV-1-FS and vprAGM-FS (Fig. 1A) containing frameshifts at codons 64 (40, 42) and 85, respectively. The mutant constructs, vprHIV-1-FS and vprAGM-FS, are unable to cause G2 arrest (data not shown).
Transfection of vprHIV-1 produced detectable cell cycle arrest in G2 both in human (SKBR3) and African green monkey (Cos-7) cells (Fig. 2). In contrast, vprAGM induced cell cycle arrest in Cos-7 cells but did not induce any detectable change in the cell cycle profile of SKBR3 cells (Fig. 2). We have obtained similar results when expressing vprAGM and vprHIV-1 in a variety of human cell lines, including osteosarcoma, leukemia/lymphoma, fibroblast-like, epidermal, colon carcinoma, rhabdomyosarcoma, and retinoblastoma cells (data not shown).
Relationship between transactivation and G2 arrest.
The ability of vprHIV-1 to induce G2 arrest has been linked to another property of vpr, transactivation of the viral promoter, the LTR. It was shown that the transcriptional activity of the LTR was increased by 5- to 10-fold (18, 20) during the G2 phase compared to that during the G1 phase. Consequently, infected cells that express vprHIV-1 are capable of producing 5- to 10-fold-larger amounts of viral progeny than are cells infected with vpr-deleted viruses (18, 20). A direct result of this enhanced transcriptional activity is that HIV-1 strains expressing functional vpr alleles are capable of spreading in culture with faster kinetics than are isogenic strains with mutations in vpr (13, 27, 37).
We wished to investigate the potential relationship between G2 arrest and transactivation in the context of vprAGM. Because vprAGM is unable to induce cell cycle arrest in human cells, we hypothesized that vprAGM would also be unable to induce transcriptional transactivation of the SIVagm LTR. The transactivation abilities of HIV-1 and SIVagm vpr genes were tested using luciferase reporter vectors (Fig. 1B). Luciferase reporter vectors were cotransfected with a mammalian expression vector expressing vpr or a truncated version of vpr (vprHIV-1-FS or vprAGM-FS) or with a control plasmid, pCMV-thy (Fig. 1A).
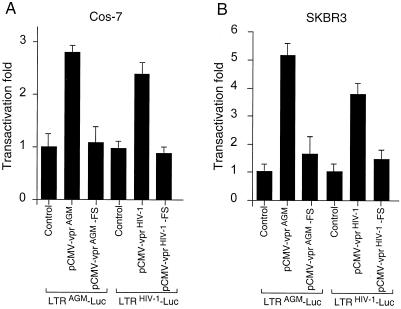
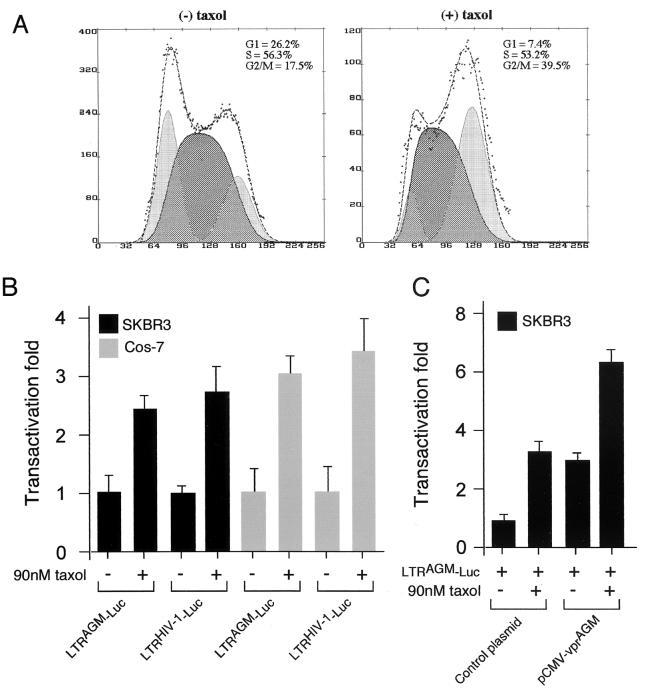
We first tested whether transactivation of the viral LTR by vprAGM would behave with species specificity as did induction of G2 arrest. In African green monkey cells, cotransfection of vprAGM with LTRAGM-Luc produced a significant level of transactivation, as did vprHIV-1 when cotransfected with LTRHIV-1-Luc (Fig. 3A). We then performed a parallel experiment with human cells. As expected, vprHIV-1 increased the level of expression of LTR-LucHIV-1 in human cells (Fig. 3B). Surprisingly, despite its inability to cause cell cycle arrest in human cells, vprAGM produced a significant level of transactivation upon the LTRAGM-Luc construct. Thus, it appears that vprAGM is able to induce transactivation of the SIVagm LTR independently of the induction of G2 arrest.
