Abstract
Glioblastoma (GBM) is the most prevalent primary malignant tumor of the nervous system. In this study, we utilized pathomics analysis to explore the expression of CD40LG and its predictive value for the prognosis of GBM patients. We analyzed the expression differences of CD40LG in GBM tissue and normal brain tissue, along with performing survival prognosis analysis. Additionally, histopathological sections of GBM were used to screen for pathological features. Subsequently, SVM and LR pathomics models were constructed, and the models’ performance was evaluated. The pathomics model was employed to predict CD40LG expression and patient prognosis. Furthermore, we investigated the potential molecular mechanisms through enrichment analysis, WGCNA analysis, immune correlation analysis, and immune checkpoint analysis. The expression level of CD40LG was significantly increased in GBM. Multivariate analysis demonstrated that high expression of CD40LG is a risk factor for overall survival (OS) in GBM patients. Five pathological features were identified, and SVM and LR pathomics models were constructed. Model evaluation showed promising predictive effects, with an AUC value of 0.779 for the SVM model, and the Hosmer–Lemeshow test confirmed the model’s prediction probability consistency (P > 0.05). The LR model achieved an AUC value of 0.785, and the Hosmer–Lemeshow test indicated good agreement between the LR model’s predicted probabilities and the true value (P > 0.05). Immune infiltration analysis revealed a significant correlation between the pathomics score (PS) and the degree of infiltration of activated DC cells, T cells CD4 naïve, Macrophages M2, Macrophages M1, and T cells CD4 memory resting. Our results demonstrate that the pathomics model exhibits predictability for CD40LG expression and GBM patient survival. These findings can be utilized to assist neurosurgeons in selecting optimal treatment strategies in clinical practice.
Subject terms: CNS cancer, Tumour biomarkers
Introduction
Glioblastoma (GBM) is the most prevalent malignant tumor of the central nervous system1. Currently, surgery supplemented by radiotherapy and chemotherapy is the primary treatment for GBM. Despite substantial understanding of the molecular pathogenesis and biology of this tumor, patients with GBM still face a poor prognosis, with a median survival of approximately 15 months2,3. Molecular markers such as IDH and MGMT are used for predicting the sensitivity and prognosis of GBM patients to TMZ chemotherapy4,5. However, the detection of these markers is time-consuming and expensive. Therefore, there is a need to develop an efficient, cost-effective method for predicting GBM, aiming to improve patient prognosis and optimize treatment.
The CD40LG gene encodes a protein expressed on the surface of T cells6, and a defect in this gene results in incapacity for immunoglobulin class switching, associated with Hyper-IgM syndrome7. Upon interaction with its cognate receptor CD40, which is constitutively expressed by B cells and antigen-presenting cells, CD40L plays a crucial role in enhancing B-cell activation and promoting T-cell-dependent humoral immune responses8. Studies indicate that CD40LG is closely related to the biological development of lung cancer and breast cancer9,10. CD40/CD40L expression correlates with survival in glioblastoma patients, and enhancement of CD40 signaling enhances vaccination efficacy in glioma models11. Treatment with CD40L blockers inhibits GBM infiltration when combined with mesenchymal stem-like cells in the GBM microenvironment12.
Histopathology is the gold standard for tumor diagnosis, providing prognostic predictions. However, differences in judgments among pathologists may impact prognosis assessment13. The introduction of the concept of pathomics may address this issue. Pathomics involves the conversion of pathological images into high-fidelity, high-throughput mineable data using artificial intelligence, covering quantitative features such as texture, morphological characteristics, edge gradients, and biological traits, and is used for quantitative pathological diagnosis, molecular expression, and disease prognosis14.
This study screened the pathological features of H&E slices of glioblastoma based on the above factors, constructed pathological models based on the screened features, predicted the expression level of CD40LG, and assessed prognosis through these pathological features. Simultaneously, it further explored the internal molecular mechanisms of pathomics and investigated the relevant molecular pathways and their correlation with immune infiltration.
Materials and methods
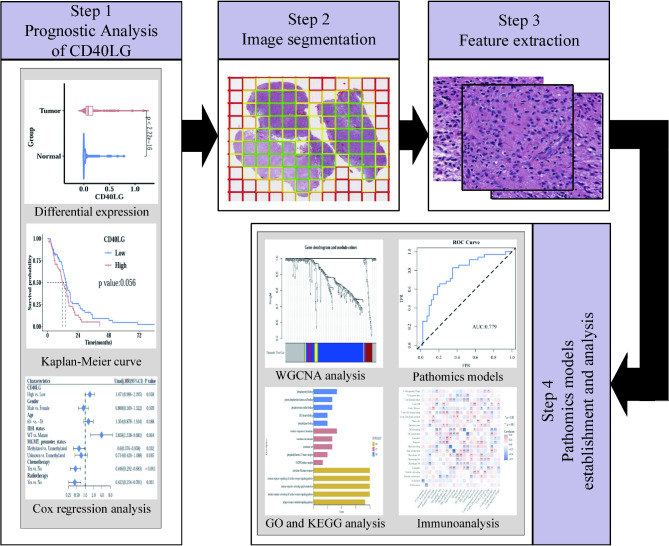
The overall flowchart of prognosis analysis of CD40LG, image features extraction and pathomics models establishment was presented in Fig. 1. The details of each part were described in the following sections.
Fig. 1.
The workflow of prognostic analysis, feature extraction and construction of pathomics models.
Patients
The clinical data of 599 GBM patients was obtained from the TCGA database. The criteria for exclusion are as follows: 1. Patients who were not first treated at the clinic; 2. Patients with missing survival data or survival time of less than one month; 3. Patients with ‘Unknown’ IDH status; 4. Patients with non-primary solid tumors and no RNA-seq data.
Additionally, pathological images of 389 GBM patients were obtained from the TCGA database. The criteria for exclusion are as follows: patients with poor pathological image quality and no overlap with clinical data.
Identification of CD40LG as a differentially expressed gene
RNA-seq data was downloaded for GBM and corresponding normal tissues from The Cancer Genome Atlas (TCGA) database and the Genotype Tissue Expression (GTEx) database. R software version 3.6.6 was used for assessing differential CD40LG gene expression between non-paired normal and cancerous tissues. A significance level of p < 0.05 was applied to determine statistical significance. The data was normalized using the Log2 (value + 1) transformation. Expression analysis was conducted using the Wilcoxon rank sum test.