FIG. 3.
Transactivation of vprHIV-1 and vprAGM on the viral LTR. Cos-7 (A) and SKBR3 (B) cells were transiently transfected with a luciferase reporter construct (LTRAGM-Luc or LTRHIV-1-Luc) and a vpr expression construct. The structures of all constructs, except the control plasmid, are shown in Fig. 1. The control plasmid was pCMV-thy. At 48 h posttransfection, the cells were lysed in 100 μl of lysis buffer and a 20-μl volume was used to measure luciferase activity. Experiments were normalized to light units measured with control plasmid, to which an arbitrary value of 1 was assigned. All experiments were performed in triplicate, and standard deviations are shown as error bars.
Effect of caffeine on transactivation by vprHIV-1 and vprAGM.
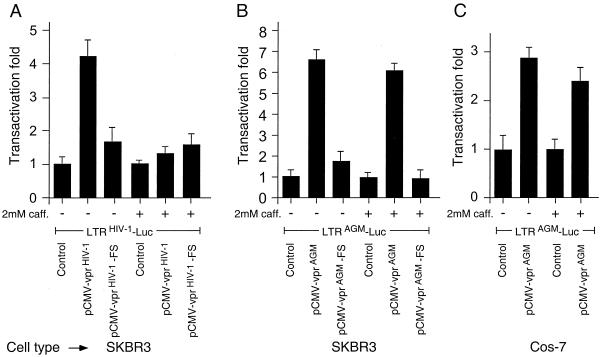
Caffeine, a methylxanthine, inhibits DNA damage-induced (36) and vprHIV-1-induced (43) cell cycle arrest. To further examine whether transactivation by vprAGM is independent of cell cycle blockade, we performed an experiment in which caffeine was used to relieve G2 arrest and the cells were analyzed for transactivation (Fig. 4). Incubation with 2 mM caffeine inhibited vprHIV-1-induced G2 arrest as well as vprAGM-induced G2 arrest (Fig. 2, bottom panels). Treatment with caffeine led to nearly complete (94%) inhibition of vprHIV-1 transactivation effect (Fig. 4A). In contrast, treatment with caffeine had no significant effect on the transactivation by vprAGM in human cells (Fig. 4B) or in African green monkey kidney cells (Fig. 4C). Thus, it appears that vprAGM can induce transactivation of the viral LTR by a mechanism which is independent of the induction of cell cycle arrest, and it represents an important mechanistic difference from the proposed mode of action of vprHIV-1 (18).
FIG. 4.
Effect of caffeine on transactivation. SKBR3 (A and B) or Cos-7 (C) cells were transiently transfected with vprHIV-1 (A) or vprAGM (B and C) vectors and 2 mM caffeine was added to the medium 1 h later; at 72 h posttransfection, the cells were lysed and analyzed for luciferase activity as described in the legend to Fig. 3.
Effect of taxol on transactivation of the HIV-1 and SIVagm LTRs.
It has been suggested that the transactivation effect induced by vprHIV-1 is a result of a higher transcriptional level of the viral LTR in the G2 phase of the cell cycle (18). According to this hypothesis, one would expect that drug-induced G2 arrest would produce similar transactivation of the LTR. To test this idea, cells were treated with a chemotherapeutic agent, taxol, which specifically induces cell cycle arrest at the G2/M transition by inhibiting the formation of the mitotic spindle (19, 24, 31). Treatment of SKBR3 cells with taxol induced G2 arrest (Fig. 5A) and transactivation of the HIV-1 LTR (Fig. 5B) in human cells. Similarly, taxol induced G2 arrest in Cos-7 cells (data not shown) and increased activity of the SIVagm LTR (Fig. 5B).
FIG. 5.
Induction of G2/M arrest by taxol leads to LTR transactivation. (A) SKBR3 cells were incubated in medium containing 90 nM taxol [(+) taxol] or medium alone [(−) taxol] and were assayed for cell cycle profile after 48 h. (B) Cells were transfected with the indicated reporter constructs; at 1 h posttransfection they were incubated in medium containing taxol or medium alone, and at 72 h they were lysed and analyzed for luciferase activity. (C) SKBR3 cells were transfected with the indicated plasmids and treated with 90 nM taxol or medium alone; at 72 h posttransfection they were lysed and analyzed for luciferase activity.
Thus, it appears that the SIVagm LTR transcriptional activity can be stimulated by two different mechanisms or pathways, one that is dependent on cell cycle manipulation and one that is independent of it. If this is true, one would expect that treatment with taxol and expression of vprAGM may have additive effects on LTR transactivation. To investigate this possibility, we co-transfected LTRAGM-Luc with vprAGM into human cells and treated the cells with taxol (Fig. 5C). If the SIVagm LTR is responsive to cell cycle manipulation, the extent of transactivation by vprAGM should be enhanced by taxol. Addition of taxol potentiated the transcriptional activity of the LTR in human cells (Fig. 5C). This indicates that the SIVagm LTR can be transactivated in part by cell cycle manipulation and in part by a cell cycle-independent pathway.
Taken together, the above results suggest that the relationship between transactivation and induction of cell cycle arrest differs in SIVagm and HIV-1 vpr genes. vprHIV-1 produces increased activity of the viral promoter through induction of G2 arrest. Abrogation of G2 arrest dramatically inhibits such transactivation by vprHIV-1. On the other hand, vprAGM induces transactivation of the viral LTR by cell cycle-dependent and -independent mechanisms.
Induction of apoptosis by vpr and its relationship to cell cycle perturbation.
Cells that are induced to arrest in G2 by either vprHIV-1 expression or infection with HIV-1 appear to be unable to enter the cell cycle again and, instead, undergo apoptosis (6, 49, 50). Because of the structural and functional similarities between vprHIV-1 and vprAGM, we wished to investigate the role of vprAGM in induction of apoptosis. In particular, we designed experiments to determine (i) whether expression of vprAGM leads to apoptosis; (ii) whether apoptosis by vprAGM is subject to species specificity, as is G2 arrest induction; and (iii) whether induction of apoptosis, for both vprHIV-1 and vprAGM, is linked to the induction of G2 arrest.
For our apoptosis studies, we used a lentivirus transduction method which we described previously (49). This delivery system has important advantages with respect to infection with full-length HIV or transfection procedures (34, 35). First, lentivirus vectors are able to deliver a small subset of viral genes. Second, lentivirus vectors using vesicular stomatitis virus glycoprotein G (VSVG) can be produced at high titers, such that high levels of infection can be achieved. Finally, we have developed lentivirus vectors (49) that express a marker, GFP, for analysis of transduction at the single-cell level (Fig. 6).
FIG. 6.
Viral vectors used for transduction of vprAGM, vprHIV-1, and mHSA. (A) Lentivirus transfer vectors were constructed by incorporating the indicated genes into the defective vector, D102; the vpr open reading frame was eliminated by mutating the translational start codon, which overlaps with vif; an XbaI restriction endonuclease site was introduced for convenient cloning of desired genes into this region of the genome; an EcoRI site was a present and was used as the downstream cloning site; discontinuous lines depict splicing donors (SD) and acceptors (SA). (B) The lentivirus packaging construct, pCMVΔR8.2-Δvpr, was described previously (4).
A defective HIV-1-derived vector, named D102 (Fig. 6A), was used for construction of three lentivirus vectors expressing vprHIV-1, vprAGM, or a control gene, encoding mHSA. All the splicing signals were left intact in D102. Only the splicing donors and acceptors involved in the generation of vpr and nef (GFP) mRNAs are shown in Fig. 6.
Infectious vectors were produced by cotransfection of one of the three transfer vectors derived from D102, plus the packaging construct, pCMVΔR8.2-Δvpr (Fig. 6), plus a VSV glycoprotein G expression vector, HCMV-VSVG (3, 7).
Target cells were transduced with lentivirus vectors, and the cells were analyzed at 72 h posttransduction for levels of infection and apoptosis. The levels of infection were assessed by measuring the levels of GFP expression and were between 80 and 95%. The levels of apoptosis were measured by TUNEL (Fig. 7). This method is based on the ability of terminal transferase to extend 3′-hydroxyl (3′-OH) ends of chromosomal breaks. Apoptotic cells, containing nicked DNA, incorporate significant amounts of labeled nucleotides, and these cells stain dark brown. The DNA from nonapoptotic cells is mostly nonfragmented and lacks free 3′-OH ends, and therefore these cells do not stain positive in the TUNEL method.
FIG. 7.