Survival prognosis analysis
Kaplan–Meier (KM) curves were utilized in the survival analysis to assess the relationship between CD40LG expression and overall survival (OS) of GBM patients. The criteria for “high” and “low” expressions in the KM analysis were based on the tumor cohort median. Additionally, Univariate and multivariate Cox regression analyses were performed to identify potential prognostic factors for overall survival (OS) in patients with GBM. The analyses were performed using the “survival (v3.2.1)”, “survminer (v3.3.3)” and “ggplot2 (v3.3.3)” R packages, where p < 0.05 was considered statistically significant.
Processing of histopathological images
Hematoxylin and Eosin (H&E) stained histopathological images (20 × or 40 × magnification) in SVS format were downloaded from the TCGA (https://tcga-data.nci.nih.gov/tcga/) database. Pathological image processing and segmentation: use the OTSU algorithm (https://opencv.org/) to obtain the tissue area of the pathological section. The 40 × image was divided into multiple 1024 × 1024-pixel sub-images, while the 20 × image was likewise resampled to 1024 × 1024 pixels after being divided into multiple 512 × 512-pixel sub-images. Sub-images with insufficient quality, such as contamination, indistinct images, or a vacant area exceeding 50%, were discarded. Ten sub-images were chosen at random for subsequent analysis from each pathology image.
Pathomics features extraction
PyRadiomics is an open-source Python library designed for extracting a large number of engineered features from medical images. It enables researchers and clinicians to automatically extract quantitative features from image data, which can be used for disease diagnosis, prognosis prediction, treatment response evaluation, and other medical researches. PyRadiomics supports various image formats and offers flexible feature extraction configurations, covering multiple categories of features such as shape, intensity, and texture15.
We standardized each sub-image using the PyRadiomics open-source package (https://pyradiomics.readthedocs.io/en/latest/). Ninety-three original features, including first-order and second-order features, were extracted from each sub-image. Additionally, high-order features (Wavelet: LL, LH, HL, HH) were also extracted, yielding a total of 465 features. After characterizing 10 sub-images from each patient’s pathological image, we calculated the average value of these characteristics for each sample, which served as the pathology characteristic for follow-up data analysis. Finally, we applied the mRMR (Maximum relevance, minimum redundancy) and RFE (Recursive feature elimination) algorithms to further filter the best characteristics.
Establishment and evaluation of pathomics models
We utilized the R language, specifically the “caret” and “stats” packages, to model the selected pathomics features using the support vector machine (SVM) and logistic regression (LR) algorithms, respectively. These models were used to predict gene expression. We assessed the model performance using various evaluation metrics, including ROC curve, PR curve, AUC, accuracy (ACC), sensitivity (SEN), specificity (SPE), positive predictive value (PPV), negative predictive value (NPV), calibration curve, and Brier score. The AUC values of the two models were compared using the Delong test. Finally, the pathomics score (PS) of the intersection sample is calculated through the pathomics model, the PS is combined with the corresponding clinical data, and the cutoff value of the PS is calculated through the survminer package and divided into binary variables.
Immune cell infiltration and enrichment analysis based on pathomics score
To explore the molecular mechanism underlying expression differences between high and low expression groups of PS genes, we used the R package “clusterProfiler” to perform genes enrichment analysis (GSEA) on genes differentially expressed between high and low PS groups in the KEGG (c2.cp.kegg.v7.5.1.symbols.gmt) and Hallmark (h.all.v7.5.1.symbols.gmt) gene sets. A p-value of less than 0.05 was considered statistically significant. We uploaded the gene expression matrix of GBM samples to the CIBERSORTx database (https://cibersortx.stanford.edu/) to calculate the immune cell infiltration of each sample, and we analyzed the correlation between PS and the extent of immune cell infiltration using the R package “corrplot”. Moreover, wilcox.test was used to analyze the difference in immune checkpoint expression between high and low PS groups.
In addition, WGCNA was used to identify BCR signaling pathway gene sets related to prognosis, an expression similarity matrix was constructed based on the obtained BCR signaling pathway gene expression data, and the scale independence and scale independence of the network with different power values (1 ~ 10) were tested. Finally, the module with the highest correlation with clinical characteristics is regarded as the key module, and this module is selected to identify relevant hub genes. Finally, GO and KEGG enrichment analysis were performed on the hub genes related to the BCR signaling pathway.
Results
Patients
Out of the clinical data of 599 GBM patients, 557 patients who were initially diagnosed and treated were screened, 29 patients with missing survival data and survival time less than 1 month were deleted, 11 patients with IDH status “Unknown” were excluded, and 290 patients with non-primary solid tumors with no RNA-seq data were eliminated, and 127 were eventually included. Out of 389 GBM pathological images, 311 cases with poor pathological image quality and no intersection with clinical data were eliminated, and 78 were eventually included.
Expression and prognostic analysis of CD40LG
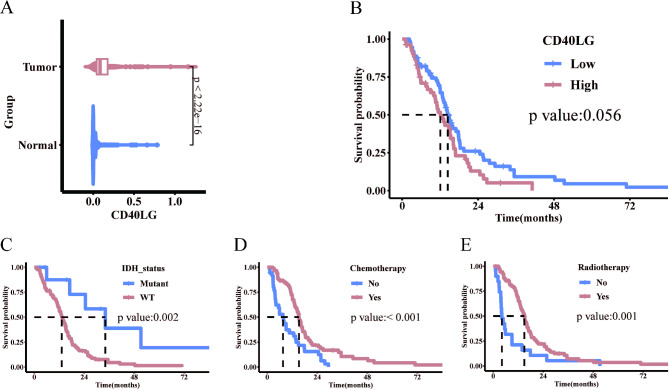
Differences in CD40LG expression between GBM (from the TCGA database) and normal tissues (from GTEx database) were compared. The analysis revealed a noticeable upregulation of CD40LG expression in GBM (Fig. 2A). Based on the expression of CD40LG using a cut-off value of 0.1237, the 127 patients were categorized into a CD40LG high expression group (n = 73) and a low expression group (n = 54). Notably, there were no significant differences in clinical and pathological characteristics between these two patient groups (Table 1). Kaplan–Meier survival curves demonstrated that the CD40LG low expression group had a median survival time of 14.4 months, while the CD40LG high expression group had a median survival time of 12.1 months. High CD40LG expression was closely associated with OS deterioration, with a borderline significant p-value of 0.056 (Fig. 2B). Additionally, the IDH mutation group exhibited significantly higher survival rates compared to the IDH wild-type group (Fig. 2C). Furthermore, both the chemotherapy group and radiotherapy group demonstrated significantly higher survival rates than the no chemotherapy and no radiotherapy groups (Fig. 2D,E).
Fig. 2.