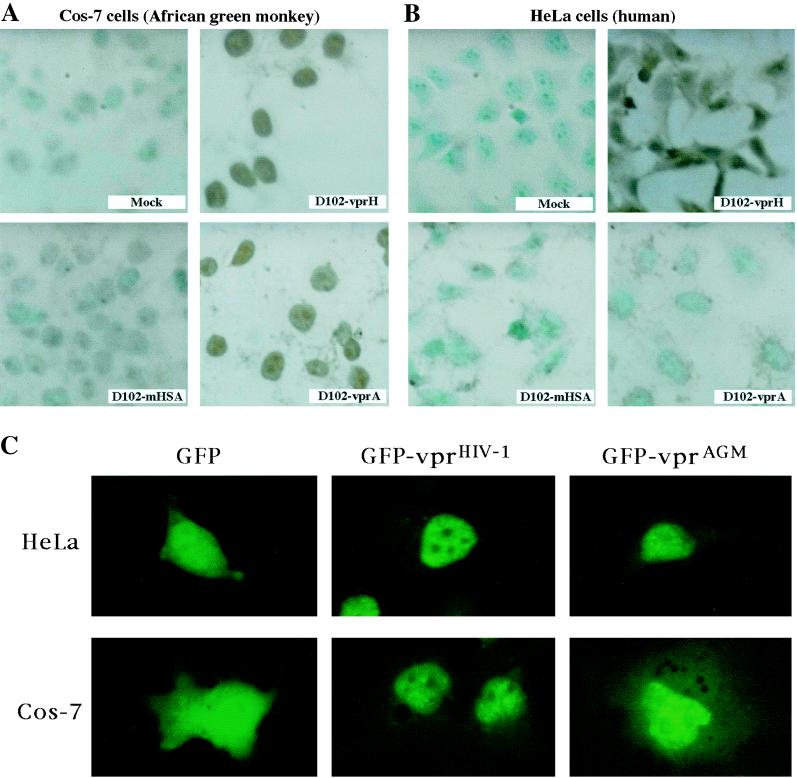
Induction of apoptosis by and subcellular localization of vprHIV-1 and vprAGM. (A) Apoptosis induction in Cos-7 cells. The cells were either mock infected or infected with one of the indicated lentivirus vectors and assayed for apoptosis at 72 h postinfection, using the TUNEL method; levels of infection were quantitated by measuring the frequency of GFP-positive cells and were between 80 and 95% for all experiments. (B) Apoptosis induction in HeLa cells. Experiments were performed in parallel with those in panel A. (C) Subcellular localization of GFP-vpr fusion proteins. Expression vectors bearing indicated fusion proteins were transiently transfected into HeLa or Cos-7 cells and visualized using fluorescence microscopy after 24 h.
The vector D102-vprH, which drives the expression of vprHIV-1 (Fig. 6), induced significant levels of apoptosis in both Cos-7 (Fig. 7A) and HeLa (Fig. 7B) cells. The vector D102-vprA, which directs the expression of vprAGM, induced significant levels of apoptosis in Cos-7 cells (Fig. 7A) but did not induce detectable apoptosis in HeLa cells (Fig. 7B). As a negative control, we used another lentivirus vector, D102-mHSA, that was engineered to express mHSA (25, 29). Transduction with D102-mHSA did not induce detectable apoptosis in either cell type (Fig. 7). The levels of apotosis were quantitated as percentages of cells staining positive in the TUNEL method (Fig. 8). Levels of apoptosis were confirmed by nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI) (49) in parallel samples (data not shown). The ability of vprHIV-1 and vprAGM to induce apoptosis was also tested in the human CD4-positive lymphocyte cell line Sup-T1 (Fig. 8C), and the results further supported the notion that induction of apoptosis by vprAGM is species specific.
FIG. 8.
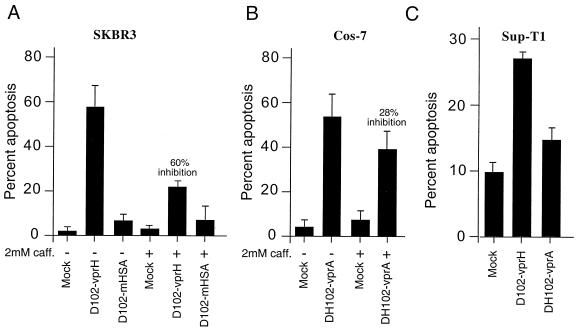
Effect of caffeine on the induction of apoptosis by vprHIV-1 and vprAGM. Infections and apoptosis measurements were performed as described in the legend to Fig. 7; TUNEL-positive (apoptotic) cells were visually counted, and the levels of apoptosis were expressed as a percentage of the total cells counted; the percentage of apoptotic cells in mock infections was always lower than 5%. All experiments were done in triplicate, and standard deviations are shown. (A) SKBR3 cells. (B) Cos-7 cells. (C) Sup-T1 cells.
It is not clear whether vprHIV-1 and vprAGM are able to induce apoptosis directly. In certain instances, apoptosis is thought to be a secondary effect of an abnormal delay in the G2 phase. In addition, recent observations in various laboratories suggested that the sensitivity of cycling cells to apoptosis is increased during the G2/M phase and G2/M arrest (30, 48, 56). If vpr induces apoptosis directly, we predict that inhibition of G2 arrest will have no effect on the level of apoptosis. Conversely, if the induction of apoptosis by vpr is dependent on the induction of G2 arrest, we would expect that alleviation of G2 arrest (i.e., by incubation with caffeine) would diminish the level of apoptosis. In a study on the effects of caffeine and radiation (36), radiation-induced G2 arrest and apoptosis decreased when the cells were treated with caffeine.
Treatment with caffeine led to a 60% decrease in the level of apoptosis induced by vprHIV-1 in human cells (Fig. 8A). This degree of inhibition was statistically significant (P < 0.05; Student's t test). Because abrogation of G2 arrest led to a dramatic decrease in the amount of apoptosis, we conclude that G2 arrest is a necessary step in the full induction of apoptosis by vprHIV-1.
Treatment of vprAGM-expressing cells with caffeine showed a different response from the one we observed with vprHIV-1. While caffeine was able to inhibit the cell cycle arrest in Cos-7 cells (Fig. 2), it produced only a moderate degree of inhibition of apoptosis (Fig. 8B), which was not statistically significant (P > 0.05; Student's t test).
Nuclear localization of vprHIV-1 and vprAGM in human versus African green monkey cells.
In view of the above results and since vprHIV-1 and vprAGM naturally localize to the cell nucleus (8, 15, 33, 51, 53), we hypothesized that species-specific differences in the functions of these proteins may be a consequence of potential differences in subcellular localization.
In an effort to compare the subcellular localization properties of vprHIV-1 and vprAGM and how localization may relate to their function in human versus African green monkey kidney cells, we constructed GFP-vpr fusion proteins and studied their localization by transient transfection followed by fluorescence microscopy (Fig. 7C). The study of subcellular localization of proteins via fusion with GFP offers the significant advantage that fluorescence may be studied in living cells, in the absence of fixation or staining procedures which may lead to artifactual patterns of distribution. In the fusion proteins, we incorporated a flexible linker (10) between the fusion partners to prevent steric hindrance. GFP displayed a diffuse pattern of distribution and did not selectively accumulate in the nucleus or the cytoplasm in human HeLa and simian Cos-7 cells. As previously reported, GFP-vprHIV-1 selectively localized to the nucleus and only very faint fluorescence could be detected in the cytoplasm. GFP-vprAGM appeared to localize in the nucleus, with a pattern that was indistinguishable from that of GFP-vprHIV-1. A previous report indicated that while both vprHIV-1 and vprAGM selectively localized to the nucleus of human cells, vprHIV-1 was evenly distributed whereas vprAGM displayed a punctate distribution (51). Our observations with GFP-vpr fusion proteins suggest that both HIV-1 and African green monkey kidney Vpr proteins display a punctate nuclear distribution. Thus, subcellular localization appears not to be the determinant behind the failure of vprAGM to induce cell cycle arrest and apotosis in human cells.
DISCUSSION
All known primate lentiviruses contain one or two genes (vpr and/or vpx) that are considered vpr homologs. The various genes in the vpr family are structurally related because they have predicted amino acid sequence homology and encode products that are virion-encapsidated (54). These genes are involved in multiple aspects of the biology of primate lentiviruses, although their precise roles in viral replication and modulation of host functions are still unclear.
We and others previously reported that species-specific factors modulate the induction of G2 arrest by members of the vpr family (42, 51). SIVagm and SIVsyk vpr genes are capable of arresting African green monkey kidney cells but are unable to do so in human cells. In contrast, HIV-1, HIV-2, and SIVsm vpr genes function in both simian and human cell types, although SIVsm Vpr functions more efficiently in simian cells than it does in human cells. These differences could not be explained on the basis of differential protein stability or subcellular localization (51). The species-specific cell cycle arrest differences between SIVagm and HIV-1 Vpr proteins indicate that these proteins may signal through cellular pathways that are divergent among primates.
We first compared vprAGM and vprHIV-1 at the level of transactivation. We wished to ascertain whether transactivation by vprAGM is species specific, as is its ability to induce G2 arrest. Surprisingly, vprAGM was able to induce transactivation in human cells, despite its inability to induce cell cycle alteration in these cells. This observation suggests that vprAGM may exert transactivation by a mechanism that is, at least in part, independent of cell cycle manipulation. To corroborate this observation, we resorted to the use of caffeine, a known inhibitor of vpr-induced (43) and DNA damage-induced (36) G2 arrest. Caffeine potently inhibited the cell cycle arrest by vprAGM in African green monkey kidney cells but produced little or no inhibition of its transactivation effect in either monkey or human cells.