Expression and prognostic analysis of CD40LG. (A) CD40LG expression in GBM and normal tissues (GBM = 166, Normal = 1157). (B) Overall survival curves of CD40LG in GBM (High = 73, Low = 54). (C-E) GBM survival curves under different IDH (Mutant = 9, WT = 118), chemotherapy (No = 36, Yes = 91) and radiotherapy status (No = 20, Yes = 107).
Table 1.
The clinical and pathological characteristics in the CD40LG low and high groups.
| Variables | Total (n = 127) | Low (n = 73) | High (n = 54) | p |
|---|---|---|---|---|
| Gender, n (%) | 0.98 | |||
| Female | 41 (32) | 23 (32) | 18 (33) | |
| Male | 86 (68) | 50 (68) | 36 (67) | |
| Age, n (%) | 0.636 | |||
| ~ 59 | 56 (44) | 34 (47) | 22 (41) | |
| 60 ~ | 71 (56) | 39 (53) | 32 (59) | |
| IDH_status, n (%) | 0.077 | |||
| Mutant | 9 (7) | 8 (11) | 1 (2) | |
| WT | 118 (93) | 65 (89) | 53 (98) | |
| MGMT_promoter_status, n (%) | 0.536 | |||
| Unmethylated | 59 (46) | 35 (48) | 24 (44) | |
| Methylated | 41 (32) | 25 (34) | 16 (30) | |
| Unknown | 27 (21) | 13 (18) | 14 (26) | |
| Chemotherapy, n (%) | 0.204 | |||
| No | 36 (28) | 17 (23) | 19 (35) | |
| Yes | 91 (72) | 56 (77) | 35 (65) | |
| Radiotherapy, n (%) | 1 | |||
| No | 20 (16) | 11 (15) | 9 (17) | |
| Yes | 107 (84) | 62 (85) | 45 (83) | |
Univariate and multivariate analyses in GBM
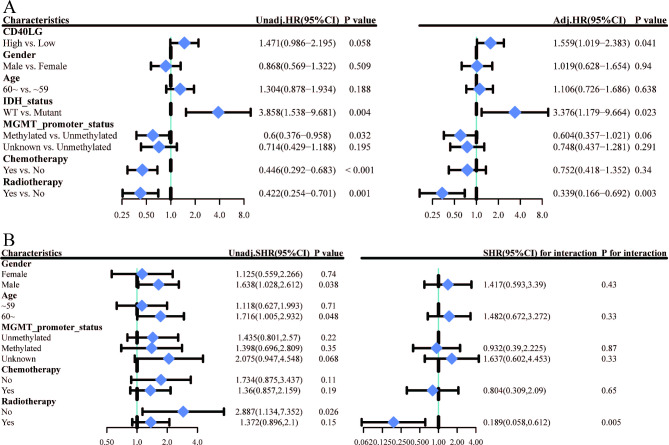
In univariate analysis, high expression of CD40LG was identified as a risk factor for overall survival (OS) (HR = 1.471, 95% CI: 0.986–2.195, P = 0.058). Additionally, IDH wild type was associated with an increased risk of OS (HR = 3.858, 95% CI: 1.538–9.681, P = 0.004), while MGMT methylation was found to be a protective factor for OS (HR = 0.6, 95% CI: 0.376–0.958, P = 0.032). Moreover, chemotherapy (HR = 0.446, 95% CI: 0.292–0.683, P < 0.001) and radiotherapy (HR = 0.442, 95% CI: 0.254–0.701, P = 0.001) were both protective factors for OS. After adjusting for multiple factors, high expression of CD40LG remained a risk factor for OS (HR = 1.559, 95% CI: 1.019 − 2.383, P = 0.041), as well as IDH wild type (HR = 3.376, 95% CI: 1.179 − 9.664, P = 0.023). Conversely, radiotherapy was observed to be a significant protective factor for OS (HR = 0.339, 95% CI: 0.166 − 0.692, P = 0.003) (Fig. 3A).
Fig. 3.
Univariate and Multivariate Analyses in GBM. (A) Univariate and multivariate Cox regression analyses of clinical features of GBM associated with OS. (B) Univariate COX regression for subgroup analysis.
To further investigate the impact of CD40LG on patient prognosis across different subgroups, we conducted exploratory subgroup analysis using univariate Cox regression. In the subgroup analysis, elevated CD40LG was identified as a significant risk factor for OS in the subgroup that did not receive radiotherapy (HR = 2.887, 95% CI: 1.134–7.352, P = 0.026). However, in the subgroup that received radiotherapy, elevated CD40LG was not significantly associated with OS (HR = 1.372, 95% CI: 0.896–2.1, P = 0.15). The interaction test yielded a p-value of 0.005, suggesting a significant interaction between the "association between CD40LG and patient OS" and the receipt of radiotherapy (Fig. 3B).
Model establishment and evaluation
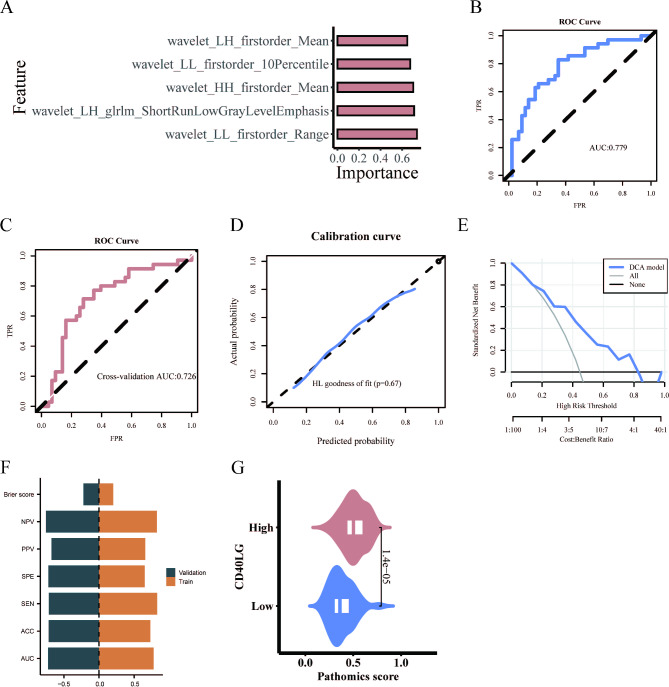
Initially, 465 original and high-order features were screened. Then, the mRMR algorithm was used to select the top 20 features, followed by the RFE algorithm to further narrow down to 5 features.
The selected pathomics features were modeled through the SVM algorithm. The results show the importance of the five features in the SVM algorithm (Fig. 4A). Subsequently, the model’s performance was evaluated. The ROC curve revealed an AUC value of 0.779 (Fig. 4B), while the tenfold cross-validation yielded an AUC of 0.726 (Fig. 4C). The calibration curve and Hosmer–Lemeshow goodness-of-fit test indicated that the pathomics prediction model accurately predicted gene expression probability (P > 0.05) (Fig. 4D). DCA further confirmed the model’s clinical applicability (Fig. 4E). The training set had an ACC of 0.731, SEN of 0.829, SPE of 0.651, PPV of 0.659, NPV of 0.824, and a Brier score of 0.202. The cross-validation had an ACC of 0.718, SEN of 0.714, SPE of 0.721, PPV of 0.676, NPV of 0.756, and a Brier score of 0.221 (Fig. 4F). Additionally, the SVM pathomics model provided a PS for predicting gene expression levels. The Wilcoxon test revealed a significant difference in PS distribution between high and low gene groups (p < 0.001). The CD40LG high expression group exhibited higher PS value (Fig. 4G).
Fig. 4.
SVM model establishment and evaluation. (A) Five features in the SVM algorithm. (B) Predictive performance of SVM model. (C) Predictive performance of the tenfold cross-validation. (D) Calibration curve analysis. (E) Decision curve analysis. (F) The ACC, SEN, SPE, PPV, NPV and Brier score of the train and validation sets. (G) Prediction of CD40LG expression by pathomic score.
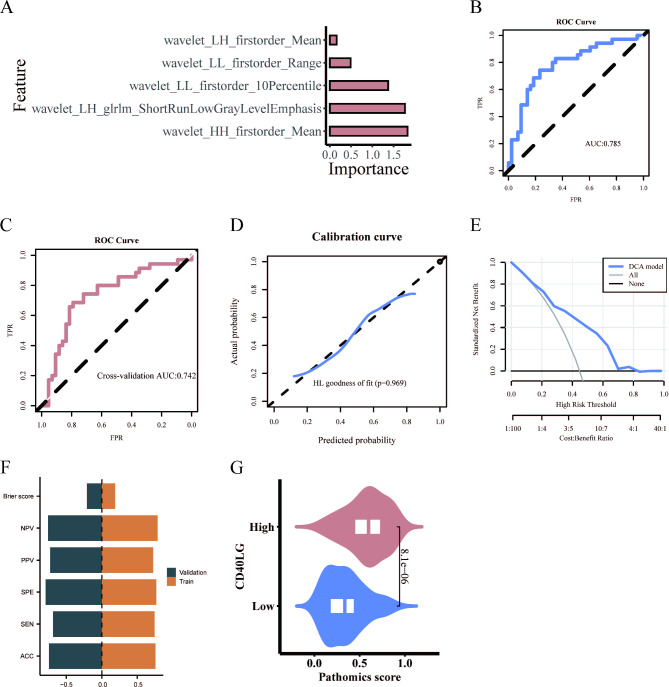
The selected pathomics features were used to build a binary classification model for gene expression prediction using logistic regression algorithm. Figure 5A illustrates the significance of pathological features in the LR algorithm. The performance of the LR model was assessed, demonstrating its effectiveness: the AUC value of the model was 0.785 according to the ROC curve (Fig. 5B), and the tenfold internal cross-validation yielded an AUC value of 0.742 (Fig. 5C). The calibration curve and Hosmer–Lemeshow goodness-of-fit test confirmed the model’s accurate prediction of high CD40LG expression (P > 0.05) (Fig. 5D). Furthermore, DCA demonstrated the model’s clinical practicality (Fig. 5E). The training set had an ACC of 0.756, SEN of 0.743, SPE of 0.767, PPV of 0.722, NPV of 0.786, and a Brier score of 0.188. The cross-validation had an ACC of 0.744, SEN of 0.686, SPE of 0.791, PPV of 0.727, NPV of 0.756, and a Brier score of 0.21 (Fig. 5F). Similarly, the LR pathomics model calculated the PS for predicting gene expression levels. The Wilcoxon test revealed significant differences in PS distribution between high and low gene groups (p < 0.001). The CD40LG high expression group exhibited higher PS value (Fig. 5G).
Fig. 5.
LR model establishment and evaluation. (A) Five features in the LR algorithm. (B) Predictive performance of LR model. (C) Predictive performance of the tenfold cross-validation. (D) Calibration curve analysis. (E) Decision curve analysis. (F) The ACC, SEN, SPE, PPV, NPV and Brier score of the train and validation sets. (G) Prediction of CD40LG expression by pathomic score.
Finally, the DeLong test was performed to compare the AUC values of the SVM omics model and the LR omics model before and after validation. The results showed no statistical difference between the two models (p = 0.549 for the training set and p = 0.844 for cross-validation). As the LR omics model had a higher AUC value compared to the SVM omics model, it was chosen for further analysis.
Enrichment analysis between high and low PS groups
We utilized the R package “survminer” to obtain the predicted value of the LR pathomics model. Patients were stratified into high PS expression (n = 47) and low expression (n = 31) groups based on the PS cutoff value of 0.3484. Subsequently, a baseline information table for each clinical variable was generated using the PS Low/High grouping. The analysis revealed no significant differences in the distribution of clinical variables between the high and low PS groups (p > 0.05) (Table 2).
Table 2.
The clinical variables between the low and high PS groups.