The transactivation effects reported here and elsewhere for the various primate vpr genes are modest compared with the transactivation effects exerted by other, more classical viral transactivators (e.g., HIV-1 tat or human T-cell leukemia virus tax). The importance of vpr-induced transactivation, however, is underscored by two observations. First, HIV-1, HIV-2, SIVmac, and SIVagm vpr genes are able to transactivate their respective LTRs (39). This observation establishes the conservation of this function through evolution. Second, experiments in which the abilities of tat and vpr to transactivate were tested in parallel indicated that they were not mutually exclusive but were synergistic (28, 47). Thus, coexpression of the two genes provided a multiplicative effect on the promoter activity of the LTR.
We also wished to examine whether the ability of vprHIV-1 to cause apoptosis would be paralleled by vprAGM. Indeed, vprAGM is capable of inducing apoptosis in African green monkey kidney cells. This induction of apoptosis was not observed when vprAGM was expressed in human cells, and therefore apoptosis appears be determined by species-specific factors.
Abrogation of the cell cycle effect by caffeine treatment suppressed the levels of apoptosis by vprAGM, as it did for vprHIV-1. The potential link between the induction of G2 arrest and the onset of apoptosis is likely to be complex, since multiple signaling connections have been proposed. Perhaps the area of radiation-induced DNA damage has produced the clearest results to date. DNA damage is a natural signal that may induce cell cycle arrest and apoptosis. Cell cycle arrest following cellular insult allows for the repair of damaged DNA to protect the organism from the repercussions of mutation (21). Cell cycle block can occur before DNA replication, at the G1/S checkpoint, or before chromosome segregation, at the G2/M checkpoint (21, 38). Many DNA-damaging agents, including certain antineoplastic drugs, exert their effect at the G2/M phase (9) and, ultimately, commit the cell to death (32). The effects of vpr, specifically the G2 block, may be mediated along similar pathways to the effects of genotoxic agents (43).
It was initially thought that the cell cycle perturbation function of vprHIV-1 would require that the protein be localized in the nucleus. This was supported by experiments by Di Marzio et al. (15), who failed to find vprHIV-1 mutants that could cause G2 arrest in the absence of nuclear localization. Later mutagenesis experiments demonstrated that nuclear localization was not a requisite for vprHIV-1 to function as a cell cycle inhibitor (33, 53). We tested whether the subcellular localization of vprAGM might explain the species specificity of the effects of this protein. We found that in both human and African green monkey kidney cells a GFP-vprAGM fusion protein is also found in the nucleus. Thus, subcellular localization does not explain the inability of vprAGM to cause G2 arrest and apoptosis in human cells.
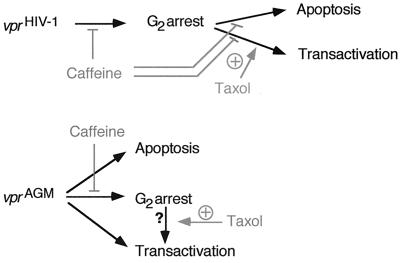
Our results point out important functional differences between the HIV-1 and SIVagm vpr genes (Fig. 9). HIV-1 vpr appears to induce G2 arrest as a primary effect, whereas transactivation and apoptosis appear to be downstream effects of G2 arrest. Thus, inhibition of G2 arrest will abrogate such downstream effects. SIVagm vpr induces these three effects independently. Thus, abrogation of vprAGM-induced G2 arrest has no effect on the onset of apoptosis or transactivation of the LTR.
FIG. 9.
Proposed model to summarize the relationships among various functions of vprHIV-1 and vprAGM. The observed effects of drugs are depicted in grey. Caffeine inhibits vprHIV-1 G2 arrest and also its downstream effects, apoptosis and transactivation. For vprAGM, caffeine inhibits G2 arrest but not apoptosis or transactivation. Transactivation by vprAGM is due, in part, to a cell cycle-independent mechanism, and therefore, caffeine has little or no effect on transactivation. However, the SIVagm LTR is also responsive to cell cycle arrest since taxol can induce transactivation. Because treatment with caffeine does not significantly affect transactivation, the significance of G2 arrest for vprAGM is uncertain (question mark).
Taken together, our results suggest that while the multiple functions of vpr are conserved among various primate lentiviruses, the mechanisms leading to the execution of such functions are divergent.