| Variables | Total (n = 78) | Low (n = 31) | High (n = 47) | p |
|---|---|---|---|---|
| Gender, n (%) | 0.547 | |||
| Female | 21 (27) | 10 (32) | 11 (23) | |
| Male | 57 (73) | 21 (68) | 36 (77) | |
| Age, n (%) | 0.458 | |||
| ~ 59 | 40 (51) | 18 (58) | 22 (47) | |
| 60 ~ | 38 (49) | 13 (42) | 25 (53) | |
| IDH_status, n (%) | 0.107 | |||
| Mutant | 7 (9) | 5 (16) | 2 (4) | |
| WT | 71 (91) | 26 (84) | 45 (96) | |
| MGMT_promoter_status, n (%) | 0.237 | |||
| Methylated | 20 (26) | 10 (32) | 10 (21) | |
| Unknown | 26 (33) | 7 (23) | 19 (40) | |
| Unmethylated | 32 (41) | 14 (45) | 18 (38) | |
| Chemotherapy, n (%) | 0.659 | |||
| No | 21 (27) | 7 (23) | 14 (30) | |
| Yes | 57 (73) | 24 (77) | 33 (70) | |
| Radiotherapy, n (%) | 0.507 | |||
| No | 10 (13) | 5 (16) | 5 (11) | |
| Yes | 68 (87) | 26 (84) | 42 (89) | |
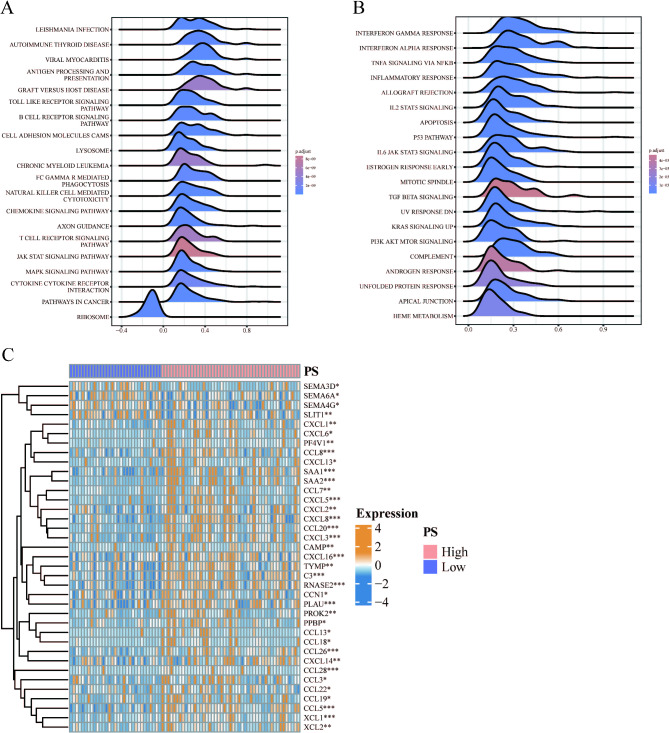
GSEA was performed on differentially expressed genes between the high and low PS groups, and the top 20 pathways were visualized using KEGG16–18 and Hallmark gene set analyses. The KEGG gene set analysis revealed significant enrichment of differential genes in the B cell receptor signaling pathway, chemokine signaling pathway, and other signaling pathways among the high and low PS groups (Fig. 6A). Similarly, Hallmark gene set analysis demonstrated significant enrichment of differential genes in signaling pathways such as KRAS SIGNALING and PI3K AKT MTOR SIGNALING between the PS high and low groups (Fig. 6B). Additionally, the differential expression of chemokines between the high and low PS groups was analyzed using the Wilcoxon test, revealing significantly increased expression of CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CXCL13, CXCL14, and CXCL16 in the PS high group (Fig. 6C).
Fig. 6.
Enrichment analysis of differential genes between high and low PS groups. (A) Gene set enrichment analysis in the KEGG gene set. (B) Gene set enrichment analysis in the Hallmark gene set. (C) Differential expression of chemokines between high and low PS groups. (*p < 0.05, **p < 0.01, ***p < 0.001).
Identification of the Hub Genes in WGCNA network
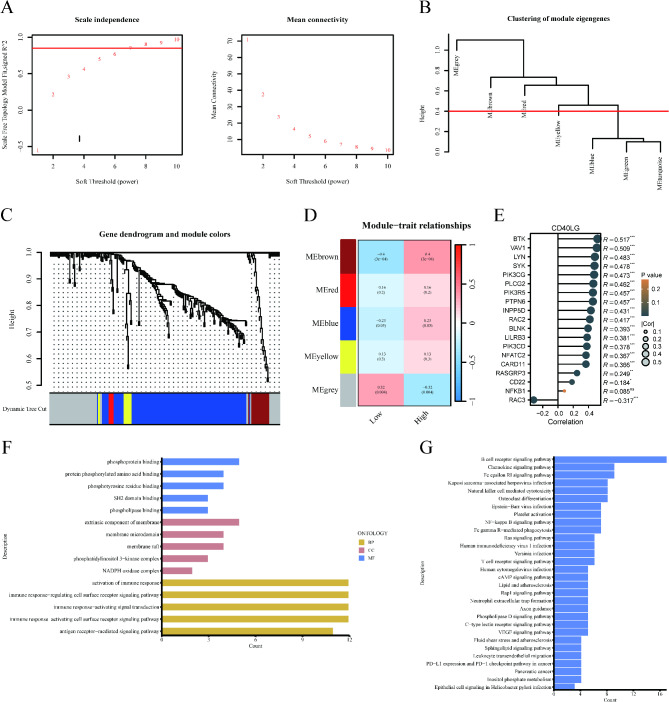
Furthermore, WGCNA was used to identify BCR signaling pathway gene sets associated with prognosis. The optimal soft threshold power, determined from the scale-free network, was 8 (Fig. 7A). Based on the average connected hierarchical clustering and the optimal soft threshold power, five modules were identified (Fig. 7B). BCR signaling pathway genes were allocated to these 5 modules (Fig. 7C). Pearson correlation coefficient analysis revealed a close relationship between the brown module and the prognosis of GBM patients (Fig. 7D). Consequently, the brown module was identified as the key module. To investigate the relationship between CD40LG and the 19 genes in the brown module, we performed a correlation analysis. The results showed that, except for NFKB1, CD40LG expression was significantly correlated with the expression of the other 18 genes (Fig. 7E). To further validate the potential functions of the 19 hub genes related to the BCR signaling pathway within the brown module, we performed GO and KEGG analyses. GO (BP) enrichment analysis revealed a significant enrichment of the activation of immune response among the 19 hub genes within the brown module (Fig. 7F). Similarly, the KEGG enrichment analysis indicated a significant enrichment of the NF-kappa B signaling pathway among the 19 hub genes in the brown module (Fig. 7G).
Fig. 7.
Identification of Hub genes in the BCR pathway by WGCNA network. (A) Scale independence and mean connectivity. (B) Clustering of module eigengenes. (C) Gene dendrogram and modules. (D) Relationships between module eigengenes and histopathological image feature. (E) The relationship between CD40LG and the 19 genes in the brown module. (F, G) GO and KEGG analysis of hub genes. (*p < 0.05, **p < 0.01, ***p < 0.001, ns no significance).
Immune correlation analysis
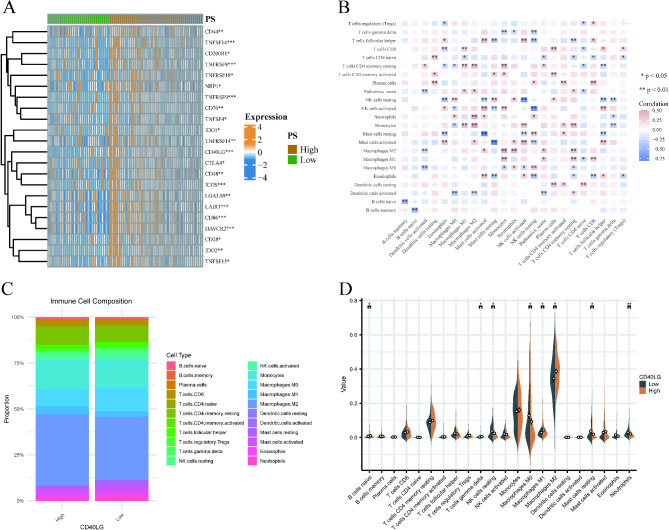
Differential analysis of immune checkpoint expression between the high and low PS groups revealed significantly increased levels of immune checkpoints, including CD86 and HAVCR2, in the PS high group (Fig. 8A).
Fig. 8.
Immune correlation analysis. (A) Differential analysis of immune checkpoint expression between high and low PS groups. (B) The correlation between the PS and the abundance of immune cells. (C) The proportions of immune cells in the high and low CD40LG expression groups. (D) The differences in infiltration of various immune cell types between the high and low CD40LG expression groups. (*p < 0.05, **p < 0.01, ***p < 0.001).
Additionally, to explore the relationship between PS and immune cell abundance, we analyzed immune cell infiltration in GBM. The results demonstrated a significant negative correlation between PS and the degree of infiltration of activated DC cells (cor = -0.313, p = 0.005) and T cell CD4 naïve (cor = -0.269, p = 0.017). Furthermore, a significant positive correlation was observed between PS and the infiltration degree of macrophage M2 (cor = 0.289, p = 0.01), macrophage M1 (cor = 0.239, p = 0.036), and T cell CD4 resting memory (cor = 0.266, p = 0.019) (Fig. 8B). Furthermore, to explore the relationship between CD40LG and the immune microenvironment, we utilized the CIBERSORT algorithm to analyze the proportions of immune cells in the high and low CD40LG expression groups. In the high-expression group, Macrophages M2 accounted for the highest proportion (38.68%), followed by Monocytes at 16.07%. Similarly, in the low-expression group, Macrophages M2 had the highest proportion (34.44%), followed by Monocytes (15.32%) (Fig. 8C). We also examined the differences in infiltration of various immune cell types between the high and low CD40LG expression groups. The results indicated that B cells naïve, T cells gamma delta, Macrophages M1, and Macrophages M2 had significantly higher infiltration in the high-expression group, whereas NK cells resting, Macrophages M0, Mast cells resting, and Neutrophils exhibited significantly higher infiltration in the low-expression group (Fig. 8D).
Discussion
Glioblastoma, a highly aggressive and frequently diagnosed malignant primary brain tumor in adults19,20, is characterized by its invasive nature, rapid growth, and resistance to treatment21. CD40L, a member of the TNF superfamily (TNFSF), and its receptor CD40, are being extensively studied as promising targets for immunotherapy. Previous studies have demonstrated a high expression of CD40L in cancer is related to an increase in proiferation and poor prognosis22,23. Moreover, studies have found that high expression of CD40 is associated with lower survival rates in glioma patients. Cell experiments have further demonstrated that knocking down CD40LG or using a CD40LG neutralizing antibody can reduce the invasiveness of GBM cells12.
Our findings conclusively demonstrate the upregulation of CD40LG in glioblastoma, which is correlated with a dismal prognosis and represents a risk factor for overall survival (OS) in patients with this disease. Consequently, we envision the utility of CD40LG as a predictive biomarker for patient prognosis. Nevertheless, the current clinical detection method for CD40LG is both time-consuming and labor-intensive, failing to provide a rapid and efficient means for its quantification.
Pathology serves as the gold standard for GBM diagnosis. Consequently, we employed H&E colored images to construct pathology models utilizing the SVM and LR algorithms. These models demonstrated favorable predictive capability for CD40LG expression levels. Given the routine use of H&E-stained images in clinical practice, our pathomics model holds significant potential for practical application. The rapid advancement of machine learning algorithms and advanced image analysis methodologies has fostered considerable promise for the application of artificial intelligence in the detection of histological features, particularly in predicting clinical outcomes24,25. High-throughput omics, including radiomics and pathomics, are progressively gaining traction in the field of precision medicine for automated diagnosis and prognosis prediction 26–28. For instance, Amin Zadeh Shirazi et al. successfully employed a deep convolutional neural network approach to discern pathological features linked to survival in glioblastoma 29. Our research shares similarities with this study; however, we employed two algorithms to construct two distinct models, thereby further validating the stability and predictive performance of our model. Various methodologies have been employed in numerous studies to develop predictive models for the clinical outcomes of patients with glioma. Cui et al. demonstrated the potential of radiomics analysis in accurately predicting survival outcomes in individuals diagnosed with low-grade glioma, reporting an AUC value of 0.709 30. Meanwhile, Yan et al. developed a deep learning feature based on diffusion tensor imaging (DTI) to forecast OS in individuals diagnosed with invasive glioma 31. Nonetheless, few studies have explored the use of pathomics and genomics in predicting the prognosis of GBM. Notably, our research stratified patients into high and low groups based on their PS, demonstrating the effectiveness of predicting CD40LG expression.