ACKNOWLEDGMENTS
We thank M. J. Renda and E. Klimatcheva for critical reading of the manuscript. We also thank B. J. Rimel and Don Nguyen for valuable assistance with the construction of vprAGM-FS and the D102 lentivirus vector, respectively. I. S. Y. Chen and D. S. An kindly provided the packaging construct, pCMVΔR8.2-Δvpr.
This work was supported by NIH research grants to V.P. (R29-AI41407) and H.A.G. (5RO1-MH56838).
REFERENCES
- 1.Accola M A, Bukovsky A A, Jones M S, Göttlinger H G. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J Virol. 1999;73:9992–9999. doi: 10.1128/jvi.73.12.9992-9999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, R. W, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akkina R K, Walton R M, Chen M L, Li Q X, Planelles V, Chen I S Y. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An D S, Morizono K, Li Q X, Mao S H, Lu S, Chen I S Y. An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication. J Virol. 1999;73:7671–7677. doi: 10.1128/jvi.73.9.7671-7677.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 6.Banki K, Hutter E, Gonchoroff N J, Perl A. Molecular ordering in HIV-induced apoptosis. Oxidative stress, activation of caspases, and cell survival are regulated by transaldolase. J Biol Chem. 1998;273:11944–11953. doi: 10.1074/jbc.273.19.11944. [DOI] [PubMed] [Google Scholar]
- 7.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell B J, Hirsch V M. Vpr of simian immunodeficiency virus of African green monkeys is required for replication in macaque macrophages and lymphocytes. J Virol. 1997;71:5593–5602. doi: 10.1128/jvi.71.7.5593-5602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabner B A. Biological basis for cancer treatment. Ann Intern Med. 1993;118:633–637. doi: 10.7326/0003-4819-118-8-199304150-00011. [DOI] [PubMed] [Google Scholar]
- 10.Challita-Eid P M, Abboud C N, Morrison S L, Penichet M L, Rosell K E, Poles T, Hilchey S P, Planelles V, Rosenblatt J D. A RANTES-antibody fusion protein retains antigen specificity and chemokine function. J Immunol. 1998;161:3729–3736. [PubMed] [Google Scholar]
- 11.Chang L, Chen C, Urlacher V, Lee T. Differential apoptosis effects of primate lentiviral vpr and vpx in mammalian cells. J Biomed Sci. 2000;7:322–333. doi: 10.1007/BF02253252. [DOI] [PubMed] [Google Scholar]
- 12.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen E A, Terwilliger E F, Jalinoos Y, Proulx J, Sodroski J G, Haseltine W A. Identification of HIV-1 vpr product and function. J Acquired Immune Defic Syndr. 1990;3:11–18. [PubMed] [Google Scholar]
- 14.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 15.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouchier R A, Meyer B E, Simon J H, Fischer U, Albright A V, Gonzalez-Scarano F, Malim M H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 19.Gotaskie G E, Andreassi B F. Paclitaxel: a new antimitotic chemotherapeutic agent. Cancer Pract. 1994;2:27–33. [PubMed] [Google Scholar]
- 20.Gummuluru S, Emerman M. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J Virol. 1999;73:5422–5430. doi: 10.1128/jvi.73.7.5422-5430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 22.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, KewalRamani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz S B. Taxol (paclitaxel): mechanisms of action. Ann Oncol. 1994;5(Suppl. 6):S3–S6. [PubMed] [Google Scholar]
- 25.Jamieson B D, Zack J A. In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J Virol. 1998;72:6520–6526. doi: 10.1128/jvi.72.8.6520-6526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from west African. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jowett J B, Planelles V, Poon B, Shah N P, Chen M L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashanchi F, Agbottah E T, Pise-Masison C A, Mahieux R, Duvall J, Kumar A, Brady J N. Cell cycle-regulated transcription by the human immunodeficiency virus type 1 Tat transactivator. J Virol. 2000;74:652–660. doi: 10.1128/jvi.74.2.652-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kay R, Takei F, Humphries R K. Expression cloning of a cDNA encoding M1/69-J11d heat-stable antigens. J Immunol. 1990;145:1952–1959. [PubMed] [Google Scholar]
- 30.Ling Y H, Tornos C, Perez-Soler R. Phosphorylation of Bcl-2 is a marker of M phase events and not a determinant of apoptosis. J Biol Chem. 1998;273:18984–18991. doi: 10.1074/jbc.273.30.18984. [DOI] [PubMed] [Google Scholar]