Most studies primarily focus on the model itself without considering the underlying molecular mechanisms associated with it. Consequently, we performed WGCNA and functional enrichment analysis on the differentially expressed genes between high and low PS groups. The results of the enrichment analysis were generally consistent with our expectations, with a main focus on the B cell receptor signaling pathway, chemokine signaling pathway, PI3K signaling pathway, and NF-κB pathway. This finding is also consistent with previous literature reports. Studies have demonstrated that the CD40/CD40L system acts as a co-stimulatory mechanism that enhances the immune response and can induce inflammation by upregulating adhesion molecules and promoting the synthesis of various cytokines and chemokines, including IL-1, TNF-α, IL-832. The transmission of a typical CD40/CD40L signal is facilitated by the recruitment of TRAF, followed by the activation of NF-κB. This process is further supported by additional signals, such as PI3K/AKT, MAPK, and JAK3/STAT33. We conducted an analysis of the relationship between PS and immune infiltration and found a significant association between PS and various immune cells, including DC cells, T cells CD4 naïve, Macrophages, T cells CD4 resting memory, and others. Our results are consistent with those of previous studies. The primary expression of CD40L occurs on activated T cells and platelets. Casamayor-Palleja et al. reported that following stimulation of T cells via the TCR and costimulatory ligands such as CD28, preformed CD40L is rapidly upregulated on the cell surface. This upregulation occurs within minutes and peaks after 6 h, followed by a gradual decrease in the surface expression of CD40L over the next 24 h34. CD40L interacts with CD40, which is present on various antigen-presenting cells (APCs) such as B cells, dendritic cells (DCs), and monocytes35. We used our pathohistological model to predict CD40LG expression, and our immune infiltration analyses indicated that PS was associated with the infiltration of certain immune cells, specifically DC cells. The studies indicate that DCs are essential components of the innate immune system, playing a critical role in coordinating antitumor responses. Specifically, their primary function is to initiate antitumor T cell responses by activating T cell receptors (TCRs)36. The interaction between CD40 and CD40L activates dendritic cells (DCs), allowing them to secrete the proinflammatory cytokine interleukin-12 (IL-12), which is critical in supporting CD4 + T cell helper responses and priming CD8 + cytotoxic T cells37–39. In vivo, temporary stimulation of the CD40 signaling pathway by soluble CD40 ligand (sCD40L) has a suppressive effect on human breast cancer cells40, demonstrating the tumor-suppressing potential of the CD40/CD40L interaction in various types of tumors. This justifies the use of CD40L + CAR T cells in treating various types of cancer. Research has shown that using a mouse model of immunocompetent lymphoma, CD40L + CAR T cells can activate dendritic cells (DCs) in vivo, leading to the recruitment of non-CAR T cells capable of recognizing tumor cells, ultimately resulting in a more potent and effective anti-tumor response41.
Despite the potential anti-tumor effect of the interaction between CD40L and CD40, it remains unclear why CD40LG, which is highly expressed in GBM, does not exert the same effect and is associated with a poor tumor prognosis. Researches have shown that tumor-associated macrophages play a critical role in tumor progression, cancer immunosuppression, and treatment resistance42–44. Tumor-associated macrophages constitute the primary population of non-tumor cells in GBM, making up half of the cells in GBM tumors45,46. This indicates that tumor-associated macrophages, particularly the M2 macrophages, are a major source of immunosuppression in GBM.
Our immune infiltration results further support this finding, showing that the proportion of M2 macrophage infiltration is significantly higher in the CD40LG high-expression group compared to the CD40LG low-expression group.
Previous studies have demonstrated the effectiveness of chimeric antigen receptor T (CAR-T) cell therapy in treating GBM. However, when CAR-T cells reach the tumor, they face numerous challenges within the microenvironment, including tumor-derived soluble substances and cytokines, immunosuppressive immune cells, as well as physical and metabolic disturbances47,48. The cytokine network within the glioblastoma (GBM) microenvironment includes molecules such as prostaglandin E2, IL-6, IL-10, and TGF-β, which have been shown to inhibit T cell proliferation and the development of effector responses49. In comparison to tumors collected before therapy, the expression levels of these immunosuppressive factors were consistently higher in surgical samples obtained from patients who had received EGFRvIII CAR-T cell infusions50. Regulatory T cells (Tregs) make up approximately 30% of the infiltrating lymphocytes in GBM51. Through their role in suppressing T cell responses, these Tregs pose a significant challenge to the development of immunotherapeutic strategies for GBM. Hence, we infer that these immunosuppressive factors in the GBM microenvironment hinder the function of CD40L, thereby impeding its tumor-suppressive effect. The efficacy of CD40L + CAR-T cell therapy in treating GBM remains unexplored and warrants further investigation.
Our study demonstrates that CD40LG is associated with a poor prognosis in GBM patients. Additionally, we developed a pathomics model to assess CD40LG. This model can effectively predict the expression of CD40LG and consequently forecast the prognosis of GBM patients. Simultaneously predicting the prognosis of GBM patients from dual perspectives enhances prediction accuracy, timeliness, and alleviates the economic burden on patients. Concurrently, analysis of the intrinsic molecular mechanisms within pathomics revealed that differentially expressed molecules were predominantly enriched in immune-related pathways and correlated with immune infiltration in GBM. Our findings are derived from bioinformatics and pathomics analyses, requiring validation through additional experimental research.
Acknowledgements
We are grateful to all the members of our study.
Author contributions
ZL: conceived and designed the research. WL: analyzed data and wrote the manuscript. JX: helped perform the analysis and edit the manuscript. CZ, XD and JY helped perform the analysis. XL and JH: download data and collate references. All authors reviewed the manuscript.
Data availability
The datasets analyzed during the current study are available in the Genotype-Tissue Expression (https://commonfund.nih.gov/GTEx) and The Cancer Genome Atlas (http://cancergenome.nih.gov) repositories.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributed equally.
References
- 1.Miller, K. D. et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J. Clin.71, 381–406. 10.3322/caac.21693 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Tan, A. C. et al. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin.70, 299–312. 10.3322/caac.21613 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med.352, 987–996. 10.1056/NEJMoa043330 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Verdugo, E., Puerto, I. & Medina, M. A. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. (Lond)42, 1083–1111. 10.1002/cac2.12361 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med.360, 765–773. 10.1056/NEJMoa0808710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cyster, J. G. & Allen, C. D. C. B cell responses: Cell interaction dynamics and decisions. Cell177, 524–540. 10.1016/j.cell.2019.03.016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weller, S. et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc. Natl. Acad. Sci. USA98, 1166–1170. 10.1073/pnas.98.3.1166 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laman, J. D., Claassen, E. & Noelle, R. J. Functions of CD40 and its ligand, gp39 (CD40L). Crit. Rev. Immunol.37, 371–420. 10.1615/CritRevImmunol.v37.i2-6.100 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Wang, C. et al. Tumor cell-associated exosomes robustly elicit anti-tumor immune responses through modulating dendritic cell vaccines in lung tumor. Int. J. Biol. Sci.16, 633–643. 