- 31.Long B H, Fairchild C R. Paclitaxel inhibits progression of mitotic cells to G1 phase by interference with spindle formation without affecting other microtubule functions during anaphase and telephase. Cancer Res. 1994;54:4355–4361. [PubMed] [Google Scholar]
- 32.Lowe S W, Ruley H E, Jacks T, Housman D E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 33.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 36.Ning S, Knox S J. G2/M-phase arrest and death by apoptosis of HL60 cells irradiated with exponentially decreasing low-dose-rate gamma radiation. Radiat Res. 1999;151:659–669. [PubMed] [Google Scholar]
- 37.Ogawa K, Shibata R, Kiyomasu T, Higuchi I, Kishida Y, Ishimoto A, Adachi A. Mutational analysis of the human immunodeficiency virus vpr open reading frame. J Virol. 1989;63:4110–4114. doi: 10.1128/jvi.63.9.4110-4114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 39.Philippon V, Matsuda Z, Essex M. Transactivation is a conserved function among primate lentivirus Vpr proteins but is not shared by Vpx. J Hum Virol. 1999;2:167–174. [PubMed] [Google Scholar]
- 40.Planelles V, Bachelerie F, Jowett J B M, Haislip A, Xie Y, Banooni P, Masuda T, Chen I S Y. Fate of the human immunodeficiency virus type 1 provirus in infected cells: a role for vpr. J Virol. 1995;69:5883–5889. doi: 10.1128/jvi.69.9.5883-5889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planelles V, Haislip A, Withers-Ward E S, Stewart S A, Xie Y, Shah N P, Chen I S Y. A new reporter system for detection of retroviral infection. Gene Ther. 1995;2:369–376. [PubMed] [Google Scholar]
- 42.Planelles V, Jowett J B M, Li Q X, Xie Y, Hahn B, Chen I S Y. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poon B, Jowett J B, Stewart S A, Armstrong R W, Rishton G M, Chen I S Y. Human immunodeficiency virus type 1 vpr gene induces phenotypic effects similar to those of the DNA alkylating agent, nitrogen mustard. J Virol. 1997;71:3961–3971. doi: 10.1128/jvi.71.5.3961-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J Biol Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 45.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawaya B E, Khalili K, Gordon J, Taube R, Amini S. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J Biol Chem. 2000;275:35209–35214. doi: 10.1074/jbc.M005197200. [DOI] [PubMed] [Google Scholar]
- 48.Scatena C D, Stewart Z A, Mays D, Tang L J, Keefer C J, Leach S D, Pietenpol J A. Mitotic phosphorylation of Bcl-2 during normal cell cycle progression and Taxol-induced growth arrest. J Biol Chem. 1998;273:30777–30784. doi: 10.1074/jbc.273.46.30777. [DOI] [PubMed] [Google Scholar]
- 49.Shostak L D, Ludlow J, Fisk J, Pursell S, Rimel B J, Nguyen D, Rosenblatt J D, Planelles V. Roles of p53 and caspases in the induction of cell cycle arrest and apoptosis by HIV-1 vpr. Exp Cell Res. 1999;251:156–165. doi: 10.1006/excr.1999.4568. [DOI] [PubMed] [Google Scholar]
- 50.Stewart S A, Poon B, Jowett J B, Chen I S Y. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J Virol. 1997;71:5579–5592. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subbramanian R A, Kessous-Elbaz A, Lodge R, Forget J, Yao X J, Bergeron D, Cohen E A. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J Exp Med. 1998;187:1103–1111. doi: 10.1084/jem.187.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subbramanian R A, Yao X J, Dilhuydy H, Rougeau N, Bergeron D, Robitaille Y, Cohen E A. Human immunodeficiency virus type 1 Vpr localization: nuclear transport of a viral protein modulated by a putative amphipathic helical structure and its relevance to biological activity. J Mol Biol. 1998;278:13–30. doi: 10.1006/jmbi.1998.1685. [DOI] [PubMed] [Google Scholar]
- 54.Tristem M, Marshall C, Karpas A, Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe N, Yamaguchi T, Akimoto Y, Rattner J B, Hirano H, Nakauchi H. Induction of M-phase arrest and apoptosis after HIV-1 vpr expression through uncoupling of nuclear and centrosomal cycle in HeLa cells. Exp Cell Res. 2000;258:261–269. doi: 10.1006/excr.2000.4908. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto K, Ichijo H, Korsmeyer S J. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G2/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan X, Matsuda Z, Matsuda M, Essex M, Lee T H. Human immunodeficiency virus vpr gene encodes a virion-associated protein. AIDS Res Hum Retroviruses. 1990;6:1265–1271. doi: 10.1089/aid.1990.6.1265. [DOI] [PubMed] [Google Scholar]