10.7150/ijbs.38414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan, M. et al. CD40LG as a prognostic molecular marker regulates tumor microenvironment through immune process in breast cancer. Int. J. Gen. Med.14, 8833–8846. 10.2147/IJGM.S336813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chonan, M. et al. CD40/CD40L expression correlates with the survival of patients with glioblastomas and an augmentation in CD40 signaling enhances the efficacy of vaccinations against glioma models. Neuro Oncol.17, 1453–1462. 10.1093/neuonc/nov090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, S. M. et al. Glioblastoma-educated mesenchymal stem-like cells promote glioblastoma infiltration via extracellular matrix remodelling in the tumour microenvironment. Clin. Transl. Med.12, e997. 10.1002/ctm2.997 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Bent, M. J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol.120, 297–304. 10.1007/s00401-010-0725-7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barisoni, L., Lafata, K. J., Hewitt, S. M., Madabhushi, A. & Balis, U. G. J. Digital pathology and computational image analysis in nephropathology. Nat. Rev. Nephrol.16, 669–685. 10.1038/s41581-020-0321-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res.77, e104–e107. 10.1158/0008-5472.CAN-17-0339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30. 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951. 10.1002/pro.3715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592. 10.1093/nar/gkac963 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young, R. M., Jamshidi, A., Davis, G. & Sherman, J. H. Current trends in the surgical management and treatment of adult glioblastoma. Ann. Transl. Med.3, 121. 10.3978/j.issn.2305-5839.2015.05.10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrom, Q. T. et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol.21, v1–v100. 10.1093/neuonc/noz150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furnari, F. B. et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev.21, 2683–2710. 10.1101/gad.1596707 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa, K. et al. Up-regulation of CD40 with juxtacrine activity in human nonsmall lung cancer cells correlates with poor prognosis. Cancer113, 530–541. 10.1002/cncr.23618 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Pang, X. et al. Expression of CD40/CD40L in colon cancer, and its effect on proliferation and apoptosis of SW48 colon cancer cells. J. BUON22, 894–899 (2017). [PubMed] [Google Scholar]
- 24.Klimov, S. et al. A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res.21, 83. 10.1186/s13058-019-1165-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis, D. N. et al. Computational pathology: An emerging definition. Arch Pathol Lab Med138, 1133–1138. 10.5858/arpa.2014-0034-ED (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lambin, P. et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol.14, 749–762. 10.1038/nrclinonc.2017.141 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Cao, R. et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics10, 11080–11091. 10.7150/thno.49864 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, Y. et al. Development and validation of a deep learning CT signature to predict survival and chemotherapy benefit in gastric cancer: A multicentre, retrospective study. Ann. Surg.274, e1153–e1161. 10.1097/SLA.0000000000003778 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Zadeh Shirazi, A. et al. A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma. Br. J. Cancer125, 337–350. 10.1038/s41416-021-01394-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi, Y. S. et al. Machine learning and radiomic phenotyping of lower grade gliomas: Improving survival prediction. Eur. Radiol.30, 3834–3842. 10.1007/s00330-020-06737-5 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Yan, J. et al. Deep learning features from diffusion tensor imaging improve glioma stratification and identify risk groups with distinct molecular pathway activities. EBioMedicine72, 103583. 10.1016/j.ebiom.2021.103583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brugnolo, F. et al. Highly Th2-skewed cytokine profile of beta-lactam-specific T cells from nonatopic subjects with adverse drug reactions. J. Immunol.163, 1053–1059 (1999). [PubMed] [Google Scholar]
- 33.Tang, T. et al. Molecular basis and therapeutic implications of CD40/CD40L immune checkpoint. Pharmacol. Ther.219, 107709. 10.1016/j.pharmthera.2020.107709 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casamayor-Palleja, M., Khan, M. & MacLennan, I. C. A subset of CD4+ memory T cells contains preformed CD40 ligand that is rapidly but transiently expressed on their surface after activation through the T cell receptor complex. J. Exp. Med.181, 1293–1301. 10.1084/jem.181.4.1293 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kooten, C. & Banchereau, J. Functions of CD40 on B cells, dendritic cells and other cells. Curr. Opin. Immunol.9, 330–337. 10.1016/s0952-7915(97)80078-7 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Banchereau, J. & Steinman, R. M. Dendritic cells and the control of immunity. Nature392, 245–252. 10.1038/32588 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Caux, C. et al. Activation of human dendritic cells through CD40 cross-linking. J. Exp. Med.180, 1263–1272. 10.1084/jem.180.4.1263 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cella, M. et al. Ligation of CD40 on dendritic cells triggers production of high levels of interleukin-12 and enhances T cell stimulatory capacity: T-T help via APC activation. J. Exp. Med.184, 747–752. 10.1084/jem.184.2.747 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoenberger, S. P., Toes, R. E., van der Voort, E. I., Offringa, R. & Melief, C. J. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature393, 480–483. 10.1038/31002 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Tong, A. W. et al. Growth-inhibitory effects of CD40 ligand (CD154) and its endogenous expression in human breast cancer. Clin. Cancer Res.7, 691–703 (2001). [PubMed] [Google Scholar]
- 41.Kuhn, N. F. et al. CD40 ligand-modified chimeric antigen receptor T cells enhance antitumor function by eliciting an endogenous antitumor response. Cancer Cell35, 473–488. 10.1016/j.ccell.2019.02.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard, J. W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer4, 71–78. 10.1038/nrc1256 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Ruffell, B. & Coussens, L. M. Macrophages and therapeutic resistance in cancer. Cancer Cell27, 462–472. 10.1016/j.ccell.2015.02.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostuni, R., Kratochvill, F., Murray, P. J. & Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol.36, 229–239. 10.1016/j.it.2015.02.004 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Morantz, R. A., Wood, G. W., Foster, M., Clark, M. & Gollahon, K. Macrophages in experimental and human brain tumors. Part 2: Studies of the macrophage content of human brain tumors. J. Neurosurg.50, 305–311. 10.3171/jns.1979.50.3.0305 (1979). [DOI] [PubMed] [Google Scholar]
- 46.Hambardzumyan, D., Gutmann, D. H. & Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci.19, 20–27. 10.1038/nn.4185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newick, K., O’Brien, S., Moon, E. & Albelda, S. M. CAR T cell therapy for solid tumors. Annu. Rev. Med.68, 139–152. 10.1146/annurev-med-062315-120245 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Mirzaei, R., Sarkar, S. & Yong, V. W. T cell exhaustion in glioblastoma: Intricacies of immune checkpoints. Trends Immunol.38, 104–115. 10.1016/j.it.2016.11.005 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Hao, C. et al. Cytokine and cytokine receptor mRNA expression in human glioblastomas: Evidence of Th1, Th2 and Th3 cytokine dysregulation. Acta Neuropathol.103, 171–178. 10.1007/s004010100448 (2002). [DOI] [PubMed] [Google Scholar]
- 50.O’Rourke, D. M. et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med.10.1126/scitranslmed.aaa0984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs, J. F. et al. Regulatory T cells and the PD-L1/PD-1 pathway mediate immune suppression in malignant human brain tumors. Neuro Oncol.11, 394–402. 10.1215/15228517-2008-104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the Genotype-Tissue Expression (https://commonfund.nih.gov/GTEx) and The Cancer Genome Atlas (http://cancergenome.nih.gov) repositories.